Abstract
Wound infections are common medical problems in sub-Saharan Africa but data on the molecular epidemiology are rare. Within this study we assessed the clonal lineages, resistance genes and virulence factors of Gram-negative bacteria isolated from Ghanaian patients with chronic wounds. From a previous study, 49 Pseudomonas aeruginosa, 21 Klebsiella pneumoniae complex members and 12 Escherichia coli were subjected to whole genome sequencing. Sequence analysis indicated high clonal diversity with only nine P. aeruginosa clusters comprising two strains each and one E. coli cluster comprising three strains with high phylogenetic relationship suggesting nosocomial transmission. Acquired beta-lactamase genes were observed in some isolates next to a broad spectrum of additional genetic resistance determinants. Phenotypical expression of extended-spectrum beta-lactamase activity in the Enterobacterales was associated with blaCTX-M-15 genes, which are frequent in Ghana. Frequently recorded virulence genes comprised genes related to invasion and iron-uptake in E. coli, genes related to adherence, iron-uptake, secretion systems and antiphagocytosis in P. aeruginosa and genes related to adherence, biofilm formation, immune evasion, iron-uptake and secretion systems in K. pneumonia complex. In summary, the study provides a piece in the puzzle of the molecular epidemiology of Gram-negative bacteria in chronic wounds in rural Ghana.
Keywords: wounds, Gram-negative bacteria, colonization, infection, clonal lineages, resistance genes, virulence factors
1. Introduction
The microbiology of chronic infected wounds, also on a molecular level, is poorly understood in sub-Saharan Africa (SSA) [1]. However, studies highlight the importance of antibiotic resistant Gram-negative bacteria [2,3,4,5,6].
From other parts in the world, in particular from industrialized countries, information on the microbiology and the role of biofilm-forming microorganisms causing such infections are well established [7,8,9,10].
In chronic wounds, Pseudomonas aeruginosa is amongst the most frequently isolated Gram-negative bacteria, associated with biofilm formation [11,12]. Tightly adhering biofilms pose a challenge in the diagnosis of P. aeruginosa using standard culturing methods [13].
In comparison, the role of Enterobacterales in chronic wounds has been much less characterized [14,15,16,17]. Studies have shown that geography seems to play a role in the estimation of their etiological relevance [18]. It was shown that skin colonization with Gram-negative bacteria is frequent in resource-limited (sub)tropical settings [19,20,21], in contrast to skin colonization of individuals from industrialized countries, where Gram-positive bacteria dominate [19]. Temperature and moisture have been discussed as likely reasons for the difference seen [22].
Isolation of potentially pathogenic bacteria from non-sterile sites like wounds does not necessarily indicate clinical relevance, which poses challenge to clinical interpretation.
In a recent study that focused on the overall bacterial composition of chronic wound infections in Ghana, from which the isolates for the present molecular analysis were taken, Enterobacterales and Pseudomonas aeruginosa constituted the majority of isolated bacterial strains [23]. A moderate proportion of ESBL-positive Enterobacterales suggests lower frequencies of antibiotic resistance [23] than what was recorded from other Ghanaian hospitals [5,24].
Within this study, we aim at characterizing clonal lineages, resistance-associated genetic elements and virulence genes of P. aeruginosa, the Klebsiella pneumoniae complex and Escherichia coli, which were recently isolated from chronic wounds of Ghanaian adult patients [23]. The molecular epidemiology of dominating clonal lineages and associated resistance genes will be assessed. Further, analysis of highly abundant virulence factors will be conducted.
2. Results
2.1. Clustering Based on Core Genome Multilocus Sequence Typing (cgMLST) Results
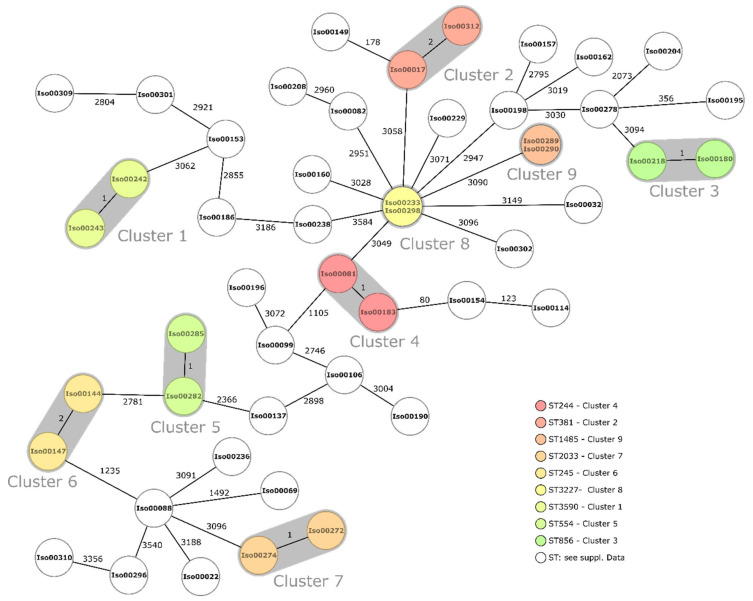
Of the 49 P. aeruginosa analyzed, a total of nine clusters comprising isolates without any recorded differences (n = 2) or with one or two alleles difference (n = 7) were found, suggesting closely related phylogeny (Figure 1). In addition to the clusters, 31 singletons with differences ranging from 80 to 3584 alleles were observed. MLST sequence types (ST) are indicated in Figure 1 and Table A1 and Table A2. Cluster sequence types included the following: ST244, ST245, ST381, ST554, ST856, ST1485, ST2033, ST3227 and ST3590.
Figure 1.
Minimum spanning tree of P. aeruginosa based on 3867 targets (core genome). Isolate numbers are found within the nodes, and numbers between nodes indicate the number of different alleles. Isolates within clusters are colored based on MLST sequence type (ST). The ST types of white nodes are indicated in Table A1.
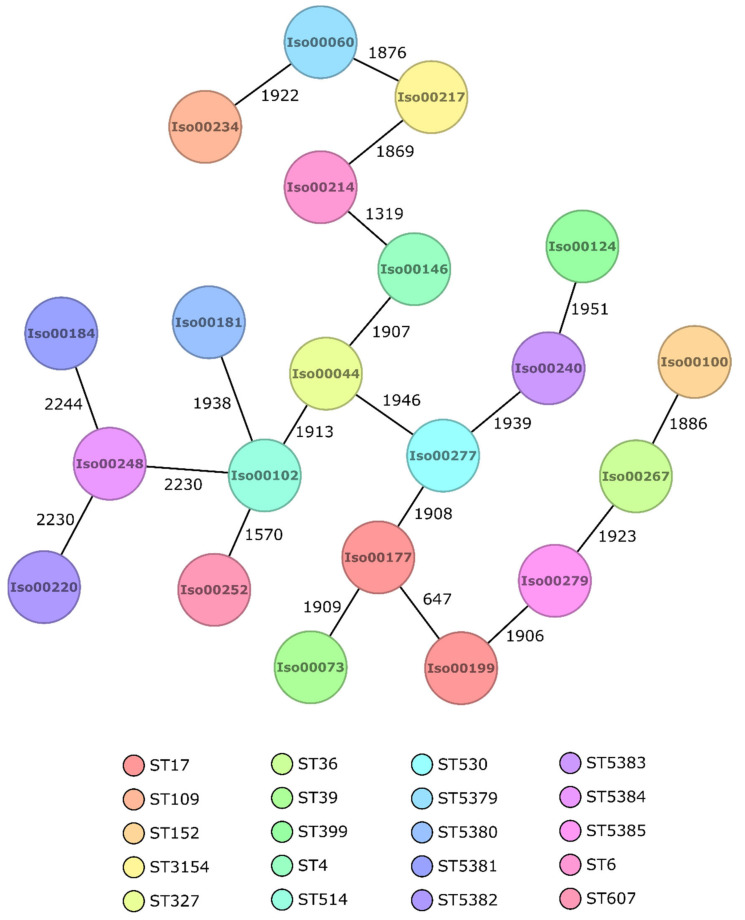
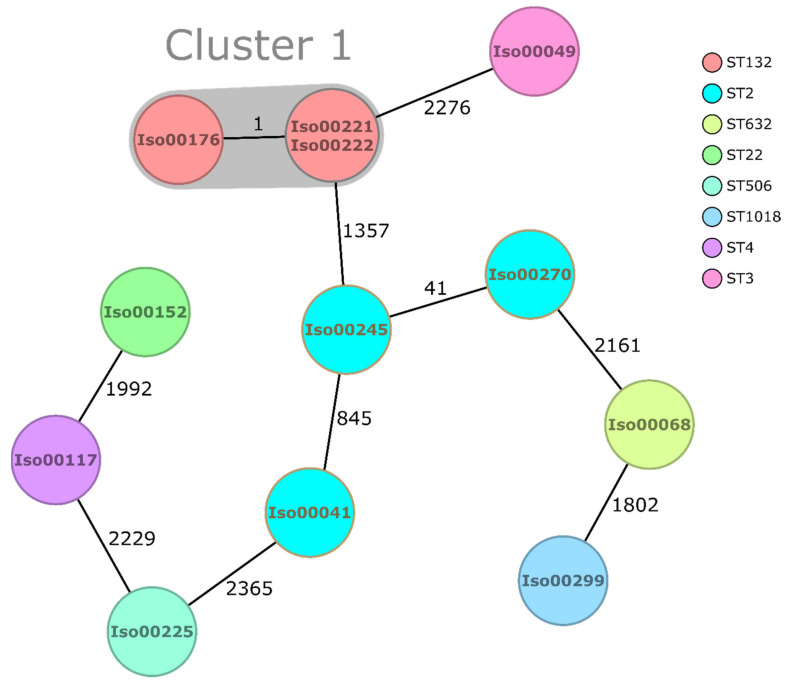
No clusters were identified among the 21 assessed K. pneumonia complex members, which were all singletons with differences ranging from 647 to 2244 alleles. K. pneumoniae complex sequence types are summarized in Figure 2. From the 12 E. coli isolates, three isolates in a cluster of close phylogenetic relationship were found (1× no allelic differences, 1 × 1 allele difference) (Figure 3). In addition to the cluster observed, nine singletons with differences ranging from 41 to 2365 alleles were recorded. The sequence type of the cluster was ST132 (Pasteur MLST scheme). Sequence types of all E. coli isolates are illustrated in Figure 3.
Figure 2.
Minimum spanning tree of K. pneumoniae complex based on 2358 targets (core genome). Isolate numbers are found within the nodes, and the numbers between the nodes indicate the number of different alleles. Colors demonstrate the MLST sequence type of the isolates.
Figure 3.
