Abstract
Biocides are frequently applied as disinfectants in animal husbandry to prevent the transmission of drug-resistant bacteria and to control zoonotic diseases. Concerns have been raised, that their use may contribute to the selection and persistence of antimicrobial-resistant bacteria. Especially, extended-spectrum β-lactamase- and AmpC β-lactamase-producing Escherichia coli have become a global health threat. In our study, 29 ESBL-/AmpC-producing and 64 NON-ESBL-/AmpC-producing E. coli isolates from three German broiler fattening farms collected in 2016 following regular cleaning and disinfection were phylogenetically characterized by whole genome sequencing, analyzed for phylogenetic distribution of virulence-associated genes, and screened for determinants of and associations between biocide tolerance and antibiotic resistance. Of the 30 known and two unknown sequence types detected, ST117 and ST297 were the most common genotypes. These STs are recognized worldwide as pandemic lineages causing disease in humans and poultry. Virulence determinants associated with extraintestinal pathogenic E. coli showed variable phylogenetic distribution patterns. Isolates with reduced biocide susceptibility were rarely found on the tested farms. Nine isolates displayed elevated MICs and/or MBCs of formaldehyde, chlorocresol, peroxyacetic acid, or benzalkonium chloride. Antibiotic resistance to ampicillin, trimethoprim, and sulfamethoxazole was most prevalent. The majority of ESBL-/AmpC-producing isolates carried blaCTX-M (55%) or blaCMY-2 (24%) genes. Phenotypic biocide tolerance and antibiotic resistance were not interlinked. However, biocide and metal resistance determinants were found on mobile genetic elements together with antibiotic resistance genes raising concerns that biocides used in the food industry may lead to selection pressure for strains carrying acquired resistance determinants to different antimicrobials.
Keywords: Escherichia coli, biocide tolerance, antibiotic resistance, biocide determinants, virulence, food safety
1. Introduction
Escherichia coli is a gram-negative, non-sporulating facultative anaerobe, a widespread gut commensal of vertebrates, and a versatile pathogen [1]. Pathogenic E. coli are categorized as intestinal pathogenic (InPEC) or extraintestinal pathogenic E. coli (ExPEC) [2]. The latter colonize the gut of healthy hosts without causing disease but by entering extraintestinal sites ExPEC can lead to urinary tract infections, meningitis, skin infections, or sepsis [3]. In addition to affecting humans, avian pathogenic E. coli (APEC), the avian pathotype of ExPEC, causes severe economic losses to the poultry industry and may represent a zoonotic risk [4]. Multidrug-resistant bacteria (particularly those producing extended-spectrum ß-lactamases (ESBL) and/or AmpC ß-lactamases (AmpC)) are a growing threat to food safety [5,6]. ESBL-/AmpC-producing E. coli from healthy hosts were classified as commensal strains but recent investigations indicated that they also show characteristics of ExPEC or ExPEC-like strains [3,7]. Humans can be exposed to ESBL-/AmpC-producing pathogens via human-to-human transmission, food, animal, and environmental sources [8]. A high prevalence of ESBL-/AmpC-producing Enterobacteriaceae was previously demonstrated on broiler farms [9,10,11]. Recent studies suggested that contaminated broiler chicken farms might play an important role in the transmission of ESBL-/AmpC-producing Enterobacteriaceae into the environment [12,13]. Luyckx et al. detected E. coli in broiler houses following hygiene measures, highlighting drain holes or floor cracks as critical locations for cleaning and disinfection (C&D) [14,15]. Biocides like quaternary ammonium compounds (QACs), aldehydes, oxidizing agents, organic acids, and cresols are widely used in animal husbandry and food processing plants to prevent microbial growth. However, concerns have been raised that the continued exposure to biocides in industrial settings including food production environments may trigger mechanisms that alter both biocide and antibiotic susceptibility and select for antimicrobial-resistant strains [16,17]. E. coli uses multiple pathways to overcome environmental stresses. Acid stress, for instance, is counteracted by a range of physiological, metabolic, and proton-consuming acid resistance mechanisms [18]. Biocide tolerance is a multifactorial process and can include several mechanisms such as target modification [19], biofilm formation [20], changes of cell envelope permeability [21], or the activity of efflux pumps [22]. Proteins involved in tolerance to quaternary ammonium compounds (QACs) include members of the small multidrug resistance (SMR) efflux family such as SugE(c), SugE(p), EmrE, YdgE/YdgF, QacE, QacE∆1, QacF, QacG, QacH, and QacI as well as members of the major facilitator superfamily (MFS) such as MdfA [23,24,25,26].
So far, laboratory methods to investigate biocide susceptibility are not standardized [27,28] and to the best of our knowledge, only one study evaluated epidemiological cutoffs (ECOFFs) for E. coli to a limited set of biocidal compounds [29]. As little is known about the link between biocide selection pressure and antibiotic resistance in E. coli field isolates in Germany we aimed to characterize a commensal E. coli study population including ESBL-/AmpC-producing and NON-ESBL-/AmpC-producing E. coli from broiler fattening farms following cleaning and disinfection. Because of the widespread use of disinfectants in hygiene processes, we assumed a high selective pressure in the investigated farm environment. We tested susceptibilities to seven biocides frequently used in farm hygiene and to antibiotics relevant for human and veterinary medicine. In addition, we characterized the genetic diversity of the E. coli strains including ExPEC associated virulence genes, and looked for associations between biocide tolerance, antibiotic resistance, and the presence of putative genetic determinants of antimicrobial resistance.
2. Materials and Methods
2.1. E. coli Isolates
A panel of 93 E. coli isolates collected in 2016 from three broiler fattening farms after cleaning and disinfection measures were investigated (Table S1). The isolates originated from surfaces of grounds, walls, and equipment such as air inlets, drains, door handles, tractors (for food and litter), electric cables, feeding and drinking troughs from four barns. E. coli were isolated from swab samples on MacConkey agar with and without cefotaxime. Species identification and differentiation of ESBL-/AmpC-producing E. coli were performed as previously described [30]. In brief, MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) was applied to suspicious isolates for species identification. Beta-lactamase genes blaCTX-M, blaSHV, blaTEM, and CIT-type pAmpC genes were identified using a multiplex real-time PCR [31] as well as Sanger sequencing [30]. Isolates were selected from different sources to obtain a highly diverse study population including ESBL-/AmpC- and NON-ESBL-/AmpC-producing E. coli (farm 1: barn 1, n = 27 including 13 AmpC-producing E. coli; barn 2, n = 15 including five ESBL-producing E. coli; farm 2: barn 3, n = 21 including three ESBL-producing E. coli; farm 3: barn 4, n = 30 including eight ESBL-producing E. coli). C&D protocols applied in the barns comprised dry cleaning, wet cleaning, and two disinfection steps. During dry cleaning, bedding and feed were removed. For wet cleaning all-purpose cleaners were used. Disinfection was carried out using formaldehyde-based disinfectants followed by either chlorocresol-based disinfectants (barns 1 and 4) or lime solutions (barns 2 and 3).
2.2. Whole Genome Sequencing
E. coli isolates were cultivated on sheep blood agar. A single colony was transferred into Miller’s lysogeny broth (LB) (Merck KGaA, Darmstadt, Germany) and incubated at 37 °C for 19 ± 1 h with shaking at 150 rpm. DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). Whole-genome sequencing (WGS) libraries were prepared with the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Paired-end sequencing (2 × 301 cycles) was performed using the MiSeq Reagent v3 600-cycle Kit (Illumina) on an Illumina MiSeq benchtop sequencer. Raw fastq data were trimmed and assembled using the AQUAMIS pipeline (https://gitlab.com/bfr_bioinformatics/AQUAMIS (accessed on 9 July 2018)) based on trimmomatic (version 0.36.), fastp (version 0.19.5), unicycler (version 0.4.4), spades (version 3.11.1), pilon (version 1.22), mash (version 2.1), and quast (version 4.6.3).
2.3. Phylogenetic Analysis
For phylogenetic analysis, multilocus-sequence typing (MLST) was performed using WGS data. The classical MLST scheme defined by alleles of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA, database hosted at the University of Warwick) was applied. MLST types were determined using the MLST 2.0 webtool of the Center for Genomic Epidemiology (http://www.genomicepidemiology.org (accessed on 6 October 2018)) [32]. For phylogroup assignment, a multiplex PCR was conducted as described previously [33] with minor modifications. The total reaction mixture of 25 µL contained 0.2 µM of each primer (except for TspE4C2.1b (0.4 µM) and TspE4C2.2b (0.4 µM)), 12.5 µL of DreamTaq Green PCR Mastermix (Thermo Fisher Scientific, Schwerte, Germany), 5 µL of PCR Water and 2.5 µL of the template DNA. An initial denaturation step of 3 min at 94 °C was followed by 33 PCR cycles with 30 s of denaturation at 94 °C, primer binding for 30 s at 57 °C, and 1 min of elongation at 72 °C, as well as a final elongation step of 5 min at 72 °C. Isolates belonging to phylogroups A and C or E and D were not further differentiated and assigned to phylogroup A/C or E/D, respectively. Furthermore, we determined genetic relatedness between E.coli isolates with ParSNP v1.0 [34]. The maximum-likelihood tree was calculated by FastTree2 [35] and visualized with EMBL interactive tree of life, iTOL v4 (https://itol.embl.de/, accessed on 20 September 2019).
