Abstract
In this study, we show that Acinetobacter baumannii ATCC 19606 harbors two sets of ohrR-ohr genes, respectively encoded in chromosomal DNA and a pMAC plasmid. We found no significant difference in organic hydroperoxide (OHP) resistance between strains with or without pMAC. However, a disk diffusion assay conducted by exposing wild-type, ∆ohrR-C, C represented gene on chromosome, or ∆ohr-C single mutants, or ∆ohrR-C∆ohr-C double mutants to tert-butyl hydroperoxide (tBHP) found that the ohrR-p-ohr-p genes, p represented genes on pMAC plasmid, may be able to complement the function of their chromosomal counterparts. Interestingly, ∆ohr-C single mutants generated in A. baumannii ATCC 17978, which does not harbor pMAC, demonstrated delayed exponential growth and loss of viability following exposure to 135 μg of tBHP. In a survival assay conducted with Galleria mellonella larvae, these mutants demonstrated almost complete loss of virulence. Via an electrophoretic mobility shift assay (EMSA), we found that OhrR-C was able to bind to the promoter regions of both chromosomal and pMAC ohr-p genes, but with varying affinity. A gain-of-function assay conducted in Escherichia coli showed that OhrR-C was not only capable of suppressing transformed ohr-C genes but may also repress endogenous enzymes. Taken together, our findings suggest that chromosomal ohrR-C-ohr-C genes act as the major system in protecting A. baumannii ATCC 19606 from OHP stresses, but the ohrR-p-ohr-p genes on pMAC can provide a supplementary protective effect, and the interaction between these genes may affect other aspects of bacterial viability, such as growth and virulence.
Keywords: Acinetobacter baumannii, Ohr, OhrR, organic hydroperoxide, antibiotic resistance, bacterial viability, bacterial virulence
1. Introduction
Bacteria are often exposed to reactive oxygen species (ROS) and other organic hydroperoxides (OHPs) produced by host phagocytic cells [1] as part of the immune response [2,3]. OHPs produced by free radical-catalyzed oxidation of polyunsaturated fatty acids (PUFAs) from the host cell [2] are highly toxic molecules that can generate organic free radicals and induce oxidative injury to bacterial cell components. Bacteria have therefore evolved several strategies to protect themselves against oxidative stress. One such strategy is to produce enzymes that can directly detoxify OHPs by transforming them into unreactive alcohols. Ohr (organic hydroperoxide resistance protein) is a thiol peroxidase that is central to the bacterial response against OHPs [4,5], and it acts to neutralize OHPs via a redox-active disulfide bond formed by two conserved cysteines at the catalytic site [6]. Two homologs of Ohr, respectively termed OhrA and OhrB, have been identified in Bacillus subtilis [7].
The expression of Ohr enzymes is suppressed by OhrR (organic hydroperoxide resistance regulator) [8]. OhrR belongs to the MarR superfamily of transcription factors and is found in both Gram-negative and Gram-positive bacteria [9]. The oxidation of a highly-conserved cysteine in the N-terminus of OhrR upon exposure to OHPs disrupts its DNA binding activity [10], leading to the derepression of ohr gene. In B. subtilis, the oxidation of the sole cysteine residue in OhrR allows the regulator to retain its DNA-binding activity, and the derivative from an additional oxidation is required for derepression [11,12]. In Burkholderia thailandensis, OhrR oxidation results in the formation of a reversible disulfide bond between conserved cysteines in the N-terminus and C-terminus of separate monomers resulting for attenuation of DNA binding in vitro [13].
Acinetobacter baumannii, a type of gammaproteobacteria of the Moraxellaceae family, is widely found in soil, water, animals, and food items, and has gained increasing attention in recent years due to its role in nosocomial infections [14,15]. A. baumannii can induce a vast range of serious infections, including pneumonia, bloodstream infections, urinary tract infections, wound and burn infections, and secondary meningitis [15,16,17,18]. According to genomic data from the National Center for Biotechnology Information (NCBI), as of January 2021, genome assemblies for at least 4914 strains of A. baumannii have been deposited, and high genomic diversity between strains has been observed. The most studied A. baumannii strain, ATCC 17978, was isolated from a four-month-old infant with fetal meningitis, while the ATCC 19606 strain used in this study was isolated from urine [19]. Plasmid availability is one of the most important distinctions between these two strains, as A. baumannii ATCC 19606 harbors pMAC, a 9540-base pair (bp) plasmid that contains 11 annotated open reading frames (ORFs) [20]. ORF8 and ORF9 have been respectively annotated as ohrA and ohrR, and this has been confirmed through functional analysis in Escherichia coli [20].
In this study, we identified the presence of another ohrR-C-ohr-C gene cluster in the chromosomal DNA of A. baumannii ATCC 19606 through in silico analysis, and subsequently investigated the function and significance of this newly-discovered set of ohr-c genes. Our results show that chromosomal ohrR-C-ohr-C may play a greater role in resistance to OHPs, and this may have implications for the study of ohr genes in other bacterial species, as well as the development of novel antibacterial therapies against A. baumannii infections.
2. Material and Methods
2.1. Bacterial Strains and Culturing Conditions
A. baumannii ATCC 19606 and E. coli strains were grown in LB medium [21] at 37 °C with shaking. All of the strains were listed in Table 1. LB medium with 1.5% agar was prepared for solid cultures. Antibiotics were used at different concentrations according to the cultured strains.
Table 1.
Plasmids and bacterial strains used in this study.
| Plasmid | Description | Antibiotic Resistance (µg/mL) | Reference/Source |
|---|---|---|---|
| pK18mobsecB | Suicide vector for homologous recombination | Km50 | [23] |
| pK18dohrR | pK18mobsecB contains the upstream and downstream regions of ohrR-C | Km50 | This study |
| pK18dohr | pK18mobsecB contains the upstream and downstream regions of ohr-C | Km50 | This study |
| pK18dohrRohr | pK18mobsecB contains the upstream and downstream regions of ohrR-C-ohr-C | Km50 | This study |
| pQE80L | Expression vector with colE1 origin for His-tag fusion protein purification | Amp50 | Qiagen |
| pOhrR-C | ohrR-C gene with N-terminal-fused His-tag in pQE80L | Amp50 | This study |
| pOhrR-C-Pohr-ohr-C | ohr-C and ohrR-C gene promoter was cloned downstream of pOhrR-C | Amp50 | This study |
| Strain | Description | Reference/Source | |
| E. coli DH5α | F−, supE44, hsdR17, recA1, gyrA96, endA1, thi-1, relA1, deoR, λ− | ATCC 53868 | |
| E. coli S17-1λpir | thi-, thr , leu , tonA , lacY , supE , recA::RP4-2-Tc::Mu, Smr, lpir | [23] | |
| Acinetobacter baumannii ATCC 19606 | Primary strain used in this study | [19] | |
|
Acinetobacter baumannii ATCC 17978 |
Most studied strain to date | [14] | |
| ∆ohrR | Marker-less ohrR-c deletion mutant | This study | |
| ∆ohr | Marker-less ohr-c deletion mutant | This study | |
| ∆ohrRohr | Marker-less ohrR-c-ohr-c deletion mutant | This study | |
Amp: ampicillin; Km: kanamycin.
The growth curve of each strain was determined according to culture turbidity, using optical density measurements at 600 nm (OD600). Viable bacteria counts for each strain were determined using the drop plate method, as previously described [22]. At each time point, 5 μL of 10-fold serially diluted bacteria was dropped on LB agar plates containing ampicillin to establish viable bacteria counts. Bacterial growth curves were determined every three hours for the first 12 h, after which bacterial viability was assessed every 12 h, until 48 h had elapsed from the initial drop.
2.2. Markerless Mutant Generation
Both ohr-C and ohrR-C mutants were generated by markerless gene deletion. The upstream and downstream DNA fragments of the respective genes were amplified by PCR, using the primers listed in Table 2, and were cloned into the BamHI and HindIII sites of plasmid pK18mobsecB (Table 1) [23] using the Gibson assembly cloning kit (New England Biolabs; Ipswich, MA, USA). The resulting plasmid was transformed into E. coli S17-1λpir to produce a donor for conjugation with A. baumannii. This plasmid DNA was maintained in the chromosomal region near ohr-C through the first homologous recombination event, and it was selected using the kanamycin resistance gene. Excision of the resulting plasmid DNA by the second crossover event was facilitated by selection on medium containing 10–20% sucrose but without kanamycin. The deletion mutant was obtained without any markers and was subsequently confirmed by PCR analysis.
Table 2.