Minimum spanning tree of E. coli based on 2513 targets (core genome). Isolate numbers are found within the nodes, and the numbers between the nodes indicate the number of different alleles. Colors demonstrate the Pasteur sequence type of the isolates.
2.2. Identified Molecular Resistance Mechanisms in Correlation to Previous Phenotypic Antibiotic Resistance
Table 1 summarizes acquired antimicrobial resistance determinants for E. coli and acquired genes mediating tolerance to disinfectants. Data for P. aeruginosa and K. pneumoniae are presented in Table A1 and Table A2. Table A3, Table A4, Table A5, Table A6, Table A7 and Table A8 summarize the phenotypic resistance results as previously recorded [23].
Table 1.
Analysis of antimicrobial resistance determinants, ordered by strain and MLST type, of the assessed E. coli isolates. ST = Sequence type.
| Sample ID | ST-Type | Acquired Resistance Determinants Against | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta lacatams | Sulfonamids | Trimethoprim | Makrodlids | Tetracyclins | Fluoroquinolones | Chloramphenicol | Aminoglycosides | Efflux pumps | Amino acid exchanges due to point mutations | Disinfectant resistance genes * | ||
| 041 | ST 2 | blaOXA-1, blaTEM-1B, blaCTX-M-15 | sul1 | dfrA17 | mph(A) | tet(B) | aac(6′)-Ib-cr, aac(6′)-Ib-cr |
catB3,
catA1 |
aac(3)-IId, aac(6′)-Ib-cr, aadA5, aac(6′)-Ib-cr | mdf(A) | parE p.S458A, gyrA p.S83L, gyrA p.D87N, parC p.S80I | sitABCD, qacE |
| 049 | ST 3 | bla TEM-1B | sul2, sul1, | dfrA12 | mph(A) | tet(A) | aadA2, aph(3″)-Ib, aph(6)-Id | mdf(A) |
sitABCD-like, qacE |
|||
| 068 | ST 632 | bla TEM-1B | sul3 | dfrA12 | tet(A) | cmlA1 | aadA1, aadA2 | mdf(A)-like | parE p.S458A, gyrA p.S83L, gyrA p.D87N, parC p.S80I | |||
| 117 | ST 4 | bla TEM-1B | sul1, sul2 | dfrA7 | tet(A) | catA1 | aph(6)-Id, aph(3 ″)-Ib | mdf(A)-like | sitABCD-like, qacE | |||
| 152 | ST 22 | blaCARB-2, blaTEM-1B | sul1 | dfrA1 | ere(B) | tet(B) | catA1 | aadA1, aadA2b | mdf(A)-like | gyrA p.S83L | qacE, sitABCD | |
| 176 | ST 132 | bla TEM-1B | sul1 | dfrA7 | tet(A) | catA1 | aph(3 ″)-Ib, aph(6)-Id | mdf(A) | qacE, sitABCD | |||
| 221 | ST 132 | bla TEM-1B | sul1 | dfrA7 | tet(A) | catA1 | aph(6)-Id, aph(3 ″)-Ib | mdf(A) | qacE, sitABCD | |||
| 222 | ST 132 | bla TEM-1B | sul1, sul2 | dfrA7 | tet(A) | catA1 | aph(3 ″)-Ib, aph(6)-Id | mdf(A) | qacE, sitABCD | |||
| 225 | ST 506 | blaTEM-1D, blaCTX-M-15 | sul1, sul2 | dfrA17 | mph(A) | tet(A) | catA1 | aadA5, aph(6)-Id, aph(3″)-Ib | mdf(A)-like | gyrA p.S83L, parE p.I529L | sitABCD-like, qacE | |
| 245 | ST 2 | bla TEM-1B | sul1 | dfrA12 | mph(A) | tet(B) | qepA4 (neu) | catA1 | aadA2, aac(3)-IId | mdf(A) | parE p.S458A, gyrA p.S83L, gyrA p.D87N, parC p.S80I | qacE |
| 270 | ST 2 | bla CTX-M-15 | tet(B) | catA1 | mdf(A) | gyrA p.S83L, gyrA p.D87N, parE p.S458A, parC p.S80I | ||||||
| 299 | ST 1018 | bla TEM-1B | sul3 | dfrA14 | tet(A) | qnrS1 | mdf(A) | |||||
* sitABCD = peroxides resistance, qacE = quaternary ammonium compounds resistance.
In the present study, phylogenetically identical or almost identical isolates also carried the same resistomes. All E. coli strains harbored acquired beta-lactamase genes with the majority coding for small spectrum beta-lactamases such as blaTEM-1 or blaOXA-1. Only four strains carried the gene for an ESBL, in all cases blaCTX-M-15. Among the K. pneumoniae complex strains, two belonged to the species K. variicola, one to the species K. quasipneumoniae and the remaining to the species K. pneumoniae sensu stricto as reflected by intrinsic blaLEN, blaOKP and blaSHV-1 like, respectively. Genes coding for ESBL (blaCTX-M-15) were found solely in four out of 18 K. pneumoniae sensu stricto strains that also displayed resistance to oxyimino cephalosporins. In addition, several K. pneumoniae complex strains harbored blaTEM-1, single strains also contained blaOXA-1 and blaSCO-1.
With respect to P. aeruginosa, only one strain harbored acquired beta-lactamase genes (blaTEM-1 and blaSCO-1). Increased minimum inhibitory concentrations (MICs) for carbapenems as observed in some P. aeruginosa strains were neither explained by matching acquired carbapenemase genes nor by full sequence analysis of the oprD gene. The associated amino acid sequences are shown in Figure A1. As indicated, the complete oprD gene was found in all 49 P. aeruginosa isolates; there was no evidence of protein truncation by premature stop of translation. The 49 isolates could be divided into 7 subgroups according to the protein sequence of the oprD protein, which differ in a total of 30 individual amino acid exchanges and in a single 12aa/10aa-stretch. Therefore, genotypic assessment could not identify the reason for the single carbapenem-resistant P. aeuroginosa isolate 088 (ST 1682).
Other frequently detected resistance genes in P. aeruginosa were the fosfomycin resistance gene fosA, the chloramphenicol resistance gene catB7, the aminoglycoside resistance gene aph(3′)-IIb and the fluoroquinolone-resistance gene crpP. In the Klebsiella pneumoniae complex isolates, single amino acid exchanges and the fosmomycin resistance gene fosA were frequent. Various fluoroquinolone resistance genes and disinfectant tolerance mediating genes also quantitatively dominated. Finally, a broad spectrum of acquired genes causing resistance to the assessed classes of antimicrobial drugs and tolerance to disinfectants was observed in the E. coli strains.
2.3. Identified Molecular Virulence Mechanisms
Table 2 summarizes the analysis of virulence-related genes in E. coli (without genes mediating enteropathogenicity). Data for P. aeruginosa and K. pneumoniae are presented in Table A9 and Table A10.
Table 2.
Analysis of virulence determinants, ordered by strain and MLST type, of the assessed E. coli isolates. ST = Sequence type.
| Sample ID | ST-Type | Pathogenicity Factor Groups | |||||
|---|---|---|---|---|---|---|---|
| Adherence | Invasion | Toxin | Immune Evasion | Iron Uptake | Protease | ||
| 041 | ST 2 | fdeC | aslA, ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, | |||
| 049 | ST 3 | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU, kpsS-like, ompA | chuS, chuU, chuV, chuW, chuY, entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG | ||||
| 068 | ST 632 | ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG | ||||
| 117 | ST 4 | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU; kpsS-like, ompA | hlyB, hlyC, hlyD, | tcpC | chuA, chuS, chuT, chuU, chuV, chuW, chuX, chuY, entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, hlyA, iroN, | pic, sat, vat | |
| 152 | ST 22 | sfaB, sfaC, sfaD, sfaE, sfaF, sfaG, sfaH, sfaS, sfaX, sfaY | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU; kpsS-like, ompA | cnf1; hlyA, hlyB, hlyC, hlyD, | tcpC | chuA, chuS, chuT, chuU, chuV, chuW, chuX, chuY, entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, iroN, | vat |
| 176 | ST 132 | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU; kpsS-like, ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, | sat | |||
| 221 | ST 132 | aslA, kpsC, kpsD, kpsE, kpsM, kpsU; kpsS-like, ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, | sat | |||
| 222 | ST 132 | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU; kpsS-like, ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, | sat | |||
| 225 | ST 506 | aslA, kpsC, kpsD, kpsE, kpsF, kpsM, kpsU; kpsS-like, ompA | chuA, chuS, chuT, chuU, chuV, chuW, chuX, chuY, entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepC, fepD, fepG, | sat | |||
| 245 | ST 2 | aslA, ompA | entA-like, entB, entC, entE, entF, entS, fepB, fepC, fepD, fepG | ||||
| 270 | ST 2 | aslA, ompA | entA-like, entB, entC, entE, entS, fepA, fepB, fepC, fepD, fepG | ||||
| 299 | ST 1018 | ompA | entA-like, entB, entC, entE, entF, entS, fepA, fepB, fepD, fepG | ||||
The virulence-associated gene exoU, which has been described in association with the P. aeruginosa high-risk clone ST 135 [25], was recorded three times, associated with ST 135 (sample ID 296), ST 532 (sample ID 310) and ST 2483 (sample ID 22), respectively. Based on a Kleborate assessment, a positive virulence score was calculated for 7 out of 21 K. pneumoniae strains, comprising the known high-risk clones ST 17 (sample IDs 177, 199) and ST 152 (sample ID 100) [26], next to the clones ST 4 (sample ID 146), ST 6 (sample ID 214), ST 36 (sample ID 267) and ST 39 (sample ID 73), respectively. With focus on some important virulence associated genes in Klebsiella spp., ybt genes were detected in the abovementioned 7 samples, iroE was recorded in all 21 strains, while clb or rpmA genes were not detected.
Iron-uptake-related genes were numerous in all analyzed bacterial strains. For P. aeruginosa and K. pneumoniae, various secretion system-associated genes were found. Immune evasion-related genes were highly abundant in K. pneumoniae but not in E. coli isolates. Adherence-related genes were numerous in P. aeruginosa and in K. pneumoniae but not in E. coli.
Numerous invasion-associated genes were detected in E. coli, antiphagocytosis-associated genes were found in P. aeruginosa, and biofilm-associated genes in K. pneumoniae.
Less frequently detected were: toxin genes in E. coli and K. pneumoniae, protease genes in E. coli and P. aeruginosa, regulation genes in P. aeruginosa and K. pneumoniae, biosurfactant and pigment genes in P. aeruginosa and nutrition factor, efflux pumps and serum resistance genes in K. pneumoniae.
