Abstract
Acinetobacter baumannii is a Gram-negative ESKAPE microorganism that poses a threat to public health by causing severe and invasive (mostly nosocomial) infections linked with high mortality rates. During the last years, this pathogen displayed multidrug resistance (MDR), mainly due to extensive antibiotic abuse and poor stewardship. MDR isolates are associated with medical history of long hospitalization stays, presence of catheters, and mechanical ventilation, while immunocompromised and severely ill hosts predispose to invasive infections. Next-generation sequencing techniques have revolutionized diagnosis of severe A. baumannii infections, contributing to timely diagnosis and personalized therapeutic regimens according to the identification of the respective resistance genes. The aim of this review is to describe in detail all current knowledge on the genetic background of A. baumannii resistance mechanisms in humans as regards beta-lactams (penicillins, cephalosporins, carbapenems, monobactams, and beta-lactamase inhibitors), aminoglycosides, tetracyclines, fluoroquinolones, macrolides, lincosamides, streptogramin antibiotics, polymyxins, and others (amphenicols, oxazolidinones, rifamycins, fosfomycin, diaminopyrimidines, sulfonamides, glycopeptide, and lipopeptide antibiotics). Mechanisms of antimicrobial resistance refer mainly to regulation of antibiotic transportation through bacterial membranes, alteration of the antibiotic target site, and enzymatic modifications resulting in antibiotic neutralization. Virulence factors that may affect antibiotic susceptibility profiles and confer drug resistance are also being discussed. Reports from cases of A. baumannii coinfection with SARS-CoV-2 during the COVID-19 pandemic in terms of resistance profiles and MDR genes have been investigated.
Keywords: Acinetobacter baumannii, antibiotic, resistance, antimicrobial, multidrug resistant, extensive drug resistant, pandrug resistant, next-generation sequencing, CRAB
1. Introduction
Antimicrobial or antibiotic resistance (AMR) has emerged as a substantial and triggering phenomenon with increasing costs for healthcare systems worldwide. In recent years it has been related to significant morbidity, mortality, and increased cost due to both prolonged length of hospitalization and treatment. Although in the last decades, there are new antimicrobial agents in our armamentarium, resistance seems to be an expanding problem with geometric evolution. Data from multicenter studies in the last decades have demonstrated that both community-acquired and nosocomial AMR are increasing alongside the increased number of older patients with primary or secondary immunodeficiencies [1,2].
Acinetobacter baumannii belongs to the Moraxellaceae family and is a Gram-negative bacterium that predominantly causes nosocomial infections. These infections are diverse and may include hospital-acquired and ventilator-associated pneumonia (HAP, VAP), urinary tract infections, meningitis, bacteremia, and gastrointestinal and skin/wound infections [3,4,5,6,7].
A. baumannii is one of the ESKAPE organisms (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.), that pose a global threat to human health and a therapeutic challenge due to emerging and constantly increasing resistance. Carbapenem resistant A. baumannii (CRAB) was ranked in 2018 by WHO as number one priority for antibiotic research and development. Carbapenem was chosen as a marker, because carbapenem resistance is usually associated with a broad range of co-resistance to other antibiotic classes [8].
The overall prevalence of multidrug-resistant strains in patients with A. baumannii HAP and VAP is estimated to be 79.9%, ranging from 56.5% in Argentina and 61.8% in Taiwan to 100% in Central America, Pakistan, Lebanon, Qatar, and Croatia, while its overall mortality can be as high as 56.2% [9]. The patterns of carbapenem resistance differ throughout Europe and also within the countries of the Arab League. Increased incidence of carbapenem resistant A. baumannii isolates has been observed in Northern and Eastern Europe as well as in the Levant countries of the Arab League (Iraq, Jordan, Lebanon, Palestinian territories, and Syria) [10,11].
Carbapenems were the preferred treatment of multidrug resistant (MDR) A. baumannii infections, but their prior use has led to increased incidence of carbapenem resistance during the last years [12]. Polymyxins are now widely used as the antibiotics of choice for MDR A. baumannii infections, although they were initially avoided due to systemic toxicities (nephrotoxicity and neurotoxicity) [13,14]. Extensive drug resistant (XDR) A. baumannii is called an isolate resistant to three or more classes of antimicrobials (penicillins and cephalosporins—including inhibitor combinations, fluoroquinolones, and aminoglycosides, resistant to carbapenems in most of cases), while pandrug resistant (PDR) A. baumannii is an XDR isolate resistant to polymyxins and tigecycline. Lately, extensively drug-resistant isolates have been led to the discovery of novel antimicrobials and the introduction of new treatment approaches [15,16].
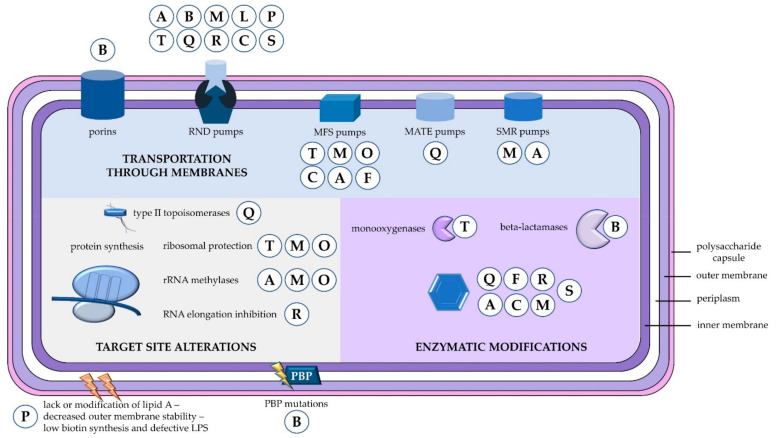
Mechanisms of antibiotic resistance can be categorized into three groups. First, resistance can be achieved by reducing membrane permeability or increasing efflux of the antibiotic and thus preventing access to the target. Second, bacteria can protect the antibiotic target through genetic mutation or post-translational modification, and last, antibiotics can be directly inactivated by hydrolysis or modification [17]. The mechanisms of A. baumannii antibiotic resistance based on this classification are summarized in Figure 1. One of the most important weapons in the armoury of Acinetobacter is its impressive genetic plasticity, facilitating rapid genetic mutations and rearrangements as well as integration of foreign determinants carried by mobile genetic elements. Of these, insertion sequences are considered one of the key forces shaping bacterial genomes and ultimately evolution [1,2]. Additionally, A. baumannii can form biofilms and thus prolong its survival on medical devices, such as ventilators in intensive care units (ICUs) [18]. However, the relationship between biofilm formation and antibiotic resistance still remains unclear [19,20]. In the present review, we report data on A. baummannii mechanisms of resistance to different classes of antibiotics and future perspectives for early identification of resistant genes.
Figure 1.
Mechanisms of antibiotic resistance in A. baumannii. Antibiotic resistance can be conferred through three main mechanisms, i.e., control of antibiotic transportation through membranes (reduction of porin permeability or increased efflux), modification of antibiotic targets, and enzymatic inactivation of the antibiotics. A = aminoglycosides; B = beta-lactams; C = chloramphenicol; F = fosfomycin; L = lincosamides; M = macrolides; MATE = multidrug and toxic compound extrusion; MFS = major facilitator superfamily; O = oxazolidinones; P = polymyxins; PBP = penicillin binding protein; Q = fluoroquinolones; R = rifamycins; RND = resistance-nodulation-division; S = diaminopyrimidines and sulfonamides; SMR = small multidrug resistance family; T = tetracyclines.
2. Resistance to Beta-Lactams
Beta-lactams comprise of penicillins, cephalosporins, carbapenems, monobactams, and beta-lactamase inhibitors. Beta-lactam antibiotics are structurally similar to the d-Ala-d-Ala moiety of peptidoglycan, where penicillin-binding proteins bind, and thus they inhibit the transpeptidation, which is the last step in peptidoglycan synthesis [21]. A. baumannii is nowadays intrinsically resistant to penicillins and cephalosporins [22]. Resistance to beta-lactam antibiotics can be conferred through all the aforementioned mechanisms, which is inactivation by hydrolysis, increase of efflux, decrease of influx, and protection of the antibiotic target.
2.1. Beta-Lactamases
Beta-lactamases are enzymes that catalyze the hydrolysis of beta-lactam antibiotics and can be divided into four classes based on sequence motifs and differences in hydrolytic mechanism [23,24].
2.1.1. Class A
Class A beta-lactamases mediate resistance to penicillin, cephalosporins, monobactams, and carbapenems. These lactamases may have narrow spectrum, or they can acquire extended spectrum antibiotic activity through point mutations. Narrow spectrum lactamases are active, mostly against penicillins, and can be inhibited by clavulanic acid [25], while extended spectrum beta-lactamases (ESBLs) can hydrolyze extended spectrum cephalosporins, like ceftazidime, ceftriaxone, cefotaxime, as well as aztreonam [26]. In addition, they are broadly distributed among Gram-negative bacteria with the help of plasmids and other mobile genetic elements [24]. Periodic surveillance of ESBL producing strains and detection of the respective genes (for example blaTEM-92, blaSHV, blaGES-11, blaGES-14, blaPER-1, blaPER-7, and blaVEB-1) can be of use in the clinical setting [27,28,29,30,31]. Other notable members of this class are the extended spectrum cefotaximases (CTX-M) and the Klebsiella pneumoniae carbapenemases (KPC) [32,33].
2.1.2. Class B
Class B or metallo-beta-lactamases (MBLs) are encoded by mobile DNA (plasmids, integrons) and catalyze the hydrolysis of virtually all beta-lactamases (including carbapenems), but not monobactams, conferring multidrug-resistance. These enzymes require either zinc or another heavy metal for the catalysis and are further classified in three subclasses (B1, B2, and B3) based on sequence diversity and differences in the structure of their active sites. Moreover, four types of MBLs have been described in A. baumannii, namely IMP, VIM, NDM, and SIM [34]. The detection of MBL-producing organisms with the help of phenotypic methods, such as double disc synergy test, MBL E-test, and combined disk test, has been challenging [35,36]; however, molecular methods and in particular next generation sequencing are going to shed some light on their detection [37,38,39]. Phenotypic methods are not sensitive enough and thus do not detect all MBL producing strains [36]. With the help of PCR, blaVIM-1 was detected in 14.3% of A. baumannii isolates characterized as MBL negative by E-test, highlighting the importance of introducing molecular methods into every-day practice in order to detect these hidden MBLs [40]. Recently, López et al. demonstrated that the expression of NDM lactamases does not compromise bacterial growth and is favored over other MBLs due to the lack of fitness cost leading to its worldwide dissemination among Gram-negative bacteria [41].
2.1.3. Class C
Class C beta-lactamases are chromosomally encoded cephalosporinases (acinetobacter-derived cephalosporinase, ADC), inherent to all A. baumannii. Overexpression of these lactamases can be induced by the insertion of ISAba1 and ISAba125 sequences upstream of the encoding gene blaADC (formerly known as blaAmpC), which appear to be stronger promoters than the intrinsic promoter [42,43,44,45,46]. Insertions sequences (IS) are short transposable elements that are repeated multiple times throughout the genome, and thus are difficult to detect. An open-source bioinformatics pipeline (panISa) that utilizes whole genome sequencing (WGS) data as input has been recently developed in order to overcome this problem [47]. Several ADC variants have been described, many of which exhibit extended spectrum antibiotic resistance [48,49]. For example, ADC-30 provides resistance not only to cephalosporins, but also to carbapenems and sulbactam [50,51]. Finally, phosphoproteomic analysis revealed that dephosphorylation of ADC may lead to imipenem resistance in clinical isolates [52].
2.1.4. Class D
Class D beta-lactamases, also called oxicillinases (OXA) or carbapenem-hydrolyzing class D β-lactamases (CHDLs), can inactivate all beta-lactams (mainly OXA-10 family) and comprise the main mechanism of carbapenem resistance. These enzymes are serine-dependent just like Class A and C beta-lactamases [53]. In addition, class D beta-lactamases usually cannot be inhibited by clavulanic acid, sulbactam, and tazobactam [54]. There are several blaOXA genes, including blaOXA-51, blaOXA-23, blaOXA-24, blaOXA-58, blaOXA-143, and blaOXA-235. The genes encoding these enzymes can be found on both the chromosome as well as the plasmids. Moreover, Wong et al. have recently confirmed that carbapenem resistance in clinical isolates of A. baumannii is mediated by over-expression of either OXA-23 or OXA-51 through insertion of ISAba1 in their promoter region [55]. With the help of molecular techniques (PCR, sequencing), reservoirs of carbapenemase-producing A. baumannii have been isolated from companion animals and pigeons, highlighting as a result the importance of global surveillance [56,57].
2.2. Outer Membrane Proteins
Antibiotic resistance due to beta-lactamases can be significantly enhanced when they collaborate with outer membrane proteins (OMPs). The low permeability outer membrane protein A (OmpA, 40 kDa) is the main non-specific porin in A. baumannii and has mainly a structural role [58,59]. It is speculated that OmpA takes part in the transport of antibiotics out of the periplasmatic space [60]. On the contrary, Iyer et al. demonstrated that OmpA selectively enables the uptake of small molecules like sulbactam, imipenem, and ETX2514 [61]. In mutant strains lacking OmpA (ΔompA and OmpA-like domain deletion), instability of the outer membrane and increased susceptibility to antibiotics (including penicillins and cephalosporins) has been observed [58,60,62]. Finally, Zhong et al. have recently shown that OmpA C-terminal domain is also responsible for anchoring beta-lactamases (Oxa23, GES-11) in the periplasmatic space, which may further explain the increased susceptibility in case of OmpA loss [63].
Mussi et al. have vigorously studied the carbapenem-associated outer membrane protein (CarO, 29 kDa) in carbapenem sensitive and resistant A. baumannii [64,65,66,67]. They have demonstrated that CarO serves as an uptake channel for L-ornithine and possibly carbapenems [66]. In carbapenem-resistant clinical isolates of A. baumannii insertion, elements can disrupt the expression of the chromosomal CarO gene and lead to complete loss of CarO [65,68,69]. In addition, CarO gene mutations can induce significant structural modifications in CarO and thus reduce outer membrane permeability and eventually cause drug resistance [70]. However, there are also studies that question the role of CarO in carbapenem resistance and suggest that other porin-mediated mechanisms might exist [71,72,73,74].
Srinivasan et al. studied the physiological functions of another OMP, AbuO, and demonstrated its role in multidrug resistance. AbuO is involved in the active efflux of multiple antibiotics, including ceftriaxone and meropenem [75]. Moreover, decreased expression of a different 33-36 kDa OMP is associated with carbapenem resistance [76,77]. Lastly, there have been contradicting studies regarding the involvement of the outer membrane carboxylate channel (Occ; formerly OprD) family in carbapenem resistance [78,79].
