Abstract
Marine sponges commonly host a repertoire of bacterial-associated organisms, which significantly contribute to their health and survival by producing several anti-predatory molecules. Many of these compounds are produced by sponge-associated bacteria and represent an incredible source of novel bioactive metabolites with biotechnological relevance. Although most investigations are focused on tropical and temperate species, to date, few studies have described the composition of microbiota hosted by Antarctic sponges and the secondary metabolites that they produce. The investigation was conducted on four sponges collected from two different sites in the framework of the XXXIV Italian National Antarctic Research Program (PNRA) in November–December 2018. Collected species were characterized as Mycale (Oxymycale) acerata, Haliclona (Rhizoniera) dancoi, Hemigellius pilosus and Microxina sarai by morphological analysis of spicules and amplification of four molecular markers. Metataxonomic analysis of these four Antarctic sponges revealed a considerable abundance of Amplicon Sequence Variants (ASVs) belonging to the phyla Proteobacteria, Bacteroidetes, Actinobacteria and Verrucomicrobia. In particular, M. (Oxymycale) acerata, displayed several genera of great interest, such as Endozoicomonas, Rubritalea, Ulvibacter, Fulvivirga and Colwellia. On the other hand, the sponges H. pilosus and H. (Rhizoniera) dancoi hosted bacteria belonging to the genera Pseudhongella, Roseobacter and Bdellovibrio, whereas M. sarai was the sole species showing some strains affiliated to the genus Polaribacter. Considering that most of the bacteria identified in the present study are known to produce valuable secondary metabolites, the four Antarctic sponges could be proposed as potential tools for the discovery of novel pharmacologically active compounds.
Keywords: Antarctica, Demospongiae, marine biotechnology, metataxonomics, microbiota
1. Introduction
The Antarctic region comprises ice shelves, waters and all the island territories in the Southern Ocean, covering about 10% of the total world ocean’s area. The Antarctic is characterized by low temperature and scarce availability of nutrients, together with a high seasonality in terms of light conditions. Due to the extreme environmental conditions, Antarctic fauna has developed several physiological and behavioural adaptations, leading to the evolution of unique characteristics [1]. For instance, a longer period of larval development or parental care has been observed in Antarctic invertebrates, including sponges [2,3,4]. Moreover, marine invertebrate communities living in this area have been subjected to a wide temporal and biogeographic isolation [5,6] dating back to about 140 million years ago when the Antarctic continent separated from Gondwana [7,8]. This event has promoted the development of specific traits, which make Antarctic organisms extremely diverse from those living in other southern hemisphere seas [9,10,11].
Sponges are sessile and filter-feeder organisms, belonging to the phylum Porifera, which represent, in terms of abundance and biomass, the major component of the Antarctic zoobenthos [12], with a total number of 400 known species [13]. Through their aquiferous system, they are able to capture several microorganisms (including bacteria, yeasts, microalgae) from surrounding water and harbour a huge microbial community within their body [14,15,16]. Sponges normally establish a strong interaction with their bacterial hosts due to several benefits that improve their fitness and survival, including nutritional supply, transport of waste products, and molecules that confer chemical and mechanical defence [17,18,19].
Although a good knowledge is available on sponge fauna, the Antarctic region covers an extraordinarily wide area that makes some zones almost unknown to the scientific community [20,21]. Until now, a few studies have investigated the composition of microbial communities living within Antarctic sponges [22,23,24,25,26,27,28,29,30]. Some of these studies have demonstrated that Antarctic sponges are mostly dominated by Proteobacteria and Bacteroidetes [23,26,29,31]. Interestingly, species composition has been found to be strictly specific, probably regulated by several bioactive molecules and quorum sensing [14,31,32,33,34,35]. Since several studies have revealed that symbiotic bacteria are able to produce bioactive metabolites (reviewed by Brinkmann et al. [36]), studying the species composition of microbiomes could shed light on the possible biotechnological applications of sponges. This scientific question becomes much more attractive when addressed to Antarctic species, which are still largely unknown, and might reserve a great potential considering that they have undergone incredible adaptations. Interactions between sponges and microorganisms may occur in many forms, representing these microorganisms’ food sources, pathogens/parasites, or mutualistic symbionts [37,38,39,40]. Microbial associates can represent up to 40% of sponge tissue volume. Furthermore, the diversity in types of interactions may be matched by the phylogenetic diversity of microbes that occur within host sponges.
In the present work, we aimed to enlarge the yet scant knowledge on the bacterial communities inhabiting Antarctic sponges. In particular, we collected four sponges from two different sites of Tethys Bay (Victoria Land, Antarctica) in the framework of the XXXIV Italian National Antarctic Research Program (PNRA) expedition. Victoria Land (Tethys Bay) belongs to the Antarctic Specially Protected Area n. 161 [41] (ASPA 161; https://www.ats.aq/devAS/Meetings/Measure/688 accessed on 29 January 2021). Sponge species were characterized by morphological and molecular analysis. Metagenomic DNA extraction and Illumina MiSeq analysis were applied in order to identify the associated communities living within the analysed sponges. More than five hundred bacterial isolates were phylogenetically identified to establish whether the associated bacterial communities were host-specific. By relying on Amplicon Sequence Variants (ASVs) data, the biotechnological potential of sponge specimens was also considered.
2. Results
2.1. Species Identification
2.1.1. Morphological Analysis
The four sponge specimens belonged to the class Demospongiae and the following two orders: Haplosclerida with three species, Hemigellius pilosus (Kirkpatrick, 1907), Microxina sarai (Calcinai and Pansini, 2000), and Haliclona (Rhizoniera) dancoi (Topsent, 1901), and Poecilosclerida with one species, Mycale (Oxymycale) acerata (Kirkpatrick, 1907) (Table 1).
Table 1.
Sites, sample IDs, species identification, MNA code, geographic coordinates, sampling method in meters (m) and depth.
| Site | Sample ID | Sponge Taxonomy | MNA code | Sampling Method | Sampling Depth (m) | Coordinates |
|---|---|---|---|---|---|---|
| 1 | B4 | Mycale (Oxymycale) acerata (Kirkpatrick, 1907) | 13264 | SCUBA | 26 | 74°42.067’S 164°02.518’E |
| 2 | C6 | Haliclona (Rhizoniera) dancoi (Topsent, 1901) | 13265 | SCUBA | 28 | 74°40.537’S 164°04.169’E |
| 2 | D4 | Hemigellius pilosus (Kirkpatrick, 1907) | 13266 | SCUBA | 28 | 74°40.537’S 164°04.169’E |
| 2 | D6 | Microxina sarai (Calcinai & Pansini, 2000) | 13267 | SCUBA | 28 | 74°40.537’S 164°04.169’E |
2.1.2. Molecular Analysis
BLAST similarity search totally agreed with the morphological identification obtained for B4 and D4 samples. Molecular analysis confirmed B4 species as M. (Oxymycale) acerata, with CO1 primers that were the most specific (98% of pairwise identity) in comparison to 18S, 28S and ITS molecular markers. Similarly, CO1 also appeared to be the best molecular marker for the identification of the sponge D4, with a highest sequence similarity to H. pilosus (98% sequence identity). Regarding sample C6, molecular markers identified the genus corresponding to Haliclona, with the most striking result achieved using the 18S marker (92% similarity to Haliclona sp.). Unfortunately, it was not possible to identify this sponge at the species level, because there are no other available sequences on GenBank for H. (Rhizoniera) dancoi. Similarly, the results achieved with sample D6 were partially unclear, since several genera at low-sequence similarity were observed from BLAST outputs. In fact, the sequences of M. sarai, identified by spicule observations, are still not uploaded in GenBank (see Tables S1–S4; details on the alignments are reported in Figures S1–S3).
