Abstract
Seed development needs the coordination of multiple molecular mechanisms to promote correct tissue development, seed filling, and the acquisition of germination capacity, desiccation tolerance, longevity, and dormancy. Heat stress can negatively impact these processes and upon the increase of global mean temperatures, global food security is threatened. Here, we explored the impact of heat stress on seed physiology, morphology, gene expression, and methylation on three stages of seed development. Notably, Arabidopsis Col-0 plants under heat stress presented a decrease in germination capacity as well as a decrease in longevity. We observed that upon mild stress, gene expression and DNA methylation were moderately affected. Nevertheless, upon severe heat stress during seed development, gene expression was intensively modified, promoting heat stress response mechanisms including the activation of the ABA pathway. By analyzing candidate epigenetic markers using the mutants’ physiological assays, we observed that the lack of DNA demethylation by the ROS1 gene impaired seed germination by affecting germination-related gene expression. On the other hand, we also observed that upon severe stress, a large proportion of differentially methylated regions (DMRs) were located in the promoters and gene sequences of germination-related genes. To conclude, our results indicate that DNA (de)methylation could be a key regulatory process to ensure proper seed germination of seeds produced under heat stress.
Keywords: heat stress, seed, DNA methylation, seed development, seed germination
1. Introduction
The accumulative anthropomorphic greenhouse gas emissions causing climate change have a diverse and mainly negative impact on life on Earth. The mean temperatures are increasing worldwide (0.35 °C from 1979–2003) and the range in diurnal temperature is higher, since the minimum temperature increases much faster than the maximum temperature rate (1.13 °C from 1979–2003) [1,2,3]. The effect of global warming induced-heat stress on plants is strong and diverse. It can promote an advance in flowering time [4,5,6,7], modifications in plant architecture [8,9,10], decrease in grain yield [11], decreased in seed dormancy [12,13], and the shift of plant establishment in higher altitudes [14], as a decrease in crop production such as maize and wheat [15]. All of those effects pose a great threat to agricultural safety and ecological diversity. Concerning plant life cycle transitions, both the timing of germination and reproduction are determined by temperature [16,17]. Therefore, the control of important seed traits has the potential to be seriously impacted by climate change [18]. Studies have shown that heat stress can impact seed development and reduce seed size [19,20]. Along with physiological observations, the plants’ molecular responses to heat stress has been extensively studied in the past decades. Heat stress factors (HSF) and heat shock proteins (HSP), calcium channels, phytohormones, chaperones, and other secondary metabolites have an important role in heat stress responses [21,22]. Moreover, transcriptional reprogramming enables plants to cope with heat stress. This reprogramming upregulates specific proteins such as kinases and transcription factors along with direct stress protection genes such as detoxifying enzymes and osmoprotectants. In contrast, growth-related genes are generally repressed [23]. Gene transcription is connected to its epigenetic state. Therefore, gene expression regulation is based on chromatin modifications (e.g., histone acetylation, methylation, phosphorylation, and ubiquitylation) and DNA modifications (e.g., cytosine methylation) [24,25]. These modifications can be triggered by developmental or environmental factors and as a consequence, they modulate chromatin architecture without changing the genomic sequences, but only the accessibility of the transcriptional machinery to specific genome regions [26,27].
However, the understanding of epigenetic regulation of heat stress response is still dawning. DNA methylation is the addition of a methyl group to a DNA cytosine base, forming a 5-methylcytosine (5mC). This process occurs in three sequence contexts in plants: the symmetric CG and CHG contexts and the asymmetric CHH context, where H stands for A, T, or C. DNA methylation can be divided into (i) de novo methylation, (ii) maintenance of methylation, and (iii) demethylation processes, in which different proteins are involved. In plants, de novo methylation is catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), which is regulated by the RNA-directed DNA methylation (RdDM) pathway [28]. In the RdDM pathway, the RNA polymerase IV (Pol IV) transcribes single-stranded RNA (ssRNAs) that are later synthesized into double-stranded RNA intermediates (dsRNAs) by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2). These dsRNAs are precursors for RNase III-class DICER-LIKE 3 (DCL3) to process into 24-nt interfering RNAs (siRNAs) that will be incorporated into ARGONAUTE 4 (AGO4) to be base-paired with Pol V transcripts, resulting in the recruitment of DRM2 and DNA base methylation [28]. Maintenance of DNA methylation is dependent on the sequence context as CG methylation is maintained by METHYLTRANSFERASE 1 (MET1) and DECREASE IN DNA METHYLATION 1 (DDM1), while CHG is maintained by CHROMOMETHYLASE 3 (CMT3), and CHH by DRM2 [29]. On the other hand, plant DNA demethylation is regulated by four bifunctional 5-methylcytosine glycosylases REPRESSOR OF SILENCING 1 (ROS1), DEMETER (DME), DME-like 2 (DML2), and DML3, which remove the 5-methylcytosine from DNA through the base excision repair pathway [30]. In ros1 mutants, all DNA methylation contexts were hypermethylated, suggesting that ROS1 demethylates all DNA methylation contexts [31]. DNA methylation regulates gene expression, normally by inhibiting the transcription of genes and also maintains genome stability against transposable elements [29,32]. Heat stress can affect DNA methylation as was demonstrated in Arabidopsis thaliana leaves with the increase of global methylation and homologous recombination frequency [33]. The methylation processes protein, DRM2, NUCLEAR RNA POLYMERASE D1(NRPD1), and NRPE1 were also shown to be upregulated upon heat stress and could contribute to an increase in DNA methylation as a heat stress response [34]. The RNA-dependent DNA methylation pathway (RdDM) is partially responsible for the transcriptional response to temperature stress [23]. Nevertheless, heat stress-induced global DNA methylation changes appear to be species and tissue-dependent. Global DNA methylation was found increased in Quercus suber under heat stress [35] and in Brassica napus [36], but decreased in Gossypium hirsutum [37] and Oryza sativa [20]. Furthermore, investigating heat stress impact on transgenerational memory showed phenotypic changes over generations [33,38,39,40]. Heat stress ancestral exposure over two consecutive generations increased fitness in the F3 heat-treated Arabidopsis plants [41]. This epigenetic inheritance could promote genomic flexibility and adaptive advantages for plant fitness under heat stress conditions [42].
Thus far, the majority of heat stress studies have focused on vegetative tissues with short-term heat stress events. Given the importance of seeds for agriculture, we aimed to evaluate the impact of different heat stress intensities during the whole seed development in A. thaliana as mimicking global warming conditions. In our study, we evaluated the impact of mild and severe heat stress on embryo growth and morphology; seed germination and longevity; gene expression and DNA methylation.
2. Material and Methods
Biological Material and Heat Treatment
A. thaliana wild-types Columbia-0 (Col-0) and Wassilewskija (WS) and homozygous insertional mutants (T-DNA) cmt3 (in WS background) [43] and ros1-4 (SALK_135293 in Col-0 background) [44] were used in this work. Plants were grown in square pots of 6 × 6 × 6 cm filled to the top with Tray substrate (Klasmann-Deilmann, Bourgoin Jallieu, France). Plants were kept at long-day conditions in a growth chamber with controlled humidity (60–70%) and an average temperature of 23 °C (24 °C/22 °C day/night) with a 16 h-light/8 h-dark photoperiod with a light intensity of 150 mmol m2s−1. Plants received watering three days per week on the base of their pots, for capillarity absorption by the substrate.
For heat stress treatment during seed development, as soon as the first flower buds appeared on the base of the leaf rosette, plants were transferred to growth chambers with the same light, photoperiod, humidity, and watering conditions, but with different temperature ranges: 21 °C (22 °C/20 °C day/night), 25 °C (26 °C/24 °C day/night) or 27 °C (28 °C/26 °C day/night), or 29 °C (30 °C/28 °C day/night). A parallel set of control plants were continually grown in the 23 °C growth.
A resumed view of this study experiments, conditions and sampling time-points is available in Supplementary Figure S1.
3. Physiological and Phenotypic Characterizations
For water content analysis, seeds were sampled at 4, 6, 8, 10, 12, and 14 DAF (days after fertilization), which represent the embryo stages of globular (4 DAF), heart (6 DAF), bent (8 DAF) and green mature (14 DAF) on standard growth conditions. Three replicates of 50 seeds were used for the determination of fresh weight and dry weight after two days of drying at 96 °C.
For the following experiments, fully matured seeds from all genotypes were equilibrated at 44% of relative humidity (RH) using a saturated solution of K2CO3 at 20 °C for three days, and then used for the physiological analysis. Col-0 plants at 29 °C heat stress treatment produced insufficient seed number for all the experiments to be performed.
Protein content was characterized using a CHNS elemental analyzer, which measured the percentage (w/w) of nitrogen using flash combustion of the sample based on the “Dumas” method. Mature seeds were ground in liquid nitrogen and dried in an oven at 90 °C for 48 h. Then, triplicates of approximately 5 mg of powder were analyzed using an Elementar Vario Micro cube analyzer (Germany). Protein content was estimated from the nitrogen content using the Jones Factor [45], which is used to convert the nitrogen concentration into total protein content.
For the germination assay, dormancy was released by incubating seeds at cold for 48 h. The germination capacity was then accessed by germinating triplicates of 100 dried seeds on Whatman paper No. 1 imbibed with 1 mL of autoclaved water in 3 cm diameter Petri dishes at 20 °C under a 16 h/8 h photoperiod for eight days. Germination was scored every two days as seeds exhibited a protruded radicle.