2.4. Biocide Susceptibility Testing
2.4.1. Biocides
Susceptibility of the E. coli isolates was tested against the two biocides formaldehyde (FA, Carl Roth, Karlsruhe, Germany) and chlorocresol (p-chloro-m-cresol, PCMC, Merck KGaA) used for C&D on the farms under study and five biocides commonly applied in farm hygiene, namely the quaternary ammonium compounds benzalkonium chloride (BAC, Sigma Aldrich, Steinheim, Germany) and didecyldimethylammonium chloride (DDAC, Merck KGaA), hydrogen peroxide (HP, Carl Roth), peroxyacetic acid (PAA, VWR, Dresden, Germany), and acetic acid (AA, Carl Roth). Biocides were serially diluted in 2-fold steps just before the experiment using standardized hard water as defined in EN 1276. The following final concentration ranges were tested: 320 to 5 mg/L BAC, 40 to 0.3 mg/L DDAC, 640 to 5 mg/L FA, 1024 to 8 mg/L HP, 2000 to 16 mg/L PAA, 16,384 to 128 mg/L AA, and 4000 to 63 mg/L PCMC.
2.4.2. Minimum Inhibitory Concentration (MIC)
Biocide MICs were determined using broth microdilution. Overnight cultures grown on tryptic soy agar (TSA; Merck KGaA) were adjusted to about 106 CFU/mL in 2-fold concentrated tryptic soy broth (TSB; Merck KGaA). In 96-well microtiter plates (Greiner Bio-One, Frickenhausen, Germany), 50 µL of the bacterial suspension was added to 50 µL of the double-concentrated biocide solution. Plates were incubated at 37 °C for 20 ± 2 h. Optical density at 595 nm (OD595) was measured after 5 s of shaking using the Mithras2 multimode reader (Berthold Technologies, Bad Wildbad, Germany; Software MikroWin 2010 v5.18, German UI). Bacterial growth was compared to a negative control (microtiter well containing biocide solution and tryptic soy broth, Thermo Fisher Scientific) and a ΔOD595 nm of 0.08 was applied as the cut-off value. The MIC was defined as the lowest concentration of a biocide at which no growth was observed. Three independent experiments were performed on different days and the median was considered as the final MIC.
2.4.3. Minimum Bactericidal Concentration (MBC)
The MBC of each strain and biocide was determined by broth microdilution according to Knapp et al., with minor modifications [28]. Dey-Engley neutralizing broth (Sigma-Aldrich) was used to quench biocidal effects for MBC testing. Neutralizer efficacy and toxicity were tested before [36]. The MBC was defined as the lowest concentration of a biocide, which revealed no visible colonies after subculture on tryptic soy agar (TSA, Thermo Fisher Scientific). The reference strain E. coli ATCC 25922 was used as internal quality control in both MIC and MBC tests and showed comparable results throughout the experiments.
2.4.4. Determination of MIC95/MBC95
To distinguish between biocide susceptible isolates and isolates with reduced susceptibility, the MIC (or MBC) that encompassed 95% of all MIC (or MBC) values in the distribution was designated as MIC95 (or MBC95).
2.5. Antibiotic Susceptibility Testing
Antibiotic susceptibility was determined by broth microdilution using the Sensititre system with EUVSEC/EUVSEC2 plates (Thermo Fisher Scientific) in concordance with the decision 2013/652/EU of the European Union. The following antimicrobial substances were used: Ampicillin, AMP; Azithromycin, AZI; Cefepime, FEP; Cefoxitin, FOX; Ceftazidime, TAZ; Cefotaxime, FOT; Cefotaxime/Clavulanic acid, F/C; Ceftazidime/Clavulanic acid, T/C; Chloramphenicol, CHL; Ciprofloxacin, CIP; Colistin, COL; Ertapenem, ETP; Gentamicin, GEN; Imipenem, IMI; Meropenem, MERO; Nalidixic acid, NAL; Sulfamethoxazole, SMX; Temocillin, TRM; Tetracycline, TET; Tigecycline, TGC; Trimethoprim, TMP. We followed CLSI guidelines and defined resistance using epidemiological cut-offs according to EUCAST.
2.6. Statistical Analysis
Spearman rank coefficients (Rho) were calculated to investigate the correlation of MICs or MBCs between tested biocides and antibiotics using SPSS (IBM SPSS Statistics, Version 21, IBM corp., Armonk, NY, USA). Data were tested for normal distribution by the Kolmogorov-Smirnov test. For comparative analysis between two groups of isolates (e.g., ESBL-/AmpC- versus NON-ESBL-/AmpC-producing isolates) the Mann-Whitney-test was applied. Statistically significant differences between antimicrobial resistance or distribution of virulence determinants in different genetic lineages were tested using the chi2 test and Fisher’s exact test. p-values < 0.05 were considered to be significant.
2.7. In Silico Screening for Biocide and Metal Tolerance Determinants at Protein Level
WGS data of the E. coli isolates under study were screened for the presence of 753 experimentally confirmed biocide- and metal-resistance proteins recorded in the BacMet database [37] (Antibacterial Biocide and Metal Resistance Genes database; http://bacmet.biomedicine.gu.se/, BacMet version 2, last updated on 9 December 2017, accessed on 5 December 2018) as described before [38].
2.8. Detection of Biocide Tolerance and Virulence Determinants at Nucleotide Level
The presence of genes conferring biocide tolerance was determined as previously described [38]. The genomes of all isolates were screened for genes encoding for small multidrug resistance (SMR) transporters, i.e., qacEΔ1, qacE, qacF, qacH, qacI, qacG, emrE, sugE(c), sugE(p), ydgE, ydgF, and for the multidrug efflux pump gene mdfA of the major facilitator superfamily (MFS). In addition, we screened for genes involved in formaldehyde and acid tolerance. An overview of the investigated genes and corresponding accession numbers is given in Table S2. A minimum sequence identity (%ID) threshold of 80% and a minimum length of 80% of the target gene were defined for the detection of biocide determinants except for qacEΔ1 and qacE (100%ID and 100% minimum length).
In addition, we screened for the presence of 49 virulence genes typically associated with ExPEC including fitness factors that are found in pathogenic and commensal strains (Table S2). Virulence-associated genes (VAGs) were chosen from public databases contained in the E. coli functional genotyping plugin (version 1.01) of Bionumerics or from previously published reports [7,39,40]. A minimum sequence identity (%ID) threshold of 90% and a minimum length of 60% of the target gene were used for the identification of VAGs.
2.9. Identification of Antibiotic Resistance Genes
Acquired antibiotic resistance determinants and chromosomal mutations leading to antibiotic resistance were identified using ResFinder 3.0 (Center for Genomic Epidemiology, http://www.genomicepidemiology.org, accessed on 11 January 2019 [41]).
2.10. Accession Numbers of Whole-Genome Sequences
Genome sequence data of the strains under study have been deposited at the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/, accessed on 9 March 2021)) under accession numbers JAFMWT000000000-JAFMVF000000000 (see Table S1).