2.2. Metataxonomic Data Analysis
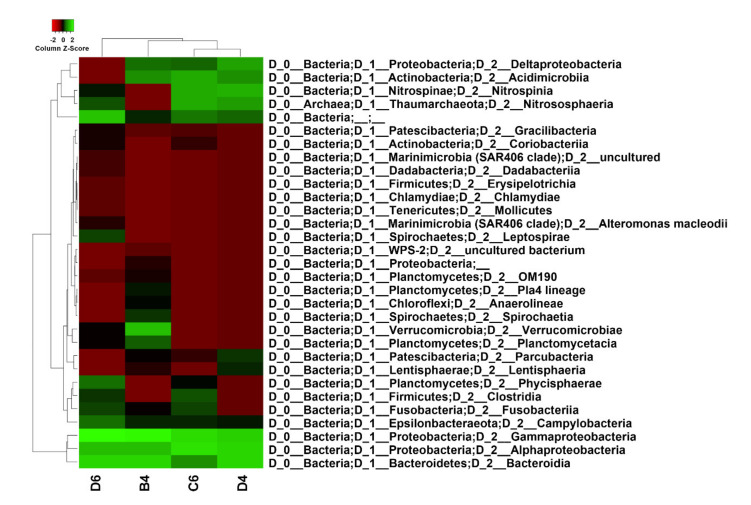
ASVs analysis was conducted considering those reporting a percentage of confidence ≥ 75%. The sponge M. (Oxymycale) acerata (B4) hosted the greatest abundance of bacterial taxa (250 ASVs), while H. pilosus (D4), H. (Rhizoniera) dancoi (C6) and M. sarai (D6) showed 47, 55 and 120 ASVs, respectively (Tables S5–S8). Overall, concerning the taxonomic profiling, sponge samples were all dominated by Gammaproteobacteria, Alphaproteobacteria and Bacteroidia (Figure 1).
Figure 1.
Heatmap of taxon relative abundance using taxonomic profiling, showing that sponge samples were all dominated by Gammaproteobacteria, Alphaproteobacteria and Bacteroidia. Sample code: B4 = M. (Oxymycale) acerata; D4 = H. pilosus, D6 = M. sarai, C6 = H. (Rhizoniera) dancoi. Scaling was done by column and clustering was performed using average linkage method and Manhattan distance measurement. Values were normalized as Log10.
In addition, M. (Oxymycale) acerata, H. pilosus and H. (Rhizoniera) dancoi revealed an abundance of both Deltaproteobacteria and Acidimicrobiia. Manhattan algorithm indicated that M. sarai clustered separately in comparison to the others, with H. pilosus and H. (Rhizoniera) dancoi resulting as the most similar in terms of species structure and abundance (Figure 1).
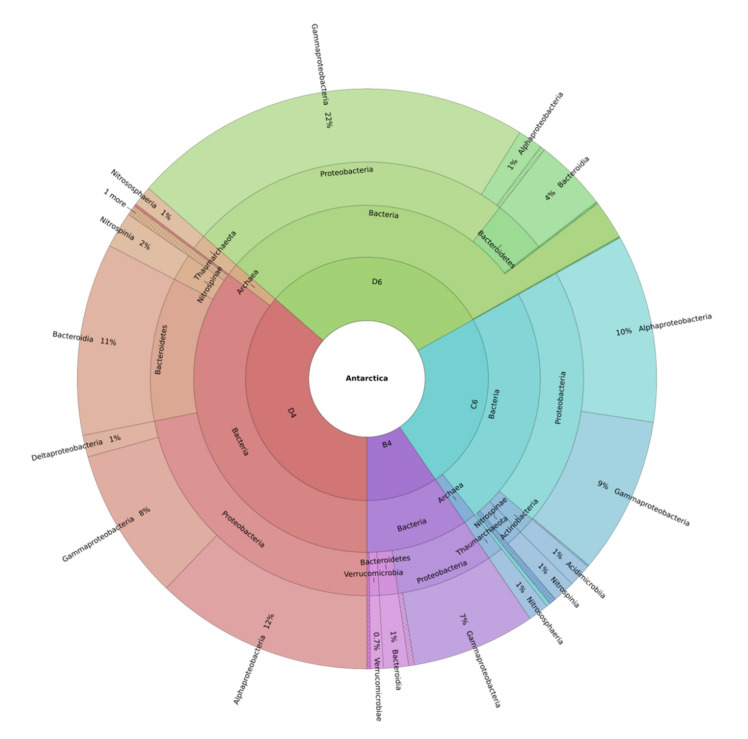
More specifically, a high relative frequency of Gammaproteobacteria and Bacteroidia were found in M. (Oxymycale) acerata (61.5% and 19%, respectively) and M. sarai (71% and 14%, respectively) (Figure 2; see also Figure S4).
Figure 2.
Krona Plot at the class level. Gammaproteobacteria of the genus Endozoicomonas were identified from M. (Oxymycale) acerata as well as Gammaproteobacteria belonging to the genus Colwellia and some bacterial strains classified as Fulvivirga and Ulvibacter, two genera included into Bacteroidetes. Bacteria of the family Rhodobacteraceae (class Alphaproteobacteria) were identified in H. (Rhizoniera) dancoi and H. pilosus. M. sarai was the only species showing a relative abundance of Polaribacter, an additional species grouped into the Bacteroidetes phylum. Sample code: B4 = M. (Oxymycale) acerata; D4 = H. pilosus, D6 = M. sarai, C6 = H. (Rhizoniera) dancoi. “1 more” corresponds to Acidicrodobiia group, which is present in trace levels.
On the contrary, a lower percentage (5–7%) of Alphaproteobacteria was detected in both species. In addition, M. (Oxymycale) acerata revealed 2–7% of bacteria belonging to Acidimicrobiia and Verrucomicrobiae classes, while lower percentages (~1%) of other bacterial phyla (Proteobacteria, Epsilonbacteraeota, Planctomycetes) together with 7% of an unknown phylum were recorded in M. sarai (Figure S4).
As reported above (Figure 1), the sponges H. (Rhizoniera) dancoi and H. pilosus revealed a similar composition in bacterial species distribution. In fact, a high abundance of Alphaproteobacteria (44% in H. (Rhizoniera) dancoi and 33.2% in H. pilosus) and Gammaproteobacteria (37% in H. (Rhizoniera) dancoi and 24% H. pilosus) was observed in both species. Moreover, lower percentages (0.5–5%) of additional taxa were recorded, including the Nitrospinia, Nitrososphaeria, Acidimicrobiia and Deltaproteobacteria groups. A huge difference was detected for bacteria belonging to the class Bacteroidia, since a higher relative abundance was found in H. pilosus (14%) in comparison to H. (Rhizoniera) dancoi (2%) (Figure S4).
3. Discussion
In the present study, we analyzed the species composition and abundance of the associated microbiota from four Antarctic sponges, M. (Oxymycale) acerata, H. (Rhizoniera) dancoi, H. pilosus and M. sarai, collected from Tethys Bay (Victoria Land, Antarctica). In particular, the associated community of M. (Oxymycale) acerata, collected from site 1 (Table 1), was similar to H. (Rhizoniera) dancoi and H. pilosus (Figure 1), retrieved from site 2 (Table 1). Interestingly, at this latter site, we collected the sponge M. sarai, whose species abundance was found statistically different by Manhattan clustering analysis (Figure 1).
As reported in most investigations focusing on sponge-associated bacteria [42,43,44,45,46,47], taxonomic profiling showed that Proteobacteria and Bacteroidetes dominated the four Antarctic sponges (Figure 2; Tables S5–S8). Previous studies identified these bacterial groups from M. (Oxymycale) acerata and other Antarctic species by metagenomic approaches [23,27,28,30,31,32,48]. These bacteria were frequently found to be the dominant bacterial phyla in marine ecosystems [49]. In particular, Proteobacteria showed different functions in host, including nitrogen fixation, and were involved in host defense mechanisms [50]. Furthermore, some bacteria were described as highly specialized hydrocarbon degrading microorganisms [51,52] and their wide distribution may be due to a strong positive interaction in environments where bacteria represent a fundamental source of nutrients, such as the case of Antarctica. This finding could be corroborated by results revealing that these bacteria are able to adapt to extreme environments, including polar habitats [53,54,55,56]. Concerning their biotechnological potential, genome-mining approaches reported several biosynthetic gene clusters (BGCs) encoding for bioactive molecules from marine Proteobacteria (reviewed by Buijs et al. [57]). However, there is no direct 100% correlation between the presence of a certain BGC, a bacterial genus and a bioactive metabolite. BGCs can be silent in certain conditions and, hence, methods should be developed to unlock their silent potential [58], to observe the production of a particular compound and induce the desired bioactivity. The most common approach known to discover new metabolites is the “OSMAC” (one strain many compounds) approach. The term OSMAC was coined for the first time by Zeeck and co-workers [59], indicating the ability of single strains to produce different metabolites when cultivated under different conditions. Examples are the use of different culturing strategies to trigger the production of secondary metabolites such as changing culturing conditions (e.g., nutrients or light exposure), mimicking environmental stressors and co-culturing with other species.
On the whole, several species belonging to Gammaproteobacteria and Alphaproteobacteria isolated from sponges and soils showed antibacterial, antiviral, antifungal and antiprotozoal activities that make them suitable tools in drug discovery research fields [36,43,60,61,62,63]. In particular, the Gammaproteobacteria of the genus Endozoicomonas, identified from M. (Oxymycale) acerata in the present work (Figure 2; Table S5), was found to induce antimicrobial activities [64,65].
