Abstract
Antiseptic use for body decolonization is the main activity applied to prevent healthcare-associated infections, including those caused by S. aureus. Consequentially, tolerance to several antiseptics such as chlorhexidine gluconate (CHG) has developed. This study aimed to estimate the prevalence of CHG tolerance among S. aureus strains in Israel and to evaluate factors that may affect this tolerance. Furthermore, it tested the associations between phenotypic and genotypic CHG tolerance. S. aureus strains (n = 190) were isolated from clinical samples of patients admitted to various medical institutions in Israel. Phenotypic susceptibility to CHG was assessed by determining minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). Genotypic tolerance was detected using real-time PCR for detection of qac A/B genes. MIC for the antibiotic mupirocin was determined using the Etest method. Presence of the Panton–Valentine Leucocidin (pvl) toxin, mecA and mecC genes was detected using an eazyplex® MRSAplus kit (AmplexDiagnostics GmbH, Gars, Germany). CHG tolerance was observed in 13.15% of the isolates. An association between phenotypic and genotypic tolerance to CHG was observed. Phenotypic tolerance to CHG was associated with methicillin resistance but not with mupirocin resistance. Additionally, most of the CHG-tolerant strains were isolated from blood cultures. In conclusion, this work shed light on the prevalence of reduced susceptibility to CHG among S. aureus strains in Israel and on the characteristics of tolerant strains. CHG-tolerant strains were more common than methicillin-resistant ones in samples from invasive infections. Further research should be performed to evaluate risk factors for the development of CHG tolerance.
Keywords: chlorhexidine, reduced susceptibility, healthcare-associated infections, S. aureus, genotypic and phenotypic susceptibility
1. Introduction
Staphylococcus aureus (S. aureus) is one of the most common causes of human diseases, ranging from skin and soft tissue infections to pneumonia, meningitis, and sepsis [1]. S. aureus infections and, specifically, methicillin-resistant S. aureus (MRSA) infections are frequent both in the community and in healthcare institutions (nosocomial infections), are associated with high morbidity and mortality rates, and incur high medical costs [2]. Magill et al. reported that S. aureus is the second most common cause of healthcare-associated infections in the United States [3]. It was estimated that 40–60% of all nosocomial S. aureus infections are due to MRSA [4].
One of the main preventive activities against healthcare-associated infections is the use of antiseptics and biocides for hand and body decolonization [5] in order to reduce the bacterial load of pathogens from the colonized or infected body [6]. One example of such an antiseptic agent is chlorhexidine gluconate (CHG). Chlorhexidine (1,6-bis(4-chlorophenylbiguanido)hexane) is active against a wide range of fungi, some viruses [7,8,9,10], and gram-positive and gram-negative bacteria, especially MRSA and vancomycin-resistant enterococci [11]. It is extensively utilized because of its long-lasting bactericidal effect and its satisfactory tolerability and safety profiles [6].
However, the massive use of CHG has led to the development of reduced susceptibility to CHG among bacteria in general and in S. aureus specifically (reviewed in [6,12]). CHG antiseptic tolerance is mediated by efflux pumps that are encoded by several genes, such as qac A, qac B, and smr [6,13]. Interestingly, strains that show phenotypic tolerance to CHG do not always carry tolerance genes and vice versa. One of the most important findings regarding these genes is their association with resistance to antimicrobial agents [6]. This phenomenon is explained by either cross-resistance or co-resistance. Cross-resistance means that one efflux pump can remove both biocides and antibiotics. Co-resistance occurs when tolerance genes and antibiotic resistance genes are colocalized on the same mobile genetic element [14].
The prevalence of reduced susceptibility to CHG among S. aureus strains in Israel is unknown. This study aimed to assess its prevalence and to search for factors that may be associated with CHG tolerance. Furthermore, it assessed the possible association between phenotypic tolerance and qac A/B presence. We found that 13.15% of our strains were CHG-tolerant; phenotypic tolerance was associated with genotypic tolerance to CHG. Some bacterial characteristics were associated with CHG tolerance.
2. Results
2.1. Characteristics of Clinical S. aureus Isolates
This study included 92 MRSA and 98 methicillin-sensitive S. aureus (MSSA) isolates recovered from blood cultures (35.3% isolates) and wound cultures (64.7% isolates) (Table 1). Thirty-nine (20.5%) isolates carried the gene for the pvl toxin; most (28/39, 71.8%) of these pvl-positive isolates were MRSA ones (p = 0.001). All MRSA isolates contained the mecA gene. None of the isolates carried the mecC gene (data not shown).
Table 1.
Characteristics of clinical isolates.
| Characteristic | MSSA n = 98 n (%) |
MRSA n = 92 n (%) |
Total Isolates n = 190 n |
p-Value 1 |
|---|---|---|---|---|
| Sample source | 0.865 | |||
| Blood cultures | 34 (50.7) | 33 (49.3) | 67 | |
| Wounds | 64 (52) | 59 (48) | 123 | |
| Presence of pvl | 0.001 2 | |||
| Yes | 11 (28.2) | 28 (71.8) | 39 | |
| No | 87 (57.6) | 64 (42.4) | 151 | |
| Susceptibility to mupirocin | <0.001 | |||
| Resistant | 2 (11.1) | 16 (88.9) | 18 | |
| Sensitive | 96 (59.25) | 76 (40.75) | 162 | |
| Mupirocin’s MIC (mg/L) | 0.023 | |||
| Mean | 15.3 | 178.75 | 94.45 | |
| Median (Q1, Q3) | 0.125 (0.94, 0.19) | 0.19 (0.102, 0.865) | 0.125 (0.094, 0.25) |
1 Chi-squared test, Mann–Whitney test. 2 Bold values indicate statistical significance.
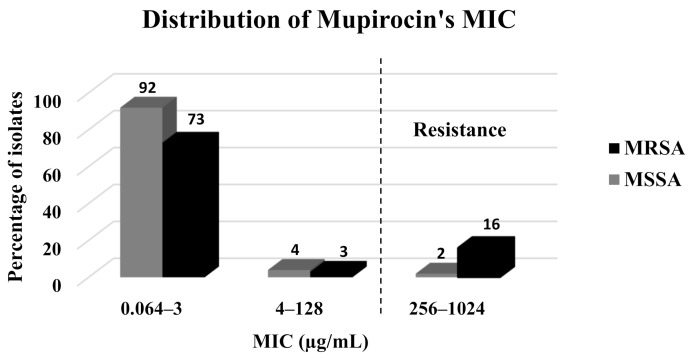
Mupirocin resistance was observed in 9.5% of the isolates and was significantly more prevalent among the MRSA isolates (16/92, 17.4%) as compared to the MSSA isolates (2/98, 2.05%) (p < 0.001) (Table 1, Figure 1). In addition, the mean and median MICs were significantly higher among the MRSA isolates (15.3 mg/L and 0.19 mg/L, respectively) as compared to the MSSA isolates (178.75 mg/L and 0.125 mg/L, respectively) (p = 0.023) (Table 1).