3. Results
3.1. Phylogenetic Diversity and Virulence-Associated Genes
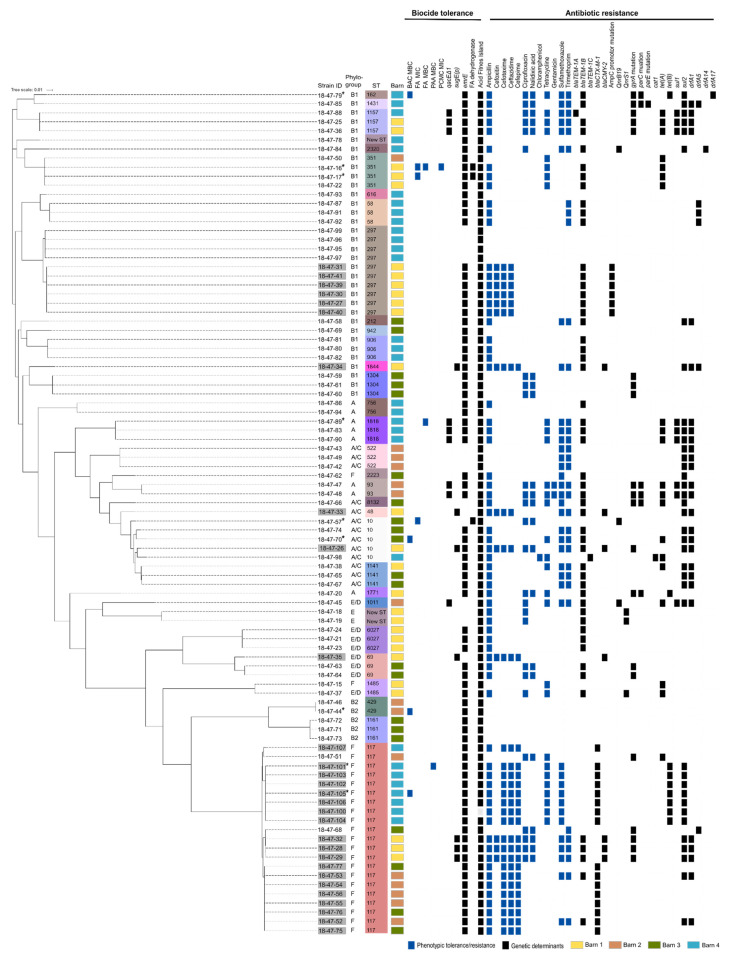
PCR-based phylotyping of the 93 E. coli isolates revealed seven different banding patterns associated with phylogroups A (n = 8), A/C (n = 13), B1 (n = 34), B2 (n = 5), E (n = 2), E/D (n = 7), and F (n = 24). E. coli isolates belonged to 30 known and two unknown multilocus sequence types (STs). The most prevalent STs were ST117 (n = 21; two NON-ESBL-/AmpC-producing E. coli from barns 2 and 3, 19 ESBL-/AmpC-producing E. coli from all barns) and ST297 (n = 10; six AmpC-producing E. coli from barn 1, and four NON-ESBL-/AmpC-producing E. coli from barn 4) (Figure 1).
Figure 1.
Phylogenetic tree of 93 E. coli isolates from broiler fattening farms including their phenotypic biocide tolerance and antibiotic resistance as well as the distribution of biocide tolerance and antibiotic resistance-conferring genes. An asterisk marks biocide tolerant strains. Reduced susceptibility to biocides and antibiotic resistance are indicated for each isolate as blue squares, tolerance, and resistance-conferring genes as black squares. Further information on ESBL-/AmpC-producing E. coli phenotype (grey shaded strain ID) and multilocus sequence type (ST) are provided. The affiliation to different barns are highlighted in yellow (barn 1), orange (barn 2), green (barn 3), and blue (barn 4). BAC = Benzalkonium chloride, FA = Formaldehyde, PCMC = Chlorocresol (p-chloro-m-cresol).
Up to 27 ExVAGs (VAGs associated with ExPEC) (55%) were detected in ST117 strains (phylogroup F), up to 23 ExVAGs (47%) in ST429 (phylogroup B2), and up to 20 ExVAGs (41%) in ST69 (phylogroup E/D) (Table S1). All isolates were positive for fimH (type 1 fimbriae), feoB (ferrous iron transporter, protein B), and ompA (outer membrane protein A). The iss (increased serum survival protein) and fimA (type 1 fimbriae) genes were present in 77 (83%) and 74 (80%) isolates, respectively. Twenty-one VAGs were significantly associated with phylogroup F. Certain genetic determinants such as papC, papEF, papG-allele II (P fimbriae formation), ireA (iron-responsive element), and hlyE (hemolysin E) exclusively occurred in isolates belonging to ST117 of phylogroup F, whereas vat (vacuolating autotransporter toxin) was present in ST429 (phylogroup B2) and some ST117 (phylogroup F) isolates. Iron capture systems were frequently represented in the genomes, but the number of encoding genes varied considerably among isolates from 1 to 11. Iron uptake systems were most prevalent in ST117 and ST429 isolates.
3.2. Susceptibility to Biocides
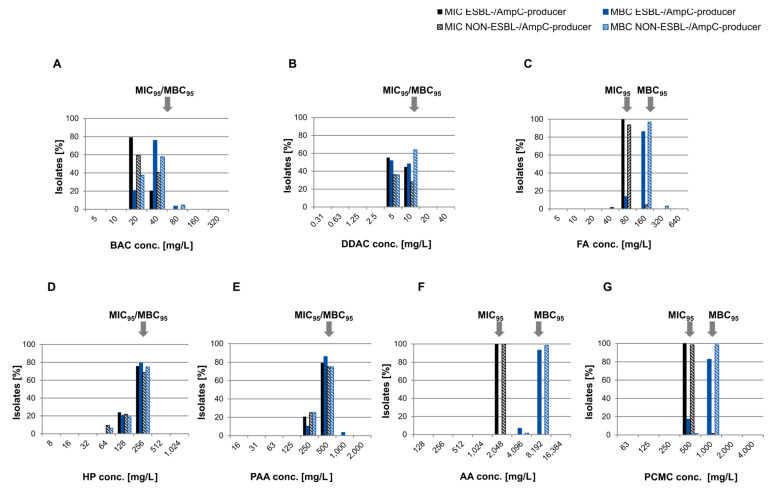
MIC and MBC data showed non-normal, unimodal distributions ranging between one and three dilution steps for all biocides (Figure 2). MIC and/or MBC values above MIC95/MBC95 indicated isolates with reduced susceptibility to the tested biocides.
Figure 2.
MIC and MBC distributions of ESBL-/AmpC-producing and NON-ESBL-/AmpC-producing E. coli isolates for common biocides used in farm hygiene. Black bars = MIC ESBL-/AmpC-producing E. coli, black striped = MIC NON-ESBL-/AmpC-producing E. coli, blue bars = MBC ESBL-/AmpC-producing E. coli, blue striped = MBC NON-ESBL-/AmpC-producing E. coli. Arrows mark MIC95 and MBC95 representing cut-off values for isolates with reduced susceptibility. (A) BAC = Benzalkonium chloride, (B) DDAC = Didecyldimethylammonium chloride, (C) FA = Formaldehyde, (D) HP = Hydrogen peroxide, (E) PAA = Peracetic acid, (F) AA = Acetic acid, (G) PCMC = Chlorocresol (p-chloro-m-cresol).
These biocide-tolerant isolates were found in all barns (barn 1: n = 2; barn 2: n = 1, barn 3: n = 2, barn 4, n = 4), and mostly originated from transitions between wall and floor as well as from cracks and crevices in the ground (Table S1). An individual NON-ESBL-/AmpC-producing E. coli isolate (ST351) displayed elevated MIC (160 mg/L) and MBC (320 mg/L) values of FA and an elevated MIC of PCMC (1000 mg/L). Furthermore, three NON-ESBL-/AmpC-producing E. coli showed either an elevated MIC (160 mg/L, n = 2, ST10, ST351) or MBC value (320 mg/L, n = 1, ST1818) of FA. Increased MBCs were also detected for PAA (1000 mg/L, n = 1, ESBL-producing E. coli, ST117) and BAC (80 mg/L, n = 4, three NON-ESBL-/AmpC-producing E. coli, ST10, ST162, ST429, and one ESBL-producing E. coli, ST117) (Figure 1).
3.3. Susceptibility to Antibiotics
All isolates were sensitive to carbapenems (ETP, IMI, MERO), COL and TGC. Antibiotic resistance to AMP (100% ESBL-/AmpC-producing E. coli, 63% NON-ESBL-/AmpC-producing E. coli), SMX (52% ESBL-/AmpC-producing E. coli, 36% NON-ESBL-/AmpC-producing E. coli), and TMP (28% ESBL-/AmpC-producing E. coli, 39% NON-ESBL-/AmpC-producing E. coli) were most common in both groups (Figure 1). Thirty-four isolates (37%) were resistant to at least one antibiotic in three or more classes and therefore defined as multidrug-resistant (MDR). Two isolates from barn 2 were resistant to antibiotics in five substance classes including aminoglycosides, ß-lactams, fluoroquinolones, sulfonamides, and tetracyclines.