Always M. (Oxymycale) acerata showed a relative abundance of a Gammaproteobacteria belonging to the genus Colwellia (Figure 2; Table S5), which is extremely interesting since it was recently proposed as a useful tool for the bioremediation of nitrogen pollutants [66]. Previous investigations also demonstrated that a sponge-associated Colwellia sp. produces several extracellular polymeric substances (EPSs) with potential use in the production of cosmeceutical and nutraceutical ingredients [35,67].
M. (Oxymycale) acerata also revealed some bacterial strains classified as Fulvivirga and Ulvibacter, two genera included into Bacteroidetes, the second most abundant phylum found in the samples under analysis (Figure 2; Table S5). Genome-mining approaches coupled to chemical analyses revealed the presence of some amine acylated desferrioxamine siderophores from Fulvivirga sp. with anticancer properties [68]. Similarly, Ulvibacter species, already observed in Antarctic habitats [69], belong to the family Flavobacteriaceae, whose biotechnological applications are well-documented. In fact, several polysaccharide-digesting enzymes together with antibiotics and other bioactive compounds, such as quercetin (known for its antioxidant, anti-inflammatory, chemopreventive properties), were isolated [70,71].
The sequencing of 16S regions revealed that M. (Oxymycale) acerata was the sole species hosting a certain abundance of Rubritalea strains (phylum Verrucomicrobia) (Figure 2; Table S5). This bacterial group was already observed in other sponge species, from which some BGCs encoding for PKSs (polyketide synthases) were identified [42,72,73,74,75,76,77]. Verrucomicrobia, coupled with Planctomycetes and Chlamydiae, was classified in the PVC (Planctomycetes, Verrucomicrobia, and Chlamydiae) superphylum, which is known to include a wide number of species with biotechnological potential [78,79,80]. The finding of these bacteria within the symbiotic community of M. (Oxymycale) acerata may be extremely attractive since several bioactive molecules, such as carotenoids and squalene, were found in several bacteria belonging to the genus Rubritalea [72,73,74,81,82,83,84]. The potential capability to produce biotechnologically relevant compounds was also demonstrated by genomic analyses, revealing some genes involved in defense mechanisms mediated by toxin-antitoxin systems from sponge-associated verrucomicrobial strains [77].
ASV’s data showed bacteria of the family Rhodobacteraceae (class Alphaproteobacteria) in H. (Rhizoniera) dancoi and H. pilosus (Figure 2; Tables S6 and S7). Several genera were recognized as a huge source of novel bioactives, especially Pseudovibrio species living in seawater and through symbiotic relationships with sponges, tunicates and corals [85,86]. For example, H. pilosus specifically hosted the genus Roseobacter, which was also studied for its antimicrobial properties [87,88]. The hydrocarbon-degrading Gammaproteobacteria of the genus Pseudohongiella, with potential use in the bioremediation of anthropogenic contaminants [89,90], were also revealed in H. (Rhizoniera) dancoi and H. pilosus (Figure 2; Tables S6 and S7).
Less abundant members living within H. pilosus and H. (Rhizoniera) dancoi belonged to the classes Nitrospinia (phylum Nitrospinae) and Nitrosophaeria (phylum Thaumarchaeota) (Figure 2; Tables S6 and S7). Recent investigations already reported low percentages of Nitrospinia from H. pilosus, H. (Rhizoniera) dancoi and other Antarctic species [26,29]. Concerning the capability to produce molecules with biotechnological potential, very little information is available so far. In a recent study, BLASTp search against the Integrated Microbial Genomes (IMG) database identified a Pseudoalteromonas luteoviolacea gene encoding for a l-amino acid oxidase (LAAO) with antimicrobial properties in the genome of a strain belonging to the phylum Nitrospinae [91].
The sponges under analysis had low percentages of Acidimicrobiia (phylum Actinobacteria), except for M. sarai (Tables S5–S8). According to our results, this bacterial class was recently reported from H. (Rhizoniera) dancoi, H. pilosus and other Antarctic sponges by metagenomic analysis [27,28]. Acidimicrobiia were widely observed in marine sponges, particularly from tropical species [63,92,93,94,95,96,97]. Similar to Proteobacteria, several studies demonstrated the great biotechnological potential of Actinobacteria, especially those belonging to the Streptomyces genus. In fact, several bioactive compounds with antimicrobial, antiviral, antiparasitic, antiprotozoal and antitumor effects have been described [98,99,100,101,102,103,104,105,106,107]. Moreover, genomic analyses revealed some BGCs encoding for secondary metabolites, such as PKS I and III, NRPS (nonribosomal peptides), terpene and bacteriocin gene clusters from a sponge-derived Actinobacteria showing antimicrobial activities [108,109,110,111,112].
ASV’s analysis of the three sponges, M. (Oxymycale) acerata, H. pilosus and H. (Rhizoniera) dancoi, displayed Deltaproteobacteria (Figure 2; Tables S5–S7) belonging to the phylum Proteobacteria, that, as mentioned above, produce interesting bioactive metabolites [57]. For instance, H. pilosus exhibited some strains included into the genus Bdellovibrio (Figure 3; Table S7), which is an obligate predator of other Gram-negative bacteria that was proposed for possible biotechnological applications toward medicinal, agricultural and industrial fields [113,114,115].
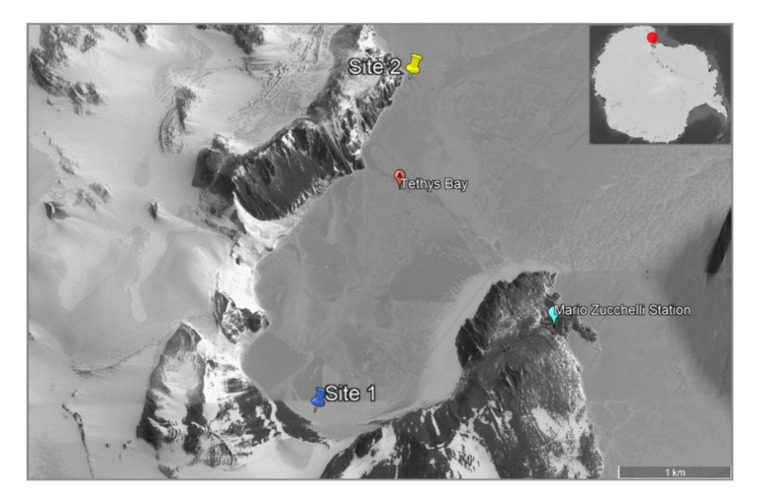
Figure 3.
Map of Tethys Bay (Victoria Land, Antarctica). The collection sites were reported as blue (site 1) and yellow (site 2) icons. Scale bar = 1 km.
M. sarai was the only species showing a relative abundance of Polaribacter, an additional species grouped into the Bacteroidetes phylum (Figure 2; Table S8). Some data demonstrated that these cold-adapted bacteria produced interesting EPSs molecules with protective effects on human skin and anti-aging properties [116,117].
4. Materials and Methods
4.1. Sponge Collection
Four sponge samples, reported as B4, C6, D4 and D6, were collected by scuba divers in November–December 2018 at two sites of Tethys Bay: 1) B4 at 26 metres of depth (74°42.067′ S, 164°02.518′ E) and 2) C6, D4 and D6 at 28 metres of depth (74°40.537′ S, 164°04.169′ E) (Figure 3). Samples were immediately washed at least three times with filter-sterilized natural seawater to remove transient and loosely attached bacteria and/or debris [14,27]. Firstly, a small fragment of each sponge was preserved in 70% ethanol for taxonomic identification. Specimens were then placed into individual sterile tubes and kept in RNAlater© at −20° C until transported to the Stazione Zoologica Anton Dohrn (Naples, Italy).
Sponge slides of spicules are deposited at the Italian National Antarctic Museum (MNA, Section of Genoa, Italy). The MNA voucher codes of the sponges investigated in the present work are reported in Table 1.
4.2. Morphological Analysis of Spicules
The taxonomic identification was conducted at the species level. Small fragments of each sponge were heat-dissolved in nitric acid, rinsed in water and dehydrated in ethanol. Then, spicules were mounted on slides for microscopic analyses, following standard methods [118]. The skeletal architecture was examined under light microscope and hand-cut sections of sponge portions were made as described in Hooper [119].
The taxonomic classification follows the updated nomenclature reported in the World Porifera Database (WPD) [120].
