Abstract
Chlamydia (C.) psittaci is the causative agent of avian chlamydiosis and human psittacosis. In this study, we extracted single-nucleotide polymorphisms (SNPs) from the whole genome sequences of 55 C. psittaci strains and identified eight major lineages, most of which are host-related. A combined PCR/high-resolution melting (HRM) assay was developed to screen for eight phylogenetically informative SNPs related to the identified C. psittaci lineages. The PCR-HRM method was validated on 11 available reference strains and with a set of 118 field isolates. Overall, PCR-HRM clustering was consistent with previous genotyping data obtained by ompA and/or MLST analysis. The method was then applied to 28 C. psittaci-positive samples from animal or human cases. As expected, PCR-HRM typing results from human samples identified genotypes linked to ducks and pigeons, a common source of human exposure, but also to the poorly described Mat116-like genotype. The new genotyping method does not require time-consuming sequencing and allows a quick identification of the source of infection.
Keywords: Chlamydia psittaci, SNP, PCR-high-resolution melting (HRM), psittacosis, avian chlamydiosis, genotyping
1. Introduction
Chlamydia (C.) psittaci, an agent from the Chlamydiaceae family, is commonly isolated from a wide range of birds worldwide [1]. This species is typically associated with infection in humans in close contact with birds, following inhalation of aerosolized infectious particles originating from dry feces and respiratory secretions. Pet bird owners and breeders, pet shop and zoo employees, poultry workers, veterinarians, laboratory technicians, and wildlife workers are particularly at risk [2]. C. psittaci-infected humans usually exhibit non-specific signs, including fever, headache, myalgia and non-productive cough. Misdiagnosis and/or inappropriate antibiotic-based treatment can also result in death due to atypical pneumonia. Avian species belonging to the orders Psittaciformes, Galliformes, Anseriformes, and Columbiformes are common sources of infection [3]. In birds, the clinical presentation may vary considerably and is influenced by the pathogen (genotypes of C. psittaci) and the host (species, age, health and immunological status). Alongside subclinical infections, severe respiratory, digestive and ocular forms are described [2].
The different C. psittaci typing methods developed over time have revealed a strain diversity within this species and a close association between serotypes/genotypes and bird groups. The initially described eight serovars (A to F for avian strains, WC and M56 for mammalian strains), based on the use of monoclonal antibodies [4,5,6] were found to be equivalent with ompA-based genotypes. Later, the ompA sequence analysis from a large panel of isolates led to the introduction of new C. psittaci provisional genotypes (1V, 6N, Mat116, R54, YP84 and CPX0308) [7]. Recently, 1V strain was re-affiliated to the C. abortus species [8], based on MLST analysis of seven conserved household genes [9]. By this method, the studied C. psittaci strains are classified into four main groups (I to IV), in connection with their avian hosts (parrot, duck, pigeon, turkey), the WC C. psittaci mammal strain being apart. Knowledge about the genotypes circulating in wildlife (e.g., sea birds and birds of prey) remains limited.
Genotyping based on ompA is commonly used, but resolution is rather low and since the gene is a hot spot for mutations and recombination [10,11], it may give misleading results. MLST provides higher discrimination and is robust, but requires time-consuming sequencing of seven gene targets. With the increase in number of whole genome sequences (WGS) available for C. psittaci strains [12,13,14,15,16], it is now possible to apply single nucleotide polymorphisms (SNPs) for the establishment of a phylogenetic tree pointing out true relationship between C. psittaci isolates. In our study, we aimed to develop a highly discriminative and user-friendly method where the work comprises three parts: (i) to identify clusters, based on SNP analysis, (ii) to compare those with MLST- and ompA-based topologies and (iii) to develop a rapid typing tool based on the identification of relevant SNPs by PCR/high-resolution melting (HRM) technology for straightforward genotyping of C. psittaci from isolates and clinical samples.
2. Materials and Methods
2.1. Bacterial Strains and Isolates
Strains and/or DNA samples were obtained from Anses (France), Friedrich-Loeffler-Institut (Germany), Veterinary Research Institute (Poland), University of Bordeaux (France) and Uppsala University Hospital (Sweden) (Table 1, Supplementary Data 1). Chlamydial strains were propagated in the yolk sac of chicken embryos or by cell culture as previously described [17,18]. DNA from strains or samples collected for diagnostic purposes was extracted using the QIAamp DNA Mini Kit (QIAGEN, Courtaboeuf, France). All samples included in this study tested positive for C. psittaci using the PCR system developed by Pantchev et al. [19].