Seed longevity was evaluated by using accelerated (or artificial) aging. Seeds were incubated in high humidity chambers (75% of RH from a saturated solution of NaCl) at 35 °C in the dark. At various intervals of aging, 100 seeds were retrieved and imbibed with 1 mL of autoclaved water for germination assay to calculate the survival percentage of aged seed lots. Then, we modeled the seed longevity curve, characterized by the loss of seed viability along different aging time. A longevity indicator was assessed as the aging-time required for the seed batch to lose 50% of germination and called P50. Two-way ANOVA, followed by Tukey’s multiple comparison test, was used to determine the statistical significance of different time-points and/or genotypes. The exact statistical analysis parameters used are described in each related figure legend. Graphical creations and statistical analyses were performed using Prism 8.0.1 (GraphPad).
4. Morpho-Anatomical Measurements
For evaluation of embryo development at different temperatures, Col-0 seeds were collected at different embryo stages: globular, transition, heart, torpedo, walking stick, and mature. Seeds were cleared using the pseudo-Schiff propidium iodide staining as detailed [46].
Mature and dried seeds were used to measure seed width and length. SEM (scanning electron microscopy) pictures of seeds were acquired with the Phenom Pro Desktop SEM (Phenom-World) and all the samples were directly imaged, without sample preparation or modification. For all measurements, mature dried seeds were obtained from four plants per replicate, and three replicates from three independent experiments were used for these measurements. Two-way ANOVA followed by Tukey’s multiple comparison test was used to determine the statistical significance of different time-points and/or genotypes. The exact statistical analysis parameters used are described in each related figure legend. Graphical creations and statistical analysis were performed using Prism 8.0.1 (GraphPad).
For evaluation of embryo survival, mature and dried seeds were incubated using a 1% solution of 2, 3, 5 triphenyl tetrazolium chloride (Tetrazolium Red- TTZ- Merck, Kenilworth, NJ, USA) in a 50 mM phosphate buffer (pH 7.0) for 24 h at 30 °C as previously described [19]. ros1-4 mutant seeds were separated between normal-shaped and wrinkled-shaped seeds. After 24 h, seeds were crushed between two glass laminas for embryo release to get in contact with the TTZ solution. Samples were then incubated at 25 °C for two hours before macroscopic observation and imagery under a stereomicroscope Olympus SZX16.
5. Total RNA Isolation and Transcriptomic Sequencing
Col-0 seeds were collected at embryo stages: heart, bent and mature, and developed at 23 °C, 25 °C, and 27 °C temperatures. ros1-4 and Col-0 seeds grown at 23 °C were sampled at the mature stage (dried seeds) and germination stages II and IV, which was achieved after drying the seeds for three days, then dried seeds were imbibed with water and incubated at 20 °C light for germination [47]. All samples were ground with micro-pistons and liquid nitrogen. Seed powders at germination stages II and IV were incubated in NucleoSpin® RNA Plant and Fungi Kit lysis buffer with 1% of polyvinylpyrrolidone (PVP-40), followed by incubation at room temperature for 10 min and centrifugation at 8000× g for 5 min before transferring the supernatant to extract RNA, as described previously [48]. Total RNA was extracted from 100 harvested dried seeds in three biological replicates using the NucleoSpin® RNA Plant and Fungi Kit (Macherey-Nagel, Düren, Germany), according to the manufacturer instructions. RNA quantity and quality were measured using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA). cDNA library preparation and single-end sequencing (SE50, 20 M) were outsourced to the Beijing Genomics Institute (BGI, https://www.bgi.com, accessed on 4 July 2020) using the DNBseq sequencing technology. After quality control, high-quality reads were mapped on Arabidopsis reference transcriptome version 11 (Araport11) using quasi-mapping alignment and quantification methods of Salmon algorithm v.1.2 [49]. For gene expression analysis, raw RNA-Seq data were first normalized as transcripts per kilobase million (TPM). Transcripts with an average above 0 TPM in at least one developmental stage/tissue and with a coefficient of variation of log2 TPM > 0.05 among all developmental stages were retained for further analysis, resulting in 27,587 genes. Differentially expressed genes (DEGs) were determined using DESeq2 package (v1.22.2) [50], in which genes with log2 FC >0 or <0 and p-adjusted value of <0.05 for multiple testing with the Benjamini–Hochberg procedure, which controls false discovery rate (FDR), were considered as differentially expressed. Gene annotation and GO terms were assigned according to the Araport11 annotation version of the Arabidopsis genome. Over representation analysis (ORA) using GO enrichment terms was performed using the Clusterprofiler [51] package in RStudio (version 1.3.1073) by applying an adjusted p-value cut-off of <0.5 obtained from the Bonferroni procedure.
Data mining for transcriptomics of Col-0 seeds from fresh seeds to seed imbibition for 48 h was obtained from the Gene Expression Omnibus (GEO, GSE94459) as a publicly available dataset [52]. Raw data were mapped/quantified using Salmon with the same parameters described above. ImpulseDE2 algorithm [53], available in R, was used to unravel differentially expressed genes along the germination phase using the ‘case-only’ differential expression analysis and an adjusted p-value cutoff of 1%.
6. DNA Isolation and Whole-Genome Bisulfite Sequencing
Col-0 seeds were collected at embryo stages bent and mature, and developed at 23 °C, 25 °C, and 27 °C average temperature. Two biological replicates were used for each stage and condition. Genomic DNA was extracted from the samples using the NucleoSpin® DNA Food Kit (Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions, then quantified using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA). DNA samples were sent to the BGI and library construction; bisulfite treatment using a ZYMO EZ DNA Methylation-Gold kit and paired-end sequencing using an Illumina Hiseq 2500 (PE100 20M) were outsourced to this company. Sequencing quality was verified by FastQC and clean reads were subsequently mapped to the A. thaliana reference genome index sequence version 10 (TAIR10) using Methylpy software [54]. Chloroplast genomic sequence from A. thaliana was used as an unmethylated control. After mapping, deduplication of sequences was performed and cytosine methylation sites were determined and quantified using Methylpy. Each context of methylation was considered independently: CG, CHG, or CHH. Putative differentially-methylated regions (DMRs) were identified from merged differentially methylated sites (DMS) using the Methylpy ‘DMRfind’ method.
7. Results
Impact of Constant Heat Stress on A. thaliana Seeds
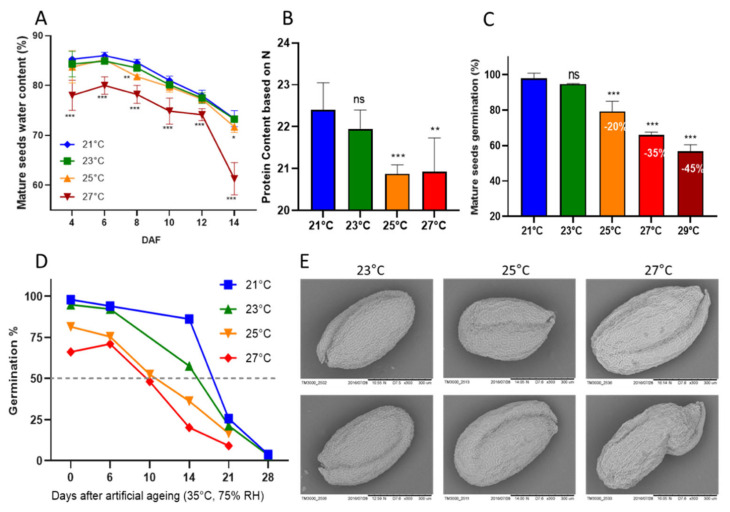
Seed physiological assays were conducted to observe which range of constant heat stress would impact seeds and how this impact could influence seed development and important seed traits such as germination capacity, longevity and seed filling. Col-0 plants were grown in optimal conditions in growth chambers until their reproductive stage (i.e., the apparition of first flowers), then moved to growth chambers with average temperatures of 21 °C, 23 °C, 25 °C, 27 °C, and 29 °C until seed maturity. Therefore, we were able to analyze seed growth and development in five temperature ranges. Our focus on this study was the evaluation of seeds, but we also observed that plants presented different phenotypes when grown at heat stress conditions. Plant biomass was increased, while the silique number and length decreased as the temperature was raised (Supplementary Figure S2). The effect of heat stress in seeds was first observed at water content measurements from 4 DAF to 14 DAF (days after fertilization). Seeds developed at 25 °C presented lower water content than seeds at 21 °C and 23 °C, and seeds at 27 °C presented a decrease in water content at all time points, from 4 DAF until 14 DAF. At 14 DAF, the water content of seeds at 27 °C was 20% less than at normal temperature (Figure 1A). Heat stress also had a negative impact on protein content. Seeds from 25 °C and 27 °C presented decreased protein content when compared to seeds from 21 °C and 23 °C (Figure 1B). When evaluating the germination capacity of mature seeds, 21 °C and 23 °C seeds presented no difference between each other, with almost 100% of seed germination. However, germination was negatively impacted by heat stress treatments. Seeds from 25 °C, 27 °C, and 29 °C presented progressively decreased seed germination, with −20% germination capacity at 25 °C, −35% at 27 °C, and −45% at 29 °C (Figure 1C). Seed longevity was accessed by artificial aging and seed germination evaluation after successive aging time-points (6, 10, 14, 21, and 28 days). The P50 for seeds from 21 °C and 23 °C seeds were of 14 days of aging. Nevertheless, for 25 °C and 27 °C, P50 was significantly decreased, with 50% of seeds incapable of germination after 10 days of aging (Figure 1D). We were also interested in evaluating the impact of constant heat stress on seed shape. By microscopically phenotyping seeds, we were able to observe the presence of wrinkled-shape seeds due to heat stress conditions. Seeds from plants at 23 °C and 25 °C presented mainly normal-shaped seeds, while seeds at 27 °C presented normal and wrinkled seeds (17%) (Figure 1E). Concerning seed development, we did not observe any effect of heat stress on seed embryogenesis. The anatomical evaluation of seed layers and embryo showed normal embryo development and growth (Supplementary Figure S3).
Figure 1.