4.3. DNA Extraction and PCR Amplification
About 10 mg of tissue were used for DNA extraction by using QIAamp® DNA Micro kit (QIAGEN), according to the manufacturer’s instructions. DNA quantity (ng/μL) was evaluated by NanoDrop spectrophotometer. PCR reactions were performed on the C1000 Touch Thermal Cycler (BioRad) in a 30 µL reaction mixture final volume including about 50–100 ng of genomic DNA, 6 µL of 5X Buffer GL (GeneSpin Srl, Milan, Italy), 3 µL of dNTPs (2 mM each), 2 µL of each forward and reverse primer (25 pmol/µL, Table 1), 0.2 µL of Xtra Taq Polymerase (5 U/µL, GeneSpin Srl, Milan, Italy), using different PCR programs for 18S and 28S, ITS and CO1 as follows:
(i) for 18S and 28S, a denaturation step at 95 °C for 2 min, [35 cycles denaturation step at 95 °C for 1 min, annealing step at 60 °C (A/B, [121]), 57 °C (C2/D2, [122]), 55 °C (18S-AF/18S-BR, NL4F/NL4R, [123]) for 1 min and 72 °C of primer extension for 2 min], a final extension step at 72 °C for 10 min;
(ii) ITS primers (RA2/ITS2.2, [121]), a first denaturation at 95 °C for 2 min, [35 cycles denaturation step at 95 °C for 1 min, annealing step at 67 °C for 1 min and 72 °C of primer extension for 2 min], a final extension step at 72 °C for 10 min.
(iii) CO1 primers (dgLCO1490/dgHCO2198, [124]), a first denaturation at 94 °C for 3 min, [35 cycles of denaturation at 94 °C for 30 sec, annealing at 45 °C for 30 sec and primer extension at 72 °C for 1 min].
Sequences of PCR primers are reported in Supporting Information (Table S9). PCR products were run on 1.5% agarose gel and the fragment length was evaluated by using 100 bp DNA ladder (GeneSpin Srl, Milan, Italy). PCR products were purified using QIAquick Gel Extraction Kit (Qiagen), according to the manufacturer’s instructions. PCR amplicons were then sequenced in both strands through Applied Biosystems (Life Technologies) 3730 Analyzer (48 capillaries). Sequences produced were ~300–650 bases long in average with more than 97.5% accuracy, starting from PCR fragments. The total 18S, 28S, ITS and CO1 region were submitted to GenBank using Basic Local Alignment Search Tool (BLAST) [125] and then aligned with highly similar sequences using MultiAlin (http://multalin.toulouse.inra.fr/multalin/ accessed on 29 January 2021) [126].
4.4. Metagenomic DNA Extraction and Illumina MiSeq Sequencing
Genomic DNA for 16S rRNA sequencing was performed from about 250 mg of tissue by using DNeasy® PowerSoil® Pro Kit (QIAGEN), according to the manufacturer’s instructions. Extractions were performed using both internal and external sponge tissue in order to obtain the whole bacterial community. DNA quantity (ng/μL) and quality (A260/280, A260/230) were evaluated by NanoDrop spectrophotometer, whereas DNA integrity was checked on 0.8% agarose gel electrophoresis in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0). 20 µL of samples (30 ng/μL final concentration) were subjected to 16S V3-V4 rRNA gene library preparation and sequencing (Bio-Fab Research, Rome, Italy). Illumina adapters overhang nucleotide sequences were added to the gene specific primer sequences targeting the V3-V4 region [127]. After 16S amplification, a PCR clean-up was done to purify the V3-V4 amplicon from free primers and primer-dimer species. A subsequent limited cycle amplification step was performed to add multiplexing indices and Illumina sequencing adapters by using a Nextera XT Index Kit. Finally, libraries were normalized and pooled by denoising processes (Table S10), and sequenced on Illumina MiSeq Platform with 2x300 bp paired-end reads. Taxonomy was assigned using “home made” Naive Bayesian Classifier trained on V3-V4 16S sequences of SILVA 132 database [128]. QIIME 2 (Quantitative Insights Into Microbial Ecology) platform [129] was used for microbiome analysis from raw DNA sequencing data. QIIME analysis workflow was performed by demultiplexing, quality filtering, chimera removal and taxonomic assignment. The full dataset of raw data has been deposited in the SRA database (submission ID: SUB8701897; BioProject ID: PRJNA687362).
4.5. Statistical Analysis
ASVs distribution and frequency in the whole dataset and for each sample are reported in the Supporting Information (Figures S5 and S6).
Heatmap was generated by using Heatmapper Software available at http://www.heatmapper.ca/ accessed on 29 January 2021 [130]. The number of features observed for each identified taxa were normalized as Log10 and scaled by column. Hierarchical clustering was applied on both rows and columns by average linkage method. For computing distance between rows and columns, Manhattan distance measurement algorithm was performed.
5. Conclusions
Our metataxonomic analysis highlights the occurrence of dominant and locally enriched microbes in the Antarctic sponges M. (Oxymycale) acerata, H. (Rhizoniera) dancoi, H. pilosus and M. sarai, characterized by morphological and molecular approaches. This can be considered a starting point in the understanding of the global Antarctic microbiome in a more complete perspective, given the scarce information in the literature for extreme environments such as the Antarctica. According to the microbial community identified, the biotechnological value should not be underestimated. In fact, our findings open new perspectives concerning the possible role of these Antarctic sponges and their symbiotic bacteria as a source of bioactive compounds. Further studies will be devoted to bioassay-guided fractionations for identifying new potential drugs useful in pharmaceutical, nutraceutical and cosmeceutical fields.
Acknowledgments
Roberta Esposito was supported by a PhD (PhD in Biology, University of Naples Federico II) fellowship funded by the Photosynthesis 2.0 project of the Stazione Zoologica Anton Dohrn.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/19/3/173/s1, Figure S1. Alignments of CO1 (PorCOI2fwd/PorCOI2rev) sequence from B4 sponge with (a) the first BLAST hit Asbestopluma lycopodium and (b) the sequence of M. acerata displaying low query cover, Figure S2: Alignment of CO1 (dgLCO1490/dgHCO2198) sequence from B4 sponge with the first BLAST hit (M. acerata), Figure S3: Alignment of CO1 (dgLCO1490/dgHCO2198) sequence from D4 sponge with the first BLAST hit (H. pilosus), Figure S4. Krona plot at increasing complexity levels. Regnum (a), phylum (b), class (c), order (d), family (e), genus (f) and species (g) were reported. Figure S5. Distribution of ASV’s frequencies, Figure S6. Distribution of ASV’s frequencies for each sample (reported as a blue bar), Table S1. BLAST results from B4 sponge (Mycale (Oxymycale) acerata). The primer names, sequence length in base pairs (bp), first hits (highlighted in bold), hits at low significance displaying the correct species (where present), query cover and identity percentages (%) were reported, Table S2. BLAST results from C6 sponge (Haliclona (Rhizoniera) dancoi). The primer names, sequence length in base pairs (bp), first hits (highlighted in bold), hits at low significance displaying the correct species (where present), query cover and identity percentages (%) were reported, Table S3. BLAST results from D4 sponge (Hemigellius pilosus). The primer names, sequence length in base pairs (bp), first hits (highlighted in bold), hits at low significance displaying the correct species (where present), query cover and identity percentages (%) were reported, Table S4. BLAST results from D6 sponge (Microxina sarai). The primer names, sequence length in base pairs (bp), first hits (highlighted in bold), query cover and identity percentages (%) were reported, Table S5. ASVs found from M. (Oxymycale) acerata with percentage of confidence ≥ 75%, Table S6. ASVs found from H. (Rhizoniera) dancoi with percentage of confidence ≥ 75%, Table S7. ASVs found from H. pilosus with percentage of confidence ≥ 75%, Table S8. ASVs found from M. sarai with percentage of confidence ≥ 75%, Table S9. Targeted region, forward and reverse names, sequences (5’→ 3’) and reference of primers pairs used for molecular characterization, Table S10. Denoising process.
Author Contributions
Molecular identification of sponges, data curation, formal analysis, N.R. and R.E.; Morphological analysis of sponges, M.B.; collection of sponge samples in the framework of XXXIV Italian National Antarctic Research Program (PNRA) expedition, G.Z.; metataxonomic analysis, M.S., F.A.; data curation, formal analysis, investigation, reviewing and editing, D.C., G.N., C.L., A.F.; funding acquisition, writing—reviewing and editing, D.G., A.I., C.V.; conceptualization, funding acquisition, supervision, writing—original draft, M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by PNRA16_00043, Cosmeceuticals and Nutraceuticals From Antarctic Biological REsources (CAN FARE). It was carried out in the framework of the SCAR Programme “Antarctic Thresholds–Ecosystem Resilience and Adaptation” (AnT-ERA).