Table 1.
PCR-high-resolution melting (HRM) clustering of reference strains, field strains and clinical samples. The host origin and available genotype and/or MLST results are shown in Supplementary Data 1.
| HRM Group | Strain/Sample | ID |
|---|---|---|
| I_Psittacine | reference strains | 6BC-04DC45, Loth, VS1 |
| field strains with typing data | 84-6461, 84-8471/1, 84-9462, 85-1173, 85-12098, 86-0191, 86-10703, 86-14356, 86-3389, 87-13654, 88-2014, 88-5558, 88-5821, 88-8795, 89-2930, 90-0057, 90-0475, 90-10445, 90-11404, 90-12937, 90-4862, 91-14273, 91-5189, 91-6047, 95-1334, 97-5075, 97-6475, 98-7627, 99-0182, 99-0923, 99-8157/1, 00-0151, 00-0476, 00-1176, 00-1268, 00-1750, 05-0949, 05-4098, 06-0372, 06-0852, C6/98, 01DC11, 03DC29, 03DC35, 04DC42, 04DC46 | |
| field strains without typing data | 96-6274, 96-12328, 96-12742, 97-822, 97-9244, 99-0313, 99-1394, 99-8888, 00-4462, 03-3227, 04-2668, 10-0485, 10-1735 | |
| clinical samples | 10-1735 *, 17-10114, 17-10090 | |
| II_Duck | reference strain | GR9 |
| field strains with typing data | 94-2306, 05-4325, 05-4461, 06-859, 06-871, 06-881, 06-889, 06-1683, 10-1398/28, 10-1400, C1/97, 07-1391, 08-2626_L3, 08-2626_L4 | |
| field strains without typing data | 04-5006, 05-553/17, 07-2962, 08-2850, 10-1393 | |
| clinical samples | 15-46D/8, 15-53D/8, 15-41/8, 15-63/3, 16-1264_MJC, 16-1264_JL1, 16-1264_JL2, 17-5203, 2008_A, 2009_A, 20-3954_C054 | |
| III_Pigeon | reference strains | CP3, Cal10, MN |
| field strains with typing data | 90-12617, 91-6568, 91-12516, 89-13210, 91-5983, 05-4036, 09-295_JF5, 09-295_JF8, 09-295_J9, 09-295_MB32, 09-295_MB33, 09-336_FA30634, 09-487_T13, 09-489_LP5, 09-489_LP7, 09-489_Mon5, 09-489_Mon13, 09-496_FA32303, 09-496_FA32311, 09-544_Van14, 09-589_S10, 09-928, 10-743_SC1, 10-881_SC22, 10-743_SC24, 10-743_SC28, 10-743_SC33, 10-743_SC42, 10-881_SC42, 10-883_EL27, 10-1048_Bat16, 01DC12, 03DC32, 09DC75, 11DC94 | |
| field strain without typing data | 10-2168 | |
| clinical samples | 15-8D/13, 15-57D/2, 10-743_SC1 *, 10-743_SC22 *, 10-743_SC24 *, 10-743_SC28 *, 10-743_SC33 *, 10-2168 *, 16-1264_VE, 20-1105_A036, 20-1105_A039 | |
| IV_Turkey | reference strains | NJ1, TT3 |
| V_Mat116 | field strain with typing data | 99DC05 |
| clinical samples | 20-1105_A041, 19-5617_I078_G2662, 19-5617_I080_K12016 | |
| VI_M56 | reference strain | M56-07DC57 |
| field strain with typing data | 16DC111 | |
| clinical samples | 12-2090_P088, 12-1950_M074 | |
| VII_VS225 | reference strain | VS225 |
| VIII_WC | reference strain | WC-07DC58 |
| field strain with typing data | Ful127 |
* linked to a field strain.