Gene-specific primers used in this study.
| Name | Sequences (5′–3′) | Function |
|---|---|---|
| ohrR MU F | GCTGTGGGTGGATATCAGGA | Construction of ∆ohrR |
| ohrR MU R | CAGTGACTCGTCTTAAAGAT | Construction of ∆ohrR |
| pk18_ohrup_F | CGAGCTCGGTACCCGGGCGG GACAACGAATAATTTTG |
Construction of ∆ohr |
| ohrup-ohrdo-R | CCAATGAAGCAAGTGAAGCA TTATTTAGCTATATTTGTACGGAGC |
Construction of ∆ohr |
| ohrup-ohrdo-F | GCTCCGTACAAATATAGCTA AATAATGCTTCACTTGCTTCATTGG |
Construction of ∆ohr |
| ohrdo-pK18-R | AACGACGGCCAGTGCCATTT ACGTCTTGCTGGTCGTG |
Construction of ∆ohr |
| pk18_ ohrRup_F | CGAGCTCGGTACCCGGGCAT CTACCCCTTTTGGCAAT |
Construction of ∆ohrR∆ohr |
| ohrRup- ohrdo-R | CCAATGAAGCAAGTGAAGCA CCTCAGAAAACTAATGGGTGCT |
Construction of ∆ohrR∆ohr |
| ohr Rup- ohr do-F | AGCACCCATTAGTTTTCTGA GGTGCTTCACTTGCTTCATTGG |
Construction of ∆ohrR∆ohr |
| nptII-F | ATGATTGAACAAGATGGATTGC | Amplification of kanamycin resistant gene |
| nptII-R | TCAGAAGAACTCGTCAAGAAG | Amplification of kanamycin resistant gene |
| gyrase-PF | AACCTATATTTGCTAGGGAG | Amplification of gyraeB promoter region |
| gyrase-PR | TACTAGAGGAATCATAAGCC | Amplification of gyraeB promoter region |
| ohr-1 up | ACCAATTTTTTTACAAAATT GACTTGTTAAAATAATTAAGTCGTG |
Amplification of ohr-c promoter region |
| ohr-1 bo | CACGACTTAATTATTTTAAC AAGTCAATTTTGTAAAAAAATTGGT |
Amplification of ohr-c promoter region |
| ohr-2 up | TACGATATATTTGCATTCAA TTTAATTTAGGAATAAAAAG |
Amplification of ohr promoter region |
| ohr-2 bo | CTTTTTATTCCTAAATTAAA TTGAATGCAAATATATCGTA |
Amplification of ohr-c promoter region |
| Ohr-3_PF | TTGACTTGTTAAAATAATTAAGTCG | Amplification of ohr-c promoter region |
| ohr_PF | ACCAATTTTTTTACAAAATT | Amplification of ohr-c promoter region |
| ohr_PR | CTTTTTATTCCTAAATTAAA | Amplification of ohr-C promoter region |
| pMAC_ohr_PF | GGCCGATATAAGCTCTATTT | Amplification of ohr-p promoter region |
| pMAC_ohr_PR | TGTATATTACCTTGCTTAAT | Amplification of ohr-p promoter region |
| ohrR _kpnI_flag-R | GGGGTACCCTTGTCGTCATCGTCTTTG TAGTCTTCAGTCACAATATTAAACG | Construction of pOhrR-C-Pohr-ohr-C |
| ohrR F | CGGGATCCATGGACCAAGACTGTCAAA A | Specific primer of chromosomal ohrR-C |
| ohrR R | GGGGTACCTTATTTAGCTATATTTGTAC | Specific primer of chromosomal ohrR-C |
| ohrR qF | TGGACCAAGACTGTCAAAATC | qRT-PCR primer for ohrR-C |
| ohrR qR | TCCCACAACACCAACATCAC | qRT-PCR primer for ohrR-C |
| ohr qF-2 | AAGCAACAGGTGGCCGTGAT | qRT-PCR and specific primer of chromosomal ohr-C |
| ohr qR-2 | ACCGACTTCACCTTCAACATACGC | qRT-PCR and specific primer of chromosomal ohr-C |
| ABpMAC_ohrR_F | TGTCCAAGAATCAGCTTTGCT | qRT-PCR and specific primer of ohrR-p |
| ABpMAC_ohrR_R | TTTGTCCAAGATCACCCACA | qRT-PCR and specific primer of ohrR-p |
| ABpMAC_ohr_F | AAAGGTGATGCAACGAATCC | qRT-PCR and specific primer of ohr-p |
| ABpMAC_ohr_R | GTCAAGGCAAATCCACCATT | qRT-PCR and specific primer of ohr-p |
| gyrB-F | GGCGGTTTATCTGAGTTTGT | qRT-PCR primer for gyrase gene of A. baumannii |
| gyrB-R | TTTGTGGAATGTTGTTTGTG | qRT-PCR primer for gyrase gene of A. baumannii |
2.3. Minimum Inhibition Concentration
The resistance of each strain to antibiotics was assessed by liquid minimum inhibition assay. Specifically, each antibiotic was inoculated into 96-well microtiter plates with 2-fold serial dilution. Each strain was cultured in 3 mL of LB medium at 37 °C overnight. Diluted overnight cultures with an OD600 of 0.1 were then inoculated into 96-well microtiter plates. Optical density was determined after overnight culture. The MIC was defined as the lowest concentration of antibiotics that inhibit bacterial proliferation [24].
2.4. Disk Diffusion Assay
The resistance of each strain to organic hydroperoxide was evaluated by disk diffusion assy. In brief, each strain was cultured in 3 mL of LB medium at 37 °C overnight. Diluted overnight cultures with an OD600 of 0.1 were then densely inoculated onto LB agar plates using a cotton swab, in order to ensure the confluent growth of each strain. Whatman filter paper discs imbued with 5 μL of 300 μM tBHP were aseptically applied to the surface of the agar plate, and the zones of growth inhibition were measured after incubation for 16 h at 37 °C.
2.5. RNA Extraction and qRT-PCR
Each strain was cultured in LB medium with agitation at 37 °C overnight. Bacteria were sub-cultured in a fresh 50 mL medium for three hours. Samples were collected for the non-treatment group and mixed with 0.1 volume of fixing solution (5% acid phenol, 95% ethanol). After centrifugation by 17,000× g at 4 °C, the supernatant was discarded, and the remaining cell pellets were stored at −80 °C for RNA extraction.
Cell pellets were thawed on ice and resuspended in 1 mL of NucleoZOL (MACHEREY-NAGEL; Düren, Germany), mixed thoroughly with 400 μL of diethyl pyrocarbonate (DEPC)-treated H2O, and then incubated at room temperature for 15 min. The supernatant was recovered after centrifugation at 17,000× g at 4 °C for 20 min, then fully mixed with 5 μL of 100% 4-bromoanisole and incubated at room temperature for 10 min. Excess protein was removed by centrifugation at 17,000× g at 4 °C for 20 min. The RNA suspension was subsequently mixed with an equal volume of isopropanol for 15 min for RNA precipitation. The RNA pellet was washed twice with ice-cold 75% ethanol and resuspended in 30 μL of DEPC-treated H2O for the following analysis.
A total of 2 μg of RNA was subsequently used to prepare cDNA after a Nanodrop spectrophotometer (NanoDrop 2000C, Thermo Fisher Scientific; Waltham, MA, USA) analysis was conducted to determine RNA concentrations. The qRT-PCR mixture contained 10× reaction buffer, 200 U of MMLV high performance reverse transcriptase (Epicentre; Madison, WI, USA), 100 mM of dithiothreitol (DTT), 2.5 mM dNTP, and 1 nM of hexamer. The reaction was conducted in a LightCycler® 480 (Roche; Basel, Switzerland). Gene-specific primers used to determine the presence and expression levels of the ohr-C, ohrR- C, ohr-p, and ohrR-p genes are listed in Table 2. The gyrase gene served as an internal control, and was amplified by PCR using the specific primers, gyrF and gyrR (Table 2) [25].
2.6. OhrR Overexpression and Purification
The PCR product of the ohrR-C gene was amplified using ohrR-c-F and ohrR-c-R primers, and cloned into the BamHI and KpnI sites of plasmid pQE80L (Qiagen, Hilden, Germany) to generate pOhrR-C. To express OhrR-C protein, E. coli DH5α(pOhrR-C) was expanded at 37 °C by inoculating a 0.5-mL overnight culture into 50 mL of LB medium containing ampicillin. Incubation was continued at 37 °C until the culture reached an OD600 of 0.6. Protein expression was induced by adding IPTG to achieve a final concentration of 1 mM. After incubation overnight at 37 °C, the cells were harvested by centrifugation at 4000× g for 15 min. The cells were then stored at −80 °C until use.
OhrR-C protein purification was performed by a method described elsewhere [26]. Frozen cells overexpressing OhrR-C were suspended in 30 mL of 1× binding buffer containing 5 mM imidazole, 0.5 mM NaCl, and 20 mM Tris-HCl (pH 8.0) and subjected to 3 cycles of freezing and thawing at –80 °C and room temperature, respectively. The thawed cells were disrupted by high pressure at 4 °C using a low-temperature cell disruptor, JNBIO JN-O2C (Guangzhou, China). The cell extract was separated from the cell debris by centrifugation at 17,000× g for 30 min at 4 °C (Avanti J-25 Centrifuge, JA25.5 rotor, Beckman Coulter; Brea, CA, USA). The OhrR-C containing cell extract was then purified by Ni-affinity chromatography (Novagen; Madison, WI, USA). Purified fractions were analyzed via 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie Brilliant Blue G-250.