Physiological characteristics of A. thaliana seeds under heat stress development. (A) Mature seed water content measured every two days for a duration of 14 days. Two-way ANOVA showing the difference between stress conditions and control conditions at each time point, therefore 25 °C and 27 °C stress treatment against 21 °C. (B) Protein content based on nitrogen content. Two-way ANOVA comparing 23 °C, 25 °C, and 27 °C against 21 °C. (C) Percentage of fresh seed germination in which the numbers inside bars represent the decrease in seed germination upon heat stress conditions compared to control temperatures. Two-way ANOVA comparing 23 °C, 25 °C, and 27 °C against 21 °C. (D) Seed longevity based on P50, in which artificial aging was imposed for 6, 10, 14, 21, and 28 days. (E) Exemplary electronic microscopy of Col-0 seeds grown at 23 °C, 25 °C, and 27 °C. The below panel of the 27 °C condition shows a wrinkled-shape seed. Asterisks represent p-value significance: * p < 0.05; ** p < 0.001; *** p < 0.0001. Standard deviation is shown in (A–C) graphs.
8. Gene Expression during Seed Development under Heat Stress
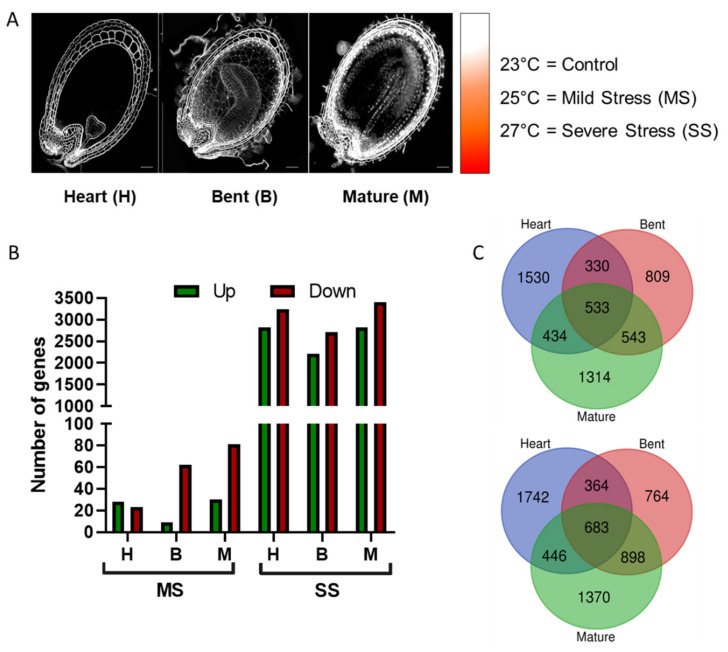
Based on the results of the physiological assays, we defined three temperatures and three embryo developmental stages for further experiments. The stages of Heart (H), Bent (B), and Mature (M) embryo were sampled from plants grown at 23 °C (control temperature), 25 °C (Mild Stress, MS), and 27 °C (Severe Stress, SS) (Figure 2A). We analyzed the dynamics of transcript abundance during seed development under control conditions and heat stress conditions by whole-transcriptome RNA-Seq analysis. Differentially expressed genes (DEGs) were observed in each stage by comparing MS versus the control and SS versus the control condition. The amount of DEGs observed at 27 °C was significantly higher than the amount of DEGs at 25 °C. At the heart stage, 27 °C seed had 16-fold more upregulated DEGs and 19-fold more downregulated DEGs than at 25 °C. The same happened at the bent stage, with 31-fold more upregulated DEGs and 22-fold more downregulated than at the MS condition, and at the mature stage, seeds on severe stress presented 9-fold more up- and 7-fold more downregulated genes than seeds on mild stress (Figure 2B). DEGs from severe stress were compared between stages and 533 genes were commonly upregulated in all stages, representing around 19–25% from the total DEGs of each stage, while 683 genes were commonly downregulated in all stages analyzed, around 20–25% of total DEGs from different stages (Figure 2C).
Figure 2.
Transcriptomics analysis of Col-0 seeds developed under heat stress conditions. (A) RNA-Seq samples were obtained during embryo developmental stages showed here in time-course order: Heart (H), Bent (B), and Mature (M). Average temperature of 23 °C was used as control against Mild Stress (MS) at 25 °C and against Severe Stress (SS) at 27 °C. (B) Scatter plot showing the quantity of differentially expressed genes (DEGs) for MS in orange and SS in red. (C) Venn diagrams of DEGs from SS at each embryo developmental stage. Upper panels show upregulated genes and lower panels show downregulated genes.
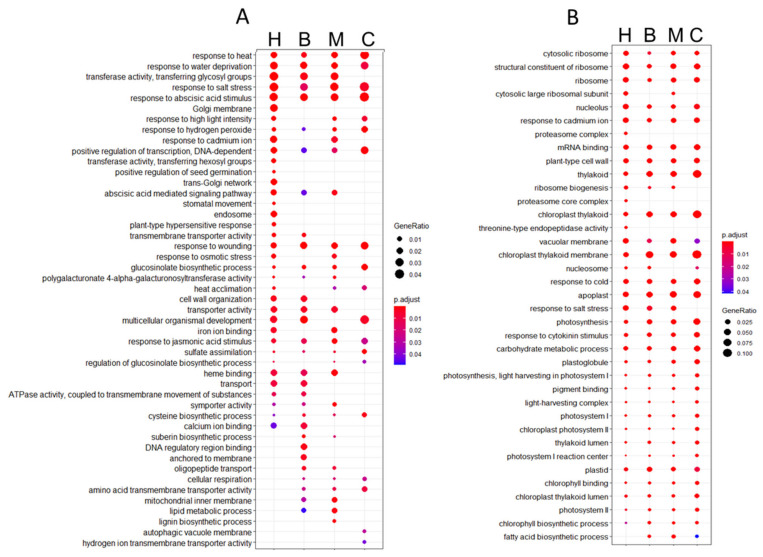
We were interested in better understanding the function of the up- and downregulated DEGs during SS since few gene changed during MS, therefore, we performed a gene set enrichment (GSEA) analysis using GO terms. The main biological functions of the upregulated DEGs were response to abiotic stress such as heat stress, water deprivation, stomatal movement, and response to hydrogen peroxide. Furthermore, secondary metabolism mechanisms were induced by severe heat stress, with the upregulation of the phytohormones abscisic acid and jasmonic acid as well as glucosinolates and suberin biosynthesis (Figure 3A). ABA signaling pathway was found upregulated in all stages upon severe stress with two related terms: ‘response to abscisic acid stimulus’ and ‘abscisic acid mediated signaling pathway’, while ‘positive regulation of germination’ terms were upregulated only during the heart stage (Figure 3A). Concerning the GO term enrichments of downregulated DEGs, ribosome biogenesis and maintenance were present for all stages, along with the decrease in the photosynthesis mechanism by repression of photosystems I and II, chlorophyll, and thylakoids (Figure 3B). The over-representation analysis showed that the downregulated DEGs were more homogeneous in their response to heat stress, but that the upregulated DEGs presented more distinct responses dependent on their development stage. Another point is that the specific DEGs of each stage, heart, bent, and mature, showed a specific response at each stage against severe heat stress (Supplementary Figure S4). The complete differential expression analysis is available in Supplementary Table S1.
Figure 3.
Gene set enrichment (GSEA) analysis of DEGs in Col-0 seeds under severe heat stress. (A) Functional enriched GO terms from upregulated differentially expressed genes. (B) Functional enriched GO terms from downregulated DEGs. The size of the dot represents the gene count. The totality of differentially up- or downregulated genes in each stage was used to perform a hypergeometric test, and the p-values were converted to false discovery rate (FDR)-corrected p-value as shown in colors, the red color being more significant than the blue color. H = heart, B = bent, M = mature, and C = common to all stages.
