Abstract
Repeated positivity and reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) is a significant concern. Our study aimed to evaluate the clinical significance of repeatedly positive testing after coronavirus disease 2019 (COVID-19) recovery. We performed a systematic literature search following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline. With available individual patient data reporting on repeatedly SARS-CoV-2 positive (RSP) patients, case reports, and case series were included in this analysis. We performed a descriptive analysis of baseline characteristics of repeatedly positive cases. We assessed the cases according to the length of their polymerase chain reaction (PCR) negative interval between the two episodes. Risk factors for the severity of second episodes were evaluated. Overall, we included 123 patients with repeated positivity from 56 publications, with a mean repeated positivity length of 47.8 ± 29.9 days. Younger patients were predominant in the delayed (>90 days) recurrent positive group. Furthermore, comparing patients with RSP intervals of below 60 and above 60 days, we found that a more severe disease course can be expected if the repeated positivity interval is shorter. Severe and critical disease courses might predict future repeatedly positive severe and critical COVID-19 episodes. In conclusion, our results show that the second episode of SARS-CoV-2 positivity is more severe if it happens within 60 days after the first positive PCR. On the other hand, the second episode’s severity correlates with the first.
Keywords: SARS-CoV-2, COVID-19, new coronavirus, polymerase chain reaction, positive, repeated, case reports, systematic review
1. Introduction
Coronavirus disease 2019 (COVID-19) pandemic affected more than 100 million patients and caused more than 2 million deaths globally when writing this report, being the most challenging healthcare crisis during the past century [1]. Active immunization for disease prevention seems to be the most feasible solution to curb the pandemic’s medical, economic, and social impact [2]. In light of this, the emerging data on repeatedly positive reverse-transcription polymerase chain reaction (PCR) samples for severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) after initial recovery from COVID-19 are of major concern [3].
The reported incidence of repeatedly positive cases among the patients who recovered from COVID-19 ranges from 2.4 to 69.2%, and the reasons for repeated positivity are unclear [3]. Multiple mechanisms have been considered, including reinfection, disease relapse, prolonged viral shedding, and laboratory or technical errors [4]. Regarding reinfection, the European Center for Disease Prevention and Control recommends whole-genome sequencing to compare the strains responsible for each episode, yet very few studies report sequencing data [5,6,7,8].
Because PCR can be positive for up to 100 days in upper respiratory tract samples, viral viability should be verified by cell culture or viral load quantification to differentiate the shedding of viral ribonucleic acid fragments from actual infection [5]. Reports have been conflicting, some of them describing a milder, others a more severe disease course at the time of second PCR positivity [9,10]. No demographic or clinical risk factors for repeated positivity have been identified, and no infections were reported among the contacts of patients with repeatedly positive tests [11]. Although infection induces the development of neutralizing antibodies in more than 90% of cases, it is unclear if and how long they provide protection [9,12].
Our study aimed to evaluate the clinical significance of repeatedly positive testing after COVID-19 recovery regarding predisposing factors for more severe symptoms and the second episode’s disease course. We also describe repeatedly positive cases’ baseline characteristics and assess them according to the length of their PCR-negative interval between the two episodes.
2. Materials and Methods
We performed a systematic literature search according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline (Table S1) [13]. The review was registered on PROSPERO under the ID number CRD42021228422 (see https://www.crd.york.ac.uk/prospero (accessed on 2 February 2021)) in advance.
We used the following PICO framework: (P): repeatedly SARS-CoV-2 positive (RSP) patients with two positive PCRs separated by a negative PCR test result, (I/C) gender, comorbidities, severity and presenting symptoms, and (O) severity of the second episode, and the time interval between repeated positivity.
2.1. Search and Selection
The MEDLINE (via PubMed), Embase, and Cochrane Central Register of Controlled Trials (CENTRAL) databases were searched until 22 November 2020 for relevant case reports, with the following search key: ((“covid 19”) OR (“coronavirus”) OR (“2019 nCoV”) OR (“SARS-cov-2”)) AND ((reinfection) OR (“second episode”) OR (“second infection”)).
With available individual patient data reporting on repeatedly COVID-19-PCR positive patients, case reports, and case series were included in this analysis. Cohort and case-control studies were excluded. A total of two review authors performed the selection by title, abstract, and full text independently. Disagreements were resolved by consensus.
2.2. Data Extraction
Relevant data, including the year of publication, name of the first author, age, gender, and existing comorbidities of the patient, and severity, symptoms, imaging, and laboratory findings of first and second episodes of the disease course, were extracted to a pre-defined Excel (Microsoft Corporation, Redmond, Washington, United States) datasheet. A total of two independent review authors performed data extraction and resolved the disagreements by consensus.
We included only nasal swab PCR-confirmed COVID-19 cases in the analysis. COVID-19 was defined as a positive PCR of SARS-nCoV-2.
Severity was assessed based on the guidelines on the Diagnosis and Treatment of COVID-19 issued by the National Health Commission of China [14]. In case of missing data regarding severity, one review author (Z.P.) classified the cases based on the patient’s symptoms following the mentioned guideline. A detailed description of the classification system is included in Table S2.
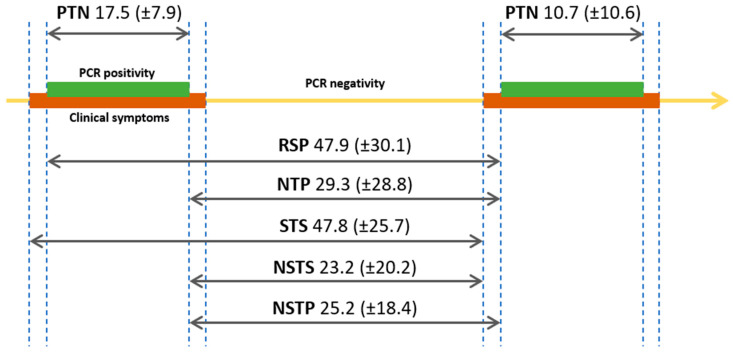
Repeated positivity was defined as two positive PCRs separated by a negative PCR test result. The RSP interval represents the interval between the two positive PCRs of each episode. Patients were classified based on the length of the RSP interval. Details of the analyzed intervals are presented in Figure 1.
Figure 1.
Summary of the analyzed time intervals. Numbers represent mean days with Scheme 2. positivity; STS: symptom to symptom interval.
2.3. Statistical Analysis
We generated and then analyzed the cohort of the included cases. Descriptive statistics were performed to characterize the RSP population. The association between categorical variables was examined with the Chi-square and Fisher’s exact tests. To assess the differences between groups t-test was applied for normally distributed variables and the Mann–Whitney U test for non-normal data. The Kruskal–Wallis test was used to compare more than two groups, with Bonferroni post-hoc adjustment. For correlation between categorical parameters, linear regression was used. A p-value of <0.05 was considered statistically significant. Statistical analyses were performed using International Business Machines Corporation (IBM)-SPSS for Windows 25 software (IBM Corp, Armonk, NY, USA).