2.7. EMSA
The DNA fragment of the ohr-C promoter and ohr-p promoter were amplified by PCR, using gene-specific primers (Table 2). EMSA was performed as previously described, with some modifications [26]. The reaction mixtures for the binding assays contained different concentrations of OhrR-C protein. The binding reactions were performed in binding reaction buffer (20 mM Tris-HCl (pH 7.5), 100 mM MgCl2, 150 mM KCl, 50 mM EDTA, 12.5% glycerol) supplemented with 250 μM DTT, 830 ng/mL poly(dI-dC) and 250 ng/mL bovine serum albumin (BSA). The reaction mixtures were incubated for 30 min at room temperature before adding 50 nM of DNA fragments. Samples were incubated for another 30 min, they were mixed with an equal volume of sample buffer (62 mM Tris-HCl, 0.01% bromophenol blue, and 10% glycerol) and loaded onto 8% non-denaturing polyacrylamide gels containing 0.5× Tris-borate-EDTA buffer. Electrophoresis was performed at 100 V for 1–1.5 h at 4 °C. Images were captured after the gels were stained with SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific; Waltham, MA, USA) for 20 min at room temperature, and imaged using the Ultra Slim LED Illuminator (MAESTROGEN; Hsinchu City, Taiwan). A DNA fragment of the gyrase gene promoter from A. baumannii was amplified by PCR, using the specific primers gyr-pF and gyr-pR (Table 2), and was used as a negative (non-specific) control.
2.8. G. Mellonella Experiments
A virulence comparison was performed for A. baumannii ATCC 19606, ATCC 17978 and the ohr-C or ohrR-C mutants of each strain. All the procedures were performed as previously described, with minor modifications [27]. Overnight cultures of each strain were washed twice with PBS (0.137 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), then diluted in PBS. Ten G. mellonella larvae were selected for the same total weight and were kept in Petri dishes without food prior to infection. Each larva was infected with 5 × 106 colony-forming units (CFU) of each strain. Bacteria in 10-mL aliquots were injected into the hemocoel of each larva via the last left proleg by a Hamilton syringe. Infected larvae were incubated at 37 °C and scored for survival (alive/dead) every 24 h. Larvae were also scored for melanization over 96 h, according to a previously described scoring method [28].
3. Results
3.1. Identification of Two Paralogous ohr-ohrR Genes in A. baumannii ATCC 19606
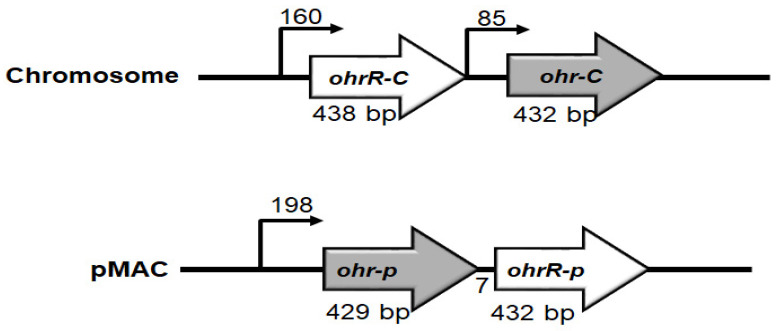
Previous research showed that the pMAC plasmid in A. baumannii ATCC 19606 contains 11 ORFs, with ORF8 and ORF9 respectively annotated as putative OhrA and OhrR enzymes that can contribute to tert-butyl hydroperoxide (tBHP) resistance [20]. Using an in silico analysis, we proceeded to identify another set of putative Ohr-C (DJ41_1043) and OhrR-C (DJ41_1042) proteins on the chromosomal DNA of A. baumannii ATCC 19606, and these shared 44.4% and 41.3% identity with ORF8 and ORF9 on pMAC. Interestingly, the chromosomal ohrR-C gene is located upstream of the chromosomal ohr-C gene, and the genetic architecture suggests that each gene possesses its own promoter regions, and can be transcribed independently in the same direction (Figure 1); however, on pMAC, ORF8 is located upstream from ORF9, and considering that the start codon of ORF9 is located only 7 bp downstream of the stop codon of ORF8, it is possible that both genes may be transcribed on the same mRNA (Figure 1).
Figure 1.
Genetic organization of ohr-ohrR genes on chromosomal DNA and pMAC. Numbers above bent arrows indicate the distance in base pairs between two genes.
We proceeded to label ORF8 as Ohr-p and ORF9 as OhrR-p, in order to distinguish them from their chromosomal counterparts. We found that Ohr-p shared 44.4% amino acid similarity with chromosomal Ohr (Ohr-C) and 41.5% similarity with OhrA of Xanthomonas campestris pv. phaseoli. Ohr-C was found to share 37.1% similarity with Ohr of E. coli (Ohr-Ec), and 56.3% similarity with OhrA of X. campestris (Ohr-Xc) (Figure S1A). Chromosomal OhrR (OhrR-C) shared 41.3% amino acid similarity with OhrR-p, and 51.5%, 48.6%, and 45.9% similarity with OhrR of X. campestris (OhrR-Xc), Pseudomonas aeruginosa (OhrR-Pa), and B. subtilis (OhrR-Bs), respectively (Figure S1B). Secondary structure analysis of OhrR-C revealed six α-helices and two β-sheets, consistent with the typical structure of a MarR protein (Figure S1B).
3.2. Role of ohr-p and ohrR-p in tBHP Resistance Is Limited
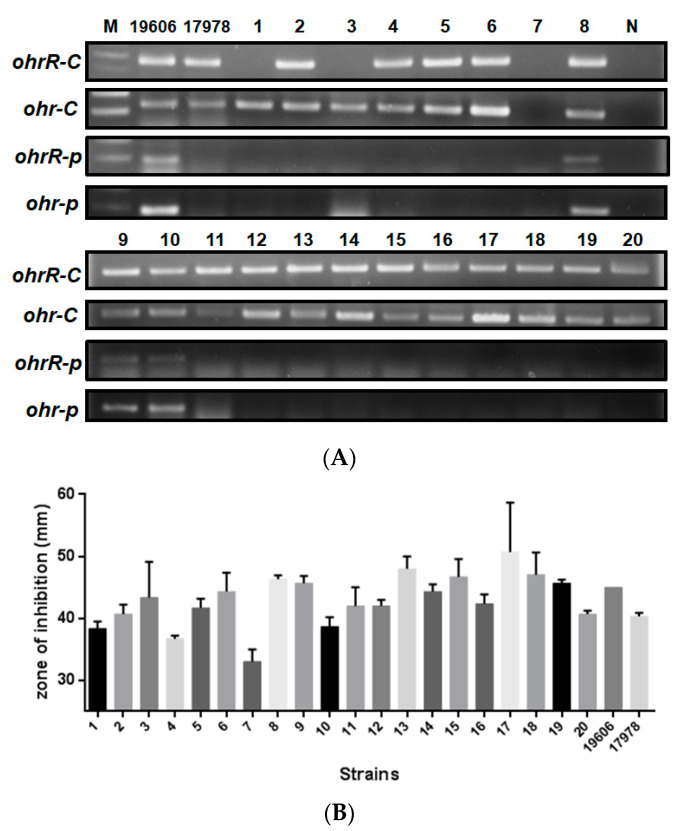
Several methods have been applied by our lab (data not shown) and others [20] to evaluate the role of pMAC in organic peroxide resistance by plasmid curing, but these efforts were not successful since we can find pMAC in bacteria after curing process. To screen for the presence of chromosomal ohrR-C-ohr-C and ohr-p-ohrR-p genes on pMAC in different Acinetobacter strains for which genomic sequences are not available but identified by 16S rRNA gene amplification and sequences analysis. Gene-specific primers were designed for gene amplification (Table 2). Subsequent PCR results revealed that most Acinetobacter spp. had chromosomal ohrR-C-ohr-C genes, except A. soli (Figure 2A, lane 7). However, ohr-p-ohrR-p genes on pMAC were only found in A. baumannii ATCC 19606 and A. baumannii ATCC 15308 (Figure 2A, lanes 8, 9, 10). Growth conditions in the presence of 135 μg of tBHP were assessed for each of these strains, using the disk diffusion assay, but no significant difference in tBHP resistance was observed between strains with or without the pMAC plasmid (Figure 2B). Different concentrations of tBHP and cumene hydroperoxide were also tested, but the results were similar to those observed with 135 μg of tBHP (data not shown). This suggests that the ohr-p-ohrR-p genes on pMAC have a limited role in tBHP resistance, and we therefore focused on elucidating the functional role of chromosomal ohrR-C-ohr-C genes in subsequent experiments.
Figure 2.
Presence of ohrR-ohr genes and tBHP resistance of 20 Acinetobacter isolates. (A) PCR products amplified by primers specific to chromosomal or pMAC ohrR-ohr genes. (B) Inhibition zones for each isolate following treatment with 135 μg of tBHP in a disk diffusion assay. Numbers 1–20 indicate the strain numbers of Acinetobacter isolates. M is a molecular weight marker. N is a negative control using ddH2O as a template. 19606 and 17978 indicate A. baumannii ATCC 19606 and ATCC 17978, respectively. These data were obtained from three independent experiments.