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Peck L.S. Antarctic marine biodiversity: Adaptations, environments and responses to change. In: Hawkins S.J., Evans A.J., Dale A.C., Firth L.B., Smith I.P., editors. Oceanography and Marine Biology: An Annual Review. Volume 56. CRC Press, Taylor & Francis Group; Boca Raton, FL, USA: 2018. pp. 2–133. [Google Scholar]
- 2.Arnaud P. Téthys. Volume 6. Station Marine d’Endoume, Centre d’Océanographie de Marseille; Marseille, France: 1974. Contribution à la bionomie marine benthique des régions antarctiques et subantarctiques; pp. 465–656. [Google Scholar]
- 3.Konecki J.T., Targett T.E. Eggs and larvae of Nototheniops larseni from the spongocoel of a hexactinellid sponge near Hugo island, Antarctic Peninsula. Polar Biol. 1989;10:197–198. doi: 10.1007/BF00238495. [DOI] [Google Scholar]
- 4.Pearse J.S., Mcclintock J.B., Bosch I. Reproduction of Antarctic benthic marine invertebrates: Tempos, modes, and timing. Integr. Comp. Biol. 1991;31:65–80. doi: 10.1093/icb/31.1.65. [DOI] [Google Scholar]
- 5.Dayton P.K. Polar benthos. In: Smith W.O., editor. Polar oceanography-Part B-Chemistry, Biology and Geology. Academic Press; London, UK: 1990. pp. 631–685. [Google Scholar]
- 6.Dayton P.K., Mordida B.J., Bacon F. Polar marine communities. Am. Zool. 1994;34:90–99. doi: 10.1093/icb/34.1.90. [DOI] [Google Scholar]
- 7.Thompson M.R.A., Crame J.A., Thompson J.W. Geological Evolution of Antarctica. Cambridge University Press; Cambridge, UK: 1991. [Google Scholar]
- 8.Tingey R. The Geology of Antarctica. Oxford University Press; Oxford, UK: 1991. [Google Scholar]
- 9.Clarke A. Is there a latitudinal diversity cline in the sea? Trends Ecol. Evol. 1992;7:286–287. doi: 10.1016/0169-5347(92)90222-W. [DOI] [PubMed] [Google Scholar]
- 10.Crame J., Clarke A. The historical component of marine taxonomic diversity gradients. In: Ormond R.F.G., Gage J.D., Angel M.V., editors. Marine Biodiversity: Patterns and Processes. Cambridge University Press; Cambridge, UK: 1997. pp. 258–273. [Google Scholar]
- 11.Gray J.S. Antarctic marine benthic biodiversity in a world-wide latitudinal context. Polar Biol. 2001;24:633–641. doi: 10.1007/s003000100244. [DOI] [Google Scholar]
- 12.McClintock J.B., Amsler C.D., Baker B.J., Van Soest R.W.M. Ecology of antarctic marine sponges: An overview. Integr. Comp. Biol. 2005;45:359–368. doi: 10.1093/icb/45.2.359. [DOI] [PubMed] [Google Scholar]
- 13.Janussen D., Downey R. V Porifera. In: De Broyer C., Koubbi P., editors. Biogeographic Atlas of the Southern Ocean. SCAR; Cambridge, UK: 2014. pp. 1–10. [Google Scholar]
- 14.Mangano S., Michaud L., Caruso C., Brilli M., Bruni V., Fani R., Lo Giudice A. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: A preliminary analysis. Res. Microbiol. 2009;160:27–37. doi: 10.1016/j.resmic.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Hentschel U., Piel J., Degnan S.M., Taylor M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012;10:641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 16.Bayer K., Kamke J., Hentschel U. Quantification of bacterial and archaeal symbionts in high and low microbial abundance sponges using real-time PCR. FEMS Microbiol. Ecol. 2014;89:679–690. doi: 10.1111/1574-6941.12369. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y.K., Lee J.H., Lee H.K. Microbial symbiosis in marine sponges. J. Microbiol. 2001;39:254–264. [Google Scholar]
- 18.Thoms C., Horn M., Wagner M., Hentschel U., Proksch P. Monitoring microbial diversity and natural product profiles of the sponge Aplysina cavernicola following transplantation. Mar. Biol. 2003;142:685–692. doi: 10.1007/s00227-002-1000-9. [DOI] [Google Scholar]
- 19.Hentschel U., Usher K.M., Taylor M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006;55:167–177. doi: 10.1111/j.1574-6941.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- 20.Calcinai B., Pansini M. Four new demosponge species from Terra Nova Bay (Ross Sea, Antarctica) Zoosystema. 2000;22:369–381. [Google Scholar]
- 21.Ríos P., Cristobo F.J., Urgorri V. Poecilosclerida (Porifera, Demospongiae) collected by the Spanish Antarctic expedition BENTART-Cah. Biol. Mar. 2004;45:97–119. [Google Scholar]
- 22.Dayton P.K., Robilliard G.A., Paine R.T., Dayton L.B. Biological accommodation in the benthic community at McMurdo Sound, Antarctica. Ecol. Monogr. 1974;44:105–128. doi: 10.2307/1942321. [DOI] [Google Scholar]
- 23.Cárdenas C.A., Font A., Steinert G., Rondon R., González-Aravena M. Temporal stability of bacterial communities in Antarctic sponges. Front. Microbiol. 2019;10:1–14. doi: 10.3389/fmicb.2019.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo Giudice A., Rizzo C. Bacteria associated with marine benthic invertebrates from polar environments: Unexplored frontiers for biodiscovery? Diversity. 2018;10:80. doi: 10.3390/d10030080. [DOI] [Google Scholar]
- 25.Lo Giudice A., Azzaro M., Schiaparelli S. Microbial symbionts of Antarctic marine benthic invertebrates. In: Castro-Sowinski S., editor. The Ecological Role of Micro-Organisms in the Antarctic Environment. Springer International Publishing; New York, NY, USA: 2019. pp. 277–296. [Google Scholar]
- 26.Steinert G., Wemheuer B., Janussen D., Erpenbeck D., Daniel R., Simon M., Brinkhoff T., Schupp P.J. Prokaryotic diversity and community patterns in antarctic continental shelf sponges. Front. Mar. Sci. 2019;6:1–15. doi: 10.3389/fmars.2019.00297. [DOI] [Google Scholar]
- 27.Savoca S., Lo Giudice A., Papale M., Mangano S., Caruso C., Spanò N., Michaud L., Rizzo C. Antarctic sponges from the Terra Nova Bay (Ross Sea) host a diversified bacterial community. Sci. Rep. 2019;9:16135. doi: 10.1038/s41598-019-52491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno-Pino M., Cristi A., Gillooly J.F., Trefault N. Characterizing the microbiomes of Antarctic sponges: A functional metagenomic approach. Sci. Rep. 2020;10:645. doi: 10.1038/s41598-020-57464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papale M., Rizzo C., Fani R., Bertolino M., Costa G., Paytuví-Gallart A., Schiaparelli S., Michaud L., Azzaro M., Lo Giudice A. Exploring the diversity and metabolic profiles of bacterial communities associated with Antarctic sponges (Terra Nova Bay, Ross Sea) Front. Ecol. Evol. 2020;8:1–16. doi: 10.3389/fevo.2020.00268. [DOI] [Google Scholar]
- 30.Sacristán-Soriano O., Pérez Criado N., Avila C. Host species determines symbiotic community composition in Antarctic sponges (Porifera: Demospongiae) Front. Mar. Sci. 2020;7:1–36. doi: 10.3389/fmars.2020.00474. [DOI] [Google Scholar]
- 31.Rodríguez-Marconi S., De La Iglesia R., Díez B., Fonseca C.A., Hajdu E., Trefault N., Webster N. Characterization of bacterial, archaeal and eukaryote symbionts from antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PLoS ONE. 2015;10:1–19. doi: 10.1371/journal.pone.0138837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster N.S., Negri A.P., Munro M.M.H.G., Battershill C.N. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 2004;6:288–300. doi: 10.1111/j.1462-2920.2004.00570.x. [DOI] [PubMed] [Google Scholar]
- 33.Mangano S., Michaud L., Caruso C., Lo Giudice A. Metal and antibiotic resistance in psychrotrophic bacteria associated with the Antarctic sponge Hemigellius pilosus (Kirkpatrick, 1907) Polar Biol. 2014;37:227–235. doi: 10.1007/s00300-013-1426-1. [DOI] [Google Scholar]
- 34.Mangano S., Caruso C., Michaud L., Lo Giudice A. First evidence of quorum sensing activity in bacteria associated with Antarctic sponges. Polar Biol. 2018;41:1435–1445. doi: 10.1007/s00300-018-2296-3. [DOI] [Google Scholar]