A subgroup analysis was performed, according to RSP intervals: (a) 1–30 days, (b) 31–60 days, (c) 61–90 days, or (d) more than 90 days between their two positive PCR positive episodes.
2.4. Data Quality
Data quality is detailed in Table S3. Data quality represents the percentage of available data for each parameter in the cohort. Parameters with a data quality under 50% were not included in our analysis.
2.5. Assessment of Risk of Bias
To assess the risk of bias of case reports and case series, we applied the Joanna Briggs Institute Critical Appraisal Checklist tools [15]. Then, two independent review authors assessed the risk of bias, and disagreements were resolved by consensus.
3. Results
The selection process is detailed in Figure S1. We identified 1612 records in three databases for evaluation. After the removal of duplicates, screening, and selection,198 full texts were assessed. Altogether, 56 articles fulfilled the eligibility criteria and were included in our systematic review and analysis.
3.1. Characteristics of the Included Studies
The main characteristics of the included studies are summarized in Tables S4 and S5. Of the 56 included studies from 18 countries, nine were case series describing 65 RSP patients, and 47 were case reports with 58 patients. In total, 123 patients with RSP were assessed in our analysis. Most of the studies were published from China (n = 22), Italy (n = 7), and the USA (n = 5).
Overall, 56% of the data required for our analyses were provided. Data were almost complete on age, gender, the severity, and symptoms of COVID-19. We could not analyze laboratory parameters due to limited data. Data were 100% complete for the interval between the two COVID-19 episodes.
3.2. Characteristics of the Cohort
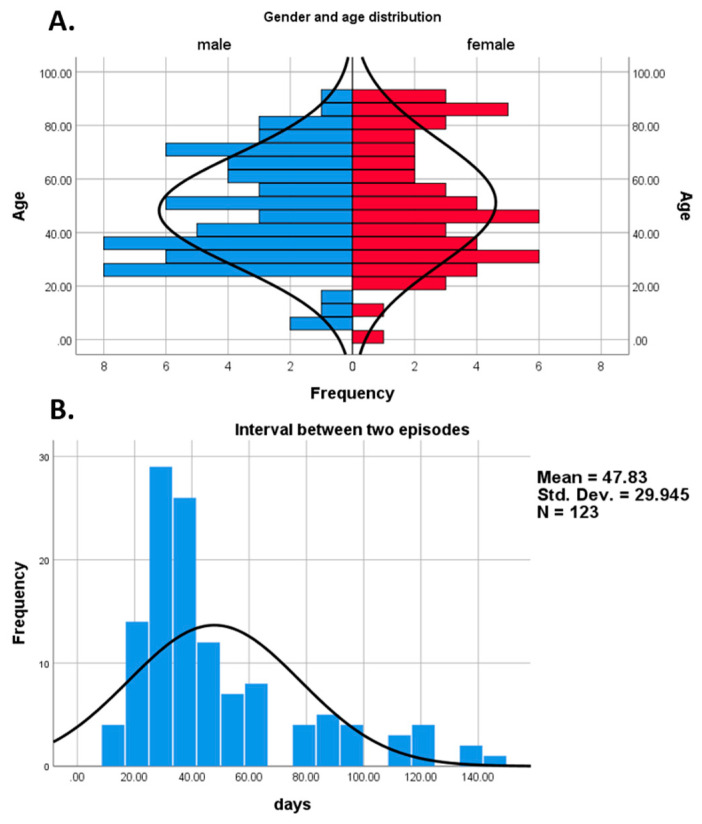
The main characteristics of the analyzed cohort are shown in Table 1. In our cohort, the mean age was 49.7 (SD 21.9, range 1–93), and 45.1% of the patients were female (n = 55/123). Age distribution based on gender is presented in Figure 2A. The most frequent comorbidities were hypertension (n = 22/82, 26.8%) and diabetes mellitus (n = 19/82, 23.2%). Overall, 66.3% (n = 57/86) of the patients had at least one comorbidity. During the first and second positive episodes, most of the patients were hospitalized (n = 95/119, 72.2% vs. n = 87/111, 70.7%, respectively).
Table 1.
Baseline characteristics of the overall analyzed population and comparison of patients with repeated positivity length under 60 with patients above 60 days.
| Parameter | Overall n/N (% Total) |
≤60 Days n/N (% Total) |
>60 Days n/N (% Total) |
p-Value |
|---|---|---|---|---|
| Total number N | 123 | 96 | 27 | |
| Female | 55/122 (45.1) | 42/96 (43.8) | 13/26 (50) | 0.570 |
| Age (mean, SD, range) | 49.7 (21.9) | 51 (22.3) | 45.2 (20.2) | 0.228 |
| Mean repeated positivity interval (SD) | 47.9 (30.1) | 34.1 (11.4) | 97 (24) | 0.001 |
| Comorbidities | ||||
| Hypertension | 22/82 (26.8) | 20/ 61 (32.8) | 2/ 21 (9.5) | 0.038 |
| Chronic heart disease | 14/84 (16.7 | 11/ 63 (17.5) | 3/ 21 (14.3) | 0.735 |
| Arrhythmia | 11/84 (13.1) | 8/ 63 (12.7) | 3/ 21 (14.3) | 0.852 |
| T2DM | 19/82 (23.2) | 15/ 61 (24.6) | 4/ 21 (19.1) | 0.604 |
| COPD | 6/82 (7.3) | 4/ 61 (6.6) | 2/ 21 (9.5) | 0.653 |
| Chronic kidney disease | 5/82 (6.1) | 5/ 61 (8.2) | 0/ 21 (0) | 0.176 |
| Chronic liver disease | 2/82 (2.4) | 1/ 61 (1.6) | 1/ 21 (4.8) | 0.424 |
| Immunosuppression | 13/82 (15.9) | 9/ 63 (14.3) | 4/ 19 (21.1) | 0.479 |
| Cancer | 3/84 (3.6) | 1/ 63 (1.6) | 2/ 21 (9.5) | 0.090 |
| Other | 31/78(39.7) | 21/ 58 (36.2) | 10/ 20 (50) | 0.277 |
| First episode | ||||
| Mean days of positivity (Mean, SD) | 17.5 (7.9) | 16.5 (6.9) | 21.1 (10) | 0.016 |
| Mild COVID-19 | 46/109 (42.2) | 32/ 82 (39) | 14/ 27 (51.9) | 0.208 |
| Moderate COVID-19 | 40/109 (36.7) | 29/ 82 (35.4) | 11/ 27 (40.7) | |
| Severe COVID-19 | 16/109 (14.7) | 14/ 82 (17.1) | 2/ 27 (7.4) | |
| Critical COVID-19 | 7/109 (6.4) | 7/ 82 (8.5) | 0/ 27 (0) | |
