Abstract
Cardiovascular diseases (CVDs) are the leading cause of mortality and morbidity worldwide leading to 31% of all global deaths. Early prediction and prevention could greatly reduce the enormous socio-economic burden posed by CVDs. Plasma lipids have been at the center stage of the prediction and prevention strategies for CVDs that have mostly relied on traditional lipids (total cholesterol, total triglycerides, HDL-C and LDL-C). The tremendous advancement in the field of lipidomics in last two decades has facilitated the research efforts to unravel the metabolic dysregulation in CVDs and their genetic determinants, enabling the understanding of pathophysiological mechanisms and identification of predictive biomarkers, beyond traditional lipids. This review presents an overview of the application of lipidomics in epidemiological and genetic studies and their contributions to the current understanding of the field. We review findings of these studies and discuss examples that demonstrates the potential of lipidomics in revealing new biology not captured by traditional lipids and lipoprotein measurements. The promising findings from these studies have raised new opportunities in the fields of personalized and predictive medicine for CVDs. The review further discusses prospects of integrating emerging genomics tools with the high-dimensional lipidome to move forward from the statistical associations towards biological understanding, therapeutic target development and risk prediction. We believe that integrating genomics with lipidome holds a great potential but further advancements in statistical and computational tools are needed to handle the high-dimensional and correlated lipidome.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-020-03715-4.
Keywords: Lipid species, Biomarkers, Genome-wide association studies, Disease risk, Pathophysiology
Introduction
Cardiovascular diseases (CVDs) are a group of complex disorders affecting heart function, vascular structure and circulatory system. Genetic and epidemiological studies have greatly improved our understanding of pathophysiology underlying the complex CVDs and have identified several risk factors for CVDs. Amongst the well-recognized predisposing factors (Fig. 1), lipid metabolism plays a central role in the development of CVDs [1, 2]. Since the landmark publications from the Framingham study [3], plasma lipids have been recognized as important predictors of future CVD events, with lipid lowering as a well-established intervention to reduce CVD risk [4]. To assess CVD risk, plasma lipids are routinely monitored by profiling total cholesterol, triglycerides, high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) (referred as “traditional lipids”). Despite these advances, CVDs remain the leading cause of mortality and morbidity worldwide [5], as the current preventive strategies are ineffective in a large proportion of the population [6].
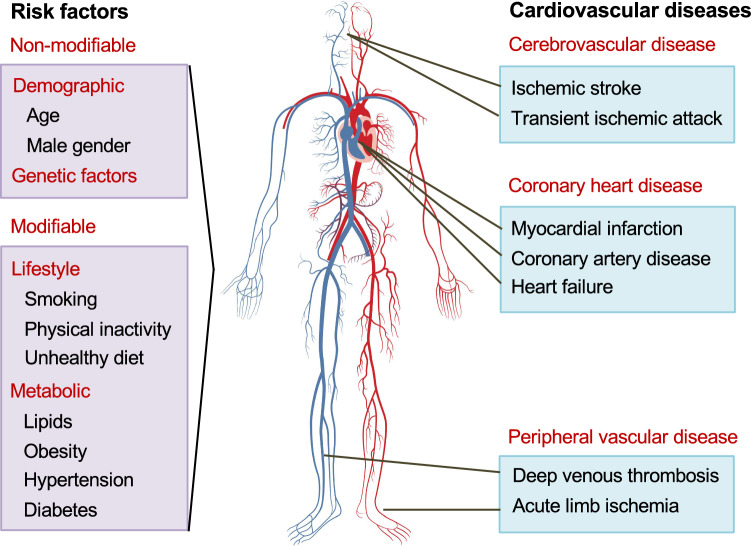
Fig. 1.
Cardiovascular diseases and their risk factors. CVDs encompass a broad range of disorders affecting the heart, brain and blood vessels. The different manifestations of CVDs include myocardial infarction, stroke and peripheral artery disease. A number of modifiable and non-modifiable risk factors have been identified that predispose individuals to CVDs. Relationships between lifestyle factors and lipids are well known and have been the target for prevention strategies
Human plasma is estimated to consist of thousands of functionally and chemically diverse molecular lipid species [7–9]. Because of the technological challenges to detect diverse yet structurally similar lipids and their isomers, efforts to understand the role of lipids in CVD pathophysiology had largely focused on traditional lipids, and to some extent on free fatty acids and lipoproteins, until last decade. Nevertheless, there have been tremendous advancements in the field of lipidomics that has facilitated the efforts to unravel the metabolic dysregulation in complex lipid-related disorders, particularly CVDs and to identify predictive biomarkers beyond traditional lipids [10, 11]. The promising findings from epidemiological studies have also led to a growing interest in understanding the genetic regulation of lipid metabolism at molecular lipid species level. Consequently, genome-wide association studies (GWAS) of lipidome profiles, have not only identified new genetic loci/genes influencing distinct molecular species but have also provided novel mechanistic insights to the known genetic loci associated with traditional lipids [12, 13].
This review presents an overview of the application of lipidomics in epidemiological and genetic studies of CVDs and their contributions to the current understanding of the field, along with a brief overview of lipidome diversity and commonly used analytical approaches. The review further discusses some new opportunities provided by integrating emerging genomics tools with the high-dimensional lipidome to move forward from the statistical associations towards therapeutic target development and personalized medicine with better prediction and prevention.
Human plasma lipidome
Lipidome, the total lipid content in a cell or tissue, is estimated to contain ~ 200,000 different molecular species with different abundance [9]. This extreme diversity arises from the extraordinary number of possible combinations of various head groups with numerous fatty acids of varying length and degree of unsaturation that are esterified to the head groups. The LIPID MAPS Initiative and the International Committee for the Classification and Nomenclature of Lipids (ICCNL) have provided a standard nomenclature that classifies lipids into eight categories—fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterols, prenols, saccharolipids and polyketides [14–16]. Lipids in each lipid category are further divided into classes and subclasses based on the head group and type of linkages [17, https://www.lipidmaps.org/].
The technological advances have tremendously helped in revealing the complexity of lipidome. The LIPID MAPS consortium revealed over 500 molecular lipid species from 6 major lipid categories, with over 200 and 160 distinct species of sphingolipids and glycerophospholipids respectively [18]. A lipidome analysis of human platelets detected over 5,600 unique lipids, with ~ 50% unidentified molecular species [19]. The major lipid categories that are commonly identified in plasma lipidomics are discussed briefly here (Fig. 2), but are reviewed in detail in [7, 15, 18, 20, 21].
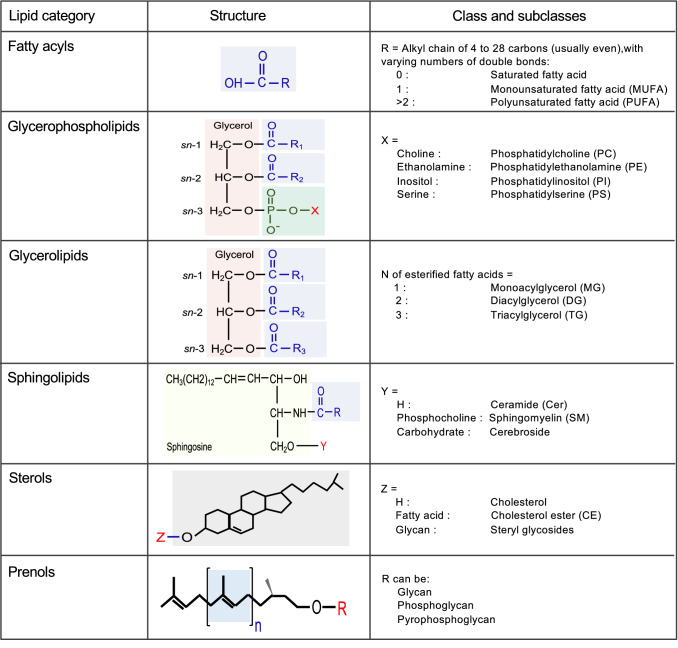
Fig. 2.
Human plasma lipidome. Six major lipid categories, of eight described by the LIPID MAPS classification system, are illustrated with their classes/subclasses and structure of representative of each lipid category. As shown, esterification of fatty acids with different backbone generates complex lipids including glycerolipids, glycerophospholipids and sphingolipds. Lipids in each lipid category are further divided into classes and subclasses based on the head group and type of linkages between the backbone and acyl chains [https://www.lipidmaps.org/]
Fatty acyls
Fatty acyls represent the most fundamental category of the lipids including fatty acids. Mostly present in esterified form with glycerol, cholesterol or other lipid components, fatty acids are carboxylic acids, often with long unbranched aliphatic chains of diverse length. Fatty acids are categorized as saturated (no carbon–carbon double bonds in aliphatic chain) and unsaturated with one (monounsaturated fatty acid-MUFA) or more double bonds (polyunsaturated fatty acid-PUFA). Human body can synthesize many of these fatty acids, except some essential fatty acids including linoleic acid (omega-6 PUFA) and alpha-linolenic acid (omega-3 PUFA). These two PUFAs are precursors for other omega-6 and omega-3 PUFAs that play crucial roles in regulating lipid metabolism and atherosclerosis [reviewed in 22, 23].
Glycerolipids
Esterification of one, two or three fatty acyls to glycerol lead to the formation of glycerolipids and are accordingly classified as monoacylglycerol (MG), diacylglycerol (DG) and triacylglycerol (TG). Glycerolipids are a large group of lipids accounting for a high proportion of total lipids in plasma. TG is the most abundant lipid class and comprises the bulk of storage fat in tissues. MGs and DGs represent intermediates in the biosynthesis and hydrolysis of TGs and function as second messengers in signal transduction processes [24, 25].
Glycerophospholipids
Also known as phospholipids, glycerophospholipids are diacylglycerides with a phosphatidyl ester attached to the terminal carbon. The terminal ester groups are mainly ethanolamine, choline, serine or inositol (Fig. 2). In addition, a number of fatty acids with varying length and unsaturation could attach to the remaining hydroxyl groups of glycerol via either acyl-, alkyl-, or alkenyl-bonds [18]. Hydrolysis of one of the fatty acids of the phospholipids by phospholipase A2 (PLA2) generates respective lysophospholipids, adding to the diversity of the lipid pool. Glycerophospholipids are the major structural component of cell membranes and are involved in various biological processes including inflammation [21].
Sphingolipids
Sphingolipids are wide-range of complex lipids defined by 18-carbon sphingoid base, usually sphingosine (SPH). Condensation of SPH and free fatty acid generates the simplest sphingolipids, ceramides which function as precursor for complex sphingolipids produced by the modification of hydroxyl group with phosphocholine (in sphingomyelins) or carbohydrates (in gangliosides) (Figs. 2, 3) [21, 26]. Sphingolipids constitute several hundreds of different species originating from the combinations of different sphingoid bases, various fatty acids that can attach to the bases and numerous carbohydrates in gangliosides. Ceramide regulates numerous cellular processes such as proliferation, differentiation, and cell signalling [27].