3. Discussion
Within this study, we aimed at filling information gaps on the molecular epidemiology of Gram-negative bacteria from chronic infected wounds in rural Ghana. Phylogenetic analyses based on core genome comparison indicated a high clonal diversity of the wound-associated isolates. Clonal clusters were restricted to nine P. aeruginosa clusters and one E. coli cluster, most likely indicating nosocomial transmission, which has most likely occurred in the wound dressing room that patients’ visit on a weekly basis.
ST 135 and ST 244, which are among the worldwide top 10 P. aeruginosa high-risk clones [25], were found among the P. aeruginosa wound isolates. In detail, one ST 135 Pseudomonas aeruginosa isolate was detected, carrying the beta-lactamase-encoding genes blaTEM-1B and blaSCO-1 and an exoU gene, next to five ST 244 without acquired beta-lactamases. Focusing on known pathogenic K. pneumoniae clones [26], two ST 17 strains, a clone reported to be associated with carbapenem-resistance, and one ST 152 strain, a clone known from the Caribbean as common carrier of multiple resistance genes, were detected. Strains carrying the ybt and iro genes were also identified as high-risk clones by the Kleborate software. From the observed E. coli ST types, none have been previously reported as being associated with pathogenic clones so far [27].
In line with the phenotypical antibiotic resistance results previously published [23], numerous acquired resistance determinants were detected in the bacterial strains under investigation. Focusing on the few observed clusters, comparable resistome compositions point towards recent nosocomial transmission. The gene blaCTX-M-15 was identified as the determinant of the detected extended spectrum beta-lactamase (ESBL) expression in ESBL positive Enterobacterales [23]. This is in line with previous reports from both human and livestock-associated ESBL positive Enterobacterales in Ghana [28,29,30,31,32,33,34]. In P. aeruginosa and K. pneumoniae, blaSCO-1, which has initially been described from an Acinetobacter baumannii isolate from Argentina [35], was observed. Beta-lactamases with high hydrolytic effects on carbapenems were lacking, the same applies to protein truncation by premature stop of translation of the oprD gene in P. aeruginosa. Accordingly, the genetic background of carbapenem resistance of a single P. aeruginosa strain could not be resolved, although downregulation of oprD expression due to mutations outside of the gene or ampC (class C betalactamase) overexpression could not be excluded as likely reasons.
Substance-specific genes and genes encoding efflux pumps mediating tolerance to disinfectants were observed in Enterobacterales. Therefore, further monitoring of the spread of disinfectant tolerance-associated genes and the effects of their abundance on disinfectant-based skin and wound decolonization strategies [36] seem advisable.
The importance of highly abundant virulence factors like iron-uptake- and secretion system-related genes in P. aeruginosa is comprehensively described in the literature [37,38]. Other genes reported in the literature like regulation-associated virulence genes, recently reported, were less frequently observed in our isolates [39,40]. However, due to lacking information on the individual etiological relevance of each isolate, any association with clinical effects remains speculative.
Further limitations of this study include a rather small sample size and the lack of a comparison strain collection containing isolates from other clinical specimens and environmental strains. Accordingly, the interpretation of the etiological relevance of individual strains remains challenging and is clearly beyond the scope of this work.
In summary, a broad spectrum of Gram-negative clones was isolated from the chronic wounds of the Ghanaian patients. Thereby, known high-risk clones [25,26,27] played only a minor role. Observed resistance patterns and mechanisms were in line with the spectrum expected from previous reports [23,28,29,30,31,32,33,34].
4. Materials and Methods
4.1. Sample Collection, Bacterial Culture and Antibiotic Susceptibility Testing
Single patient strains of P. aeruginosa, E. coli and K. pneumoniae complex were isolated from patients ≥15 years with an infected chronic wound at the Outpatient Department (OPD) of the Agogo Presbyterian Hospital, in the Asante Akim North District of rural Ghana. Patients typically visit the wound dressing room of the OPD on a weekly basis. Sampling was performed from January 2016 to November 2016. Sample collection and microbiological investigations were reported previously [23]. Antibiotic susceptibility was tested by the disk diffusion method and interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines v.6.0 (http://www.eucast.org (accessed on 15 January 2016)). Bacterial strains and antibiotic susceptibility were confirmed using the VITEK2 System. Those data have been published before [23].
4.2. DNA Isolation and Whole Genome Sequencing
Bacterial DNA was isolated using the MasterPure Complete DNA and RNA Purification Kit (LGC standards GmbH, Wesel, Germany) and sent for whole genome sequencing (WGS) to BGI Europe, Denmark, Copenhagen. A BGISEQ-500 device was used for sequencing, generating 2 × 150 bp paired-end reads with an aimed coverage of 100×. Original raw data were upload for public use to the short-read archive (SRA, NCBI) under the accession number PRJNA699140. Details on the strain-specific SRA accession numbers are provided in Table A11.
4.3. Whole Genome Sequencing and Data Analysis
All raw data passed quality control using FASTQC v.0.11.4 [41] and were used for further analysis. Taxonomic classification and contamination check of raw-reads was performed using KRAKEN2 v.2.0.8-beta [42]. Phylogenetic analysis based on core genome multi locus sequence typing (cgMLST) analysis was performed using the commercial software SeqSphere+ v. 7.2.0 (Ridom GmbH, Münster, Germany) [43]. The software pipeline included assessment of read data and adapter control using FASTQC followed by genome assembly using the internally provided assembler Velvet, applying default settings. The reference genomes NC_000913.3 (E. coli), NC_002516.2 (P. aeruginosa) and NC_01273.1 (K. pneumoniae species complex) were used for cgMLST analyses. Only samples with a ration of “good cgMLST targets” higher than 90% were included in the phylogenetic analysis. Novel cgMLST-based complex types (CT) were automatically assigned by the SeqSphere software. Unknown alleles and profiles of MLST genes were submitted to pubmlst.org or Institute Pasteur to establish novel sequence types (ST). Isolates were defined to be clonally identical with allele differences less than four. Moreover, raw data were assembled with SPAdes v3.13.11 [44] using the careful option. Scaffolds shorter than 500 bp or with a coverage smaller than ten were sorted out, using an in-house script. Abricate v.0.9.9 [45] was used to screen for resistance and virulence genes in SPAdes assembly files, using NCBI AMRFinderPlus [46] and VFDB [47] as reference databases (both updated 6 November 2020), respectively. Additionally, SPAdes assemblies were uploaded to ResFinder4.1 [48] to obtain WGS predicted phenotypes against different antimicrobials by using default settings (%ID > 90, minimum length > 60%) and to Kleborate to predict virulence genes in Klebsiella isolates.
4.4. Ethical Considerations
The Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology in Kumasi, Ghana, approved this study (approval number CHRPE/AP/078/16).
5. Conclusions
In conclusion, this study provides a molecular insight into the epidemiology of Gram-negative bacteria isolated from chronic wound infections from patients in rural Ghana. Epidemiological data that focus on the distribution and spread of antimicrobial resistance determinants and associated virulence factors in resource-limited settings are scarce. Although the study is a small cross-sectional assessment, which cannot replace continuous surveillance programs, it might provide a glimpse of prevailing Gram-negative bacteria isolated from wound infections in this area of Ghana. Considering the ongoing need for resistance and virulence surveillance in tropical regions, larger future studies are desirable.
Acknowledgments
We thank all patients that participated in this study and the staff at the Agogo Presbyterian Hospital. Without their support, this research study would not have been possible. We thank the team of curators pubmlst.org and the Institute Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr/ (accessed on 22 March 2021).
Appendix A
Table A1.
Analysis of antimicrobial resistance determinants, ordered by strain and MLST type, of the assessed P. aeruginosa isolates. ST = Sequence type.
| Sample ID | ST-Type | Acquired Resistance Determinants Against | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta Lacatams | Sulfonamids | Fosfomycin | Trimethoprim | Makrolides | Tetracyclinws | Fluoroquinolones | Chloramphenicol | Rifampicin | Aminoglycosides | Efflux Pumps | Amino Acid Exchanges Due to Point Mutations | Disinfectant Resistance Genes | ||
| 017 | ST 381 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 022 | ST 2483 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 032 | ST 3587 | sul1 | fosA | dfrA15 | tet(G) | catB7 | aph(3′)-IIb | |||||||
| 069 | ST 360 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 081 | ST 244 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 082 | ST 514 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 088 | ST 1682 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 099 | ST 244 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 106 | ST 1521 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 114 | ST 244 | fosA | crpP-like | catB7 | aph(3′)-IIb-like | |||||||||
| 137 | ST 3014 | fosA | crpP-like | catB7 | aph(3′)-IIb-like | |||||||||
| 144 | ST 245 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 147 | ST 245 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 149 | ST 381 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 153 | ST 704 | fosA-like | crpP-like | catB7-like | aph(3′)-IIb- like | |||||||||
| 154 | ST 244 | , | fosA | crpP-like | catB7 | aph(3′)-IIb-like | ||||||||
| 157 | ST 2616 | fosA | catB7-like | aph(3′)-IIb | ||||||||||
| 160 | ST 170 | fosA-like | aph(3′)-IIb | |||||||||||
| 162 | ST 274 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 180 | ST 856 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 183 | ST 244 | fosA | catB7 | aph(3′)-IIb-like | ||||||||||
| 186 | ST 3588 | fosA-like | catB7-like | aph(3′)-IIb-like | ||||||||||
| 190 | ST 871 | fosA | catB7-like | aph(3′)-IIb | ||||||||||
| 195 | ST 988 | fosA | crpP-like | catB7-like | aph(3′)-IIb-like | |||||||||
| 196 | ST 2475 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 198 | ST 2476 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 204 | ST 639 | fosA | crpP | catB7 | aph(3′)-IIb-like | |||||||||
| 208 | ST 132 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 218 | ST 856 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 229 | ST 270 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 233 | ST 3227 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 236 | ST 266 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 238 | ST 3589 | fosA-like | crpP-like | catB7-like | aph(3′)-IIb-like | |||||||||
| 242 | ST 3590 | fosA-like | ||||||||||||
| 243 | ST 3590 | fosA-like | catB7-like | aph(3′)-IIb-like | ||||||||||
| 272 | ST 2033 | fosA | catB7-like | aph(3′)-IIb-like | ||||||||||
| 274 | ST 2033 | fosA | catB7-like | aph(3′)-IIb | ||||||||||
| 278 | ST 988 | fosA | crpP-like | catB7-like | ||||||||||
| 282 | ST 554 | fosA | crpP-like | catB7 | aph(3′)-IIb | |||||||||
| 285 | ST 554 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 289 | ST 1485 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 290 | ST 1485 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 296 | ST 235 | blaTEM-1B, blaSCO-1 | sul1 | fosA | tet(G) | catB7-like | aph(3′)-IIb-like, aac(3)-IIa | |||||||
| 298 | ST 3227 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 301 | ST 3593 | fosA-like | catB7-like | aph(3′)-IIb-like | ||||||||||
| 302 | ST 1755 | fosA | catB7 | aph(3′)-IIb | ||||||||||
| 309 | ST 3592 | fosA like | crpP-like | catB7-like | aph(3′)-IIb-like | |||||||||
| 310 | ST 532 | sul1 | fosA | catB7-like | aph(3″)-Ib, aph(6)-Id, aph(3′)-IIb | |||||||||
| 312 | ST 381 | fosA | catB7 | aph(3′)-IIb | ||||||||||
Acquired resistance genes for macrolides, rifampicin, resistance-associated point mutations, genes for efflux pumps or genes mediating tolerance against disinfectants were not detected.
