Abstract
Stromal stiffening accompanies malignancy, compromises treatment, and promotes tumor aggression. Clarifying the molecular nature and the factors that regulate stromal stiffening in tumors should identify biomarkers to stratify patients for therapy and interventions to improve outcome. We profiled lysyl hydroxylase- and lysyl oxidase-mediated collagen crosslinks and quantified the greatest abundance of total and complex collagen crosslinks in aggressive human breast cancer subtypes with the stiffest stroma. These tissues harbor the highest number of tumor-associated macrophages (TAM), whose therapeutic ablation in experimental models reduced metastasis, and decreased collagen crosslinks and stromal stiffening. Epithelial-targeted expression of the crosslinking enzyme, lysyl oxidase, had no impact on collagen crosslinking in PyMT mammary tumors, whereas stromal cell targeting did. Stromal cells in microdissected human tumors expressed the highest level of collagen crosslinking enzymes. Immunohistochemical analysis of a cohort of breast cancer patient biopsies revealed that stromal expression of lysyl hydroxylase two, an enzyme that induces hydroxylysine aldehyde-derived collagen crosslinks and stromal stiffening, correlated significantly with disease specific mortality. The findings link tissue inflammation, stromal cell-mediated collagen crosslinking and stiffening to tumor aggression and identify lysyl hydroxylase two as a stromal biomarker.
Introduction
Extracellular matrix (ECM) accumulation accompanies the formation of solid tumors1-3. The tumor ECM is composed of interstitial collagen that is progressively reorganized and stiffened2,4. Tumor fibrosis compromises treatment and associates with poor patient prognosis5-8. Less differentiated tumors are more fibrotic, and tumor fibrosis predicts poor patient survival1,4,9. Patients with pancreatic ductal adenocarcinomas surrounded by stiff, thick collagens have a shorter survival, and invasive breast carcinomas with the stiffest stroma are the most aggressive1,2. Studies in culture and in vivo have provided evidence for a causal relationship between collagen organization, stromal stiffness and tumor progression10-14. These data underscore the need to clarify the molecular nature of tumor-associated collagens and stromal stiffness to identify biomarkers and anti-cancer therapeutics1,2,15,16.
Interstitial type I fibrillar collagen is the ECM component that regulates the tensile strength of tissues17. Collagen tensile strength is regulated by two families of enzymes: the lysyl oxidases (LOX), which regulate fibrillogenesis of newly synthesized collagen molecules through intermolecular covalent crosslinking, and the lysyl hydroxylases (LH; gene name procollagen-lysine, 2-oxoglutarate 5-dioxygenase or PLOD), which catalyze a posttranslational modification of lysine residues that specify the profile of crosslinking reaction products18-20. Human tumors express high levels of LOX and LH21. Tumor grade and patient survival associate with tissue LOX and PLOD2 mRNA20,22,23. Inhibition of LOX in MMTV-Her2/Neu mice or genetic reduction of PLOD2 in injected lung tumor cells reduce fibrosis, stromal stiffening and collagen crosslinking and decrease tumor incidence and aggression12,22. Elevating LOX or LH2-mediated collagen crosslinking stiffens the stroma and promotes malignancy and tumor aggression in tumor xenografts, implying targeting collagen crosslinking enzymes has clinical merit12,22. Nevertheless, given caveats with clinical trials targeting ECM modifiers including suboptimal activity of inhibitory treatments and off-target effects, strategies that interfere with the induction and activation of these enzymes offer attractive alternatives24.
Fibrotic tissues are inflamed, and inflammation promotes fibrosis25,26,14,26-29. Tumor inflammation promotes cancer aggression, and preventing inflammation reduces metastasis and improves anti-tumor treatment2,25,30-33. Whether inflammation promotes these phenotypes by inducing stromal stiffening remains unclear.
Results
xAAA profiling identifies collagen crosslinks and stromal stiffness associated with breast tumor aggression
To explore the role of collagen crosslinking in malignancy we developed a crosslinked amino acid analysis (xAAA) method that enables the characterization of specific collagen crosslinks in tissues across a range of collagen levels. We utilized solid phase extraction (SPE) enrichment followed by high pH amide hydrophilic chromatography (HILIC) coupled to a benchtop orbitrap (QExactive) mass spectrometer. The validated method detected all known LOX-generated crosslinks including divalent (lysinonorleucine, dihydroxy lysinonorleucine), trivalent (pyridinoline and deoxy-pyridinoline) and tetravalent (desmosine and isodesmosine) crosslinked amino acids linearly over four orders of magnitude, with calculated limits of quantification (LLOQ) in the femtomolar range (Extended Data Figure 1; Supplementary Table 1)34. The technique revealed a positive correlation between collagen crosslinking and abundance in clinical specimens with very low to very high collagen concentrations (Extended Data Figure 2). The method identified a subset of LH2-dependent hydroxylysine aldehyde (Hylald)-derived collagen crosslinks (HLCCs) crucial for the mechanical strength of tissue17,35.