Figure 1.
Distribution of mupirocin’s MIC (mg/L) among the clinical isolates.
2.2. Susceptibility to CHG among Clinical S. aureus Isolates
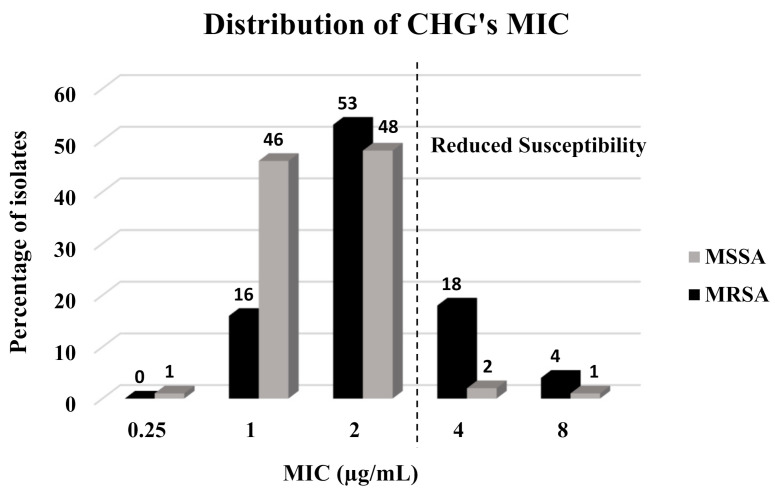
The prevalence of reduced susceptibility to CHG was 13.15% (25/190) (Table 2), with most of the tolerant isolates belonging to the MRSA group (22/25, 88%) (p < 0.001) (Table 2, Figure 2). Mean CHG’s MICs were higher and the MIC distribution was significantly different between MRSA and MSSA isolates (2.5 mg/L and 1.62 mg/L, respectively) (p < 0.001). Mean CHG MBCs were also higher and the distribution was significantly different between MRSA and MSSA isolates (2.45 mg/L and 2.23 mg/L, respectively) (p < 0.001). In parallel, out of the 25 (13.15%) isolates that carried the qac A/B gene, 21 (84%) were MRSA isolates and only four (16%) were MSSA isolates (p < 0.001).
Table 2.
Susceptibility to CHG among clinical isolates.
| Characteristic | MSSA n = 98 n (%) |
MRSA n = 92 n (%) |
Total Isolates n = 190 n |
p-Value 1 |
|---|---|---|---|---|
| Reduced susceptibility to CHG | <0.001 2 | |||
| Yes | 3 (12) | 22 (88) | 25 | |
| No | 95 (57.6) | 70 (42.4) | 165 | |
| CHG’s MIC (mg/L) | <0.001 | |||
| Mean | 1.62 | 2.5 | 2.04 | |
| Median (Q1, Q3) | 2 (1, 2) | 2 (2, 3.5) | 2 (1, 2) | |
| CHG’s MBC (mg/L) | <0.035 | |||
| Mean | 2.23 | 2.45 | 2.33 | |
| Median (Q1, Q3) | 2 (1, 2) | 2 (2, 2) | 2 (2, 2) | |
| qac A/B presence | <0.001 | |||
| Yes | 4 (16) | 21 (84) | 25 | |
| No | 94 (57) | 71 (43) | 165 |
1 Chi-squared test, Mann–Whitney test. 2 Bold values indicate statistical significance.
Figure 2.
Distribution of CHG’s MICs (mg/L) among clinical isolates.
2.3. The Associations between Phenotypic and Genotypic Tolerance to CHG
An association was found between the genotypic and phenotypic tolerance to CHG (p < 0.001) (Table 3). When only MSSA isolates were assessed, no association between the genotype and the phenotype was found (p = 0.717). In contrast, in MRSA strains, the phenotype and the genotype of CHG susceptibility were linked (p = 0.004) (Table 4).
Table 3.
Clinical and epidemiological characteristics of CHG-tolerant vs. CHG-susceptible isolates.
| Characteristic | Reduced Susceptibility to CHG | Total Isolates n = 190 n (%) |
p-Value 1 | |
|---|---|---|---|---|
| Yes n = 25 n (%) |
No n = 165 n (%) |
|||
| qac A/B presence | <0.0001 2 | |||
| Yes | 10 (40) | 15 (60) | 25 | |
| No | 15 (9) | 150 (81) | 165 | |
| Methicillin susceptibility | <0.0001 | |||
| Resistant (MRSA) | 22 (24) | 70 (76) | 92 | |
| Sensitive (MSSA) | 3 (3) | 95 (97) | 98 | |
| Mupirocin susceptibility | 0.001 | |||
| Resistant | 7 (39) | 11 (61) | 18 | |
| Sensitive | 18 (10.5) | 154 (89.5) | 172 | |
| Mupirocin’s MIC (mg/L) | 0.066 | |||
| Mean | 257.44 | 69.74 | 94.45 | |
| Median (Q1, Q3) | 0.19 (0.125, 256) | 0.064 (0.094, 0.25) | 0.125 (0.094, 0.25) | |
| pvl presence | 0.548 | |||
| Yes | 4 (10.25) | 35 (89.75) | 39 | |
| No | 21 (12.3) | 130 (87.7) | 171 | |
| Sample source | 0.020 | |||
| Blood culture | 14 (21) | 53 (79) | 67 | |
| Wounds | 11 (8.95) | 112 (91.05) | 123 | |
1 Chi-squared test, Mann–Whitney test. 2 Bold values indicate statistical significance.
Table 4.