2.2. Whole Genome Phylogenetic Analysis and Selection of PCR-HRM Markers
The genomic sequences of 55 available C. psittaci strains used for comparison are listed in Supplementary Data 2. The whole genome SNP (wgSNP) pipeline of the BioNumerics software v7.6.1 (Applied Maths, Sint-Martens-Latem, Belgium) was used in order to detect SNPs on whole genome sequences and perform cluster analyses on the resulting wgSNP matrix. The input of the wgSNP module is raw data except for the reference. Each genome file were processed with the ART-MountRainier-2016-06-05 simulation tool that generates synthetic paired-end reads with coverage of 50 [20]. These reads were aligned and mapped against the reference sequence C. psittaci 6BC (CP002549.1) using the BWA algorithm implemented in BioNumerics with minimum 90% of sequence identity. Strain-specific SNPs were identified using the BioNumerics wgSNP module and then filtered using the following conditions: minimum 5× coverage to call a SNP, removal of positions with at least one ambiguous base, one unreliable base or non-informative SNP and minimum inter-SNP distance of 25 bp. A phylogenetic tree built using RAxML version 8.2.9 with the GTRGAMMA model and 1000 bootstrap replicates based on the filtered SNP matrix (4143 SNPs) from BioNumerics [21]. These SNPs are distributed throughout the genome.
For the eight lineages identified (groups I to VIII), an SNP was randomly selected and PCR-HRM primers were designed using Primer3Plus software [22]. The post-real-time-PCR HRM analysis offers the possibility to detect sequence variation inside amplicons without the need for sequencing or sequence-specific probes. With high precision, the melt profile of the PCR products is determined using double-stranded DNA binding dyes and accurate fluorescence data acquisition over small temperature increments. This method enables the discrimination of amplicons differing in a single SNP, according to their melting temperature (Tm). The positions of the selected SNPs in the C. psittaci 6BC genome and the primer sequences used in this study are listed in Table 2 and Table 3, respectively.
Table 2.
List of selected single-nucleotide polymorphisms (SNPs) used for this study. SNPs specific to each group are in bold.
| SNP for Each Group | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP No. | Associated Group | Group I | Group II | Group III | Group IV | Group V | Group VI | Group VII | Group VIII | Position on 6BC | Gene | Locus Tag |
| (Psittacine) | (Duck) | (Pigeon) | (Turkey) | (M56) | (Mat116) | (VS225) | (WC) | |||||
| 1 | Group I_Psittacine | T | C | C | C | C | C | C | C | 126074 | lipoate-protein ligase family protein | CPSIT_RS00640 |
| 2 | Group II_Duck | T | C | T | T | T | T | T | T | 39653 | CesT family type III secretion system chaperone | CPSIT_RS00155 |
| 3 | Group III_Pigeon | A | A | G | A | A | A | A | A | 1038463 | cation-translocating P-type ATPas | CPSIT_RS04480 |
| 4 | Group IV_Turkey | C | C | C | T | C | C | C | C | 1352 | Na(+)-translocating NADH-quinone reductase subunit A | CPSIT_RS00010 |
| 5 | Group V_M56 | C | C | C | C | T | C | C | C | 961 | hemB | CPSIT_RS00005 |
| 6 | Group VI_Mat116 | T | T | T | T | T | C | T | T | 85629 | anti-sigma regulatory factor | CPSIT_RS00375 |
| 7 | Group VII_VS225 | A | A | A | A | A | A | G | A | 13908 | exodeoxyribonuclease V subunit gamma | CPSIT_RS00045 |
| 8 | Group VIII_WC | G | G | G | G | G | G | G | A | 722388 | YqgE/AlgH family protein | CPSIT_RS03140 |
Table 3.