3.4. In Silico Analysis of Determinants Conferring Biocide and Metal Tolerance
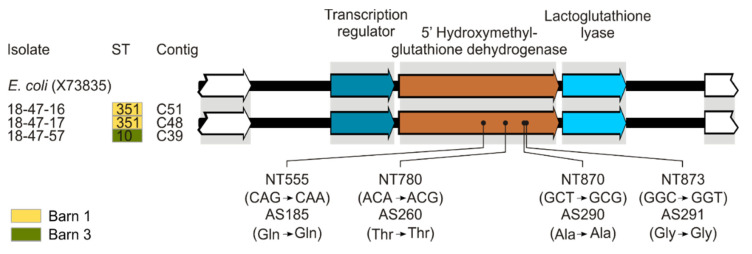
Out of 753 proteins potentially conferring biocide or metal tolerance 249 were identified in our study population (Table S3). Four tolerance determinants were exclusively present in three isolates with increased MIC values of FA (18-47-16 (ST351), 18-47-17 (ST351), and 18-47-57 (ST10). Three of these determinants belonged to an arsenic resistance operon whereas the other one was annotated as nickel/cobalt efflux transporter NcrC that is involved in nickel and cobalt resistance. All isolates under study harbored glutathione- and NAD-dependent formaldehyde dehydrogenase with ≥80% nucleotide identity to the reference (Genbank Acc. No. X73835) found in the formaldehyde-tolerant strain Escherichia coli VU3695 [19]. The three isolates with reduced susceptibility to formaldehyde harbored an additional formaldehyde dehydrogenase with 99.6% identity to X73835. Sequence analysis revealed only synonymous mutations compared to the reference (Figure 3).
Figure 3.
Glutathione-dependent formaldehyde dehydrogenases of E. coli isolates compared to the plasmid-encoded reference X73835. The alignment was created using Bionumerics and adjusted by CorelDraw Graphic Suite 3.0 (version 17) for better interpretation. Relevant CDS (arrows) were labeled by protein function based on RAST annotation.
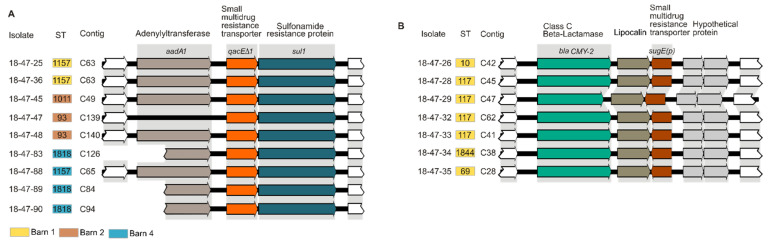
Genes of the E. coli acid fitness island were found in all but one isolate of the study population. SMR efflux pump genes sugE(c), ydgE, and ydgF and the MFS efflux pump gene mdfA were always present. We could not detect genes encoding the QAC-specific efflux determinants QacE, QacG, QacF, QacI, and QacH. Seventy-nine isolates (85%) carried emrE. The SMR efflux pump gene qacE∆1 was detected in nine NON-ESBL-/AmpC-producing E. coli isolates (10%) of ST93 (n = 2), ST1011 (n = 1), ST1157 (n = 3), and ST1818 (n = 3) taken at different sampling sites in the barns 1, 2 and 4 (Figure 4A). SugE(p) was detected in seven plasmid-mediated AmpC β-lactamase-(pAmpC-)producing E. coli isolates (8%) from barn 1 (ST117 (n = 3), ST10 (n = 1), ST48 (n = 1), ST69 (n = 1), ST1844 (n = 1)) (Figure 4B). However, the presence of efflux determinants was not associated with reduced susceptibility to tested biocides.
Figure 4.
Colocalization of biocide tolerance determinants and antibiotic resistance genes. (A) SMR efflux pump encoding gene qacE∆1 located between aminoglycoside (aadA1) and sulfonamide (sul1) resistance genes on the same contig. (B) SMR efflux pump encoding gene sugE(p) located downstream of class C beta-lactamase. The alignment was created using Bionumerics and adjusted by CorelDraw Graphic Suite 3.0 (version 17) for better interpretation. Relevant CDS (arrows) were labeled by protein function based on RAST annotation.
3.5. In Silico Analysis of Antibiotic Resistance Gene Profiles
Phenotypic antibiotic resistance could be attributed to known genetic resistance determinants except for gentamicin (Figure 1). Identified determinants responsible for beta-lactam antibiotic resistance were blaTEM-1A (n = 1, ST1157, barn 4), blaTEM-1B (n = 52, 25 STs from all barns), blaTEM-1C (n = 1, ST10, barn 4), blaCTX-M-1 (n = 16, ST117, barns 2, 3, and 4) and blaCMY-2 (n = 7, ST10, ST48, ST69, ST117, ST1844, barn 1) as well as ampC promotor mutations (n = 6, ST297, barn 1). Target mutations of gyrA (n = 20, 10 STs from all barns), parC (n = 6, ST93, ST162, ST1431, ST1771, ST8132, from all barns) and/or parE (n = 1, ST1431, barn 4) as well as the resistance genes qnrB19 (n = 3, ST10, ST1011, ST2320, barns 2, 3, and 4) and qnrS1 (n = 3, ST1485, unknown ST, barn 1) were found in quinolone resistant isolates. Chloramphenicol resistance could be attributed to the presence of cat1 (n = 1, ST10, barn 4). All tetracycline resistant isolates were positive for tet(A) (n = 20, 10 STs from all barns) or tet(B) (n = 9, ST117, ST162, ST1771, barns 1 and 4). In sulfonamide and trimethoprim resistant isolates the resistance genes sul1 (n = 9, ST93, ST1011, ST1157, ST1818, barns 1, 2, and 4) and sul2 (n = 38, 16 STs from all barns) as well as drfA1 (n = 27, 12 STs from all barns), drfA5 (n = 6, ST58, ST117, ST1431, ST1844, barns 1, 3, and 4), drfA14 (n = 1, ST2320, barn 4) and/or drfA17 (n = 1, ST162, barn 4) were present.
3.6. Association Between Reduced Biocide Susceptibility and Antibiotic Resistance and Co-occurrence of Antimicrobial Resistance Genes
Antibiotic resistance was not significantly associated with reduced susceptibility to biocides. There was also no significant difference between isolates from different barns. In addition, ESBL-/AmpC-producing isolates were in general not less susceptible to biocides than NON-ESBL-/AmpC-producing isolates. On the contrary, a higher proportion of NON-ESBL-/AmpC-producing E. coli showed reduced susceptibility in terms of MBCs of FA and PCMC compared to ESBL-/AmpC-producing E. coli (Figure 2). Interestingly, several isolates carried biocide and metal tolerance genes on mobile genetic elements closely linked to antibiotic resistance genes. For example, eight qacE∆1-positive isolates carried qacE∆1, sul1, and aadA1 on the same contig (Figure 4A). These determinants could be found downstream of an integron-integrase (intI1) gene in four out of nine isolates verifying their localization on a class 1 integron. The same element carried a mercury-resistance operon. Similarly, all sugE(p)-positive isolates (n = 7) carried sugE(p) and blaCMY-2 on the same contig (Figure 4B). Sequence data revealed genes associated with conjugal transfer and transcription in close proximity indicating plasmid localization of sugE(p) and blaCMY-2.
4. Discussion
Our study aimed at investigating (i) the phylogenetic diversity and virulence determinants, (ii) potential relationships between susceptibilities to biocides and antibiotics, and (iii) genetic determinants of biocide tolerance and antibiotic resistance of E. coli isolates from German broiler fattening farms. The study population consisted of 93 isolates sampled after C&D. Most of the field isolates belonged to phylogroup B1 and F. While phylogroup B1 and A mainly comprise commensals or intestinal pathogens [42], phylogroup F are frequently associated with ExPECs in humans, companion animals, and birds [43,44,45]. Furthermore, ExPEC strains are closely related to avian pathogenic E. coli suggesting poultry as a reservoir of zoonotic APEC strains [39,46]. APEC can cause avian colibacillosis, which threatens poultry flocks worldwide. Three of the STs detected in our study, ST10, ST48, and ST117 have been previously linked to APEC strains [47,48,49] and were also isolated from human patients [50,51,52], emphasizing a zoonotic risk. ST297, which is known to be highly prevalent in environmental and food samples, and ST69 were also found in our study population and can be pathogenic for poultry and humans [53]. In general, our data revealed a broad heterogeneity of E. coli isolates on German broiler fattening farms with variable numbers of virulence-associated genes involved in adhesion, iron uptake, and cytotoxic activity. ST117 (phylogroup F) and ST429 (phylogroup B2) carried the highest number of iron uptake-related genes. Similarly, Projahn et al. observed a high prevalence of determinants involved in iron acquisition in ST117 isolates collected during the years 2014 and 2015 from German broiler meat production chains [7]. E. coli can survive extreme acid stress [54] making use of amino acid-dependent and independent resistance mechanisms [55]. One of the amino acid-dependent systems, encoded by 12 genes of the acid fitness island, is highly conserved in E. coli and was found in virtually all isolates of our study population.