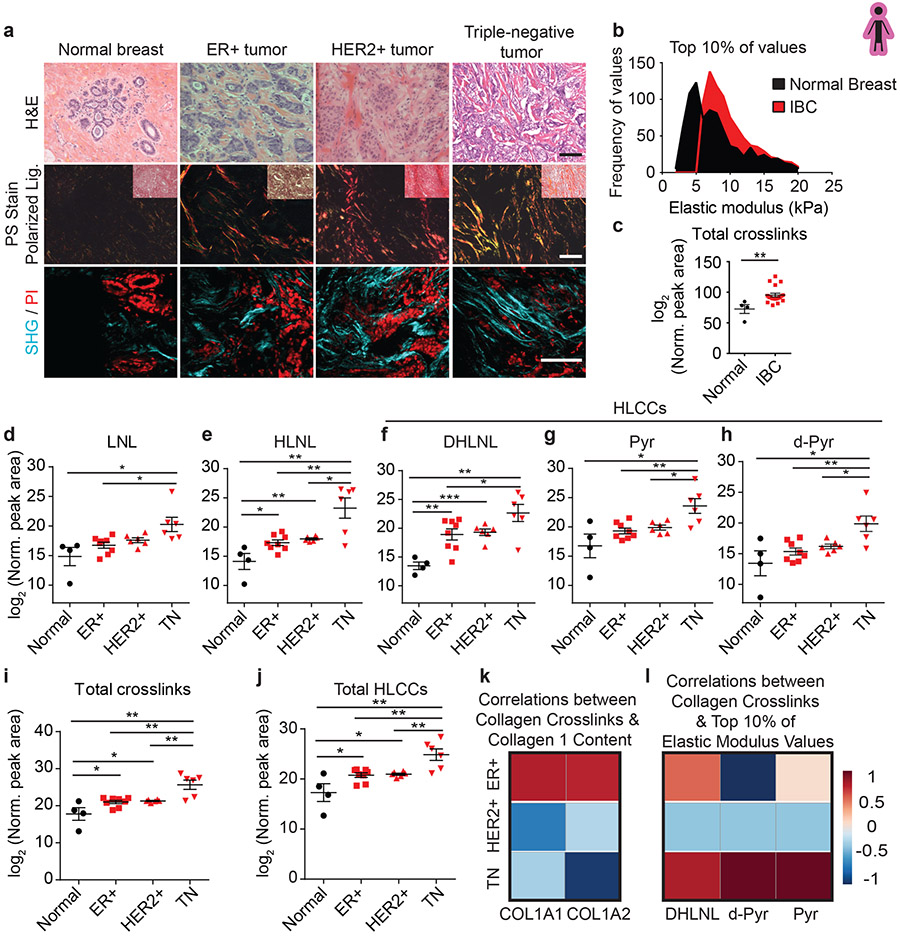
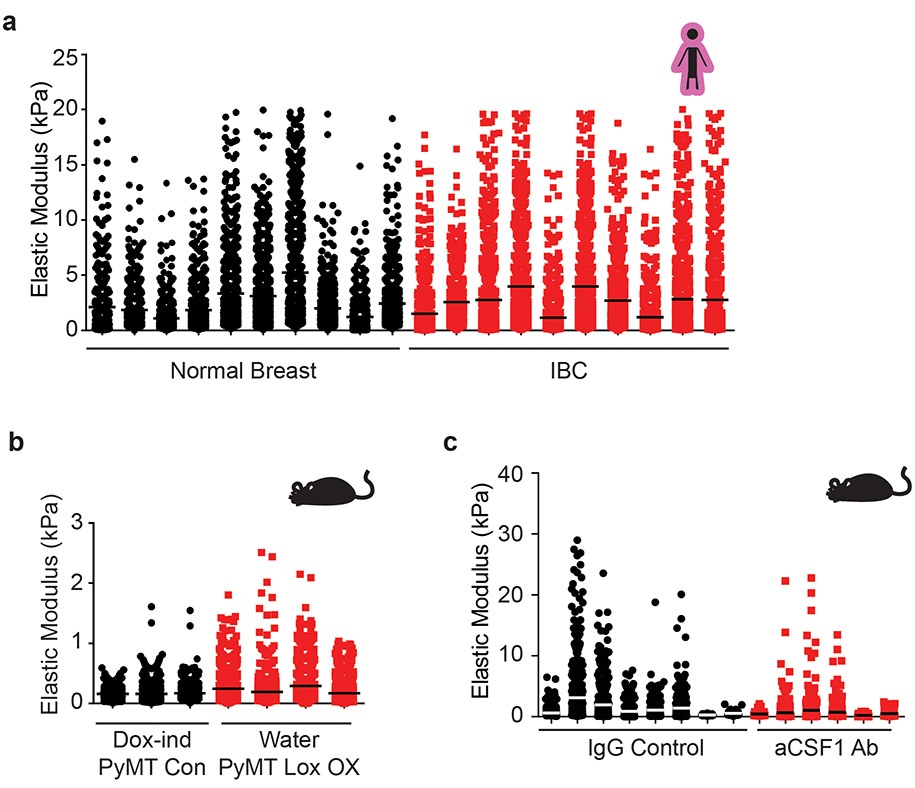
We obtained biospecimens of normal human breast tissue (N=10; age 22-58) and human tumor biopsies representing early stage (stage 1-2) invasive breast cancers (IBC) excised from mastectomy specimens. Molecular subtyping subdivides human breast tumors by estrogen receptor (ER+) and epidermal growth factor receptor two (HER2+) status and ER/PR/HER2-negative (triple negative; TN). Accordingly, we chose human breast tumor biopsies that represented ER+ (n = 8; age 42-71); HER2+ (n = 6; age 40-76) and TN (n = 6; age 50-71). H&E stained tissue confirmed the presence of normal glandular structures in the normal controls and invasive breast cancer in the tumor specimens (Figure 1a). Polarized light imaging of Picrosirius-stained (PS) tissue revealed that the normal breast stroma had little fibrillar collagen, whereas stromal tissue in all patients with IBCs contained abundant fibrillar collagen (Figure 1a) that second harmonic generation (SHG) imaging indicated was thicker and more linearized (Figure 1a). Polarized light microscopy and two-photon imaging revealed that the level of fibrosis in the tissue was higher in HER2+ compared to the ER+ breast tumors, and increased further in the TN tumors, consistent with reports that TN tumors contain abundant aligned collagen fibers (Figure 1a)2. Atomic force microscopy (AFM) microindentation revealed an increase in the elastic modulus of the stroma at the invasive front of the IBC tissues (Figure 1b, Extended Data Figure 8a)2,4,12. xAAA analysis revealed an increase in total collagen crosslinking normalized to collagen content in all IBCs (Figure 1c, Extended Data Figure 3). The findings are consistent with an association between collagen crosslinks, tissue fibrosis and stromal stiffness12. When we subdivided the IBC collagen crosslinking analysis into breast tumor subtype, the most significant increase in total collagen crosslinks was calculated to be in the TN breast tumors, which also have the highest elastic modulus of the stroma at the tumor invasive front (Figure 1i). Molecular characterization of the isolated collagen revealed that the TN tumors had a distinctive crosslink profile with a preferential enrichment of DHLNL, Pyr, and d-Pyr, all members of HLCC (Figure 1d-j). Quantification of total tissue collagen 1A1 or 1A2 showed that collagen I synthesis and crosslinking are not similarly regulated across molecular subtypes. Only in ER+ tumors did the higher total number of collagen crosslinks correlate positively with collagen levels (Figure 1k). Subtype analysis revealed that the strongest positive correlation between the level of HLCCs and tumor-associated ECM stiffness occurred in the TN subtype (Figure 1l). These findings highlight the characteristic differences in fibrosis among molecular breast tumor subtypes and suggest a role for hydroxylysine aldehyde (Hylald)-derived collagen crosslinking and its regulators in breast cancer aggression.