2.3. Efflux Pumps
Increased expression of efflux pumps contributes synergistically with beta-lactamases to antibiotic resistance [80]. Overexpression of AdeABC efflux pump is associated with A. baumannii carbapenem and cephalosporin resistance [81,82,83]. The AdeABC is a three-component efflux pump, member of the resistance–nodulation–division (RND) family. AdeB component expels antibiotics out of the cell, while AdeA is a membrane fusion protein and AdeC an outer membrane protein [84]. The substrates of AdeB can be diverse; they can range from hydrophilic to hydrophobic and can be either positively charged or neutral [83]. The expression of this efflux pump is regulated by the AdeRS two-component system. Point mutations in the adeRS operon can lead to increased expression of the pump and consequently to antibiotic resistance [85,86]. It has been shown that other efflux pumps AdeIJK and AbeM may also contribute to imipenem and cephalosporin resistance [87,88].
2.4. Penicillin-Binding Proteins
Penicillin-binding proteins (PBPs) are enzymes that catalyze the polymerization of peptidoglycan and are responsible for its insertion into the cell wall [89]. Beta-lactams bind to PBPs because they mimic their substrate. Inhibition of PBPs by beta-lactams then leads to an imbalance in cell wall metabolism and as a result to cell death [90].
The role of this resistance mechanism appears to be less significant but cannot be ignored. Although mutations in hot spot regions have been observed in PBP genes, the corresponding changes in amino acid sequences were not directly related to beta-lactam resistance [91]. However, Gehrlein et al. attributed imipenem resistance in a clone of A. baumannii strain No. 4852/88 to complex PBP alterations [92]. Moreover, Yun et al. reported increased expression of PBPs in a MDR A. baumannii strain (MIC of imipenem >128 µg/mL) as a response to imipenem exposure [93]. On the other hand, Fernández-Cuenca et al. observed reduced expression of a 73 kDa PBP [94] and Vashist et al. PBP alterations in carbapenem resistant A. baumannii [95]. Siroy et al. have also showed decreased expression of PBP1b in a clinical MDR isolate [96]. Finally, PBP3 mutants have been associated with resistance to meropenem, sulbactam, and cefiderocol [82,94,95,96].
In Table 1 are summarized the genes and the respective proteins that are involved in beta-lactam resistance retrieved from the pathogen detection microbial browser for identification of genetic and genomic elements (MicroBIGG-E), a database housed by National Center for Biotechnology Information (NCBI) [97].
Table 1.
Mechanisms of A. baumannii resistance to beta-lactams.
| Mechanism of Resistance | Element Name | Resistance | Element Symbol (Gene) | Protein Products | |
|---|---|---|---|---|---|
| Class A beta lactamases | class A broad-spectrum beta-lactamase TEM-1 | extended-spectrum | blaTEM-1 | WP_000027057.1 and others | |
| class A extended-spectrum beta-lactamase SHV-5 | blaSHV-5 | WP_011117369.1 and others | |||
| class A extended-spectrum beta-lactamase SHV-12 | blaSHV-12 | WP_002904004.1 and others | |||
| carbapenem-hydrolyzing class A beta-lactamase GES-5 | blaGES-5 | WP_012658785.1 | |||
| class A extended-spectrum beta-lactamase GES-11 | blaGES-11 | WP_001211000.1 and others | |||
| class A beta-lactamase GES-12 | blaGES-12 | WP_063860500.1 and others | |||
| inhibitor-resistant class A extended-spectrum beta-lactamase PER-1 | blaPER-1 | WP_001100753.1 and others | |||
| class A extended-spectrum beta-lactamase PER-7 | blaPER-7 | WP_032495440.1 and others | |||
| class A extended-spectrum beta-lactamase VEB-1 | blaVEB-1 | WP_000706731.1 and others | |||
| class A extended-spectrum beta-lactamase CTX-M-15 | blaCTX-M-15 | WP_000239590.1 and others | |||
| class A extended-spectrum beta-lactamase CTX-M-55 | blaCTX-M-55 | WP_015387340.1 | |||
| class A extended-spectrum beta-lactamase CTX-M-115 | blaCTX-M-115 | WP_035895532.1 | |||
| carbapenem-hydrolyzing class A beta-lactamase KPC-2 | blaKPC-2 | WP_004199234.1 and others | |||
| Class B metallo-beta-lactamases | subclass B1 metallo-beta-lactamase NDM-1 | all (except monobactams) | blaNDM-1 | WP_004201164.1 and others | |
| subclass B1 metallo-beta-lactamase IMP-1 | blaIMP-1 | WP_003159548.1 | |||
| subclass B1 metallo-beta-lactamase IMP-4 | blaIMP-4 | WP_015060105.1 | |||
| subclass B1 metallo-beta-lactamase IMP-14 | blaIMP-14 | WP_039819893.1 | |||
| subclass B1 metallo-beta-lactamase IMP-16 | blaIMP-16 | WP_063860576.1 | |||
| Class C beta-lactamases | class C extended-spectrum beta-lactamase ADC-11 | extended-spectrum | blaADC-11 | WP_001211205.1 and others | |
| class C beta-lactamase ADC-25 | blaADC-25 | WP_001211217.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-26 | blaADC-26 | WP_001211238.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-30 | blaADC-30 | WP_001211218.1 and others | |||
| cefepime-hydrolyzing class C extended-spectrum beta-lactamase ADC-33 | blaADC-33 | WP_001211220.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-52 | blaADC-52 | WP_001211232.1 and others | |||
| cefepime-hydrolyzing class C extended-spectrum beta-lactamase ADC-56 | blaADC-56 | WP_031973850.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-73 | blaADC-73 | WP_001211219.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-74 | blaADC-74 | WP_001211203.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-76 | blaADC-76 | WP_001211237.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-79 | blaADC-79 | WP_001159760.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-80 | blaADC-80 | WP_029424536.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-82 | blaADC-82 | WP_001211216.1 and others | |||
| class C beta-lactamase ADC-152 | CEPH | blaADC-152 | WP_001211233.1 and others | ||
| class C beta-lactamase ADC-156 | blaADC-156 | WP_024436624.1 and others | |||
| class C beta-lactamase ADC-162 | blaADC-162 | WP_031980335.1 and others | |||
| class C beta-lactamase ADC-176 | blaADC-176 | WP_001159761.1 and others | |||
| class C beta-lactamase ADC-182 | blaADC-182 | WP_057691114.1 and others | |||
| class C beta-lactamase ADC-212 | blaADC-212 | WP_031975357.1 and others | |||
| class C beta-lactamase ADC-222 | blaADC-222 | WP_031960999.1 and others | |||
| Class D beta-lactamases (oxicillinases) | carbapenem-hydrolyzing class D beta-lactamase OXA-23 | CAR | blaOXA-23 | WP_001046004.1 and others | |
| OXA-23 family carbapenem-hydrolyzing class D beta-lactamase OXA-239 | blaOXA-239 | WP_063862190.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-24 | CAR | blaOXA-24 | WP_012754353.1 and others | ||
| OXA-24 family carbapenem-hydrolyzing class D beta-lactamase OXA-72 | blaOXA-72 | WP_000713530.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-51 | CAR | blaOXA-51 | WP_002033109.1 and others | ||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-64 | blaOXA-64 | WP_001021788.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-65 | blaOXA-65 | WP_001021782.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-66 | blaOXA-66 | WP_001021792.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-68 | blaOXA-68 | WP_001021775.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-69 | blaOXA-69 | WP_001021779.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-71 | blaOXA-71 | WP_001021785.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-82 | blaOXA-82 | WP_001021793.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-90 | blaOXA-90 | WP_001021781.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-91 | blaOXA-91 | WP_001021776.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-94 | blaOXA-94 | WP_029424390.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-95 | blaOXA-95 | WP_031960432.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-98 | blaOXA-98 | WP_001021777.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-100 | blaOXA-100 | WP_001021795.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-104 | blaOXA-104 | WP_024433915.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-120 | blaOXA-120 | WP_004738885.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-223 | blaOXA-223 | WP_001022758.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-259 | blaOXA-259 | WP_001021784.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-371 | blaOXA-371 | WP_063862738.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-402 | blaOXA-402 | WP_001021789.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-58 | CAR | blaOXA-58 | WP_002002480.1 and others | ||
| OXA-58 family carbapenem-hydrolyzing class D beta-lactamase OXA-96 | blaOXA-96 | WP_063864543.1 | |||
| OXA-134 family carbapenem-hydrolyzing class D beta-lactamase OXA-235 | CAR | blaOXA-235 | WP_000854009.1 and others | ||
| OXA-134 family carbapenem-hydrolyzing class D beta-lactamase OXA-237 | blaOXA-237 | WP_000854010.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-143 | CAR | blaOXA-143 | WP_063861042.1 | ||
| OXA-143 family carbapenem-hydrolyzing class D beta-lactamase OXA-253 | blaOXA-253 | WP_032495764.1 | |||
| Efflux pumps | multidrug efflux RND transporter AdeABC outer membrane channel subunit AdeC | CEPH, CAR | adeC | WP_000047249.1 and others | |
| Acinetobacter baumannii efflux resistant AdeR | adeR_A91V | WP_039198290.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeR | adeR_P56S | WP_088753133.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeR | adeR_P116L | WP_111853508.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeS | adeS_G336S, adeS_N125K | WP_031975145.1, WP_057691178.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeS | adeS_H189Y | WP_119491814.1, WP_000837466.1, WP_046882653.1 | |||
| Penicillin-binding proteins | Acinetobacter baumannii carbapenem resistant FtsI | CAR | ftsI_A515V | WP_000227939.1 and others (penicillin-binding protein PBP3) | |
CEPH = cephalosporins; CAR = carbapenems.
3. Resistance to Aminoglycosides
According to MicroBIGG-E, resistance to aminoglycosides (AG) in A. baumannii can result through three distinct mechanisms (Table 2) [97]: aminoglycoside-modifying enzymes (AMEs) that weaken AG binding capacity, target site alteration by 16S rRNA methyltransferases, and limited AG uptake subsequent to loss of permeability or overactivity of efflux pumps. AMEs are further categorized as acetyl-, adenyl-, and phospho-transferases, depending on the site of AG modification (primarily by N-acetylation, O-nucleotidylation, and O-phosphorylation, respectively). A. baumannii displays intrinsic resistance to many antibiotics, while it seems to have acquired resistance to AGs in 19% to 31% of cases [98]. AGs are protein synthesis inhibitors that exert their action after crossing bacterial cell wall and by disturbing peptide elongation at the 30S ribosomal subunit. Genes conferring resistance can be transported by means of integrons, gene cassettes, transposons, and conjugated elements. Beyond the molecular level, and at cellular level, AG resistance genes can be transferred by means of mobilizable or conjugative plasmids, natural transformation, or transduction [99]. More specifically, AG resistance genes can be located in: (i) plasmid: aac(3)-Ia, aac(3)-IVa, AAC(6’)-Ia family, aac(6’)-Ib, aac(6’)-Ib-cr, aac(6′)-Ih, aac(6’)-IIc, ant(2’’)-Ia, ant(3’’)-Ia, aadA1, aadA2, aadA5, aadA13, aadA16, aph(3’)-VIa, aph(3’)-VIb, aph(3’’)-Ib, aph(4)-Ia, aph(6)-Id, and armA; (ii) integron: aac(3)-Ia, AAC(6’)-Ia family, aac(6’)-Ib, aac(6’)-Ib’, aac(6’)-Ib3, aac(6’)-Ib-cr, aac(6’)-IIc, ant(2’’)-Ia, ant(3’’)-Ia, aadA1, aadA2, aadA5, aadA11, aadA13, and aadA16; (iii) transposon: aac(3)-Ia, AAC(6’)-Ia family, aac(6’)-Ib, aac(6’)-Ib-cr, ant(3’’)-Ia, aadA1, aadA5, aph(3’)-Ia, aph(3’)-IIa, aph(3’’)-Ib, and armA; (iv) integrative conjugative element: aph(6)-Id, and aph(3’’)-Ib; (v) chromosome: aac(2′)-Ib, and aph(3’’)-Ib; or (vi) chromosomal genomic island: aph(6)-Id [100].
Table 2.
Mechanisms of A. baumannii resistance to aminoglycosides.