Clinical and epidemiological characteristics of CHG-tolerant isolates vs. CHG-susceptible MSSA and MRSA isolates.
| Characteristic | Reduced Susceptibility to CHG among MSSA Isolates (n = 98) |
p-Value 1 | Reduced Susceptibility to CHG among MRSA Isolates (n = 92) |
p-Value 1 | ||
|---|---|---|---|---|---|---|
| Yes (n = 3) | No (n = 95) | Yes (n = 22) | No (n = 70) | |||
| qac A/B presence | 0.717 | 0.004 2 | ||||
| Yes | 0 | 4 (100) | 10 (47.6) | 11 (52.4) | ||
| No | 3 (3.2) | 91 (96.8) | 12 (16.9) | 59 (83.1) | ||
| Mupirocin susceptibility | <0.0001 | 0.161 | ||||
| Resistant | 1 (50) | 1 (50) | 6 (37.5) | 10 (62.5) | ||
| Sensitive | 2 (2.1) | 94 (97.9) | 16 (21.1) | 60 (78.9) | ||
| Mupirocin’s MIC (mg/L) | 0.846 | 0.243 | ||||
| Mean | 85.4 | 13.1 | 280.9 | 146.6 | ||
| Median (Q1, Q3) | 0.125 (0.094, 0.19) | 0.22 (0.125, 776) | 0.125 (0.101, 0.25) | |||
| pvl presence | 0.532 | 0.152 | ||||
| Yes | 0 | 11(100) | 4 (14.3) | 24 (85.7) | ||
| No | 3 (3.4) | 84 (96.6) | 18 (28.1) | 46 (71.9) | ||
| Sample source | 0.960 | 0.009 | ||||
| Blood culture | 1 (2.9) | 33 (97.1) | 13 (39.4) | 20 (60.6) | ||
| Wounds | 2 (3.1) | 62 (96.9) | 9 (15.3) | 50 (84.7) | ||
1 Chi-squared test, Mann-Whitney test. 2 Bold values indicate statistical significance.
2.4. The Associations between Reduced Susceptibility to CHG and Methicillin Resistance
The prevalence rate of reduced susceptibility to CHG among MRSA isolates was 8-fold higher than the prevalence rate among MSSA isolates (24% of the MRSA isolates were CHG-tolerant as compared to 3% of the MSSA isolates) (p < 0.0001) (Table 3).
2.5. The Associations between Reduced Susceptibility to CHG and Mupirocin Resistance
Since CHG is used for body decolonization, a possible association between CHG tolerance and resistance to mupirocin, which is an antibiotic that is used superficially, was considered. Most (61%) mupirocin-resistant isolates were susceptible to CHG. The rate of reduced CHG susceptibility was significantly lower among mupirocin-sensitive isolates as compared to mupirocin-resistant isolates (10.5% vs. 39%, respectively, p = 0.001) (Table 3). It should be noted that the average mupirocin MIC was higher among isolates with a reduced CHG susceptibility (257.44 mg/L) as compared to susceptible isolates (69.74 mg/L). However, the difference in the distribution of mupirocin MIC was not statistically significant (p = 0.066).
There was no association between CHG tolerance and mupirocin resistance among MRSA isolates. In addition, no significant difference was seen in the distribution of mupirocin MIC for CHG-tolerant vs. CHG-susceptible MRSA isolates (Table 4). In contrast, among the MSSA isolates, while 50% of the mupirocin-resistant isolates were CHG tolerant, only 2.1% of the mupirocin-sensitive isolates were tolerant (p = 0.000) (Table 4). However, the difference in the distribution of the mupirocin’s MIC was not statistically significant between the CHG-tolerant isolates as compared to the CHG-susceptible MSSA isolates (p = 0.846).
2.6. The Association between Reduced Susceptibility to CHG and the pvl Toxin Presence
No association was found between reduced susceptibility to CHG and the pvl toxin presence (p = 0.548, Table 3) neither in MRSA nor in MSSA isolates (Table 4).
2.7. The Association between Reduced Susceptibility to CHG and the Sample Source
As mentioned above, 67 isolates were originally isolated from blood cultures and 123 isolates were isolated from wounds. The highest percentage of CHG-tolerant isolates was found among isolates from blood cultures (21%). An association between the sample source and the tolerance to CHG was identified (p = 0.020, Table 3). However, no such association was found when analyzing only the MSSA isolates (Table 4). Among the MRSA isolates, there was an association between the sample source and CHG tolerance; reduced susceptibility to CHG was found in 39.4% and 15.3% of the blood culture isolates and the wound isolates, respectively (p = 0.009) (Table 4).
3. Discussion
The rates of phenotypic tolerance to CHG vary between countries, ranging from 0.06% to 35% [15,16,17,18]. In addition, the median CHG’s MIC values differ in different countries [15,16,17,18]. Similarly, the rate of genotypic tolerance differs between different countries (reviewed in [6]). There are several possible reasons for these differences, including different infection control and intervention programs and different protocols for CHG use. In the current study performed on clinical isolates collected in Israel, 13.15% of the clinical S. aureus strains exhibited reduced susceptibility to CHG. In parallel, out of 190 isolates, 13.15% harbored the qac A/B genes.
Although some qac A/B-negative strains were phenotypically tolerant to CHG and some qac A/B-positive strains were phenotypically CHG-susceptible, an association was found between phenotypic and genotypic tolerance to CHG, particularly among the MRSA strains. It is not surprising to find CHG-tolerant S. aureus isolates that lack the qac A/B genes, since there are over 11 genes encoding the efflux pumps that mediate tolerance to biocides [6]; the current work tested for the presence of only two. On the other hand, the presence of these genes does not necessarily provide phenotypic tolerance as was seen with antibiotic resistance genes. This may be due to carriage of the gene without its expression. Various studies have reported on this discrepancy between phenotypic and genotypic tolerance [16,17,18,19]. It should be noted that none of the previous studies found a statistically significant association between genotypic and phenotypic susceptibility to CHG.