Figure 1: Hydroxylysine collagen crosslink abundance correlates with human breast cancer aggression.
(a) Representative images of normal breast tissue (n = 4) and invasive tumors diagnosed as ER+ tumor (n = 8), HER2+ tumor (n = 6), and TN tumor (n = 6). Brightfield images of human breast tissue stained with hematoxylin and eosin (H&E). Polarized light images of picrosirius red (PS) stained human breast tissue with an inset brightfield image. Two photon second harmonic generation (SHG) images of human breast tissue revealing collagen organization (turquoise) and propidium iodide (PI; red) stained nuclei. Scale bar, 100 μm. (b) The distribution of the top 10% of elastic modulus values of normal breast tissue (n = 10) and invasive breast carcinoma (IBC; n = 10) measured by AFM microindentation. Statistical analysis was performing using two-tailed Mann-Whitney U test (****p < 0.0001). (c) Scatter plot with mean ± SEM of total collagen crosslinks in normal breast tissue (n = 4) and in IBC tissues (n = 19). Statistical analysis was performed using two-tailed Mann-Whitney U test (**p = 0.0016). (d-j) Scatter plots showing individual and mean values ± SEM of the levels of each crosslink, total abundance, and total HLCCs measured in normal breast tissues (n = 4), and in ER+ (n = 8), HER2+ (n = 6) and TN (n = 6) breast tumors. Statistical analysis of crosslinks was performed using one-way ANOVA test for overall analysis and two-tailed unpaired t-test without adjustment for multiple comparisons was used for individual comparisons. (d) Overall **p = 0.0051, Normal-TN *p = 0.0228, ER+-TN *p = 0.0116, HER2+-TN p = 0.0612. (e) Overall **p = 0.0039, Normal-ER+ *p = 0.0187, Normal-HER2+ **p = 0.0083, Normal-TN **p = 0.0054, ER+-TN **p = 0.0027, HER2+-TN *p = 0.0124. (f) Overall ***p = 0.0003, Normal-ER+ **p = 0.0041, Normal-HER2+ ***p = 0.0002, Normal-TN **p = 0.0014, ER+-TN *p = 0.0475, HER2+-TN p = 0.06. (g) Overall **p = 0.002, Normal-TN *p = 0.016, ER+-TN **p = 0.0043, HER2+-TN *p = 0.0191. (h) Overall **p = 0.003, Normal-TN *p = 0.0212, ER+-TN **p = 0.0019, HER2+-TN *p = 0.0186. (i) Overall ***p = 0.0001, Normal-ER+ *p = 0.0329, Normal-HER2+ *p = 0.0315, Normal-TN **p = 0.0049, ER+-TN **p = 0.0019), HER2+-TN **p = 0.0057. (j) Overall ***p = 0.0002, Normal-ER+ *p = 0.028, Normal-HER2+ **p = 0.0083, Normal-TN **p = 0.0053, ER+-TN **p = 0.0032), HER2+-TN **p = 0.0072. (k,l) Heat maps of Spearman correlation coefficients indicating correlations between levels of total collagen crosslinks and collagen I content (k) and between levels of each HLCC and the top 10% of elastic modulus measurements (l) stratified by tumor subtype. All n values represent biologically independent human tissue specimens.
Collagen crosslinking correlates with stromal LOX and LH2
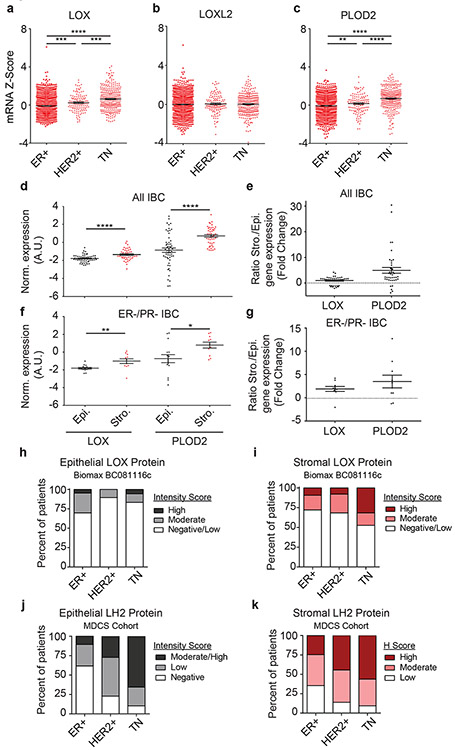
We examined associations between the levels of enzymes implicated in regulating collagen crosslinking and breast tumor subtype. We analyzed publicly available human breast cancer gene expression array data (n = 1904) for the genes coding these enzymes and evaluated their correlation to breast cancer subtype. Bioinformatics analyses revealed an increase in the collagen crosslinking enzyme LOX, but not lysyl oxidase like two (LOXL2), in the HER2 and TN tumor subtypes (Figure 2a-b). LOX and PLOD2, the major regulators of HLCC accumulation, were particularly high in TN tumors (Figure 2a,c).
Figure 2: LOX and PLOD2 are enriched in TNBC and predominantly expressed by stromal cells.