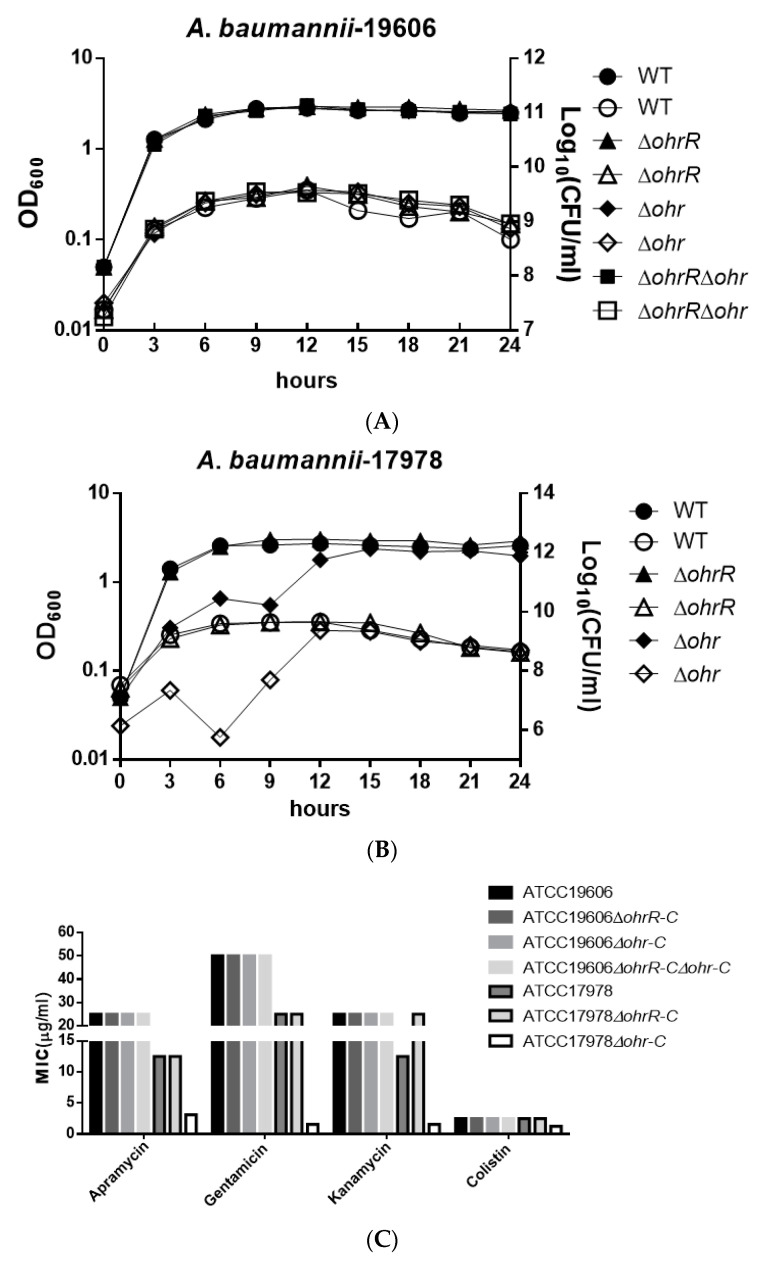
3.3. Chromosomal ohr-C Plays an Important Role in Bacterial Proliferation
To understand the importance of chromosomal ohrR-C-ohr-C genes for A. baumannii ATCC 19606, we constructed single and double ohrR-C and ohr-C mutants by markerless allelic exchange. Bacteria used in these experiments were all cultured in lysogeny broth (LB), and the optical density and viable count of cultures was determined every three hours over a 24-h period, in order to obtain growth curves for each strain. Results revealed no significant difference in growth curves for wild-type and mutant A. baumannii ATCC 19606 (Figure 3A). In addition, ohrR-C and ohr-C single mutants were also generated in A. baumannii ATCC 17978, and growth curves showed that the ohr-C mutant underwent a delay in entering the exponential growth phase during the first 12 h, but eventually reached comparable levels of growth with wild-type after entering the stationary phase (Figure 3B). These results suggest an important role for chromosomal ohr-C in overcoming organic peroxide stress during the exponential growth phase for A. baumannii ATCC 17978. However, we did not observe similar results with ohr-C single and ohrR-C-ohr-C double mutants of A. baumannii ATCC 19606 cultured in LB medium for 24 h, implying that the ohr-p-ohrR-p genes on pMAC may act to reduce organic peroxide stress (Figure 3A).
Figure 3.
Growth curve of different A. baumannii strains in LB medium and minimum inhibition concentration (MIC) of different strains treated by different antibiotics. Growth curve of (A) A. baumannii ATCC 19606 and (B) A. baumannii ATCC 17978 wild-type and mutant strains. The Y-axis at left represents OD600, while the Y-axis at right represents viable cell count. Filled symbols represent the optical density and empty symbols indicate the viable cell counts of each strain. These data were obtained from three independent experiments. (C) MIC of different strains.
Antibiotic testing was used to determine the minimum inhibition concentration of different strains, and the results showed that there were no significant differences between A. baumannii ATCC 19606 strains. However, loss of ohrR-C increased kanamycin resistance in A. baumannii ATCC 17978, while the ohr-C mutant became more susceptible to antibiotics tested (Figure 3C). This suggests that the ohr-p-ohrR-p genes on pMAC may enhance resistance by mitigating organic peroxide stress generated during antibiotic treatment.
3.4. Gene Expression of Chromosomal ohr-C Can Be Induced by tBHP
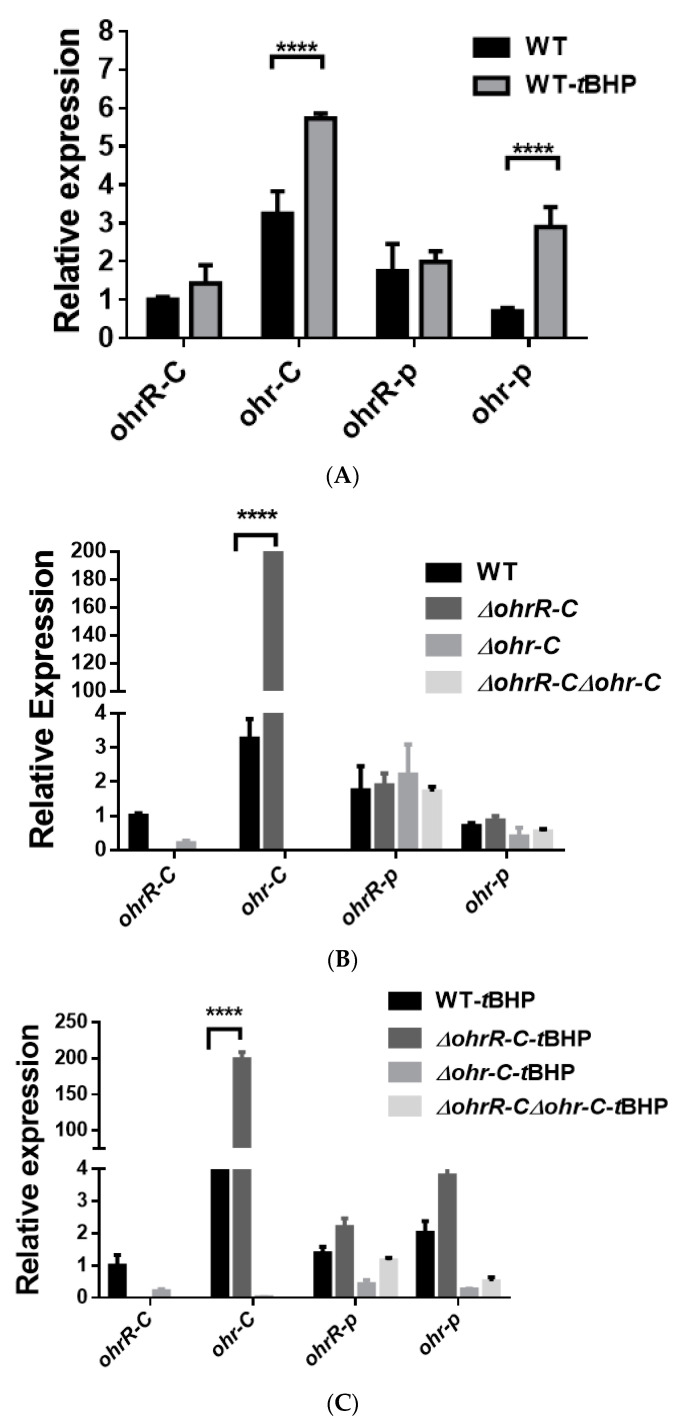
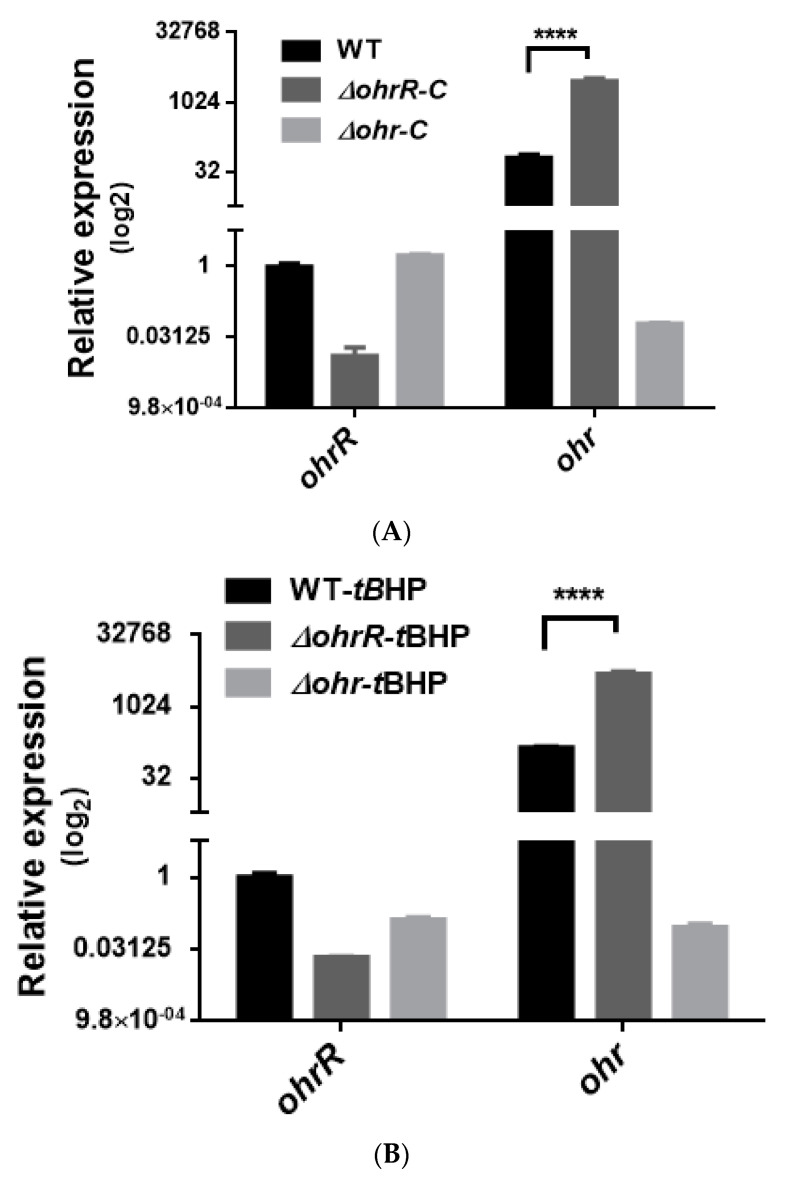
To assess gene expression levels for chromosomal ohrR-C-ohr-C and pMAC ohr-p-ohrR-p genes, quantitative reverse transcription PCR (qRT-PCR) was conducted to analyze gene expression in bacteria with or without tBHP treatment. A. baumannii wild type, ohrR-C, ohr-C single and double mutants were sub-cultured in LB medium at an initial OD600 of 0.05 for 3 h, and then treated with 200 μM of tBHP for 20 min prior to acid phenol fixation. Samples were stored at −80 °C if RNA extraction was not performed immediately. Gene-specific primers (Table 2) were used to perform qRT-PCR, and the results showed that in the presence of tBHP, ohr-C and ohr-p expression respectively increased by 1.76-fold and 4.12-fold over untreated controls (Figure 4A). In ∆ohrR-C mutants, ohr-C expression increased more than 100-fold over wild-type. Both ohrR-p and ohr-p did not demonstrate significantly elevated gene expression