9. Severe Heat Stress Effect on Methylation-Related Mutants
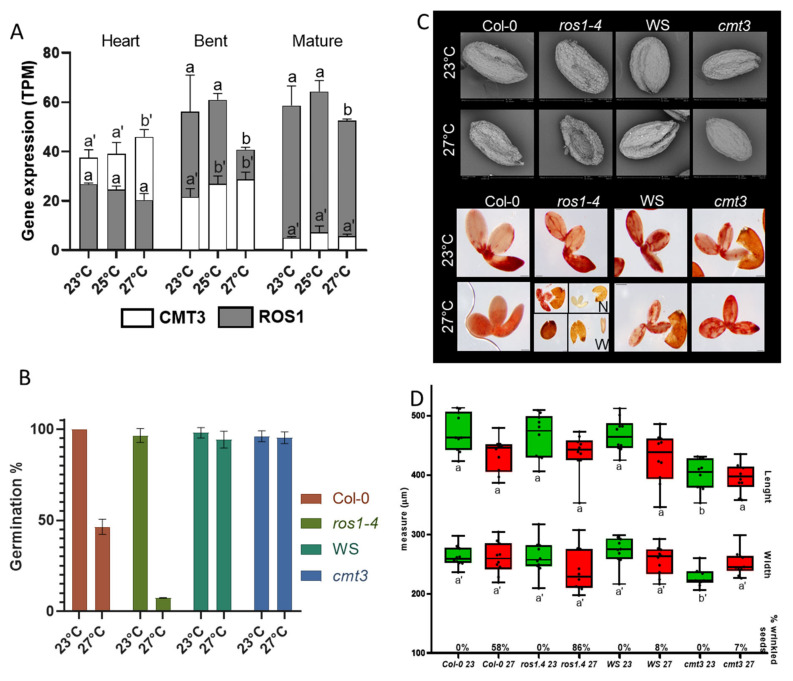
Our transcriptomic results also showed differences in the expression of genes related to chromatin organization and DNA methylation on seeds upon heat stress. The expression of CMT3 and ROS1 genes was affected by mild and severe heat stress in the embryo stages analyzed (Figure 4A). CMT3 expression increased in the heart stage at 27 °C, and at the bent stage, its expression increased at mild and severe stress. However, at the mature stage, CMT3 gene expression remained stable upon mild and severe stress. For the demethylase gene ROS1, severe heat stress dramatically impacted expression at the bent and mature stages (Figure 4A). ROS1 was also differentially expressed (downregulated) in all developmental stages upon severe stress. Therefore, to tackle the role of these genes on seeds under severe heat stress development, we decided to further investigate both gene mutants. We produced seeds from cmt3 and ros1-4 mutant plants grown at 23 °C and 27 °C for anatomical and physiological assays. First, we evaluated their seed germination rate. At 23 °C, mutants cmt3 and ros1-4 seeds presented no difference from its background, WS, and Col-0, respectively. Upon severe heat stress, seed germination was not affected for both WS and cmt3 genotypes. However, for Col-0 and ros1-4, severe heat stress decreased seed germination. Col-0 germination decreased in half while on the loss of function mutant ros1-4, severe heat stress caused an almost complete loss of germination capacity (Figure 4B). To better understand the consequences of severe heat stress on these mutants’ seeds, we observed and measured seeds with electronic microscopy (Figure 4C,D). From all the genotypes analyses, ros1-4 under 27 °C presented the most drastic phenotype. A total of 86% of ros1-4 seeds were wrinkled and deformed, some with the aspect of ‘empty seeds’ (Figure 4C,D). Concerning seed measurements, there was no significant difference in length and width for ros1-4 at 27 °C, nonetheless, there was a significant difference between cmt3 length and width at 23 °C when compared to its background and 7% of its seeds produced at 27 °C displayed the wrinkled phenotype (Figure 4D). By observing ros1-4 seed phenotype upon severe heat stress, we raised the hypothesis that ros1-4 embryos were defected or lost their viability. To observe embryo survival, we used the tetrazolium red to formazan assay. In this assay, living tissue or cells containing dehydrogenase enzymes thanks to hydrogen released during cell respiration, will reduce tetrazolium chloride to formazan, a reddish, water-insoluble compound [55]. Therefore, living cells will be red-stained, indicating normal cell respiration. cmt3 displayed red-colored embryos at control and upon severe heat stress. ros1-4 seeds at 23 °C also displayed normal anatomic and reddish embryos. Under 27 °C, ros1-4 seeds were divided between normal and wrinkled-shape for preventing erroneous interpretation of results. Normal-shaped seeds presented viable embryos as well as non-viable embryos, while the majority of wrinkled seeds were empty or presented embryos with arrested development, as seen in Figure 4C.
Figure 4.
Evaluation of CMT3 and ROS1 expression and physiology of cmt3 and ros1-4 mutants. (A) Gene expression in TPM of CMT3 and ROS1 at heart, bent, and mature stages of embryo development during control temperature (23 °C), MS (25 °C) and SS (27 °C). CMT3 is represented by white bars while ROS1 is represented by grey bars. Standard deviation is shown. Two-way ANOVA comparing differences in gene expression at 23 °C, 25 °C, and 27 °C for each genotype at the same stage. Letters represent statistically significant differences with a p-value < 0.001. Letters a or b used for ROS1 and letters a’ or b’ used for CMT3. (B) Seed germination for wild-type (Col-0 or WS) and mutant (ros1-4 or cmt3) genotypes at 23 °C (green) and 27 °C (red) average temperature. (C) Panels show embryos and seed coats from wild-types (Col-0 or WS) and mutant (ros1-4 or cmt3) genotypes at 23 °C and 27 °C average temperature. The upper panel is composed of electronic microscopy photos for phenotype observations and the lower panel shows embryo survival evaluation with TTZ. ros1-4 at 27 °C was divided into normal-shaped seeds (N) or wrinkled-shaped seeds (W). (D) Seed measurements of length and width in micrometers. Boxes and whiskers showing minimum and maximum with median line from all observations. Green bars represent temperature at 23 °C and red bars represent the temperature at 27 °C (SS). Two-way ANOVA comparing difference size between wild-types (Col-0 or WS) and mutants (ros1-4 or cmt3), respectively, at 23 °C wild-type against 23 °C mutant, and 27 °C wild-type against 27 °C mutant, therefore green against green and red against red. Letters represent statistically significant differences with a p-value < 0.001. Letters a or b used for length measures and letters a’ or b’ used for width measures. Percentage of wrinkle-shaped seeds for each genotype at 23 °C and 27 °C is shown at the base of the x-axis.
10. Whole-Genome DNA Methylation of Seeds under Heat Stress
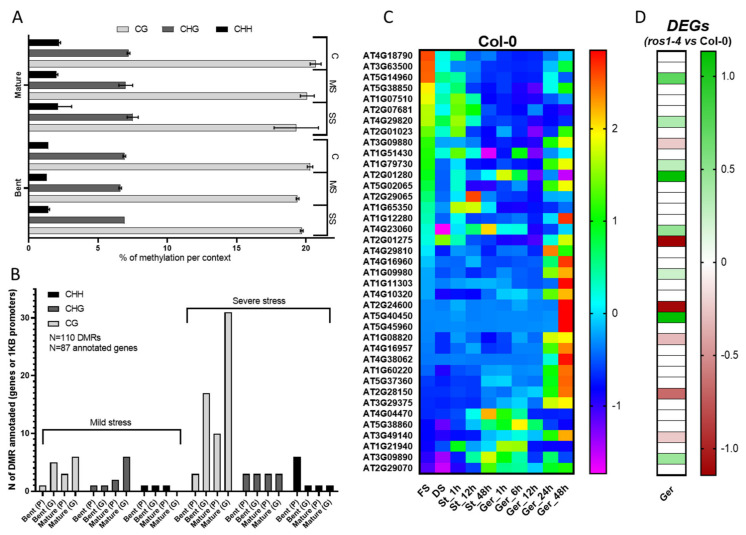
To understand the precise role of DNA methylation on seed response to heat stress, we performed bisulfite sequencing of the genomic DNA isolated from Col-0 seeds produced at 23 °C, 25 °C, and 27 °C average temperature, therefore the control, mild, and severe heat stress. We used the stages of bent and mature embryo for this analysis. About 20 million high-quality read pairs were generated for each sample and mapped uniquely to the Arabidopsis genome (TAIR10). The percentage of methylcytosines identified in each context did not change upon mild nor severe stress (Figure 5A). At the bent stage, % of methylated cytosines without heat stress was of 19.7% for CG, 6.9% for CHG, and 1.4% for CHH, while at mild heat stress CG = 19.4, CHG = 6.6, and CHH = 1.3, and at severe stress, CG = 20.3, CHG = 6.9, and CHH = 1.4. The same tendency was observed at the mature stage of seeds produced at 23 °C (CG = 19.3, CHG = 7.5, CHH = 2.1), 25 °C (CG = 20.1, CHG = 7, CHH = 2), and 27 °C (CG = 20.7, CHG = 7.2, CHH = 2.2) with a similar percentage of methylcytosines per context and per heat stress (Figure 5A). We identified differentially methylated regions (DMRs) upon mild heat stress and upon severe heat stress when comparing heat-stressed samples to the control samples. DMRs were identified in gene sequences and 1 Kb promoter gene regions. A total of 110 DMRs were identified, and from those, 87 unique genes were annotated (Figure 5B). Severe stress induced more DNA methylation changes than mild stress, with 33 DMRs at the bent and 49 DMRs at the mature stage, against 10 DMRs at the bent and 18 DMRs at the mature stage upon mild stress. The mature stage had a greater total amount of DMRs (67) when compared to the bent stage (43). Interestingly, DMRs were also found in mitochondrial DNA, with 13 DMRs present upon mild stress and 1 DMR upon severe heat stress. From these, five DMRs were localized at mitochondrial coding or promoter gene regions. A complete list of the genes related to the DMRs and their contexts are shown in Supplementary Table S2.
Figure 5.
Methylome analysis and DMRs related to germination genes. (A) Percentage of cytosine methylation per context (CG, CHG, CHH) on bent and mature embryo stages of Col-0 seeds grown under the Control temperature (C = 23 °C) or Mild Stress (MS = 25 °C), or Severe Stress (SS = 27 °C). (B) Amount of DMRs between control condition and MS or control condition and SS in all contexts (CH, CHG, CHH) for bent and mature stages at 1 Kb promoter regions (P) and gene sequences (G). (C) Normalized expression (TPM) heat-map of 39 genes differentially expressed during seed germination and containing a DMR at mature stage with severe heat stress (MethDEGs). Expression in Col-0 seeds at conditions: fresh seed (FS); dried seed (DS); stratified and imbibed seeds at 4 °C (St) for 1 h, 12 h and 48 h; stratified and imbibed seeds at 20 °C light for germination (Ger) for 1 h, 6 h, 12 h, 24 h, and 48 h. (D) Differential expression analysis of MethDEGs between ros1-4 and Col-0, comparing dry seeds against two germination stages (II and IV), whereas green indicates upregulated genes and red indicates downregulated genes in ros1-4.