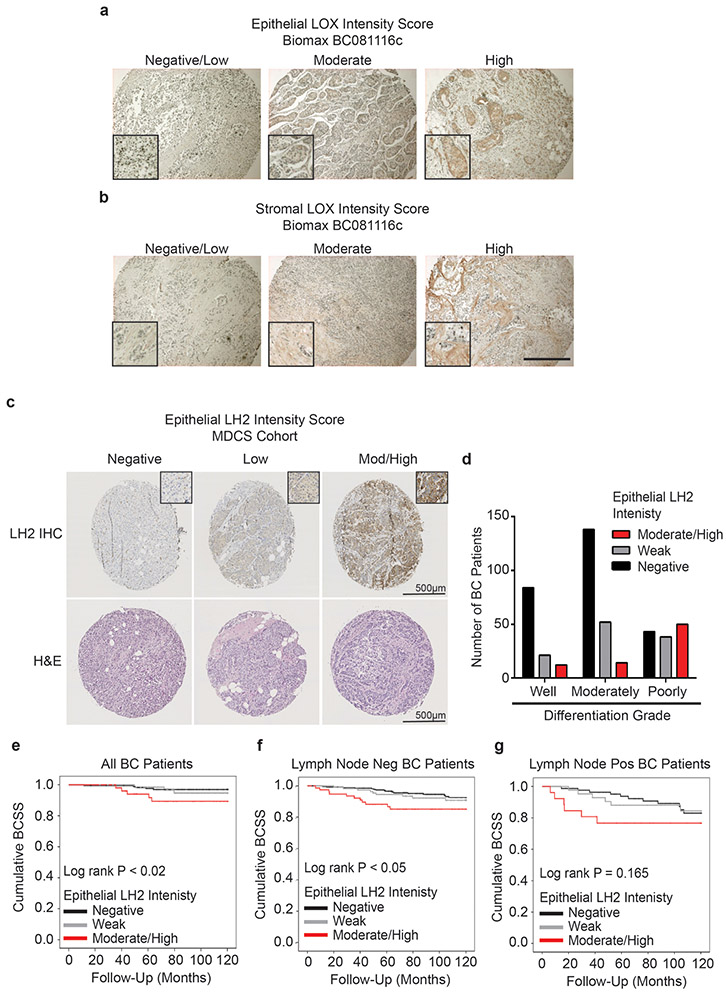
(a-c) Gene expression analysis of LOX (a), LOXL2 (b), and PLOD2 (c) stratified by ER+ (n = 1355), HER2+ (n = 127), and triple negative (TN; n = 299) subtypes. Gene expression is plotted as a scatter plot of mRNA z scores with the mean ± SEM. Statistical analysis was performed using one-way ANOVA for overall analysis and two-tailed unpaired t-test without adjustment for multiple comparisons was used for individual comparisons. (a) Overall ****p < 0.0001, ER+-HER2+ ***p = 0.0001, ER+-TN ****p < 0.0001, HER2+-TN ***p = 0.0008. (b) Overall p = 0.7984, ER+-HER2+ p = 0.5435, ER+-TN p = 0.7198, HER2+-TN p = 0.7505. (c) Overall ****p < 0.0001, ER+-HER2+ **p = 0.01, ER+-TN ****p < 0.0001, HER2+-TN ****p < 0.0001. (d) Scatter plot of individual and mean values ± SEM comparing LOX (n = 47) and PLOD2 (n = 57) gene expression in microdissected epithelial and stromal compartments of human invasive breast carcinomas. Statistical analysis was performed using two-tailed Mann-Whitney U test ****p < 0.0001. (e) Scatter plot of individual and mean values ± SEM of LOX and PLOD2 gene expression fold change from (d) in stromal cells relative to epithelial cells. (f-g) Restriction of the stromal/epithelial gene expression analysis in (d) and (e) to estrogen receptor (ER) negative and progesterone receptor (PR) negative samples. (LOX n = 11, PLOD2 n = 15). Statistical analysis was performed using two-tailed Mann-Whitney U test **p = 0.002, *p = 0.0204. (h) Bar plot showing distribution of epithelial LOX staining intensity among human patient tumors stratified by molecular subtype. (i) Bar plot showing distribution of stromal LOX staining intensity among human patient tumors stratified by molecular subtype. LOX scoring of Biomax BC081116c TMA was performed by a researcher (ER+ n = 43, HER2+ n = 38, TN n = 18 epithelial, 19 stromal). (j) Bar plot showing distribution of epithelial LH2 staining intensity among human patient tumors stratified by molecular subtype. (k) Bar plot showing distribution of stromal LH2 H Score among human patient tumors stratified by molecular subtype. LH2 scoring of patient samples from the MDCS cohort was performed by a pathologist. See Supplementary Tables 2 and 3 for patient information. All n values represent biologically independent human tissue specimens.
To identify the tissue origins of LOX and LH2 in IBCs, we used laser capture microdissection to isolate regions of tumor epithelium and stroma36,37. Gene expression analysis of the stromal and epithelial compartments revealed that the stromal cells in the IBCs expressed more LOX and PLOD2 than adjacent epithelium. The relationship was more evident in the breast tissue from women with ER−/PR− IBCs (Figure 2d-g). LOX IHC performed on Biomax BC081116c tissue microarray (TMA) revealed higher protein levels in the IBC stroma compared to the epithelium (Figure 2h-i, Extended Data Figure 10a-b). Stromal LOX intensity was the most elevated in TN tumor samples as compared to the IBC epithelium (Figure 2i). LH2 IHC performed on samples from the Malmö Diet and Cancer Study (MDCS) patient cohort revealed higher LH2 levels in breast tumor stroma compared to the epithelium with the highest levels quantified in the stroma of HER2+ and TN tumors (Figure 2j-k).
The findings suggest LOX and LH2 enzymes likely generate the increased level and complexity of collagen crosslinking in aggressive human breast tumors. The data implicate stromal cell LOX and LH2 as likely contributors to HLCC enrichment and IBC aggression.