Escherichia coli can survive hygiene measures, persist over a long period of time, and spread throughout the barns of broiler chicken farms [14,15,56,57]. Overall, phenotypic biocide susceptibility testing did not prove tolerance to disinfectants within our study population since MIC and MBC values of the biocides tested were well below in-use concentrations. Modal MIC values of E. coli determined for BAC [58,59,60,61,62], DDAC [63,64], FA [58,60,61], AA [58,65], PAA [66] and PCMC [67,68] in previous studies were similar to our results. In contrast, modal MIC and MBC values of HP reported for avian pathogenic E. coli differed by two dilution steps (64 versus 256 mg/L) [61]. So far, breakpoints to distinguish between biocide susceptible and tolerant isolates are missing. Morrissey et al. [29] suggested ECOFFs for the most commonly applied biocides such as BAC, chlorhexidine, triclosan, and sodium hypochlorite considering various species including E. coli. According to published MICs (>64 mg/L) and MBCs (>128 mg/L) of BAC, none of our E. coli isolates could be defined as tolerant. However, MIC values of biocides are difficult to compare across studies because experimental conditions have not yet been harmonized. In this context, Slipski et al. compared different antimicrobial susceptibility test methods (broth, agar spot colony, and pegged lid biofilms) and showed that the mode of bacterial growth significantly influenced QAC tolerant phenotypes related to SMR over-expression [69]. Thus, standardized methods are urgently needed.
Based on the MIC95/MBC95 values determined, nine isolates from our study population showed reduced susceptibility to at least one biocide (Figure 1 and Figure 2). Six of these isolates were taken from transitions between floor and wall or cracks and crevices. These are well-known critical locations in broiler houses because they are difficult to clean and disinfect [14,15], and exposure to subinhibitory concentrations of biocides in such niches is very likely. Three out of the nine isolates showed elevated MICs of formaldehyde and one isolate additionally had an elevated MIC of chlorocresol. The most widespread bacterial pathway for formaldehyde detoxification involves a glutathione-dependent dehydrogenase catalyzing the reversible formation of S-formylglutathione and NADH from formaldehyde, glutathione, and NAD [70]. Enzymatic degradation of formaldehyde by a plasmid-encoded variant of the enzyme has been previously described as a formaldehyde resistance mechanism in E. coli [19,71,72,73]. In our study, the plasmid-encoded variant of the formaldehyde dehydrogenase was exclusively present in isolates displaying elevated MICs of formaldehyde (160 mg/L) indicating that this enzyme may contribute to the observed phenotype. Interestingly, genes involved in arsenic and nickel/cobalt resistance were also uniquely detected in these formaldehyde tolerant isolates.
SMR efflux pumps are known to confer resistance to a variety of substances, including QACs and antibiotics [23,24,26,74,75,76,77], and are commonly found in E. coli [59,64,78,79]. Since QACs are frequently used for cleaning and disinfection in the food industry, strains armed with appropriate biocide tolerance mechanisms have an increased ability to persist in food processing environments. Not only drugs and toxic metabolites are expelled from bacterial cells by multidrug efflux pumps, molecules that may be important for cell communication, biofilm formation, and osmoregulation or protection of the cell are also released [76,80].
In our study, all isolates harbored the putative QAC tolerance conferring genes sugE(c), ydgE, ydgF, and mdfA, while qacE, qacF, qacG, qacH, and qacI were absent. These results are in line with previous findings on the prevalence of ydgE/ydgF (87–100%), mdfA (86–100%), and qac genes (0–18%) in E. coli isolates from different sources [64,79]. The SMR transporters emrE, qacE∆1, and sugE(p) were detected in varying frequencies within our study population. Nevertheless, our data were similar to those obtained from other epidemiological studies on E. coli isolated from poultry meat, meat products, and farms in Germany [81], the United States [79], and China [64]. The contribution of qacE∆1 as a partially functional derivative of qacE [82] on QAC tolerance is controversially discussed [83]. As described before [81,84], we were not able to show an association between the presence of qacE∆1 and reduced QAC susceptibility. The SMR efflux pump SugE has its role in QAC tolerance [26,64] with a rather narrow substrate specificity, including cetyltrimethyl ammonium, cetyldimethyl ammonium, cetylpyridinium, and cetrimide cations [69,74], which may explain the phenotypic susceptibility to BAC and DDAC of isolates carrying sugE(p) in our study.
Antibiotic resistance profiles were generally consistent with zoonoses monitoring data of commensal E. coli from broiler fattening farms in Germany, 2016 [85]. However, 8.3% colistin-resistant isolates were reported in the national monitoring program, whereas colistin resistance was not found in our study population. A significant number of isolates showed resistance to three or more classes of antibiotics including critically important antimicrobials as classified by the World Health Organization such as quinolones and 3rd generation cephalosporins [86]. With the exception of gentamicin, all phenotypic resistances could be traced back to genetic determinants. Different mechanisms are known to confer gentamicin resistance. Most common are enzymes modifying the drug by acetylation (aminoglycoside acetyltransferase, AAC), adenylation (adenylate aminoglycoside nucleotidyltransferase, ANT) or phosphorylation (aminoglycoside phosphotransferase, APH) [87,88]. Mutations in the ribosomal target have also been described [89], but could not be confirmed in our isolates. According to clinical breakpoints of CLSI, E. coli is supposed to be resistant to gentamicin if MIC ≥ 16 mg/L [90]. As our isolates had MIC values below the clinical but above epidemiological cut-off (ECOFF 2 mg/L), these isolates may have developed resistance. Within the EU, gentamicin is not authorized for use in poultry [91] and resistance is rarely found in conventional broiler stocks in Germany (1.3% in 2016) [85].
In vitro studies showed that antibiotic cross-resistance can occur during bacterial exposure to subinhibitory concentrations of biocides like QACs [92], biguanides [93], and phenolic compounds [94]. The E. coli isolates in our study revealed no association between phenotypic biocide tolerance and antibiotic resistance as described before [60,95]. On the contrary, FA and PCMC killed ESBL-/AmpC-producing E. coli at slightly lower concentrations than NON-ESBL-/AmpC-producing E. coli. Similarly, lower MICs of DDAC were reported for ESBL-/AmpC-producing E. coli in another study [81].
The biocide tolerance determinants qacE∆1 and sugE(p) were located on mobile genetic elements in close proximity to the antibiotic resistance genes sul1 and blaCMY-2, respectively. QacE∆1 is common in enteric bacteria and is typically associated with the presence of class 1 integrons that carry the sulfonamide resistance determinant sul1 explaining why all qacE∆1 positive isolates showed co-resistance to sulfamethoxazole [96]. On the same genetic element, several mercury resistance genes were observed, which frequently occur on plasmids together with antibiotic resistance genes and the qacE∆1 gene [97]. Furthermore, multiple gene cassettes can be arranged in tandem within these elements conferring additional resistance to ß-lactams, tetracycline, gentamicin as well as aminoglycosides [59,64,79]. Worldwide, blaCMY-2 is associated with pAmpC-producing E. coli from poultry [98]. The genetic element, blaCMY-2-blc-sugE, has already been found in IncK plasmids of E. coli isolated from humans in Spain and poultry in Norway and Switzerland [99,100,101]. Plasmids carrying sugE(p) and blaCMY-2 antibiotic resistance genes have been detected in various STs of E. coli [99,101,102] and may be spread by conjugative transfer to different reservoirs. Even though isolates carrying qacE∆1 or sugE(p) did not show reduced susceptibility to the QACs investigated in our study, the use of QACs in broiler fattening farms may provide selection pressure to strains that carry genes encoding resistance to clinically important antibiotics [64].
5. Conclusions
Our study revealed a high genetic diversity of E. coli isolates from German broiler fattening farms including genotypes characteristic of ExPEC strains. Our findings support the hypothesis that poultry farm environments may act as a reservoir of human ExPEC and could play a role in the spread of facultative pathogenic E. coli. While the overall prevalence of biocide tolerant strains was low, the detection of isolates carrying formaldehyde tolerance determinants and at the same time showing a reduced MIC to the compound indicates that the use of disinfectants could have provided selection pressure. The QAC tolerance determinants qacE∆1 and sugE(p) were both located on mobile genetic elements in close proximity to antibiotic resistance genes. In this case, disinfectants may simultaneously select strains with acquired resistance to other antimicrobials. Whether disinfectants can be a driver of antibiotic resistance in zoonotic pathogens from stable to table has to be clarified to assess the consumer risks related to hygiene measures.