| Hospitalization | 95/119 (77.2) | 79/ 92 (85.9) | 16/ 27 (59.3) | 0.002 |
| Pneumonia | 46/62 (74.2) | 39/51 (76.5) | 7/11 (63.3) | 0.452 |
| Fever | 76/108 (70.4) | 60/ 81 (74.1) | 16/ 27 (59.3) | 0.144 |
| Cough | 67/108 (62) | 52/ 81 (64.2) | 15/ 27 (55.6) | 0.423 |
| Dyspnea | 28/105 (26.7) | 20/ 78 (25.6) | 8/ 27 (29.6) | 0.686 |
| Arthromyalgia | 20/108 (18.5) | 15/ 81 (18.5) | 5/ 27 (18.5) | 1.000 |
| Headache | 13/108 (12) | 9/ 81 (11.1) | 4/ 27 (14.8) | 0.609 |
| General cold symptoms | 19/108 (17.6) | 15/ 81 (18.5) | 4/ 27 (14.8) | 0.662 |
| Asthenia | 14/109 (12.8) | 11/ 82 (13.4) | 3/ 27 (11.1) | 0.756 |
| Gastrointestinal symptoms | 20/109 (18.3) | 20/ 82 (24.4) | 0/ 27 (0) | 0.005 |
| Second episode | ||||
| Mean days of positivity (Mean, SD) | 10.7 (10.6) | 10.6 (10.7) | 10.9 (10.9) | 0.928 |
| Mild COVID-19 | 59/97 (60.8) | 38/ 71 (53.5) | 21/ 26 (80.8) | 0.039 |
| Moderate COVID-19 | 22/97 (22.7) | 17/ 71 (23.9) | 5/ 26 (19.2) | |
| Severe COVID-19 | 7/97 (7.2) | 7/ 71 (9.9) | 0/ 26 (0) | |
| Critical COVID-19 | 9/97 (9.3) | 9/ 71 (12.7) | 0/ 26 (0) | |
| Hospitalization | 87/111 (70.7) | 73/ 88 (83) | 14/ 23 (60.9) | 0.043 |
| Pneumonia | 51/61 (83.6) | 49/53 (92.5) | 2/8 (25) | 0.001 |
| Fever | 30/100 (30) | 24/ 74 (32.4) | 6/ 26 (23.1) | 0.371 |
| Cough | 29/97 (29.9) | 24/ 71 (33.8) | 5/ 26 (19.2) | 0.165 |
| Dyspnea | 27/100 (27) | 24/ 74 (32.4) | 3/ 26 (11.5) | 0.039 |
| Arthromyalgia | 19/99 (19.2) | 14/ 73 (19.2) | 5/ 26 (19.2) | 1.000 |
| Headache | 10/100 (10) | 7/ 74 (9.5) | 3/ 26 (11.5) | 0.717 |
| General cold symptoms | 16/100 (16) | 12/ 74 (16.2) | 4/ 26 (15.4) | 1.000 |
| Asthenia | 8/100 (8) | 5/ 74 (6.8) | 3/ 26 (11.5) | 0.425 |
| Gastrointestinal symptoms | 9/100 (9) | 8/ 74 (10.8) | 1/ 26 (3.8) | 0.439 |
COPD: chronic obstructive pulmonary disease; COVID-19: Coronavirus disease 2019; SD: standard deviation; T2DM: type 2 diabetes mellitus.
Figure 2.
Age distribution of patients with repeatedly SARS-CoV-2 positivity (A), and the histogram of repeated positivity lengths (B).
Despite the high hospitalization rate, most of the cases were mild and moderate (n = 86/109, 78.9% and n = 81/97, 82.7%, respectively), and only few patients developed a critical condition (n = 7/109, 6.4% and n = 9/97, 9.3%, respectively) during the first and second episodes. The most frequent symptoms were fever (n = 76/108, 70.4%), cough (n = 67/108, 62%), and dyspnea (n = 28/105, 26.7%) during the first episode, compared to the second episode, where these symptoms were present in a smaller proportion (n = 30/100, 30%; n = 29/97, 29.9%; n = 27/100, 27%, respectively). Pneumonia was present in similar proportion during the first and second episodes (n = 46/62, 74.2% vs. n = 51/61, 83.6%); however, the data quality was borderline low for this parameter in the two episodes (50.41 and 49.59%, respectively). Immunoglobulin G was positive in 86.1% (n = 31/36) and 94.2% (n = 49/52) of the patients after the first and second episode, respectively.
Asymptomatic patients were present in 6.12% (n = 6/98) and 41.94% (n = 39/93) during the first and second episodes, respectively. A total of three cases presented symptoms between the two episodes, although they presented with negative COVID-19 PCR test results. Overall, out of the 123 patients only three deaths occurred during the second PCR positive episode, and three patients were in the hospital on reporting.
3.3. Length of Positivity and Intervals between Episodes
The first episode’s duration was significantly longer than the second episode’s (17.53 ± 7.86 vs. 10.71 ± 10.63 days, respectively; p = 0.001). The length of RSP for each patient was calculated, the mean interval being 47.9 ± 30.1 days (Figure 2B). The mean length of the rest of the analyzed intervals is presented in Figure 1 and Figure S2. The mean and median length for the negative to positive (NTP) interval was 29.3 (±28.8 SD) days, and 17 (10.75-37.25 Q1-Q3) days, respectively.