Stromal crosslinking enzymes regulate fibrosis and collagen crosslinking
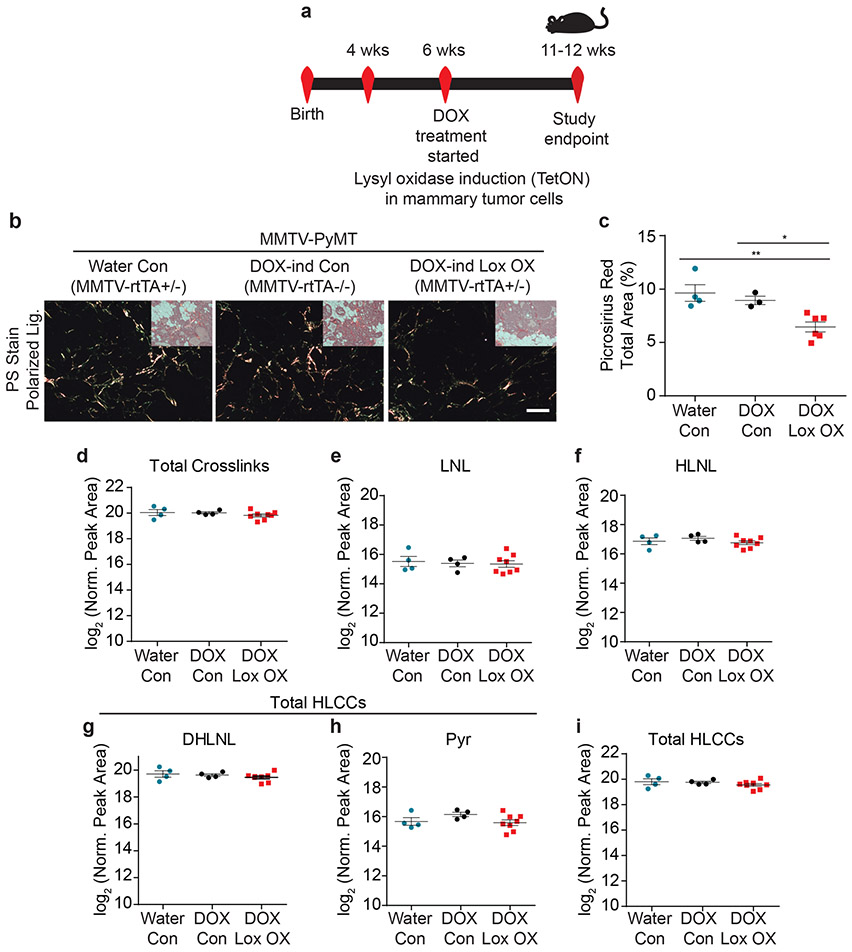
We next created genetically engineered mouse models (GEMMs) in which we targeted fibroblast versus epithelial-specific expression of mouse LOX. We first generated GEMMs in which we targeted luminal epithelial-specific expression of mouse LOX using the MMTV-rtTA promoter (Epithelial LOX overexpression [OX]) (Figure 3a, Extended Data Figure 4a-d). We then validated the LOX OX mouse model confirming ectopic LOX expression and elevated cleaved LOX protein in the mammary tumor epithelium following LOX induction (Extended Data Figure 4e). Thereafter, we crossed the LOX OX mice into the MMTV-PyMT, a spontaneous mammary tumor model, to enhance LOX expression in the mammary tumor epithelium (PyMT epithelial LOX OX). Once tumors formed, we assayed the mammary tissue for tissue fibrosis compared to the mammary glands from age-matched PyMT control mice (PyMT). We did not detect any increase in either the level or extent of mammary tumor fibrosis in the PyMT epithelial LOX OX GEMM including differences in the quantity of type I fibrillar collagen, or interstitial collagen organization (Figure 3b-c). xAAA crosslinking analysis revealed that the levels of collagen crosslinks between the PyMT control and PyMT epithelial LOX OX were indistinguishable (Figure 3d-i). However, we detected an increase in fibrillar collagen, collagen crosslinks and stromal stiffness in the PyMT mammary tumors compared to age-matched wildtype FVB mammary glands (Figure 4c,f-k). Thus, tumor epithelial LOX may not be the primary driver of collagen crosslinking, stromal stiffening and tissue fibrosis in spontaneous mammary tumors.
Figure 3: Epithelial-derived collagen crosslinking enzymes fail to induce collagen crosslinking.
(a) Schematic depicting the experimental strategy used to induce epithelial Lox overexpression. (b) Polarized light images with brightfield inset of picrosirius red stained murine mammary tissues. Scale bar is 100 μm. (c) Scatter plot showing mean ± SEM of percent area of picrosirius red staining per field of view in PyMT controls (Water n = 4, DOX n = 3) and PyMT epithelial Lox overexpression (n = 6). Statistical analysis was performed using Kruskal-Wallis one-way ANOVA *p = 0.010 and two-tailed Mann-Whitney U-test without adjustment for multiple comparisons for individual comparisons **p = 0.0095, *p = 0.0238. (d) Scatter plot showing individual and mean values ± SEM of total tissue collagen crosslinks in PyMT controls (Water n = 4, DOX n = 4) and PyMT epithelial Lox overexpression (n = 8). (e-h) Scatter plots showing individual and mean values ± SEM for each LCC and HLCC collagen crosslink measured in PyMT control (Water n = 4, DOX n = 4) and PyMT epithelial Lox overexpression (n = 8) tumor tissue. (i) Scatter plot showing individual and mean values ± SEM of total HLCCs. Statistical analyses for crosslinking data were performed using one-way ANOVA for overall comparison and unpaired t-test without adjustment for multiple comparisons for individual comparisons. (d) Overall p = 0.8077. (e) Overall p = 0.899. (f) Overall p = 0.4286. (g) Overall p = 0.4586. (h) Overall p = 0.2128. (i) Overall p = 0.4325. All n values represent biologically independent mouse tissue specimens.