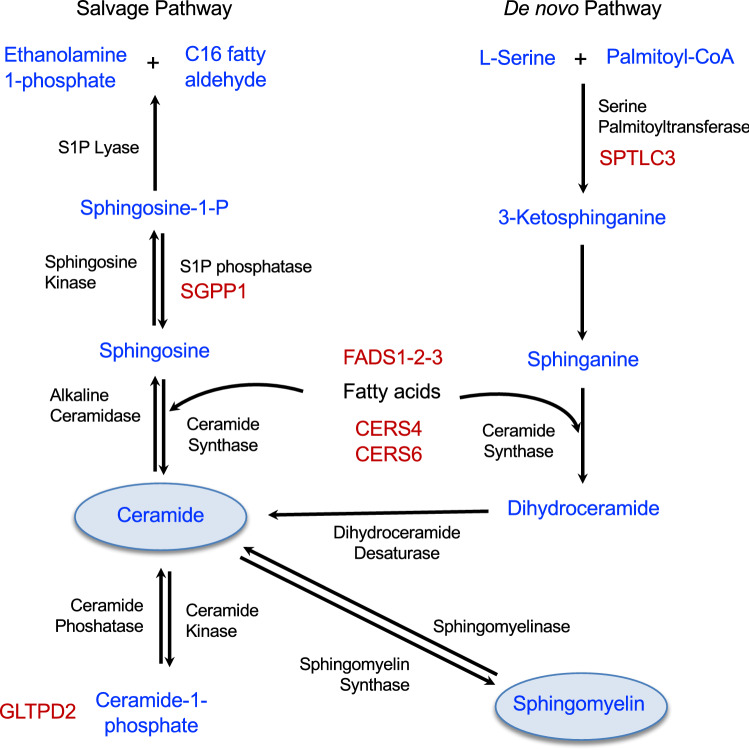
Fig. 3.
Role of sphingolipid associated loci in major sphingolipid metabolic pathways. Most of the sphingolipid associated loci contain genes that code for enzymes (highlighted in red font) involved in sphingolipid metabolic pathways. SGPP1 codes for a S1P phosphatase that catalyzes the degradation of sphingosine-1-P to sphingosine to facilitate ceramides synthesis catalyzed by ceramide synthases (CERS1-6) including CERS4 and CERS6. SPTLC3 gene encodes a subunit of the serine palmitoyltransferase complex which catalyzes the rate-limiting step of de novo pathway in sphingolipid biosynthesis. FADS1-2-3 locus encodes enzymes that regulate the desaturation of fatty acids and have important role in generation of unsaturated ceramides. GLTPD2 codes for glycolipid transfer protein domain-containing protein 2 and has putative role in transfer of ceramide-1-phosphate
Sterol lipids
Sterols typically have a sterol nucleus composed of four tightly fused carbon rings and a hydroxyl group attached to the first ring [28]. Cholesterol, the well-known and widely measured lipid, is the simplest and most abundant sterol in plasma, accounting for more than 99% of all plasma sterols. Cholesterol exits in both free and esterified forms as cholesterol esters, mainly in association with lipoproteins.
Prenols
Prenols are synthesized from five-carbon isoprene units that can be combined in wide variety of polymeric units and configuration to make diverse products [21]. Prenols include vitamins A, E and carotenoids and are essential for immune system or regulatory functions in the brain [21]. Prenols are understudied in the current lipidomic technologies.
Analytical methods in lipidomics
Of the hundreds of thousands of distinct lipid structures that are estimated to occur in nature, only a small fraction of lipids has been identified so far, highlighting the lack of knowledge and importance of development of high-throughput screening methods for lipid identification and profiling. There are two main approaches in lipidomics: (a) targeted approach that focuses on detection of known lipids using pre-existing knowledge and (b) non-targeted approach that screens all the lipid species without preselection. Non-targeted approach provides large coverage but is limited by the complexity of data processing and identification of lipids from large number of signals. Given the structural diversity of lipid species, no single analytical method could capture the entire lipidome, hence many different methods have been employed [reviewed in 29–35]. Here we briefly discuss two methods—Nuclear Magnetic Resonance (NMR) spectroscopy and Mass spectrometry (MS), that have been used commonly in the epidemiological and genetic studies of lipidome in the context of CVDs.
Lipidome analysis using NMR spectrometry is based on the measurement of magnetic spin of nuclei (1H, 13C, 15N and 31P) contained in the lipids. NMR can efficiently and accurately quantify density, size and particle number of different lipoprotein subclasses along with their total lipid content (e.g., total TGs, total phospholipids, total cholesteryl esters, total sphingolipids in HDLs, LDLs, and very-low-density lipoprotein (VLDL)) [35]. Identification of individual lipid species within lipoprotein subclasses is difficult using NMR. As lipid composition of various lipoprotein subclasses varies considerably, NMR has been applied to measure lipid content in lipoproteins to examine their relationship with CVDs [36, 37] and to determine their genetic determinants [38–42].
MS, on the other hand, provides higher resolution of molecular composition of lipidome [reviewed in 31–34]. MS is either coupled with prior chromatographic separation such as gas chromatography (GC–MS) and liquid chromatography (LC–MS), or involve direct infusion of lipid extract (shotgun lipidomics). LC–MS which provides excellent separation efficiency, high sensitivity and strong specificity, is one of the most important and widely used methods for lipidomics research (Table 1, Supplementary Table 2). Liquid chromatography separates lipids based on their physiochemical properties, i.e., polar head group, carbon chain length, number of double bonds. After chromatographic separation, the isolated lipids are ionized that are detected using a mass analyser [31]. GC–MS provides limited coverage of the lipidome and hence, is restricted to studies focused on specific lipid classes and fatty acids quantifications [43–45]. In recent times, shotgun lipidomics has gain popularity due to its relative simplicity of operation and short run times to quantify hundreds of lipids [32–34] and has been applied in several large-scale studies [12, 46, 47]. Shotgun lipidomics technology directly infuses lipids extract into an electrospray ionization mass spectrometer for the detection of lipids, without chromatographic separation. Shotgun lipidomics has lower sensitivity than LC–MS and hence many of the low abundant lipid species are not captured in shotgun lipidomic approaches.
Table 1.
Major findings of lipidomics-based epidemiological studies of CVDs
| Study Reference, Cohort | Platform | Samples | Lipids analysed |
|---|---|---|---|
| Ceramides as prognostic markers | |||
| Sigruener et al. [50], LURIC study | Shotgun MS | 3316 (768 incident CVD; 484 mortality) | 38 PCs, 15 LPCs, 30 PC Os, 31 PEs, 24 PE Os, 33 SMs, 7 Cers |
| Cheng et al. [51], ATHEROREMO-IVUS | LC–MS | 581 (underwent angiography) | 8 lipids (CEs and Cers) identified in LURIC study |
| Laaksonen et al. [52], Corogene, SPUM-ACS, BECAC | LC–MS | 3377 (mostly CVD patients) | 4 Cers (C16:0, C18:0, C24:0, C24:1) |
| Havulinna et al. [54], FINRISK2002 | LC–MS | 8101 healthy subjects (813 incident MACE) | 4 Cers (C16:0, C18:0, C24:0, C24:1) |
| Wang et al. [53], PREDIMED | LC–MS | 1017 (230 incident cases; 787 random samples) | 4 Cers (C16:0, C22:0, C24:0, C24:1) |
| Anroedh et al. [46], ATHEROREMO-IVUS | Shotgun MS | 581 (underwent angiography) | 10 lipids (CEs and Cers) identified in LURIC study |
| Alshehry et al. [55], ADVANCE | Targeted LC–MS | 3779 (case–control) | Lipidome-wide (310 lipid species) |
| Paynter et al. [56], WHI, PREDIMED | LC–MS | 944 (472 incident CHD; 472 controls); 627 (312 incident CHD; 315 controls) | Lipidome-wide (217 lipids) |
| Poss et al. [57], Utah population | LC–MS/MS | 674 (462 CAD patients; 212 controls) | 32 Sphingolipids |
| Opposite effects of MUFA and PUFA containing phospholipids | |||
| Sigruener et al. [50], LURIC study | Shotgun MS | 3316 (768 incident CVD; 484 mortality) | 38 PCs, 15 LPCs, 30 PC Os, 31 PEs, 24 PE Os, 33 SMs, 7 Cers |
| Alshehry et al. [55], ADVANCE | Targeted LC–MS | 3779 (case–control) | Lipidome-wide (310 lipid species) |
| Wang et al. [69], PREDIMED | LC–MS | 1017 (230 incident CVD; 787 random samples) | Lipidome-wide (200 lipids) |
| Wurtz et al. [36], FINRISK, SABRE, BWHHS | Targeted NMR metabolomics | 7256 (800 CVD events); 2622 (573 CVD events); 3563 (368 CVD events) | 14 lipoprotein subclasses and fatty acid composition |
| Razquin et al. [68], PREDIMED | LC–MS | 983 (case–control) | Lipidome-wide (202 lipids) |
| Mundra et al. [66], LIPID, ADVANCE | LC–MS | 5991; 3779 | Lipidome-wide (342 lipids) |
| TG species and risk of CVDs | |||
| Fernandez et al. [47], MDC study | Shotgun MS | 427 (211 incident CVD; 216 controls) | 85 lipids (TGs, DGs, CEs, SMs, PC Os, LPCs, PEs, PE Os) |
| Stegemann et al. [65], Bruneck study | Shotgun MS | 685 (90 incident CVD) | 135 lipids (PCs, LPCs, CEs, SMs, PSs, PEs, LPEs, TGs) |
| Alshehry et al. [55], ADVANCE | Targeted LC–MS | 3779 (case–control) | Lipidome-wide (310 lipid species) |
| Razquin et al. [68], PREDIMED | LC–MS | 983 (case–control) | Lipidome-wide (202 lipids) |
| Wang et al. [69], PREDIMED | LC–MS | 1017 (230 incident CVD; 787 random samples) | Lipidome-wide (200 lipids) |
CAD coronary artery disease, CE cholesteryl ester, Cer ceramide, CHD coronary heart disease, DG diacylglyceride, LPC lysophosphatidylcholine, LPE lysophosphatidylethanolamine, LC–MS liquid chromatography–mass spectrometry, MACE major adverse cardiovascular event, MS mass spectrometry, NMR nuclear magnetic resonance, PC phosphatidylcholine, PC O phosphatidylcholine–ether, PE phosphatidylethanolamine, PE O phosphatidylethanolamine–ether, SM sphingomyelin, TG triacyglycerol
Lipidomics in CVD risk prediction
With the advances in high-throughput lipidomics technologies, several studies were undertaken to perform in-depth examination of role of distinct lipid species in CVDs. The studies and their major findings are described in Supplementary Table 1. Taken together, findings from these studies suggest: (1) ceramides as prognostic markers for CVDs, (2) opposite effects of saturated or MUFA containing lipids and PUFA containing lipids on the risk of future CVD events or death, and (3) distinct role of TG species based on carbon content (Table 1).
Ceramides as prognostic markers for CVDs
Though first regarded as inert components of cell membrane, sphingolipids have emerged as important bioactive molecules owing to their wide-range of biological functions. Of particular note is the link between ceramides and CVDs mediated through atherosclerotic processes by promoting LDL infiltration into blood vessel wall, aggregation of LDL in arterial plaque and accumulation of cholesterol in macrophages [48, 49]. Findings from in vitro and in vivo animal studies have also supported their role in cardiometabolic disorders including atherosclerosis, and heart failure [27]. In this regard, lipidomics of human plasma in large cohorts not only reinforced the role of ceramides in CVD manifestations, but has also pointed to the distinct ceramide species which are independent predictors of future CVD events or death.