PCR primers used for the PCR-HRM analysis.
| SNP No. | Associated Group | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon Size (bp) |
|---|---|---|---|---|
| 1 | Group I_Psittacine | gacccaacgagatttctgga | cccaaagacatttgccttaca | 94 |
| 2 | Group II_Duck | gcgatctcgtcaagatacgtg | ttggtatccgaagaaggaggt | 94 |
| 3 | Group III_Pigeon | cttctttcttgcaggaactccag | atccgaaagctgctgacgtc | 101 |
| 4 | Group IV_Turkey | aagaaccctaacatgcacgc | ggcgatgaaaatccctgttgt | 81 |
| 5 | Group V_M56 | tgatgtgttgcatcgagtga | ccactgacttgataggctgct | 63 |
| 6 | Group VI_Mat116 | cgcttcttggtatgcataggag | agaacaactcaaaacattcccaa | 100 |
| 7 | Group VII_VS225 | aagggagtcagaagaagagaaaa | actaatgctacgagtaaccacg | 63 |
| 8 | Group VIII_WC | tgaacaggaatgcaaaagca | tgggtttagaaatagctgacga | 119 |
2.3. PCR-HRM Assay
For samples from human or animal origin with a low DNA content (Cq higher than 33 with the C. psittaci real-time PCR), a pre-amplification step was done to increase the amount of DNA template (using the Perfecta® pre-amplification kit (Quantabio) and a mix of the eight set of primers for 15 cycles).
PCR-HRM amplifications were performed on the ViiA7™ Real-Time PCR System (Life Technologies, Carlsbad, CA, USA) using the LightCycler® 480 High Resolution Melting Master Mix (Roche Diagnostics, Roche, Switzerland). The reaction mixture consisted of 0.2 μM of each primer, 1 × LightCycler® 480 HRM master mix and 2.5 mM MgCl2 in an 18-μL final volume. The following parameters were used: 10 min at 95 °C were followed by 40 cycles consisting of 10 s at 95 °C, 10 s at 60 °C and 20 s at 72 °C. Samples were next heated to 95 °C for 30 s, cooled down to 65 °C for 1 min and heated from 65 °C to 88 °C at a rate of 1 °C/s with 25 acquisitions/°C. HRM data were analyzed by the ViiA7™ Software (version 1.2.1). Synthetic oligonucleotide templates were PCR amplified and used as controls for each marker (dilution of 10−7 from a 100 µM solution) in HRM analysis.
2.4. ompA and MLST Typing
ompA sequencing using primers 3GPB and 5GPF was performed as previously described [23]. The seven housekeeping genes of the MLST method [9], namely gatA, oppA, hflX, gidA, enoA, hemN and fumC, were amplified and sequenced using primers and conditions described on the Chlamydiales MLST website (http://pubmlst.org/chlamydiales/http://mlst.ucc.ie/, accessed on 1 February 2020). Sequencing of both DNA strands was performed by Eurofins (Reichenwalde, Germany) and numbers for alleles and sequence types (STs) were assigned in accordance with the Chlamydiales MLST Database.
2.5. In-Silico MLST Typing
Assemblies of the 55 C. psittaci strains were downloaded from NCBI and a script (https://github.com/tseemann/mlst, accessed on 1 February 2020) was used to scan contig files against the PubMLST chlamydiales scheme. The resulting table was used to build a tree in BioNumerics using the parameter “categorical values” to calculate the similarity matrix and UPGMA to reconstruct the tree.
3. Results and Discussion
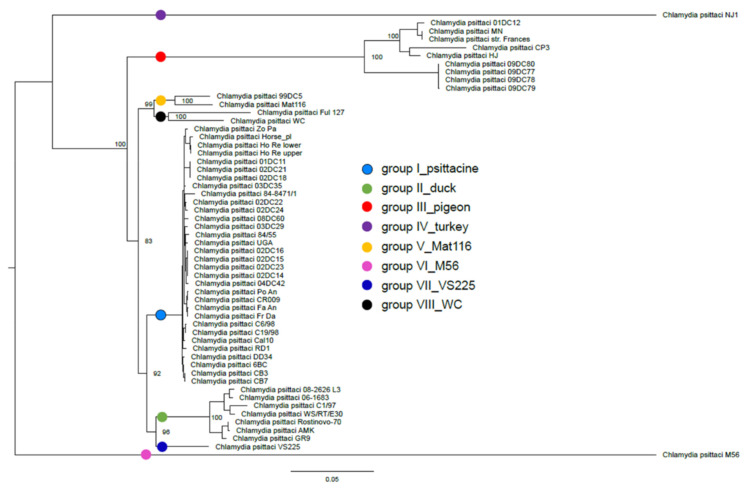
The availability of complete genome sequences of a large panel of C. psittaci strains allowed the establishment of phylogenetic relationships between C. psittaci isolates. Using a large number of SNPs scattered through the genomes of 55 strains, the construction of a SNP-based tree led to the identification of eight distinct lineages (Figure 1), all correlating with the currently defined genotypes. Indeed, in this tree, most of the strains are distributed into three main groups: strains isolated from psittacine birds (genotype A), ducks (genotypes C and E/B), or pigeons (genotypes B and E). This clustering is consistent with the MLST clustering described by Pannekoek et al. [9], with strains of C. psittaci grouped mainly according to the bird groups they infect. The other five groups included the more anecdotal genotypes, such as NJ1 associated with turkeys (genotype D), VS225 (genotype F), Mat116 (previously proposed without distinct host species assignments [7]), as well as the mammalian genotypes related to the WC or M56 strains. These eight groups were named: group I_psittacine, group II_duck, group III_pigeon, group IV_turkey, group V_Mat116, group VI_M56, group VII_VS225, and group VIII_WC.