- 35.Caruso C., Rizzo C., Mangano S., Poli A., di Donato P., Finore I., Nicolaus B., di Marco G., Michaud L., Lo Giudice A. Production and biotechnological potential of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol. 2018;84:e01624-17. doi: 10.1128/AEM.01624-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkmann C.M., Marker A., Kurtböke D.I. An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity. 2017;9:40. doi: 10.3390/d9040040. [DOI] [Google Scholar]
- 37.Taylor M.W., Radax R., Stegar D., Wagner M. Sponge-associated microorganisms: Evolution, evology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster N.S., Taylor M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 39.Thacker R.W., Freeman C.J. Advances in Marine Biology. Volume 62. Academic Press; Cambridge, MA, USA: 2012. Sponge-microbe symbioses. Recent advances and new directions; pp. 57–111. [DOI] [PubMed] [Google Scholar]
- 40.Björk J.R., Díez-Vives C., Coma R., Ribes M., Montoya J.M. Specificity and temporal dynamics of complex bacteria–sponge symbiotic interactions. Ecology. 2013;94:2781–2791. doi: 10.1890/13-0557.1. [DOI] [PubMed] [Google Scholar]
- 41.Hughes K.A., Pertierra L.R., Walton D.W.H. Area protection in Antarctica: How can conservation and scientific research goals be managed compatibly? Environ. Sci. Policy. 2013;31:120–132. doi: 10.1016/j.envsci.2013.03.012. [DOI] [Google Scholar]
- 42.Kennedy J., Codling C.E., Jones B.V., Dobson A.D.W., Marchesi J.R. Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase genes from the sponge metagenome. Environ. Microbiol. 2008;10:1888–1902. doi: 10.1111/j.1462-2920.2008.01614.x. [DOI] [PubMed] [Google Scholar]
- 43.Dupont S., Carré-Mlouka A., Descarrega F., Ereskovsky A., Longeon A., Mouray E., Florent I., Bourguet-Kondracki M.L. Diversity and biological activities of the bacterial community associated with the marine sponge Phorbas tenacior (Porifera, Demospongiae) Lett. Appl. Microbiol. 2014;58:42–52. doi: 10.1111/lam.12154. [DOI] [PubMed] [Google Scholar]
- 44.Karimi E., Ramos M., Gonçalves J.M.S., Xavier J.R., Reis M.P., Costa R. Comparative metagenomics reveals the distinctive adaptive features of the Spongia officinalis endosymbiotic consortium. Front. Microbiol. 2017;8:1–16. doi: 10.3389/fmicb.2017.02499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slaby B.M., Hackl T., Horn H., Bayer K., Hentschel U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017;11:2465–2478. doi: 10.1038/ismej.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Najafi A., Moradinasab M., Nabipour I. First record of microbiomes of sponges collected from the Persian Gulf, using tag pyrosequencing. Front. Microbiol. 2018;9:1–18. doi: 10.3389/fmicb.2018.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey M.A., Andreassend S.K., Bracegirdle J., Brown A., Keyzers R.A., Ackerley D.F., Northcote P.T., Owen J.G. Metagenomic exploration of the marine sponge Mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. MBio. 2020;11:1–16. doi: 10.1128/mBio.02997-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S., Zhang W., Ding W., Wang M., Fan S., Yang B., McMinn A., Wang M., Xie B.B., Qin Q.L., et al. Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics. Microbiome. 2020;8:47. doi: 10.1186/s40168-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens H., Stübner M., Simon M., Brinkhoff T. Phylogeny of Proteobacteria and Bacteroidetes from oxic habitats of a tidal flat ecosystem. FEMS Microbiol. Ecol. 2005;54:351–365. doi: 10.1016/j.femsec.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Bibi F., Alvi S.A., Al-Sofyani A., Naseer M.I., Yasir M., Azhar E.I. Pyrosequencing reveals sponge specific bacterial communities in marine sponges of Red Sea, Saudi Arabia. Saudi J. Biol. Sci. 2020;27:67–73. doi: 10.1016/j.sjbs.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joye S.B., Kleindienst S., Gilbert J.A., Handley K.M., Weisenhorn P., Overholt W.A., Kostka J.E. Responses of microbial communities to hydrocarbon exposures. Oceanography. 2016;29:136–149. doi: 10.5670/oceanog.2016.78. [DOI] [Google Scholar]
- 52.McGenity T.J. Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. Springer International Publishing; New York, NY, USA: 2019. [Google Scholar]
- 53.Hamdan L.J., Coffin R.B., Sikaroodi M., Greinert J., Treude T., Gillevet P.M. Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 2013;7:685–696. doi: 10.1038/ismej.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams T.J., Cavicchioli R. Marine metaproteomics: Deciphering the microbial metabolic food web. Trends Microbiol. 2014;22:248–260. doi: 10.1016/j.tim.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Shivaji S., Reddy G.S.N., Chattopadhyay M.K. Bacterial biodiversity, cold adaptation and biotechnological importance of bacteria occurring in Antarctica. Proc. Ind. Natl. Sci. Acad. USA. 2017;83:327–352. doi: 10.16943/ptinsa/2017/48956. [DOI] [Google Scholar]
- 56.Picazo A., Rochera C., Villaescusa J.A., Miralles-Lorenzo J., Velázquez D., Quesada A., Camacho A. Bacterioplankton community composition along environmental gradients in lakes from Byers peninsula (Maritime Antarctica) as determined by next-generation sequencing. Front. Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buijs Y., Bech P.K., Vazquez-Albacete D., Bentzon-Tilia M., Sonnenschein E.C., Gram L., Zhang S. Da Marine Proteobacteria as a source of natural products: Advances in molecular tools and strategies. Nat. Prod. Rep. 2019;36:1333–1350. doi: 10.1039/C9NP00020H. [DOI] [PubMed] [Google Scholar]
- 58.Reen F., Gutiérrez-Barranquero J., Dobson A., Adams C., O’Gara F. Mar. Drugs. Vol. 13. 2015. Emerging Concepts Promising New Horizons for Marine Biodiscovery and Synthetic Biology; pp. 2924–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 60.Santos O.C.S., Pontes P.V.M.L., Santos J.F.M., Muricy G., Giambiagi-deMarval M., Laport M.S. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res. Microbiol. 2010;161:604–612. doi: 10.1016/j.resmic.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Lee L.H., Cheah Y.K., Nurul Syakima A.M., Shiran M.S., Tang Y.L., Lin H.P., Hong K. Analysis of Antarctic protobacteria by PCR fingerprinting and screening for antimicrobial secondary metabolites. Genet. Mol. Res. 2012;11:1627–1641. doi: 10.4238/2012.June.15.12. [DOI] [PubMed] [Google Scholar]
- 62.Desriac F., Jégou C., Balnois E., Brillet B., Le Chevalier P., Fleury Y. Antimicrobial peptides from marine proteobacteria. Mar. Drugs. 2013;11:3632–3660. doi: 10.3390/md11103632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bibi F., Yasir M., Al-Sofyani A., Naseer M.I., Azhar E.I. Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EASaudi J. Biol. Sci. 2020;27:1139–1147. doi: 10.1016/j.sjbs.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haber M., Ilan M. Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J. Appl. Microbiol. 2014;116:519–532. doi: 10.1111/jam.12401. [DOI] [PubMed] [Google Scholar]
- 65.Rua C.P.J., Trindade-Silva A.E., Appolinario L.R., Venas T.M., Garcia G.D., Carvalho L.S., Lima A., Kruger R., Pereira R.C., Berlinck R.G.S., et al. Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera Brasiliensis. PeerJ. 2014;2014:1–14. doi: 10.7717/peerj.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J., Meng Y., Shi Y., Lin X. Complete genome sequences of Colwellia sp. Arc7-635, a denitrifying bacterium isolated from Arctic seawater. Curr. Microbiol. 2019;76:1061–1065. doi: 10.1007/s00284-019-01714-8. [DOI] [PubMed] [Google Scholar]