Relationship between distinct ceramide species, particularly Cer(d18:1/16:0), Cer(d18:1/18:0) and Cer(d18:1/24:1), and CVD mortality was first suggested by the LURIC study [50], which was supported by report from ATHEROREMO-IVUS study [51]. This led to a growing interest in these ceramide species among the researchers and several studies focusing on them added to the supporting evidences of their relationship with secondary CVD outcomes [46, 52, 53] and future CVD events in healthy individuals [54]. The findings were also corroborated by lipidome-wide studies [55–58] and a large meta-analysis of seven cohort studies with over 29,800 individuals [59]. On the contrary, a recent study by Seah et al. that explored association of 79 sphingolipid species with CVDs in a Chinese ethnic population did not support the role of ceramides in CVD [60]. Although reasons such as difference in quantification methods and statistical power could not be ruled out, this study may point to the population-specific effect of lipid species on CVD risk. The meta-analysis by Mantovani et al. also suggested that associations may be stronger for ceramides with long acyl chain and for those with unsaturated acyl chain [59]. On the similar lines, Lemaitre earlier showed associations of higher plasma levels of shorter sphingolipids (C16 acyl chain) with increased risk of heart failure, whereas higher levels of longer sphingolipids (C20-24 acyl chains) with decreased risk of heart failure [61].
Clinical utility of prediction scores based on ceramide species has been proposed by several independent studies (Table 2). Laaksonen et al. [52] showed that CERT score based on Cer(d18:1/16:0), Cer(d18:1/18:0) and Cer(d18:1/24:1) and their ratios with Cer(d18:1/24:0) predict cardiovascular death in patients with stable CAD and acute coronary syndromes beyond LDL-C. It was further shown that the prognostic value of CERT score could be improved by adding phosphatidylcholine (PC) species [62]. Subsequently, the predictive value of ceramide-based scores have been repeatedly confirmed by many independent studies including the Framingham Heart study [63], Mayo clinic study [64] and FINRISK2002 [54]. All these strong evidences provided basis for the ceramide-based clinical test recommended by the Mayo clinic [https://news.mayocliniclabs.com/ceramides-miheart/] to assess risk of adverse clinical outcomes in CAD patients. A recent study showed that in addition to ceramides, sphingomyelin species (SM) could be important predictor of CVD and proposed a new risk score termed as the sphingolipid-inclusive CAD (SIC) risk score which included dihydro-Cer(d18:0/18:0), Cer(d18:1/18:0), Cer(d18:1/22:0), Cer(d18:1/24:0), SM(d18:0/24:1), SM(d18:1/24:0), SM(d18:1/18:0) and sphingosine [57]. Authors showed that the SIC risk score provides strong prediction value and outperform other measures including LDL-C and CERT score. Thus, through lipidomics-based studies, plasma ceramides have emerged as promising new diagnostic or prognostic marker for CVD with clinical application.
Table 2.
Lipidome-based prediction scores for CVD risk and death
| Prediction score/Study | Components | C-statistics |
|---|---|---|
| CERT1, Laaksonen et al. [52] | Cer(d18:1/16:0), Cer (d18:1/18:0), Cer (d18:1/24:1), Cer (d18:1/16:0)/Cer(d18:1/24:0), Cer (d18:1/18:0)/Cer(d18:1/24:0), Cer (d18:1/24:1)/Cer(d18:1/24:0) | 0.80 for CVD death |
| CERT2, Hilvo et al. [62] | Cer(d18:1/24:1)/Cer(d18:1/24:0), Cer(d18:1/16:0)/PC 16:0/22:5, Cer(d18:1/16:0)/PC 14:0/22:6, PC 16:0/16:0 | 0.76 for CVD death |
| SIC, Poss et al. [57] | Cer(d18:1/18:0), Cer(d18:1/24:0), SM(d18:1/24:0), SM(d18:0/24:1)/SM(d18:1/18:0), Cer(d18:1/18:0)/Cer(d18:1/22:0), Sphingosine | 0.79 for risk of CAD |
| Mundra et al. [66] | PC O-34:2, PC 38:5, PI 38:3, PC O-36:1, GM3(d18:1/16:0), LPI 18:2, PE 38:6 | 0.65 for CVD events |
| Alshehry et al. [55] | PC O-36:1, CE 18:0, PE O-36:4, PC 28:0, LPC 20:0, PC 35:4, LPC 18:2 | 0.70 for CVD events |
| Alshehry et al. [55] | PC O 36:1, DG 16:0_22:5, SM 34:1, PC O-36:5 | 0.76 for CVD death |
C-statistic is a standard measure of the predictive accuracy of a model)
SIC sphingolipid-inclusive CAD risk score, CER Ceramide, DG diacylglyceride, GM monosialated ganglioside, LPC lysophosphatidylcholine, LPI lysophosphatidylinositol, PC phosphatidylcholine, PCO phosphatidylcholine-ether, PE phosphatidylethanolamine, PEO phosphatidylethanolamine-ether, SM sphingomyelin
Opposite effects of MUFA and PUFA containing phospholipids
Many distinct phospholipid species have been consistently identified as risk factor for CVDs in lipidomics-based studies (Supplementary Table 1) [47, 50, 55, 56, 65–69]. Recently, alterations in phospholipids levels in patients with ischemic cardiomyopathy have been shown suggesting the changes in metabolic profiles during progression from ischemic heart disease to ischemic cardiomyopathy [70]. Addition of phospholipids to the base model of traditional risk factors also improved CVD risk prediction (Table 2). Bruneck study showed that addition of a phosphatidylethanolamine species PE 36:5 and two other lipid species to a model including conventional risk factors increased prediction value [65]. Similarly, LIPID study [66] and ADVANCE trial [55] showed that addition of phospholipid species to the traditional risk factors improved prediction of CVD events and mortality (Table 2). Although clinical utility of lysophospholipids has also been suggested by Ganna et al. [67], there have been inconsistent reports on the direction of their effect on CVDs [47, 55, 65].
An interesting observation that emerged from these studies is that the phospholipids have opposite effects on CVD risk based on the degree of unsaturation of their acyl chains. It was first observed in the LURIC study that phospholipids with saturated and monounsaturated fatty acyl chains were positively associated with risk of CVD, while polyunsaturated phospholipids were inversely associated with the CVD risk [50]. Later, several lipidome-wide investigations including ADVANCE trial, LIPID study, PREDIMED trial, WHI study and Bruneck study provided consistent findings (Table 1). On the similar lines, Wurtz et al. showed that MUFAs levels increase cardiovascular risk, while higher omega-6 and omega-3 PUFAs lower the risk [36]. Consistently, network-based analysis of lipidome data in PREDIMED study also showed that the lipid species are clustered based on degree of unsaturation and that the cluster containing phospholipids with more double bonds was associated with decreased risk of CVD [69]. Thus, the lipidomics-based studies have suggested that there are two subgroups of phospholipids based on the degree of unsaturation that have opposite effects on CVD pathophysiology.
Distinct role of TG species in CVDs
The routine clinical risk assessment quantifies the total mass of triglycerides, however, their contribution to the development of CVD has been debatable as clinical trials of lowering TG with fibrates provided inconsistent results [71, 72], whereas genetic evidence supported the causal role [73]. The apparent inconsistency is not surprising due to the large number of functionally diverse TG species in circulation that have varied effects (opposite directions or different magnitude) on CVD risk, as revealed by high-resolution lipidomics. The MDC study first identified association of five TG species including TG 48:1, TG 48:2, TG 48:3, TG 50:3 and TG 50:4 with adverse CVD outcome, after adjusting for Framingham risk factors [47]. Later, many TG species were found to be associated with CVD risk over a 10-year observation period in the Bruneck study [65], but associations were more pronounced for TGs of lower carbon number and double-bond content (saturated and MUFAs). Similarly, ADVANCE study found inverse association of TG 56:6 with recurrent CVD incidence and CVD mortality [55] whereas PREDIMED trial showed that short TGs were associated with increased risk of CVD [68]. These findings were further supported by the network and cluster analysis of lipidome in PREDIMED trial which showed that saturated TGs cluster consisting mainly of DGs and TGs with saturated fatty acids was associated with increased CVD risk [69]. These studies clearly suggest that abnormalities in different TG molecular species levels could have different pathological consequences, which might not be detected in enzymatic measurement of total triglycerides, as evident from a study that found decrease in a TG species without observed change in total triglycerides [74].
Genetic regulation of lipidome
Despite the expected influence of dietary intake on the circulatory lipids, contribution of genetic factors in endogenous regulation of lipid metabolism is well recognized. Studies using pedigree information and genetic data have shown that 10−60% of the variation in plasma levels of circulatory lipid species is contributed by the genetic factors, with considerable variation across lipid categories [12, 75–77]. For example, in general, sphingolipids have higher heritability than glycerophospholipids, with ceramides having highest estimated SNP-based heritability (35–40%) and phosphatidylinositols with the least heritability (11–31%) [12]. Interestingly, genetic mechanisms do not regulate all human plasma lipid species belonging to a lipid class in the same way [12, 75, 76], as also observed in mice lipidomics studies [78, 79]. Rather, it seems to depend on the length and degree of unsaturation of the acyl chains. For instance, lipids containing polyunsaturated fatty acids have higher heritability compared to other lipid species [12]. It is also reported that phosphatidylcholine species (PCs) with larger number of carbon atoms have lower heritability estimates, while PCs with a larger number of double bonds have higher heritability [80].
Although over 400 genomic loci are now known to influence the plasma traditional lipid levels [81, 82], their effects on detailed lipid metabolism at molecular levels are not completely known. As epidemiological studies have demonstrated that distinct molecular species (e.g., TGs and PCs) have different or opposite effect on disease outcomes, genetic variants/loci could potentially have different effect on functionally diverse lipid species. To ascertain that, several genome-wide association studies have been performed with individual lipid species (Table 3) [12, 38–45, 83–100]. These studies have not only identified several new loci/genes contributing to lipid metabolism, but also provided novel mechanistic insights to the known loci identified for traditional lipids. The major findings of these GWASs are discussed here. Figure 4 illustrates the genes identified for different lipid categories and their overlap. The list of all the genetic variants reported to be associated in these studies is provided in Supplementary Table 3.
Table 3.