in all mutants (Figure 4B). In ∆ohrR-C mutants, tBHP induced 50-fold higher chromosomal ohr-C expression over wild-type strains, indicating that OhrR-C has a repressive effect on ohr-C expression, but little effect on ohr-p expression (Figure 4C). As for the expression of ohr-p-ohrR-p genes on pMAC in the presence of tBHP, neither ohr-p nor ohrR-p gene expression was strongly induced in ohr-C single mutants or ohrR-C-ohr-C double mutants (Figure 4C). These results indicate that even with the loss of ohrR-C-ohr-C, the ohr-p genes on pMAC will not be upregulated to overcome the effects of tBHP treatment. To further clarify the role of chromosomal ohrR-C-ohr-C, mutants were generated in a strain without pMAC (A. baumannii ATCC 17978), and qRT-PCR was conducted to assess gene expression with or without tBHP treatment. The results were similar to those observed for A. baumannii ATCC 19606, in which ohr-C expression was induced by tBHP (Figure 5A) and repressed by OhrR-C (Figure 5B).
Figure 4.
Transcriptional expression of chromosomal ohrR-C-ohr-C and pMAC ohr-p-ohrR-p genes in different strains of A. baumannii ATCC 19606, as quantified by qRT-PCR. (A) Relative expression of chromosomal ohrR-C-ohr-C and pMAC ohr-p-ohrR-p genes in wild-type strains cultured in the presence (WT-tBHP) or absence (WT) of 200 μM of tBHP for 20 min. Relative expression of chromosomal ohrR-C-ohr-C and pMAC ohr-p-ohrR-p genes in ohrR-C mutant (∆ohrR-C), ohr-C mutant (∆ohr-C), and ohrR-C-ohr-C double mutant (∆ohrR-C∆ohr-C) strains compared with wild-type strains in the absence (B) and presence (C) of tBHP. These data were obtained from three independent experiments. Each sample was normalized using gyrase gene expression as an internal control. The expression of ohrR-C gene in wild type was determined as 1 for comparison. Multiple-way ANOVA was used to determine the significance of each phase. **** indicates p < 0.001.
Figure 5.
Transcriptional expression of chromosomal ohrR-C-ohr-C genes in different strains of A. baumannii ATCC 17978, as quantified by qRT-PCR. Relative expression of chromosomal ohrR-C-ohr-C genes in ohrR-C mutant (∆ohrR-C) and ohr-C mutant (∆ohr-C) strains compared with wild-type strains in the absence (A) and presence of 200 μM of tBHP for 20 min (B). Each sample was normalized using gyrase gene expression as an internal control. The expression of ohrR-C gene in wild type was determined as 1 for comparison. These data were obtained from three independent experiments. Multiple-way ANOVA was used to determine the significance of each phase. **** indicates p < 0.0001.
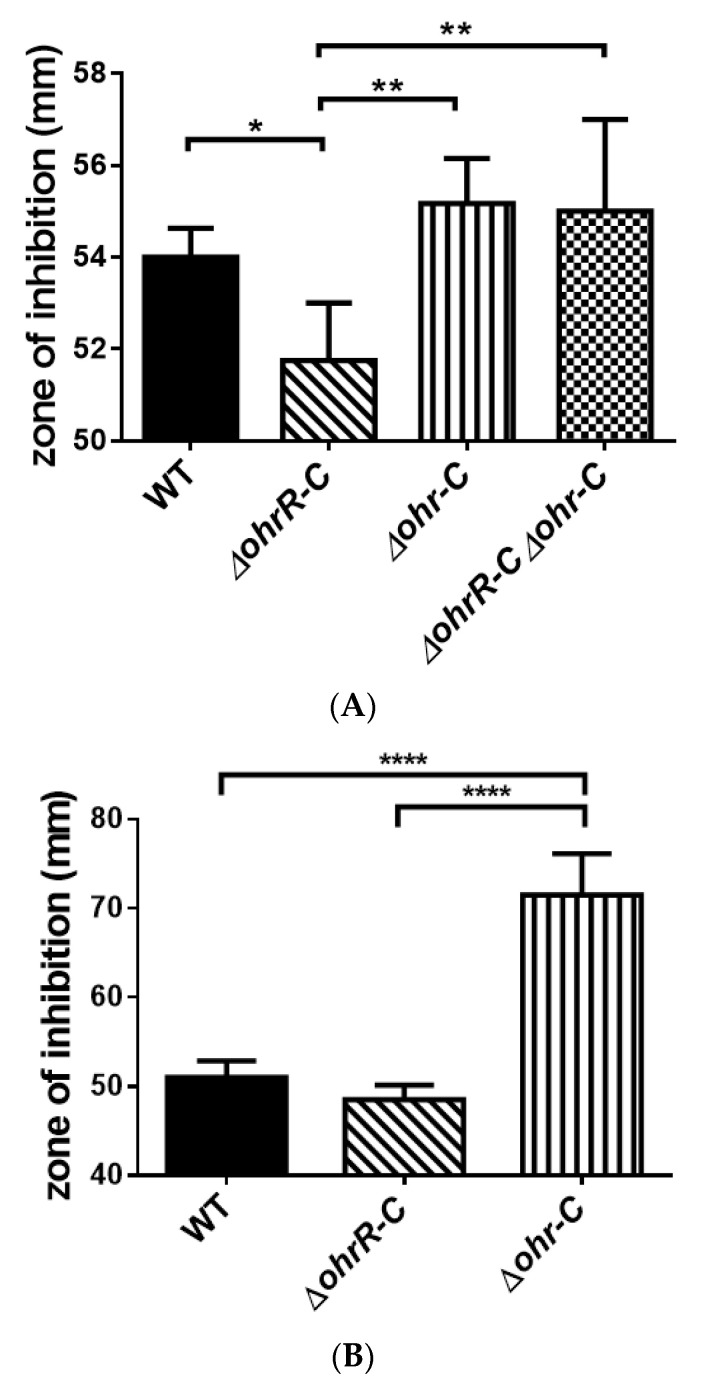
A disk diffusion assay was conducted to assess tBHP resistance for each strain of A. baumannii 19606. The zone of inhibition was diminished in ohrR-C mutants as compared with wild-type (Figure 6A), and this observation concurs with the qRT-PCR results. A previous study has shown that the ohr-p-ohrR-p genes on pMAC contribute to organic peroxide resistance in A. baumannii 19606 [20], and to better ascertain the role of the pMAC genes, a chromosomal ohrR-C-ohr-C double mutant was generated by markerless allelic exchange, to create strains with ohr-p-ohrR-p only. We found that pMAC alone did not contribute to tBHP resistance (Figure 6A). Moreover, a disk diffusion assay conducted with ohrR-C and ohr-C single mutants of A. baumannii ATCC 17978 found that the ohr-C single mutant had reduced resistance against tBHP (Figure 6B). These results further confirm the importance of ohr-C to tBHP resistance.