Given that germination was affected by heat stress and that ros1-4 presented a strong germination phenotype upon severe stress, we compared our DMR annotated genes with germination-related genes identified by transcriptomics analysis [52]. The result showed that 69% of the differentially methylated genes (39 out of 87 genes) were also differentially expressed genes during seed germination, therefore potentially involved in the germination process. To examine the influence of differential methylation on differential gene expression, we analyzed the expression profiles of these 39 methylated and differentially expressed genes (MethDEGs) between successive stages of seed development germination from fresh seed, dry seed, and stratified seeds at 4 °C and seeds incubated at 20 °C light for germination from 1 h to 48 h (Figure 5C). The complete list of the MethDEGs and their annotation is available in Supplementary Table S2. We observed two gene clusters: (i) genes with accumulated transcripts on mature seed, that progressively decreased in expression along with seed germination process (upper heat map), and (ii) genes that were induced upon stratification and seed imbibition, with an increased expression during seed germination with seed coat rupture and radicle expansion (middle-bottom heat map) (Figure 5C). We also performed RNA-Seq with Col-0 and ros1-4 seed grown at 23 °C, using mature dried seeds, and two stages of germination: stage II (seed coat rupture and radicle point) and stage IV (seed coat rupture and radicle 0.5 mm expansion). A total of 84% (32 genes) of the MethDEGs were found to be differentially expressed in ros1-4 during germination when comparing the mature stage of ros1-4 against its samples at stage II or against stage IV of germination. We also performed a differential expression analysis between Col-0 and ros1-4 (Figure 5D). On the ros1-4 mutant that showed decreased germination and decreased heat stress resistance, we observed that 14 MethDEGs were differentially expressed, in germination stages II or IV. From those genes, eight were upregulated and six were downregulated in the mutant (Figure 5D). From the upregulated genes, ELF7 (EARLY FLOWERING 7—AT1G79730), DEL2 (DP-E2F-like transcription factor—AT5G14960), and BRF3 (TFIIIB-related factor—AT2G01280) genes are interesting because they were already shown to be required for seed dormancy, proper plant cell proliferation and elongation, and thermotolerance. In detail, ELF7 is required for the expression of the flowering repressor FLC [56] and is also part of the Polymerase II Associated Factor 1 Complex (PAF1C), which interacts with Reduced Dormancy 2 factor (RDO2). A study with elf7 mutant showed that seed dormancy was reduced, suggesting the role of this gene as a negative regulator of seed germination [57]. DEL2 promotes cell proliferation and represses cell elongation in the root meristem and its altered expression affects the expression of several cell-cycle regulators [58]. As cell elongation is the first step in the germination stage, before cell division [59,60], DEL2 might repress radicle emergence. Moreover, the BRF3 gene, which is part of the core unit of the RNA Pol III transcription complex [61], was shown to negatively regulate the thermotolerance in Arabidopsis [62]. The upregulation of these genes in ros1-4 during germination could be part of the answer for the decrease in ros1-4 seed germination upon heat stress. The complete differential expression analysis between ros1-4 and Col-0 is available in Supplementary Table S1.
11. Discussion
Constant Heat Stress Does Not Affect Seed Embryogenesis but Negatively Impacts Seed Germination and Longevity
Here, we aimed to verify the impact of constant heat stress on seed development. To perform the experiments with A. thaliana, we based our temperature selection on global warming projections. Initial projections estimated an increase of 1 °C to 2 °C on average global surface temperature, but recent projections foresee an increase by up to 4.8 °C by the year 2100, depending on the level of greenhouse gas emissions [63]. Along with these gloomy data, climate change is projected to impose more extreme weather conditions, with the rise in global temperature including longer and hotter summers that possibly will severely disrupt plant growth and productivity [64].
We were keen to investigate if heat stress could disrupt seed embryogenesis, which is the developmental process that transforms a single totipotent cell, the zygote, into a mature embryo, by producing and developing the embryo first tissues precursors and first stem cells [65]. We observed that seed embryogenesis was not affected upon our heat stress assays. One explanation might be the strength of the embryogenesis cell pattern in A. thaliana. Most flowering plants present apparently random and disordered embryogenic cell divisions while a few present a highly regular and predictable pattern such as the case of the Brassicaceae family [66,67]. This is why most of the knowledge on plant embryogenesis comes from Arabidopsis research, again with the advantages of small genome size, rapid life cycle, and amenability to genetic transformation [65]. Our assays showed that the majority of Col-0 seeds at severe heat stress still developed correctly and this fact could also be due to the plants’ mechanism of basal thermotolerance, that is when plant survival above optimal temperatures is assured. Basal thermotolerance must be distinguished from acquired thermotolerance, which is survival to otherwise lethal heat stress, both being physiologically and molecularly different processes [68].
Even if we did not observe anatomical abnormalities during embryogenesis, we observed reduced silique number and length as temperature increased. As silique length is correlated with the number of seeds per silique [69,70], it is probable that less efficient fertilization leads to fewer seeds and shorter siliques. This was the case for heat stress assays in A. thaliana and in Lycopersicon esculentum, which negatively affected flower fertilization [70,71,72,73]. Flower infertility might also occur when heat stress inhibits anther dehiscence, thereby reducing pollen release and pollen viability by disrupting male meiosis [73,74,75]. Heat stress can also have an impact on the female gametes with malformation of the ovule and accessory tissues as well as a decrease in stigma receptivity [76,77]. In our assays, all plants were grown at control conditions until bolting, then plants were transferred to heat stress conditions as described. Nevertheless, as Arabidopsis flowers continuously appear, later-developed flowers might have been more stressed than the first-developed flowers. This could be the explanation for shorter and fewer siliques, with less seeds.
Regarding heat stress induced impairment of seed essential traits, heat stress reduced germination by 30% to 50% and longevity by half (Figure 1D). The negative impact of heat stress on germination has already been reported [78,79,80]. These major impacts on seed germination and longevity can have disastrous consequences for plant species survival since these are key processes to ensure species propagation [81,82,83]. For promoting seed germination, two phytohormones are key players: gibberellic acid (GA) and abscisic acid (ABA) [84]. During seed germination, ABA biosynthesis is repressed while GA increases [85]. The contrary is true to dormancy in which ABA content is high and GA is low [86]. We observed that upon severe heat stress, the ABA pathway was induced in all stages, and as the regulation of plant process upon abiotic stress has a modulated activity depending on the water status in the environment [85], the extremely low water content on seeds upon severe heat stress might have disrupted the regulation of germination process.
12. Heat Stress Induces Impaired Cell Development and Metabolic Imbalance
Our study had the objective of characterizing the transcriptomic changes that occur on seeds when they face constant heat stress during development. We observed that the transcriptional changes due to heat stress was significant when plants were grown in severe heat stress conditions and that DEGs upon severe heat stress from all the stages analyzed were mainly related to abiotic stress response and the repression of photosynthesis machinery. These results reinforced previous works that showed that a myriad of cell processes is affected by heat stress, since even enzyme function can be disrupted by temperature and provoke metabolic imbalance [87,88] or even promote programmed cell death [89,90]. Moreover, a variety of membrane-linked processes are impacted due to changes in membrane fluidity and permeability [91,92]. By promoting membrane and protein damage, heat-induced oxidative stress is generated by the production of active oxygen species [93,94,95,96]. Taking together all the possible damage, heat stress in plants is characterized by impaired translocation of assimilates and reduced carbon gain and reduced photosynthesis [93]. Heat stress influences plant growth, productivity, and development, being more related to impaired development. However, soon after the stress period, plants will reinitiate their developmental and growth program and end the transcriptional response to stress, which will provide successful recovery.
13. DNA Demethylation Is Partially Responsible for Ensuring Seed Germination under Heat Stress Conditions
We also aimed to evaluate if heat stress during seed growth was capable of inducing epigenetic changes. The epigenetic regulation of heat stress response varies, from DNA methylation [20,35,37], to histone modifications [20,35,37,97,98] and histone variants [99,100], and also microRNAs and siRNAs [40,101,102]. Nevertheless, these epigenetic responses are dependent on the heat intensity and duration. To better understand the dynamics of DNA methylation on embryogenesis stages upon heat stress, we chose to closely examine two stages that represent the middle and the end of embryogenesis, the bent and mature stages, respectively. When analyzing the epigenetic changes on the DNA, we observed that global DNA methylation levels were not modified by heat stress. The Arabidopsis genome-wide methylation levels were 24% in CG, 6.7% in CHG, and 1.7% in CHH contexts [103]. We detected a slight difference from each context against data from leaves [103], but not between stages or upon mild or severe heat stress. This could indicate that different tissues have different DNA methylation levels and also that methylation is stably maintained throughout the genome upon constant heat stress, like a basal DNA-methylation thermotolerance.
In plants, the DNA methylation homeostasis is regulated by DNA methylation and also by demethylation processes [27,30] in a way that active DNA methylation and demethylation are balanced in the cells [104]. Since we observed the differential expression of CMT3 and ROS1 upon heat stress, we aimed to investigate if maintenance of methylation by CMT3 and/or DNA demethylation by ROS1 could have a role in heat stress response on seeds. During the mature embryo stage, cell division is arrested, and therefore it suggests that DNA replication and maintenance of DNA methylation by MET1 during replication is also stopped. Therefore, the creation of DMRs during this stage is hypothesized to be due to DNA demethylation processes [104]. Recent studies have shown that ROS1 is active in developing seeds during late embryogenesis by promoting differentially methylated regions in seed-development-related genes, more specifically for the demethylation of endosperm-specific methylated regions [104,105]. At the mature embryo stage, we observed an increase in ROS1 expression in normal conditions and a decrease in CMT3 expression, however, severe heat stress repressed ROS1 expression. Furthermore, DML3 expression increased upon severe heat stress, while other demethylases, DML2 and DME were repressed as well as methylases DRM1, CMT2. RDM1, RDM4, and CMT3 remained stably low. The same was detected by a recent study that showed that while DME, DML2, and DML3 were repressed, ROS1 expression was induced in the maturation stages [104]. In addition to CMT3 expression, a study in rice seeds showed repression of its expression with 48 h of moderate heat stress (34 °C) after fertilization, which was proposed to cause differences in the DNA methylation levels of Fertilization-Independent Endosperm1 (OsFIE1), a member of Polycomb Repressive Complex2 (PRC2) [20].