Table A2.
Analysis of antimicrobial resistance determinants, ordered by strain and MLST type, of the assessed K. pneumoniae isolates. ST = Sequence type.
| Sample ID | ST-Type | Acquired Resistance Determinants Against | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta Lacatams | Sulfonamids | Fosfomycin | Trimethoprim | Macrolides | Tetracyclines | Fluoroquinolones | Chloramphenicol | Rifampicin | Aminoglycosides | Efflux Pumps | Amino Acid Exchanges Due to Point Mutations | Disinfectant Resistance Genes * | ||
| 044 | ST 327 | fosA | oqxB, oqxA | ompK37 p.I70M, ompK37 p.I128M,ompK37 p.I128M, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB, oqxA | |||||||||
| 060 | ST 5379 | bla TEM-1C | sul1, sul2 | fosA | dfrA12 | mph(A) | oqxA, oqxB, qnrS1 | catA2-like | aph(6)-Id, aph(3″)-Ib, aph(3′)-Ia, aadA2, aac(3)-IIa | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.T184P, ompK37 p.I70M, ompK37 p.I128M | oqxA, qacE, oqxB | |||
| 073 | ST 39 | blaTEM-1B , bla CTX-M-15 | sul1, | fosA | dfrA27 | erm(B), mph(A) | tet(D) | oqxB, oqxA, aac(6′)-Ib-cr, qnrB2, aac(6′)-Ib-cr | catA2-like | ARR-3 | aac(6′)-Ib-cr, aadA16, aac(3)-IIa, aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M, ompK37 p.N230G, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S | oqxB, oqxA, qacE | |
| 100 | ST 152 | blaCTX-M-15 , bla OXA-1 , bla TEM-1B | sul2, sul1 | fosA | dfrA1, dfrA27 | mph(A) | tet(D) | aac(6′)-Ib-cr, oqxB, qnrB6, oqxA, aac(6′)-Ib-cr | catB3, catA1, catB3 | ARR-3 | aac(3)-IIa, aph(6)-Id, aph(3″)-Ib, aadA1, aadA16, aph(3′)-Ia, aac(6′)-Ib-cr, aac(6′)-Ib-cr | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.G189T, ompK36 p.F198Y, ompK36 p.F207Y, ompK36 p.A217S, ompK36 p.T222L, ompK36 p.D223G, ompK36 p.E232R, ompK36 p.N304E, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M, ompK37 p.N230G | oqxB, oqxA | |
| 102 | ST 514 | fosA | tet(C) | oqxB, oqxA | catA1 | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S, ompK37 p.I70M, ompK37 p.I128M, ompK37 p.N230G, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB, oqxA | |||||||
| 124 | ST 399 | fosA | oqxA, oqxB | catA1 | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.G189T, ompK36 p.F198Y, ompK36 p.F207Y, ompK36 p.A217S, ompK36 p.T222L, ompK36 p.D223G, ompK36 p.E232R, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M | oqxA, oqxB | ||||||||
| 146 | ST 4 | sul2 | fosA | tet(D) | oqxA, oqxB | catA2-like | oqxA, oqxB | |||||||
| 177 | ST 17 | sul1, sul2 | fosA | dfrA15 | tet(A) | oqxA, oqxB-like | catA1 | aadA1, aph(3″)-Ib, aph(6)-Id | ompK37 p.I70M, ompK37 p.I128M, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK36 p.N49S, ompK36 p.L59V,ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232RompK36 p.T254S | qacE, oqxB-like, oqxA | ||||
| 181 | ST 5380 | fosA | oqxA, oqxB | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S,ompK37 p.I70M, ompK37 p.I128M, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxA, oqxB | |||||||||
| 184 | ST 5381 | fosA | oqxA-like, oqxB-like | ompK37 p.I70M, ompK37 p.I128M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191Q, ompK36 p.F198Y,ompK36 p.A217S, ompK36 p.N218H, ompK36 p.Q227N, ompK36 p.L229V, ompK36 p.N304E, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxA-like, oqxB-like | |||||||||
| 199 | ST 17 | blaCTX-M-15 , bla TEM-1B | sul2, sul1 | fosA-like | dfrA16 | oqxA, oqxB | aadA2b, aac(3)-IIa | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.T86V, ompK36 p.S89T, ompK36 p.D91K, ompK36 p.A93S, ompK36 p.L191Q, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.Q227N, ompK36 p.L229V, ompK36 p.E232R,ompK36 p.H235D, ompK36 p.T254S | oqxA, oqxB, qacE | |||||
| 214 | ST 6 | sul1 | fosA-like | dfrA14 | oqxB-like, oqxA | catA1 | aph(3′)-Ia | ompK37 p.I70M, ompK37 p.I128M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.G189T, ompK36 p.F198Y, ompK36 p.F207Y, ompK36 p.A217S, ompK36 p.T222L, ompK36 p.D223G, ompK36 p.E232R, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB- like, oqxA | |||||
| 217 | ST 3154 | blaSCO-1 , bla TEM-1B | sul1, sul2 | fosA | dfrA12, dfrA14 | tet(A) | oqxA, oqxB-like | catA2-like | aph(6)-Id, aph(3″)-Ib, aac(3)-IIa, aadA2 | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.G189T, ompK36 p.F198Y, ompK36 p.F207Y, ompK36 p.A217S, ompK36 p.T222L, ompK36 p.D223G, ompK36 p.E232R, ompK36 p.N304E | oqxA, qacE, oqxB-like | |||
| 220 | ST 5382 | fosA-like | oqxB-like, oqxA-like | catA1 | ompK37 p.I70M, ompK37 p.I128M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191Q, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.Q227N, ompK36 p.L229V, ompK36 p.N304E, acrR p.P161R, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB-like, oqxA-like | ||||||||
| 234 | ST 109 | fosA | oqxA, oqxB-like | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S, acrR p.P161R, acrR p.G164A, acrR p.F172S,acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK37 p.I70M, ompK37 p.I128M | oqxA, oqxB-like | |||||||||
| 240 | ST 5383 | fosA-like | tet(D) | oqxA, oqxB-like | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK37 p.I70M, ompK37 p.I128M, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I,acrR p.K201M | oqxA, oqxB-like | ||||||||
| 248 | ST 5384 | fosA-like | tet(A) | oqxB-like, oqxA-like | catA1 | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F198Y, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK37 p.I70M, ompK37 p.I128M, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB-like, oqxA-like | |||||||
| 252 | ST 607 | bla TEM-1B | sul2, sul1 | fosA-like | dfrA7 | tet(A) | oqxB-like, oqxA | catA1 | aph(3″)-Ib, aph(6)-Id | ompK37 p.I70M, ompK37 p.I128M, ompK37 p.N230G, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R, ompK36 p.T254S, acrR p.P161R,acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxB-like, oqxA, qacE | |||
| 267 | ST 36 | blaCTX-M-15 , bla TEM-1B | sul2, sul1 | fosA | dfrA27 | tet(D) | aac(6′)-Ib-cr, oqxA, oqxB | catA2-like | ARR-3 | aph(6)-Id, aph(3″)-Ib, aac(6′)-Ib-cr, aadA16, aac(3)-IIa, aph(6)-Id | ompK36 p.N49S, ompK36 p.L59V, ompK36 p.T184P, ompK37 p.I70M, ompK37 p.I128M, ompK37 p.N230G, acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M | oqxA, qacE, oqxB | ||
| 277 | ST 530 | bla TEM-35 | sul2 | fosA-like | dfrA14 | tet(D) | oqxA, oqxB-like | aph(3″)-Ib, aph(6)-Id | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.L191S, ompK36 p.F207W, ompK36 p.A217S, ompK36 p.N218H, ompK36 p.D224E, ompK36 p.L228V, ompK36 p.E232R,ompK36 p.T254S, ompK37 p.I70M, ompK37 p.I128M | oqxA, oqxB-like | ||||
| 279 | ST 5385 | fosA | oqxA, oqxB | acrR p.P161R, acrR p.G164A, acrR p.F172S, acrR p.R173G, acrR p.L195V, acrR p.F197I, acrR p.K201M, ompK36 p.N49S, ompK36 p.L59V, ompK36 p.T184P, ompK37 p.I70M, ompK37 p.I128M | oqxA, oqxB | |||||||||
* qacE = quaternary ammonium compounds resistance and oqxB and oqxA = efflux pumps mediating resistance against disinfectants.
Table A3.
Phenotypic resistance the P. aeruginosa strains. Data are missing for strains 198, 218 and 312, due to loss during subcultivation. MIC = minimum inhibitory concentration. N.a. = value missing due to loss of strain or failed reaction.