To identify risk factors that may be associated with CHG tolerance, we first tested resistance to methicillin. The prevalence of CHG tolerance and the average CHG’s MIC were higher among MRSA strains as compared to MSSA strains. These findings align with previous reports, in which MRSA strains were less susceptible to CHG as compared to MSSA strains [20,21,22]. These observations are concerning in light of the common use of CHG as a disinfectant, particularly for MRSA infections. On the one hand, the clinical significance of the high CHG’s MIC among MRSA isolates remains unknown, as CHG is used at much higher concentrations than the tested MICs [23]. On the other hand, there is some evidence of its clinical impact; for example, one study reported on the spread of an MRSA strain with an elevated MIC following the use of a standard CHG disinfection protocol in an intensive care unit (ICU) [24]. In addition, there are subpopulations of staphylococci with heterogeneous tolerance to CHG [6]. Therefore, the phenomenon of reduced susceptibility of S. aureus, specifically of MRSA strains, to CHG should be further investigated.
As with the phenotypic tolerance, the prevalence of genotypic tolerance was also higher among the MRSA strains (22.8%) versus the MSSA strains (4.1%). The reported prevalence of qac A/B genes in MRSA isolates varies across different geographic areas, from 0.9% in the USA [25] to 83.3% in Malaysia [26] (reviewed in [12]). MSSA isolates show a qac A/B prevalence of 2–12% [27,28,29]. As already mentioned, these differences may result from diverse decolonization and prevention policies and different protocols for CHG use, among other factors.
The mupirocin resistance rate was 9.5% among all isolates, 2.05% among MSSA isolates, and 17.4% among MRSA isolates. Most previous studies analyzed mupirocin resistance rates among MRSA samples only; these rates ranged between 0% and 65% (reviewed in [7,30]). This broad range may result from the diverse mupirocin treatment protocols for MRSA infections (including different frequencies of mupirocin use), different study populations, different study settings (community vs. hospital, clinical isolates vs. colonizing isolates), among other factors. A recent systematic review reported on a higher prevalence of mupirocin resistance among MRSA isolates as compared to MSSA isolates (13.8% and 7.6%, respectively) [31] as shown in our work. A possible explanation for this finding is that mupirocin is more frequently used against MRSA infections than for MSSA infections.
Regarding the association between mupirocin resistance and CHG tolerance, we surprisingly found an opposite association, i.e., most (72%) of the CHG-tolerant strains were susceptible to mupirocin. One possible explanation is that mupirocin and CHG are not administered simultaneously. Thus, the treated strain develops resistance/tolerance either to mupirocin or to CHG, but not to both. While no reports of assessment of this association were found in the literature, one study did report on coexistence of a low level of resistance to mupirocin in MRSA with qac A/B genes, which was associated with elevated risk for persistent carriage following decolonization [32]. Thus, even though no association was found between CHG tolerance and mupirocin resistance in the current study, it is important to continue monitoring S. aureus isolates and their susceptibility to mupirocin and CHG.
The presence of the pvl toxin was the third tested characteristic. The pvl toxin creates pores in leukocyte membranes and is associated with increased bacterial virulence. In the current work, 20.5% of the strains carried the pvl toxin gene. A comparison with other studies concluded that pvl prevalence among S. aureus isolates varies between geographical areas or the type of patients (e.g., children vs. adults) [33], from 3% to 75% [34,35,36]. Interestingly, the majority of pvl-positive strains in our study were MRSA, an observation that aligns with earlier reports [33,37,38,39]. For example, a study performed in Alaska in 2000 reported that while no MSSA isolates carried the pvl genes, 92% of MRSA isolates had the toxin [39]. However, several studies found the opposite pattern, where the pvl toxin was more common among MSSA strains [40,41,42]. These discrepancies may derive from different characteristics of study cohorts, including patient type and age, sample sources, and sample size. Although we hypothesized that pvl presence may be linked to CHG tolerance, no such association was found. No previous studies investigated the relationships between pvl presence and CHG tolerance. Thus, further research should be performed on this aspect.
The last characteristic that we tested was the sample source and its possible association with CHG tolerance. The CHG tolerance rate was higher among the strains isolated from blood cultures as compared to wounds. This association was also seen among the MRSA strains. To the best of our knowledge, this is the first evidence of an association between phenotypic CHG tolerance in S. aureus and the invasiveness of the condition. These observations strengthen two previous reports that analyzed S. aureus strains isolated from children and showed that CHG’s genotypic tolerance was associated with a higher rate of invasive infection and with central venous catheters [43,44]. This is likely related to the fact that invasive S. aureus strains have the ability to form biofilms that are impermeable to antibiotics and biocides. Therefore, it is reasonable that strains causing invasive infections such as bacteremia are more tolerant to antiseptics.
The study had several limitations; first, no data regarding patient exposure to CHG were collected; second, other data regarding risk factors for CHG tolerance, such as patients’ underlying diseases, previous hospital admissions, presence of central venous lines, etc., should have been collected.
In summary, this work shed light on the prevalence of reduced CHG susceptibility among S. aureus strains in Israel and on the characteristics of tolerated strains. It can be concluded that CHG-tolerant strains are more often isolated from invasive infections and are more likely methicillin-resistant. Additionally, phenotypic tolerance to CHG is associated with the presence of the qac A/B genes. This is the first time that such a study was conducted in Israel. Further research should be performed to evaluate risk factors for the development of CHG tolerance.
4. Materials and Methods
4.1. Bacterial Strains
The study included 190 S. aureus isolates (98 MSSA and 92 MRSA isolates) from human blood and wound cultures. Ninety-two strains (52 MSSA and 40 MRSA strains) were isolated from blood and wound cultures of patients (age range: 0 to 100 years) admitted to the Padeh Poriya Medical Centre between January 2018 and December 2019. These isolates were recovered as part of the routine diagnosis workup at the clinical microbiology laboratory. Briefly, colonies suspected to be S. aureus-positive were isolated on blood agar (BD Diagnostics, Sparks, MD, USA) and on a selective chromogenic growth medium ChromagarTM MRSA/MSSA (Hy Laboratories Ltd., Rehovot, Israel), and then incubated at 37 ± 1 °C for 18–24 h. Confirmatory tests, including gram staining, a catalase test, and a rapid agglutination test, for simultaneous detection of the fibrinogen affinity antigen (clumping factor), protein A, and capsular polysaccharides (Pastorex™ Staph Plus, BIO RAD, Marnes-la-Coquette, France) were performed on each isolate. Final identification of S. aureus was performed using the Bruker Biotyper system (Bruker Daltonics, Bremen, Germany), which is based on the matrix-assisted laser desorption ionization–time of flight (MALDI–TOF) technique. Methicillin resistance testing was performed according to the routine laboratory protocol, by testing the isolate susceptibility to cefoxitin.