Figure 1.
Maximum likelihood SNP-based tree determined from 55 C. psittaci whole genome sequences. The tree was built using RAxML version 8.2.9 with the GTRGAMMA model and 1000 bootstrap replicates. The eight distinct lineages determined in this study (group I_psittacine, group II_duck, group III_pigeon, group IV_turkey, group V_Mat116, group VI_M56, group VII_VS225, and group VIII_WC) are represented by coloured circles. Bootstrap values indicate the stability of the branches and the scale bar represents the number of substitution per site.
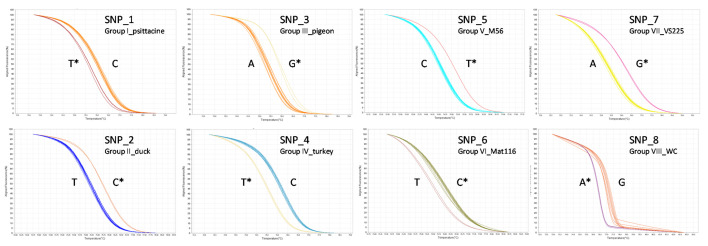
Based on this clustering, a specific SNP for each of these eight groups (named SNP1 to 8) was selected and primers designed for a specific amplification by PCR. The HRM curves obtained for the eight C. psittaci targeted SNPs are shown in Figure 2. All SNPs allowed a clear distinction between amplicons from the different targeted groups. Synthetic oligonucleotides corresponding to PCR amplified fragments were used as template controls for each marker, as well as DNA from an initial set of 11 reference strains of C. psittaci (except Mat116, not available). All these samples clustered in their intended group and all yielded amplicons producing a single melting peak, with Tm values depending on the SNP carried by the amplicon. On average, differences in Tm values of about 0.3 to 1.1 °C were observed between the paired amplicons specific of each group (Supplementary Data 3).
Figure 2.
Normalized melting curves obtained with the eight SNP markers for reference C. psittaci strains and the respective synthetic controls. The SNP specific to the targeted group is noted by an *.
The developed PCR-HRM method was then applied to 118 DNA preparations from C. psittaci strains isolated from different avian or animal hosts. Results are summarized in Table 1. All strains isolated from psittacine birds, ducks, or pigeons clustered in their respective group, except 91-5983, isolated from a psittacine that clustered in the pigeon group (group III). In line with this result, this strain was previously typed as genotype E by PCR-restriction fragment length polymorphism (RFLP) and microarray [24], a genotype commonly associated with pigeons. A strain of unknown origin (87-1365), clustered in the psittacine group (group I_psittacine) and was previously characterized as genotype A. Most of the strains included in this study were genotyped with the microarray test and/or PCR-RFLP and/or ompA sequencing (n = 94/117), and identical clustering results were obtained for all of them (Table 1, Supplementary Data 1). Field strains without preliminary genotyping clustered into the group corresponding to their bird host (group I_psittacine (n = 13), group II_duck (n = 5) or group III_pigeon (n = 1)). It is interesting to note that the group I_psittacine also includes strains of C. psittaci isolated from non-avian hosts (rabbit, pig, tick, rat) indicating a certain extent of variability in host tropism of C. psittaci strains. Shared grazing and mixed farming may also explain the detection of a duck genotype (group II_duck) in a ruminant (C1/97) or a pigeon genotype (group III_pigeon) in a pig (01DC12).