Genome-wide association studies with lipidome and the identified loci
| Study [Ref] | Lipids included | Study population | Loci identified |
|---|---|---|---|
| Gieger et al. [83] | 85 sphingolipids, 208 phospholipids | German (N = 284) | FADS, LIPC, PLEK |
| Hicks et al. [84] | 33 Sphingolipids | European (N = 4400) | SPTLC3, LASS4, SGPP1, ATP10D, FADS1-3 |
| Illig et al. [85] | PCs, SMs | German (N = 1809), British (N = 422) | FADS1, ELOVL2, PLEKHH1, SYNE2, SPTLC3 |
| Lemaitre et al. [43] | n-3 PUFAs (ALA, EPA, DPA, DHA) | European (N = 8866) | FADS1, ELOVL2, GCKR |
| Suhre et al. [86] | Fatty acids, PCs, LPCs, MGs, | German (N = 1768), British (N = 1052) | FADS1, SCD, CYP4A, SLCO1B1, PDXDC1, ELOVL2 |
| Nicholson et al. [38] | Phospholipids | British (N = 211) | None identified for lipids |
| Kettunen et al. [40] | Phospholipids and CEs content in lipoproteins | Finnish (N = 8330) | PPP1R11, ALB, FCGR2B, ANGPTL3, LPL, APOA1-C3-A4-A5, APOE-C1-C2, LDLR, PCSK9, MLX1PL, CETP, LIPC, PLTP, LIPG, CPT1A, ABCA1, FADS1-2–3, PDXDC1 |
| Demirkan et al. [87] | PCs and SMs | European (N = 4034) | ABHD3, AGPAT1, ALG1, ALG14, APOA5, APOE-C1-C2-C4, ATP10D, CDH8, CDK17, CNTNAP4, DLG2, ELOVL2, GCKR, ILKAP, ITGA9, KCNH7, KLF12, LASS4, LIPC, LPAR2, OR8I2, PAPD7, PAQR9, PLD2, PCDH20, PDXDC1, PKD2L1, PLEKHH1, PNLIPRP2, SGPP1, SPTLC3, SYT9, ZNF600, FADS1-2-3 |
| Wu et al. [44] | FAs 16:0, 16:1n-7, 18:0, 18:1n-9 | European (N = 8961) | ALG14, LPGAT1, FADS1-2-3, PKD2L1, HIF1AN, GCKR, rs6722456 |
| Rhee et al. [88] | PCs, TGs, LPCs, SMs, LPEs, CEs, DGs, | American (European ancestry) (N = 2076) | FADS1-2-3, LIPC, GCKR, APOA1/C3/A4/A5, SLCO1B1, SYNE2, PDE4D, SEC61G, rs6593086, GNAL, BX18, NTAN1 |
| Yu et al. [89] | FAs, lysolipids | African American (N = 1260) | PKD2L1 |
| Shin et al. [90] | FAs, lysolipids | European (N = 7824) | FADS-1-2-3, APOA5, SLCO1B1, LIPC, MBOAT7, SPTLC3 |
| Ried et al. [91] | Phospholipids | European (N = 1809 + 843) | CPS1, MYL1, MSH4, FNIP2, RABGGTB, SNORD45A, SNORD45B, SNORD45C, C4orf46, PPID, FTFDH, SLC16A9, FADS1-2-3, MIR1908, MIR611, BEST1, RAB3IL1, FEN1, C11orf9, DAGLA, SYNE2, SGPP1, SLC22A4, P4HA2, PDLIM4, SLC22A5, ACADS, CABP1, MLEC, POP5, UNC119B, SPPL3, OASL, SLC22A1, IGF2R, MFSD2A, MYCL1, PDCD6IP, IL3, C12orf75, INTS8, DKFZp68601327 |
| Draisma et al. [92] | PCs, LPCs, SMs, | European (N = 7478 + 1182) |
ABHD3, AGPS, APOE-C1-C2-C4, CERS4, ACSL1, CETP, ELOVL2, FADS1-2-3, GATAD2A, GCKR, MFSD2A, PDXDC1, SCD, SGPP1, SPTLC3, TMEM229B, APOA1/C3/A4/A5 |
| Mozaffarian et al. [93] | Trans FAs from isolated phospholipids | European (N = 8013) + non-European (N = 1082 + 669 + 657) | FADS1-2-3 |
| Kettunen et al. [41] | Lipids content in Lipoproteins, plasma FAs | European (N = 24,925) | PCSK9, SORT1, USF1, GALNT2, APOB, ABCG5, LPA, HMGCR, DNAH11, MLX1PL, ABCA1, LPL, TRIB1, TTC39B, ABO, CILP, ZNF259, SCARB1, LIPC, CETP, LIPG, ANGPTL4, LDLR, APOE, MAFB, PLTP |
| Rhee et al. [94] | PCs, TGs, LPCs, SMs, LPEs, CEs, DGs, | European-American FHS (N = 2076) + ARIC (N = 1528) | None identified for lipids (Exome array) |
| Yu et al. [95] | FAs, lysolipids | African-American (1361 + 508) | None identified for lipids (LoF analysis) |
| Hu et al. [45] | MUFAs | Meta-analysis of Chinese and European cohorts (N = 3521 + 12,020) | FADS1-2-3, PKD2L1, GCKR, HIF1AN, LPCAT3 |
| Long et al. [96] | PCs, LPCs, SMs, Fas | British (N = 1960) | LIPC, APOA5, CERS4, FAAH/CYP4A11, FADS1-2-3, MBOAT7, PDXDC1, PIGH, SCD, SGPP1 |
| Yousri et al. [97] | PCs, LPCs, SMs, Fas | Qatari (N = 614 + 382) | ACADS, FADS2 |
| Feofanova et al. [98] | FAs, Lysolipids | European American (N = 1552) + African American (N = 1872) | ALMS1P, FADS1-2-3, PKD2L1, SLCO1B1, ELOVL2, PKD2L1, TMC4, LIPC |
| Tabassum et al. [12] | PCs, LPCs, PEs, CEs, TGs, SM, Cer | Finnish (N = 2045) |
KLHL17, KAZN, ABCG5/8, VWA3B, ZNF385D, SYT1, PAQR9, PDHA2, ABLIM2, DDX43, TNFAIP3, SHTN1, COL26A1, PTPRN2, BLK, LPL, CD33, COL5A1, MAF, FADS2, APOA5, MIR100HG, DHRS12, SYNE2, LIPC, GALNT16, GATM, GLTPD2, RBFOX3, CERS4, ROCK1 |
| Yazdani et al. [99] | FAs, lysolipids | European American (N = 1376) | MYO1A, PDE4DIP, EGFL8, CCDC154, ZNF211, GPR97, CLEC4C, MAP10, KIAA1755, CD300C |
| Demirkan et al. [13] | TGs, PCs, LPCs, PEs, SMs | European (N = 5537) | GCKR, FADS1, APOA, SGPP1, TMEM229B, LIPC, PDXDC1, CETP, CERS4, SPTLC3 |
| Lotta et al. [100] | PCs, LPCs, SMs | European (N = 86,507) | CERS6 |
| Coltell et al. [42] | PUFAs | European (N = 426) | FADS1-2-3 |
The genetic loci identified for lipid species in different GWASs are listed. Details of the associations between genetic variants and lipids are provided in Supplementary Table 3
ALA α-Linolenic acid, CE cholesteryl ester, Cer ceramide, DG diacylglyceride, DPA docosapentaenoic acid, DHA docosahexaenoic acid, EPA eicosapentaenoic acid, FA fatty acid, LPC lysophosphatidylcholine, LPE lysophosphatidylethanolamine, MUFA monounsaturated fatty acid, PC phosphatidylcholine, PC O phosphatidylcholine-ether, PE phosphatidylethanolamine, PE O phosphatidylethanolamine-ether, PUFA polyunsaturated fatty acid, SM sphingomyelin, TG triacyglycerol
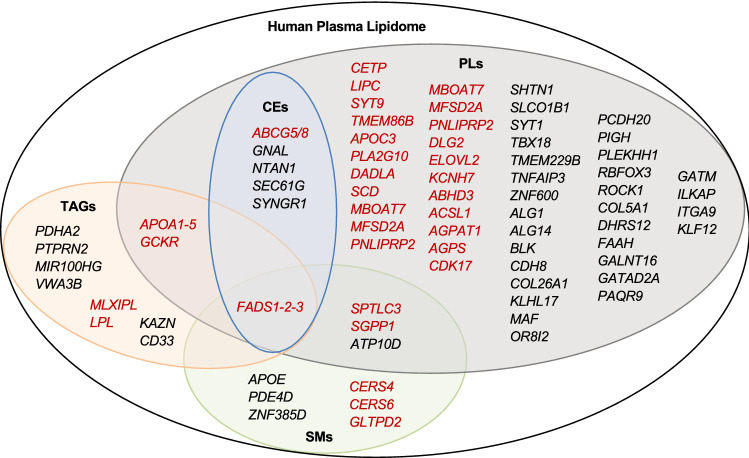
Fig. 4.
Genetic loci identified in GWAS with lipidome. Genetic loci identified for different lipid classes and their overlap are shown. Genes highlighted in red font have direct role in lipid metabolic pathways. SMs: Sphingomyelins; PLs: Phospholipids; CEs: Cholesteryl esters; TGs: Triacylglycerides
Sphingolipids
The first genetic investigation of sphingolipids was performed by Geiger et al. in 2008 [83], that included 85 sphingolipids and 284 individuals of the KORA study. The study identified two loci for sphingolipids-PLEK and ANKRD30A at genome-wide significance that did not stand multiple testing correction. Later, large-scale studies for 33 sphingolipids including over 4400 subjects from 5 diverse European populations identified 7 loci- ATP10D, FADS1-2-3, SGPP1, CERS4, SPTLC3, APOE and GLTPD2-PLD2 [84, 87]. As most of the loci identified by Hicks et al. [84] contains genes encoding enzymes involved in sphingolipids/ceramide synthesis (Fig. 3), and none of the genes involved in ceramide degradation or signaling was identified, authors speculated that the plasma levels of ceramide are primarily regulated by genes involved in ceramide production. Given the prominent roles of these genes in sphingolipid metabolic pathways, association of these genes were subsequently replicated in many studies [12, 85, 92, 95]. Furthermore, new loci were also discovered by studies with larger sample sizes including PDE4D and CERS6 [88, 100]. A recent metabolomics-based GWAS in 7 European cohorts with total sample size of over 7,000 that included 14 sphingomyelins confirmed previous associations of SPTLC3, APOE, SGPP1, CERS4 with sphingomyelins, but no new locus was identified [13].
From these studies, 6 genes (SPTLC3, CERS4, CERS6, SGPP1, GLTPD2 and FADS1-2-3) with direct role in ceramide metabolism have emerged as prominent regulator of plasma levels of ceramides. The rate of ceramide synthesis is regulated by the first step of de novo pathway, which is catalyzed by serine palmitoyltransferase (SPT). SPTLC3 codes for a subunit of the SPT complex which catalyses the condensation of serine with palmitoyl-CoA (Fig. 3). The increased expression or activity of SPTLC3 could result in increased ceramide production by increasing the influx of sphinganine in de novo pathway (Fig. 3). Several cis-eQTLs (expression quantitative trait) of SPTLC3 (GTEx v7) are found to be associated with plasma levels of ceramides and sphingomyelins (Supplementary Table 3), suggesting that genetic effect of SPTLC3 variants is mediated by regulating the expression of the gene. SPT product sphinganine is then metabolized to dihydroceramides by the addition of different acyl-chains by ceramide synthases (CerS) [101], which is subsequently converted to ceramides and sphingomyelins. CerS also catalyzes the conversion of sphingosine to ceramide in the salvage pathway. Six isoforms of CerS (CerS1-6) exist in humans with different preferences for specific fatty acids. CerS4 has high selectivity towards long acyl chains (C18-C20) while CerS6 has preference for short acyl chains (C14-C16) [102]. Consistently, association of variants in CERS4 that encode CerS4, with ceramides and SMs containing C18-C20 acyl chains have been identified repeatedly in many studies (SupplementaryTable 3).
FADS1-2-3 locus encodes three enzymes that regulate desaturation of fatty acids and production of unsaturated fatty acids. Unsaturated ceramides are synthesized by the incorporation of unsaturated fatty acids into sphingosine/sphinganine bases. Association of variants in the FADS1-2-3 locus with unsaturated ceramides levels suggests crucial role of fatty acid desaturases in generation of unsaturated ceramides and sphingolipid metabolic pathways [84]. On the other hand, SGPP1 codes for sphingosine-1-phosphate phosphohydrolase 1 that play important role in salvage pathway. SGPP1 belongs to the phosphatase super-family that converts sphingosine-1-phosphate to sphingosine that is readily metabolized to ceramide. Enhanced SGPP1 activity could lead to elevated sphingolipid levels by shifting the stochiometric balance towards sphingosine and ceramide production. Consistently, variants in SGPP1 have shown association with circulating sphingomyelins, mainly containing C14-C16 and C22-C24 acyl chains. GLTPD2 codes for glycolipid transfer protein domain-containing protein 2 and has putative role in transfer of ceramide-1-phosphate. Thus, findings from the major GWASs on sphingolipids suggest that the plasma levels of ceramides and sphingomyelins are primarily driven by the genes involved in sphingolipid metabolism, particularly ceramide biosynthesis (Fig. 3).