After physiological analysis of both cmt3 and ros1-4 mutants, ros1-4 presented an interesting phenotype, with seed germination dramatically affected by severe stress, and with deformed seeds and undeveloped embryos. In our study, we did not observe the influence of CMT3 on heat stress resistance. This might be due to the potential functional redundancy of chromomethylases - or by its heat stress resistant background, WS [74]. The ros1-4 mutant had a Col-0 ecotype background with T-DNA insertion in the ROS1 gene, proved to cause complete loss of function of ROS1 [106]. As de novo methylation is established through the RdDM pathway, ROS1 antagonizes RdDM and RdDM-independent DNA methylation. This mechanism was shown to prevent the spread of DNA methylation from transposable elements to protein-coding genes [31,107,108]. At the same time, ROS1 expression is positively regulated by proximal RdDM-dependent TE methylation [109,110]. Seed establishment is largely influenced by the amount and diversity of compounds stored during seed maturation [111], whereas germination capacity is not completely dependent on seed reserve accumulation, since the germination of mutants deficient in lipid reserve mobilization pathways is only slightly affected [112,113,114,115,116]. Therefore, germination efficiency must be controlled by other yet unknown mechanisms, one on those being the DNA demethylation by ROS1. Here, we observed a set of germination-related genes that were differentially expressed upon severe heat stress and were also differentially methylated at seed maturity. When analyzing their expression pattern, we identified the remodeling of the transcriptome upon the steps culminating in germination. These changes were also observed in previous works that investigated the seed transition from fresh, dry seed stage, seed stratification, germination, and post germination, following exposure to light [52]. Studies have shown that during the seed maturation process, a diverse set of molecules such as proteins, lipids, sugars, and transcripts will be accumulated for use upon germination [115,117,118,119]. Upon seed imbibition, a metabolic switch happens and the metabolites accumulated during seed maturation are consumed by being mobilized and/or degraded, also with that, germination-associated gene expression programs already start during seed imbibition [83]. Thus, we can also witness these two clusters of gene expressions in the germination-related MethDEGs.
Here, we showed that mother-plants grown under heat-stress generated seeds that had DMRs located on germination-related genes. These DMRs could be conserved in future generations. Many works have presented the inheritance of epigenetic changes caused by heat stress by the next generations and that this transgenerational inheritance was maintained for at least three generations in a genotypic and phenotypic manner [33,38,39,40,98]. The epigenetic memory can increase plant fitness to provide better heat stress responses and therefore contribute to plant evolutionary adaptation [41,42]. Additionally, ROS1 was shown to decrease its expression by heat stress and that this decrease was maintained in heat-stressed A. thaliana F1 descendants, which also presented earlier-bolting as well as fewer and larger leaves [39].
Remarkably, five mitochondrial genes presented DMRs upon heat stress. These genes were all found to be highly expressed in seed development, imbibition, and germination. Given that mitochondria are responsible for the respiratory chain, these genes could be important actors during seed germination, and different DNA methylation status caused by heat stress could change their expression and affect seed germination. Studies have shown the role and versatility of mitochondria structures during seed germination. At the dry seed stage, there are promitochondria, which are larger, isodiametric structures that will eventually develop into mitochondria upon seed imbibition. Later, upon germination, mitochondria undergo variations in morphology along with cell differentiation and cell division in the course of early root development [120,121]. The bioenergetic reactivation of mitochondria is immediately observed upon seed imbibition and the reactivation of mitochondrial dynamics occurs after transfer to germination conditions [122]. Besides, as mitochondria are the main source of reactive oxygen species (ROS) production, they are strongly exposed to oxidative damage [123], and the continuous heat stress could affect them greatly, also in an epigenetic manner, as we observed in our work. Further studies on these genes could clarify their role on heat stress resistance in seeds.
Taken together, our results showed a partial control of heat stress response on seeds by the demethylation of germination-related genes. This is a finding of great importance because seed germination and plantlet establishment are the base of global food production, and ensuring food security in the climate change era is one of the major challenges in the coming decades. Plant breeding strategies focused on selecting varieties with increased resistance to abiotic stresses, like heat stress, will play a central role in confronting this challenge.
Acknowledgments
The authors sincerely thank all of the SEED team at INRAE, IRHS from Angers, more specifically Joseph Ly Vu and Benoit Ly Vu for advice regarding plant care and physiological assays. The authors gratefully acknowledge the technical platforms ANAN and iMAC of the SFR 4207 QUASAV.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/3/457/s1, Figure S1: Schematic organization of samples, experiments, and ecotypes used in this work, Figure S2: Col-0 plant phenotype under heat stress conditions. (A) Photos of Col-0 plants showing increased biomass related to increase in temperature. (B) Photos of Col-0 plants showing decreased silique number related to the increase in temperature. (C) Average silique number per branch at 21 °C, 23 °C, 25 °C, 27 °C, and 29 °C average temperatures. (D) Average silique length at 21 °C, 23 °C, 25 °C, 27 °C, and 29 °C average temperatures. Standard deviations are indicated in both C and D graphs, Figure S3: Microscopy of Col-0 embryo development under heat stress. Embryo stages showed are globular, transition, heart, torpedo, walking stick, and mature at 21 °C, 23 °C, 25 °C, and 27 °C average temperature, Figure S4: Gene set enrichment analysis (GSEA) of specific DEGs in Col-0 seeds under severe heat stress. (A) Enriched GO terms from upregulated differentially expressed genes. (B) Enriched GO terms from downregulated differentially expressed genes. The size of the dot represents the gene count. The totality of differentially up- or downregulated genes specific to each stage was used to perform a hypergeometric test, and the p-values from the tests were converted to false discovery rate (FDR)-corrected p-value as shown in colors, the red color being more significant than the blue color. H = heart, B = bent, M = mature, and C = common to all stages, Table S1: Deseq2 summary data from RNA-Seq with Col-0 seeds during development under different ranges of heat stress. Table S2: Deseq2 summary data from RNA-Seq with ros1-4 and Col-0 seeds at maturation and germination stages.
Author Contributions
J.M., D.W. and W.X. contributed equally to this work; Conceptualization, J.M., D.W. and J.V.; Methodology, J.M., D.W., W.X. and J.V.; Validation, J.M. and J.V.; Formal Analysis, J.M., J.V., W.X. and D.W.; Investigation, J.M., D.W., W.X. and J.V.; Data Curation, J.M. and J.V.; Writing—Original Draft Preparation, J.M.: Writing—Review & Editing, J.V., D.W., W.X. and J.M.; Visualization, J.M. and J.V.; Supervision, J.V.; Project Administration, J.V.; Funding Acquisition, J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was part of the EpiBASIS project funded by the University of Angers and by the DEswitch project funded by ANR (ANR-19-CE20-0027-01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167245, accessed on 22 March 2021). Supplementary Data: RNA-Seq and methylome data are available at GSE167245 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167245, accessed on 22 March 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Venterea R.T. Climate Change 2014: Mitigation of Climate Change. J. Environ. Qual. 2014;38:837. doi: 10.2134/jeq2008.0024br. [DOI] [Google Scholar]
- 2.Walther G., Post E., Convey P., Menzel A., Parmesank C., Beebee T.J.C., Fromentin J., Hoegh-Guldberg O., Bairlein F. Ecological response to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 3.Teixeira E.I., Fischer G., Van Velthuizen H., Walter C., Ewert F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013;170:206–215. doi: 10.1016/j.agrformet.2011.09.002. [DOI] [Google Scholar]
- 4.Penuelas J., Rutishauser T., Filella I. Phenology Feedbacks on Climate Change. Science. 2009;324:887–888. doi: 10.1126/science.1173004. [DOI] [PubMed] [Google Scholar]
- 5.Fitter A.H., Fitter R.S.R. Rapid changes in flowering time in British plants. Science. 2002;296:1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:0980–0989. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Cheng R., Spokas K.A., Palmer A.A., Borevitz J.O. Genetic variation for life history sensitivity to seasonal warming in Arabidopsis thaliana. Genetics. 2014;196:569–577. doi: 10.1534/genetics.113.157628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahid A., Gelani S., Ashraf M., Foolad M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 9.Patel D., Franklin K.A. Temperature-regulation of plant architecture. Plant Signal. Behav. 2009;4:577–579. doi: 10.4161/psb.4.7.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoun M., Ouellet F. Growth temperature affects inflorescence architecture in Arabidopsis thaliana. Botany. 2013;91:642–651. doi: 10.1139/cjb-2013-0011. [DOI] [Google Scholar]
- 11.Long S.P., Ort D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010;13:240–247. doi: 10.1016/j.pbi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Donohue K. Completing the cycle: Maternal effects as the missing link in plant life histories. Philos. Trans. R. Soc. B Biol. Sci. 2009;364:1059–1074. doi: 10.1098/rstb.2008.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Z., Footitt S., Finch-Savage W.E. The effect of temperature on reproduction in the summer and winter annual Arabidopsis thaliana ecotypes Bur and Cvi. Ann. Bot. 2014;113:921–929. doi: 10.1093/aob/mcu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pauli H., Gottfried M., Dullinger S., Abdaladze O., Akhalkatsi M., Alonso J.L.B., Coldea G., Dick J., Erschbamer B., Calzado R.F., et al. Recent Plant Diversity Changes on Europe’s Mountain Summits. Science. 2012;336:353–355. doi: 10.1126/science.1219033. [DOI] [PubMed] [Google Scholar]