The other 98 strains (46 MSSA and 52 MRSA strains) were randomly collected at the S. aureus National Reference Centre, Israel Ministry of Health, Jerusalem, Israel. These isolates were originally isolated from wound and blood cultures of patients (age range: 0 to 100 years) admitted to various medical institutions in Israel between May 2015 and February 2018.
4.2. Antibiotic Susceptibility Testing (AST)
Susceptibility to mupirocin was assessed using the Etest method that is used to determine the MIC, i.e., the minimal concentration (mg/L) of a given antibiotic that inhibits growth of a particular bacterium under specific experimental conditions. All isolates were grown on blood agar (Hy Laboratories Ltd., Rehovot, Israel) at 37 ± 1 °C for 18–24 h. Several colonies were then suspended in saline to a turbidity of 0.5 McFarland. The suspensions were seeded on Mueller–Hinton agar plates (Hy Laboratories Ltd., Rehovot, Israel), after which an Etest strip of mupirocin (bioMérieux, Durham, NC, United States) was added to each agar plate. All plates were incubated at 37 ± 1 °C for 18–24 h. Mupirocin susceptibility was determined in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2018 guidelines [45].
4.3. Phenotypic Susceptibility to CHG
4.3.1. Broth Microdilution for MIC Determination
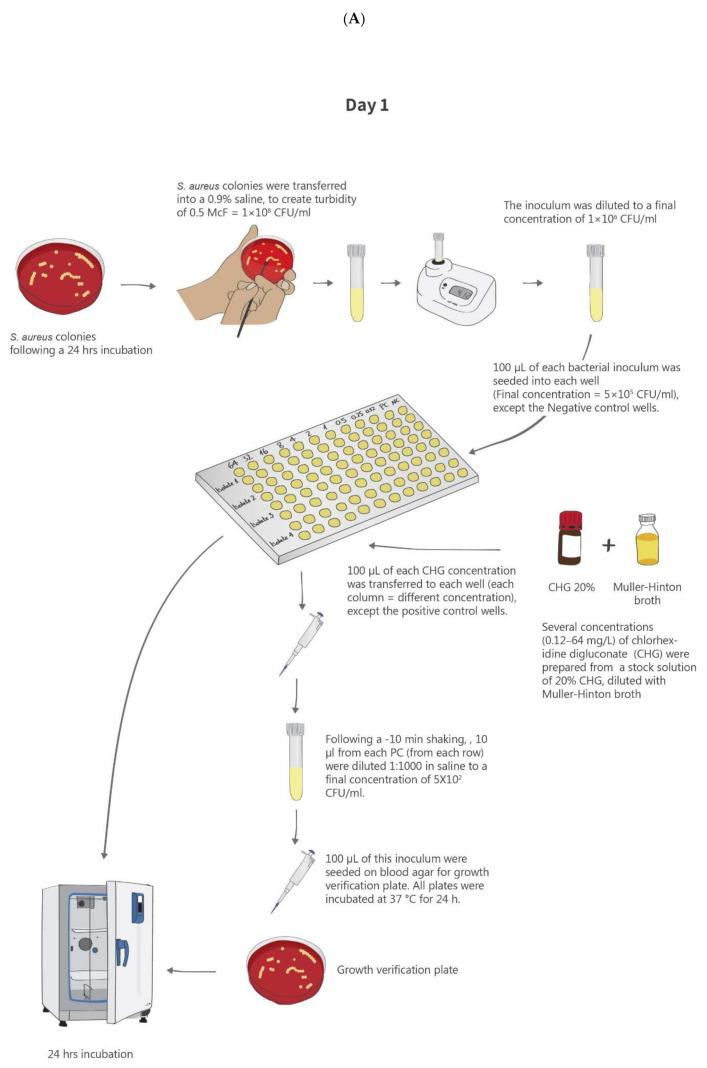
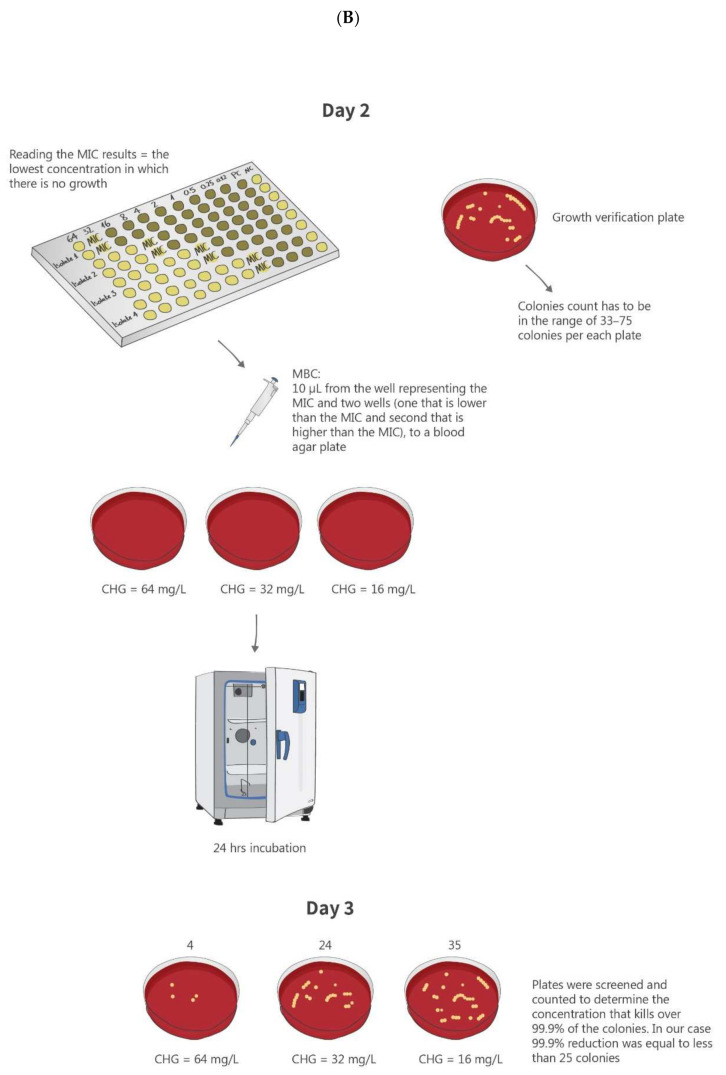
The MIC was determined using the broth microdilution method as previously described [46] and as recommended by the American Society for Microbiology (ASM) guidelines [47]. Figure 3 summarizes the methods used for CHG susceptibility testing.