| Element Name and Symbol | Resistance | Gene | Protein Products | |
|---|---|---|---|---|
| Aminoglycoside acetyltransferases | Aminoglycoside 2’-N-acetyltransferase AAC(2’)-Ib | GEN, TOB, DIB, NET [101] | aac(2′)-Ib | WP_001159732.1 † |
| Aminoglycoside 3-N-acetyltransferase | GEN | aac(3) | WP_195206917.1 | |
| AAC(3)-I family aminoglycoside 3-N-acetyltransferase | GEN | aac(3)-I | HAV6561382.1, WP_069597335.1 | |
| Aminoglycoside N-acetyltransferase AAC(3)-Ia | AST, GEN, SIS [102] | aac(3)-Ia | WP_002089484.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(3)-IId | GEN | aac(3)-IId | WP_000557454.1, WP_126562472.1 | |
| Aminoglycoside N-acetyltransferase AAC(3)-IIe | GEN | aac(3)-IIe | WP_000557452.1, WP_002063884.1, WP_033107705.1, WP_095530619.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(3)-Iva | APR, GEN, TOB | aac(3)-IVa | WP_001199192.1 | |
| Aminoglycoside 6’-N-acetyltransferase | all | aac(6’) | HAV4276337.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-31 | all | aac(6’)-31 | WP_044424439.1 | |
| Aminoglycoside 6’-N-acetyltransferase AAC(6’)-33 | all | aac(6’)-33 | WP_015059044.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase | AMI, KAN, TOB, putatively against all | aac(6’) | EGY2236091.1, EGY5968849.1, WP_088756823.1, WP_088774065.1, WP_140976846.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase AacA16 | all | aacA16 | WP_001109644.1 | |
| Aminoglycoside 6’-N-acetyltransferase AacA34 | all | aacA34 | WP_052285801.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase AacA43 | KAN, TOB | aacA43 | WP_024437351.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ian | AMI, KAN, TOB, putatively against all | aac(6’)-Ian or aacA57-2 | WP_000960976.1 | |
| AAC(6’)-Ib family aminoglycoside 6’-N-acetyltransferase | AMI, DIB, GEN, ISE, KAN, NET, SIS, TOB [102] | aac(6’)-Ib | WP_063840280.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib’ | GEN | aac(6’)-Ib’ | WP_014454105.1 | |
| AAC(6’)-Ighjkrstuvwx family aminoglycoside N-acetyltransferase | AMI, KAN, TOB | aac(6’)-I | WP_169109636.1, WP_150956588.1, WP_005288246.1, WP_005243483.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib3 | AMI, KAN, TOB | aac(6’)-Ib3 | WP_032488579.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib4 | GEN | aac(6’)-Ib4 | WP_003159191.1 | |
| aminoglycoside N-acetyltransferase AAC(6’)-Ih | AMI, KAN, TOB | aac(6’)-Ih | WP_016541245.1 † | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Il | AMI, KAN, TOB | aac(6’)-Il | WP_156193962.1 | |
| AAC(6’)-II family aminoglycoside 6’-N-acetyltransferase AacA35 | GEN, KAN, TOB | aacA35 | WP_024437054.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-IIc | GEN, KAN, TOB | aac(6’)-IIc | WP_149959345.1, WP_149938250.1 | |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr | AMI, KAN, TOB, QUI | aac(6’)-Ib-cr | WP_185936887.1 | |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr5 | AMI, KAN, TOB, QUI | aac(6’)-Ib-cr5 | WP_063840321.1 | |
| Aminoglycoside adenyltransferases | Aminoglycoside nucleotidyltransferase ANT(2’’)-Ia | DIB, GEN, KAN, SIS, TOB [102] | ant(2’’)-Ia | WP_000381802.1, and others |
| ANT(3’’)-I family aminoglycoside nucleotidyltransferase | STR, SPE | ant(3’’) | WP_038350223.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA | STR, SPE | ant(3’’)-Ia | WP_001205725.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA1 | STR | aadA1 | WP_001206316.1, and others | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA2 | STR | aadA2 | WP_001206356.1, WP_001261740.1, and others | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA5 | STR | aadA5 | WP_000503573.1, WP_000503574.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA11 | STR | aadA11 | WP_048608579.1, HAV4466908.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA13 | STR | aadA13 | WP_001424636.1 † | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA16 | STR | aadA16 | WP_001749984.1, WP_185936919.1 | |
| ANT(3’’)-II family aminoglycoside nucleotidyltransferase | STR, SPE | ant(3’’)-II | WP_005240470.1 | |
| Aminoglycoside nucleotidyltransferase ANT(3’’)-IIa | STR, SPE | ant(3’’)-IIa | WP_001279062.1, WP_001279061.1,WP_001112625.1, and others | |
| Aminoglycoside nucleotidyltransferase ANT(3’’)-IIc | STR, SPE | ant(3’’)-IIc | WP_005281276.1 | |
| Aminoglycoside phosphotransferases | APH(3’) family aminoglycoside O-phosphotransferase | all | aph(3’) | WP_196077463.1 |
| Aminoglycoside O-phosphotransferase APH(3’)-Ia | GEN, KAN, NEO, PAR, LIV, RIB [102] | aph(3’)-Ia | WP_000018326.1, and others | |
| APH(3’)-II family aminoglycoside O-phosphotransferase | KAN | aph(3’)-II | WP_000262467.1 | |
| Aminoglycoside O-phosphotransferase APH(3’)-IIa | KAN | aph(3’)-IIa | WP_000572405.1, WP_171502934.1, and others | |
| APH(3’)-VI family aminoglycoside O-phosphotransferase | AMI, KAN | aph(3’)-VI | WP_014386410.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’)-VIa | AMI, KAN | aph(3’)-VIa | WP_000422636.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’)-VIb | AMI, KAN | aph(3’)-VIb | WP_000422633.1, WP_000422632.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’’)-Ib | STR | aph(3’’)-Ib | WP_001082319.1, and others | |
| Aminoglycoside O-phosphotransferase APH(4)-Ia | HYG | aph(4)-Ia | WP_185218783.1 | |
| Aminoglycoside O-phosphotransferase APH(6)-Id | STR | aph(6)-Id | WP_000480968.1, and others | |
| Target mutation: 16S rRNA methylase genes | ArmA family 16S rRNA (guanine(1405)-N(7))-methyltransferase | GEN | armA | WP_000359986.1, and others |
| 16S rRNA (guanine(1405)-N(7))-methyltransferase RmtB1 | all | rmtB and rmtB1 | WP_012372818.1 | |
| RmtE family 16S rRNA (guanine(1405)-N(7))-methyltransferase | all | rmtE | WP_120494548.1 | |
| Efflux pump overactivity | Multidrug efflux MFS transporter AmvA | Putatively against all | amvA | WP_001170321.1, and others |
| Multidrug efflux RND transporter AdeABC outer membrane channel subunit AdeC | adeC | WP_000047249.1, and others | ||
| Multidrug efflux RND transporter periplasmic adaptor subunit AdeD | adeD | WP_002119008.1, WP_039254548.1 | ||
| Multidrug efflux RND transporter permease subunit AdeE | adeE | WP_002118518.1, WP_039254549.1 | ||
| Efflux system DNA-binding response regulator transcription factor AdeR | adeR | WP_032002707.1, EGY8404952.1,WP_020752724.1 | ||
| Two-component sensor histidine kinase AdeS two-component sensor histidine kinase | adeS | WP_031975145.1, and others | ||
| Multidrug efflux SMR transporter EmrE | emrE | WP_109847152.1 |
AMI = amikacin; APR = apramycin; AST = astromicin; DIB = dibekacin; DNA = deoxyribonucleic acid; GEN = gentamicin; HYG = hygromycin; ISE = isepamicin; KAN = kanamycin; LIV = lividomycin; MFS = major facilitator superfamily; NEO = neomycin; NET = netilmicin; PAR = paromomycin; QUI = quinolone; RIB = ribostamycin; RNA = ribonucleic acid; RND = resistance/nodulation/cell division family; SIS = sisomicin; SMR = small multidrug resistance family; SPE = spectinomycin; STR = streptomycin; TOB = tobramycin; † = Identified via NCBI Identical Protein Groups, but with no respective mention in MicroBIGG-E.
AME genes aph(3’)-VIa, aph(3’)-VIb, and aph(3’)-VI seem to be significantly more prevalent among amikacin and kanamycin resistant isolates, while isolates harboring aac(6’)-Ian, aac(6’)-Ib, aac(6’)-Ib3, aac(6’)-I, and aac(6’)-Il genes exhibit resistance against amikacin, kanamycin, and tobramycin [81,87]. Conversely, resistance to gentamycin should not be taken for granted in the presence of the above genes, while it is mainly mediated by 16S rRNA methylation (armA gene), 3-N- and 6’-N-acetylation of gentamycin, and by 2’’-O-nucleotidylation (ant(2’’)-Ia gene; correlated originally with resistance to kanamycin and tobramycin [97,103]. Unlike amikacin, gentamicin, and tobramycin that lack 3’-hydroxyl, groups usually retain activity in the presence of O-phospotransferases due to their inability to accept phosphate [104]. Resistance to AGs is increasing, and target site alteration seems to play a pivotal role with armA and rmtB1 genes to be detected with plentiful of coexisting resistance genes [99].
Although AMEs remain the principal procedure by which A. baumannii evades elimination by AGs, efflux is also an emerging problem with AG use. Seven different gene products (Table 2) result in effective efflux of AGs, including pumps, permeases, periplasmic adaptors, and two component systems (TCSs) [105]. Of note, gentamicin and netilmicin are effectively cleared by AdeABC and AbeM pumps, but efflux is significantly weaker as regards more hydrophilic AGs, like amikacin and kanamycin [106]. Changes in membrane lipids and porin expression are only minor mechanisms of AG resistance and are still under investigation [99]. Recent case reports have indicated the usefulness of next-generation sequencing (NGS) in the prompt diagnosis and appropriate treatment of A. baumannii infections by early identification of resistance genes [107].
4. Resistance to Tetracyclines
Tetracycline antibiotics bind to the 30S ribosomal subunit and thereby inhibit protein synthesis by deterring the start of translation [108]. Resistance to tetracycline antibiotics is attributed to three main mechanisms: (i) efflux dependent on ATP, (ii) inactivation of tetracyclines by enzymes, and (iii) ribosomal protection proteins (RPPs) [109]. Table 3 describes resistance mechanisms that are unique for this antibiotic group.
Table 3.
Tetracycline-specific mechanisms of A. baumannii resistance.
| Element name and symbol | Resistance | Gene | Protein products |
|---|---|---|---|
| Tetracycline efflux MFS transporter Tet(39) | DOX, TET [110,111] | tet(39) | WP_004856455.1, and others |
| Tetracycline efflux MFS transporter Tet(A) | DOX, MIN, TET, TIG * | tet(A) | WP_000804064.1, and others |
| tetracycline efflux MFS transporter Tet(B) | DOX, MIN, TET [111] | tet(B) | WP_001089072.1, and others |
| Tetracycline efflux MFS transporter Tet(C) | TET [112] | tet(C) | WP_000841448.1 |
| Tetracycline efflux MFS transporter Tet(D) | TET [113] | tet(D) | WP_024436252.1 |
| Tetracycline efflux MFS transporter Tet(G) | DOX, MIN, TET | tet(G) | WP_001257840.1 |
| Tetracycline efflux MFS transporter Tet(H) | OXY, TET [114] | tet(H) | WP_006248867.1 |
| Tetracycline resistance ribosomal protection protein Tet(M) | TET, MIN | tet(M) | WP_000691727.1 |
| Tetracycline-inactivating monooxygenase Tet(X) | all | tet(X) | WP_024160783.1, and others |
DOX = doxycycline; MFS = major facilitator superfamily; MIN = minocycline; OXY = oxytetracycline; TET = tetracycline; TIG = tigecycline; * = only when RND-pumps AdeABC and AdeIJK are present.
Two types of efflux pumps that require energy are responsible for tetracycline resistance in A. baumannii. The resistance/nodulation/cell division (RND) family-type pumps are constitutive non-specific pumps originating from adeA, adeB, and adeC genes, which encode periplasmic adaptor subunits, permease subunits, and outer membrane pump elements, respectively [83]. RND pumps, and predominantly AdeABC, can effectively eliminate tetracyclines, while correspondingly, they mediate a substantial elevation of minimum inhibitory concentrations (MICs) for tigecyline, minocycline, and tetracycline [106]. RND pump AdeIJK seems to have a minor role in tetracycline resistance against A. baumannii, but it can act synergistically with other overexpressed efflux pumps (like AdeABC and AcrAB-TolC) and result in tigecycline resistance [115]. The second category refers to tetracycline major facilitator superfamily (MFS) efflux pumps: TetA and TetB [116]. TetA seems to lead efflux of tigecycline into the periplasm, and subsequently, RND pumps drive to elimination through the outer membrane [117].
TetM/TetW/TetO/TetS family tetracycline resistance RPPs abolish the inhibitory effect of tetracycline on protein synthesis by a non-covalent modification of the ribosomes [118]. Though rare, resistance to minocycline in A. baumannii has been attributed to ribosomal protection and tetM gene. The latter gene product mediates tetracycline release from its ribosomal binding site by a GTP-dependent mechanism, enabling the continuation of translation despite the presence of tetracycline [119].
Recent transcriptomic studies have shown that A. baumannii can swiftly become resistant to tigecycline [120]. Tigecycline has been a powerful tool against A. baumannii in the clinician’s armamentarium until recently, when various plasmid-mediated tet(X) gene variants emerged. Tet(X3), Tet(X4), and Tet(X5) are monooxygenases that can inactivate all tetracyclines even tigecycline and the recently authorized eravacycline and omadacycline [121,122].
5. Resistance to Fluoroquinolones
Quinolones are bactericidals with a broad spectrum that are characterized by a bicyclic core formation bearing resemblance to 4-quinolone. Quinolone antibiotics are mostly fluoroquinolones displaying efficacy against both Gram-negative and Gram-positive pathogens [123]. Regarding their mechanism of action, quinolone antibiotics interrupt DNA replication by averting bacterial DNA from loosening and being cloned. Quinolones exert their action by inhibiting the ligase activity of the type II topoisomerases, DNA gyrase, and topoisomerase IV, which normally induce supercoiling in collaboration with DNA nucleases. Disrupting ligase activity, bacteria remain with double-stranded DNA breaks and thus are led to cell death. Notably, quinolones primarily affect gyrase activity, while toxicity against topoisomerase IV is secondary (i.e., there is no proof of sole parC mutations without concomitant alterations in gyrA) [123]. Quinolone resistance occurs via three different mechanisms: (i) target mutations in gyrase and topoisomerase IV, which weaken the respective quinolone–enzyme interactions; (ii) plasmid-borne resistance mediated by Qnr proteins, the AMEs AAC(6′)-Ib-cr and AAC(6′)-Ib-cr5, and by plasmid-encoded efflux pumps; and (iii) chromosome-derived resistance resulted by either low expression of porins or overexpression of chromosome-encoded efflux pumps [123,124,125]. A recent review reported resistance of A. baumannii to fluoroquinolones between 50% and 73% of cases, while the respective resistance in developing countries during the last years displayed a marked increase reaching 75% to 97.7% [98,126]. Table 4 lists all recorded quinolone-specific resistance mechanisms for A. baumannii.
Table 4.
Quinolone-specific mechanisms of A. baumannii resistance.
| Element Name and Symbol | Resistance | Gene | Protein Products |
|---|---|---|---|
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr | CIP, NOR | aac(6’)-Ib-cr | WP_185936887.1 |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr5 | CIP, NOR | aac(6’)-Ib-cr5 | WP_063840321.1 |
| A. baumannii quinolone resistant GyrA (DNA gyrase subunit A) | CIP, LEV | gyrA_S81L | WP_000116444.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_E88K | WP_000202265.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_S84F | WP_000202250.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_S84L | WP_000202252.1, and others |
| QnrA family quinolone resistance pentapeptide repeat protein | CIP | qnrA | HAV5951840.1 |
| QnrB family quinolone resistance pentapeptide repeat protein | CIP | qnrB | WP_185936934.1 |
| Quinolone resistance pentapeptide repeat protein QnrB19 | CIP | qnrB19 | WP_012954666.1 |
| QnrS family quinolone resistance pentapeptide repeat protein | CIP, * | qnrS | WP_147508156.1 |
CIP = ciprofloxacin; LEV = levofloxacin; NOR = norfloxacin; * = resistance to CIP, LEV, NOR, and enrofloxacin in the presence of rmtB [127].
Quinolone resistance occurring secondary to overactive RND pumps has been long documented and is quite common [128]. Mutations in the TCS elements regarding both regulator (AdeR, primarily with polymorphisms D20N, A91V, A136V, and P116L) and sensor (AdeS, mainly due to polymorphisms G30D, A94V, G103D, G186V, and T153M) of AdeABC pump result in higher fluoroquinolone efflux. Mutations in adeR and adeS seem to trigger overexpression of the AdeABC efflux system (especially of the permease subunit adeB gene) and are associated with resistance to ciprofloxacin, norfloxacin, and ofloxacin [128]. Beyond AdeABC, efflux systems AdeIJK and AdeFGH are also utilized by A. baumannii to remove fluoroquinolones from the cell and result in high MICs. In addition, contribution of the multidrug and toxic compound extrusion (MATE) transporters AbeM and AbeS to A. baumannii resistance against quinolones is disputable. AbeM pump primarily affects the hydrophilic fluoroquinolones, norfloxacin and ciprofloxacin, rather than the hydrophobic ones (such as ofloxacin). In general, these non-RND efflux pumps confer low-level resistance to fluoroquinolones, even though some studies suggest otherwise [124,129].
Quinolone resistance-determining regions (QRDRs) refer mainly to alteration of target sites in gyrase (Ser83Leu, Gly81Asp, and Ser81Leu mutations preventing quinolones from binding its alpha-subunit) and topoisomerase IV (mutations Ser80Leu, Glu84Lys, and Gly78Cys, and Ser84Leu in its subunit C). Although a single point mutation in DNA gyrase is usually not enough for resistance to fluoroquinolones in A. baumannii (maybe only against levofloxacin; single parC mutations link with ciprofloxacin resistance), concurrent mutations within QRDR regions of the gyrA and parC genes are linked with significantly higher level of quinolone resistance [130,131]. Alterations in gyrB and parE genes are of minor significance [126].