| Sample ID | Piperacillin | Piperacillin/Tazobactam | Ceftrazidime | Cefepime | Imipenem | Meropenem | Gentamicin | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | |
| 17 | ≤4 | S | ≤4 | S | ≤1 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S |
| 22 | ≤4 | S | ≤4 | S | ≤1 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S |
| 32 | ≤4 | S | 8 | S | 2 | S | 2 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 69 | ≤4 | S | ≤4 | S | 4 | S | 2 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 81 | ≤4 | S | 8 | S | 2 | S | ≤1 | S | 1 | S | 1 | S | ≤1 | S |
| 82 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 88 | ≥128 | R | ≥128 | R | ≥64 | R | 32 | R | ≥16 | R | 4 | I | ≤1 | S |
| 99 | 8 | S | 8 | S | 4 | S | 2 | S | 2 | S | 1 | S | ≤1 | S |
| 106 | ≤4 | S | 8 | S | 2 | S | 2 | S | 2 | S | 1 | S | ≤1 | S |
| 114 | ≤4 | S | 8 | S | 2 | S | ≤1 | S | 2 | S | 1 | S | ≤1 | S |
| 137 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | 2 | S | ≤1 | S |
| 144 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | 1 | S | ≤1 | S |
| 147 | 8 | S | ≤4 | S | 4 | S | 8 | S | 2 | S | 0.5 | S | 4 | S |
| 149 | 8 | S | 8 | S | 4 | S | 2 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S |
| 153 | ≤4 | S | 8 | S | 2 | S | ≤1 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 154 | 64 | R | ≤4 | S | ≤1 | S | ≤1 | S | 2 | S | 0.5 | S | ≤1 | S |
| 157 | 16 | S | 8 | S | 4 | S | 4 | S | 2 | S | ≤0.25 | S | 2 | S |
| 160 | ≥128 | R | 32 | R | 16 | R | 32 | R | 8 | I | 8 | I | 8 | R |
| 162 | 64 | R | 32 | R | 8 | S | 8 | S | 2 | S | 1 | S | 2 | S |
| 180 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 183 | 8 | S | 8 | S | 4 | S | 2 | S | 2 | S | 0.5 | S | ≤1 | S |
| 186 | 16 | n.a. | n.a. | S | 4 | S | 2 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 190 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | 0.5 | S | ≤1 | S |
| 195 | 8 | S | 8 | S | 4 | S | ≤1 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 196 | ≤4 | S | ≤4 | S | 2 | S | ≤1 | S | 2 | S | 0.5 | S | ≤1 | S |
| 198 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 204 | 8 | S | 8 | S | 2 | S | ≤1 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 208 | 8 | S | 8 | S | 4 | S | 2 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 218 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 229 | 8 | S | 8 | S | 4 | S | 2 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 233 | ≥128 | R | ≥128 | R | 32 | R | 8 | S | 8 | I | 4 | I | ≤1 | S |
| 236 | 16 | S | 16 | S | 4 | S | 2 | S | 2 | S | 0.5 | S | 2 | S |
| 238 | 8 | S | 16 | S | 4 | S | 2 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 242 | 8 | S | 8 | S | 4 | S | 2 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 243 | 16 | S | 8 | S | 4 | S | 2 | S | 1 | S | ≤0.25 | S | ≤1 | S |
| 272 | 64 | R | 64 | R | 8 | S | 4 | S | 2 | S | 1 | S | ≤1 | S |
| 274 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | 0.5 | S | ≤1 | S |
| 278 | ≤4 | S | ≤4 | S | 2 | S | ≤1 | S | 2 | S | ≤0.25 | S | ≤1 | S |
| 282 | ≤4 | S | 8 | S | ≤1 | S | ≤1 | S | 2 | S | 1 | S | ≤1 | S |
| 285 | ≤4 | S | ≤4 | S | ≤1 | S | ≤1 | S | 2 | S | 1 | S | ≤1 | S |
| 289 | ≤4 | S | 8 | S | ≤1 | S | ≤1 | S | 2 | S | 1 | S | ≤1 | S |
| 290 | 8 | S | 8 | S | ≤1 | S | ≤1 | S | 2 | S | 1 | S | ≤1 | S |
| 296 | ≥128 | R | 64 | R | 4 | S | 8 | S | 1 | S | 1 | S | ≥16 | R |
| 298 | ≥128 | R | ≥128 | R | ≥64 | R | 8 | S | 8 | I | 4 | I | ≤1 | S |
| 301 | 16 | S | 8 | S | 4 | S | 2 | S | 2 | S | 0.5 | S | ≤1 | S |
| 302 | 8 | S | 8 | S | 4 | S | ≤1 | S | 2 | S | 0.5 | S | ≤1 | S |
| 309 | 16 | S | 8 | S | 4 | S | 4 | S | 2 | S | 1 | S | ≤1 | S |
| 310 | 32 | R | 16 | S | 4 | S | 4 | S | 2 | S | 1 | S | ≤1 | S |
| 312 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
Table A4.
Phenotypic resistance of P. aeruginosa strains. Data are missing for strains 198, 218 and 312 due to loss during subcultivation. MIC = minimum inhibitory concentration. N.a. = value missing due to loss of strain or failed reaction.
| Sample ID | Ciprofloxacin | Moxifloxacin | Aztreonam | Amikacin | Tobramycin | Fosfomycin | Colistin | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | |
| 17 | ≤0.25 | S | 1 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 22 | ≤0.25 | S | 1 | R | 2 | I | ≤2 | S | ≤1 | S | 128 | R | 1 | S |
| 32 | ≤0.25 | S | 2 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 69 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 81 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 82 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 88 | 2 | R | ≥8 | R | 32 | R | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 99 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 106 | 2 | R | 1 | R | 8 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 114 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 137 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 144 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 147 | ≤0.25 | S | 2 | R | 4 | I | 8 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 149 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 153 | ≤0.25 | S | 2 | R | 4 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 154 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 157 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 160 | 1 | R | ≥8 | R | ≥64 | R | 16 | I | ≤1 | S | 64 | R | ≤0.5 | S |
| 162 | 0.5 | S | 2 | R | 32 | R | 4 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 180 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 183 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 186 | ≤0.25 | S | 1 | R | 16 | I | ≤2 | S | ≤1 | S | 32 | R | ≤0.5 | S |
| 190 | ≤0.25 | S | 1 | R | 16 | I | 4 | S | ≤1 | S | 32 | R | ≤0.5 | S |
| 195 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 64 | R | ≤0.5 | S |
| 196 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 64 | R | ≤0.5 | S |
| 198 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 204 | ≤0.25 | S | 1 | R | 2 | I | ≤2 | S | ≤1 | S | 32 | R | 2 | S |
| 208 | ≤0.25 | S | 1 | R | 8 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 218 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 229 | ≤0.25 | S | 1 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 233 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 236 | ≤0.25 | S | 1 | R | 16 | I | 8 | S | ≤1 | S | 64 | R | ≤0.5 | S |
| 238 | ≤0.25 | S | 2 | R | 8 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 242 | ≤0.25 | S | 1 | R | 8 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 243 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | 64 | R | ≤0.5 | S |
| 272 | ≤0.25 | S | 2 | R | 32 | R | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 274 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | ≥256 | R | ≤0.5 | S |
| 278 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 282 | ≤0.25 | S | 1 | R | 2 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 285 | ≤0.25 | S | 1 | R | 2 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 289 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 290 | ≤0.25 | S | 0.5 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 296 | ≥4 | R | ≥8 | R | 32 | R | ≤2 | S | ≥16 | R | 64 | R | ≤0.5 | S |
| 298 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 301 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 302 | ≤0.25 | S | 1 | R | 4 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 309 | ≤0.25 | S | 1 | R | 4 | I | ≤2 | S | ≤1 | S | ≤16 | R | ≤0.5 | S |
| 310 | ≤0.25 | S | 2 | R | 16 | I | ≤2 | S | ≤1 | S | 128 | R | ≤0.5 | S |
| 312 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
Table A5.
Phenotypic resistance of the Klebsiella strains. MIC = minimum inhibitory concentration. ESBL = signal in phenotypic testing for extended-spectrum beta-lactamases.
| Sample ID | ESBL | Ampicillin | Ampicillin/Sulbactam | Piperacillin/Tazobactam | Cefuroxime | Cefuroxime Axetil | Cefpodoxime | Cefotaxime | Ceftrazidime | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | ||
| 44 | negative | ≥32 | R | 16 | R | ≤4 | S | 4 | I | 4 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 60 | negative | ≥32 | R | 16 | R | ≤4 | S | ≤1 | I | ≤1 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 73 | positive | ≥32 | R | ≥32 | R | ≥128 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 8 | R |
| 100 | positive | ≥32 | R | ≥32 | R | ≥128 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 16 | R |
| 102 | negative | ≥32 | R | ≤2 | I | 8 | S | 8 | I | 8 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 124 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 146 | negative | ≥32 | R | ≤2 | I | 8 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 177 | positive | ≥32 | R | ≥32 | R | ≥128 | R | 2 | I | 2 | S | ≤0.25 | S | ≤1 | S | ≤1 | I |
| 181 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 184 | negative | 16 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 199 | positive | ≥32 | R | ≥32 | R | 8 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 16 | R |
| 214 | negative | ≥32 | R | ≤2 | I | ≤4 | S | ≤1 | I | ≤1 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 217 | negative | ≥32 | R | ≥32 | R | ≥128 | R | 4 | I | 4 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 220 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 4 | I | 4 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 234 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 240 | negative | ≥32 | R | ≤2 | I | ≤4 | S | ≤1 | I | ≤1 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 248 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 252 | negative | ≥32 | R | 16 | R | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 267 | positive | ≥32 | R | ≥32 | R | 32 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 16 | R |
| 277 | negative | ≥32 | R | ≥32 | R | 64 | R | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 279 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 2 | I | 2 | I | ≤0.25 | S | ≤1 | S | ≤1 | S |
Table A6.
Phenotypic resistance of the Klebsiella strains. MIC = minimum inhibitory concentration. ESBL = signal in phenotypic testing for extended-spectrum beta-lactamases.
| Sample ID | ESBL | Ertapenem | Imipenem | Meropenem | Gentamicin | Ciprofloxacin | Moxifloxacin | Tigecycline | Trimethoprim/Sulfamethoxazole | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | ||
| 44 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
| 60 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | 1 | R | 2 | R | ≤0.5 | S | ≥320 | R |
| 73 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | 1 | R | 2 | R | ≤0.5 | S | ≥320 | R |
| 100 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | ≥4 | R | ≥8 | R | ≤0.5 | S | ≥320 | R |
| 102 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | 0.5 | R | 4 | R | ≤20 | S |
| 124 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
| 146 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | 1 | S | ≤20 | S |
| 177 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | 2 | I | ≥320 | R |
| 181 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
| 184 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
| 199 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 214 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 217 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | ≤0.25 | S | ≤0.25 | S | 2 | I | ≥320 | R |
| 220 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | 0.5 | R | 1 | S | ≤20 | S |
| 234 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | 0.5 | R | 1 | S | ≤20 | S |
| 240 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
| 248 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | 2 | I | ≤20 | S |
| 252 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | 1 | S | ≥320 | R |
| 267 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 277 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | 1 | S | ≥320 | R |
| 279 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≤20 | S |
Table A7.
Phenotypic resistance of Escherichia coli strains. MIC = minimum inhibitory concentration. ESBL = signal in phenotypic testing for extended-spectrum beta-lactamases.
| Sample ID | ESBL | Ampicillin | Ampicillin/Sulbactam | Piperacillin/Tazobactam | Cefuroxime | Cefuroxime Axetil | Cefpodoxime | Cefotaxime | Ceftrazidime | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | ||
| 41 | positive | ≥32 | R | ≥32 | R | 64 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 16 | R |
| 49 | negative | ≥32 | R | 16 | R | ≤4 | S | 4 | I | 4 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 68 | negative | ≥32 | R | 16 | R | ≤4 | S | ≤1 | I | ≤1 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 117 | negative | ≥32 | R | ≥32 | R | 64 | R | 4 | I | 4 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 152 | negative | ≥32 | R | ≥32 | R | ≥128 | R | 4 | I | 4 | S | 0.5 | S | ≤1 | S | ≤1 | S |
| 176 | negative | ≥32 | R | ≥32 | R | ≤4 | I | 2 | I | 2 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 221 | negative | ≥32 | R | ≥32 | R | ≤4 | I | 2 | I | 2 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 222 | negative | ≥32 | R | ≥32 | R | ≤4 | I | 4 | I | 4 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
| 225 | positive | ≥32 | R | ≥32 | R | ≤4 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | 16 | R |
| 245 | positive | ≥32 | R | ≥32 | R | 16 | I | 16 | R | 16 | R | 1 | S | 2 | I | ≤1 | S |
| 270 | positive | ≥32 | R | 16 | R | ≤4 | R | ≥64 | R | ≥64 | R | ≥8 | R | ≥64 | R | ≥64 | R |
| 299 | negative | ≥32 | R | ≤2 | I | ≤4 | S | 4 | I | 4 | S | ≤0.25 | S | ≤1 | S | ≤1 | S |
Table A8.