Glycerolipids
Only a few GWASs have included molecular TG species [12, 13, 88, 94]. The study by Rhee et al. in over 2,000 participants from Framingham Heart Study (FHS) included 46 TG species and revealed association of FADS1-2-3, GCKR and APOA1-5 loci with several TG species [88]. These genes with direct role in triglyceride metabolism were subsequently replicated in other GWASs [12, 94]. The lead GCKR variant rs1260326, a missense variant (L446P) associated with TG species, is established as the likely causal variant through functional studies [103, 104]. In another study by Rhee et al. that focused only on the rare variants, no additional variants for TGs could be identified [94]. However, recent studies have suggested association of common variants at new loci for TGs. Our previous study suggested new signals for TGs at genetic variants in or near KAZN, VWA3B, ABLIM2, PDHA2, PTPRN2, LPL, APOA5, CD33 and MIR100HG at genome-wide significance that did not remain significant after multiple testing correction [12]. Also, Demirkan et al. identified association between a new locus MLXIPL and TGs (TG 48:1 and TG 50:1) [13].
Interestingly, similar to epidemiological finding that different TG species have different effect on CVD risk, findings from the genetic studies revealed different patterns of association of TG species with genetic variants. GCKR demonstrated a stronger association with TGs of relatively lower carbon content (TG 48:2, TG 48:3, TG 50:3, TG 50:2, TG 50:4) [13, 88], while APOA1/A5 and LPL have stronger effect on medium length TG species (TG 54:4, TG 52:3, TG 52:4) [12, 88]. On the other hand, FADS1-2-3 associate with TGs in a fatty acid saturation specific manner, with the direction of effect differed at the extremes of TG carbon content, and strongest association with TGs of relatively higher carbon and double bond content such as TG 58:10 and TG 58:11 [13, 88]. Such a pattern of association was also observed in one of the loci identified in GWASs for enzymatically measured triglycerides-CILP2 that had different effect sizes across different TG species and was mainly associated with the unsaturated TGs [13]. These findings suggest that genes involved in TG metabolism have species-specific effect that apparently depend on the length of acyl chains. Further light on this was provided by our previous study which showed that a genetic variant at LPL locus (rs11570891) increases the expression and enzymatic activity of LPL [12]. We further showed that the increased LPL enzymatic activity had stronger effect on medium length TGs than other TGs. Taken together, GWAS findings suggest that genetic regulation of TGs is determined by their carbon content and degree of unsaturation and further reinforce that such effects might not be detected by enzymatic measurement of total triglycerides.
Phospholipids
A number of genetic loci have been associated with plasma levels of distinct phospholipids species, including genes with direct role in phospholipid metabolism (Table 3; Fig. 4). In the KORA study that included 208 phospholipid species, Geiger et al. [83] identified association of phospholipids with FADS1-2-3 and LIPC. Later, Illig et al. [85] identified five loci for phospholipids-FADS1-2-3, ELOVL2, PLEKHH1, SYNE2 and SPTLC3 in a larger dataset. Further, a comprehensive genetic investigation of phospholipids with 57 PCs, 20 lyso PCs, 27 PEs, 15 plasmalogens in over 4000 samples identified 25 loci at genome-wide significance [87]. In the pathway analysis, 13 genes (KCNH7, AGPAT1, PNLIPRP2, SYT9, FADS2, DAGLA, DLG2, APOA1, APOC3, ELOVL2, CDK17, LIPC and PLA2G10) located in 11 loci from the 25 loci were mapped to the glycerophospholipid metabolism pathway [87]. Several additional loci for phospholipids were discovered as illustrated in Fig. 4 and listed in Supplementary Table 3. Here we discuss two examples that highlight the potential of lipidomics in identifying new lipid modifying genes and providing mechanistic insights to the known lipid loci.
MBOAT7 encodes a lysophosphatidylinositol acyltransferase that incorporates arachidonic acid (C20:4) into lysophosphatidylinositol (LPI) to generate phosphatidylinositols (PI) [105]. The activity of MBOAT7 regulates the levels of free arachidonic acid and its availability for eicosanoid production which mediates pro-inflammatory signalling [106]. Consistent with its biochemical function, Shin et al. identified association of variants in MBOAT7 with the ratio of arachidonate (20:4n6) to 1-arachidonoylglycerophophoinositol [90]. Later, its association with PI species was confirmed in other studies [12, 96]. MBOAT7 variants also increase the susceptibility to liver disorders including liver cirrhosis and non-alcoholic fatty liver disease (NAFLD) by inducing a reduction in its expression in liver [107–109]. Notably, our previous study also suggested association of MBOAT7 variant with venous thromboembolism [12]. The example of MBOAT7 further exemplifies that lipidome-based GWAS could identify new genes with prominent role in lipid metabolism that could not be detected through GWAS of traditional lipids.
Fatty acid desaturase (FADS) gene cluster has been consistently reported to be associated with omega-3 and omega-6 fatty acids levels with inverse effects on different PUFAs [43, 44, 83, 87]. The FADS gene cluster contains genes coding for three key enzymes in PUFA metabolism-FADS1 (delta-5 desaturase), FADS2 (delta-6 desaturase) and FADS3 (delta-9 desaturase). The delta-6 desaturation by FADS2 generates gamma-linolenic acid (C18:3 n-6) and stearidonic acid (C18:4 n-3) from linoleic acid (C18:2 n-6) and alpha-linolenic acid (C18:3 n-3) respectively, that by elongation yield dihomo-gamma-linolenic acid (C20:3 n-6) and eicosatetraenoic acid (C20:4 n-3) [110]. Further, delta-5 desaturation of dihomo-gamma-linolenic acid by FADS1 generates arachidonic acid (C20:4 n-6) and eicosapentaenoic acid (C20:5 n-3). Genetic variants in FADS1 and FADS2 genes are associated with the increased levels of phospholipids with three or less double bonds while with the decreased levels of phospholipids with four or more double bonds [12, 83]. We previously showed that a variant in FADS2 increases FADS2 expression while reduces the expression of FADS1 that explain the inverse relationship of FADS2 variants with lipids containing different polyunsatureated fatty acids (PUFAs) [12]. The association of FADS1-2-3 locus with the reduced levels of lipids containing arachidonic acid may also explain its assocition with reduced risk of atherosclerotic CVD outcomes-peripheral artery disesae (PAD) and aterial embolism and thrombosis [12]. The example of FADS1-2-3 along with other known lipid genes such as LPL, GCKR (discussed above) highlights how detailed lipidomic profile could provide the mechanistic understanding of effects of well-established lipid loci.
Sterols
Sterol lipids including CEs are not represented well in the GWASs with lipidome or metabolome, and only two studies have reported association of CEs with genetic variants [12, 88]. Rhee et al. [88] identified four loci associated with CE species- FADS1-2-3, GNAL, NTAN1 and SEC61G. We found previously association of three loci with CEs-ABCG5/8, FADS2 and SYNGR1. ABCG5/G8 codes for ABC cholesterol transporters G5 and G8, that have been associated with total cholesterol, LDL-C and cholesterol esters in LDL. However, our study revealed a novel association of ABCG5/G8 variant with a specific CE species- CE 20:2;0. All of the identified loci for CEs overlap with the loci associated with phospholipids (Fig. 4).
Discussion
It is apparent from epidemiological and genetic studies that lipidomics has great potential in revealing new biology not captured by traditional lipids and lipoprotein measurements. Lipid species measurements, like other intermediate phenotypes, increases statistical power to detect genetic associations and hence provide opportunity to discover new lipid loci [12, 111]. In an analysis with over 500 known genetic variants for traditional lipids, we previously showed that as compared to traditional lipids, associations with detailed molecular lipids are several orders of magnitudes stronger for the variants in or near genes involved in lipid metabolism such as FADS1-2-3, LIPC, ABCG5/8, SGPP1, SPTLC3 [12]. This demonstrates the prospects of lipidomics in identifying lipid-modulating variants, particularly the ones with direct role in lipid metabolism. Consistently, GWASs with distinct lipid species discovered many new genes with direct role in lipid metabolic pathways and provided new insights into the genomic loci associated with traditional lipids. For instance, detailed TG profiles revealed that the total triglycerides associated loci such as GCKR, FADS1-2-3, LPL, APOA5 drive association of distinct TG species depending on number of carbon atoms and degree of unsaturation, as discussed above.
Integrating lipidomics and genomics: opportunities beyond GWAS
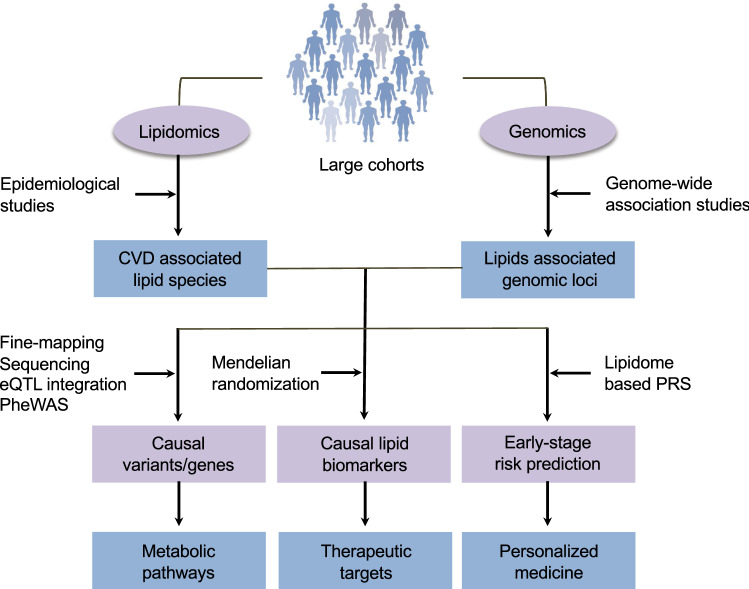
After the success of the GWASs in identifying new genomic loci associated with lipid species, one of the next challenges is to translate these findings towards predictive and personalized medicine. Emerging tools in genomics provides many new opportunities in this direction [112]. This is demonstrated by the success of genetic studies guiding the development of antibodies targeting PCSK9 (proprotein convertase subtilisin/kexin type 9) to treat hyperlipidemia and CVD [113]. Here we discuss approaches that could be used to translate the statistical associations identified in epidemiological studies and GWASs to biological understanding, drug target identification and disease risk characterization (Fig. 5).
Fig. 5.
Approaches to move beyond GWAS. New opportunities and prospects of application of genomics to translate findings from lipidomics to develop better predictive and preventive strategies are illustrated
Inferring causality towards drug targets development
Epidemiological studies have provided strong evidence of association of several distinct lipid species with CVD outcomes. However, it cannot be inferred from simple observational studies which of these associated lipid species have causal effect on CVDs. Nonetheless, genomics provides an alternative approach to infer causality using the intrinsic properties of the genome, i.e., the random assortment of alleles at conception, in a statistical framework referred as Mendelian randomization (MR) [114–116]. MR is a routinely used genetic tool in observational studies which uses genetic variants as proxies for exposure variable (risk factors) to infer whether the increased or decreased risk factor causes the disease [41, 117, 118]. First described in 1986 [119], many methods and approaches in MR analysis have been developed that allow use of GWAS summary statistic data for risk factor and disease of interest, either from one dataset (one-sample MR) or two datasets (two-sample MR) [114]. Development of two-sample MR methods using the existing and publicly available GWAS summary data has made MR analysis increasingly easy and popular.