The strain 99DC05 isolated from a horse in Germany [25] clustered in group V_Mat116 and Ful127, a strain recently isolated from a fulmar (Procellariidae) [26], clustered in group VIII_WC. The MLST sequences of these two strains are both close to the MLST ST24, corresponding to the reference strain 6BC (group I_psittacine), with only two mutations in the hflX gene for Ful127 and one mutation in the enoA gene for 99DC05. In this case, the SNP-based typing generates a different topology than MLST analysis (Supplementary Data 4). Our study was conducted on a large set of SNPs (4143 SNPs) distributed through the genome of C. psittaci strains whereas only seven housekeeping genes were analyzed for MLST. It is likely that WGS analysis will help, in the near future, to refine the classification of different genotypes.
In a second step, the PCR-HRM method was applied to samples of animal or human origin. Of the 17 animal samples, seven were tissues from which strains included in the study had been isolated (six from pigeons (10-743_SC1, 10-743_SC22, 10-743_SC24, 10-743_SC28, 10-743_SC33 and 10-2168) and one from a psittacine (10-1735)). Identical clustering results were obtained. Other samples from psittacine birds (17-10114 and 17-10090), duck/waterfowl birds (15-46D/8, 15-53D/8, 15-41/8, 15-63/3) or a hooded crow (15-8D/13) gave a consistent clustering with the bird groups, confirmed for most of them by ompA sequencing results when available, except for 15-57D/2. Indeed, this sample came from a black-headed gull and was ompA-genotyped as Mat116-like [8], whereas the PCR-HRM analysis clustered it in group II_pigeon. This discordant result could be due to the well-known heterogeneity of the ompA gene being a hot spot for mutations and recombination events [11].
Furthermore, PCR-HRM typing confirmed that two ruminant samples (12-2090_P088 and 12-1950_M074) belonged to group VI_M56, in agreement with previous ompA analysis. Interestingly, these two samples were isolated from cases of cattle abortion illustrating that this mammalian genotype of C. psittaci can be associated with reproductive failure in cattle.
When human cases are suspected, it is important to obtain information on the source of infection to avoid the spread of the infection in animals, but also to potentially prevent further human exposure. It is often difficult to determine the origin of infection outside a workplace or a family context, with a clear and identified exposure to birds. Indeed, strains of C. psittaci are hosted by a variety of birds, ranging from domestic birds, to exotic and wild birds, and direct or indirect exposures can be linked to a variety of activities. The low amount of chlamydial DNA in human samples, especially when non-invasive samplings are performed (throat swabs, nasopharyngeal aspirates), is a limitation for a sequence-based genotyping that also requires time. In this study, the PCR-HRM typing scheme was applied to 20 human samples after a pre-amplification step, but only 13 were successfully typed. The limit of amplification for typing, determined from the synthetic control templates, was estimated at 5 × 104 copies per µL. The seven unamplified samples were very weakly positive with Cq > 38 in the C. psittaci real time PCR, which explains the failure to generate amplicons for PCR-HRM. Six of the successfully amplified samples were collected from duck breeders (16-1264_MJC, 16-1264_JL1, 16-1264_JL2, 2008_A, 2009_A, and 20-3954_C054) and genotyped as group II_duck by PCR-HRM) and three other samples (16-1264_VE, 20-1105_A036, and 20-1105_A039) were genotyped as group III_pigeon by PCR-HRM, in line with the suspected infection source (dust exposure, cleaning of attics). These bird groups are among the common sources of human infection [3].
In particular, the PCR-HRM method was applied to the sample 17-5203 from a deceased patient whose infection source was not clearly established at the time of hospitalization, as the person had both raised poultry and hunted game shortly before the onset of clinical signs. PCR-HRM analysis identified the group II_duck genotype, presumably from wildlife, as all backyard birds tested negative. This result was confirmed by analysis of the ompA (genotype C) and MLST (ST28) results.