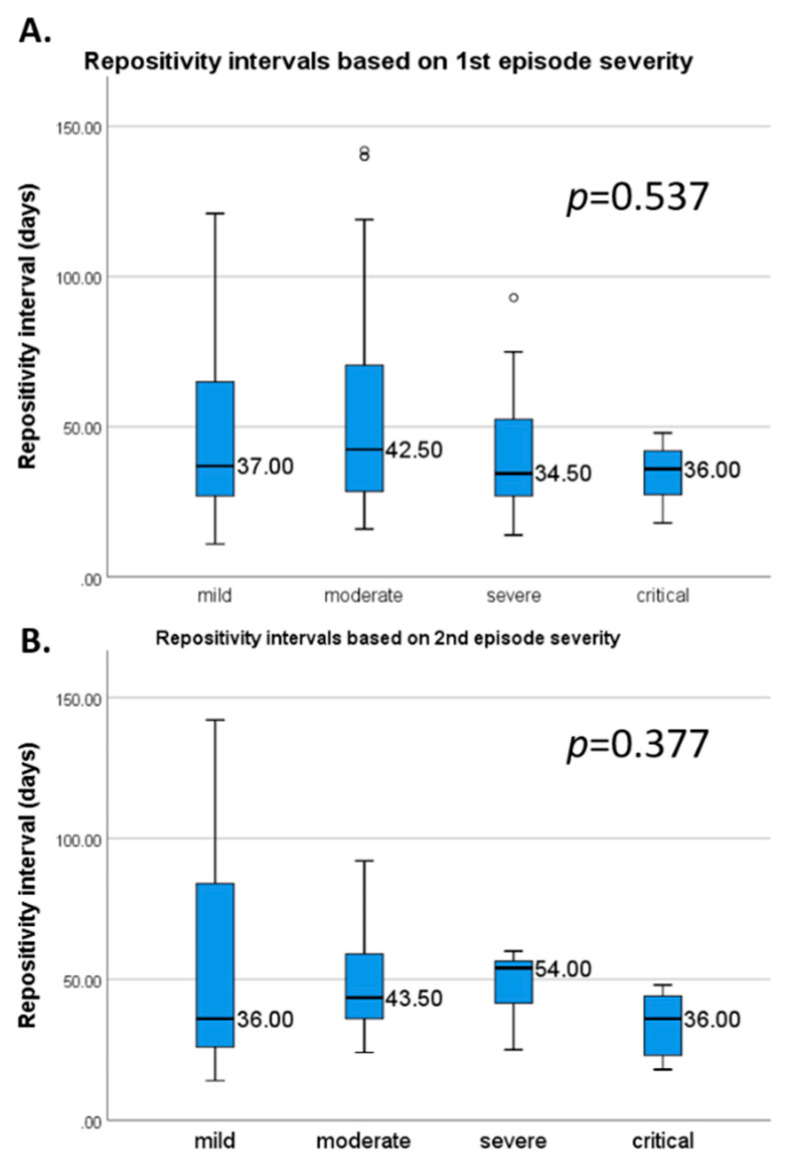
We found that younger patients might have a second episode later than older patients, although without statistical significance (R = 0.064, p = 0.486, Figure S3). Median RSP intervals based on the first- and second-episodes severity are presented in Figure 3. Mean RSP intervals for each parameter are shown in Table 2. Only the hospitalization during the first episode (44.2 ± 29.3 vs. 64.6 ± 29.8 days, respectively; p = 0.001) and the presence of pneumonia on imaging examinations (35.3 ± 12.9 vs. 70.8 ± 40.9 days, respectively; p = 0.012) during the second episode was associated with a shorter RSP interval. On the other hand, only the presence of headache during the first episode was associated with a longer mean RSP interval (63.8 ± 38.8 vs. 47.6 ± 30.1 days, respectively; p = 0.042).
Figure 3.
Median repeated positivity lengths based on the first (A) and second episode severity (B). Error bars represent 1.5 times the interquartile range.
Table 2.
Mean repeated positivity length based on the listed parameters.
| Parameter | Number with and without Assessed Parameter | Mean RSP (SD) Parameter Present | Mean RSP (SD) Parameter Absent | p-Value |
|---|---|---|---|---|
| Female | 55 | 47.7 (31.6) | − | 0.651 |
| Male | 67 | 46.7 (26.9) | − | |
| Comorbidities | ||||
| Hypertension | 22 vs. 60 | 44.8 (24.8) | 55.8 (32.5) | 0.379 |
| Chronic heart disease | 14 vs. 70 | 46.8 (20.2) | 53.6 (32.3) | 0.773 |
| Arrhythmia | 11 vs. 73 | 49.2 (23.3) | 53 (31.7) | 0.974 |
| T2DM | 19 vs. 63 | 46.2 (22.6) | 54.9 (32.8) | 0.271 |
| COPD | 6 vs. 76 | 61.8 (39.8) | 52.2 (30.3) | 0.345 |
| Chronic kidney disease | 5 vs. 77 | 46.6 (9.6) | 53.3 (31.7) | 0.651 |
| Chronic liver disease | 2 vs. 80 | 58 (38.2) | 52.7 (30.9) | 0.939 |
| Immunosuppression | 13 vs. 69 | 52.9 (33.7) | 51.6 (30.3) | 0.899 |
| Cancer | 3 vs. 81 | 72.7 (23.1) | 51.7 (30.7) | 0.123 |
| Other | 31 vs. 47 | 53.8 (27.3) | 52.7 (32.5) | 0.434 |
| First episode | ||||
| Mild COVID-19 | 46 | 51.3 (32.9) | − | 0.537 |
| Moderate COVID-19 | 40 | 54 (34.7) | − | |
| Severe COVID-19 | 16 | 41.1 (20.6) | − | |
| Critical COVID-19 | 7 | 34.4 (11.4) | − | |
| Hospitalization | 95 vs. 24 | 44.2 (29.3) | 64.6 (29.8) | 0.001 |
| Pneumonia | 46 vs. 16 | 42.8 (28.2) | 42.2 (28.9) | 0.342 |
| Fever | 76 vs. 32 | 48.1 (31.4) | 53.1 (32.1) | 0.638 |
| Cough | 67 vs. 41 | 47.7 (32.6) | 52.6 (30) | 0.209 |
| Dyspnea | 28 vs. 77 | 48.4 (25.6) | 50.7 (34) | 0.873 |
| Arthromyalgia | 20 vs. 88 | 46.7 (20.8) | 50.2 (33.6) | 0.687 |
| Headache | 13 vs. 95 | 63.8 (38.8) | 47.6 (30.1) | 0.042 |
| General cold symptoms | 19 vs. 89 | 53.6 (35.9) | 48.7 (30.7) | 0.475 |
| Asthenia | 14 vs. 95 | 46.2 (28) | 49.8 (32.1) | 0.942 |
| Gastrointestinal symptoms | 20 vs. 89 | 33.8 (10.5) | 52.8 (33.6) | 0.061 |
| Second episode | ||||
| Mild COVID-19 | 59 | 55.1 (37.8) | − | 0.377 |
| Moderate COVID-19 | 22 | 48.5 (18.7) | − | |
| Severe COVID-19 | 7 | 47.9 (13.1) | − | |
| Critical COVID-19 | 9 | 33.9 (11.4) | − | |
| Hospitalization | 87 vs. 24 | 44 (27.9) | 56.4 (37) | 0.148 |
| Pneumonia | 51 vs. 10 | 35.3 (12.9) | 70.8 (41) | 0.012 |
| Fever | 30 vs. 70 | 48.9 (26.1) | 51.9 (33) | 0.798 |
| Cough | 29 vs. 68 | 47.2 (26) | 53.5 (33.3) | 0.750 |
| Dyspnea | 27 vs. 73 | 48.3 (25) | 52 (33.1) | 0.661 |
| Arthromyalgia | 19 vs. 80 | 53 (28.2) | 50.6 (32) | 0.344 |
| Headache | 10 vs. 90 | 60.8 (35.3) | 49.9 (30.5) | 0.186 |
| General cold symptoms | 16 vs. 84 | 58.8 (36.6) | 49.5 (29.9) | 0.220 |
| Asthenia | 8 vs. 92 | 65.3 (38.1) | 49.8 (30.3) | 0.155 |
| Gastrointestinal symptoms | 9 vs. 91 | 39.1 (15.2) | 52.2 (32) | 0.511 |
COPD: chronic obstructive pulmonary disease; COVID-19: Coronavirus disease 2019; SD: standard deviation; RSP: repeatedly severe acute respiratory syndrome coronavirus 2 positivity; T2DM: type 2 diabetes mellitus. Minus sign means no data was available
3.4. Comparing Patients with below and above 60 Days of RSP
Based on Figure 2B, we compared patients with less than 60 days between RSP episodes with those with more than 60 days. A summary of the findings is included in Table 1. The mean RSP interval in the below 60 days group was 34.1 ± 11.4 days (n = 96), while in the above 60 days group was 97 ± 24 days (n = 27), with a significant difference between the groups (p = 0.001). The two groups were not different regarding age (51 ± 22.3 vs. 45.2 ± 20.2 years; p = 0.228) and gender (female n = 42/96, 43.8% vs. n = 13/26, 50%; p = 0.570). Similarly, most of the listed parameters were comparable between the two groups.