Plasmid-mediated quinolone resistance (PMQR) has been recently identified as a clinical problem against A. baumannii infections, but generally confers still low-level (≤10-fold) resistance [123,132]. Qnr genes qnrAI (the first PMQR gene to be identified), qnrB, qnrB19, and qnrS encode members of the pentapeptide-repeat protein family (homologous to McbG and MfpA proteins), which originally inhibit gyrase action through competition with DNA for binding, leading to a decrease of DNA binding with topoisomerase and thus protecting enzyme–DNA complexes from the action of quinolones [97,132]. Due to the aforementioned DNA homology, these pentapeptide repeat-containing Qnr proteins can also interact with gyrase and topoisomerase IV and prevent quinolones from cleavage, leading eventually to aggregation and subsequent accumulation of double-stranded DNA breaks that would be, in this case, lethal to A. baumannii [133]. PMQR by AMEs AAC(6′)-Ib-cr and AAC(6′)-Ib-cr5 is attributed practically to mutant variants of 6’ aminoglycoside acetyltransferase-Ib gene (“-cr” implying ciprofloxacin resistance; W102R and D179Y mutations) that encode enzymes acetylating the C7 unsubstituted nitrogen in the piperazine ring of norfloxacin and ciprofloxacin [134]. The third category of plasmid-borne quinolone resistance refers to efflux pumps, but no such case has been documented for A. baumannii so far. QepA (quinolone efflux pumps A1 and A2; contributing to resistance to norfloxacin, enrofloxacin, levofloxacin, and ciprofloxacin among rmtB-positive E. Coli) and OqxAB (quinoxaline-di-N-oxide olaquindox) are the first described pumps of this category [127,133].
Chromosome-derived resistance to fluoroquinolones associates either with low influx rhythm by downregulated or dysfunctional porins, or with efflux pumps that are overactive. In fact, porins are virulence factors of Gram-negative bacteria that regulate cellular permeability through the outer membrane and are merely linked with carbapenem resistance (e.g., in Pseudomonas aeruginosa isolates with low OprD expression). Porin genes like ompA, omp25, omp33, oprC, oprD, oprW, dcap-like, and carO have been implicated in this resistance mechanism. Low relative expression of Omp25 and CarO porins seems to correlate with resistant A. baumannii strains, but resistance to quinolones has not been justified [111,135]. In the case of fluoroquinolones, chromosome-encoded efflux pumps (described in Table 2) and porin alterations alone do not seem to render significant clinical resistance to A. baumannii [123,129].
6. Resistance to Macrolides—Lincosamides—Streptogramin Antibiotics
Macrolide antibiotics are of little use in A. baumannii infections. Azithromycin, but no other macrolide, seems to inhibit mucin production, suggesting efficacy against ventilator-associated pneumonia. Azithromycin for nosocomial pneumonia due to A. baumannii for pediatric and adult patients in intensive care units (ICUs) is used in combination with other antibiotics. In the latter case, azithromycin’s mechanism of action differentiates from its established way of action (inhibiting translation and protein biosynthesis by attaching to the 50S subunit): suppression of ERK/JNK pathway phosphorylation (between extracellular signal-regulated kinases and c-Jun N-terminal kinases) and nuclear translocation of NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) [136]. According to MicroBIGG-E database, resistance to macrolides in A. baumannii is attributed to (i) three 23S rRNA (adenine(2058)-N(6))-methyltransferases, encoded by erm(B), erm(C), and erm(F) genes), (ii) ABC-F type ribosomal protection protein Msr(E) or msr(E), and (iii) two macrolide 2’-phosphotransferases encoded by mph(A) and mph(E) [97]. The first two classes lead to resistance by modification of the target site, while the third class results in macrolide inactivation. In particular, Mph(A) and Mph(E) seem to confer resistance to erythromycin, clarithromycin, azithromycin, and oleandomycin, but only in the presence of specific regulatory proteins [137].
Additional literature search revealed several references to efflux pumps involved in macrolide resistance in A. baumannii infections. WGS confirmed recently the potential role of multidrug efflux MFS transporter AmvA (encoded by amvA) in erythromycin resistance [138,139]. Another WGS study identified transferable genetic elements in macrolide-resistant A. baumannii strains encoding the proton motive macrolide efflux MFS transporter and ribosomal protection protein mef(E)/mel (originally found in resistant streptococci) in an operon of MEGA-element (a DNA fragment named after the macrolide efflux genetic assembly). Unlike other mechanisms discussed above, MefE pumps do not result in higher MICs for lincosamides or streptogramins [140,141,142]. The tripartite MacA–MacB–TolC transporter in A. baumannii intersects inner and outer membranes so as to actively extrude macrolides. Main components of this transmembrane machine are the ATPase MacB, belonging to the ABC superfamily, and the membrane fusion protein MacA, conferring synergistically resistance to azithromycin and roxithromycin [143]. In addition, detection of the SMR family transporter AbeS (homologue of E. Coli EmrE) links with erythromycin resistance [144]. Another pump involved in macrolide-resistant A. baumannii is AdeABC efflux pump, regulated by base substitutions in the adeRS operon. Conversely, inactivation of the latter operon negatively affects biofilm formation and prompts decreased expression of AdeABC [145].
Lincosamides compounds lincomycin, clindamycin, and pirlimycin are of limited use as single antimicrobial agents in A. baumannii infections. Their mechanism of action resembles that of macrolides, by binding the ribosomal 50S subunit and specifically its 23S portion to a site corresponding to the peptidyl–transferase center. Consequently, plasmid- and transposon-derived 23S rRNA methyltransferases belonging to the erm family (erythromycin ribosome methylase) elicit combined resistance to macrolides, lincosamides, and streptogramins B (i.e., MLSB phenotype) [146]. Although resistance to lincosamides can occur through ribosomal modification, efflux, and drug inactivation, reports from A. baumannii strains are limited to efflux pumps. RND efflux pumps AdeABC, AdeFGH, and AdeIJK have been documented to significantly increase MICs of all lincosamides in several A. baumannii isolates [129,147,148,149]. Far from pumps, WGS revealed the link between ABC transporter Msr(E) and macrolide phosphotransferase mph(E) with MLSB phenotype [150,151,152].
7. Resistance to Polymyxins
As mentioned before, Gram-negative bacteria, such as A. baumannii, bear a semi-permeable outer membrane for insertion of essential elements and clearance of toxic compounds. Lipopolysaccharides (LPSs) reside on the outer surface and contain a negatively charged hydrophobic lipid A, which in turn interacts with the cationic non-ribosomal lipopeptides of polymyxins B and E (widely known as colistin). This interaction results in destabilization of the outer membrane, uptake of the polymyxins into the periplasm, and increased permeability by disrupting the integrity of both outer and inner membranes. Although the detailed mechanism of action is unknown, the hydrophobic tail of polymyxins seems to be crucial for the induction of membrane damage, suggesting a detergent-like mode of action [153].
Mechanisms of resistance to polymyxins in A. baumannii include (i) drug target alteration by LPS lipid A modification subsequent to mutations in the pmrCAB operon and mcr genes; (ii) mutations of lpxA, lpxC, and lpxD genes -encoding acyltransferases essential to lipid A biosynthesis and associated with lipid A deficiency; (iii) lpsB, lptD, and vacJ expression associated with permeability defects and osmotic resistance of the outer membrane, subsequently leading to markedly elevated MICs for polymyxins; (iv) insufficient concentration of cofactors constitutional for LPS formation, like biotin, which are essential for susceptibility to polymyxins; and (v) efflux pumps [153,154,155,156].
Operon pmrCAB includes pmrC gene encoding a phosphoethanolamine (PEA) transferase, along with pmrA and pmrB that encode the PmrA/PmrB TCS. Mutations in the latter TCS (and especially of pmrA) induce the overexpression of pmrC. PEA residue addition to the 4′-phosphate group site of lipid A hepta-acylated form is the most commonly reported modification in polymyxin-resistant A. baumannii, resulting in removal of negative charges and thus lowering the affinity of LPS to this drug class. The addition of 4-amino-L-arabinose and/or galactosamine to the phosphate groups of lipid A and to residues within the core oligosaccharide acts similarly. Likewise, PmrA/PmrB TCS can lead to polymyxin resistance by upregulating the transcription of NaxD deacetylase and therefore by modifying LPS lipid A by means of the deacetylated β-galactosamine. In addition, mutations in miaA gene (encoding the posttranscriptional regulator tRNA dimethylallyl diphosphate transferase) seem to act synergistically with mutations in pmrA towards producing polymyxin-resistant phenotypes [157,158]. In the same context, mutations in the pmrC paralogues eptA-1 and eptA-2 have been associated with increased expression and colistin resistance [153,159]. Other PEA transferases that affiliate with polymyxin-resistance are encoded by mcr-1, mcr-4, and mcr-4.3 genes. Noteworthily, resistance emerging from MCR PEA transferases that was thought to be originally chromosomal (and therefore limiting its rapid distribution and dissemination) has been recently reported to be also carried by plasmids [160,161,162].
Biotin, which is essentially involved in fatty acid synthesis, contributes in a dose-dependent manner to LPS synthesis. Deletions in lpsB, a gene encoding a glycosyltransferase involved in biotin synthesis, have been associated with colistin resistance [163].
A recent WGS study on colistin-resistant isolates of A. baumannii identified linkage with vacJ, zndP, pldA, ttg2C, and pheS genes. VacJ is linked with the Vps-VacJ ABC transporter system, which is in charge of preserving LPS and phospholipids at the outer and inner leaflet of the outer membrane respectively. Correspondingly, PldA is designated to remove phospholipids in the outer leaflet of the outer membrane, while ZndP is a zinc-dependent peptidase A, upstream of PldA, that also has a pivotal role in the outer membrane processing. Ttg2C is an efflux ABC transporter upregulated in response to phenol exposure (an organic solvent that solubilizes the cell wall, acting equivalently with polymyxins). Moreover, mutations in pheS that encodes phenylalanine-tRNA ligase subunit alpha seem to play a role in colistin resistance [164]. Beyond the two aforementioned ABC transporters, and as stated before in text, overexpressed RND efflux pump MexAB-OprM correlates with colistin resistance [156,165].
8. Resistance to Others
8.1. Resistance to Amphenicols—Oxazolidinones
A. baumannii usually acquires resistance to amphenicols through five distinct mechanisms: (i) chloramphenicol O-acetyltransferases: type A-1, type A-2 CatII, type B-2 CatB11, type B-3 CatB3, and type B-3 CatB8 encoded by catA1, catA2, catB11, catB3, and catB8 genes, respectively; (ii) bifunctional type B-3 chloramphenicol O-acetyltransferase CatB8/aminoglycoside N-acetyltransferase AAC(6’)-Ib or catB8/aac(6’)-Ib’ found in A. pittii; (iii) chloramphenicol efflux MFS transporters: CmlA/FloR family (cml), CmlA family (cmlA), CmlA1, CmlA5, CmlA6, and CmlB1; (iv) chloramphenicol/florfenicol efflux MFS transporters FloR and FloR2; and (v) multidrug efflux pumps [97]. All the above mechanisms confer resistance to chloramphenicol; the second class confers combined resistance to both chloramphenicol and gentamicin, while the fourth class confers resistance to chloramphenicol and florfenicol [166]. Specifically, the presence of cmlA5 induces resistance to chloramphenicol and thiamphenicol, while craA identification associates with loss of susceptibility to chloramphenicol, thiamphenicol, and florfenicol. Of note, single-component MFS family chloramphenicol/H+ antiporter CraA (a homologue of E. Coli MdfA and MdtM pumps) can act in synergy with RND AdeABC, AdeFGH, and AdeIJK transporters towards the amphenicol resistance phenotype, particularly when chloramphenicol acetyltransferases are absent [167]. In the same context, MDR A. baumannii strain 5075 (Ab5075) chloramphenicol resistance has been linked with efflux by an inner membrane permease encoded by ABUW_0982 gene [168]. Moreover, a recent study identified 8 MDR A. baumannii isolates bearing tet(X5) in plasmids and two isolates with tet(X6) in their chromosomes that displayed resistance to tigecycline, tetracycline, ciprofloxacin, trimethoprim/sulfamethoxazole, and florfenicol [169].
Although decreased membrane permeability has been mainly attributed to carbapenem-resistance in A. baumannii, low expression of porin OmpA has been associated with chloramphenicol, aztreonam, and nalidixic acid resistance [60]. Another gene, abrp, encoding a C13 family peptidase has been linked with decreased cell membrane permeability, faster cell growth rate, and decreased susceptibility to chloramphenicol, tetracycline, minocycline, doxycycline, tigecycline, and fosfomycin [170]. Conversely, overexpression of global regulator SoxR led to increased susceptibility to chloramphenicol, by downregulating abeS, abeM, adeJ, and adeG, although adeB and craA expression remained intact [171]. Additionally, deletion of valine–glycine repeat G (vgrG gene), a component of the Type VI Secretion System (T6SS), which is considered as a crucial virulence factor of A. baumannii, led to reduced resistance to chloramphenicol [172].
Rather bizarre combinations of colistin with anti-Gram-positive antibiotics, like the oxazolidinone linezolid, have demonstrated efficacy and have been utilized in cases of MDR A. baumannii [173]. Protein biosynthesis inhibition is mediated by different mechanisms as regards oxazolidinones, chloramphenicol, and lincosamides, but their target sites on the 23S ribosomal RNA subunit may be overlapping. The most prevalent mechanisms for linezolid resistance are multidrug efflux pumps, base substitutions in domain V of the ribosomal RNA 23S (conferring resistance to oxazolidinones, macrolides, lincosamides, streptogramins, phenicols, pleuromutilins, and glycopeptide antibiotics), and/or the presence of a transmissible Cfr(B) 23S ribosomal RNA methyltransferase (conferring resistance to oxazolidinones, streptogramins, phenicols, and lincosamides). Interestingly, plenty of microorganisms carry multiple gene copies that encode the 23S rRNA subunit, while the observed linezolid resistance corresponds to the ratio of wild type to mutant 23S rRNA [156,174]. According to the Comprehensive Antibiotic Resistance Database (CARD), resistance to linezolid can also occur by mutations in the 50S ribosomal subunit P-site (which attaches the peptidyl-tRNA to the developing polypeptide chain), by the MFS antibiotic efflux pump LmrS (capable of extruding oxazolidinones, macrolides, phenicols, aminoglycosides, and diaminopyrimidine antibiotics), ClcD (a cfr-like 23S ribosomal RNA methyltransferase linked with resistance to oxazolidinones, lincosamides, streptogramins, phenicols, and pleuromutilins), and ABC-F ATP-binding cassette ribosomal protection protein encoded by poxtA (conferring resistance to oxazolidinones, tetracyclines, macrolide antibiotic, lincosamides, streptogramins, phenicols, and pleuromutilins) [156].