Interestingly, analysis of human samples recently collected in France (20-1105-A041) and in Sweden (19-5617_I078_G2662, 19-5617_I080_K12016) revealed the group V_Mat116 genotype, a genotype poorly described so far and also isolated from a horse in Germany (strain 99DC05). While the original Mat116 strain was isolated from an unspecified psittacosis outbreak in Japan (Genbank CP002744), the French and Swedish human cases are likely to have wild birds as the probable origin, since no contact with poultry or psittacine birds has been identified.
The PCR-HRM typing tool presented in this study should be considered as a scheme under development, which should be enriched with sequencing data as genomes are contributed and which may then require an update of the SNP and corresponding primer panels. Indeed, during the development work of this study, the PCR-HRM analysis performed with the initially established set of primers (not shown), did not allow affiliation of the recently isolated strain Ful127 to one of the eight determined groups and a new SNP marker for the group VIII_WC had to be implemented.
In summary, our findings show that this first set of PCR-HRM markers can be used as a new typing method providing discrimination between C. psittaci isolates from diverse hosts. Moreover, as additional chlamydial genome sequences become available, it will be possible to search for new SNP markers that can be used for further strain discrimination. Given the zoonotic risks associated with C. psittaci infection, a prompt source determination is crucial for a good sanitary management of cases and prevention of human exposure.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/3/625/s1, Supplementary Data 1. PCR-HRM clustering of reference strains, field strains and clinical samples from human and animal origin. Available genotype and/or MLST results are indicated. Supplementary Data 2. NCBI accession numbers of the 55 C. psittaci strains used in this study. Supplementary Data 3. Melting temperature (Tm) values for each SNP. Values were determined for the respective controls tested 6 times. Supplementary Data 4. MLST-based tree determined from ST of the 55 C. psittaci strains used for the SNP determination. The tree was built using BioNumerics software with the parameter “categorical values” to calculate the similarity matrix and UPGMA to reconstruct the tree.
Author Contributions
F.V., R.A. and K.L. conceived the study; B.d.B., O.P., M.S.-C., B.H. and C.S. provided strains and samples from their collections; F.V., R.A. and K.L. conducted analyses and compiled resulting data sets for the manuscripts. K.L., C.S. and B.H. wrote the manuscript. All authors have read, edited and approved the final manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Detailed data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaleta E.F., Taday E.M.A. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 2003;32:435–461. doi: 10.1080/03079450310001593613. [DOI] [PubMed] [Google Scholar]
- 2.Balsamo G., Maxted A.M., Midla J.W., Murphy J.M., Wohrle R., Edling T.M., Fish P.H., Flammer K., Hyde D., Kutty P.K., et al. Compendium of Measures to Control Chlamydia psittaci Infection Among Humans (Psittacosis) and Pet Birds (Avian Chlamydiosis) J. Avian Med. Surg. 2017;31:262–282. doi: 10.1647/217-265. [DOI] [PubMed] [Google Scholar]
- 3.Hogerwerf L., Roof I., de Jong M.J.K., Dijkstra F., van der Hoek W. Animal sources for zoonotic transmission of psittacosis: A systematic review. BMC Infect. Dis. 2020;20:192. doi: 10.1186/s12879-020-4918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen A.A. Serotyping of Chlamydia psittaci isolates using serovar-specific monoclonal antibodies with the microimmunofluorescence test. J. Clin. Microbiol. 1991;29:707–711. doi: 10.1128/JCM.29.4.707-711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen A.A. Two new serovars of Chlamydia psittaci from North American birds. J. Vet. Diagn. Investig. 1997;9:159–164. doi: 10.1177/104063879700900209. [DOI] [PubMed] [Google Scholar]
- 6.Vanrompay D., Butaye P., Sayada C., Ducatelle R., Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res. Microbiol. 1997;148:327–333. doi: 10.1016/S0923-2508(97)81588-4. [DOI] [PubMed] [Google Scholar]