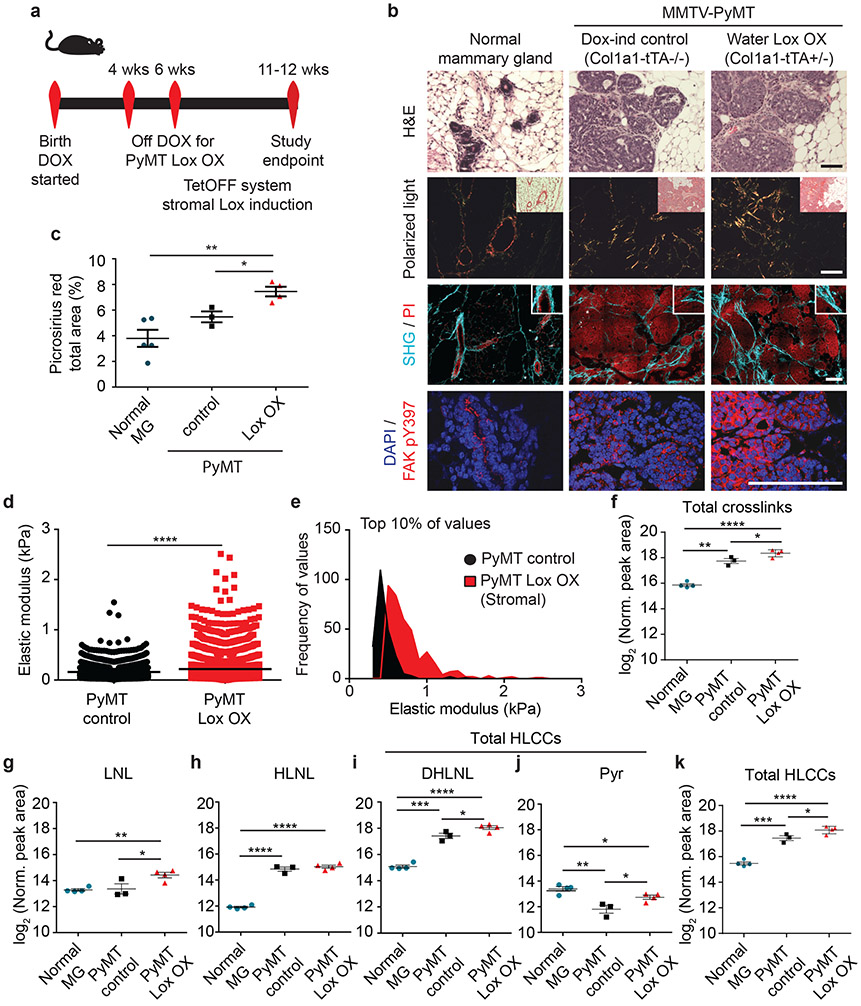
Figure 4: Stromal-derived LOX regulates collagen crosslinking and stiffening.
(a) Schematic depicting the experimental strategy. (b) Representative images of normal murine mammary gland (n = 4) and PyMT Control (n = 3) and stromal Lox overexpressing (PyMT Lox OX; n = 4 on) tumors with brightfield images of H&E stain, polarized light images with brightfield inset of picrosirius red (PS) stain, second harmonic generation (SHG) images (turquoise) and propidium iodide (PI; red) stained nuclei, and confocal images of pY397FAK stain (red) and DAPI (blue; nuclei). Scale bars are 100 μm. (c) Scatter plot of individual values with mean ± SEM picrosirius red staining of normal mammary gland (n = 5), PyMT Control (n = 3), and PyMT Lox OX (n = 4) by percent area per field of view. Statistical analysis was performed using ordinary one-way ANOVA **p = 0.0034 for overall relationship and two-tailed unpaired t-test without adjustment for multiple comparisons for individual groups **p = 0.003 *p = 0.018. (d) Scatter plot showing individual values and mean of the top 10% of elastic modulus measurements performed by AFM microindentation on the mammary stroma from PyMT Control (n = 3) compared to PyMT Lox OX (n = 4). Statistical analyses were performed using two-tailed Mann-Whitney U test (****p < 0.0001). (e) Histogram showing the distribution of the top 10% of elastic modulus measurements by AFM in PyMT Control and Lox OX tumors. Statistical analysis was performed using two-tailed Mann-Whitney U test (****p < 0.0001). (f) Scatter plots showing individual and mean values ± SEM of total collagen crosslink abundance in the normal mammary gland (n = 4) as compared to glands with doxycycline-treated PyMT control tumors (n = 3) and PyMT Lox OX tumors (n = 4). (g-j) Scatter plots showing individual and mean values ± SEM of LCC and HLCC crosslinks quantified in normal mammary gland (n = 4), PyMT Control tumors (n = 3) and PyMT Lox OX tumors (n = 4). (k) Scatter plot showing individual and mean values ± SEM of total HLCCs in normal mammary gland (n = 4), PyMT Control tumors (n = 3) and PyMT Lox OX tumors (n = 4). Statistical analyses for crosslinking data were performed using one-way ANOVA for overall comparison and two-tailed unpaired t-test without adjustment for multiple comparisons for individual comparisons. (f) Overall ****p < 0.0001, Normal-Control **p = 0.0019, Normal-Lox OX ****p < 0.0001, Control-Lox OX *p = 0.0236. (g) Overall *p < 0.0125, Normal-Lox OX **p = 0.0027, Control-Lox OX *p = 0.0494. (h) Overall ****p < 0.0001, Normal-Control ****p < 0.0019, Normal-Lox OX ****p < 0.0001. (i) Overall ****p < 0.0001, Normal-Control ***p = 0.0001, Normal-Lox OX ****p < 0.0001, Control-Lox OX *p = 0.0473. (j) Overall **p = 0.0025, Normal-Control **p = 0.005, Normal-Lox OX *p = 0.0356, Control-Lox OX *p = 0.0318. (k) Overall ****p < 0.0001, Normal-Control ***p = 0.0005, Normal-Lox OX ****p < 0.0001, Control-Lox OX *p = 0.0251. All n values represent biologically independent mouse tissue specimens.