Figure 6.
Disk diffusion assay for different strains of A. baumannii. (A) ATCC 19606 and (B) ATCC 17978 disk diffusion assay results. The zone of inhibition was determined after bacteria were treated with 135 μg of tBHP for 12 h. These data were obtained from three independent experiments. Multiple-way ANOVA was used to determine the significance of each phase. * indicates p < 0.05; ** indicates p < 0.01; **** indicates p < 0.0001.
3.5. Binding of OhrR-C to the Promoter of the ohr-C and ohr-p Genes
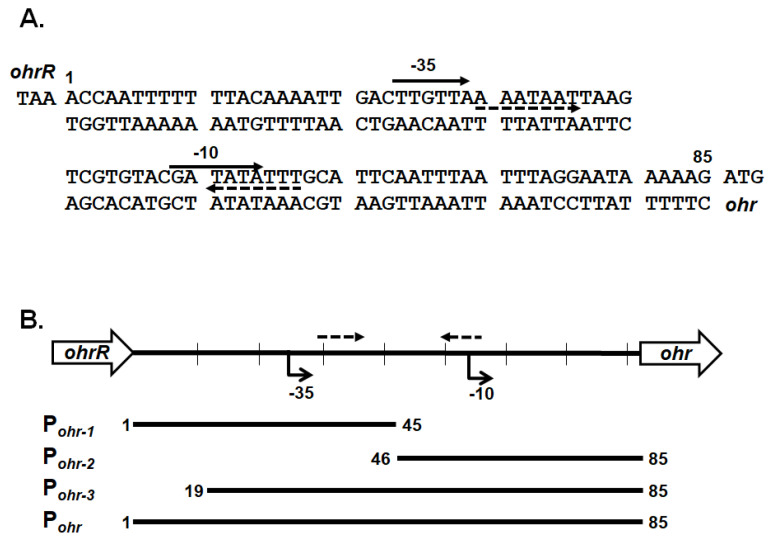
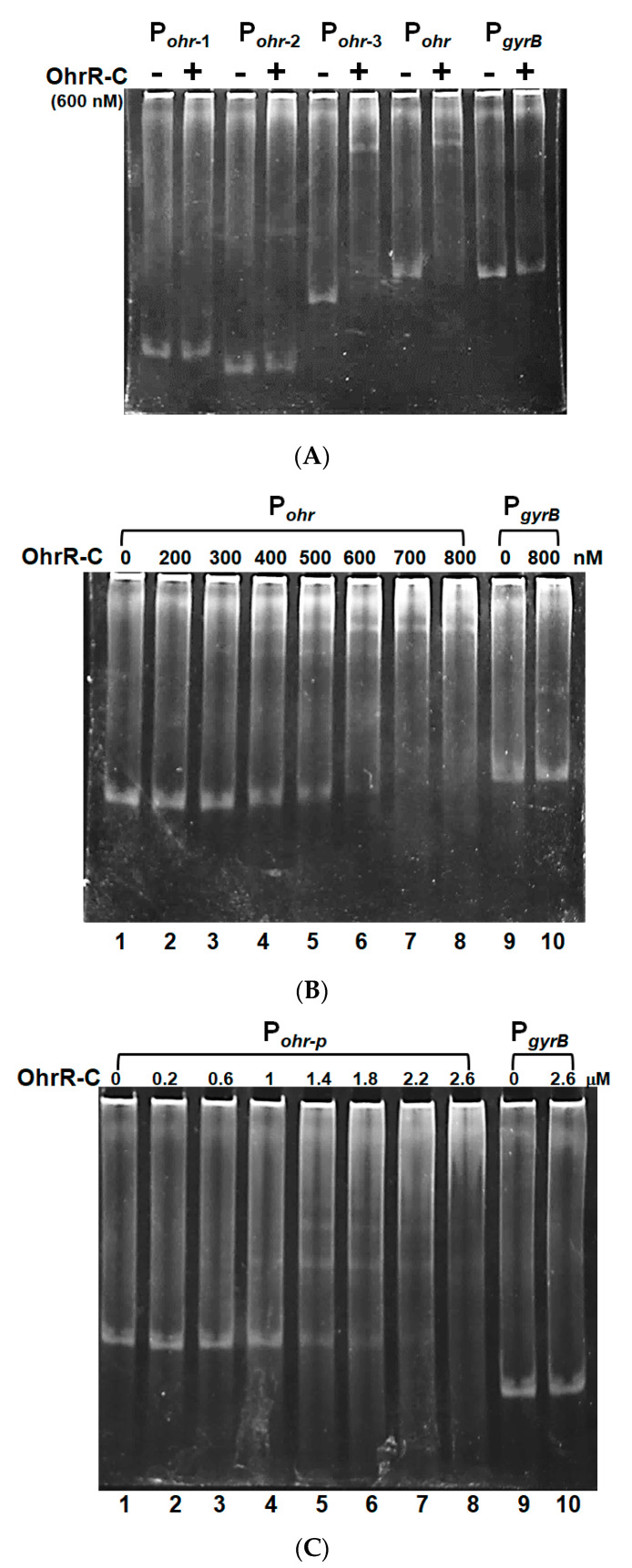
To confirm that chromosomal OhrR-C suppressed ohr-C gene expression, EMSA was conducted to ascertain the direct binding of Ohr-C to the promoter region of the ohr-C gene. The putative OhrR-C box of A. baumannii, AAATXAT-14-ATXTATTT (Figure 7) contains an AT-rich motif that has been found in most of the DNA-binding sequences of OhrR-C homologs [7,13,29,30,31]. The entire 85-bp putative promoter region of the ohr-C gene was split into different fragments (Figure 7). Pohr-1 and Pohr-2 divided this region into two parts, each covering half of the promoter region. Pohr-3 is 18-bp shorter than Pohr (Figure 8A). We incubated 50 nM of DNA probes with recombinant OhrR-C purified from an E. coli overexpression strain. The results showed that only Pohr-3 and Pohr could be bound by 600 nM of OhrR-C, with a mobility shift in electrophoresis observed (Figure 8A). We then used different concentrations of OhrR-C to interact with Pohr. Results revealed that a mobility shift could be observed with just 400 nM of OhrR-C mixed with DNA (Figure 8B, lane 4). Increasing OhrR-C concentration to 600 nM led to a significant mobility shift (Figure 8B, lane 6). The DNA/protein complexes became trapped in electrophoresis wells when the amount of OhrR-C exceeded 800 nM (Figure 8B, lane 8).
Figure 7.
Genetic organization and intergenic sequences for chromosomal ohrR-C and ohr-C genes. (A) Intergenic sequences between the chromosomal ohrR-C and ohr-C genes. The numbers indicate positions relative to the stop codon of ohrR. Arrows with dashed lines indicate the putative inverted repeats. (B) Genetic organization and relative position of probes for EMSA.
Figure 8.
OhrR-C binds specifically to the chromosomal ohrR-ohr intergenic region and the ohr-p promoter region on pMAC. (A) OhrR-C (600 nM) incubated with different probes (50 nM). (B) Pohr incubated with different concentrations of OhrR-C (0–800 nM). PgyrB is a gyrase gene promoter that was incubated with or without 800 nM of OhrR-C, to serve as a control. (C) Pohr-p interaction with different concentrations of OhrR-C (0–2600 nM). PgyrB is a gyrase gene promoter that was also incubated with or without 2600 nM of OhrR-C, to serve as a control. Concentrations of OhrR-C are indicated in the top row.
Recombinant chromosomal OhrR-C was also used to test binding capability with the promoter region of ohr-p on pMAC. A mobility shift was observed when 1 μM of OhrR-C was mixed with DNA (Figure 8C, lane 4). Increasing OhrR-C concentration to 1.4 μM led to a significant mobility shift (Figure 8C, lane 5), and the DNA/protein complexes became trapped in the well when the amount of OhrR-C exceeded 2.6 μM (Figure 8C, lane 8). Taken together, recombinant chromosomal OhrR-C showed less affinity for the promoter region of ohr-p on pMAC, compared with chromosomal ohr-C.