Phenotypic resistance of Escherichia coli strains. MIC = minimum inhibitory concentration. ESBL = signal in phenotypic testing for extended-spectrum beta-lactamases.
| Sample ID | ESBL | Ertapenem | Imipenem | Meropenem | Gentamicin | Ciprofloxacin | Moxifloxacin | Tigecycline | Trimethoprim/Sulfamethoxazole | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | ||
| 41 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≥16 | R | ≥4 | R | ≥8 | R | ≤0.5 | S | ≥320 | R |
| 49 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 68 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≥4 | R | ≥8 | R | ≤0.5 | S | ≥320 | R |
| 117 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 152 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | 2 | S | 1 | R | 2 | R | ≤0.5 | S | ≥320 | R |
| 176 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 221 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 222 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.5 | S | ≥320 | R |
| 225 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | 0.5 | I | 1 | R | ≤0.5 | S | ≥320 | R |
| 245 | positive | ≤0.5 | S | 0.5 | S | ≤0.25 | S | ≥16 | R | ≥4 | R | ≥8 | R | ≤0.5 | S | ≥320 | R |
| 270 | positive | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ≥4 | R | ≥8 | R | ≤0.5 | S | ≤20 | S |
| 299 | negative | ≤0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤1 | S | 0.5 | I | 2 | R | ≤0.5 | S | ≥320 | R |
Figure A1.
Clustal omega multiple alignment of oprD proteins—one example for the 7 detected subgroups.
Table A9.
Analysis of virulence determinants, ordered by strain and MLST type, of the assessed P. aeruginosa isolates. ST = Sequence type.
| Sample ID | ST-Type | Pathogenicity Factor Groups | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adherence | Anti-Phagocytosis | Biosurfactant | Iron Uptake | Pigment | Protease | Toxin | Regulation | Secretion System | ||
| 017 | ST 381 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algP/algR3 algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 022 | ST 2483 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 032 | ST 3587 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 069 | ST 360 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 081 | ST 244 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW like, xcpX, xcpY, xcpZ |
| 082 | ST 514 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 088 | ST 1682 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 099 | ST 244 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 106 | ST 1521 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 114 | ST 244 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW like, xcpX, xcpY, xcpZ |
| 137 | ST 3014 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 144 | ST 245 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 147 | ST 245 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 149 | ST 381 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimV, pilA, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 153 | ST 704 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | plcH | lasI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 154 | ST 244 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW like, xcpX, xcpY, xcpZ |
| 157 | ST 2616 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilA, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 160 | ST 170 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilA, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 162 | ST 274 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilA, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 180 | ST 856 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 183 | ST 244 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW like, xcpX, xcpY, xcpZ |
| 186 | ST 3588 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 190 | ST 871 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 195 | ST 988 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 196 | ST 2475 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 198 | ST 2476 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 204 | ST 639 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 208 | ST 132 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 218 | ST 856 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 229 | ST 270 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 233 | ST 3227 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 236 | ST 266 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 238 | ST 3589 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | plcH | lasI, rhlI | xcpP, xcpQ, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 242 | ST 3590 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3 like, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 243 | ST 3590 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 272 | ST 2033 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 274 | ST 2033 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 278 | ST 988 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 282 | ST 554 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 285 | ST 554 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilA like, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 289 | ST 1485 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 290 | ST 1485 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 296 | ST 235 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW like, xcpX, xcpY, xcpZ |
| 298 | ST 3227 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 301 | ST 3593 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 302 | ST 1755 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 309 | ST 3592 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimT, fimU, fimV, pilB, pilD, pilE, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilV, pilW, pilX pilY1, pilY2, pilC, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdE | phzM, phzS | aprA, lasA | plcH | lasI, rhlI | xcpP, xcpQ, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 310 | ST 532 | waaA, waaC, waaF, waaG, waaP, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, fpvA pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA | phzM, phzS | aprA lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
| 312 | ST 381 | waaA, waaC, waaF, waaG, waaP, wzy, wzz, chpA, chpB, chpC, chpD, chpE, fimV, pilB, pilD, pilF, pilG, pilH, pilI, pilK, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, pilC like, xcpA/pilD | alg44, alg8, algA, algB, algC, algD, algE, algF, algG, algI, algJ, algK, algL, algP/algR3, algQ, algR, algU, algW, algX, algZ, mucA, mucB, mucC | rhlA, rhlB | fptA, pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchH, pchI, pchR, pvdA, pvdD, pvdE | phzM, phzS | aprA, lasA | toxA, plcH | lasI, rhlI | xcpP, xcpQ, xcpR, xcpS, xcpT, xcpU, xcpV, xcpW, xcpX, xcpY, xcpZ |
Table A10.
Analysis of virulence determinants, ordered by strain and MLST type, of the assessed K. pneumoniae isolates. ST = Sequence type.
| Sample ID | ST-Type | Pathogenicity Factor Groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adherence | Biofilm Formation | Efflux Pump | Immune Evasion | Iron Uptake | Nutritional Factor | Regulation | Secretion System | Serum Resistance | Toxin | ||
| 044 | ST 327 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entD, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE like | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 060 | ST 5379 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | impA/tssA, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | |||
| 073 | ST 39 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA, sciN/tssJ, tle1, tli1, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 100 | ST 152 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entD, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG | |||
| 102 | ST 514 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 124 | ST 399 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkF, mrkH, mrkJ | acrA, acrB | cpsACP, galF, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | glf, wbbM, wbbN, wbbO, wzm, wzt | |||
| 146 | ST 4 | fimA, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 177 | ST 17 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entD, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 181 | ST 5380 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gmd like, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB, | impA/tssA, sciN/tssJ, tle1, tli1, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 184 | ST 5381 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entD like, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE like | rcsA, rcsB | impA/tssA, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | |||
| 199 | ST 17 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB like, manC, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | |||
| 214 | ST 6 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 217 | ST 3154 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 220 | ST 5382 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB like, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, | allA, allB, allC, allD, allR, allS | rcsA, rcsB | |||
| 234 | ST 109 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB, | impA/tssA, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 240 | ST 5383 | fimC, fimD, fimF, fimG, fimH, fimI, fimK | mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | glf, wbbM, wbbN, wbbO, wzm, wzt | |||
| 248 | ST 5384 | mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | |||
| 252 | ST 607 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, wza like, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 267 | ST 36 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE, irp1, irp2, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX | rcsA, rcsB | impA/tssA, sciN/tssJ, tle1, tli1, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 277 | ST 530 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, manB, manC, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB | impA/tssA like, sciN/tssJ, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | glf, wbbM, wbbN, wbbO, wzm, wzt | ||
| 279 | ST 5385 | fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | acrA, acrB | cpsACP, galF, gnd, ugd, wza, wzi | entA, entB, entC, entE, entF, fepA, fepB, fepC, fepD, fepG, fes, ybdA, iroE | rcsA, rcsB, | impA/tssA, sciN/tssJ, tle1, tli1, tssF, tssG, vasE/tssK, vgrG/tssI, vipA/tssB, vipB/tssC | wzm, wzt | ||
Table A11.
Details on the strain-specific short-read archive (SRA) accession numbers.