- 15.Lobell D.B., Schlenker W., Costa-Roberts J. Climate Trends and Global Crop Production Since 1980. Science. 2011;333:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 16.Donohue K., Rubio De Casas R., Burghardt L., Kovach K., Willis C.G. Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 2010;41:293–319. doi: 10.1146/annurev-ecolsys-102209-144715. [DOI] [Google Scholar]
- 17.Burghardt L.T., Edwards B.R., Donohue K. Multiple paths to similar germination behavior in Arabidopsis thaliana. New Phytol. 2016;209:1301–1312. doi: 10.1111/nph.13685. [DOI] [PubMed] [Google Scholar]
- 18.Walck J.L., Hidayati S.N., Dixon K.W., Thompson K., Poschlod P. Climate change and plant regeneration from seed. Glob. Chang. Biol. 2011;17:2145–2161. doi: 10.1111/j.1365-2486.2010.02368.x. [DOI] [Google Scholar]
- 19.Righetti K., Vu J.L., Pelletier S., Vu B.L., Glaab E., Lalanne D., Pasha A., Patel R.V., Provart N.J., Verdier J., et al. Inference of Longevity-Related Genes from a Robust Coexpression Network of Seed Maturation Identifies Regulators Linking Seed Storability to Biotic Defense-Related Pathways. Plant Cell. 2015;27:2692–2708. doi: 10.1105/tpc.15.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folsom J.J., Begcy K., Hao X., Wang D., Walia H. Rice Fertilization-Independent Endosperm1 Regulates Seed Size under Heat Stress by Controlling Early Endosperm Development. Plant Physiol. 2014;165:238–248. doi: 10.1104/pp.113.232413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClung C.R., Davis S.J. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 2010;20:R1086–R1092. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 22.Bokszczanin K.L., Fragkostefanakis S. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 2013;4:315. doi: 10.3389/fpls.2013.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova O.V., Dinh H.Q., Aufsatz W., Jonak C. The RdDM Pathway Is Required for Basal Heat Tolerance in Arabidopsis. Mol. Plant. 2013;6:396–410. doi: 10.1093/mp/sst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonasio R., Tu S., Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibney E.R., Nolan C.M. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 26.Lukens L.N., Zhan S. The plant genome’s methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 2007;10:317–322. doi: 10.1016/j.pbi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Chinnusamy V., Zhu J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matzke M.A., Mosher R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 29.Chan S.W.L., Henderson I.R., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Zhu J.-K. Active DNA Demethylation in Plants and Animals. Cold Spring Harb. Symp. Quant. Biol. 2012;77:161–173. doi: 10.1101/sqb.2012.77.014936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang K., Lang Z., Zhang H., Zhu J.-K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants. 2016;2:16169. doi: 10.1038/nplants.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 33.Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., Hollander J., Meins F., Kovalchuk I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE. 2010;5:e9514. doi: 10.1371/annotation/726f31b5-99c4-44e9-9cd6-b8d66b3f6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naydenov M., Baev V., Apostolova E., Gospodinova N., Sablok G., Gozmanova M., Yahubyan G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015;87:102–108. doi: 10.1016/j.plaphy.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Correia B., Valledor L., Meijón M., Rodriguez J.L., Dias M.C., Santos C., Cañal M.J., Rodriguez R., Pinto G. Is the Interplay between Epigenetic Markers Related to the Acclimation of Cork Oak Plants to High Temperatures? PLoS ONE. 2013;8:e53543. doi: 10.1371/journal.pone.0053543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao G., Li J., Li H., Li F., Xu K., Yan G., Chen B., Qiao J., Wu X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 2014;64:125–133. doi: 10.1270/jsbbs.64.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min L., Li Y., Hu Q., Zhu L., Gao W., Wu Y., Ding Y., Liu S., Yang X., Zhang X. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 2014;164:1293–1308. doi: 10.1104/pp.113.232314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suter L., Widmer A. Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS ONE. 2013;8:e80819. doi: 10.1371/journal.pone.0080819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Migicovsky Z., Yao Y., Kovalchuk I. Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 2014;9:e27971. doi: 10.4161/psb.27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong S.H., Liu J.Z., Jin H., Lin L., Li Q., Chen Y., Yuan Y.X., Wang Z.Y., Huang H., Qi Y.J., et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:9171–9176. doi: 10.1073/pnas.1219655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittle C.A., Otto S.P., Johnston M.O., Krochko J.E. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany. 2009;87:650–657. doi: 10.1139/B09-030. [DOI] [Google Scholar]
- 42.Liu J., Feng L., Li J., He Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015;6:267. doi: 10.3389/fpls.2015.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López Sánchez A., Stassen J.H.M., Furci L., Smith L.M., Ton J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016;88:361–374. doi: 10.1111/tpj.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones D.B. Factors for converting percentages of nitrogen in foods and feeds into percentages of protein. Br. Food J. 1932 [Google Scholar]
- 46.Xu W., Fiume E., Coen O., Pechoux C., Lepiniec L., Magnani E. Endosperm and nucellus develop antagonistically in arabidopsis seeds. Plant Cell. 2016;28:1343–1360. doi: 10.1105/tpc.16.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maia J., Dekkers B.J.W., Provart N.J., Ligterink W., Hilhorst H.W.M. The re-establishment of desiccation tolerance in germinated arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE. 2011;6:e29123. doi: 10.1371/journal.pone.0029123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanai M., Mano S., Nishimura M. An efficient method for the isolation of highly purified RNA from seeds for use in quantitative transcriptome analysis. J. Vis. Exp. 2017;2017:4–9. doi: 10.3791/55008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narsai R., Gouil Q., Secco D., Srivastava A., Karpievitch Y.V., Liew L.C., Lister R., Lewsey M.G., Whelan J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017;18:172. doi: 10.1186/s13059-017-1302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer D.S., Theis F.J., Yosef N. Impulse model-based differential expression analysis of time course sequencing data. Nucleic Acids Res. 2018;46:e119. doi: 10.1093/nar/gky675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz M.D., He Y., Whitaker J.W., Hariharan M., Mukamel E.A., Leung D., Rajagopal N., Nery J.R., Urich M.A., Chen H., et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De França-Neto J.B., Krzyzanowski F.C. Tetrazolium: An important test for physiological seed quality evaluation. J. Seed Sci. 2019;41:359–366. doi: 10.1590/2317-1545v41n3223104. [DOI] [Google Scholar]
- 56.Berr A., Xu L., Gao J., Cognat V., Steinmetz A., Dong A., Shen W.H. Set Domain Group25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y., Geyer R., van Zanten M., Carles A., Li Y., Hörold A., van Nocker S., Soppe W.J.J. Identification of the Arabidopsis reduced dormancy 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE. 2011;6:e22241. doi: 10.1371/journal.pone.0022241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sozzani R., Maggio C., Giordo R., Umana E., Ascencio-Ibañez J.T., Hanley-Bowdoin L., Bergounioux C., Cella R., Albani D. The E2FD/DEL2 factor is a component of a regulatory network controlling cell proliferation and development in Arabidopsis. Plant Mol. Biol. 2010;72:381–395. doi: 10.1007/s11103-009-9577-8. [DOI] [PubMed] [Google Scholar]
- 59.Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sliwinska E., Bassel G.W., Bewley J.D. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 2009 Aug 20;60:3587–3594. doi: 10.1093/jxb/erp203. [DOI] [PubMed] [Google Scholar]
- 61.Willis I., Schmidt P., Soll D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989;8:4281–4288. doi: 10.1002/j.1460-2075.1989.tb08614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu C., Liu X., Li X., Huo P., Ge J., Hou Y., Yang W., Zhang J., Zhang L., Zhao D., et al. BRF Negatively Regulates Thermotolerance Defect of fes1a in Arabidopsis. Front. Plant Sci. 2020;11:171. doi: 10.3389/fpls.2020.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pachauri R., Meyer L. Climate change 2014: Synthesis report. Geneva Intergov. Panel Clim. Chang. 2015;16:408. [Google Scholar]
- 64.Dolzblasz A., Dołzbłasz S. Arabidopsis high temperature stress research. Acta Soc. Bot. Pol. 2018;87:3. doi: 10.5586/asbp.3594. [DOI] [Google Scholar]
- 65.Ten Hove C.A., Lu K.J., Weijers D. Building a plant: Cell fate specification in the early arabidopsis embryo. Development. 2015;142:420–430. doi: 10.1242/dev.111500. [DOI] [PubMed] [Google Scholar]
- 66.Palovaara J., De Zeeuw T., Weijers D. Tissue and Organ Initiation in the Plant Embryo: A First Time for Everything. Annu. Rev. Cell Dev. Biol. 2016;32:47–75. doi: 10.1146/annurev-cellbio-111315-124929. [DOI] [PubMed] [Google Scholar]
- 67.Boscá S., Knauer S., Laux T. Embryonic development in arabidopsis thaliana: From the zygote division to the shoot meristem. Front. Plant Sci. 2011;2:93. doi: 10.3389/fpls.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkindale J., Hall J.D., Knight M.R., Vierling E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malabarba J., Buffon V., Mariath J.E.A., Gaeta M.L., Dornelas M.C., Margis-Pinheiro M., Pasquali G., Revers L.F. The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine. J. Exp. Bot. 2017;68:1493–1506. doi: 10.1093/jxb/erx025. [DOI] [PubMed] [Google Scholar]
- 70.Bac-Molenaar J.A., Fradin E.F., Becker F.F.M., Rienstra J.A., van der Schoot J., Vreugdenhil D., Keurentjes J.J.B. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis Thaliana. Plant Cell. 2015;27:1857–1874. doi: 10.1105/tpc.15.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zinn K.E., Tunc-Ozdemir M., Harper J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010;61:1959–1968. doi: 10.1093/jxb/erq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warner R.M., Erwin J.E. Naturally occurring variation in high temperature induced floral bud abortion across Arabidopsis thaliana accessions. Plant Cell Environ. 2005;28:1255–1266. doi: 10.1111/j.1365-3040.2005.01361.x. [DOI] [Google Scholar]