Figure 3.
Schematic of the methods used for CHG susceptibility testing. (A). Determination of minimum inhibitory concentration (MIC) for CHG. (B). Determination of minimum bactericidal concentration (MBC) for CHG.
Bacterial Inoculum
All isolates were seeded on blood agar plates (BD Diagnostics, Sparks, MD, United States) and incubated at 37 °C for 24 h prior to the experiment. Isolated colonies from each culture were suspended in 0.9% saline to create a 0.5 McFarland turbidity, which is equivalent to approximately 1–2 × 108 colony-forming units (CFU)/mL. This stock was then diluted with 0.9% saline to a final concentration of 1–2 × 106 CFU/mL.
Two strains were used for quality control: S. aureus ATCC strain 29213, which is recommended by the American Society for Microbiology (ASM) guidelines [47], and the ATCC strain 700699, which has reduced susceptibility to CHG [48].
CHG Preparation
Serial dilutions of a primary CHG stock (200 mg/mL; Sigma-Aldrich, Rehovot, Israel) using the Mueller–Hinton broth (MHB) (Hy Laboratories Ltd., Rehovot, Israel) were prepared in order to create 10 final concentrations (0.12, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 µg/mL).
For each CHG concentration and for each isolate, the following positive and negative controls were prepared: the positive control (PC) included the bacterial inoculum in the MHB without CHG and the negative control (NC) included CHG with the saline, without the bacterial inoculum. Each CHG concentration and control was tested in duplicates.
Determination of the MIC
The assay was performed in 96-well polypropylene plates (Sigma-Aldrich, Rehovot, Israel). The bacterial inoculum (100 µL) was seeded at the same concentration (final concentration of 5 × 106 CFU/mL) to all wells, aside from the NC wells. A different concentration of CHG was added to each column of wells, except for the PC wells. Quality control strains were added to the dedicated wells. Following seeding, the 96-well plates were incubated at 37 °C for 24 h. Thereafter, controls were checked, and then the 96-well plates were screened for presence of bacterial growth. Growth was indicated by a single turbidity dot. The MIC was defined as the lowest CHG concentration that inhibited bacterial growth. When duplicate wells presented different results, the test was repeated. Reduced susceptibility to CHG was defined according to the epidemiological cut-off of the MIC ≥ 4 (mg/L) [27,45].
4.3.2. Minimum Bactericidal Concentration (MBC) Determination
MBC is the concentration that kills ≥99.9% of cells. In order to determine the MBC as recommended by the ASM guidelines [47], 10 µL samples from the well representing the MIC and two wells treated with the CHG concentrations just below and just above the MIC were transferred to a blood agar plate. The agar plate was incubated at 37 °C for 24 h. The plates were screened for colonies, which were counted to determine the MBC. The MBC was determined as the concentration in which fewer than 25 colonies grew [47].
4.4. Genotypic Susceptibility to CHG
Molecular detection of qac A/B genes was performed using the real-time polymerase chain reaction (PCR) technique. To this end, DNA was extracted from bacteria using a GenEluteTM Bacterial Genomic kit (Sigma-Aldrich, Rehovot, Israel) as per the manufacturer’s instructions. PCR was performed using the primers and a probe as previously described [49] under the following conditions: 50 °C for 2 min, 95 °C for 20 s, and 40 cycles of 95 °C for 35 s, 60 °C for 30 s.
4.5. Panton–Valentine Leucocidin (pvl), mecC, and mecA Detection
Presence of the Panton–Valentine leucocidin (pvl) gene was detected using an eazyplex® MRSAplus kit (AmplexDiagnostics GmbH, Germany) which is based on the rapid isothermal amplification reaction. This qualitative molecular test detects the presence of the pvl toxin gene and identifies MRSA isolates by detection of the mecA and mecC genes. When the mecA, mecC, or pvl genes are present in the detected S. aureus isolate, specific amplification products are generated. Due to the binding of a fluorescence dye to these double-stranded DNA amplification products, presence of the corresponding genes is visualized by real-time fluorescence. The procedure was performed according to the manufacturer’s instructions.
4.6. Statistical Analysis
Univariate tests were applied to analyze the differences in the phenotypic and genotypic characteristics and between CHG susceptibility of MSSA isolates vs. MRSA isolates.
Comparisons between groups were made using the chi-squared or non-parametric Wilcoxon–Mann–Whitney rank-sum test for independent samples for the categorical and continuous variables, respectively.
Statistical significance was determined with the p-value < 0.05. The data were analyzed using SPSS version 25.