- 67.Casillo A., Parrilli E., Sannino F., Mitchell D.E., Gibson M.I., Marino G., Lanzetta R., Parrilli M., Cosconati S., Novellino E., et al. Structure-activity relationship of the exopolysaccharide from a psychrophilic bacterium: A strategy for cryoprotection. Carbohydr. Polym. 2017;156:364–371. doi: 10.1016/j.carbpol.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z.J., Zhou H., Zhong G., Huo L., Tang Y.J., Zhang Y., Bian X. Genome mining and biosynthesis of primary amine-acylated desferrioxamines in a marine gliding bacterium. Org. Lett. 2020;22:939–943. doi: 10.1021/acs.orglett.9b04490. [DOI] [PubMed] [Google Scholar]
- 69.Choi T.H., Lee H.K., Lee K., Cho J.C. Ulvibacter antarcticus sp. nov., isolated from Antarctic coastal seawater. Int. J. Syst. Evol. Microbiol. 2007;57:2922–2925. doi: 10.1099/ijs.0.65265-0. [DOI] [PubMed] [Google Scholar]
- 70.McBride M.J. The Family Flavobacteriaceae. In: Rosenberg E., De Long E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. Springer-Verlag; Berlin/Heidelberg, Germany: 2014. pp. 1–1028. [Google Scholar]
- 71.Enisoglu-Atalay V., Atasever-Arslan B., Yaman B., Cebecioglu R., Kul A., Ozilhan S., Ozen F., Cata T. Chemical and molecular characterization of metabolites from Flavobacterium sp. PLoS ONE. 2018;13:e0205817. doi: 10.1371/journal.pone.0205817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheuermayer M., Gulder T.A.M., Bringmann G., Hentschel U. Rubritalea marina gen. nov., sp. nov., a marine representative of the phylum “Verrucomicrobia”, isolated from a sponge (Porifera) Int. J. Syst. Evol. Microbiol. 2006;56:2119–2124. doi: 10.1099/ijs.0.64360-0. [DOI] [PubMed] [Google Scholar]
- 73.Kasai H., Katsuta A., Sekiguchi H., Matsuda S., Adachi K., Shindo K., Yoon J., Yokota A., Shizuri Y. Rubritalea squalenifaciens sp. nov., a squalene-producing marine bacterium belonging to subdivision 1 of the phylum “Verrucomicrobia”. Int. J. Syst. Evol. Microbiol. 2007;57:1630–1634. doi: 10.1099/ijs.0.65010-0. [DOI] [PubMed] [Google Scholar]
- 74.Yoon J., Matsuo Y., Matsuda S., Adachi K., Kasai H., Yokota A. Rubritalea spongiae sp. nov. and Rubritalea tangerina sp. nov., two carotenoid- and squalene-producing marine bacteria of the family Verrucomicrobiaceae within the phylum “Verrucomicrobia”, isolated from marine animals. Int. J. Syst. Evol. Microbiol. 2007;57:2337–2343. doi: 10.1099/ijs.0.65243-0. [DOI] [PubMed] [Google Scholar]
- 75.Cuc N.T.K., Dat T.T.H., Hong T.T., Cuong P.V. Phylogenetic diversity of microorganisms associated with three marine sponges from Mien Trung Sea of Vietnam. J. Sci. Technol. 2017;55:168–177. doi: 10.15625/0866-708X/55/2/8577. [DOI] [Google Scholar]
- 76.Kaluzhnaya O., Itskovich V. Diversity of potential producers of bioactive metabolites having polyketide nature in the Baikal sponge community of Rezinkovia echinata. Limnol. Freshw. Biol. 2020;2020:423–428. doi: 10.31951/2658-3518-2020-A-3-423. [DOI] [Google Scholar]
- 77.Sizikov S., Burgsdorf I., Handley K.M., Lahyani M., Haber M., Steindler L. Characterization of sponge-associated Verrucomicrobia: Microcompartment-based sugar utilization and enhanced toxin–antitoxin modules as features of host-associated Opitutales. Environ. Microbiol. 2020;22:4669–4688. doi: 10.1111/1462-2920.15210. [DOI] [PubMed] [Google Scholar]
- 78.Wagner M., Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 79.DeVos D.P., Ward N.L. Mind the PVCs. Environ. Microbiol. 2014;16:1217–1221. doi: 10.1111/1462-2920.12349. [DOI] [PubMed] [Google Scholar]
- 80.Spring S., Bunk B., Spröer C., Schumann P., Rohde M., Tindall B.J., Klenk H.P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016;10:2801–2816. doi: 10.1038/ismej.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoon J., Matsuo Y., Matsuda S., Adachi K., Kasai H., Yokota A. Rubritalea sabuli sp. nov., a carotenoid- and squalene-producing member of the family Verrucomicrobiaceae, isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2008;58:992–997. doi: 10.1099/ijs.0.65540-0. [DOI] [PubMed] [Google Scholar]
- 82.Yoon J., Matsuda S., Adachi K., Kasai H., Yokota A. Rubritalea halochordaticola sp. nov., a carotenoid producing verrucomicrobial species isolated from a marine chordate. Int. J. Syst. Evol. Microbiol. 2011;61:1515–1520. doi: 10.1099/ijs.0.025031-0. [DOI] [PubMed] [Google Scholar]
- 83.Shindo K., Asagi E., Sano A., Hotta E., Minemura N., Mikami K., Tamesada E., Misawa N., Maoka T. Diapolycopenedioic acid xylosyl esters A, B, and C, novel antioxidative glyco-C30-carotenoic acids produced by a new marine bacterium Rubritalea Squalenifaciens. J. Antibiot. 2008;61:185–191. doi: 10.1038/ja.2008.28. [DOI] [PubMed] [Google Scholar]
- 84.Shindo K., Misawa N. New and rare carotenoids isolated from marine bacteria and their antioxidant activities. Mar. Drugs. 2014;12:1690–1698. doi: 10.3390/md12031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choudhary A., Naughton L.M., Dobson A.D.W., Rai D.K. High-performance liquid chromatography/electrospray ionisation mass spectrometric characterisation of metabolites produced by Pseudovibrio sp. W64, a marine sponge derived bacterium isolated from Irish waters. Rapid Commun. Mass Spectrom. 2018;32:1737–1745. doi: 10.1002/rcm.8226. [DOI] [PubMed] [Google Scholar]
- 86.Romano S. Ecology and biotechnological potential of bacteria belonging to the genus Pseudovibrio. Appl. Environ. Microbiol. 2018;84:e02516-17. doi: 10.1128/AEM.02516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brinkhoff T., Bach G., Heidorn T., Liang L., Schlingloff A., Simon M. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 2004;70:2560–2565. doi: 10.1128/AEM.70.4.2560-2565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bruhn J.B., Gram L., Belas R. Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 2007;73:442–450. doi: 10.1128/AEM.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cerqueda-García D., García-Maldonado J.Q., Aguirre-Macedo L., García-Cruz U. A succession of marine bacterial communities in batch reactor experiments during the degradation of five different petroleum types. Mar. Pollut. Bull. 2020;150:110775. doi: 10.1016/j.marpolbul.2019.110775. [DOI] [PubMed] [Google Scholar]
- 90.Peng C., Tang Y., Yang H., He Y., Liu Y., Liu D., Qian Y., Lu L. Time- and compound-dependent microbial community compositions and oil hydrocarbon degrading activities in seawater near the Chinese Zhoushan Archipelago. Mar. Pollut. Bull. 2020;152:110907. doi: 10.1016/j.marpolbul.2020.110907. [DOI] [PubMed] [Google Scholar]
- 91.Andreo-Vidal A., Sanchez-Amat A., Campillo-Brocal J.C. The Pseudoalteromonas luteoviolacea L-amino acid oxidase with antimicrobial activity is a flavoenzyme. Mar. Drugs. 2018;16:499. doi: 10.3390/md16120499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Montalvo N.F., Mohamed N.M., Enticknap J.J., Hill R.T. Novel actinobacteria from marine sponges. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2005;87:29–36. doi: 10.1007/s10482-004-6536-x. [DOI] [PubMed] [Google Scholar]
- 93.Zhang H., Zhang W., Jin Y., Jin M., Yu X. A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2008;93:241–248. doi: 10.1007/s10482-007-9196-9. [DOI] [PubMed] [Google Scholar]
- 94.Xin Y., Huang J., Deng M., Zhang W. Culture-independent nested PCR method reveals high diversity of actinobacteria associated with the marine sponges Hymeniacidon perleve and Sponge sp. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2008;94:533–542. doi: 10.1007/s10482-008-9270-y. [DOI] [PubMed] [Google Scholar]
- 95.Selvin J., Gandhimathi R., Kiran G.S., Priya S.S., Ravji T.R., Hema T.A. Culturable heterotrophic bacteria from the marine sponge Dendrilla nigra: Isolation and phylogenetic diversity of actinobacteria. Helgol. Mar. Res. 2009;63:239–247. doi: 10.1007/s10152-009-0153-z. [DOI] [Google Scholar]