With the improved understanding of the genetic architecture of lipidome and availability of large-scale GWAS summary statistics data both for lipidome and CVD, MR provides an excellent avenue to infer the causal role of the CVD associated lipid species. Efforts at this front have been limited so far, most likely due to the highly correlated nature of the high-dimensional lipidome profiles. Ganna et al. [67] used summary stats from CARDIoGRAMplusC4D to reveal causal effect of MG 18:2 on CHD risk. On the other hand, a recent study performed a MR analysis to investigate the causal relationship between PC 38:3 and P wave duration (PWD, an indicator of atrial conduction) [120]. The study found that PC 38:3 and PWD association is not causal and is mediated by BMI. Larger efforts are required to highlight the lipid species with therapeutic potential from the large number of associations for follow-up investigations, as exhibited in a recent MR analysis [121]. The study performed GWAS on untargeted plasma metabolome with ~ 11,000 metabolites in ~ 10,000 individuals and used two-sample MR approach to assess the causal effect of both identified and unidentified metabolites on 45 common diseases. The study provided evidence of causal effect of 31 metabolites on at least one of the 5 diseases- CHD, schizophrenia, bipolar disorder, rheumatoid arthritis, primary sclerosing cholangitis. Among the causal metabolites, 19 metabolites were causal for coronary heart disease and 6 of which were found to be associated with incident CHD. Recent development of multivariate MR methods provides a powerful tool to incorporate high-dimensional data like lipidome profiles in MR analysis [122–124]. Application of these approaches to infer the causality of CVD associated lipid species is one of the ways to move forward in the direction of drug target development.
Refining GWAS signals to causal variants to reveal metabolic networks
Although GWASs have been successful in identifying new genomic loci associated with lipid species, the associated variant, in most cases, does not cause the trait or disease itself but serve as surrogate for neighbouring SNPs in a large genomic region that are in linkage disequilibrium (LD) with it. Because of the complex LD patterns among the SNPs, pinpointing causal variants from the associated variants is a challenging task. However, statistical fine-mapping approaches allow the refinement of the trait-associated regions to identify genetic variants with likely causal influence on the trait [125, 126]. The fine-mapping approach in lipidomics could not only help in refining the genomic loci but the multidimensional association data may also help in highlighting the metabolite-specific effects and hence reveal new metabolic networks. For instance, Gallois et al. [127] performed fine-mapping of LIPC region which suggested that there are at least three distinct sites with metabolite-specific variants within the gene. The study showed that large HDL and triglyceride in lipoproteins are influenced by all the three sites in LIPC. But interestingly, intermediate density lipoproteins (IDLs) and fatty acids are mostly influenced by two sites and very small VLDL (very low-density lipoprotein) are influenced by only one of sites in the gene [127]. On the similar note, earlier Tukiainen et al. [111] showed that LIPC region has opposite associations between the lipid measures of larger and smaller HDL particles.
Another commonly used approach is targeted sequencing of the trait-associated region that allow identification of rare coding or loss-of-function (LoF) variants with putative causal effects in the region [128–132]. Exome or whole genome sequencing allow the identification of full spectrum of variants, including rare and loss-of-function variants that may have direct functional effects than the common variants [99, 133, 134]. Due to the limited sample sizes, the sequencing efforts have been so far less successful in lipidomics and only a few rare or LoF variants influencing lipid levels could be discovered [96–99]. Long et al. identified seven rare variants in four genes (ACADS, CRAT, DMGDH, ETFDH) involved in fatty acid metabolism [96]. Thus, larger sequencing efforts are required to better understand the potential role of rare coding variants involved in lipidomic variation.
Further insight to the putative causal variant could also be provided by integrating GWAS results with other “omics” data such as transcriptome, proteome and epigenome. About 80% of the genetic variants identified by the GWASs lie in the non-coding regions, thus exploring the association with gene expression levels (eQTLs), protein levels (pQTLs) and epigenetic changes such as DNA methylation in relevant tissues could provide information about the biological effects of the variants and putative causal genes. Also, utilizing the phenome-wide association (PheWAS) data for thousands of clinical outcomes from biobanks such as UK Biobank (https://www.ukbiobank.ac.uk/) and FinnGen (https://www.finngen.fi/en) could further provide mechanistic insights. To facilitate the efforts in this direction, colocalization tools have been developed in recent years that can integrate multi-omics datasets such as GWAS, eQTLs, pQTLs and PheWAS data using statistical methods [135–137]. The colocalization analysis by Franceschini et al. provided evidence for the role of novel genes in the subclinical measures such as carotid intima-media thickness (cIMT) and carotid plaque formation and provided insights into the regulatory mechanisms linking atherosclerosis and clinical outcomes [137].
Predicting CVD risk using lipidome-based genetic risk scores
Early prediction and prevention could greatly reduce the enormous socio-economic burden of the CVDs [138]. The intensity of risk management regime is generally guided by the risk estimates of the individuals [139], which are not always precise [140]. Moreover, existing clinical risk assessment tools, that typically include traditional plasma lipids, identify individuals with high CVD risk at a stage when atherosclerotic events have already developed. Polygenic risk scores (PRS), a weighted sum of the number of risk alleles carried by an individual, has shown potential in early prediction, but currently have limited clinical utility [141–144]. Thus, the quest for better and early-stage prediction scores to maximize the benefits of risk management has been the focus of the CVD research, but have provided limited success so far [145, 146].
One of the challenges in early CVD detection and prevention is the heterogeneity owing to the diverse pathological conditions that are preceded by atherosclerotic and metabolic events developing at young age [147], resulting in different CVD subtypes. Hence, individuals’ predisposition to different subtypes is influenced by multiple independent risk factors that need to be understood and incorporated in prediction algorithms to guide appropriate and personalized interventions. Traditional lipid profiling that measures HDL-C, LDL-C, triglycerides and total cholesterol, does not reflect precise molecular perturbations in lipid metabolism underlying CVD subtypes. Moreover, a PRS based on genetic loci for CVD represents a combination of genetic risk factors acting through different pathways, whose roles may vary in different CVD subtypes. In such scenario, individuals would respond very differently to the same risk management strategies. As revealed by genetic studies of lipidome that many genomic loci have lipid species specific effects, integrating information of genetic variants of lipidomic measures in PRS algorithms could provide more specific and sensitive CVD risk stratification than those based on CVD variants. Our proposition is that the risk prediction model should incorporate information on perturbations in individual's lipidome profile and their genetic determinants. Thus, the next challenge is to develop predictive tools to incorporate the genetic data on high-dimensional lipidome profiles.
Conclusion
The increasing global burden of CVDs highlights the pressing need for better personalized prediction and prevention strategies. One key step is to open new therapeutic opportunities by understanding the causal roles of lipid metabolism at molecular lipid species/sub-species resolution in heterogenous CVDs etiologies and their regulation by genetic and lifestyle factors. To this end, the technological advances in lipidomics and other omics technologies have led to a tremendous progress in the CVD research field in last two decades. Lipidomics has not only provided a closer look at the lipid metabolic perturbations in CVDs, but has improved our understanding of the genetic control of lipid metabolism. Further improvement in technologies will continue to improve our understanding of CVDs. However, the ultimate goal of a personalized translational research is to find the right intervention (target biomarker) for right individual (CVD subtype) at the right time (at early stage). Integrating the emerging genomics tools with the high-dimensional lipidome holds a great potential in moving towards this goal. We discussed some of the commonly used approaches in translational research that could be employed in lipidomics-based studies, however further advancements in statistical and computational tools would be required to deal with the high-dimensional and correlated structure of the lipidome profiles.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Sari Kivikko and Ulla Tuomainen for management assistance.
Author contributions
RT performed the literature search and data analysis; RT and SR had the idea and wrote the manuscript.
Funding
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. SR was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (Grant No 312062), the Finnish Foundation for Cardiovascular Research, the Sigrid Juselius Foundation and University of Helsinki HiLIFE Fellow and Grand Challenge grants.
Code availability
Not applicable.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Availability of data and material
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rubina Tabassum, Email: rubina.tabassum@helsinki.fi.
Samuli Ripatti, Email: samuli.ripatti@helsinki.fi.
References
- 1.O'Donnell MJ, Xavier D, Liu L, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. 2010;376:112–123. doi: 10.1016/S0140-6736(10)60834-3. [DOI] [PubMed] [Google Scholar]
- 2.Yusuf S, Joseph P, Rangarajan S, et al. Modifiable risk factors, cardiovascular disease, and mortality in 155 722 individuals from 21 high-income, middle-income, and low-income countries (PURE): a prospective cohort study. Lancet. 2019;395:795–808. doi: 10.1016/S0140-6736(19)32008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannel WB, Dawber TR, Friedman GD, Glennon WE, Mcnamara PM. Risk factors in coronary heart disease. an evaluation of several serum lipids as predictors of coronary heart disease; the Framingham study. Ann Intern Med. 1964;61:888–899. doi: 10.7326/0003-4819-61-5-888. [DOI] [PubMed] [Google Scholar]
- 4.Arnett DK, Blumenthal RS, Albert MA, et al. 2019 ACC/AHA Guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Global Burden of Disease Causes of Death Collaborators (2017) Global, regional, and national age-specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2016;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akyea RK, Kai J, Qureshi N, Iyen B, Weng SF. Sub-optimal cholesterol response to initiation of statins and future risk of cardiovascular disease. Heart. 2019;13:975–981. doi: 10.1136/heartjnl-2018-314253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quehenberger O, Dennis EA. The human plasma lipidome. N Engl J Med. 2011;365:1812–1823. doi: 10.1056/NEJMra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci USA. 2019;106:2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M. Informatics and computational strategies for the study of lipids. Mol Biosyst. 2008;4:121–127. doi: 10.1039/b715468b. [DOI] [PubMed] [Google Scholar]
- 10.Lydic TA, Goo YH. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clin Transl Med. 2018;7:4. doi: 10.1186/s40169-018-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 12.Tabassum R, Rämö JT, Ripatti P, et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun. 2019;10:4329. doi: 10.1038/s41467-019-11954-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demirkan A, Pool R, Deelen J, Beekman M, Liu J, Harms AC, Vaarhorst A, Hagenbeek FA, Willemsen G, Verhoeven A, Amin N, van Dijk KW, Hankemeier T, Boomsma DI, Slagboom E, van Duijn CM. Genome-wide association study of plasma triglycerides, phospholipids and relation to cardio-metabolic risk factors. BioRxiv. 2019 doi: 10.1101/621334. [DOI] [Google Scholar]