Author Contributions
Conceptualization, M.A., C.S., K.A.-S., and A.P.; data curation, M.A.; formal analysis, M.A., C.S., K.A.-S., and A.P.; investigation, M.A., K.A.-S., and A.P.; methodology, M.A., C.S., T.L., Z.H., M.B., A.R., K.A.-S., and A.P.; project administration, M.A. and A.P.; supervision, M.A., K.A.-S., and A.P.; validation, M.A., C.S., T.L., Z.H., and M.B.; visualization, M.A., K.A.-S., and A.P.; writing—original draft preparation, M.A., K.A.-S., and A.P.; writing—review and editing, M.A., C.S., T.L., Z.H., M.B., A.R., K.A.-S., and A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tatarkiewicz J., Staniszewska A., Bujalska-Zadrożny M. New agents approved for treatment of acute staphylococcal skin infections. Arch. Med. Sci. 2016;12:1327–1336. doi: 10.5114/aoms.2016.59838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purrello S.M., Garau J., Giamarellos E., Mazzei T., Pea F., Soriano A., Stefani S. Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 2016;7:178–186. doi: 10.1016/j.jgar.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., et al. Multistate Point-Prevalence Survey of Health Care–Associated Infections. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köck R., Becker K., Cookson B., E Van Gemert-Pijnen J., Harbarth S., Kluytmans J., Mielke M., Peters G., Skov R.L., Struelens M.J., et al. Methicillin-resistant Staphylococcus aureus (MRSA): Burden of disease and control challenges in Europe. Euro Surveill. 2010;15:19688. doi: 10.2807/ese.15.41.19688-en. Erratum in: Euro Surveill.2010, 15, 19694. [DOI] [PubMed] [Google Scholar]
- 5.Calfee D.P. Goldman-Cecil Medicine. 25th ed. Elsevier Saunders; Philadelphia, PA, USA: 2015. Prevention and control of health care-associated infections; pp. 1861–1863. [DOI] [Google Scholar]
- 6.Horner C., Mawer D., Wilcox M. Reduced susceptibility to chlorhexidine in staphylococci: Is it increasing and does it matter? Antimicrob. Chemother. 2012;67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 7.Williamson D.A., Carter G.P., Howden B.P. Current and Emerging Topical Antibacterials and Antiseptics: Agents, Action, and Resistance Patterns. Clin. Microbiol. Rev. 2017;30:827–860. doi: 10.1128/CMR.00112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugo W.B., Longworth A.R. Some aspects of the mode of action of chlorhexidine. J. Pharm. Pharmacol. 1964;16:655–662. doi: 10.1111/j.2042-7158.1964.tb07384.x. [DOI] [PubMed] [Google Scholar]
- 9.Denton G.W. Chlorhexidine. In: Block S.S., editor. Disinfection, Sterilization, and Preservation. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2001. pp. 321–336. [Google Scholar]
- 10.Kampf G., Kramer A. Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin. Microbiol. Rev. 2004;17:863–893. doi: 10.1128/CMR.17.4.863-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soto-Hernandez J.L. Chlorhexidine bathing and infections in critically ill patients. JAMA. 2015;313:1863. doi: 10.1001/jama.2015.3533. [DOI] [PubMed] [Google Scholar]
- 12.Kampf G. Acquired resistance to chlorhexidine—Is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 2016;94:213–227. doi: 10.1016/j.jhin.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Madden G.R., Sifri C.D. Antimicrobial Resistance to Agents Used for Staphylococcus aureus Decolonization: Is There a Reason for Concern? Curr. Infect. Dis. Rep. 2018;20:26. doi: 10.1007/s11908-018-0630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonnell G., Russell A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999;12:147–179. doi: 10.1128/CMR.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes C., Ferguson J. Phenotypic chlorhexidine and triclosan susceptibility in clinical Staphylococcus aureus isolates in Australia. Pathology. 2017;49:633–637. doi: 10.1016/j.pathol.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Sheng W.-H., Wang J.-T., Lauderdale T.-L., Weng C.-M., Chen D., Chang S.-C. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: Emphasis on chlorhexidine susceptibility. Diagn. Microbiol. Infect. Dis. 2009;63:309–313. doi: 10.1016/j.diagmicrobio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Longtin J., Seah C., Siebert K., McGeer A., Simor A., Longtin Y., Low D.E., Melano R.G. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob. Agents Chemother. 2011;55:2999–3001. doi: 10.1128/AAC.01707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGann P., Kwak Y.I., Summers A., Cummings J.F., Waterman P.E., Lesho E.P. Detection of qacA/B in clinical isolates of methicillin-resistant Staphylococcus aureus from a regional healthcare network in the eastern United States. Infect. Control Hosp. Epidemiol. 2011;32:1116–1119. doi: 10.1086/662380. [DOI] [PubMed] [Google Scholar]
- 19.Wang J.-L., Sheng W.-H., Chen D., Chen M.-L., Chen Y.-C., Chang S.-C. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J. Antimicrob. Chemother. 2008;62:514–517. doi: 10.1093/jac/dkn208. [DOI] [PubMed] [Google Scholar]
- 20.Akinkunmi E.O., Lamikanra A. Susceptibility of community associated methicillin resistant Staphylococcus aureus isolated from faeces to antiseptics. J. Infect. Dev. Ctries. 2012;6:317–323. doi: 10.3855/jidc.1609. [DOI] [PubMed] [Google Scholar]
- 21.Wootton M., Walsh T.R., Davies E.M., Howe R.A. Evaluation of the effectiveness of common hospital hand disinfectants against methicillin-resistant Staphylococcus aureus, glycopeptide-intermediate S. aureus, and heterogeneous glycopeptide-intermediate S. aureus. Infect. Control Hosp. Epidemiol. 2009;30:226–232. doi: 10.1086/595691. [DOI] [PubMed] [Google Scholar]
- 22.Hasanvand A., Ghafourian S., Taherikalani M., Jalilian F.A., Sadeghifard N., Pakzad I. Antiseptic resistance in methicillin sensitive and methicillin resistant Staphylococcus aureus isolates from some major hospitals, Iran. Recent Pat. Anti-Infect. Drug Discov. 2015;10:105–112. doi: 10.2174/1574891X10666150623093259. [DOI] [PubMed] [Google Scholar]
- 23.Smith K., Gemmell C.G., Hunter I.S. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J. Antimicrob. Chemother. 2008;61:78–84. doi: 10.1093/jac/dkm395. [DOI] [PubMed] [Google Scholar]
- 24.Batra R., Whiteley C., Wyncoll D., Cooper B.S., Patel A.K., Edgeworth J.D. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 2010;50:210–217. doi: 10.1086/648717. [DOI] [PubMed] [Google Scholar]