- 96.Sun W., Dai S., Jiang S., Wang G., Liu G., Wu H., Li X. Culture-dependent and culture-independent diversity of Actinobacteria associated with the marine sponge Hymeniacidon perleve from the South China Sea. Antonie Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2010;98:65–75. doi: 10.1007/s10482-010-9430-8. [DOI] [PubMed] [Google Scholar]
- 97.Li L., Zhang D., Tang W.Z., Lin H.W., Lu Y.H. Geodermatophilus marinus sp. nov., isolated from the marine sponge Leucetta chagosensis. Int. J. Syst. Evol. Microbiol. 2019;69:2966–2971. doi: 10.1099/ijsem.0.003478. [DOI] [PubMed] [Google Scholar]
- 98.Dita S.F., Budiarti S.R.I., Lestari Y. Sponge-associated Actinobacteria: Morphological character and antibacterial activity against pathogenic bacteria. J. Sumberd. Hayati. 2017;3:21–26. [Google Scholar]
- 99.Xu D., Han L., Li C., Cao Q., Zhu D., Barrett N.H., Harmody D., Chen J., Zhu H., McCarthy P.J., et al. Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Front. Microbiol. 2018;9:1–9. doi: 10.3389/fmicb.2018.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kasanah N., Triyanto T. Bioactivities of halometabolites from marine Actinobacteria. Biomolecules. 2019;9:225. doi: 10.3390/biom9060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu T., Wu S., Zhang R., Wang D., Chen J., Zhao J. Diversity and antimicrobial potential of Actinobacteria isolated from diverse marine sponges along the Beibu Gulf of the South China Sea. FEMS Microbiol. Ecol. 2019;95:1–10. doi: 10.1093/femsec/fiz089. [DOI] [PubMed] [Google Scholar]
- 102.Phuong T.V., Thi N., Dung P., Diep C.N. Diversified collection of novel sponge-associated actinobacteria of Ha Tien sea, Kien Giang province, Vietnam. Int. J. Innov. Eng. Technol. 2019;14:33–41. [Google Scholar]
- 103.Puttaswamygowda G.H., Olakkaran S., Antony A., Purayil A.K. Present status and future perspectives of marine actinobacterial metabolites. In: Buddolla V., editor. Recent Developments in Applied Microbiology and Biochemistry. Academic Press; Cambridge, CA, USA: 2019. pp. 307–319. [Google Scholar]
- 104.Santos J.D., Vitorino I., De La Cruz M., Díaz C., Cautain B., Annang F., Pérez-Moreno G., Martinez I.G., Tormo J.R., Martín J.M., et al. Bioactivities and extract dereplication of Actinomycetales isolated from marine sponges. Front. Microbiol. 2019;10:1–11. doi: 10.3389/fmicb.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baig U., Dahanukar N., Shintre N., Holkar K., Pund A., Lele U., Gujarathi T., Patel K., Jakati A., Singh R., et al. Phylogenetic diversity and activity screening of cultivable actinobacteria isolated from marine sponges and associated environments from the western coast of India. bioRxiv Prepr. Serv. 2020:1–36. doi: 10.1101/2020.01.09.901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gohain A., Manpoong C., Saikia R., De Mandal S. Actinobacteria: Diversity and biotechnological applications. In: De Mandal S., Bhatt P., editors. Recent Advancements in Microbial Diversity. Elsevier Inc.; Berlin/Heidelberg, Germany: 2020. pp. 217–231. [Google Scholar]
- 107.Kamala K., Sivaperumal P., Kamath S.M., Thilagaraj W.R., Rajaram R. Marine actinobacteria as a source for emerging biopharmaceuticals. In: Se-Kwon K., editor. Encyclopedia of Marine Biotechnology. John Wiley & Sons Ltd.; Hoboken, NJ, USA: 2020. pp. 2095–2105. [Google Scholar]
- 108.Jiang S., Sun W., Chen M., Dai S., Zhang L., Liu Y., Lee K.J., Li X. Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2007;92:405–416. doi: 10.1007/s10482-007-9169-z. [DOI] [PubMed] [Google Scholar]
- 109.Jiang S., Li X., Zhang L., Sun W., Dai S., Xie L., Liu Y., Lee K.J. Culturable actinobacteria isolated from marine sponge Iotrochota sp. Mar. Biol. 2008;153:945–952. doi: 10.1007/s00227-007-0866-y. [DOI] [Google Scholar]
- 110.Schneemann I., Nagel K., Kajahn I., Labes A., Wiese J., Imhoff J.F. Comprehensive investigation of marine actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010;76:3702–3714. doi: 10.1128/AEM.00780-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selvin J., Sathiyanarayanan G., Lipton A.N., Al-Dhabi N.A., Arasu M.V., Kiran G.S. Ketide Synthase (KS) domain prediction and analysis of iterative type II PKS gene in marine sponge-associated actinobacteria producing biosurfactants and antimicrobial agents. Front. Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang X., Kong F., Zhou S., Huang D., Zheng J., Zhu W. Streptomyces tirandamycinicus sp. nov., a novel marine sponge-derived actinobacterium with antibacterial potential against Streptococcus Agalactiae. Front. Microbiol. 2019;10:1–11. doi: 10.3389/fmicb.2019.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Madhusoodanan J. Probing predatory bacteria as an antibacterial remedy. Proc. Natl. Acad. Sci. USA. 2019;116:22887–22890. doi: 10.1073/pnas.1917513116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Williams L.E., Cullen N., Degiorgis J.A., Martinez K.J., Mellone J., Oser M., Wang J., Zhang Y. Variation in genome content and predatory phenotypes between Bdellovibrio sp. NC01 isolated from soil and B. bacteriovorus type strain HD100. Microbiology. 2019;165:1315–1330. doi: 10.1099/mic.0.000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bratanis E., Andersson T., Lood R., Bukowska-Faniband E. Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front. Microbiol. 2020;11:1–14. doi: 10.3389/fmicb.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun M.L., Zhao F., Shi M., Zhang X.Y., Zhou B.C., Zhang Y.Z., Chen X.L. Characterization and biotechnological potential analysis of a new exopolysaccharide from the Arctic marine bacterium Polaribacter sp. SMSci. Rep. 2015;5:1–12. doi: 10.1038/srep18435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun M.L., Zhao F., Zhang X.K., Zhang X.Y., Zhang Y.Z., Song X.Y., Chen X.L. Improvement of the production of an Arctic bacterial exopolysaccharide with protective effect on human skin cells against UV-induced oxidative stress. Appl. Microbiol. Biotechnol. 2020;104:4863–4875. doi: 10.1007/s00253-020-10524-z. [DOI] [PubMed] [Google Scholar]
- 118.Rützler K. Sponges in coral reefs. In: Stoddart D.R., Johannes R.E., editors. Coral reefs: Research Methods, Monographs on Oceanographic Methodology. UNESCO; Paris, France: 1978. pp. 299–313. [Google Scholar]
- 119.Hooper J.N.A. ‘Sponguide’. Guide to sponge collection and identification. [(accessed on 17 March 2021)]; Available online: https://www.academia.edu/34258606/SPONGE_GUIDE_GUIDE_TO_SPONGE_COLLECTION_AND_IDENTIFICATION_Version_August_2000.
- 120.Van Soest R.W.M., Boury-Esnault N., Hooper J.N.A., Rützler K., de Voogd N.J., Alvarez B., Hajdu E., Pisera A.B., Manconi R., Schönberg C., et al. World Porifera Database. [(accessed on 17 March 2021)]; Available online: http://www.marinespecies.org/porifera/
- 121.Schmitt S., Hentschel U., Zea S., Dandekar T., Wolf M. ITS-2 and 18S rRNA gene phylogeny of Aplysinidae (Verongida, Demospongiae) J. Mol. Evol. 2005;60:327–336. doi: 10.1007/s00239-004-0162-0. [DOI] [PubMed] [Google Scholar]
- 122.Chombard C., Boury-Esnault N., Tillier S. Reassessment of homology of morphological characters in Tetractinellid sponges based on molecular data. Syst. Biol. 1998;47:351–366. doi: 10.1080/106351598260761. [DOI] [PubMed] [Google Scholar]
- 123.Dohrmann M., Janussen D., Reitner J., Collins A.G., Wörheide G. Phylogeny and evolution of glass sponges (Porifera, Hexactinellida) Syst. Biol. 2008;57:388–405. doi: 10.1080/10635150802161088. [DOI] [PubMed] [Google Scholar]
- 124.Meyer C.P., Geller J.B., Paulay G. Fine scale endemism on coral reefs: Archipelagic differentiation in turbinid gastropods. Evolution (N. Y). 2005;59:113–125. doi: 10.1111/j.0014-3820.2005.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 125.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 126.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.