On the other hand, hypertension was more frequent in the below 60 days group (n = 20/61, 32.8% vs. n = 2/21, 9.5%; p = 0.038). During the first episode, hospitalization (n = 79/119, 85.9 vs. n = 16/27, 59.3%; p = 0.002) and gastrointestinal symptoms (n = 20/82, 24.4% vs. n = 0/27, 0%; p = 0.005) were more frequent in the below 60 days group; however, the length of the episode was longer in the above 60 days group (16.5 ± 6.9 vs. 21.1 ± 10 days, p = 0.016).
During the second episode, hospitalization was similarly more frequent in the below 60 days group (n = 73/88, 83% vs. n = 14/23, 60.9%, p = 0.043). Furthermore, pneumonia (n = 49/53, 92.5% vs n = 2/8, 25%, p = 0.001) and dyspnea (n = 24/74, 32.4% vs n = 3/26, 11.5%, p = 0.039) were also more frequent in the below 60 days group. We found a significant difference in severity, severe (n = 7/71, 9.9% vs. n = 0/26, 0%), and critical (n = 9/71, 12.7% vs. n = 0/26, 0%; p = 0.039) cases being more frequent in the below 60 days group.
3.5. Comparison Based on Intervals
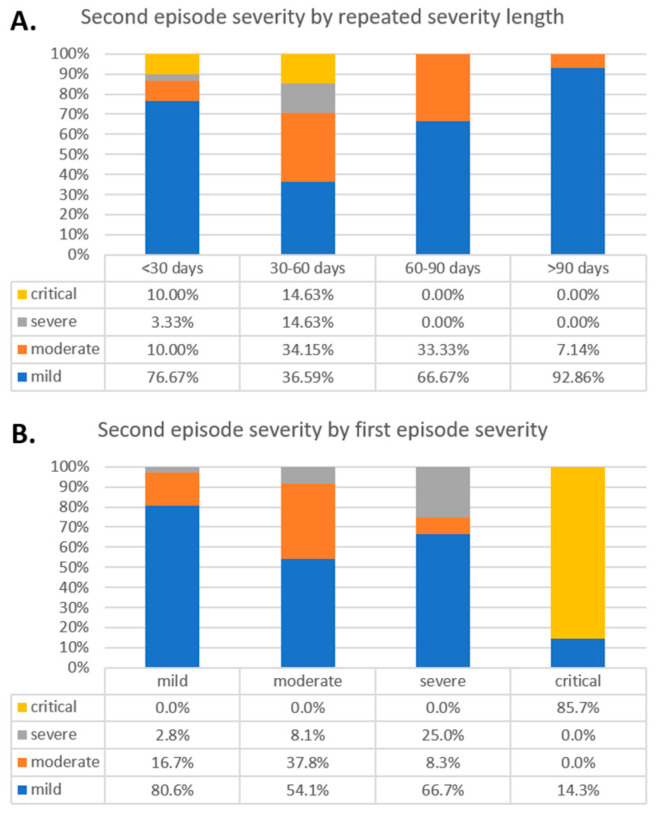
We divided RSP patients into four groups (below 30, 30–60, 60–90, and above 90 days), based on the length of the RSP intervals. The summary of findings is included in Table 3. The severity distributions based on the selected intervals are presented in Figure 4A. The critical and severe second episodes were more frequent in the below 30- and 30–60-day interval (p = 0.005), compared to the above 60 groups. Similarly, the difference was present regarding the first episode’s severity; however, the difference was non-significant (p = 0.797).
Table 3.
Characteristics of the analyzed patients based on the repeated positivity intervals.
| Parameter | ≤30 Days | 31 to 60 Days | 61 to 90 Days | >90 Days | p-Value |
|---|---|---|---|---|---|
| Total number | 42 | 54 | 12 | 15 | |
| Female | 22/42 (52.4) | 20/54 (37) | 5/12 (41.7) | 8/14 (57.1) | 0.363 |
| Age (mean, SD, range) | 43.9 (22.9) | 56.4 (20.5) | 55.9 (20.3) | 36.6 (16.1) | 0.001 |
| Mean repeated positivity interval (SD) | 24.1 (4.9) | 41.9 (8.5) | 75.5 (9.3) | 114.1 (17) | 0.001 |
| Comorbidities | |||||
| Hypertension | 31.25% | 33.33% | 11.11% | 8.33% | 0.118 |
| Chronic heart disease | 17.65% | 17.39% | 33.33% | 0.00% | 0.238 |
| Arrhythmia | 11.76% | 13.04% | 33.33% | 0.00% | 0.166 |
| T2DM | 25.00% | 24.44% | 33.33% | 8.33% | 0.557 |
| COPD | 0.00% | 8.89% | 11.11% | 8.33% | 0.651 |
| Chronic kidney disease | 0.00% | 11.11% | 0.00% | 0.00% | 0.223 |
| Chronic liver disease | 0.00% | 2.22% | 11.11% | 0.00% | 0.314 |
| Immunosuppression | 17.65% | 13.04% | 42.86% | 8.33% | 0.199 |
| Cancer | 0.00% | 2.17% | 22.22% | 0.00% | 0.015 |
| Other | 37.50% | 35.71% | 66.67% | 36.36% | 0.377 |
| First episode | |||||
| Mean days of positivity (Mean, SD) | 13.5 (5.3) | 19.3 (7) | 23.3 (9.8) | 19.2 (10.2) | 0.001 |
| Mild COVID-19 | 40.00% | 38.30% | 50.00% | 53.33% | 0.797 |
| Moderate COVID-19 | 37.14% | 34.04% | 41.67% | 40.00% | |
| Severe COVID-19 | 17.14% | 17.02% | 8.33% | 6.67% | |
| Critical COVID-19 | 5.71% | 10.64% | 0.00% | 0.00% | |
| Hospitalization | 95.0% | 78.8% | 58.3% | 60.0% | 0.005 |
| Pneumonia | 63.0% | 91.7% | 75.0% | 57.1% | 0.452 |
| Fever | 69.44% | 77.78% | 58.33% | 60.00% | 0.422 |
| Cough | 69.44% | 60.00% | 50.00% | 60.00% | 0.641 |
| Dyspnea | 22.86% | 27.91% | 41.67% | 20.00% | 0.569 |
| Arthromyalgia | 16.67% | 20.00% | 33.33% | 6.67% | 0.349 |
| Headache | 2.78% | 17.78% | 8.33% | 20.00% | 0.147 |
| General cold symptoms | 16.67% | 20.00% | 16.67% | 13.33% | 0.941 |
| Asthenia | 10.81% | 15.56% | 8.33% | 13.33% | 0.884 |
| Gastrointestinal symptoms | 24.32% | 24.44% | 0.00% | 0.00% | 0.045 |
| Second episode | |||||
| Mean days of positivity (Mean, SD) | 10.1 (10.5) | 11.2 (11.1) | 5.7 (6) | 16.1 (12.7) | 0.238 |
| Mild COVID-19 | 76.67% | 36.59% | 66.67% | 92.86% | 0.005 |
| Moderate COVID-19 | 10.00% | 34.15% | 33.33% | 7.14% | |
| Severe COVID-19 | 3.33% | 14.63% | 0.00% | 0.00% | |
| Critical COVID-19 | 10.00% | 14.63% | 0.00% | 0.00% | |
| Hospitalization | 82.5% | 83.3% | 55.6% | 64.3% | 0.135 |
| Pneumonia | 87.5% | 96.6% | 50.0% | 0.0% | 0.001 |
| Fever | 25.81% | 37.21% | 33.33% | 14.29% | 0.417 |
| Cough | 33.33% | 34.15% | 25.00% | 14.29% | 0.505 |
| Dyspnea | 16.13% | 44.19% | 8.33% | 14.29% | 0.011 |
| Arthromyalgia | 12.90% | 23.81% | 33.33% | 7.14% | 0.257 |
| Headache | 3.23% | 13.95% | 8.33% | 14.29% | 0.423 |
| General cold symptoms | 12.90% | 18.60% | 8.33% | 21.43% | 0.778 |
| Asthenia | 3.23% | 9.30% | 8.33% | 14.29% | 0.487 |
| Gastrointestinal symptoms | 6.45% | 13.95% | 8.33% | 0.00% | 0.521 |