| Sample ID | Percentage of Good Targets (SeqSphere+) | Average Coverage (Assembled) (SeqSphere+) | Approximated Genome Size (Megabases) (SeqSphere+) | Species (Kraken2) | Sequence Type | Complex Type (SeqSphere+) | SRA Accession |
|---|---|---|---|---|---|---|---|
| Iso00017 | 99.4 | 105 | 6.7 | Pseudomonas aeruginosa | 381 | 1791 | SRR13617317 |
| Iso00022 | 99.4 | 102 | 6.9 | Pseudomonas aeruginosa | 2483 | 1792 | SRR13617316 |
| Iso00032 | 99.2 | 106 | 6.6 | Pseudomonas aeruginosa | 3587 | 1793 | SRR13617305 |
| Iso00041 | 99.4 | 97 | 5.0 | Escherichia coli | 2 (Pasteur) | 11349 | SRR13617294 |
| Iso00044 | 99.7 | 116 | 5.1 | Klebsiella pneumoniae | 327 | 5462 | SRR13617283 |
| Iso00049 | 98.7 | 94 | 5.2 | Escherichia coli | 3 (Pasteur) | 11350 | SRR13617272 |
| Iso00060 | 99.6 | 112 | 5.3 | Klebsiella pneumoniae | 5379 | 5463 | SRR13617261 |
| Iso00068 | 99.6 | 109 | 4.9 | Escherichia coli | 632 (Pasteur) | 11351 | SRR13617250 |
| Iso00069 | 99.6 | 104 | 6.8 | Pseudomonas aeruginosa | 360 | 1794 | SRR13617239 |
| Iso00073 | 99.4 | 104 | 5.8 | Klebsiella pneumoniae | 39 | 5464 | SRR13617236 |
| Iso00081 | 98.7 | 108 | 6.6 | Pseudomonas aeruginosa | 244 | 1795 | SRR13617315 |
| Iso00082 | 99.4 | 112 | 6.3 | Pseudomonas aeruginosa | 514 | 1796 | SRR13617314 |
| Iso00088 | 97.8 | 105 | 6.8 | Pseudomonas aeruginosa | 1682 | 1797 | SRR13617313 |
| Iso00099 | 99.4 | 106 | 6.6 | Pseudomonas aeruginosa | 244 | 1798 | SRR13617312 |
| Iso00100 | 99.2 | 108 | 5.5 | Klebsiella pneumoniae | 152 | 5465 | SRR13617311 |
| Iso00102 | 99.2 | 111 | 5.4 | Klebsiella pneumoniae | 514 | 5466 | SRR13617310 |
| Iso00106 | 99.5 | 110 | 6.4 | Pseudomonas aeruginosa | 1521 | 1799 | SRR13617309 |
| Iso00114 | 99.4 | 105 | 6.7 | Pseudomonas aeruginosa | 244 | 1800 | SRR13617308 |
| Iso00117 | 99.2 | 95 | 5.3 | Escherichia coli | 4 (Pasteur) | 11352 | SRR13617307 |
| Iso00124 | 99.4 | 112 | 5.3 | Klebsiella pneumoniae | 399 | 5467 | SRR13617306 |
| Iso00137 | 99.4 | 110 | 6.4 | Pseudomonas aeruginosa | 3014 | 1801 | SRR13617304 |
| Iso00144 | 99.6 | 109 | 6.5 | Pseudomonas aeruginosa | 245 | 1802 | SRR13617303 |
| Iso00146 | 99.4 | 110 | 5.5 | Klebsiella pneumoniae | 4 | 5468 | SRR13617302 |
| Iso00147 | 99.5 | 108 | 6.6 | Pseudomonas aeruginosa | 245 | 1802 | SRR13617301 |
| Iso00149 | 99.6 | 104 | 6.9 | Pseudomonas aeruginosa | 381 | 1803 | SRR13617300 |
| Iso00152 | 99.4 | 98 | 5.2 | Escherichia coli | 22 (Pasteur) | 11353 | SRR13617299 |
| Iso00153 | 98.5 | 111 | 6.4 | Pseudomonas aeruginosa | 704 | ? | SRR13617298 |
| Iso00154 | 99.4 | 102 | 7.0 | Pseudomonas aeruginosa | 244 | 1805 | SRR13617297 |
| Iso00157 | 99.6 | 114 | 6.3 | Pseudomonas aeruginosa | 2616 | 1806 | SRR13617296 |
| Iso00160 | 99.2 | 115 | 6.2 | Pseudomonas aeruginosa | 170 | 1807 | SRR13617295 |
| Iso00162 | 99.1 | 111 | 6.5 | Pseudomonas aeruginosa | 274 | 1808 | SRR13617293 |
| Iso00176 | 99.0 | 98 | 5.1 | Escherichia coli | 132 (Pasteur) | 11354 | SRR13617292 |
| Iso00177 | 99.6 | 108 | 5.5 | Klebsiella pneumoniae | 17 | 5469 | SRR13617291 |
| Iso00180 | 99.8 | 110 | 6.5 | Pseudomonas aeruginosa | 856 | 1809 | SRR13617290 |
| Iso00181 | 99.9 | 107 | 5.6 | Klebsiella pneumoniae | 5380 | 5470 | SRR13617289 |
| Iso00183 | 99.5 | 107 | 6.7 | Pseudomonas aeruginosa | 244 | 1795 | SRR13617288 |
| Iso00184 | 98.3 | 104 | 5.6 | Klebsiella variicola subsp. variicola | 5381 | 5471 | SRR13617287 |
| Iso00186 | 98.7 | 113 | 6.3 | Pseudomonas aeruginosa | 3588 | 1810 | SRR13617286 |
| Iso00190 | 99.7 | 114 | 6.3 | Pseudomonas aeruginosa | 871 | 1811 | SRR13617285 |
| Iso00195 | 99.5 | 111 | 6.5 | Pseudomonas aeruginosa | 988 | 1812 | SRR13617284 |
| Iso00196 | 99.5 | 101 | 7.1 | Pseudomonas aeruginosa | 2475 | 1813 | SRR13617282 |
| Iso00198 | 99.6 | 112 | 6.4 | Pseudomonas aeruginosa | 2476 | 1814 | SRR13617281 |
| Iso00199 | 99.4 | 108 | 5.6 | Klebsiella pneumoniae | 17 | 5472 | SRR13617280 |
| Iso00204 | 99.5 | 104 | 6.9 | Pseudomonas aeruginosa | 639 | 1815 | SRR13617279 |
| Iso00208 | 99.7 | 109 | 6.5 | Pseudomonas aeruginosa | 132 | 1816 | SRR13617278 |
| Iso00214 | 99.7 | 108 | 5.5 | Klebsiella pneumoniae | 6 | 5473 | SRR13617277 |
| Iso00217 | 99.8 | 104 | 5.7 | Klebsiella pneumoniae | 3154 | 5474 | SRR13617276 |
| Iso00218 | 99.7 | 109 | 6.5 | Pseudomonas aeruginosa | 856 | 1809 | SRR13617275 |
| Iso00220 | 97.8 | 110 | 5.4 | Klebsiella quasipneumoniae subsp. similipneumoniae | 5382 | 5475 | SRR13617274 |
| Iso00221 | 99.0 | 94 | 5.1 | Escherichia coli | 132 (Pasteur) | 11354 | SRR13617273 |
| Iso00222 | 99.0 | 96 | 5.1 | Escherichia coli | 132 (Pasteur) | 11354 | SRR13617271 |
| Iso00225 | 99.1 | 99 | 5.2 | Escherichia coli | 506 (Pasteur) | 11355 | SRR13617270 |
| Iso00229 | 99.6 | 109 | 6.5 | Pseudomonas aeruginosa | 270 | 1817 | SRR13617269 |
| Iso00233 | 97.8 | 114 | 6.1 | Pseudomonas aeruginosa | 3227 | 1818 | SRR13617268 |
| Iso00234 | 99.7 | 111 | 5.5 | Klebsiella pneumoniae | 109 | 5476 | SRR13617267 |
| Iso00236 | 99.7 | 112 | 6.4 | Pseudomonas aeruginosa | 266 | 1819 | SRR13617266 |
| Iso00238 | 98.7 | 108 | 6.6 | Pseudomonas aeruginosa | 3589 | 1820 | SRR13617265 |
| Iso00240 | 98.9 | 112 | 5.4 | Klebsiella pneumoniae | 5383 | 5477 | SRR13617264 |
| Iso00242 | 98.9 | 111 | 6.4 | Pseudomonas aeruginosa | 3590 | 1821 | SRR13617263 |
| Iso00243 | 98.9 | 111 | 6.4 | Pseudomonas aeruginosa | 3590 | 1821 | SRR13617262 |
| Iso00245 | 99.3 | 107 | 4.8 | Escherichia coli | 2 (Pasteur) | 11356 | SRR13617260 |
| Iso00248 | 97.2 | 108 | 5.5 | Klebsiella quasivariicola | 5384 | 5478 | SRR13617259 |
| Iso00252 | 99.6 | 112 | 5.3 | Klebsiella pneumoniae | 607 | 5479 | SRR13617258 |
| Iso00267 | 99.6 | 103 | 5.7 | Klebsiella pneumoniae | 36 | 5480 | SRR13617257 |
| Iso00270 | 99.2 | 100 | 4.9 | Escherichia coli | 2 (Pasteur) | 11358 | SRR13617256 |
| Iso00272 | 99.5 | 109 | 6.5 | Pseudomonas aeruginosa | 2033 | 1822 | SRR13617255 |
| Iso00274 | 99.4 | 109 | 6.5 | Pseudomonas aeruginosa | 2033 | 1822 | SRR13617254 |
| Iso00277 | 99.4 | 109 | 5.5 | Klebsiella pneumoniae | 530 | 5481 | SRR13617253 |
| Iso00278 | 99.6 | 110 | 6.5 | Pseudomonas aeruginosa | 988 | 1823 | SRR13617252 |
| Iso00279 | 99.7 | 111 | 5.5 | Klebsiella pneumoniae | 5385 | 5482 | SRR13617251 |
| Iso00282 | 99.3 | 108 | 6.6 | Pseudomonas aeruginosa | 554 | 1824 | SRR13617249 |
| Iso00285 | 99.3 | 109 | 6.5 | Pseudomonas aeruginosa | 554 | 1824 | SRR13617248 |
| Iso00289 | 99.6 | 112 | 6.3 | Pseudomonas aeruginosa | 1485 | 1825 | SRR13617247 |
| Iso00290 | 99.7 | 113 | 6.3 | Pseudomonas aeruginosa | 1485 | 1825 | SRR13617246 |
| Iso00296 | 99.7 | 106 | 6.7 | Pseudomonas aeruginosa | 235 | 1826 | SRR13617245 |
| Iso00298 | 97.8 | 116 | 6.1 | Pseudomonas aeruginosa | 3227 | 1818 | SRR13617244 |
| Iso00299 | 99.3 | 108 | 4.6 | Escherichia coli | 1018 (Pasteur) | 11357 | SRR13617243 |
| Iso00301 | 98.6 | 112 | 6.3 | Pseudomonas aeruginosa | 3593 | 1827 | SRR13617242 |
| Iso00302 | 99.6 | 113 | 6.3 | Pseudomonas aeruginosa | 1755 | 1828 | SRR13617241 |
| Iso00309 | 98.6 | 109 | 6.5 | Pseudomonas aeruginosa | 3592 | 1829 | SRR13617240 |
| Iso00310 | 99.3 | 105 | 6.8 | Pseudomonas aeruginosa | 532 | 1830 | SRR13617238 |
| Iso00312 | 99.4 | 106 | 6.7 | Pseudomonas aeruginosa | 381 | 1791 | SRR13617237 |
Author Contributions
U.L., D.D. and J.M. designed and coordinated this study. T.T., F.P. and S.T. performed bioinformatic analysis. M.L. supported the management of this study. A.J. managed the data collection. H.F., D.D. and U.L. wrote the first draft of this manuscript. K.O. conducted and supervised fieldwork. C.W.A. and K.T. conducted and supervised lab work. M.K. and S.S. supported the interpretation of the results, writing and editing the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by institutional funds of the Bernhard Nocht Institute for Tropical Medicine (BNITM).
Institutional Review Board Statement
The study was conducted according to guidelines of the Declaration of Helsinki. The Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology in Kumasi, Ghana, approved this study (approval number CHRPE/AP/078/16).
Informed Consent Statement
Informed consent was obtained from all study participants.