Acknowledgments
Our work was intramurally funded by the German Federal Institute for Risk Assessment (BfR grant no. 1322-674). We thank Anna-Louisa Hauffe for excellent technical assistance and our colleagues of the BfR sequencing platform (4SZ) for their support. We further thank Katrin Boll, Anja Blasse, and their colleagues from the Institute for Animal Hygiene and Environmental Health of Freie Universität Berlin for excellent assistance in sampling and monitoring of the poultry farms and subsequent E. coli strain isolation and characterization.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/3/651/s1, Table S1: Supplementary information of investigated isolates, Table S2: Biocide tolerance genes and virulence-associated genes (VAGs) screened in this study, Table S3: In silico analysis of determinants conferring biocide tolerance.
Author Contributions
Conceptualization, A.R., M.N., S.A.D., and R.D.; methodology, A.R., M.P., and C.R.; software, A.R. and J.A.H.; validation, A.R., S.V., M.P., J.A.H., S.A.D., and R.D.; formal analysis, A.R., S.V., J.A.H., and R.D.; investigation, A.R., S.V., M.P., J.A.H., and R.D.; resources, U.R., C.R., S.A.D. and R.D.; data curation, A.R., S.V., M.P., and J.A.H.; writing—original draft preparation, A.R., S.V., S.A.D., and R.D.; writing—review and editing, all authors; visualization, A.R., and J.A.H.; supervision, M.N., S.A.D., and R.D.; project administration, M.N., S.A.D. and R.D.; funding acquisition, S.A.D. and R.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tenaillon O., Skurnik D., Picard B., Denamur E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 2.Smith J.L., Fratamico P.M., Gunther N.W. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007;4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 3.Vila J., Saez-Lopez E., Johnson J.R., Romling U., Dobrindt U., Canton R., Giske C.G., Naas T., Carattoli A., Martinez-Medina M., et al. Escherichia coli: An old friend with new tidings. FEMS Microbiol. Rev. 2016;40:437–463. doi: 10.1093/femsre/fuw005. [DOI] [PubMed] [Google Scholar]
- 4.Mellata M. Human and avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013;10:916–932. doi: 10.1089/fpd.2013.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EFSA Panel on Biological Hazards (BIOHAZ) Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011;9:2322. doi: 10.2903/j.efsa.2011.2322. [DOI] [Google Scholar]
- 6.Kaesbohrer A., Bakran-Lebl K., Irrgang A., Fischer J., Kämpf P., Schiffmann A., Werckenthin C., Busch M., Kreienbrock L., Hille K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019;233:52–60. doi: 10.1016/j.vetmic.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Projahn M., Daehre K., Semmler T., Guenther S., Roesler U., Friese A. Environmental adaptation and vertical dissemination of ESBL-/pAmpC-producing Escherichia coli in an integrated broiler production chain in the absence of an antibiotic treatment. Microb. Biotechnol. 2018;11:1017–1026. doi: 10.1111/1751-7915.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mughini-Gras L., Dorado-Garcia A., van Duijkeren E., van den Bunt G., Dierikx C.M., Bonten M.J.M., Bootsma M.C.J., Schmitt H., Hald T., Evers E.G., et al. Attributable sources of community-acquired carriage of Escherichia coli containing beta-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet. Health. 2019;3:e357–e369. doi: 10.1016/S2542-5196(19)30130-5. [DOI] [PubMed] [Google Scholar]
- 9.Friese A., Schulz J., Laube H., von Salviati C., Hartung J., Roesler U. Faecal occurrence and emissions of livestock-associated methicillin-resistant Staphylococcus aureus (laMRSA) and ESBL/AmpC-producing E. coli from animal farms in Germany. Berl. Muench. Tieraerztl. Wochenschr. 2013;126:175–180. [PubMed] [Google Scholar]
- 10.Huijbers P.M., Graat E.A., Haenen A.P., van Santen M.G., van Essen-Zandbergen A., Mevius D.J., van Duijkeren E., van Hoek A.H. Extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: Prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 2014;69:2669–2675. doi: 10.1093/jac/dku178. [DOI] [PubMed] [Google Scholar]
- 11.Laube H., Friese A., von Salviati C., Guerra B., Käsbohrer A., Kreienbrock L., Roesler U. Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 2013;79:4815–4820. doi: 10.1128/AEM.00856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daehre K., Projahn M., Semmler T., Roesler U., Friese A. Extended-Spectrum Beta-Lactamase-/AmpC Beta-Lactamase-Producing Enterobacteriaceae in Broiler Farms: Transmission Dynamics at Farm Level. Microb. Drug Resist. 2018;24:511–518. doi: 10.1089/mdr.2017.0150. [DOI] [PubMed] [Google Scholar]
- 13.Laube H., Friese A., von Salviati C., Guerra B., Rosler U. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol. 2014;172:519–527. doi: 10.1016/j.vetmic.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Luyckx K., Dewulf J., Van Weyenberg S., Herman L., Zoons J., Vervaet E., Heyndrickx M., De Reu K. Comparison of sampling procedures and microbiological and non-microbiological parameters to evaluate cleaning and disinfection in broiler houses. Poult. Sci. 2015;94:740–749. doi: 10.3382/ps/pev019. [DOI] [PubMed] [Google Scholar]
- 15.Luyckx K.Y., Van Weyenberg S., Dewulf J., Herman L., Zoons J., Vervaet E., Heyndrickx M., De Reu K. On-farm comparisons of different cleaning protocols in broiler houses. Poult. Sci. 2015;94:1986–1993. doi: 10.3382/ps/pev143. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert P., McBain A.J. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 2003;16:189–208. doi: 10.1128/CMR.16.2.189-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SCENIHR Opinion on the Assessment of the Antibiotic Resistance Effects of Biocides. [(accessed on 21 January 2021)]; Available online: https://ec.europa.eu/health/archive/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf.
- 18.Kanjee U., Houry W.A. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 2013;67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 19.Kummerle N., Feucht H.H., Kaulfers P.M. Plasmid-mediated formaldehyde resistance in Escherichia coli: Characterization of resistance gene. Antimicrob. Agents Chemother. 1996;40:2276–2279. doi: 10.1128/AAC.40.10.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bas S., Kramer M., Stopar D. Biofilm Surface Density Determines Biocide Effectiveness. Front. Microbiol. 2017;8:2443. doi: 10.3389/fmicb.2017.02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denyer S.P., Maillard J.Y. Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. J. Appl. Microbiol. 2002;92:35S–45S. doi: 10.1046/j.1365-2672.92.5s1.19.x. [DOI] [PubMed] [Google Scholar]
- 22.Levy S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 2002;92:65S–71S. doi: 10.1046/j.1365-2672.92.5s1.4.x. [DOI] [PubMed] [Google Scholar]
- 23.Bay D.C., Rommens K.L., Turner R.J. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim. Biophys. Acta. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Bay D.C., Turner R.J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 2009;9:140. doi: 10.1186/1471-2148-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R., Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 1997;179:2274–2280. doi: 10.1128/JB.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He G.X., Zhang C., Crow R.R., Thorpe C., Chen H., Kumar S., Tsuchiya T., Varela M.F. SugE, a new member of the SMR family of transporters, contributes to antimicrobial resistance in Enterobacter cloacae. Antimicrob. Agents Chemother. 2011;55:3954–3957. doi: 10.1128/AAC.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fessler A.T., Schug A.R., Geber F., Scholtzek A.D., Merle R., Brombach J., Hensel V., Meurer M., Michael G.B., Reinhardt M., et al. Development and evaluation of a broth macrodilution method to determine the biocide susceptibility of bacteria. Vet. Microbiol. 2018;223:59–64. doi: 10.1016/j.vetmic.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Knapp L., Amezquita A., McClure P., Stewart S., Maillard J.Y. Development of a protocol for predicting bacterial resistance to microbicides. Appl. Environ. Microbiol. 2015;81:2652–2659. doi: 10.1128/AEM.03843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrissey I., Oggioni M.R., Knight D., Curiao T., Coque T., Kalkanci A., Martinez J.L. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE. 2014;9:e86669. doi: 10.1371/journal.pone.0086669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Projahn M., Daehre K., Roesler U., Friese A. Extended-Spectrum-Beta-Lactamase- and Plasmid-Encoded Cephamycinase-Producing Enterobacteria in the Broiler Hatchery as a Potential Mode of Pseudo-Vertical Transmission. Appl. Environ. Microbiol. 2017;83:e02364-16. doi: 10.1128/AEM.02364-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roschanski N., Fischer J., Guerra B., Roesler U. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS ONE. 2014;9:e100956. doi: 10.1371/journal.pone.0100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomsen M.C., Ahrenfeldt J., Cisneros J.L., Jurtz V., Larsen M.V., Hasman H., Aarestrup F.M., Lund O. A Bacterial Analysis Platform: An Integrated System for Analysing Bacterial Whole Genome Sequencing Data for Clinical Diagnostics and Surveillance. PLoS ONE. 2016;11:e0157718. doi: 10.1371/journal.pone.0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clermont O., Christenson J.K., Denamur E., Gordon D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 34.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knapp L., Rushton L., Stapleton H., Sass A., Stewart S., Amezquita A., McClure P., Mahenthiralingam E., Maillard J.Y. The effect of cationic microbicide exposure against Burkholderia cepacia complex (Bcc); the use of Burkholderia lata strain 383 as a model bacterium. J. Appl. Microbiol. 2013;115:1117–1126. doi: 10.1111/jam.12320. [DOI] [PubMed] [Google Scholar]