COPD: chronic obstructive pulmonary disease; COVID-19: Coronavirus disease 2019; SD: standard deviation; T2DM: type 2 diabetes mellitus.
Figure 4.
Severity based on repeated positivity intervals (A), and second episode’s severity based on the first episode severities (B).
Among the groups, the mean age was the highest (56.4 ± 20.5 years) in the 30 to 60 days group, and the lowest in the above 90 days group (36.6 ± 16.1 years), with a significant difference between groups (p = 0.001). Regarding comorbidities, cancer was also more frequent in the 60 to 90 days group (22.2%, p = 0.015). During the first episode, hospitalization was higher in the below 30 and 30–60-days group (95 and 78.8%, respectively; p = 0.005). Gastrointestinal symptoms were present only in the below 30 and 30–60-days group (p = 0.045), with no other significant differences regarding symptoms. Lastly, during the second episode, pneumonia, and dyspnea were most frequent in the between 30–60-days group (96.6 and 44.2%, respectively).
3.6. The Severity of the First and Second Episodes
According to the first episode’s severity, the severity of the second episode is presented in Figure 4B. After mild and moderate first episodes, most of the second episodes were similarly mild or moderate (97.3 and 91.9%, respectively). During the second episode, severe cases were more common if the first episode was severe (25%). Lastly, critical cases tended to stay critical (85.7%). We did not find significant difference in the analyzed intervals based on first- and second-episode severity.
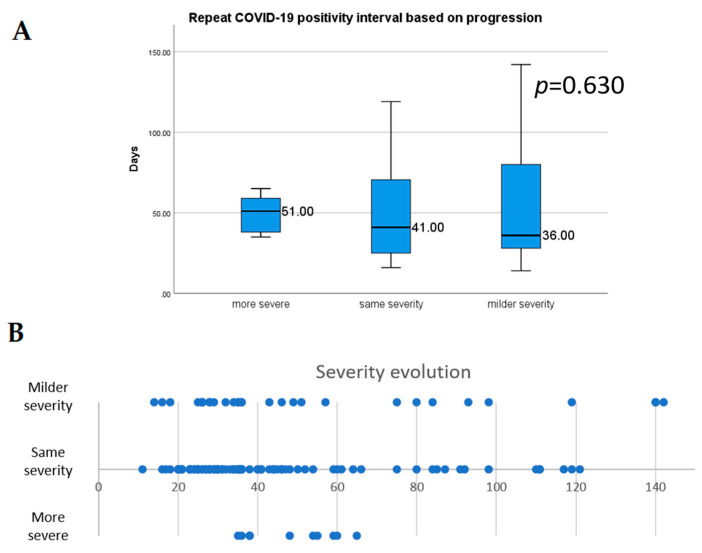
In Figure 5, the evolution of severity is presented. The mean RSP interval was similar when comparing progression direction (p = 0.630); in the case of a worse severity during the second episode compared to the first episode, the mean RSP interval was 48.8 ± 11.3 days, for a similar severity, it was 50.7 ± 30.8 days, and in the case of a less severe course the mean was 55.4 ± 39 days.
Figure 5.
Median repeated positivity length based on progression (A), and distribution of the progression of the second episode (B). Error bars represent 1.5 times the interquartile range.
Lastly, the mean and median RSP interval for asymptomatic patients are summarized in Table S6. RSP interval was higher in asymptomatic patients.
3.7. Risk of Bias Assessment
The summary of the risk of bias assessment is shown in Tables S8 and S9. We waived the scoring of the statistical analysis domain in case series since it was not a factor of interest.
4. Discussion
Our analysis aimed to systematically review case reports and case series, with individual patient data reporting on repeated COVID-19 positivity. We found that repeatedly positive patients did not differ from patients who had only one episode of COVID-19. In a meta-analysis of more than 3000 patients with a single episode of COVID-19, patients were predominantly male, similarly to our findings, while the same symptoms dominated in our analyzed cohort (fever, cough, and dyspnea) [16]. We generally see a lower prevalence of the analyzed symptoms during the second episode, which correlates with the higher proportion of asymptomatic or mild cases. The proportion of pneumonia during each episode was high, although data was present only in 50% of the cases.
Based on the correlation between the age and RSP length, younger patients might have a second episode later than older patients. Similarly, in our cohort younger patients were predominant in the delayed (>90 days) repeated positivity group, which are more likely to represent actual reinfections [17]. Unfortunately, serology and viral viability data were scarce in our cohort. Nevertheless, knowing that milder cases are more prevalent among youth, our data suggest that younger patients might be more prone to reinfection because of a lack of efficacious active immunization after a less severe first episode of disease [18,19].
Shorter RSP interval was associated with increased number of pneumonia and longer hospital stay during the second episode. Furthermore, comparing patients with RSP intervals of below 60 and above 60 days, we found that a more severe disease course can be expected if the RSP interval is shorter. These cases can probably be explained by a continued and aggravated disease course with false-negative PCR results midpoint in their illness rather than genuine reinfection [17]. Furthermore, the higher number of pneumonias in the case of below 30 days RSP group also strengthens this theory. Lastly, in our cohort the severe and critical disease courses might be predictive of future severe and critical episodes.