- 25.Mc Gann P., Milillo M., Kwak Y.I., Quintero R., Waterman P.E., Lesho E. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2013;77:270–272. doi: 10.1016/j.diagmicrobio.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Shamsudin M.N., Alreshidi M.A., Hamat R.A., Alshrari A.S., Atshan S.S., Neela V. High prevalence of qacA/B carriage among clinical isolates of methicillin-resistant Staphylococcus aureus in Malaysia. J. Hosp. Infect. 2012;81:206–208. doi: 10.1016/j.jhin.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Alam M.M., Kobayashi N., Uehara N., Watanabe N. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb. Drug Resist. 2003;9:109–121. doi: 10.1089/107662903765826697. [DOI] [PubMed] [Google Scholar]
- 28.Opacic D., Lepsanovic Z., Sbutega-Milosevic G. Distribution of disinfectant resistance genes qacA/B in clinical isolates of meticillin-resistant and -susceptible Staphylococcus aureus in one Belgrade hospital. J. Hosp. Infect. 2010;76:266–267. doi: 10.1016/j.jhin.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Mayer S., Boos M., Beyer A., Fluit A.C., Schmitz F.-J. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 2001;47:896–897. doi: 10.1093/jac/47.6.896. [DOI] [PubMed] [Google Scholar]
- 30.Khoshnood S., Heidary M., Asadi A., Soleimani S., Motahar M., Savari M., Saki M., Abdi M. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed. Pharmacother. 2019;109:1809–1818. doi: 10.1016/j.biopha.2018.10.131. [DOI] [PubMed] [Google Scholar]
- 31.Dadashi M., Hajikhani B., Darban-Sarokhalil D., van Belkum A., Goudarzi M. Mupirocin resistance in Staphylococcus aureus: A systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 2019;20:238–247. doi: 10.1016/j.jgar.2019.07.032. [DOI] [PubMed] [Google Scholar]
- 32.Lee A.S., Macedo-Vinas M., François P., Renzi G., Schrenzel J., Vernaz N., Pittet D., Harbarth S. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: A case–control study. Clin. Infect. Dis. 2011;52:1422–1430. doi: 10.1093/cid/cir233. [DOI] [PubMed] [Google Scholar]
- 33.Bastidas C.A., Villacrés-Granda I., Navarrete D., Monsalve M., Coral-Almeida M., Cifuentes S.G. Antibiotic susceptibility profile and prevalence of mecA and lukS-PV/lukF-PV genes in Staphylococcus aureus isolated from nasal and pharyngeal sources of medical students in Ecuador. Infect. Drug Resist. 2019;12:2553–2560. doi: 10.2147/IDR.S219358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran G.J., Krishnadasan A., Gorwitz R.J., Fosheim G.E., McDougal L.K., Carey R.B., Talan D.A. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 35.Del Giudice P., Blanc V., Durupt F., Bes M., Martinez J.-P., Counillon E., Lina G., Vandenesch F., Etienne J. Emergence of two populations of methicillin-resistant Staphylococcus aureus with distinct epidemiological, clinical and biological features, isolated from patients with community-acquired skin infections. Br. J. Dermatol. 2006;154:118–124. doi: 10.1111/j.1365-2133.2005.06910.x. [DOI] [PubMed] [Google Scholar]
- 36.Tristan A., Bes M., Meugnier H., Lina G., Bozdoğan B., Courvalin P., Reverdy M.-E., Enright M.C., Vandenesch F., Etienne J. Global distribution of panton valentine leukocidin–positive methicillin-resistant. Emerg. Infect. Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David M.Z., Daum R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baggett H.C., Hennessy T.W., Rudolph K., Bruden D., Reasonover A., Parkinson A., Sparks R., Donlan R.M., Martinez P., Mongkolrattanothai K., et al. Community-onset methicillin-resistant Staphylococcus aureus associated with antibiotic use and the cytotoxin Panton-Valentine leukocidin during a furunculosis outbreak in rural Alaska. J. Infect. Dis. 2004;2189:1565–1573. doi: 10.1086/383247. [DOI] [PubMed] [Google Scholar]
- 39.Goudarzi M., Navidinia M., Beiranvand E., Goudarzi H. Phenotypic and molecular characterization of methicillin-resistant Staphylococcus aureus clones carrying the panton-valentine leukocidin genes disseminating in Iranian hospitals. Microb. Drug Resist. 2018;24:1543–1551. doi: 10.1089/mdr.2018.0033. [DOI] [PubMed] [Google Scholar]
- 40.Tajik S., Najar-Peerayeh S., Bakhshi B. Hospital clones of Panton-Valentine leukocidin positive and methicillin-resistant Staphylococcus aureus are circulating in the community of Tehran. J. Glob. Antimicrob. Resist. 2019;22:177–181. doi: 10.1016/j.jgar.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Ahmadpour M., Havaei S.A., Poursina F., Havaei S.R., Ruzbahani M. Detection of panton-valentine leukocidin gene isoforms of Staphylococcus aureus isolates in Al-Zahra Hospital, Isfahan-Iran. Adv. Biomed. Res. 2017;6:93. doi: 10.4103/2277-9175.211798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vali L., Dashti A.A., Mathew F., Udo E.E. Characterization of heterogeneous MRSA and MSSA with reduced susceptibility to chlorhexidine in Kuwaiti hospitals. Front. Microbiol. 2017;8:1359. doi: 10.3389/fmicb.2017.01359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNeil J.C., Hultén K.G., Mason E.O., Kaplan S.L. Impact of health care exposure on genotypic antiseptic tolerance in Staphylococcus aureus infections in a pediatric population. Antimicrob. Agents Chemother. 2017;61:e00223-17. doi: 10.1128/AAC.00223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeil J.C., Kok E.Y., Vallejo J.G., Campbell J.R., Hulten K.G., Mason E.O., Kaplan S.L. Clinical and molecular features of decreased chlorhexidine susceptibility among nosocomial Staphylococcus aureus isolates at Texas Children’s Hospital. Antimicrob. Agents Chemother. 2016;60:1121–1128. doi: 10.1128/AAC.02011-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.EUCAST: Clinical Breakpoints—Breakpoints and Guidance. [(accessed on 8 January 2020)]; Available online: http://www.eucast.org/clinical_breakpoints/
- 46.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 47.Garcia L.S., editor. Clinical Microbiology Procedures Handbook. American Society for Microbiology Press; Washington, DC, USA: 2010. [Google Scholar]
- 48.Panda S., Kar S., Choudhury R., Sharma S., Singh D.V. Development and evaluation of hexaplex PCR for rapid detection of methicillin, cadmium/zinc and antiseptic-resistant staphylococci, with simultaneous identification of PVL-positive and -negative Staphylococcus aureus and coagulase negative staphylococci. FEMS Microbiol. Lett. 2014;352:114–122. doi: 10.1111/1574-6968.12383. [DOI] [PubMed] [Google Scholar]
- 49.Otter J.A., Patel A., Cliff P.R., Halligan E.P., Tosas O., Edgeworth J.D. Selection for qacA carriage in CC22, but not CC30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J. Antimicrob. Chemother. 2013;68:992–999. doi: 10.1093/jac/dks500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.