8.2. Resistance to Glycopeptide and Lipopetide Antibiotics
Zeocin is a formulation of phleomycin D1, a glycopeptide antibiotic of the bleomycin family. Resistance to zeocin is linked with phleomycin/bleomycin binding protein Ble-Sh (ble-Sh gene), while resistance to bleomycin is usually conferred by the binding protein Ble-MBL (metallo-beta-lactamase-associated ble gene) [175,176,177].
The outer membrane of Gram-negative bacteria is impenetrable to large glycopeptide molecules, and therefore A. baumannii displays intrinsic resistance to vancomycin. However, VanD-type vancomycin resistance histidine kinase VanS (vanS-D gene) has been reported in A. baumannii infection [97]. Contrariwise, colistin combination with glycopeptides (vancomycin, teicoplanin, and telavancin) and lipopeptides (like daptomycin) has been successfully employed in MDR A. baumannii infections [178]. Studies on glycopeptide- and lipopeptide-resistant A. baumannii isolates are lacking.
8.3. Resistance to Rifamycins
Resistance to rifampin (also known as rifabicin) in A. baumannii infections has been linked with mutations in the rpoB gene, which encodes rifamycin sensitive beta-subunit of RNA polymerase and averts RNA elongation just after adding the first nucleotides. Beyond rifampin, RpoB associates with resistance to all rifamycins (rifabutin, rifaximin, and rifapentine) [179]. Enzymatic modification by rifampin ADP-ribosyltransferases Arr, Arr-2, Arr-3, and Arr-4 impels to inactivation of rifamycins’ 23-OH position using NAD+ [97,180]. As mentioned above in text, RND multidrug efflux pumps AdeIJK and AcrAB-TolC are also potential mechanisms of rifamycin-resistant isolates [156]. Conversely, fhuE gene encodes an outer membrane protein transporter that is involved with iron acquisition and is upregulated in low iron conditions and seems to associate with increased susceptibility to rifabutin (200-fold more potent than rifampin). The previous finding was confirmed by successful in vivo treatment of extreme drug resistant A. baumannii with rifabutin/colistin combination [181].
8.4. Resistance to Fosfomycin
Intrinsic fosfomycin resistance in A. baumannii occurs predominantly as a result of two mechanisms: (i) fosfomycin efflux MFS transporter AbaF encoded (abaF gene); (ii) fosfomycin resistance glutathione transferases FosLL, FosA3, and FosA3/FosA4 family (encoded by fos, fosA3, and fosA, respectively), and FosB1/FosB3 family fosfomycin resistance bacillithiol transferase (encoded by fosB) [97,182]. Disruption of abaF has displayed an increase in fosfomycin susceptibility and a decrease in biofilm formation and virulence, suggesting a major role for this pump [183]. Of note, FosA impels the binding of glutathione to Fosfomycin C1 and thereby renders it inactive, while FosB opens enzymatically the epoxide rind of Fosfomycin after employing either bacillithiol or L-cysteine [156].
Fosfomycin sensitive pyruvyl transferase MurA has been recently proposed as a potential drug target for MDR A. baumannii. This transferase, whih has no counterparts in eukaryotes, catalyzes the pathway of peptidoglycan biosynthesis and is indispensable for cell integrity, while it is highly sensitive to inhibition from fosfomycin [184].
8.5. Resistance to Diaminopyrimidines—Sulfonamides
Antifolate antibiotics exert their action by inhibiting purine metabolism, and thereby DNA and RNA synthesis. Trimethoprim is a dihydrofolate reductase (DHFR) inhibitor (blocking tetrahydrofolic acid formation by dihydrofolic acid, an essential step of folate biosynthesis, which in turn is involved in the generation of many nucleotides and amino acids), while sulfonamides are known dihydropteroate synthase (DHPS) inhibitors (halting the conversion of para-aminobenzoate or PABA to dihydropteroate; including sulfamethoxazole, commonly used in combination with trimethoprim, and sulfonamides) [185]. Resistance against diaminopyrimidines in A. baumannii infections is mainly conferred by trimethoprim-resistant dihydrofolate reductases DfrA1, DfrA5, DfrA7, DfrA10, DfrA12, DfrA14, DfrA16, DfrA17, DfrA19, DfrA20, DfrA27, and DfrB1 (encoded by dfrA1, dfrA5, dfrA7, dfrA10, dfrA12, dfrA14, dfrA16, dfrA17, dfrA19, dfrA20, dfrA27, and dfrB1, respectively) [97]. A recent study of A. baumannii in turkey and chicken raw meat revealed that prevalence of dfrA was as high as 71% [186]. The latter antibiotic resistance gene seems to be a typical part of cassettes belonging to class I integron [187]. RND efflux pump MexAB-OprM consists of MexA (the membrane fusion protein), MexB (the carrier located in the inner membrane), and OprM (the transporter positioned in the outer membrane). MexAB-OprM associates with lack of susceptibility to most antibiotics, including trimethoprim and even colistin, when expressed in high levels [156,165]. As mentioned above in text, efflux pumps AdeIJK and LmrS confer resistance to trimethoprim [148]. Another pump, the RND plasmid-borne OqxAB, links with resistance to trimethoprim, quinolones, tetracyclines, glycylcycline, and nitrofurans [188,189].
In general, A. baumannii is intrinsically resistant to sulfonamides in approximately 71.3% of isolates [98]. Resistance, in these cases, is being materialized through two main mechanisms: (i) sulfonamide-resistant dihydropteroate synthases encoded by sul1 and sul2, and (ii) RND efflux pump MexAB-OprM with or without MexR mutations [156,190].
9. SARS-CoV-2 and Resistant A. baumannii Coinfections
During the COVID-19 pandemic, coinfection with A. baumannii secondary to SARS-CoV-2 infections has been reported multiple times in literature. An isolate with OXA-23 has been responsible for an outbreak in COVID-19 ICUs of a tertiary Japanese hospital [191]. Incidence of secondary infections (mainly lower respiratory tract infections) attributed to A. baumannii has been reported to be as high as 1% of hospitalized COVID-19 patients in an Italian hospital [192]. The same incidence (1%) has been reported by an early study in hospitalized patients from Wuhan, China [193]. A concurrent study from Wuhan reported A. baumannii coinfection in one out of 69 hospitalized patients (1.4%) with COVID-19 [194]. In addition, a recent study from a French ICU calculated coinfection with A. baumannii at 1.1% (1 out of 92; susceptible to third generation cephalosporins) in severe SARS-CoV-2 pneumonia patients [195]. Higher incidence of MDR A. baumannii coinfection has been documented in a study from Egypt (2.7%; seven out of 260 COVID-19 hospitalized patients; susceptible only to tigecycline and fluoroquinolones; bearing resistance genes NDM-1, TEM, and CTX-M) [196]. A recent study in hospitalized COVID-19 patients from Spain showed that coinfection with A. baumannii was apparent in 2.4% of hospitalized patients (17 out of 712; 16 out of these 17 patients in ICU) and it has been the strongest determinant of mortality with OR 9.329 (95% CI: 2.289 to 38.02; p = 0.002; higher than sex, bacteriemia, and number of comorbidities) [197]. A study from an Iranian ICU reported coinfection with MDR A. baumannii in 17 out of 19 COVID-19 patients (89.5%; no MBL-producing strain; 52% resistance rate to colistin), while all these patients died [198]. A. baumannii was detected in 20% of samples acquired from COVID-19 patients in an ICU from Beijing, China. All of these cases were identified during late ICU admission [199]. Of interest, a recent study from a Mexican ICU assigned to COVID-19 patients revealed clonal dispersion of MDR AdeABCRS+ A. baumannii that was only susceptible to gentamicin, nitrofurans, and phenicols [200]. Steroids and long ICU stays, but not MDR infections, have been associated with higher mortality in an Italian retrospective analysis of 32 COVID-19 ICU patients. In the latter cohort, four patients have been coinfected with MDR A. baumannii (12.5%) [201]. CRAB outbreaks in COVID-19 patients seem to be effectively treated with a pre-optimized two-phage cocktail [202]. This finding is important, because OXA-23 CRAB infections have become a serious threat against SARS-CoV-2 infected patients in ICUs [203].
10. Conclusions
As justified above in text, A. baumannii may acquire antibiotic resistance through several distinct mechanisms: by altering the antibiotic target site, by controlling the passage of antibiotics through its membranes, and by enzymatic modification of antibiotics, rendering them neutralized. Secondary to innate mechanisms of antibiotic resistance that are de facto conferred by genes, A. baumannii may facilitate antibiotic resistance through various mechanisms linked with its virulence: outer membrane proteins (like porins), cell envelope factors (like LPS and the capsule around its bacterial surface), specific enzymes (like phospholipases C and D, and glycan-specific adamalysin-like protease CpaA), quorum sensing, and biofilm formation (BfmRS TCS regulating Csu pili, Csu expression regulated by the GacSA TCS, biofilm-associated proteins BapAb, synthesis of the exopolysaccharide poly-β-1,6-N-acetylglucosamine PNAG, acyl-homoserine lactones through AbaR receptor, and AbaI autoinducer synthase), by attaining twitching motility via type IV pili, micronutrient acquisition systems (like siderophores and iron transporters FecA and FecI, ZnuABC transporter, and ZigA GTPase incorporating a zinc-scavenging system, resistance-associated macrophage protein NRAMP for manganese transportation), type II (with CpaA and lipases LipA and LipH as effectors), and type VI protein secretion systems [1].
Utilization of next-generation sequencing techniques has helped clinicians to decipher the molecular mechanisms of antibiotic resistance in PDR isolates of A. baumannii, and WGS has emerged as a powerful tool in the clinician’s armamentarium. In the future, application of WGS and other NGS techniques at diagnosis could provide useful insights on the microbiologic behavior and virulence of each case of critical A. baumannii infection. Apart from timely diagnosis, detailed mapping of the genetic background of each isolate could provide useful information as regards antibiotic treatment (in terms of precision medicine) and identification of novel therapeutic targets [102,107,204].
Of special note is the contribution of molecular methods, such as PCR and NGS, to the prompt identification of MDR A. baumannii epidemics. Specifically, the detection of integrase genes (intI1, intI2) is significantly associated with the epidemic character of A. baumannii as well as antibiotic resistance to multiple antimicrobials [205,206,207]. In the Netherlands, integrons are found in 43% of MDR and 85.7% of XDR A. baumannii strains [208], while in China and Iran, the prevalence of integrase genes in total A. baumannii isolates is higher and reaches 69.6% and 94.3%, respectively [209,210]. This correlation of integrase gene presence and antibiotic resistance highlights the importance of regular surveillance in order to prevent outbreaks of hospital acquired MDR A. baumannii infections.
Except from specific clinical scenarios, MDR isolates are usually treated successfully with synergistic therapeutic combinations with either beta-lactamase inhibitor sulbactam or colistin, while last-line schemes involve combination of colistin with rifampin or polymyxin B with tigecycline. In the first case, we aim for inhibiting bacterial DNA-dependent RNA polymerase along with disruption of the bacterial membranes, while in the latter case, we aim for inhibiting bacterial protein biosynthesis (by attaching to the 30S rRNA subunit) in combination with loss of the bacterial membranes’ integrity. Of course, tigecycline remains the first choice for MDR strains isolated in the setting of the ICU, while phages (like Bφ-C62) are an alternative against increased antibiotic resistance in MDR isolates [98]. Clinical experience has confirmed efficacy of trimethoprim/sulfamethoxazole against carbapenem-resistant isolates, while isolates resistant to both carbapenems and sulbactam are preferred to be treated with either minocycline or doxycycline [98,211]. XDR and PDR strains remain a serious clinical problem and identification of resistance mechanisms of this staggering microbe will elucidate pathogenetic mechanisms and propose new therapeutic targets and agents.