To our knowledge, this is the first systematic review and meta-analysis of case reports and case series on repeatedly SARS-CoV-2 positive patients. Previous studies aimed to assess the question of repeated positivity, with different results. One of these focused on the relapse of the disease, which is one of the potential causes of repeated positivity. It is noteworthy that none of the patients included in this study were asymptomatic at the time of relapse, which could act well to differentiate between reinfection and relapse [20]. In accordance, another study of 182 patients reported that repeated positivity during isolation after the initial episode of infection is rarely associated with the recurrence of disease if symptoms are absent [21]. Another meta-analysis concluded that to diagnose reinfection the RSP interval needs to be above 90 days and the patients must have symptoms [22]. Similarly, both the Center for disease Control and Prevention and the Health Protection Surveillance Center recommend an interval of at least 90 days to consider for the possibility of a reinfection [23,24].
Although multiple reports have described recurrent positivity after an initial episode, most of them did not differentiate between reinfection, recurrent positivity, relapse, or false positive tests. In our analysis we tried to assess the clinical importance of repeated positivity, but we cannot ignore the importance of reinfection. In our analyzed cohort we found a short NTP interval, which can correlate with long lasting resolution in some of the cases, rather than reinfection.
After more than one year since the first reported case of COVID-19, a consensus regarding reinfection by SARS-CoV-2 is still lacking. To diagnose reinfection, it requires the evidence of a new infection by a phylogenetically distinct form of the SARS-CoV-2 virus after the elimination of the previous one [7,23].
Out of the 56 articles included in our analysis, only eight described different strains during the two COVID-19 episodes (see Table S10) [6,7,25,26,27,28,29].
On the other hand, long shedding might be the most common background of repeated positivity. In a study of 38 long-term SARS-CoV-2 carrier patients, the median carrying history was 92 days after the onset of COVID-19, and the longest reported period was 118 days. During these periods, negative-positive PCR result fluctuation were observed [30]. Another study reported on 99 patients with a positive SARS-CoV-2 PCR after 4 weeks from the first positive viral test [31].
In our analyzed cohort, a high percentage of patients were asymptomatic during the second episode. Data in the literature regarding cycle threshold (Ct) value in asymptomatic patients is contradictory. There are studies that show Ct values are higher for asymptomatic patients while other show no difference by comparison with the symptomatic cases [32,33]. On the other hand, Ct value could be used as a surrogate marker to assess viral viability, because viral culture, is not widely available and requires high level biosafety [34]. The CDC recommends Ct value under 33 for cases diagnosed only by one positive PCR [23], while other authors suggest two positive tests with Ct < 35 as criteria for reinfection [17].
4.1. Strengths and Limitations
One of the strengths of our analysis is the preregistered protocol, which prevents publication bias. To our knowledge, this analysis is the most comprehensive work, also including individual patient data.
However, our work has multiple limitations. Most of the included case series and case reports are retrospective. The case series did not follow the Case Report (CARE) guidelines when reporting individual cases. Most of the studies lacked follow-up of the infected patients. To confirm a reinfection, one should assess the strain of the virus, which was not done in most of the articles. The case series themselves carry the limitation of publication bias, general overinterpretation, furthermore these are not representative for the whole COVID-19 population. In the case of case reports, mostly rare and unusual cases are reported, while being outcome oriented.
4.2. Implication for Research
We strongly emphasize the conduction of follow-up studies, with PCR testing after the convalescence of the disease. Cases with high Ct values should be considered with caution and ideally viral viability should be assessed if available to confirm reinfection. Animal model studies are also needed to assess the possibility of reinfection with the same strain. Based on these findings we might be able to create a classification, which can distinguish between false positivity, relapse, and reinfection.
4.3. Implication for Practice
From a practical point of view critical evaluation of a repeated positivity is suggested. One should assess whether it is associated with symptoms, how long the time interval between the two episodes is and whether the cycle threshold of the second PCR is too high.
5. Conclusions
In summary, our results show that the second episode of SARS-CoV-2 positivity is more severe if it happens within 60 days after the first positive PCR. On the other hand, the second episode severity correlates with the first infection.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/13/3/512/s1, Table S1: PRISMA checklist, Table S2: Classification of COVID-19 severity, Table S3: Data quality of the analyzed cohort, Table S4: Basic characteristics of the included case reports, Table S5: Basic characteristics of the included case series, Table S6: Difference of intervals organized by disease severity, Table S7: Repeated SARS-CoV-2 positive intervals based on symptomatic cases, Table S8: Risk of bias assessment for case reports, Table S9: Risk of bias assessment for case series, Figure S1: PRISMA flowchart, Figure S2: Histogram of analyzed intervals, Figure S3: Correlation analysis.
Author Contributions
P.H. and S.V. conceived the study. S.V. and L.S. wrote the protocol. F.D. and B.T. did the literature search. R.N. and S.B. screened the records and extracted the data. R.N. and B.T. assessed the quality of included studies. N.F. did the statistical analysis. S.V. prepared the tables. S.V. and L.S. wrote the first draft of this manuscript. A.P., B.E., Z.P. and P.H. supervised the manuscript and approved the submitted draft. Z.P. is the guarantor of this paper and, as an infectious disease specialist, provided the team with an expert background. All authors provided critical conceptual input, interpreted the data analysis, and critically revised and approved the manuscript’s final version.
Funding
Funding was provided by an Economic Development and Innovation Operative Program Grant (GINOP 2.3.2-15-2016-00048, GINOP-2.3.4-15-2020-00010) and by a Human Resources Development Operational Program Grant (EFOP-3.6.2-16-2017-00006), both co-financed by the European Union (European Regional Development Fund) within the framework of the Széchenyi 2020 Program. Furthermore, funding was provided by János Bolyai Research Scholarship of the Hungarian Academy of Sciences (to AP), and the ÚNKP20-3, a New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development, and Innovation Fund (for SV).
Ethical Approval: No ethical approval was required for this review, as all data were already published in peer-reviewed journals. No patients were involved in the design, conduction, or interpretation of our review. The datasets used in this study can be found in the full-text articles included in the systematic review and meta-analysis.
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO Coronavirus Disease (COVID-19) [(accessed on 2 February 2021)];2020 Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 2.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 3.Dao T.L., Hoang V.T., Gautret P. Recurrence of SARS-CoV-2 viral RNA in recovered COVID-19 patients: A narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:13–25. doi: 10.1007/s10096-020-04088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J., Peng J., Xiong Q., Liu Z., Lin H., Tan X., Kang M., Yuan R., Zeng L., Zhou P., et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMed. 2020;59:102960. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ECDC. European Centre for Disease Prevention and Control Reinfection with SARS-CoV: Considerations for Public Health Response. [(accessed on 25 January 2021)]; Available online: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-reinfection-sars-cov-2.