- 7.Sachse K., Laroucau K., Hotzel H., Schubert E., Ehricht R., Slickers P. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008;8:63. doi: 10.1186/1471-2180-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymańska-Czerwińska M., Mitura A., Niemczuk K., Zaręba K., Jodełko A., Pluta A., Scharf S., Vitek B., Aaziz R., Vorimore F., et al. Dissemination and genetic diversity of chlamydial agents in Polish wildfowl: Isolation and molecular characterisation of avian Chlamydia abortus strains. PLoS ONE. 2017;12:e0174599. doi: 10.1371/journal.pone.0174599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pannekoek Y., Dickx V., Beeckman D.S.A., Jolley K.A., Keijzers W.C., Vretou E., Maiden M.C.J., Vanrompay D., van der Ende A. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS ONE. 2010;5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunelle B.W., Sensabaugh G.F. Nucleotide and phylogenetic analyses of the Chlamydia trachomatis ompA gene indicates it is a hotspot for mutation. BMC Res. Notes. 2012;5:53. doi: 10.1186/1756-0500-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Loock M., Vanrompay D., Herrmann B., Vander Stappen J., Volckaert G., Goddeeris B.M., Everett K.D.E. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int. J. Syst. Evol. Microbiol. 2003;53:761–770. doi: 10.1099/ijs.0.02329-0. [DOI] [PubMed] [Google Scholar]
- 12.Voigt A., Schöfl G., Heidrich A., Sachse K., Saluz H.P. Full-length de novo sequence of the Chlamydophila psittaci type strain, 6BC. J. Bacteriol. 2011;193:2662–2663. doi: 10.1128/JB.00236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schöfl G., Voigt A., Litsche K., Sachse K., Saluz H.P. Complete genome sequences of four mammalian isolates of Chlamydophila psittaci. J. Bacteriol. 2011;193:4258. doi: 10.1128/JB.05382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seth-Smith H.M., Sait M., Sachse K., Gaede W., Longbottom D., Thomson N.R. Genome Sequence of Chlamydia psittaci Strain 01DC12 Originating from Swine. Genome Announc. 2013;1:e00078-12. doi: 10.1128/genomeA.00078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu J., Sun R., Wu Z., Liu S., Li D., Zhang Q., Ling Y., Gong Y., Wu R., Wu H., et al. Whole-Genome Sequences of Low-Virulence Strain CB3 and Mild Strain CB7 of Chlamydia psittaci. Genome Announc. 2014;2:e00456-14. doi: 10.1128/genomeA.00456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q., Wu Z., Sun R., Chu J., Han E., Zhang Y., Ling Y., Gong Y., Li D., Wu H., et al. Whole-Genome Sequences of Chlamydia psittaci Strain HJ, Isolated from Meat Pigeons with Severe Respiratory Distress and High Mortality. Genome Announc. 2015;3:e00035-15. doi: 10.1128/genomeA.00035-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laroucau K., de Barbeyrac B., Vorimore F., Clerc M., Bertin C., Harkinezhad T., Verminnen K., Obeniche F., Capek I., Bébéar C., et al. Chlamydial infections in duck farms associated with human cases of psittacosis in France. Vet. Microbiol. 2009;135:82–89. doi: 10.1016/j.vetmic.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 18.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Creasy H.H., Weidmann M., Myers G., Vorimore F., Vicari N., et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst. Appl. Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet. J. 2009;181:145–150. doi: 10.1016/j.tvjl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 20.Huang W., Li L., Myers J.R., Marth G.T. ART: A next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Untergasser A., Nijvee H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltenboeck B., Kousoulas K.G., Storz J. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J. Bacteriol. 1993;175:487–502. doi: 10.1128/JB.175.2.487-502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachse K., Laroucau K., Vorimore F., Magnino S., Feige J., Müller W., Kube S., Hotzel H., Schubert E., Slickers P., et al. DNA microarray-based genotyping of Chlamydophila psittaci strains from culture and clinical samples. Vet. Microbiol. 2009;135:22–30. doi: 10.1016/j.vetmic.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 25.Henning K., Sachse K., Sting R. Demonstration of Chlamydia from an equine abortion. Dtsch. Tierärztl. Wochenschr. 2000;107:49–52. [PubMed] [Google Scholar]
- 26.Wang H., Jensen J.K., Olsson A., Vorimore F., Aaziz R., Guy L., Ellström P., Laroucau K., Herrmann B. Chlamydia psittaci in fulmars on the Faroe Islands: A causative link to South American psittacines eight decades after a severe epidemic. Microbes Infect. 2020;22:356–359. doi: 10.1016/j.micinf.2020.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed data are available upon request.