Author Contributions
Conceptualization: I.K.; methodology: I.K. and E.V.; software: I.K. and E.V.; validation: E.V. and A.T.; formal analysis: I.K., E.V., Z.D.P. and A.T.; investigation: I.K. and E.V.; resources: A.T.; data curation: A.T.; writing—original draft preparation: I.K. and E.V.; writing—review and editing: I.K., E.V., Z.D.P. and A.T.; visualization: I.K.; supervision: A.T. and Z.D.P.; project administration: I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ayoub Moubareck C., Hammoudi Halat D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics. 2020;9:119. doi: 10.3390/antibiotics9030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vrancianu C.O., Gheorghe I., Czobor I.B., Chifiriuc M.C. Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of acinetobacter baumannii. Microorganisms. 2020;8:935. doi: 10.3390/microorganisms8060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy-Blitchtein S., Roca I., Plasencia-Rebata S., Vicente-Taboada W., Velásquez-Pomar J., Muñoz L., Moreno-Morales J., Pons M.J., Del Valle-Mendoza J., Vila J. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru article. Emerg. Microbes Infect. 2018;7:119. doi: 10.1038/s41426-018-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasr P. Genetics, epidemiology, and clinical manifestations of multidrugresistant Acinetobacter baumannii. J. Hosp. Infect. 2020;104:4–11. doi: 10.1016/j.jhin.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Doi Y., Murray G.L., Peleg A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015;36:85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyriakidis I., Palabougiouki M., Vasileiou E., Tragiannidis A., Stamou M., Moudiou T., Vyzantiadis T., Gombakis N., Hatzilianou M. Candidemia complicating biliary atresia in an infant with hemoglobinopathy. Turk. Pediatri. Ars. 2019;54:129–132. doi: 10.14744/TurkPediatriArs.2019.67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 9.Mohd Sazlly Lim S., Zainal Abidin A., Liew S.M., Roberts J.A., Sime F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019;79:593–600. doi: 10.1016/j.jinf.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control . Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019. ECDC; Stockholm, Sweden: 2020. [Google Scholar]
- 11.Moghnieh R.A., Kanafani Z.A., Tabaja H.Z., Sharara S.L., Awad L.S., Kanj S.S. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect. Dis. 2018;18:e379–e394. doi: 10.1016/S1473-3099(18)30414-6. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piperaki E.T., Tzouvelekis L.S., Miriagou V., Daikos G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019;25:951–957. doi: 10.1016/j.cmi.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Garnacho-Montero J., Timsit J.F. Managing Acinetobacter baumannii infections. Curr. Opin. Infect. Dis. 2019;32:69–76. doi: 10.1097/QCO.0000000000000518. [DOI] [PubMed] [Google Scholar]
- 15.Karakonstantis S., Kritsotakis E.I., Gikas A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection. 2020;48:835–851. doi: 10.1007/s15010-020-01520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulani M.S., Kamble E.E., Kumkar S.N., Tawre M.S., Pardesi K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair J.M.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 18.Pakharukova N., Tuittila M., Paavilainen S., Malmi H., Parilova O., Teneberg S., Knight S.D., Zavialov A.V. Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl. Acad. Sci. USA. 2018;115:5558–5563. doi: 10.1073/pnas.1800961115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C.H., Su P.W., Moi S.H., Chuang L.Y. Biofilm formation in Acinetobacter baumannii: Genotype-phenotype correlation. Molecules. 2019;24:1849. doi: 10.3390/molecules24101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzyściak P., Chmielarczyk A., Pobiega M., Romaniszyn D., Wójkowska-Mach J. Acinetobacter baumannii isolated from hospital-acquired infection: Biofilm production and drug susceptibility. APMIS. 2017;125:1017–1026. doi: 10.1111/apm.12739. [DOI] [PubMed] [Google Scholar]
- 21.Green D.W. The bacterial cell wall as a source of antibacterial targets. Expert. Opin. Ther. Targets. 2002;6:1–19. doi: 10.1517/14728222.6.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Dijkshoorn L., Nemec A., Seifert H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 23.Ambler R.P. The structure of beta-lactamases. Philos. Trans. R Soc. Lond. B Biol. Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 24.Tooke C.L., Hinchliffe P., Bragginton E.C., Colenso C.K., Hirvonen V.H.A., Takebayashi Y., Spencer J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019;431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L., Corvec S., Rapoport M., Mugnier P., Petroni A., Pasteran F., Faccone D., Galas M., Drugeon H., Cattoir V., et al. Identification of the Novel Narrow-Spectrum β-Lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 2007;51:2179–2184. doi: 10.1128/AAC.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghafourian S., Sadeghifard N., Soheili S., Sekawi Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015;17:11–22. doi: 10.21775/cimb.017.011. [DOI] [PubMed] [Google Scholar]
- 27.Smiline A.S.G., Vijayashree J.P., Paramasivam A. Molecular characterization of plasmid-encoded blaTEM, blaSHV and blaCTX-M among extended spectrum β-lactamases [ESBLs] producing Acinetobacter baumannii. Br. J. Biomed. Sci. 2018;75:200–202. doi: 10.1080/09674845.2018.1492207. [DOI] [PubMed] [Google Scholar]
- 28.Bonnin R.A., Nordmann P., Potron A., Lecuyer H., Zahar J.R., Poirel L. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:349–354. doi: 10.1128/AAC.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moubareck C., Brémont S., Conroy M.C., Courvalin P., Lambert T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:3579–3581. doi: 10.1128/AAC.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnin R.A., Potron A., Poirel L., Lecuyer H., Neri R., Nordmann P. PER-7, an extended-spectrum β-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:2424–2427. doi: 10.1128/AAC.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L., Menuteau O., Agoli N., Cattoen C., Nordmann P. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 2003;41:3542–3547. doi: 10.1128/JCM.41.8.3542-3547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walther-Rasmussen J., Høiby N. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can. J. Microbiol. 2004;50:137–165. doi: 10.1139/w03-111. [DOI] [PubMed] [Google Scholar]
- 33.Martinez T., Martinez I., Vazquez G.J., Aquino E.E., Robledo I.E. Genetic environment of the KPC gene in Acinetobacter baumannii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J. Med. Microbiol. 2016;65:784–792. doi: 10.1099/jmm.0.000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornaglia G., Giamarellou H., Rossolini G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 35.Shivaprasad A., Antony B., Shenoy P. Comparative evaluation of four phenotypic tests for detection of Metallo-β-lactamase and Carbapenemase production in Acinetobacter baumannii. J. Clin. Diagn. Res. 2014;8:5–8. doi: 10.7860/JCDR/2014/6447.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikonomidis A., Ntokou E., Maniatis A.N., Tsakris A., Pournaras S. Hidden VIM-1 metallo-β-lactamase phenotypes among Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 2008;46:346–349. doi: 10.1128/JCM.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moulana Z., Babazadeh A., Eslamdost Z., Shokri M., Ebrahimpour S. Phenotypic and genotypic detection of metallo-beta-lactamases in Carbapenem resistant Acinetobacter baumannii. Casp. J. Intern. Med. 2020;11:171–176. doi: 10.22088/cjim.11.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee S., Datta S., Roy S., Ramanan L., Saha A., Viswanathan R., Som T., Basu S. Carbapenem resistance in Acinetobacter baumannii and other Acinetobacter spp. causing neonatal sepsis: Focus on NDM-1 and its linkage to ISAba125. Front. Microbiol. 2016;7:1126. doi: 10.3389/fmicb.2016.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amin M., Navidifar T., Shooshtari F.S., Goodarzi H. Association of the genes encoding metallo-β-lactamase with the presence of integrons among multidrug-resistant clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2019;12:1171–1180. doi: 10.2147/IDR.S196575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Ageery S.M., Al-Hazmi S.S. Microbiological and molecular detection of VIM-1 metallo beta lactamase-producing Acinetobacter baumannii. Eur. Rev. Med. Pharmacol. Sci. 2014;18:965–970. [PubMed] [Google Scholar]
- 41.López C., Ayala J.A., Bonomo R.A., González L.J., Vila A.J. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat. Commun. 2019;10:3617. doi: 10.1038/s41467-019-11615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Héritier C., Poirel L., Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 2006;12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 43.Hamidian M., Hall R.M. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J. Antimicrob. Chemother. 2013;68:2682–2683. doi: 10.1093/jac/dkt233. [DOI] [PubMed] [Google Scholar]
- 44.Lopes B.S., Amyes S.G.B. Role of ISAba1 and ISAba125 in governing the expression of bla ADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 2012;61:1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 45.Hamidian M., Hancock D.P., Hall R.M. Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J. Antimicrob. Chemother. 2013;68:244–245. doi: 10.1093/jac/dks345. [DOI] [PubMed] [Google Scholar]
- 46.Hamidian M., Hall R.M. Tn6168, a transposon carrying an ISAba1-activated ampC gene and conferring cephalosporin resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2014;69:77–80. doi: 10.1093/jac/dkt312. [DOI] [PubMed] [Google Scholar]
- 47.Couchoud C., Bertrand X., Valot B., Hocquet D. Deciphering the role of insertion sequences in the evolution of bacterial epidemic pathogens with panisa software. Microb. Genom. 2020;6:e000356. doi: 10.1099/mgen.0.000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodríguez-Martínez J.M., Poirel L., Nordmann P. Genetic and functional variability of AmpC-type β-lactamases from Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010;54:4930–4933. doi: 10.1128/AAC.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian G.B., Adams-Haduch J.M., Taracila M., Bonomo R.A., Wang H.N., Doi Y. Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: ADC-56 confers resistance to cefepime. Antimicrob. Agents Chemother. 2011;55:4922–4925. doi: 10.1128/AAC.00704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingti B., Upadhyay S., Hazarika M., Khyriem A.B., Paul D., Bhattacharya P., Joshi S.R., Bora D., Dhar D., Bhattacharjee A. Distribution of carbapenem resistant Acinetobacter baumannii with blaADC-30 and induction of ADC-30 in response to beta-lactam antibiotics. Res. Microbiol. 2020;171:128–133. doi: 10.1016/j.resmic.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Kuo S.C., Lee Y.T., Lauderdale T.L.Y., Huang W.C., Chuang M.F., Chen C.P., Su S.C., Lee K.R., Chen T.L. Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front. Microbiol. 2015;6:231. doi: 10.3389/fmicb.2015.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai J.H., Yang J.T., Chern J., Chen T.L., Wu W.L., Liao J.H., Tsai S.F., Liang S.Y., Chou C.C., Wu S.H. Comparative phosphoproteomics reveals the role of AmpC β-lactamase phosphorylation in the clinical imipenem-resistant strain Acinetobacter baumannii SK17. Mol. Cell Proteom. 2016;15:12–25. doi: 10.1074/mcp.M115.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antunes N.T., Fisher J.F. Acquired class D β-Lactamases. Antibiotics. 2014;3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poirel L., Naas T., Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong M.H.-Y., Chan B.K.-W., Chan E.W.-C., Chen S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ß-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019;10:2809. doi: 10.3389/fmicb.2019.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ewers C., Klotz P., Leidner U., Stamm I., Prenger-Berninghoff E., Göttig S., Semmler T., Scheufen S. OXA-23 and ISAba1–OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int. J. Antimicrob. Agents. 2017;49:37–44. doi: 10.1016/j.ijantimicag.2016.09.033. [DOI] [PubMed] [Google Scholar]
- 57.Morakchi H., Loucif L., Gacemi-Kirane D., Rolain J.M. Molecular characterisation of carbapenemases in urban pigeon droppings in France and Algeria. J. Glob. Antimicrob. Resist. 2017;9:103–110. doi: 10.1016/j.jgar.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Sugawara E., Nikaido H. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol. 2012;194:4089–4096. doi: 10.1128/JB.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jyothisri K., Deepak V., Rajeswari M.R. Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett. 1999;443:57–60. doi: 10.1016/S0014-5793(98)01679-2. [DOI] [PubMed] [Google Scholar]
- 60.Smani Y., Fàbrega A., Roca I., Sańchez-Encinales V., Vila J., Pachón J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014;58:1806–1808. doi: 10.1128/AAC.02101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iyer R., Moussa S.H., Durand-Réville T.F., Tommasi R., Miller A. Acinetobacter baumannii OmpA Is a Selective Antibiotic Permeant Porin. ACS Infect. Dis. 2018;4:373–381. doi: 10.1021/acsinfecdis.7b00168. [DOI] [PubMed] [Google Scholar]
- 62.Kwon H.I., Kim S., Oh M.H., Na S.H., Kim Y.J., Jeon Y.H., Lee J.C. Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 2017;72:3012–3015. doi: 10.1093/jac/dkx257. [DOI] [PubMed] [Google Scholar]
- 63.Zhong X., Wu X., Schweppe D.K., Chavez J.D., Mathay M., Eng J.K., Keller A., Bruce J.E. In Vivo Cross-Linking MS Reveals Conservation in OmpA Linkage to Different Classes of β-Lactamase Enzymes. J Am. Soc. Mass Spectrom. 2020;31:190–195. doi: 10.1021/jasms.9b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Limansky A.S., Mussi M.A., Viale A.M. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 2002;40:4776–4778. doi: 10.1128/JCM.40.12.4776-4778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mussi M.A., Limansky A.S., Viale A.M. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 2005;49:1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mussi M.A., Relling V.M., Limansky A.S., Viale A.M. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for l-ornithine uptake. FEBS Lett. 2007;581:5573–5578. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 67.Mussi M.A., Limansky A.S., Relling V., Ravasi P., Arakaki A., Actis L.A., Viale A.M. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J. Bacteriolnal. Bacteriol. 2011;193:4736–4748. doi: 10.1128/JB.01533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong H.W., Cheong H.J., Kim W.J., Kim M.J., Song K.J., Song J.W., Kim H.S., Roh K.H. Loss of the 29-kilodalton outer membrane protein in the presence of OXA-51-like enzymes in Acinetobacter baumannii is associated with decreased imipenem susceptibility. Microb. Drug Resist. 2009;15:151–158. doi: 10.1089/mdr.2009.0828. [DOI] [PubMed] [Google Scholar]
- 69.Khorsi K., Messai Y., Ammari H., Hamidi M., Bakour R. ISAba36 inserted into the outer membrane protein gene carO and associated with the carbapenemase gene blaOXA-24-like in Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2018;15:107–108. doi: 10.1016/j.jgar.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 70.Zhu L.J., Chen X.Y., Hou P.F. Mutation of CarO participates in drug resistance in imipenem-resistant Acinetobacter baumannii. J. Clin. Lab. Anal. 2019;33:e22976. doi: 10.1002/jcla.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zahn M., D’Agostino T., Eren E., Baslé A., Ceccarelli M., Van Berg B.D. Small-molecule transport by CarO, an abundant eight-stranded β-barrel outer membrane protein from Acinetobacter baumannii. J. Mol. Biol. 2015;427:2329–2339. doi: 10.1016/j.jmb.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 72.Fonseca E.L., Scheidegger E., Freitas F.S., Cipriano R., Vicente A.C.P. Carbapenem-resistant Acinetobacter baumannii from Brazil: Role of carO alleles expression and blaOXA-23 gene. BMC Microbiol. 2013;13:1–7. doi: 10.1186/1471-2180-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Catel-Ferreira M., Coadou G., Molle V., Mugnier P., Nordmann P., Siroy A., Jouenne T., Dé E. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J. Antimicrob. Chemother. 2011;66:2053–2056. doi: 10.1093/jac/dkr267. [DOI] [PubMed] [Google Scholar]
- 74.Siroy A., Molle V., Lemaître-Guillier C., Vallenet D., Pestel-Caron M., Cozzone A.J., Jouenne T., Dé E. Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005;49:4876–4883. doi: 10.1128/AAC.49.12.4876-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Srinivasan V.B., Vaidyanathan V., Rajamohan G. AbuO, a tolc-like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrob. Agents Chemother. 2015;59:1236–1245. doi: 10.1128/AAC.03626-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clark R.B. Imipenem resistance among Acinetobacter baumannii: Association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 1996;38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 77.Tomás M.D.M., Beceiro A., Pérez A., Velasco D., Moure R., Villanueva R., Martínez-Beltrán J., Bou G. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005;49:5172–5175. doi: 10.1128/AAC.49.12.5172-5175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zahn M., Bhamidimarri S.P., Baslé A., Winterhalter M., Van Den Berg B. Structural Insights into Outer Membrane Permeability of Acinetobacter baumannii. Structure. 2016;24:221–231. doi: 10.1016/j.str.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 79.Catel-Ferreira M., Nehmé R., Molle V., Aranda J., Bouffartigues E., Chevalier S., Bou G., Jouenne T., Dé E. Deciphering the function of the outer membrane protein OprD homologue of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012;56:3826–3832. doi: 10.1128/AAC.06022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cecchini T., Yoon E.J., Charretier Y., Bardet C., Beaulieu C., Lacoux X., Docquier J.D., Lemoine J., Courvalin P., Grillot-Courvalin C., et al. Deciphering multifactorial resistance phenotypes in Acinetobacter baumannii by genomics and targeted label-free proteomics. Mol. Cellul Proteom. 2018;17:442–456. doi: 10.1074/mcp.RA117.000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang L., Sun L., Xu G., Xia T. Differential susceptibility to carbapenems due to the AdeABC efflux pump among nosocomial outbreak isolates of Acinetobacter baumannii in a Chinese hospital. Diagn. Microbiol. Infect. Dis. 2008;62:326–332. doi: 10.1016/j.diagmicrobio.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 82.Hawkey J., Ascher D.B., Judd L.M., Wick R.R., Kostoulias X., Cleland H., Spelman D.W., Padiglione A., Peleg A.Y., Holt K.E. Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb. Genom. 2018;4:e000165. doi: 10.1099/mgen.0.000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Magnet S., Courvalin P., Lambert T. Resistance-Nodulation-Cell Division-Type Efflux Pump Involved in Aminoglycoside Resistance in Acinetobacter baumannii Strain BM4454. Antimicrob. Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vila J., Martí S., Sánchez-Céspedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007;59:1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- 85.Marchand I., Marchand I., Damier-piolle L., Damier-piolle L., Courvalin P., Courvalin P., Lambert T., Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004;48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leus I.V., Weeks J.W., Bonifay V., Smith L., Richardson S., Zgurskaya H.I. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 2018;200:e00049-18. doi: 10.1128/JB.00049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hou P.F., Chen X.Y., Yan G.F., Wang Y.P., Ying C.M. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy. 2012;58:152–158. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- 88.Yoon E.J., Chabane Y.N., Goussard S., Snesrud E., Courvalin P., Dé E., Grillot-Courvalin C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio. 2015;6:e00309-15. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sauvage E., Kerff F., Terrak M., Ayala J.A., Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 90.Zapun A., Contreras-Martel C., Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 91.Cayô R., Rodríguez M.C., Espinal P., Fernández-Cuenca F., Ocampo-Sosa A.A., Pascual Á., Ayala J.A., Vila J., Martínez-Martínez L. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:5907–5913. doi: 10.1128/AAC.00459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gehrlein M., Leying H., Cullmann W., Wendt S., Opferkuch W. Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy. 1991;37:405–412. doi: 10.1159/000238887. [DOI] [PubMed] [Google Scholar]
- 93.Yun S.H., Choi C.W., Kwon S.O., Park G.W., Cho K., Kwon K.H., Kim J.Y., Yoo J.S., Lee J.C., Choi J.S., et al. Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 2011;10:459–469. doi: 10.1021/pr101012s. [DOI] [PubMed] [Google Scholar]
- 94.Fernández-Cuenca F., Martínez-Martínez L., Conejo M.C., Ayala J.A., Perea E.J., Pascual A. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003;51:565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 95.Vashist J., Tiwari V., Das R., Kapil A., Rajeswari M.R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 2011;133:332–338. [PMC free article] [PubMed] [Google Scholar]
- 96.Siroy A., Cosette P., Seyer D., Lemaître-Guillier C., Vallenet D., Van Dorsselaer A., Boyer-Mariotte S., Jouenne T., Dé E. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 2006;5:3385–3398. doi: 10.1021/pr060372s. [DOI] [PubMed] [Google Scholar]
- 97.Pathogen Detection Microbial Browser for Identification of Genetic and Genomic Elements (MicroBIGG-E)—Pathogen Detection—NCBI. [(accessed on 5 March 2021)]; Available online: https://www.ncbi.nlm.nih.gov/pathogens/microbigge/#.