- 37.Pal C., Bengtsson-Palme J., Rensing C., Kristiansson E., Larsson D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42:D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roedel A., Dieckmann R., Brendebach H., Hammerl J.A., Kleta S., Noll M., Al Dahouk S., Vincze S. Biocide tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 2019;85:e01253-19. doi: 10.1128/AEM.01253-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Siek K.E., Giddings C.W., Doetkott C., Johnson T.J., Fakhr M.K., Nolan L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- 40.Ronco T., Stegger M., Olsen R.H., Sekse C., Nordstoga A.B., Pohjanvirta T., Lilje B., Lyhs U., Andersen P.S., Pedersen K. Spread of avian pathogenic Escherichia coli ST117 O78:H4 in Nordic broiler production. BMC Genom. 2017;18:13. doi: 10.1186/s12864-016-3415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohler C.D., Dobrindt U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011;301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Bourne J.A., Chong W.L., Gordon D.M. Genetic structure, antimicrobial resistance and frequency of human associated Escherichia coli sequence types among faecal isolates from healthy dogs and cats living in Canberra, Australia. PLoS ONE. 2019;14:e0212867. doi: 10.1371/journal.pone.0212867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo S., Wakeham D., Brouwers H.J., Cobbold R.N., Abraham S., Mollinger J.L., Johnson J.R., Chapman T.A., Gordon D.M., Barrs V.R., et al. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect. 2015;17:266–274. doi: 10.1016/j.micinf.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Vangchhia B., Abraham S., Bell J.M., Collignon P., Gibson J.S., Ingram P.R., Johnson J.R., Kennedy K., Trott D.J., Turnidge J.D., et al. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology. 2016;162:1904–1912. doi: 10.1099/mic.0.000367. [DOI] [PubMed] [Google Scholar]
- 46.Maluta R.P., Logue C.M., Casas M.R., Meng T., Guastalli E.A., Rojas T.C., Montelli A.C., Sadatsune T., de Carvalho Ramos M., Nolan L.K., et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE. 2014;9:e105016. doi: 10.1371/journal.pone.0105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordoni G., Woodward M.J., Wu H., Alanazi M., Wallis T., La Ragione R.M. Comparative genomics of European avian pathogenic E. coli (APEC) BMC Genom. 2016;17:960. doi: 10.1186/s12864-016-3289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dissanayake D.R., Octavia S., Lan R. Population structure and virulence content of avian pathogenic Escherichia coli isolated from outbreaks in Sri Lanka. Vet. Microbiol. 2014;168:403–412. doi: 10.1016/j.vetmic.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 49.Mora A., Lopez C., Herrera A., Viso S., Mamani R., Dhabi G., Alonso M.P., Blanco M., Blanco J.E., Blanco J. Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111:H4-D-ST2085 and O111:H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet. Microbiol. 2012;156:347–352. doi: 10.1016/j.vetmic.2011.10.033. [DOI] [PubMed] [Google Scholar]
- 50.Oteo J., Diestra K., Juan C., Bautista V., Novais A., Perez-Vazquez M., Moya B., Miro E., Coque T.M., Oliver A., et al. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents. 2009;34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Smet A., Martel A., Persoons D., Dewulf J., Heyndrickx M., Claeys G., Lontie M., Van Meensel B., Herman L., Haesebrouck F., et al. Characterization of extended-spectrum beta-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: Emergence of CTX-M-15-producing strains causing urinary tract infections. Microb. Drug Resist. 2010;16:129–134. doi: 10.1089/mdr.2009.0132. [DOI] [PubMed] [Google Scholar]
- 52.Vincent C., Boerlin P., Daignault D., Dozois C.M., Dutil L., Galanakis C., Reid-Smith R.J., Tellier P.P., Tellis P.A., Ziebell K., et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 2010;16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heidemann Olsen R., Christensen H., Kabell S., Bisgaard M. Characterization of prevalent bacterial pathogens associated with pododermatitis in table egg layers. Avian Pathol. 2018;47:281–285. doi: 10.1080/03079457.2018.1440066. [DOI] [PubMed] [Google Scholar]
- 54.Mates A.K., Sayed A.K., Foster J.W. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 2007;189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foster J.W. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 56.Luyckx K., Van Coillie E., Dewulf J., Van Weyenberg S., Herman L., Zoons J., Vervaet E., Heyndrickx M., De Reu K. Identification and biocide susceptibility of dominant bacteria after cleaning and disinfection of broiler houses. Poult. Sci. 2016 doi: 10.3382/ps/pew355. [DOI] [PubMed] [Google Scholar]
- 57.Schwaiger K., Bauer J., Holzel C.S. Selection and persistence of antimicrobial-resistant Escherichia coli including extended-spectrum beta-lactamase producers in different poultry flocks on one chicken farm. Microb. Drug Resist. 2013;19:498–506. doi: 10.1089/mdr.2012.0257. [DOI] [PubMed] [Google Scholar]
- 58.Beier R.C., Poole T.L., Brichta-Harhay D.M., Anderson R.C., Bischoff K.M., Hernandez C.A., Bono J.L., Arthur T.M., Nagaraja T.G., Crippen T.L., et al. Disinfectant and antibiotic susceptibility profiles of Escherichia coli O157:H7 strains from cattle carcasses, feces, and hides and ground beef from the United States. J. Food Prot. 2013;76:6–17. doi: 10.4315/0362-028X.JFP-12-253. [DOI] [PubMed] [Google Scholar]
- 59.Deus D., Krischek C., Pfeifer Y., Sharifi A.R., Fiegen U., Reich F., Klein G., Kehrenberg C. Comparative analysis of the susceptibility to biocides and heavy metals of extended-spectrum beta-lactamase-producing Escherichia coli isolates of human and avian origin, Germany. Diagn. Microbiol. Infect. Dis. 2017;88:88–92. doi: 10.1016/j.diagmicrobio.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 60.Maertens H., De Reu K., Meyer E., Van Coillie E., Dewulf J. Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Vet. Res. 2019;15:310. doi: 10.1186/s12917-019-2044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oosterik L.H., Peeters L., Mutuku I., Goddeeris B.M., Butaye P. Susceptibility of avian pathogenic Escherichia coli from laying hens in Belgium to antibiotics and disinfectants and integron prevalence. Avian Dis. 2014;58:271–278. doi: 10.1637/10680-100113-RegR. [DOI] [PubMed] [Google Scholar]
- 62.Sheridan À., Lenahan M., Duffy G., Fanning S., Burgess C. The potential for biocide tolerance in Escherichia coli and its impact on the response to food processing stresses. Food Control. 2012;26:98–106. doi: 10.1016/j.foodcont.2012.01.018. [DOI] [Google Scholar]
- 63.Schwaiger K., Harms K.S., Bischoff M., Preikschat P., Mölle G., Bauer-Unkauf I., Lindorfer S., Thalhammer S., Bauer J., Hölzel C.S. Insusceptibility to disinfectants in bacteria from animals, food and humans-is there a link to antimicrobial resistance? Front. Microbiol. 2014;5:88. doi: 10.3389/fmicb.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang A., He X., Meng Y., Guo L., Long M., Yu H., Li B., Fan L., Liu S., Wang H., et al. Antibiotic and Disinfectant Resistance of Escherichia coli Isolated from Retail Meats in Sichuan, China. Microb. Drug Resist. 2016;22:80–87. doi: 10.1089/mdr.2015.0061. [DOI] [PubMed] [Google Scholar]