- 14.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. The LIPID MAPS initiative in lipidomics. Methods Enzymol. 2007;432:169–181. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 15.Fahy E, Subramaniam S, Brown HA, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Fahy E, Subramaniam S, Murphy RC, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim Biophys Acta. 2011;1811:637–647. doi: 10.1016/j.bbalip.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slatter DA, Aldrovandi M, O'Connor A, Allen SM, Brasher CJ, Murphy RC, Mecklemann S, Ravi S, Darley-Usmar V, O'Donnell VB. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metab. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. 2016;12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 21.Stephenson DJ, Hoeferlin LA, Chalfant CE. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl Res. 2017;189:13–29. doi: 10.1016/j.trsl.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokoła-Wysoczańska E, Wysoczański T, Wagner J, Czyż K, Bodkowski R, Lochyński S, Patkowska-Sokoła B. Polyunsaturated fatty acids and their potential therapeutic role in cardiovascular system disorders—a review. Nutrients. 2018;10:1561. doi: 10.3390/nu10101561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innes JK, Calder PC. Marine omega-3 (N-3) fatty acids for cardiovascular health: an update for 2020. Int J Mol Sci. 2020;21:1362. doi: 10.3390/ijms21041362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 25.Prentki M, Madiraju SR. Glycerolipid metabolism and signaling in health and disease. Endocr Rev. 2008;29:647–676. doi: 10.1210/er.2008-0007. [DOI] [PubMed] [Google Scholar]
- 26.Hannun Y, Obeid L. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers SA, Chaurasia B, Holland WL. Metabolic messengers: ceramides. Nat Metab. 2019;1:1051–1058. doi: 10.1038/s42255-019-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dufourc EJ. Sterols and membrane dynamics. J Chem Biol. 2008;1:63–77. doi: 10.1007/s12154-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tumanov S, Kamphorst JJ. Recent advances in expanding the coverage of the lipidome. Curr Opin Biotechnol. 2017;43:127–133. doi: 10.1016/j.copbio.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Züllig T, Trötzmüller M, Köfeler HC. Lipidomics from sample preparation to data analysis: a primer. Anal Bioanal Chem. 2020;412:2191–2209. doi: 10.1007/s00216-019-02241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal Chem. 2014;61:192–206. doi: 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Wang C, Han RH, Han X. Novel advances in shotgun lipidomics for biology and medicine. Prog Lipid Res. 2016;61:83–108. doi: 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu FF. Mass spectrometry-based shotgun lipidomics—a critical review from the technical point of view. Anal Bioanal Chem. 2018;410:6387–6409. doi: 10.1007/s00216-018-1252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aru V, Lam C, Khakimov B, Hoefsloot HC, Zwanenburg G, Lind MV, Schäfer H, van Duynhoven J, Jacobs DM, Smilde AK, Soren B. Quantification of lipoprotein profiles by nuclear magnetic resonance spectroscopy and multivariate data analysis. Trends Anal Chem. 2017;94:210–219. [Google Scholar]
- 36.Würtz P, Havulinna AS, Soininen P, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131:774–785. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes MV, Millwood IY, Kartsonaki C, Hill MR, Bennett DA, Boxall R, Guo Y, Xu X, Bian Z, Hu R, Walters RG, Chen J, Ala-Korpela M, Parish S, Clarke RJ, Peto R, Collins R, Li L, Chen Z, China Kadoorie Biobank Collaborative Group Lipids, lipoproteins, and metabolites and risk of myocardial infarction and stroke. J Am Coll Cardiol. 2018;71:620–632. doi: 10.1016/j.jacc.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson G, Rantalainen M, Li JV, Maher AD, Malmodin D, et al. A genome-wide metabolic QTL analysis in Europeans implicates two loci shaped by recent positive selection. PLoS Genet. 2011;7:e1002270. doi: 10.1371/journal.pgen.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inouye M, Ripatti S, Kettunen J, Lyytikäinen LP, Oksala N, Laurila PP, Kangas AJ, Soininen P, Savolainen MJ, Viikari J, Kähönen M, Perola M, Salomaa V, Raitakari O, Lehtimäki T, Taskinen MR, Järvelin MR, Ala-Korpela M, Palotie A, de Bakker PI. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012;8:e1002907. doi: 10.1371/journal.pgen.1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kettunen J, Tukiainen T, Sarin AP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettunen J, Demirkan A, Würtz P, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Comm. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coltell O, Sorlí JV, Asensio EM, Barragán R, González JI, Giménez-Alba IM, Zanón-Moreno V, Estruch R, Ramírez-Sabio JB, Pascual EC, Ortega-Azorín C, Ordovas JM, Corella D. Genome-wide association study for serum omega-3 and omega-6 polyunsaturated fatty acids: exploratory analysis of the sex-specific effects and dietary modulation in mediterranean subjects with metabolic syndrome. Nutrients. 2020;12:310. doi: 10.3390/nu12020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaitre RN, Tanaka T, Tang W, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu JH, Lemaitre RN, Manichaikul A, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Gene. 2013;6:171–183. doi: 10.1161/CIRCGENETICS.112.964619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, Tanaka T, Zhu J, et al. Discovery and fine-mapping of loci associated with MUFAs through trans-ethnic meta-analysis in Chinese and European populations. J Lipid Res. 2017;58:974–981. doi: 10.1194/jlr.P071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anroedh S, Hilvo M, Akkerhuis KM, Kauhanen D, Koistinen K, Oemrawsingh R, Serruys P, van Geuns RJ, Boersma E, Laaksonen R, Kardys I. Plasma concentrations of molecular lipid species predict long-term clinical outcome in coronary artery disease patients. J Lipid Res. 2018;59:1729–1737. doi: 10.1194/jlr.P081281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez C, Sandin M, Sampaio JL, Almgren P, Narkiewicz K, Hoffmann M, Hedner T, Wahlstrand B, Simons K, Shevchenko A, James P, Melander O. Plasma lipid composition and risk of developing cardiovascular disease. PLoS ONE. 2013;8:e71846. doi: 10.1371/journal.pone.0071846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bismuth J, Lin P, Yao Q, Chen C. Ceramide: a common pathway for atherosclerosis? Atherosclerosis. 2008;196:497–504. doi: 10.1016/j.atherosclerosis.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borodzicz S, Czarzasta K, Kuch M, Cudnoch-Jedrzejewska A. Sphingolipids in cardiovascular diseases and metabolic disorders. Lipids Health Dis. 2015;14:55. doi: 10.1186/s12944-015-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigruener A, Kleber ME, Heimerl S, Liebisch G, Schmitz G, Maerz W. Glycerophospholipid and sphingolipid species and mortality: the Ludwigshafen risk and cardiovascular health (LURIC) study. PLoS ONE. 2014;9:e85724. doi: 10.1371/journal.pone.0085724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng JM, Suoniemi M, Kardys I, Vihervaara T, de Boer SP, Akkerhuis KM, Sysi-Aho M, Ekroos K, Garcia-Garcia HM, Oemrawsingh RM, Regar E, Koenig W, Serruys PW, van Geuns RJ, Boersma E, Laaksonen R. Plasma concentrations of molecular lipid species in relation to coronary plaque characteristics and cardiovascular outcome: results of the ATHEROREMO-IVUS study. Atherosclerosis. 2015;243:560–656. doi: 10.1016/j.atherosclerosis.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Laaksonen R, Ekroos K, Sysi-Aho M, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J. 2016;37:1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang DD, Toledo E, Hruby A, et al. Plasma ceramides, Mediterranean diet, and incident cardiovascular disease in the PREDIMED Trial (Prevención con Dieta Mediterránea) Circulation. 2017;135:2028–2040. doi: 10.1161/CIRCULATIONAHA.116.024261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Havulinna AS, Sysi-Aho M, Hilvo M, Kauhanen D, Hurme R, Ekroos K, Salomaa V, Laaksonen R. Circulating ceramides predict cardiovascular outcomes in the population-based FINRISK 2002 cohort. Arterioscler Thromb Vasc Biol. 2016;36:2424–2430. doi: 10.1161/ATVBAHA.116.307497. [DOI] [PubMed] [Google Scholar]
- 55.Alshehry ZH, Mundra PA, Barlow CK, et al. Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in Type 2 diabetes mellitus. Circulation. 2016;134:1637–1650. doi: 10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed] [Google Scholar]
- 56.Paynter NP, Balasubramanian R, Giulianini F, Wang DD, Tinker LF, Gopal S, Deik AA, Bullock K, Pierce KA, Scott J, Martínez-González MA, Estruch R, Manson JE, Cook NR, Albert CM, Clish CB, Rexrode KM. Metabolic predictors of incident coronary heart disease in women. Circulation. 2018;137:841–853. doi: 10.1161/CIRCULATIONAHA.117.029468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poss AM, Maschek JA, Cox JE, Hauner BJ, Hopkins PN, Hunt SC, Holland WL, Summers SA, Playdon MC. Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J Clin Invest. 2020;130:1363–1376. doi: 10.1172/JCI131838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Carvalho LP, Tan SH, Ow GS, Tang Z, Ching J, Kovalik JP, Poh SC, Chin CT, Richards AM, Martinez EC, Troughton RW, Fong AY, Yan BP, Seneviratna A, Sorokin V, Summers SA, Kuznetsov VA, Chan MY. Plasma ceramides as prognostic biomarkers and their arterial and myocardial tissue correlates in acute myocardial infarction. JACC Basic Transl Sci. 2018;3:163–175. doi: 10.1016/j.jacbts.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mantovani A, Dugo C. Ceramides and risk of major adverse cardiovascular events: a meta-analysis of longitudinal studies. J Clin Lipidol. 2020;14:176–185. doi: 10.1016/j.jacl.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Seah JYH, Chew WS, Torta F, Khoo CM, Wenk MR, Herr DR, Choi H, Tai ES, van Dam RM. Plasma sphingolipids and risk of cardiovascular diseases: a large-scale lipidomic analysis. Metabolomics. 2020;16:89. doi: 10.1007/s11306-020-01709-8. [DOI] [PubMed] [Google Scholar]
- 61.Lemaitre RN, Jensen PN, Hoofnagle A, McKnight B, Fretts AM, King IB, Siscovick DS, Psaty BM, Heckbert SR, Mozaffarian D, Sotoodehnia N. Plasma ceramides and sphingomyelins in relation to heart failure risk. Circ Heart Fail. 2019;12:e005708. doi: 10.1161/CIRCHEARTFAILURE.118.005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilvo M, Meikle PJ, Pedersen ER, et al. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J. 2020;41:371–380. doi: 10.1093/eurheartj/ehz387. [DOI] [PubMed] [Google Scholar]
- 63.Peterson LR, Xanthakis V, Duncan MS, et al. Ceramide remodeling and risk of cardiovascular events and mortality. J Am Heart Assoc. 2018;7:e007931. doi: 10.1161/JAHA.117.007931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meeusen JW, Donato LJ, Bryant SC, Baudhuin LM, Berger PB, Jaffe AS. Plasma ceramides. Arterioscler Thromb Vasc Biol. 2018;38:1933–1939. doi: 10.1161/ATVBAHA.118.311199. [DOI] [PubMed] [Google Scholar]
- 65.Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, Menni C, Moayyeri A, Santer P, Rungger G, Spector TD, Willeit J, Kiechl S, Mayr M. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation. 2014;129:1821–1831. doi: 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- 66.Mundra PA, Barlow CK, Nestel PJ, et al. Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI Insight. 2018;3:e121326. doi: 10.1172/jci.insight.121326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ganna A, Salihovic S, Sundstrom J, et al. Large-scale Metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10:e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Razquin C, Liang L, Toledo E, et al. Plasma lipidome patterns associated with cardiovascular risk in the PREDIMED trial: a case-cohort study. Int J Cardiol. 2018;253:126–132. doi: 10.1016/j.ijcard.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang DD, Zheng Y, Toledo E, et al. Lipid metabolic networks, Mediterranean diet and cardiovascular disease in the PREDIMED trial. Int J Epidemiol. 2018;47:1830–1845. doi: 10.1093/ije/dyy198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang L, Wang L, Deng Y, Sun L, Lou B, Yuan Z, Wu Y, Zhou B, Liu J, She J. Serum lipids profiling perturbances in patients with ischemic heart disease and ischemic cardiomyopathy. Lipids Health Dis. 2020;19:89. doi: 10.1186/s12944-020-01269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubins HB, Robins SJ, Collins D, et al. Diabetes, plasma insulin, and cardiovascular disease: subgroup analysis from the Department of Veterans Affairs high-density lipoprotein intervention trial (VA-HIT) Arch Intern Med. 2002;162:2597–2604. doi: 10.1001/archinte.162.22.2597. [DOI] [PubMed] [Google Scholar]