- 98.Vázquez-López R., Solano-Gálvez S.G., Vignon-Whaley J.J.J., Vaamonde J.A.A., Alonzo L.A.P., Reséndiz A.R., Álvarez M.M., López E.N.V., Franyuti-Kelly G., Álvarez-Hernández D.A., et al. Acinetobacter baumannii resistance: A real challenge for clinicians. Antibiotics. 2020;9:205. doi: 10.3390/antibiotics9040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garneau-Tsodikova S., Labby K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. MedChemComm. 2016;7:11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramirez M.S., Tolmasky M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aínsa J.A., Martin C., Gicquel B., Gomez-Lus R. Characterization of the chromosomal aminoglycoside 2’-N- acetyltransferase gene from Mycobacterium fortuitum. Antimicrob. Agents Chemother. 1996;40:2350–2355. doi: 10.1128/AAC.40.10.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H., Wang J., Yu P., Ge P., Jiang Y., Xu R., Chen R., Liu X. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int. J. Mol. Med. 2017;39:364–372. doi: 10.3892/ijmm.2016.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sheikhalizadeh V., Hasani A., Rezaee M.A., Rahmati-Yamchi M., Hasani A., Ghotaslou R., Goli H.R. Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. 2017;23:74–79. doi: 10.1016/j.jiac.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 104.Jana S., Deb J.K. Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 2006;70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 105.De Silva P.M., Kumar A. Signal transduction proteins in Acinetobacter baumannii: Role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019;10:49. doi: 10.3389/fmicb.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu C., Bilya S.R., Xu W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019;30:100549. doi: 10.1016/j.nmni.2019.100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu A., Zhu H., Gao B., Weng H., Ding Z., Li M., Weng X., He G. Diagnosis of severe community-acquired pneumonia caused by Acinetobacter baumannii through next-generation sequencing: A case report. BMC Infect. Dis. 2020;20:45. doi: 10.1186/s12879-019-4733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chukwudi C.U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 2016;60:4433–4441. doi: 10.1128/AAC.00594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Warburton P.J., Amodeo N., Roberts A.P. Mosaic tetracycline resistance genes encoding ribosomal protection proteins. J. Antimicrob. Chemother. 2016;71:3333–3339. doi: 10.1093/jac/dkw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hua X., Pan C., Zhu L., Liu Z., Xu Q., Wang H., Yu Y. Complete genome sequence of Acinetobacter baumannii A1296 (ST1469) with a small plasmid harbouring the tet(39) tetracycline resistance gene. J. Glob. Antimicrob. Resist. 2017;11:105–107. doi: 10.1016/j.jgar.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 111.Rumbo C., Gato E., López M., Ruiz De Alegría C., Fernández-Cuenca F., Martínez-Martínez L., Vila J., Pachón J., Cisneros J.M., Rodríguez-Baño J., et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nikolakopoulou T.L., Giannoutsou E.P., Karabatsou A.A., Karagouni A.D. Prevalence of tetracycline resistance genes in Greek seawater habitats. J. Microbiol. 2008;46:633–640. doi: 10.1007/s12275-008-0080-8. [DOI] [PubMed] [Google Scholar]
- 113.Lin L., Wang S.F., Yang T.Y., Hung W.C., Chan M.Y., Tseng S.P. Antimicrobial resistance and genetic diversity in ceftazidime non-susceptible bacterial pathogens from ready-to-eat street foods in three Taiwanese cities. Sci. Rep. 2017;7:15515. doi: 10.1038/s41598-017-15627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miranda C.D., Kehrenberg C., Ulep C., Schwarz S., Roberts M.C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 2003;47:883–888. doi: 10.1128/AAC.47.3.883-888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuhan Y., Ziyun Y., Yongbo Z., Fuqiang L., Qinghua Z. Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of Acinetobacter baumannii and Klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 2016;49:165–171. doi: 10.1590/0037-8682-0411-2015. [DOI] [PubMed] [Google Scholar]
- 116.Huys G., Cnockaert M., Vaneechoutte M., Woodford N., Nemec A., Dijkshoorn L., Swings J. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 2005;156:348–355. doi: 10.1016/j.resmic.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 117.Foong W.E., Wilhelm J., Tam H.K., Pos K.M. Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J. Antimicrob. Chemother. 2020;75:1135–1139. doi: 10.1093/jac/dkaa015. [DOI] [PubMed] [Google Scholar]
- 118.Dönhöfer A., Franckenberg S., Wickles S., Berninghausen O., Beckmann R., Wilson D.N. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA. 2012;109:16900–16905. doi: 10.1073/pnas.1208037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ribera A., Ruiz J., Vila J. Presence of the tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2003;47:2310–2312. doi: 10.1128/AAC.47.7.2310-2312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li L., Hassan K.A., Tetu S.G., Naidu V., Pokhrel A., Cain A.K., Paulsen I.T. The Transcriptomic Signature of Tigecycline in Acinetobacter baumannii. Front. Microbiol. 2020;11:565438. doi: 10.3389/fmicb.2020.565438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He T., Wang R., Liu D., Walsh T.R., Zhang R., Lv Y., Ke Y., Ji Q., Wei R., Liu Z., et al. Emergence of Plasmid-Mediated High-Level Tigecycline Resistance Genes in Animals and Humans. Nat. Microbiol. 2019;4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 122.Wang L., Liu D., Lv Y., Cui L., Li Y., Li T., Song H., Hao Y., Shen J., Wang Y., et al. Novel Plasmid-Mediated tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2020;64:e01326-19. doi: 10.1128/AAC.01326-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aldred K.J., Kerns R.J., Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Su X.Z., Chen J., Mizushima T., Kuroda T., Tsuchiya T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005;49:4362–4364. doi: 10.1128/AAC.49.10.4362-4364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perez F., Hujer A.M., Hujer K.M., Decker B.K., Rather P.N., Bonomo R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zaki M.E.S., Abou ElKheir N., Mofreh M. Molecular Study of Quinolone Resistance Determining Regions of gyrA Gene and parC Genes in Clinical Isolates of Acintobacter baumannii Resistant to Fluoroquinolone. Open Microbiol. J. 2018;12:116–122. doi: 10.2174/1874285801812010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu J.H., Deng Y.T., Zeng Z.L., Gao J.H., Chen L., Arakawa Y., Chen Z.L. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob. Agents Chemother. 2008;52:2992–2993. doi: 10.1128/AAC.01686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lari A.R., Ardebili A., Hashemi A. Ader-ades mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates. Indian J. Med. Res. 2018;147:413–421. doi: 10.4103/ijmr.IJMR_644_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coyne S., Courvalin P., Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011;55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ardebili A., Lari A.R., Beheshti M., Lari E.R. Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran. J. Basic Med. Sci. 2015;18:623–626. doi: 10.22038/ijbms.2015.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.D’Souza R., Pinto N.A., Le Phuong N., Higgins P.G., Vu T.N., Byun J.H., Cho Y.L., Choi J.R., Yong D. Phenotypic and genotypic characterization of Acinetobacter spp. panel strains: A cornerstone to facilitate antimicrobial development. Front. Microbiol. 2019;10:559. doi: 10.3389/fmicb.2019.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robicsek A., Jacoby G.A., Hooper D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 133.Xiong X., Bromley E.H.C., Oelschlaeger P., Woolfson D.N., Spencer J. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: Conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 2011;39:3917–3927. doi: 10.1093/nar/gkq1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Robicsek A., Strahilevitz J., Jacoby G.A., Macielag M., Abbanat D., Chi H.P., Bush K., Hooper D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 135.Lee C.R., Lee J.H., Park M., Park K.S., Bae I.K., Kim Y.B., Cha C.J., Jeong B.C., Lee S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamada K., Morinaga Y., Yanagihara K., Kaku N., Harada Y., Uno N., Nakamura S., Imamura Y., Hasegawa H., Miyazaki T., et al. Azithromycin inhibits MUC5AC induction via multidrug-resistant Acinetobacter baumannii in human airway epithelial cells. Pulm. Pharmacol. Ther. 2014;28:165–170. doi: 10.1016/j.pupt.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 137.Cheng Y., Yang S., Jia M., Zhao L., Hou C., You X., Zhao J., Chen A. Comparative study between macrolide regulatory proteins MphR(A) and MphR(E) in ligand identification and DNA binding based on the rapid in vitro detection system. Anal. Bioanal. Chem. 2016;408:1623–1631. doi: 10.1007/s00216-015-9270-5. [DOI] [PubMed] [Google Scholar]
- 138.Chopjitt P., Kerdsin A., Takeuchi D., Hatrongjit R., Boueroy P., Akeda Y., Tomono K., Hamada S. Whole genome analysis of extensively drug-resistant Acinetobacter baumannii clinical isolates in Thailand. Infect. Disord Drug Targets. 2020;20 doi: 10.2174/1871526520999201116201911. [DOI] [PubMed] [Google Scholar]
- 139.Rajamohan G., Srinivasan V.B., Gebreyes W.A. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2010;65:1919–1925. doi: 10.1093/jac/dkq195. [DOI] [PubMed] [Google Scholar]
- 140.Lean S.S., Yeo C.C., Suhaili Z., Thong K.L. Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: Insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int. J. Antimicrob. Agents. 2015;45:178–182. doi: 10.1016/j.ijantimicag.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 141.Schroeder M.R., Lohsen S., Chancey S.T., Stephens D.S. High-Level Macrolide Resistance Due to the Mega Element [mef(E)/mel] in Streptococcus pneumoniae. Front. Microbiol. 2019;10:868. doi: 10.3389/fmicb.2019.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Munita J., Arias C. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Okada U., Yamashita E., Neuberger A., Morimoto M., Van Veen H.W., Murakami S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 2017;8:1336. doi: 10.1038/s41467-017-01399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Srinivasan V.B., Rajamohan G., Gebreyes W.A. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:5312–5316. doi: 10.1128/AAC.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Richmond G.E., Evans L.P., Anderson M.J., Wand M.E., Bonney L.C., Ivens A., Chua K.L., Webber M.A., Mark Sutton J., Peterson M.L., et al. The Acinetobacter baumannii two-component system aders regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio. 2016;7:e00430-16. doi: 10.1128/mBio.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leclercq R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 147.Cortez-Cordova J., Kumar A. Activity of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2011;37:420–424. doi: 10.1016/j.ijantimicag.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 148.Damier-Piolle L., Magnet S., Brémont S., Lambert T., Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008;52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoon E.J., Balloy V., Fiette L., Chignard M., Courvalin P., Grillot-Courvalin C. Contribution of the ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio. 2016;7:e00697-16. doi: 10.1128/mBio.00697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee Y., D’Souza R., Yong D., Lee K. Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Ann. Lab. Med. 2016;36:320–324. doi: 10.3343/alm.2016.36.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li P., Huang Y., Yu L., Liu Y., Niu W., Zou D., Liu H., Zheng J., Yin X., Yuan J., et al. Isolation and Whole-genome Sequence Analysis of the Imipenem Heteroresistant Acinetobacter baumannii Clinical Isolate HRAB-85. Int. J. Infect. Dis. 2017;62:94–101. doi: 10.1016/j.ijid.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 152.López-Durán P.A., Fonseca-Coronado S., Lozano-Trenado L.M., Araujo-Betanzos S., Rugerio-Trujillo D.A., Vaughan G., Saldaña-Rivera E. Nosocomial transmission of extensively drug resistant Acinetobacter baumannii strains in a tertiary level hospital. PLoS ONE. 2020;15:e0231829. doi: 10.1371/journal.pone.0231829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moffatt J.H., Harper M., Boyce J.D. Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 2019;1145:55–71. doi: 10.1007/978-3-030-16373-0_5. [DOI] [PubMed] [Google Scholar]
- 154.Bojkovic J., Richie D.L., Six D.A., Rath C.M., Sawyer W.S., Hu Q., Dean C.R. Characterization of an Acinetobacter baumannii lptD deletion strain: Permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 2016;198:731–741. doi: 10.1128/JB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luke N.R., Sauberan S.L., Russo T.A., Beanan J.M., Olson R., Loehfelm T.W., Cox A.D., St. Michael F., Vinogradov E.V., Campagnari A.A. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 2010;78:2017–2023. doi: 10.1128/IAI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.The Comprehensive Antibiotic Resistance Database (CARD) [(accessed on 5 March 2021)]; Available online: https://card.mcmaster.ca/ontology/35989.