Stromal fibroblast LOX is implicated in tumor progression and aggression12,14,38. Therefore, we generated mouse cohorts of PyMT GEMMs in which we restricted ectopic LOX expression to the stromal population using the Col1a1-tTA promoter (MMTV-PyMT+/−; Col1a1-tTA+/−;TetO_mLox+/−; herein denoted PyMT LOX OX) (Figure 4a, Extended Data Figure 5b). Induction of LOX in the stromal cells markedly enhanced the amount of fibrillar collagen in mammary glands from MMTV-PyMT LOX OX mice, as revealed by polarized images of Picrosirius Red stained tissue (Figure 4b-c). We also detected more and thicker linearized interstitial collagen in these glands by second harmonic generation imaging (Figure 4b). Immunostaining revealed more phosphorylated tyrosine 397 focal adhesion kinase protein (pY397FAK) in mammary epithelium of the glands in which stromal LOX was elevated (Figure 4b), reflecting the increase in elasticity measured in the stroma using AFM indentation12 (Figure 4d-e, Extended Data Figure 8b). We measured higher levels of total collagen crosslinks in the PyMT LOX OX mice as compared to the levels quantified in the MMTV-PyMT control glands (Figure 4f). The most significant increases quantified in the PyMT LOX OX glands were dihydroxy lysinonorleucine (DHLNL) and pyridinoline (Pyr) crosslinks (Figure 4g-k), which are the crosslinks generated through the HLCC pathway that promote mechanical stability and strength in skeletal tissue35,39. By contrast, we did not detect any increase in DHLNL or Pyr in the mammary glands in which ectopic LOX expression was elevated in the mammary epithelium using the MMTV promoter (Figure 3g-h). The findings imply that stromal cells are the primary regulator of interstitial collagen crosslinking, stromal stiffening and tissue fibrosis in spontaneous mammary tumors.
Tissue inflammation regulates fibrosis, collagen crosslinking and stromal stiffening
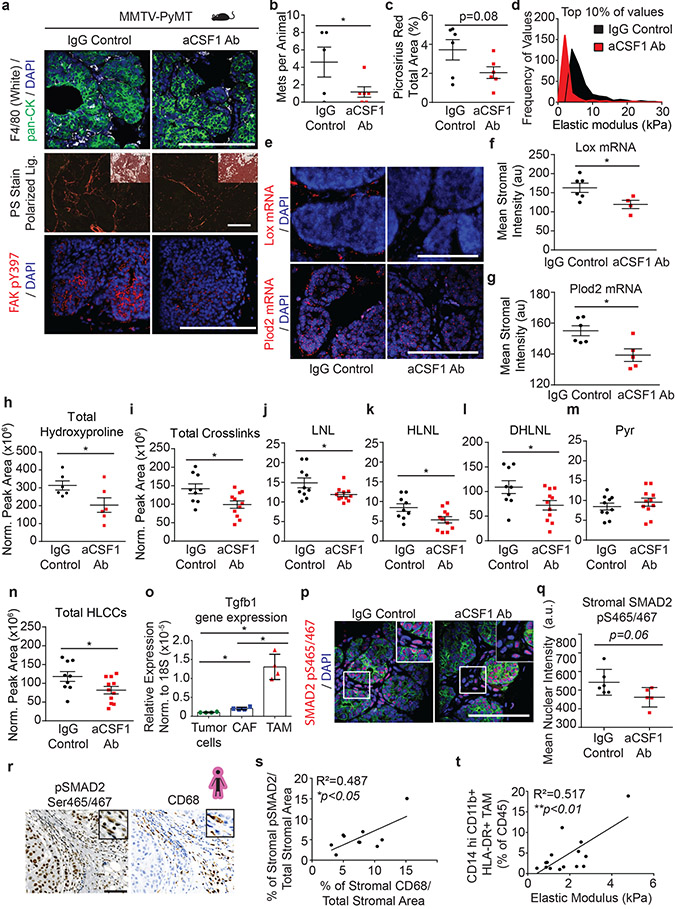
Human breast tumors are inflamed and those with the stiffest stroma harbor the highest density of macrophages2,33. Decreasing tumor-associated macrophages (TAMs) reduces lung metastasis in PyMT mice, but only when TAM depletion occurs prior to tumor invasion, when we observed fibrosis and collagen crosslinking develop (Extended Data Figure 6a-f)31,32,40,41. This raises the possibility that TAMs promote tumor metastasis by stimulating stromal-mediated collagen crosslinking and stiffening (Figures 3 & 4).