- 73.Kim S.Y., Hong C.B., Lee I. Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J. Plant Res. 2001;114:301–307. doi: 10.1007/PL00013991. [DOI] [Google Scholar]
- 74.Endo M., Tsuchiya T., Hamada K., Kawamura S., Yano K., Ohshima M., Higashitani A., Watanabe M., Kawagishi-Kobayashi M. High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 2009;50:1911–1922. doi: 10.1093/pcp/pcp135. [DOI] [PubMed] [Google Scholar]
- 75.Sato S., Kamiyama M., Iwata T., Makita N., Furukawa H., Ikeda H. Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Ann. Bot. 2006;97:731–738. doi: 10.1093/aob/mcl037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hedhly A. Sensitivity of flowering plant gametophytes to temperature fluctuations. Environ. Exp. Bot. 2011;74:9–16. doi: 10.1016/j.envexpbot.2011.03.016. [DOI] [Google Scholar]
- 77.Saini H.S., Aspinall D. Abnormal sporogenesis in wheat (Triticum aestivum L.) induced by short periods of high temperature. Ann. Bot. 1982;49:835–846. doi: 10.1093/oxfordjournals.aob.a086310. [DOI] [Google Scholar]
- 78.Schmuths H., Bachmann K., Weber W.E., Horres R., Hoffmann M.H. Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann. Bot. 2006;97:623–634. doi: 10.1093/aob/mcl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He H., De Souza Vidigal D., Basten Snoek L., Schnabel S., Nijveen H., Hilhorst H., Bentsink L. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 2014;65:6603–6615. doi: 10.1093/jxb/eru378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silva-Correia J., Freitas S., Tavares R.M., Lino-Neto T., Azevedo H. Phenotypic analysis of the Arabidopsis heat stress response during germination and early seedling development. Plant Methods. 2014;10:7. doi: 10.1186/1746-4811-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fenner M., Thompson K. The Ecology of Seeds. Volume 15. Cambridge University Press; Cambridge, UK: 2005. [Google Scholar]
- 82.Gaff D.F., Oliver M. The evolution of desiccation tolerance in angiosperm plants: A rare yet common phenomenon. Funct. Plant Biol. 2013;40:315–328. doi: 10.1071/FP12321. [DOI] [PubMed] [Google Scholar]
- 83.Fait A., Angelovici R., Less H., Ohad I., Urbanczyk-Wochniak E., Fernie A.R., Galili G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006;142:839–854. doi: 10.1104/pp.106.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacobsen J.V., Pearce D.W., Poole A.T., Pharis R.P., Mander L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 2002;115:428–441. doi: 10.1034/j.1399-3054.2002.1150313.x. [DOI] [PubMed] [Google Scholar]
- 85.Skubacz A., Daszkowska-Golec A., Szarejko I. The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016;7:1884. doi: 10.3389/fpls.2016.01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu H., Stone S.L. Regulation of ABI5 turnover by reversiblepost-translational modifications. Plant Signal. Behav. 2014;9:e27577. doi: 10.4161/psb.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vierling E. The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991;42:579–620. doi: 10.1146/annurev.pp.42.060191.003051. [DOI] [Google Scholar]
- 88.Kampinga H.H., Brunsting J.F., Stege G.J.J., Burgman P.W.J.J., Konings A.W.T. Thermal protein denaturation and protein aggregation in cells made thermotolerant by various chemicals: Roleof heat shock proteins. Exp. Cell Res. 1995;219:536–546. doi: 10.1006/excr.1995.1262. [DOI] [PubMed] [Google Scholar]
- 89.Swidzinski J.A., Sweetlove L.J., Leaver C.J. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J. 2002;30:431–446. doi: 10.1046/j.1365-313X.2002.01301.x. [DOI] [PubMed] [Google Scholar]
- 90.Vacca R.A., De Pinto M.C., Valenti D., Passarella S., Marra E., De Gara L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco bright-yellow 2 cells. Plant Physiol. 2004;134:1100–1112. doi: 10.1104/pp.103.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alfonso M., Yruela I., Almárcegui S., Torrado E., Pérez M.A., Picorel R. Unusual tolerance to high temperatures in a new herbicide-resistant D1 mutant from Glycine max (L.) Merr. cell cultures deficient in fatty acid desaturation. Planta. 2001;212:573–582. doi: 10.1007/s004250000421. [DOI] [PubMed] [Google Scholar]
- 92.Sangwan V., Örvar B.L., Beyerly J., Hirt H., Dhindsa Rajinder S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002;31:629–638. doi: 10.1046/j.1365-313X.2002.01384.x. [DOI] [PubMed] [Google Scholar]
- 93.Hall A.E. Crop Responses to Environment. CRC Press; Boca Raton, FL, USA: 2001. [Google Scholar]
- 94.Dat J.F., Foyer C.H., Scott I.M. Changes in salicylic acid and antioxidants during induced thermotolerance in mustard seedlings. Plant Physiol. 1998;118:1455–1461. doi: 10.1104/pp.118.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Larkindale J., Knight M.R. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong M., Li Y.J., Chen S.Z. Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J. Plant Physiol. 1998;153:488–496. doi: 10.1016/S0176-1617(98)80179-X. [DOI] [Google Scholar]
- 97.Singh P., Yekondi S., Chen P.W., Tsai C.H., Yu C.W., Wu K., Zimmerli L. Environmental history modulates Arabidopsis pattern-triggered immunity in a histone acetyltransferase1-dependent manner. Plant Cell. 2014;26:2676–2688. doi: 10.1105/tpc.114.123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lang-Mladek C., Popova O., Kiok K., Berlinger M., Rakic B., Aufsatz W., Jonak C., Hauser M.T., Luschnig C. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in arabidopsis. Mol. Plant. 2010;3:594–602. doi: 10.1093/mp/ssq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumar S.V., Wigge P.A. H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 101.Chen L., Ren Y., Zhang Y., Xu J., Sun F., Zhang Z., Wang Y. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene. 2012;504:160–165. doi: 10.1016/j.gene.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 102.Chellappan P., Vanitharani R., Ogbe F., Fauquet C.M. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005;138:1828–1841. doi: 10.1104/pp.105.066563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kawakatsu T., Nery J.R., Castanon R., Ecker J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017;18:171. doi: 10.1186/s13059-017-1251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsieh T.-F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. Genome-Wide Demethylation of Arabidopsis Endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Penterman J., Uzawa R., Fischer R.L. Genetic interactions between DNA demethylation and methylation in arabidopsis. Plant Physiol. 2007;145:1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y., Córdoba-Cañero D., Qian W., Zhu X., Tang K., Zhang H., Ariza R.R., Roldán-Arjona T., Zhu J.K. An AP Endonuclease Functions in Active DNA Dimethylation and Gene Imprinting in Arabidopsis. PLoS Genet. 2015;11:e1004905. doi: 10.1371/journal.pgen.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martínez-Macías M.I., Qian W., Miki D., Pontes O., Liu Y., Tang K., Liu R., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., et al. A DNA 3’ Phosphatase Functions in Active DNA Demethylation in Arabidopsis. Mol. Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qian W., Miki D., Zhang H., Liu Y., Zhang X., Tang K., Kan Y., La H., Li X., Li S., et al. A Histone Acetyltransferase Regulates Active DNA Demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lang Z., Lei M., Wang X., Tang K., Miki D., Zhang H., Mangrauthia S.K., Liu W., Nie W., Ma G., et al. The Methyl-CpG-Binding Protein MBD7 Facilitates Active DNA Demethylation to Limit DNA Hyper-Methylation and Transcriptional Gene Silencing. Mol. Cell. 2015;57:971–983. doi: 10.1016/j.molcel.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brocard-Gifford I.M., Lynch T.J., Finkelstein R.R. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003;131:78–92. doi: 10.1104/pp.011916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Katavic V., Reed D.W., Taylor D.C., Giblin E.M., Barton D.L., Zou J., MacKenzie S.L., Covello P.S., Kunst L. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995;108:399–409. doi: 10.1104/pp.108.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Focks N., Benning C. Wrinkled 1: A novel, low-seed-oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu C., Hills M.J. Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol. 2002;129:1352–1358. doi: 10.1104/pp.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Penfield S., Graham S., Graham I.A. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 2005;33:380–383. doi: 10.1042/BST0330380. [DOI] [PubMed] [Google Scholar]
- 116.Pritchard Sarah L., Charlton Wayne L., Baker A., Graham Ian A. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002;31:639–647. doi: 10.1046/j.1365-313X.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- 117.Bewley J.D. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bewley J.D., Bradford K.J., Hilhorst H.W.M., Nonogaki H. Seeds: Physiology of Development, Germination and Dormancy. 3rd ed. Springer; New York, NY, USA: 2013. [Google Scholar]
- 119.Borisjuk L., Rolletschek H., Radchuk R., Weschke W., Wobus U., Weber H. Seed Development and Differentiation: A Role for Metabolic Regulation. Plant Biol. 2004;6:375–386. doi: 10.1055/s-2004-817908. [DOI] [PubMed] [Google Scholar]
- 120.Rodríguez J.L., De Diego J.G., Rodríguez F.D., Cervantes E. Mitochondrial structures during seed germination and early seedling development in Arabidopsis thaliana. Biologia. 2015;70:1019–1025. doi: 10.1515/biolog-2015-0130. [DOI] [Google Scholar]
- 121.Law S.R., Narsai R., Whelan J. Mitochondrial biogenesis in plants during seed germination. Mitochondrion. 2014;19:214–221. doi: 10.1016/j.mito.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 122.Paszkiewicz G., Gualberto J.M., Benamar A., Macherel D., Logan D.C. Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell. 2017;29:109–128. doi: 10.1105/tpc.16.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ratajczak E., Małecka A., Ciereszko I., Staszak A.M. Mitochondria are important determinants of the aging of seeds. Int. J. Mol. Sci. 2019;20:1568. doi: 10.3390/ijms20071568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167245, accessed on 22 March 2021). Supplementary Data: RNA-Seq and methylome data are available at GSE167245 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167245, accessed on 22 March 2021).