3.6. OhrR-C Suppressed Ohr-C Expression in E. coli
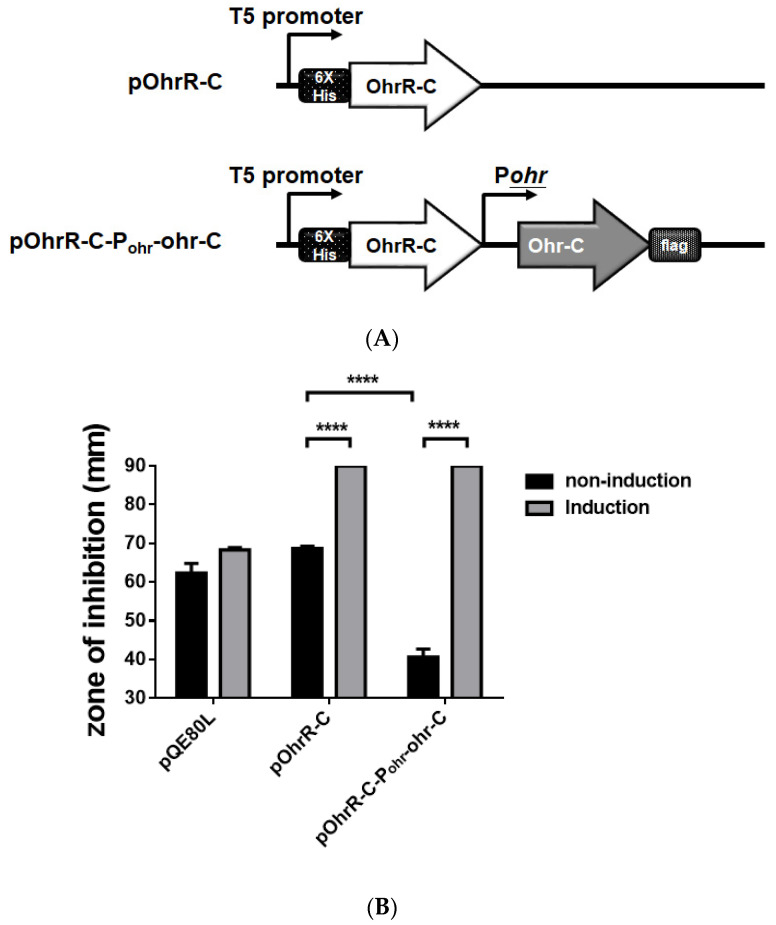
We sought to examine the role of ohrR-C-ohr-C genes by cloning them into a pQE80L plasmid, and then transformed the plasmid into E. coli to ascertain whether gain of tBHP resistance functions could be observed in the transformants. We therefore cloned ohrR-C into pQE80L under the isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible T5 promoter with N-terminal 6-His fusion, to generate the pQE80L_OhrR-C plasmid. Subsequently, ohr-C with its endogenous promoter was constructed downstream of ohrR-C, with a FLAG-tagged C-terminal, to generate the pQE80L_OhrR-C_Ohr-C plasmid (Figure 9A). Each plasmid was transformed into the E. coli DH5α strain for tBHP resistance tests. E. coli transformed with an empty pQE80L showed no difference in tBHP resistant properties, but in transformants with OhrR-C induced by 1 mM IPTG, increased tBHP sensitivity was observed, suggesting that OhrR-C may also suppress the tBHP resistance genes in E. coli. However, transformants with the ohr-C gene had reduced inhibition zones, indicative of increased tBHP resistance; but following the induction of OhrR-C expression, Ohr-C was subsequently repressed, and increased tBHP sensitivity was observed (Figure 9B). From this gain-of-function experiment, it was observed that OhrR-C was able to regulate the expression of the ohr-C gene derived from A. baumannii.
Figure 9.
Genetic organization and disk diffusion assay for the gain of function assay in E. coli. (A) Genetic organization of OhrR-C expressing strains and OhrR-C-Ohr-C expressing strains. (B) Disk diffusion assay. Different E. coli strains without (black bar) or with (gray bar) 1 mM IPTG induction. Cultures were treated with 135 μg tBHP for 12 h. Multiple-way ANOVA was used to determine the significance of each phase. **** indicates p < 0.0001.
3.7. The ohrR-C Mutant Demonstrated Decreased Virulence in Galleria mellonella
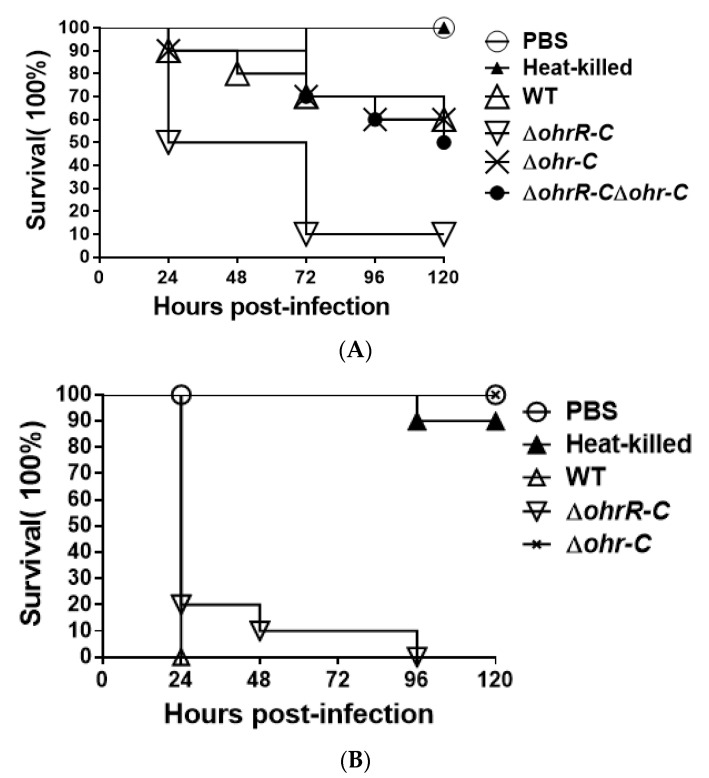
To assess the overall virulence of ohr-C and ohrR-C mutants compared with wild-type, G. mellonella larvae (n = 10) were respectively incubated with 5 × 106 colony-forming units (CFU) of each A. baumannii strain. We observed that ohrR-C mutants displayed significantly greater virulence in G. mellonella as compared to wild-type, ohr-C single mutants, and ohrR-C-ohr-C double mutants, with only 10% larvae survival at 72 h compared to 70% survival for the other three strains. However, ohr-C single mutants and ohrR-C-ohr-C double mutants retained 60% virulence in G. mellonella at 96 h, indicating that the ohr-p-ohrR-p genes on pMAC may play a role in virulence for G. mellonella (Figure 10A). A survival assay was performed with A. baumannii ATCC 17978 strains without pMAC, to better understand the importance of chromosomal ohrR-C and ohr-C for virulence against G. mellonella. The results showed that, while wild-type strains killed all infected larvae within 24 h, the ohr-C single mutants had significantly reduced virulence, even at 96 h after infection (Figure 10B). The results suggest that the chromosomal ohr-C gene plays an important role in A. baumannii virulence toward G. mellonella, and this may have implications for other hosts as well.
Figure 10.
G. mellonella survival curve following infection with A. baumannii wild-type and mutants. Larvae were infected with 5 × 106 CFU of wild-type or mutant (A) ATCC 19606 or (B) ATCC 17978 strains. PBS was used as the buffer. Heat-killed indicates wild-type bacteria treated at 100 °C for 10 min. WT, wild type bacteria; ΔohrR-C, ohrR-C mutant; Δohr-C, ohr-C mutant; ΔohrR-CΔohr-C, ohrR-C-ohr-C double mutant. The curve represents a single experiment performed with 10 larvae.
4. Discussion
The Ohr protein is a thiol-dependent peroxidase that plays a major role in the response of bacteria against organic peroxide [29]. Most bacteria harbor at least one Ohr protein [30,31]. In this study, both chromosomal and plasmid ohr genes were identified in A. baumannii 19606. The promoter regions of both ohr genes can be bound by OhrR, albeit with varying affinity. Other examples of bacteria with more than one ohr gene have previously been identified. In B. subtilis, two homologs of Ohr, OhrA and OhrB, have been identified and studied [7]. OhrR is capable of repressing ohrA expression, while ohrB is unaffected [7]. However, in P. aeruginosa, homologous OhrR and OspR have been shown to act as regulatory switches in response to oxidative stress [32]. OspR regulates gpx, which encodes a glutathione peroxidase. In an ospR mutant, in which gpx is depressed, the induction of ohr expression by oxidative stress is reduced. Similarly, in an ohrR mutant, where ohr is derepressed, organic hydroperoxide induction of gpx is reduced [32].
Several thousand strains of A. baumannii have been sequenced, and some of these have been reported to harbor plasmids [20]. Genomic sequence analysis revealed that pM1301-3 of Acinetobacter sp. M131 and a plasmid in A. baumannii ab736 had comparable length and shared high sequence similarity with pMAC of A. baumannii ATCC 19606. All of these plasmids contained ohr-p-ohrR-p genes. Multi-sequence alignment also revealed that ohr-p-ohrR-p genes can be found in pALWS1.1 of Acinetobacter lwoffii VS15 and p1AsACE of Acinetobacter schindleri ACE. These observations suggest that A. baumannii may gain these genes through horizontal gene transfer. Based on our observation of growth curves for the ohr-C mutant of A. baumannii ATCC 17978, chromosomal ohr-C not only plays a role in generating resistance to OHPs but is also associated with rapid bacterial growth (Figure 3B). However, this was not observed for the ohr-C single mutant and ohrR-C-ohr-C double mutant of A. baumannii ATCC 19606 (Figure 3A), and this may be due to the complementary effect of the ohr-p-ohrR-p genes on pMAC.