- 6.Van Elslande J., Vermeersch P., Vandervoort K., Wawina-Bokalanga T., Vanmechelen B., Wollants E., Laenen L., André E., Van Ranst M., Lagrou K., et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafaie L., Célarier T., Goethals L., Pozzetto B., Grange S., Ojardias E., Annweiler C., Botelho-Nevers E. Recurrence or Relapse of COVID-19 in Older Patients: A Description of Three Cases. J. Am. Geriatr. Soc. 2020;68:2179–2183. doi: 10.1111/jgs.16728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murchu E.O., Byrne P., Walsh K.A., Carty P.G., Connolly M., De Gascun C., Jordan K., Keoghan M., O’Brien K.K., O’Neill M., et al. Immune response following infection with SARS-CoV-2 and other coronaviruses: A rapid review. Rev. Med. Virol. 2020;n/a:e2162. doi: 10.1002/rmv.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki A. What reinfections mean for COVID-19. Lancet Infect. Dis. 2021;21:3–5. doi: 10.1016/S1473-3099(20)30783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gidari A., Nofri M., Saccarelli L., Bastianelli S., Sabbatini S., Bozza S., Camilloni B., Fusco-Moffa I., Monari C., De Robertis E., et al. Is recurrence possible in coronavirus disease 2019 (COVID-19)? Case series and systematic review of literature. Eur. J. Clin. Microbiol. Infect. Dis. 2020 doi: 10.1007/s10096-020-04057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lumley S.F., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagnier J.J., Kienle G., Altman D.G., Moher D., Sox H., Riley D., CARE Group The CARE guidelines: Consensus-based clinical case reporting guideline development. J. Med. Case Rep. 2013;7:223. doi: 10.1186/1752-1947-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J., Ji P., Pang J., Zhong Z., Li H., He C., Zhang J., Zhao C. Clinical characteristics of 3062 COVID-19 patients: A meta-analysis. J. Med. Virol. 2020;92:1902–1914. doi: 10.1002/jmv.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yahav D., Yelin D., Eckerle I., Eberhardt C.S., Wang J., Cao B., Kaiser L. Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolowska M., Lukasik Z.M., Agache I., Akdis C.A., Akdis D., Akdis M., Barcik W., Brough H.A., Eiwegger T., Eljaszewicz A., et al. Immunology of COVID-19: Mechanisms, clinical outcome, diagnostics, and perspectives-A report of the European Academy of Allergy and Clinical Immunology (EAACI) Allergy. 2020;75:2445–2476. doi: 10.1111/all.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seow J., Graham C., Merrick B., Acors S., Steel K.J.A., Hemmings O., O’Bryne A., Kouphou N., Pickering S., Galao R.P., et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.09.20148429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsayed S.M., Reddy M.K., Murthy P.M., Gupta I., Valiuskyte M., Sánchez D.F., Diaz M.A.J.C. The Possibility and Cause of Relapse After Previously Recovering From COVID-19: A Systematic Review. Cureus. 2020;12 doi: 10.7759/cureus.10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan B., Liu H.-Q., Yang Z.-R., Chen Y.-X., Liu Z.-Y., Zhang K., Wang C., Li W.-X., An Y.-W., Wang J.-C., et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci. Rep. 2020;10:11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arafkas M., Khosrawipour T., Kocbach P., Zielinski K., Schubert J., Mikolajczyk A., Celinska M., Khosrawipour V. Current meta-analysis does not support the possibility of COVID-19 reinfections. J. Med. Virol. 2021;93:1599–1604. doi: 10.1002/jmv.26496. [DOI] [PubMed] [Google Scholar]
- 23.CDC Centers for Disease Control and Prevention: Common Investigation Protocol for Investigating Suspected SARS-CoV-2 Reinfection. [(accessed on 25 January 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html.
- 24.HPSC Health Protection Surveillence Centre: Guidance on the Management of Weak Positive (High Ct Value) PCR Results in the Setting of Testing Individuals for SARS-CoV-2. [(accessed on 25 January 2021)]; Available online: https://www.hpsc.ie/a-z/respiratory/coronavirus/novelcoronavirus/guidance/outbreakmanagementguidance/
- 25.Colson P., Finaud M., Levy N., Lagier J.-C., Raoult D. Evidence of SARS-CoV-2 re-infection with a different genotype. J. Infect. 2020 doi: 10.1016/j.jinf.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman J.D., Wang K., Roltgen K., Nielsen S.C.A., Roach J.C., Naccache S.N., Yang F., Wirz O.F., Yost K.E., Lee J.-Y., et al. Reinfection with SARS-CoV-2 and Failure of Humoral Immunity: A case report. medRxiv. 2020 doi: 10.1101/2020.09.22.20192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder M., van der Vegt D.S.J.M., Oude Munnink B.B., GeurtsvanKessel C.H., van de Bovenkamp J., Sikkema R.S., Jacobs E.M.G., Koopmans M.P.G., Wegdam-Blans M.C.A. Reinfection of Severe Acute Respiratory Syndrome Coronavirus 2 in an Immunocompromised Patient: A Case Report. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao W., Wang X., Zhang G., Guo M., Ma H., Zhao D., Sun Y., He J., Liu L., Zhang K., et al. Re-detectable positive SARS-CoV-2 RNA tests in patients who recovered from COVID-19 with intestinal infection. Protein Cell. 2020 doi: 10.1007/s13238-020-00778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To K.K.-W., Hung I.F.-N., Ip J.D., Chu A.W.-H., Chan W.-M., Tam A.R., Fong C.H.-Y., Yuan S., Tsoi H.-W., Ng A.C.-K., et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q., Zheng X.-S., Shen X.-R., Si H.-R., Wang X., Wang Q., Li B., Zhang W., Zhu Y., Jiang R.-D., et al. Prolonged shedding of severe acute respiratory syndrome coronavirus 2 in patients with COVID-19. Emerg. Microbes Infect. 2020;9:2571–2577. doi: 10.1080/22221751.2020.1852058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V., Venkatakrishnan A.J., Puranik A., Kirkup C., Lopez-Marquez A., Challener D.W., Theel E.S., O’Horo J.C., Binnicker M.J., Kremers W.K., et al. Long-term SARS-CoV-2 RNA shedding and its temporal association to IgG seropositivity. Cell Death Discov. 2020;6:138. doi: 10.1038/s41420-020-00375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ra S.H., Lim J.S., Kim G.-u., Kim M.J., Jung J., Kim S.-H. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax. 2021;76:61. doi: 10.1136/thoraxjnl-2020-215042. [DOI] [PubMed] [Google Scholar]
- 33.Zhou R., Li F., Chen F., Liu H., Zheng J., Lei C., Wu X. Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mokhtar K.M. Improved RT-PCR SARS-Cov2 results interpretation by indirect determination of cut-off cycle threshold value. medRxiv. 2020 doi: 10.1101/2020.11.20.20235390. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.