Data Availability Statement
All relevant data have been provided in the paper and its Appendix A materials. Raw data are available applying the links as indicated in the methods chapter and can also be provided by the authors on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai P.S., Bebell L.M., Meney C., Valeri L., White M.C. Epidemiology of antibiotic-resistant wound infections from six countries in Africa. BMJ Glob. Health. 2018;2(Suppl. 4):e000475. doi: 10.1136/bmjgh-2017-000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mama M., Abdissa A., Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2014;13:14. doi: 10.1186/1476-0711-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moremi N., Mushi M.F., Fidelis M., Chalya P., Mirambo M., Mshana S.E. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res. Notes. 2014;7:211. doi: 10.1186/1756-0500-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kassam N.A., Damian D.J., Kajeguka D., Nyombi B., Kibiki G.S. Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a Tertiary Hospital, northern Tanzania. BMC Res. Notes. 2017;10:757. doi: 10.1186/s13104-017-3092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen H., Janssen I., Cooper P., Kainyah C., Pellio T., Quintel M., Monnheimer M., Groß U., Schulze M.H. Antimicrobial-Resistant Bacteria in Infected Wounds, Ghana, 2014. Emerg. Infect. Dis. 2018;24:916–919. doi: 10.3201/eid2405.171506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazimoto T., Abdulla S., Bategereza L., Juma O., Mhimbira F., Weisser M., Utzinger J., von Müller L., Becker S.L. Causative agents and antimicrobial resistance patterns of human skin and soft tissue infections in Bagamoyo, Tanzania. Acta Trop. 2018;186:102–106. doi: 10.1016/j.actatropica.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Høiby N., Ciofu O., Johansen H.K., Song Z.J., Moser C., Jensen P.Ø., Molin S., Givskov M., Tolker-Nielsen T., Bjarnsholt T. The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 2011;3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Percival S.L., Hill K.E., Williams D.W., Hooper S.J., Thomas D.W., Costerton J.W. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012;20:647–657. doi: 10.1111/j.1524-475X.2012.00836.x. [DOI] [PubMed] [Google Scholar]
- 9.Rahim K., Saleha S., Zhu X., Huo L., Basit A., Franco O.L. Bacterial Contributionin Chronicity of Wounds. Microb. Ecol. 2017;73:710–721. doi: 10.1007/s00248-016-0867-9. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z., Hsia H.C. The Impact of Microbial Communities on Wound Healing: A Review. Ann. Plast. Surg. 2018;81:113–123. doi: 10.1097/SAP.0000000000001450. [DOI] [PubMed] [Google Scholar]
- 11.Mulcahy L.R., Isabella V.M., Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2014;68:1–12. doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serra R., Grande R., Butrico L., Rossi A., Settimio U.F., Caroleo B., Amato B., Gallelli L., de Franciscis S. Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert. Rev. Anti. Infect. Ther. 2015;13:605–613. doi: 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 13.Kirketerp-Møller K., Jensen P.Ø., Fazli M., Madsen K.G., Pedersen J., Moser C., Tolker-Nielsen T., Høiby N., Givskov M., Bjarnsholt T. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008;46:2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler J.E., Hutchens M.P., Sadow P.M., Modi B.P., Tavakkolizadeh A., Gates J.D. Klebsiella pneumoniae necrotizing fasciitis and septic arthritis: An appearance in the Western hemisphere. Surg. Infect. 2007;8:227–232. doi: 10.1089/sur.2006.007. [DOI] [PubMed] [Google Scholar]
- 15.Dana A.N., Bauman W.A. Bacteriology of pressure ulcers in individuals with spinal cord injury: What we know and what we should know. J. Spinal Cord. Med. 2015;38:147–160. doi: 10.1179/2045772314Y.0000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washington M.A., Barnhill J.C., Duff M.A., Griffin J. Recovery of Bacteria and Fungi from a Leg Wound. J. Spec. Oper. Med. 2015;15:113–116. doi: 10.55460/DW1G-SZNG. [DOI] [PubMed] [Google Scholar]
- 17.Vyas K.S., Wong L.K. Detection of Biofilm in Wounds as an Early Indicator for Risk for Tissue Infection and Wound Chronicity. Ann. Plast. Surg. 2016;76:127–131. doi: 10.1097/SAP.0000000000000440. [DOI] [PubMed] [Google Scholar]
- 18.Wetterslev M., Rose-Larsen K., Hansen-Schwartz J., Steen-Andersen J., Møller K., Møller-Sørensen H. Mechanism of injury and microbiological flora of the geographical location are essential for the prognosis in soldiers with serious warfare injuries. Dan. Med. J. 2013;60:A4704. [PubMed] [Google Scholar]
- 19.Yun H.C., Murray C.K., Roop S.A., Hospenthal D.R., Gourdine E., Dooley D.P. Bacteria recovered from patients admitted to a deployed U.S. military hospital in Baghdad, Iraq. Mil. Med. 2006;171:821–825. doi: 10.7205/MILMED.171.9.821. [DOI] [PubMed] [Google Scholar]
- 20.Micheel V., Hogan B., Rakotoarivelo R.A., Rakotozandrindrainy R., Razafimanatsoa F., Razafindrabe T., Rakotondrainiarivelo J.P., Crusius S., Poppert S., Schwarz N.G., et al. Identification of nasal colonization with β-lactamase-producing Enterobacteriaceae in patients, health care workers and students in Madagascar. Eur. J. Microbiol. Immunol. 2015;5:116–125. doi: 10.1556/EuJMI-D-15-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frickmann H., Podbielski A., Kreikemeyer B. Resistant Gram-Negative Bacteria and Diagnostic Point-of-Care Options for the Field Setting during Military Operations. Biomed. Res. Int. 2018;2018:9395420. doi: 10.1155/2018/9395420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride M.E., Duncan W.C., Knox J.M. Physiological and environmental control of Gram negative bacteria on skin. Br. J. Dermatol. 1975;93:191–199. doi: 10.1111/j.1365-2133.1975.tb06740.x. [DOI] [PubMed] [Google Scholar]
- 23.Krumkamp R., Oppong K., Hogan B., Strauss R., Frickmann H., Wiafe-Akenten C., Boahen K.G., Rickerts V., McCormick Smith I., Groß U., et al. Spectrum of antibiotic resistant bacteria and fungi isolated from chronically infected wounds in a rural district hospital in Ghana. PLoS ONE. 2020;15:e0237263. doi: 10.1371/journal.pone.0237263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codjoe F.S., Donkor E.S., Smith T.J., Miller K. Phenotypic and Genotypic Characterization of Carbapenem-Resistant Gram-Negative Bacilli Pathogens from Hospitals in Ghana. Microb. Drug. Resist. 2019;25:1449–1457. doi: 10.1089/mdr.2018.0278. [DOI] [PubMed] [Google Scholar]
- 25.Del Barrio-Tofiño E., López-Causapé C., Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents. 2020;56:106196. doi: 10.1016/j.ijantimicag.2020.106196. [DOI] [PubMed] [Google Scholar]
- 26.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 27.Denamur E., Clermont O., Bonacorsi S., Gordon D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021;19:37–54. doi: 10.1038/s41579-020-0416-x. [DOI] [PubMed] [Google Scholar]
- 28.Eibach D., Belmar Campos C., Krumkamp R., Al-Emran H.M., Dekker D., Boahen K.G., Kreuels B., Adu-Sarkodie Y., Aepfelbacher M., Park S.E., et al. Extended spectrum beta-lactamase producing Enterobacteriaceae causing bloodstream infections in rural Ghana, 2007–2012. Int. J. Med. Microbiol. 2016;306:249–254. doi: 10.1016/j.ijmm.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Eibach D., Dekker D., Gyau Boahen K., Wiafe Akenten C., Sarpong N., Belmar Campos C., Berneking L., Aepfelbacher M., Krumkamp R., Owusu-Dabo E., et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Vet. Microbiol. 2018;217:7–12. doi: 10.1016/j.vetmic.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Labi A.K., Bjerrum S., Enweronu-Laryea C.C., Ayibor P.K., Nielsen K.L., Marvig R.L., Newman M.J., Andersen L.P., Kurtzhals J.A.L. High Carriage Rates of Multidrug-Resistant Gram-Negative Bacteria in Neonatal Intensive Care Units from Ghana. Open Forum Infect. Dis. 2020;7:ofaa109. doi: 10.1093/ofid/ofaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agyepong N., Govinden U., Owusu-Ofori A., Amoako D.G., Allam M., Janice J., Pedersen T., Sundsfjord A., Essack S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019;85:117–123. doi: 10.1016/j.ijid.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Falgenhauer L., Imirzalioglu C., Oppong K., Akenten C.W., Hogan B., Krumkamp R., Poppert S., Levermann V., Schwengers O., Sarpong N., et al. Detection and Characterization of ESBL-Producing Escherichia coli from Humans and Poultry in Ghana. Front. Microbiol. 2019;9:3358. doi: 10.3389/fmicb.2018.03358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donkor E.S., Horlortu P.Z., Dayie N.T., Obeng-Nkrumah N., Labi A.K. Community acquired urinary tract infections among adults in Accra, Ghana. Infect. Drug. Resist. 2019;12:2059–2067. doi: 10.2147/IDR.S204880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obeng-Nkrumah N., Labi A.K., Blankson H., Awuah-Mensah G., Oduro-Mensah D., Anum J., Teye J., Kwashie S.D., Bako E., Ayeh-Kumi P.F., et al. Household cockroaches carry CTX-M-15-, OXA-48- and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019;19:272. doi: 10.1186/s12866-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel L., Corvec S., Rapoport M., Mugnier P., Petroni A., Pasteran F., Faccone D., Galas M., Drugeon H., Cattoir V., et al. Identification of the novel narrow-spectrum beta-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 2007;51:2179–2184. doi: 10.1128/AAC.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Münch J., Hagen R.M., Müller M., Kellert V., Wiemer D.F., Hinz R., Schwarz N.G., Frickmann H. Colonization with Multidrug-Resistant Bacteria—On the Efficiency of Local Decolonization Procedures. Eur. J. Microbiol. Immunol. 2017;7:99–111. doi: 10.1556/1886.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornelis P., Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasannejad-Bibalan M., Jafari A., Sabati H., Goswami R., Jafaryparvar Z., Sedaghat F., Sedigh Ebrahim-Saraie H. Risk of type III secretion systems in burn patients with Pseudomonas aeruginosa wound infection: A systematic review and meta-analysis. Burns. 2020 doi: 10.1016/j.burns.2020.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Rumbaugh K.P., Griswold J.A., Hamood A.N. Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. J. Burn Care Rehabil. 1999;20(Pt 1):42–49. doi: 10.1097/00004630-199901001-00008. [DOI] [PubMed] [Google Scholar]
- 40.Colmer-Hamood J.A., Dzvova N., Kruczek C., Hamood A.N. In Vitro Analysis of Pseudomonas aeruginosa Virulence Using Conditions That Mimic the Environment at Specific Infection Sites. Prog. Mol. Biol. Transl. Sci. 2016;142:151–191. doi: 10.1016/bs.pmbts.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. [(accessed on 15 December 2020)]; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 42.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jünemann S., Sedlazeck F.J., Prior K., Albersmeier A., John U., Kalinowski J., Mellmann A., Goesmann A., von Haeseler A., Stoye J., et al. Updating benchtop sequencing performance comparison. Nature Biotechnol. 2013;31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 44.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 45.Seemann T. Abricate, Github. [(accessed on 22 March 2021)]; Available online: https://github.com/tseemann/abricate.
- 46.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.H., McDermott P.F., et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data have been provided in the paper and its Appendix A materials. Raw data are available applying the links as indicated in the methods chapter and can also be provided by the authors on reasonable request.