Both A. baumannii ATCC 19606 [33] and A. baumannii ATCC 17978 [34] harbor resistance to multiple antibiotics, but with different resistance profiles. For example, previous research [34] and our unpublished data demonstrate that A. baumannii ATCC 19606 has higher minimal inhibitory concentrations (MIC) with tigecycline (2 mg/L), imipenem (2 mg/L), and colistin (1 mg/L), as compared to A. baumannii ATCC 17978 [34]. Some bactericidal antibiotics operate through the production of highly deleterious hydroxyl radicals in bacteria [35]. Surprisingly, lipoyl groups are essential for parasite survival in host cells [36,37] and for virulence [38]. Taken together, our findings suggest that A. baumannii ATCC 19606 may have gained pMAC via horizontal gene transfer, and this strain exhibits higher MIC to antibiotics not only because of the antibiotic resistance genes encoded on pMAC, but also because two sets of ohr genes are present in the chromosomal DNA and pMAC respectively. This can increase resistance to OHPs and hydroxyl radicals from bactericidal antibiotics, thereby increasing virulence. Future research into novel antibacterial therapies that target these ohr genes may therefore be warranted.
5. Conclusions
A. baumannii has gained increasing attention in recent years due to its role in nosocomial infections. The strains examined in this study all harbor resistance to multiple antibiotics, and considering that some antibiotics act by producing hydroxyl radicals that are highly deleterious, it is possible that aside from antibiotic resistance genes present on plasmids, the presence of ohr genes on plasmids and chromosomal DNA can contribute to bacterial viability and resistance as well. Therefore, a study of the ohr genes and their functions may have important implications for understanding bacterial resistance and preventing or addressing nosocomial infections.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/3/629/s1, Figure S1: Amino acid identity of Ohr and OhrR proteins.
Author Contributions
Conceived and designed the experiment, H.-Y.S. and G.-H.L.; performed the experiment, S.-J.C. and H.-Y.S.; analyzed the data, S.-J.C. and H.-Y.S.; wrote the paper, H.-Y.S. and G.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Grant No. TCMF-SP 110-02 from Buddhist Tzu Chi Medical Foundation to Guang-Huey Lin. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Data Availability Statement
All relevant data are within the manuscript and its supplementary information.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hurst J.K. What really happens in the neutrophil phagosome? Free Radic. Biol. Med. 2012;53:508–520. doi: 10.1016/j.freeradbiomed.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayala A., Munoz M.F., Arguelles S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang F.C. Antimicrobial actions of reactive oxygen species. mBio. 2011;2:e00141-11. doi: 10.1128/mBio.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cussiol J.R., Alegria T.G., Szweda L.I., Netto L.E. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 2010;285:21943–21950. doi: 10.1074/jbc.M110.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Si Y., Guo D., Deng S., Lu X., Zhu J., Rao B., Cao Y., Jiang G., Yu D., Zhong Z., et al. Ohr and OhrR Are Critical for Organic Peroxide Resistance and Symbiosis in Azorhizobium caulinodans ORS571. Genes. 2020;11:335. doi: 10.3390/genes11030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cussiol J.R., Alves S.V., de Oliveira M.A., Netto L.E. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 2003;278:11570–11578. doi: 10.1074/jbc.M300252200. [DOI] [PubMed] [Google Scholar]
- 7.Fuangthong M., Atichartpongkul S., Mongkolsuk S., Helmann J.D. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das K., Garnica O., Dhandayuthapani S. Utility of OhrR-Ohr system for the expression of recombinant proteins in mycobacteria and for the delivery of M. tuberculosis antigens to the phagosomal compartment. Tuberculosis. 2019;116S:S19–S27. doi: 10.1016/j.tube.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Will W.R., Fang F.C. The evolution of MarR family transcription factors as counter-silencers in regulatory networks. Curr. Opin. Microbiol. 2020;55:1–8. doi: 10.1016/j.mib.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuangthong M., Helmann J.D. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.W., Soonsanga S., Helmann J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warmbold B., Ronzheimer S., Freibert S.A., Seubert A., Hoffmann T., Bremer E. Two MarR-Type Repressors Balance Precursor Uptake and Glycine Betaine Synthesis in Bacillus subtilis to Provide Cytoprotection Against Sustained Osmotic Stress. Front. Microbiol. 2020;11:1700. doi: 10.3389/fmicb.2020.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pande A., Veale T.C., Grove A. Gene regulation by redox-sensitive Burkholderia thailandensis OhrR and its role in bacterial killing of Caenorhabditis elegans. Infect. Immun. :2018. doi: 10.1128/IAI.00322-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumann P., Doudoroff M., Stanier R.Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter) J. Bacteriol. 1968;95:1520–1541. doi: 10.1128/JB.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antunes L.C., Visca P., Towner K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 16.Bergogne-Berezin E., Decre D., Joly-Guillou M.L. Opportunistic nosocomial multiply resistant bacterial infections—their treatment and prevention. J. Antimicrob. Chemother. 1993;32(Suppl. A):39–47. doi: 10.1093/jac/32.suppl_A.39. [DOI] [PubMed] [Google Scholar]
- 17.Bergogne-Berezin E., Towner K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villegas M.V., Hartstein A.I. Acinetobacter outbreaks, 1977–2000. Infect. Control Hosp. Epidemiol. 2003;24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 19.Bouvet P.J., Grimont P.A. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 1986;36:228–240. [Google Scholar]
- 20.Dorsey C.W., Tomaras A.P., Actis L.A. Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid. 2006;56:112–123. doi: 10.1016/j.plasmid.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herigstad B., Hamilton M., Heersink J. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods. 2001;44:121–129. doi: 10.1016/S0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer A., Tauch A., Jager W., Kalinowski J., Thierbach G., Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 24.Beeton M.L., Chalker V.J., Maxwell N., Kotecha S., Spiller O.B. Concurrent titration and determination of antibiotic resistance in ureaplasma species with identification of novel point mutations in genes associated with resistance. Antimicrob. Agents. Chemother. 2009;53:2020–2027. doi: 10.1128/AAC.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin H.R., Shu H.Y., Lin G.H. Biological roles of indole-3-acetic acid in Acinetobacter baumannii. Microbiol. Res. 2018;216:30–39. doi: 10.1016/j.micres.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Shu H.Y., Lin L.C., Lin T.K., Chen H.P., Yang H.H., Peng K.C., Lin G.H. Transcriptional regulation of the iac locus from Acinetobacter baumannii by the phytohormone indole-3-acetic acid. Antonie Van Leeuwenhoek. 2015 doi: 10.1007/s10482-015-0417-3. [DOI] [PubMed] [Google Scholar]
- 27.Wang-Kan X., Blair J.M.A., Chirullo B., Betts J., La Ragione R.M., Ivens A., Ricci V., Opperman T.J., Piddock L.J.V. Lack of AcrB Efflux Function Confers Loss of Virulence on Salmonella enterica Serovar Typhimurium. mBio. 2017;8:e00968-17. doi: 10.1128/mBio.00968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai C.J., Loh J.M., Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuchue T., Tanboon W., Prapagdee B., Dubbs J.M., Vattanaviboon P., Mongkolsuk S. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J. Bacteriol. 2006;188:842–851. doi: 10.1128/JB.188.3.842-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh S.Y., Shin J.H., Roe J.H. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 2007;189:6284–6292. doi: 10.1128/JB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saikolappan S., Das K., Dhandayuthapani S. Inactivation of the organic hydroperoxide stress resistance regulator OhrR enhances resistance to oxidative stress and isoniazid in Mycobacterium smegmatis. J. Bacteriol. 2015;197:51–62. doi: 10.1128/JB.02252-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atichartpongkul S., Vattanaviboon P., Wisitkamol R., Jaroensuk J., Mongkolsuk S., Fuangthong M. Regulation of Organic Hydroperoxide Stress Response by Two OhrR Homologs in Pseudomonas aeruginosa. PLoS ONE. 2016;11:e0161982. doi: 10.1371/journal.pone.0161982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi. L., Li H., Zhang C., Liang B., Li J., Wang L., Du X., Liu X., Qiu S., Song H. Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando D.M., Khan I.U., Patidar R., Lapen D.R., Talbot G., Topp E., Kumar A. Isolation and Characterization of Acinetobacter baumannii Recovered from Campylobacter Selective Medium. Front. Microbiol. 2016;7:1871. doi: 10.3389/fmicb.2016.01871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 36.O’Riordan M., Moors M.A., Portnoy D.A. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- 37.Allary M., Lu J.Z., Zhu L., Prigge S.T. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 2007;63:1331–1344. doi: 10.1111/j.1365-2958.2007.05592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith A.W., Roche H., Trombe M.C., Briles D.E., Hakansson A. Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol. Microbiol. 2002;44:431–448. doi: 10.1046/j.1365-2958.2002.02883.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its supplementary information.