- 65.Halstead F.D., Rauf M., Moiemen N.S., Bamford A., Wearn C.M., Fraise A.P., Lund P.A., Oppenheim B.A., Webber M.A. The Antibacterial Activity of Acetic Acid against Biofilm-Producing Pathogens of Relevance to Burns Patients. PLoS ONE. 2015;10:e0136190. doi: 10.1371/journal.pone.0136190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bottini C.B. Master’s Thesis. Oklahoma State University; Stillwater, OK, USA: 2012. Evaluation of Antimicrobials against Multi-Strain Cocktails of Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes Using a Kinetic Growth Inhibition Assay. [Google Scholar]
- 67.Lesser K.H. p-Chloro-m-cresol. In: Kabara J.J., editor. Cosmetic and Drug Preservation Principle and Practice. Marcel Dekker; New York, NY, USA: 1984. pp. 683–684. [Google Scholar]
- 68.Paulus W. Phenolics. In: Paulus W., editor. Directory of Microbicides for the Protection of Materials—A Handbook. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. p. 534. [Google Scholar]
- 69.Slipski C.J., Jamieson T.R., Lam A., Shing V.L., Bell K., Zhanel G.G., Bay D.C. Plasmid transmitted small multidrug resistant (SMR) efflux pumps differ in gene regulation and enhance tolerance to quaternary ammonium compounds (QAC) when grown as biofilms. bioRxiv. 2019:768630. doi: 10.1101/768630. [DOI] [Google Scholar]
- 70.Chen N.H., Djoko K.Y., Veyrier F.J., McEwan A.G. Formaldehyde Stress Responses in Bacterial Pathogens. Front. Microbiol. 2016;7:257. doi: 10.3389/fmicb.2016.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaulfers P.-M., Brandt D. Isolation of a conjugative plasmid in Escherichia coli determining formaldehyde resistance. FEMS Microbiol. Lett. 1987;43:161–163. doi: 10.1111/j.1574-6968.1987.tb02116.x. [DOI] [Google Scholar]
- 72.Kaulfers P.-M., Marquardt A. Demonstration of formaldehyde dehydrogenase activity in formaldehyde-resistant Enterobacteriaceae. FEMS Microbiol. Lett. 1991;79:335–338. doi: 10.1111/j.1574-6968.1991.tb04551.x. [DOI] [PubMed] [Google Scholar]
- 73.Dorsey C.W., Actis L.A. Analysis of pVU3695, a plasmid encoding glutathione-dependent formaldehyde dehydrogenase activity and formaldehyde resistance in the Escherichia coli VU3695 clinical strain. Plasmid. 2004;51:116–126. doi: 10.1016/j.plasmid.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Chung Y.J., Saier M.H., Jr. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J. Bacteriol. 2002;184:2543–2545. doi: 10.1128/JB.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lewinson O., Adler J., Poelarends G.J., Mazurkiewicz P., Driessen A.J.M., Bibi E. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. USA. 2003;100:1667–1672. doi: 10.1073/pnas.0435544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 77.Rotem D., Schuldiner S. EmrE, a Multidrug Transporter from Escherichia coli, Transports Monovalent and Divalent Substrates with the Same Stoichiometry. J. Biol. Chem. 2004;279:48787–48793. doi: 10.1074/jbc.M408187200. [DOI] [PubMed] [Google Scholar]
- 78.Li L., Ye L., Kromann S., Meng H. Occurrence of Extended-Spectrum β-Lactamases, Plasmid-Mediated Quinolone Resistance, and Disinfectant Resistance Genes in Escherichia coli Isolated from Ready-To-Eat Meat Products. Foodborne Pathog. Dis. 2016;14:109–115. doi: 10.1089/fpd.2016.2191. [DOI] [PubMed] [Google Scholar]
- 79.Zou L., Meng J., McDermott P.F., Wang F., Yang Q., Cao G., Hoffmann M., Zhao S. Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. J. Antimicrob. Chemother. 2014;69:2644–2649. doi: 10.1093/jac/dku197. [DOI] [PubMed] [Google Scholar]
- 80.Slipski C.J., Zhanel G.G., Bay D.C. Biocide Selective TolC-Independent Efflux Pumps in Enterobacteriaceae. J. Membr. Biol. 2018;251:15–33. doi: 10.1007/s00232-017-9992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wieland N., Boss J., Lettmann S., Fritz B., Schwaiger K., Bauer J., Hölzel C.S. Susceptibility to disinfectants in antimicrobial-resistant and -susceptible isolates of Escherichia coli, Enterococcus faecalis and Enterococcus faecium from poultry–ESBL/AmpC-phenotype of E. coli is not associated with resistance to a quaternary ammonium compound, DDAC. J. Appl. Microbiol. 2017;122:1508–1517. doi: 10.1111/jam.13440. [DOI] [PubMed] [Google Scholar]
- 82.Paulsen I.T., Littlejohn T.G., Radstrom P., Sundstrom L., Skold O., Swedberg G., Skurray R.A. The 3’ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 1993;37:761–768. doi: 10.1128/AAC.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vijayakumar R., Sandle T. A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes-special notes on pharmaceutical environmental isolates. J. Appl. Microbiol. 2019;126:1011–1022. doi: 10.1111/jam.14118. [DOI] [PubMed] [Google Scholar]
- 84.Kücken D., Feucht H., Kaulfers P. Association of qacE and qacEDelta1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol. Lett. 2000;183:95–98. doi: 10.1016/S0378-1097(99)00636-9. [DOI] [PubMed] [Google Scholar]
- 85.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). BVL-Report 12.2 Berichte zur Lebensmittelsicherheit Zoonosen-Monitoring 2016. [(accessed on 29 November 2019)]; Available online: https://www.bvl.bund.de/SharedDocs/Berichte/03_Zoonosen_Monitoring/2016_zoonosen_monitoring_bericht.pdf?__blob=publicationFile&v=5.
- 86.World Health Organization . Critically Important Antimicrobials for Human Medicine, 6th Revision. World Health Organization; Geneva, Switzerland: 2019. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 87.Davies J., Wright G.D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 88.Shaw K.J., Rather P.N., Hare R.S., Miller G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993;57:138–163. doi: 10.1128/MR.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garneau-Tsodikova S., Labby K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm. 2016;7:11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI; Wayne, PA, USA: 2019. CLSI Supplement M100. [Google Scholar]
- 91.European Medicines Agency . Reflection Paper on Use of Aminoglycosides in Animals in the European Union: Development of Resistance And Impact on Human and Animal Health. European Medicines Agency; Amsterdam, The Netherlands: 2018. [Google Scholar]
- 92.Soumet C.F., Legrandois E.P., Maris P. Resistance to phenicol compounds following adaptation to quaternary ammonium compounds in Escherichia coli. Vet. Microbiol. 2012;158:147–152. doi: 10.1016/j.vetmic.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 93.Wand M.E., Bock L.J., Bonney L.C., Sutton J.M. Mechanisms of Increased Resistance to Chlorhexidine and Cross-Resistance to Colistin following Exposure of Klebsiella pneumoniae Clinical Isolates to Chlorhexidine. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernando D.M., Xu W., Loewen P.C., Zhanel G.G., Kumar A. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 2014;58:6424–6431. doi: 10.1128/AAC.03074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wales A.D., Davies R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics. 2015;4:567–604. doi: 10.3390/antibiotics4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kazama H., Hamashima H., Sasatsu M., Arai T. Distribution of the antiseptic-resistance genes qacE and qacE delta 1 in gram-negative bacteria. FEMS Microbiol. Lett. 1998;159:173–178. doi: 10.1111/j.1574-6968.1998.tb12857.x. [DOI] [PubMed] [Google Scholar]
- 97.Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015;16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ewers C., Bethe A., Semmler T., Guenther S., Wieler L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012;18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 99.Mo S.S., Slettemeas J.S., Berg E.S., Norstrom M., Sunde M. Plasmid and Host Strain Characteristics of Escherichia coli Resistant to Extended-Spectrum Cephalosporins in the Norwegian Broiler Production. PLoS ONE. 2016;11:e0154019. doi: 10.1371/journal.pone.0154019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Porres-Osante N., Saenz Y., Somalo S., Torres C. Characterization of Beta-lactamases in Faecal Enterobacteriaceae Recovered from Healthy Humans in Spain: Focusing on AmpC Polymorphisms. Microb. Ecol. 2015;70:132–140. doi: 10.1007/s00248-014-0544-9. [DOI] [PubMed] [Google Scholar]
- 101.Seiffert S.N., Carattoli A., Schwendener S., Collaud A., Endimiani A., Perreten V. Plasmids Carrying blaCMY-2/4 in Escherichia coli from Poultry, Poultry Meat, and Humans Belong to a Novel IncK Subgroup Designated IncK2. Front. Microbiol. 2017;8:407. doi: 10.3389/fmicb.2017.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pietsch M., Irrgang A., Roschanski N., Brenner Michael G., Hamprecht A., Rieber H., Käsbohrer A., Schwarz S., Rösler U., Kreienbrock L., et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018;19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or Supplementary Materials.