- 72.Keech A, Simes RJ, Barter P, et al. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366:1849–1861. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]
- 73.Do R, Willer CJ, Schmidt EM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sysi-Aho M, Koikkalainen J, Seppänen-Laakso T, Kaartinen M, Kuusisto J, Peuhkurinen K, Kärkkäinen S, Antila M, Lauerma K, Reissell E, Jurkko R, Lötjönen J, Heliö T, Orešič M. Serum lipidomics meets cardiac magnetic resonance imaging: profiling of subjects at risk of dilated cardiomyopathy. PLoS ONE. 2011;6:e15744. doi: 10.1371/journal.pone.0015744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bellis C, Kulkarni H, Mamtani M, et al. Human plasma lipidome is pleiotropically associated with cardiovascular risk factors and death. Circ Cardiovasc Genet. 2014;7:854–863. doi: 10.1161/CIRCGENETICS.114.000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frahnow T, Osterhoff MA, Hornemann S, Kruse M, Surma MA, Klose C, Simons K, Pfeiffer AFH. Heritability and responses to high fat diet of plasma lipidomics in a twin study. Sci Rep. 2017;7:3750. doi: 10.1038/s41598-017-03965-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cadby G, Melton PE, McCarthy NS, Giles C, Mellett NA, Huynh K, Hung J, Beilby J, Dubé MP, Watts GF, Blangero J, Meikle PJ, Moses EK. Heritability of 596 lipid species and genetic correlation with cardiovascular traits in the Busselton Family Heart Study. J Lipid Res. 2020;61:537–545. doi: 10.1194/jlr.RA119000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jha P, McDevitt MT, Gupta R, Quiros PM, Williams EG, Gariani K, Sleiman MB, Diserens L, Jochem A, Ulbrich A, Coon JJ, Auwerx J, Pagliarini DJ. Systems analyses reveal physiological roles and genetic regulators of liver lipid species. Cell Syst. 2018;6:722–733.e6. doi: 10.1016/j.cels.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jha P, McDevitt MT, Halilbasic E, Williams EG, Quiros PM, Gariani K, Sleiman MB, Gupta R, Ulbrich A, Jochem A, Coon JJ, Trauner M, Pagliarini DJ, Auwerx J. Genetic regulation of plasma lipid species and their association with metabolic phenotypes. Cell Syst. 2018;6:709–721.e6. doi: 10.1016/j.cels.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hagenbeek FA, Pool R, van Dongen J, et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun. 2020;11:39. doi: 10.1038/s41467-019-13770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Surakka I, Horikoshi M, Mägi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klarin D, Damrauer SM, Cho K, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, Illig T, Suhre K. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hicks AA, Pramstaller PP, Johansson A, et al. Genetic Determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Demirkan A, van Duijn CM, Ugocsai P, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rhee EP, Ho JE, Chen MH, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu B, Zheng Y, Alexander D, et al. Genetic Determinants Influencing Human Serum Metabolome among African Americans. PLoS Genet. 2014;10:e1004212. doi: 10.1371/journal.pgen.1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ried JS, Shin SY, Krumsiek J, et al. Novel genetic associations with serum level metabolites identified by phenotype set enrichment analyses. Hum Mol Genet. 2014;23:5847–5857. doi: 10.1093/hmg/ddu301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Draisma HHM, Pool R, Kobl M, et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mozaffarian D, Kabagambe EK, Johnson CO, et al. Genetic loci associated with circulating phospholipid trans fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. Am J Clin Nutr. 2015;101:398–406. doi: 10.3945/ajcn.114.094557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rhee EP, Yang Q, Yu B, et al. An exome array study of the plasma metabolome. Nat Commun. 2016;7:12360. doi: 10.1038/ncomms12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu B, Li AH, Metcalf GA, et al. Loss-of-function variants influence the human serum metabolome. Sci Adv. 2016;2:e1600800. doi: 10.1126/sciadv.1600800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Long T, Hicks M, Yu HC et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 49:568–578 [DOI] [PubMed]
- 97.Yousri NA, Fakhro KA, Robay A, et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat Commun. 2018;9:333. doi: 10.1038/s41467-017-01972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feofanova EV, Yu B, Metcalf GA, et al. Sequence-Based Analysis of Lipid-Related Metabolites in a Multiethnic Study. Genetics. 2018;209:607–616. doi: 10.1534/genetics.118.300751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yazdani A, Yazdani A, Elsea SH, et al. Genome analysis and pleiotropy assessment using causal networks with loss of function mutation and metabolomics. BMC Genomics. 2019;20:395. doi: 10.1186/s12864-019-5772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lotta LA, Pietzner M, Stewart ID, et al. Cross-platform genetic discovery of small molecule products of metabolism and application to clinical outcomes. bioRxiv. 2020 doi: 10.1101/2020.02.03.932541. [DOI] [Google Scholar]
- 101.Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J. 2012;441:789–802. doi: 10.1042/BJ20111626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Orho-Melander M, Melander O, Guiducci C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee HC, Inoue T, Imae R, Kono N, Shirae S, Matsuda S, Gengyo-Ando K, Mitani S, Arai H. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol Biol Cell. 2008;19:1174–1184. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gijón MA, Riekhof WR, Zarini S, Murphy RC, Voelker DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283:30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luukkonen PK, Zhou Y, Hyötyläinen T, Leivonen M, Arola J, Orho-Melander M, Orešič M, Yki-Järvinen H. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 108.Meroni M, Longo M, Fracanzani AL, Dongiovanni P. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine. 2020;57:102866. doi: 10.1016/j.ebiom.2020.102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 110.Saini RK, Keum YS. Omega-3 and omega-6 polyunsaturated fatty acids: dietary sources, metabolism, and significance—a review. Life Sci. 2018;203:255–267. doi: 10.1016/j.lfs.2018.04.049. [DOI] [PubMed] [Google Scholar]
- 111.Tukiainen T, Kettunen J, Kangas AJ, et al. Detailed metabolic and genetic characterization reveals new associations for 30 known lipid loci. Hum Mol Genet. 2012;21:1444–1455. doi: 10.1093/hmg/ddr581. [DOI] [PubMed] [Google Scholar]
- 112.Kamb A, Harper S, Stefansson K. Human genetics as a foundation for innovative drug development. Nat Biotechnol. 2013;31:975–978. doi: 10.1038/nbt.2732. [DOI] [PubMed] [Google Scholar]
- 113.Hess CN, Low Wang CC, Hiatt WR. PCSK9 Inhibitors: Mechanisms of Action, Metabolic Effects, and Clinical Outcomes. Annu Rev Med. 2018;69:133–145. doi: 10.1146/annurev-med-042716-091351. [DOI] [PubMed] [Google Scholar]
- 114.Davey Smith G, Ebrahim S. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ. 2005;330:1076–1079. doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 116.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 118.Gudbjartsson DF, Thorgeirsson G, Sulem P, et al. Lipoprotein(a) concentration and risks of cardiovascular disease and diabetes. J Am Coll Cardiol. 2019;74:2982–2994. doi: 10.1016/j.jacc.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 119.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 120.Del Greco MF, Foco L, Teumer A, et al. Lipidomics, atrial conduction, and body mass index. Circ Genom Precis Med. 2019;12:e002384. doi: 10.1161/CIRCGEN.118.002384. [DOI] [PubMed] [Google Scholar]
- 121.Qin Y, Meric G, Long T, Watrous J, Burgess S, Havulinna A, Ritchie SC, Brozynska M, Jousilahti P, Perola M, Lahti L, Niiranen T, Cheng S, Salomaa V, Jain M, Inouye M. Genome-wide association and Mendelian randomization analysis prioritizes bioactive metabolites with putative causal effects on common diseases. medRxiv. 2020 doi: 10.1101/2020.08.01.20166413. [DOI] [Google Scholar]
- 122.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zuber V, Colijn JM, Klaver C, et al. Selecting likely causal risk factors from high-throughput experiments using multivariable Mendelian randomization. Nat Commun. 2020;11:29. doi: 10.1038/s41467-019-13870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, Holmes MV. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 2020;17:e1003062. doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gallois A, Mefford J, Ko A, et al. A comprehensive study of metabolite genetics reveals strong pleiotropy and heterogeneity across time and context. Nat Commun. 2019;10:4788. doi: 10.1038/s41467-019-12703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van de Ven JP, Nilsson SC, Tan PL, et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45:813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 131.Patel AP, Peloso GM, Pirruccello JP, Johansen CT, Dubé JB, Larach DB, Ban MR, Dallinge-Thie GM, Gupta N, Boehnke M, Abecasis GR, Kastelein JJ, Hovingh GK, Hegele RA, Rader DJ, Kathiresan S. Targeted exonic sequencing of GWAS loci in the high extremes of the plasma lipids distribution. Atherosclerosis. 2016;250:63–68. doi: 10.1016/j.atherosclerosis.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Service SK, Teslovich TM, Fuchsberger C, et al (2014) Re-sequencing expands our understanding of the phenotypic impact of variants at GWAS loci. PLoS Genet 10:e1004147 [DOI] [PMC free article] [PubMed]
- 133.Liu DJ, Peloso GM, Yu H, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Natarajan P, Peloso GM, Zekavat SM, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun. 2018;9:3391. doi: 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Franceschini N, Giambartolomei C, de Vries PS, et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun. 2018;9:5141. doi: 10.1038/s41467-018-07340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carter HE, Schofield D, Shrestha R. Productivity costs of cardiovascular disease mortality across disease types and socioeconomic groups. Open Heart. 2019;6:e000939. doi: 10.1136/openhrt-2018-000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Twenty Seventh Bethesda Conference Matching the intensity of risk factor management with the hazard for coronary disease events. September 14–15, 1995. J Am Coll Cardiol. 1995;27:957–1047. [PubMed] [Google Scholar]
- 140.Kent DM, Shah ND. Risk models and patient-centered evidence: should physicians expect one right answer? JAMA. 2012;307:1585–1586. doi: 10.1001/jama.2012.469. [DOI] [PubMed] [Google Scholar]
- 141.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 142.Mars N, Koskela JT, Ripatti P, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med. 2020;26:549–557. doi: 10.1038/s41591-020-0800-0. [DOI] [PubMed] [Google Scholar]
- 143.Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P, Tzoulaki I. Predictive Accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–645. doi: 10.1001/jama.2019.22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, Kundu S, Robinson-Cohen C, Psaty BM, Rich SS, Post WS, Guo X, Rotter JI, Roden DM, Gerszten RE, Wang TJ. Predictive Accuracy of a Polygenic Risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323:627–635. doi: 10.1001/jama.2019.21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.US Preventive Services Task Force, Curry SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, Doubeni CA, Epling JW Jr, Kemper AR, Kubik M, Landefeld CS, Mangione CM, Silverstein M, Simon MA, Tseng CW, Wong JB Risk assessment for cardiovascular disease with nontraditional risk factors: US Preventive Services Task Force Recommendation Statement. JAMA. 2018;320:272–280. doi: 10.1001/jama.2018.8359. [DOI] [PubMed] [Google Scholar]
- 146.Lin JS, Evans CV, Johnson E, Redmond N, Coppola EL, Smith N. Nontraditional risk factors in cardiovascular disease risk assessment: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2018;320:281–297. doi: 10.1001/jama.2018.4242. [DOI] [PubMed] [Google Scholar]
- 147.Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents, National Heart, Lung, and Blood Institute Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents: summary report. Pediatrics. 2011;128:S213–S256. doi: 10.1542/peds.2009-2107C. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.