- 157.Sun B., Liu H., Jiang Y., Shao L., Yang S., Chen D. New Mutations Involved in Colistin Resistance in Acinetobacter baumannii. mSphere. 2020;5:e00895-19. doi: 10.1128/mSphere.00895-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chin C.Y., Gregg K.A., Napier B.A., Ernst R.K., Weiss D.S. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015;59:7911–7914. doi: 10.1128/AAC.00515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gerson S., Betts J.W., Lucaßen K., Nodari C.S., Wille J., Josten M., Göttig S., Nowak J., Stefanik D., Roca I., et al. Investigation of Novel pmrB and eptA Mutations in Isogenic Acinetobacter baumannii Isolates Associated with Colistin Resistance and Increased Virulence In Vivo. Antimicrob. Agents Chemother. 2019;63:e01586-18. doi: 10.1128/AAC.01586-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gelbíčová T., Baráková A., Florianová M., Karpíšková R. Detection of colistin-resistant Acinetobacter baumannii with the mcr-4 gene. Klin. Mikrobiol. Infekc. Lek. 2019;25:4–6. [PubMed] [Google Scholar]
- 161.Martins-Sorenson N., Snesrud E., Xavier D.E., Cacci L.C., Iavarone A.T., McGann P., Riley L.W., Moreira B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020;75:60–64. doi: 10.1093/jac/dkz413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hameed F., Khan M.A., Muhammad H., Sarwar T., Bilal H., Rehman T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019;52:e20190237. doi: 10.1590/0037-8682-0237-2019. [DOI] [PubMed] [Google Scholar]
- 163.Hood M.I., Becker K.W., Roux C.M., Dunman P.M., Skaar E.P. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 2013;81:542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nhu N.T.K., Riordan D.W., Nhu T.D.H., Thanh D.P., Thwaites G., Lan N.P.H., Wren B.W., Baker S., Stabler R.A. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016;6:28291. doi: 10.1038/srep28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Paul D., Mallick S., Das S., Saha S., Ghosh A.K., Mandal S.M. Colistin Induced Assortment of Antimicrobial Resistance in a Clinical Isolate of Acinetobacter baumannii SD01. Infect. Disord. Drug Targets. 2019;20:501–505. doi: 10.2174/1871526519666190426153258. [DOI] [PubMed] [Google Scholar]
- 166.Traglia G., Chiem K., Quinn B., Fernandez J.S., Montaña S., Almuzara M., Mussi M.A., Tolmasky M.E., Iriarte A., Centrón D., et al. Genome sequence analysis of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain depicts evidence of increase genome plasticity. Sci. Rep. 2018;8:16961. doi: 10.1038/s41598-018-35377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Foong W.E., Tam H.K., Crames J.J., Averhoff B., Pos K.M. The chloramphenicol/H+ antiporter CraA of Acinetobacter baumannii AYE reveals a broad substrate specificity. J. Antimicrob. Chemother. 2019;74:1192–1201. doi: 10.1093/jac/dkz024. [DOI] [PubMed] [Google Scholar]
- 168.Karalewitz A.P.A., Millera S.I. Multidrug-resistant Acinetobacter baumannii chloramphenicol resistance requires an inner membrane permease. Antimicrob. Agents Chemother. 2018;62:e00513-18. doi: 10.1128/AAC.00513-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen C., Cui C.Y., Wu X.T., Fang L.X., He Q., He B., Long T.F., Liao X.P., Chen L., Liu Y.H., et al. Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 2021;253:108954. doi: 10.1016/j.vetmic.2020.108954. [DOI] [PubMed] [Google Scholar]
- 170.Li X., Quan J., Yang Y., Ji J., Liu L., Fu Y., Hua X., Chen Y., Pi B., Jiang Y., et al. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1371–1375. doi: 10.1007/s10096-016-2674-0. [DOI] [PubMed] [Google Scholar]
- 171.Li H., Wang Q., Wang R., Zhang Y., Wang X., Wang H. Global regulator SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in Acinetobacter baumannii. Med. Baltim. 2017;96:e7188. doi: 10.1097/MD.0000000000007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang J., Zhou Z., He F., Ruan Z., Jiang Y., Hua X., Yu Y. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS ONE. 2018;13:e0192288. doi: 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ma X.L., Guo Y.Z., Wu Y.M., Gong W.T., Sun J., Huang Z. In vivo bactericidal effect of colistin–linezolid combination in a murine model of MDR and XDR Acinetobacter baumannii pneumonia. Sci. Rep. 2020;10:17518. doi: 10.1038/s41598-020-74503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gu B., Kelesidis T., Tsiodras S., Hindler J., Humphries R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wareth G., Linde J., Hammer P., Nguyen N.H., Nguyen T.N.M., Splettstoesser W.D., Makarewicz O., Neubauer H., Sprague L.D., Pletz M.W. Phenotypic and WGS-derived antimicrobial resistance profiles of clinical and non-clinical Acinetobacter baumannii isolates from Germany and Vietnam. Int. J. Antimicrob. Agents. 2020;56:106127. doi: 10.1016/j.ijantimicag.2020.106127. [DOI] [PubMed] [Google Scholar]
- 176.Ducas-Mowchun K., De Silva P.M., Crisostomo L., Fernando D.M., Chao T.C., Pelka P., Schweizer H.P., Kumar A. Next generation of Tn7-based single-copy insertion elements for use in multi- and pan-drug-resistant strains of Acinetobacter baumannii. Appl. Environ. Microbiol. 2019;85:e00066-19. doi: 10.1128/AEM.00066-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Luna B.M., Ulhaq A., Yan J., Pantapalangkoor P., Nielsen T.B., Davies B.W., Actis L.A., Spellberg B. Selectable Markers for Use in Genetic Manipulation of Extensively Drug-Resistant (XDR) Acinetobacter baumannii HUMC1. mSphere. 2017;2:e00140-17. doi: 10.1128/mSphere.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Claeys K.C., Fiorvento A.D., Rybak M.J. A Review of Novel Combinations of Colistin and Lipopeptide or Glycopeptide Antibiotics for the Treatment of Multidrug-Resistant Acinetobacter baumannii. Infect. Ther. 2014;3:69–81. doi: 10.1007/s40121-014-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rolain J.M., Diene S.M., Kempf M., Gimenez G., Robert C., Raoult D. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan-drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob. Agents Chemother. 2013;57:592–596. doi: 10.1128/AAC.01314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Giannouli M., Di Popolo A., Durante-Mangoni E., Bernardo M., Cuccurullo S., Amato G., Tripodi M.F., Triassi M., Utili R., Zarrilli R. Molecular epidemiology and mechanisms of rifampicin resistance in Acinetobacter baumannii isolates from Italy. Int. J. Antimicrob. Agents. 2012;39:58–63. doi: 10.1016/j.ijantimicag.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 181.Luna B., Trebosc V., Lee B., Bakowski M., Ulhaq A., Yan J., Lu P., Cheng J., Nielsen T., Lim J., et al. A nutrient-limited screen unmasks rifabutin hyperactivity for extensively drug-resistant Acinetobacter baumannii. Nat. Microbiol. 2020;5:1134–1143. doi: 10.1038/s41564-020-0737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ito R., Mustapha M.M., Tomich A.D., Callaghan J.D., McElheny C.L., Mettus R.T., Shanks R.M.Q., Sluis-Cremer N., Doi Y. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio. 2017;8:e00749-17. doi: 10.1128/mBio.00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sharma A., Sharma R., Bhattacharyya T., Bhando T., Pathania R. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother. 2017;72:68–74. doi: 10.1093/jac/dkw382. [DOI] [PubMed] [Google Scholar]
- 184.Sonkar A., Shukla H., Shukla R., Kalita J., Pandey T., Tripathi T. UDP-N-Acetylglucosamine enolpyruvyl transferase (MurA) of Acinetobacter baumannii (AbMurA): Structural and functional properties. Int. J. Biol. Macromol. 2017;97:106–114. doi: 10.1016/j.ijbiomac.2016.12.082. [DOI] [PubMed] [Google Scholar]
- 185.Sánchez-Osuna M., Cortés P., Llagostera M., Barbé J., Erill I. Exploration into the origins and mobilization of di-hydrofolate reductase genes and the emergence of clinical resistance to trimethoprim. Microb. Genom. 2020;6:mgen000440. doi: 10.1099/mgen.0.000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ghaffoori Kanaan M.H., Al-Shadeedi S.M.J., Al-Massody A.J., Ghasemian A. Drug resistance and virulence traits of Acinetobacter baumannii from Turkey and chicken raw meat. Comp. Immunol. Microbiol. Infect. Dis. 2020;70:101451. doi: 10.1016/j.cimid.2020.101451. [DOI] [PubMed] [Google Scholar]
- 187.Huang C., Long Q., Qian K., Fu T., Zhang Z., Liao P., Xie J. Resistance and integron characterization of Acinetobacter baumannii in a teaching hospital in Chongqing, China. New Microbes New Infect. 2015;8:103–108. doi: 10.1016/j.nmni.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hong B.K., Wang M., Chi H.P., Kim E.C., Jacoby G.A., Hooper D.C. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Osei Sekyere J., Reta M.A. Genomic and Resistance Epidemiology of Gram-Negative Bacteria in Africa: A Systematic Review and Phylogenomic Analyses from a One Health Perspective. mSystems. 2020;5:e00897-20. doi: 10.1128/mSystems.00897-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Girija A.S.S., Vijayashree Priyadharsini J., Paramasivam A. Plasmid-encoded resistance to trimethoprim/sulfamethoxazole mediated by dfrA1, dfrA5, sul1 and sul2 among Acinetobacter baumannii isolated from urine samples of patients with severe urinary tract infection. J. Glob. Antimicrob. Resist. 2019;17:145–146. doi: 10.1016/j.jgar.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 191.Duployez C., Guern R.L., Milliere L., Caplan M., Loïez C., Ledoux G., Jaillette E., Favory R., Mathieu D., Wallet F. An outbreak can hide another. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.705. [DOI] [PubMed] [Google Scholar]
- 192.Ripa M., Galli L., Poli A., Oltolini C., Spagnuolo V., Mastrangelo A., Muccini C., Monti G., De Luca G., Landoni G., et al. Secondary infections in patients hospitalized with COVID-19: Incidence and predictive factors. Clin. Microbiol. Infect. 2021;27:451–457. doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Contou D., Claudinon A., Pajot O., Micaëlo M., Longuet Flandre P., Dubert M., Cally R., Logre E., Fraissé M., Mentec H., et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care. 2020;10:119. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ramadan H.K.-A., Mahmoud M.A., Aburahma M.Z., Elkhawaga A.A., El-Mokhtar M.A., Sayed I.M., Hosni A., Hassany S.M., Medhat M.A. Predictors of Severity and Co-Infection Resistance Profile in COVID-19 Patients: First Report from Upper Egypt. Infect. Drug Resist. 2020;13:3409–3422. doi: 10.2147/IDR.S272605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nebreda-Mayoral T., Miguel-Gómez M.A., March-Rosselló G.A., Puente-Fuertes L., Cantón-Benito E., Martínez-García A.M., Muñoz-Martín A.B., Orduña-Domingo A. Infección bacteriana/fúngica en pacientes con COVID-19 ingresados en un hospital de tercer nivel de Castilla y León, España. Enferm. Infecc. Microbiol. Clin. 2020;3 doi: 10.1016/j.eimc.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sharifipour E., Shams S., Esmkhani M., Khodadadi J., Fotouhi-Ardakani R., Koohpaei A., Doosti Z., EJ Golzari S. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020;20:646. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang S., Hua M., Liu X., Du C., Pu L., Xiang P., Wang L., Liu J. Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. 2021:104806. doi: 10.1016/j.micinf.2021.104806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Durán-Manuel E.M., Cruz-Cruz C., Ibáñez-Cervantes G., Bravata-Alcantará J.C., Sosa-Hernández O., Delgado-Balbuena L., León-García G., Cortés-Ortíz I.A., Cureño-Díaz M.A., Castro-Escarpulli G., et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries. 2021;15:58–68. doi: 10.3855/jidc.13545. [DOI] [PubMed] [Google Scholar]
- 201.Karruli A., Boccia F., Gagliardi M., Patauner F., Ursi M.P., Sommese P., De Rosa R., Murino P., Ruocco G., Corcione A., et al. Multidrug-Resistant Infections and Outcome of Critically Ill Patients with Coronavirus Disease 2019: A Single Center Experience. Microb. Drug Resist. 2021 doi: 10.1089/mdr.2020.0489. [DOI] [PubMed] [Google Scholar]
- 202.Wu N., Dai J., Guo M., Li J., Zhou X., Li F., Gao Y., Qu H., Lu H., Jin J., et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 2021:1–31. doi: 10.1080/22221751.2021.1902754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Perez S., Innes G.K., Walters M.S., Mehr J., Arias J., Greeley R., Chew D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1827–1831. doi: 10.15585/mmwr.mm6948e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fitzpatrick M.A., Ozer E.A., Hauser A.R. Utility of Whole-Genome Sequencing in Characterizing Acinetobacter Epidemiology and Analyzing Hospital Outbreaks. J. Clin. Microbiol. 2016;54:593–612. doi: 10.1128/JCM.01818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koeleman J.G.M., Stoof J., Van Der Bijl M.W., Vandenbroucke-Grauls C.M.J.E., Savelkoul P.H.M. Identification of Epidemic Strains of Acinetobacter baumannii by Integrase Gene PCR. J. Clin. Microbiol. 2001;39:8–13. doi: 10.1128/JCM.39.1.8-13.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wu T.-L., Ma L., Chang J.-C., Su L.-H., Chu C., Leu H.-S., Siu L.K. Variable Resistance Patterns of Integron-Associated Multidrug-Resistant Acinetobacter baumannii Isolates in a Surgical Intensive Care Unit. Microb. Drug Resist. 2004;10:292–299. doi: 10.1089/mdr.2004.10.292. [DOI] [PubMed] [Google Scholar]
- 207.Turton J.F., Kaufmann M.E., Glover J., Coelho J.M., Warner M., Pike R., Pitt T.L. Detection and Typing of Integrons in Epidemic Strains of Acinetobacter baumannii Found in the United Kingdom. J. Clin. Microbiol. 2005;43:3074–3082. doi: 10.1128/JCM.43.7.3074-3082.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gaur A., Prakash P., Anupurba S., Mohapatra T.M. Possible role of integrase gene polymerase chain reaction as an epidemiological marker: Study of multidrug-resistant Acinetobacter baumannii isolated from nosocomial infections. Int. J. Antimicrob. Agents. 2007;29:446–450. doi: 10.1016/j.ijantimicag.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 209.Chen J., Li H., Yang J., Zhan R., Chen A., Yan Y. Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study. Int. J. Env. Res. Public Health. 2015;12:10093–10105. doi: 10.3390/ijerph120810093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Deylam Salehi M., Ferdosi-Shahandashti E., Yahyapour Y., Khafri S., Pournajaf A., Rajabnia R. Integron-Mediated Antibiotic Resistance in Acinetobacter baumannii Isolated from Intensive Care Unit Patients, Babol, North of Iran. Biomed. Res. Int. 2017;2017:7157923. doi: 10.1155/2017/7157923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nepka M., Perivolioti E., Kraniotaki E., Politi L., Tsakris A., Pournaras S. In Vitro Bactericidal Activity of Trimethoprim-Sulfamethoxazole Alone and in Combination with Colistin against Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2016;60:6903–6906. doi: 10.1128/AAC.01082-16. [DOI] [PMC free article] [PubMed] [Google Scholar]