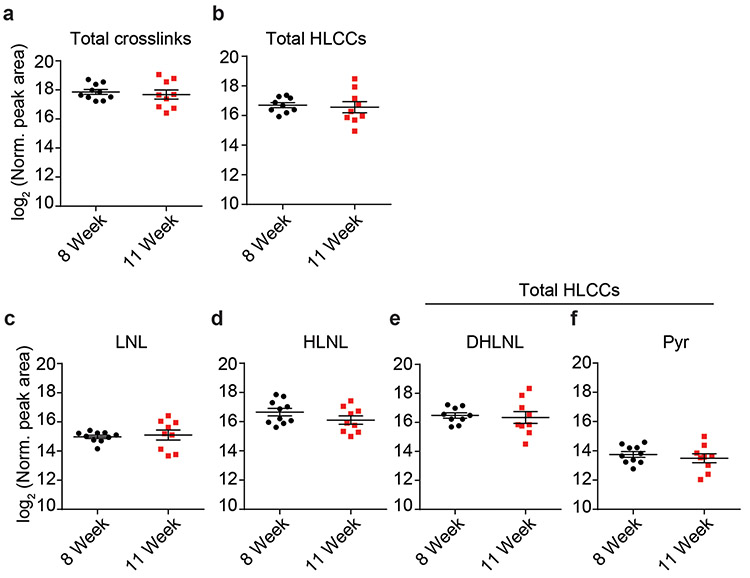
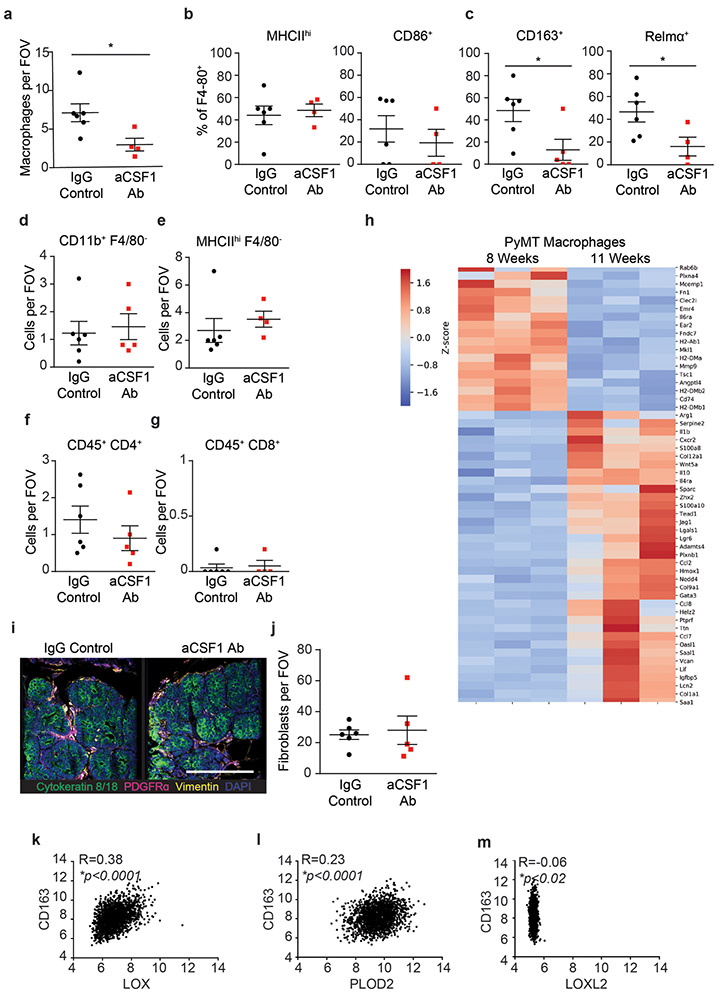
To test this, we treated PyMT mice with anti-CSF-1 antibody or a non-specific IgG control antibody prior to the onset of ductal hyperplasia, tumor cell invasion and fibrosis (four weeks of age)42. Mouse cohorts were sacrificed at eight and eleven weeks of age. F4/80 immunostaining showed TAMs were reduced in the mammary glands of both the eight-week- and eleven-week-old treated mice, (Figure 5a, Extended Data Figure 7a). The eleven-week-old treated mice demonstrated reduced lung metastasis (Figure 5b)31. Polarized images of PS stained mammary tissue revealed lower levels of total fibrillar collagen in the stroma of the eight-week-old anti-CSF1 antibody treated group, despite similar fibroblast density in both groups (Figure 5a,c; Extended Data Figure 7i-j). AFM microindentation of the anti-CSF1 antibody treated group demonstrated that the stroma from both eight-week-old (Figure 5d, Extended Data Figure 8c), and eleven-week-old (not shown) mammary glands was softer and the epithelium had lower integrin mechanosignaling, when compared to the control IgG antibody treated group, as revealed by less intense staining for pY397FAK (Figure 5a). In situ hybridization demonstrated reduced levels of Lox mRNA and Plod2 mRNA that was restricted to stromal cells in the eight-week-old, anti-CSF1 treated mice (Figure 5e-g)14. The decrease in stromal Lox and Plod2 with TAM depletion was accompanied by a reduction in collagen content and in total collagen crosslinks and HLCCs as well as divalent LOX-mediated collagen crosslinks (Figure 5h-n). We failed to detect different levels of trivalent Pyr crosslinks in 8-week-old control versus TAM-ablated mouse tumor tissues, likely because trivalent crosslinking is a slow process and the development of these crosslinks takes time to mature. The findings suggest a TAM subset exists that promotes metastasis by stimulating stromal cell expression of LOX and LH2 to induce collagen crosslinking, stromal stiffening and fibrosis.
Figure 5: Tumor infiltrating macrophages secrete TGFb to activate stromal-mediated collagen crosslinking.
(a) Representative images of PyMT tumors from 8-week-old mice treated with anti-CSF1 blocking antibody or IgG1 control. IgG1 treated (n = 6) and anti-CSF1 treated (n = 5) PyMT tumors stained for pan-cytokeratin (green), F4/80 (white) and DAPI (blue). Polarized light images with brightfield inset of IgG1 treated (n = 6) and anti-CSF1 treated (n = 6) PyMT tumors stained with picrosirius red (PS). IgG1 treated (n = 6) and anti-CSF1 treated (n = 5) PyMT tumors stained pY397FAK (red) and DAPI (blue). Scale bars are 100 μm. (b) Scatter plot with individual values and mean ± SEM of metastatic colonies in lung tissues from 11-week-old IgG1 treated (n = 5) and anti-CSF1 treated (n = 6) mice via PyMT IHC. Statistical analysis was performed using two-tailed unpaired t-test *p = 0.0363. (c) Scatter plot with individual values and mean ± SEM of PS staining by percent area per field of view in 8-week-old mice treated with anti-CSF1 blocking antibody (n = 6) or IgG1 control (n = 6). Statistical analysis was performed using two-tailed unpaired t-test p = 0.08. (d) Histogram of the top 10% of elastic modulus measurements by AFM microindentation in PyMT IgG1 treated (n = 7) and anti-CSF1 treated (n = 5) tumors. Statistical analysis was performed using two-tailed Mann-Whitney U test ****p < 0.0001. (e) Representative images of Lox and Plod2 mRNA in situ hybridization in IgG1 treated (n = 6) and anti-CSF1 treated (n = 5) PyMT tumors and DAPI (blue). Scale bars are all 100μm. (f-g) Scatter plot with individual values and mean ± SEM of stromal Lox (f) or stromal Plod2 (g) mRNA in situ hybridization mean stromal intensity in 8-week-old mice treated with anti-CSF1 (n = 5) or IgG1 control (n = 6). Statistical analyses were performed using two-tailed Mann-Whitney U test (f) *p = 0.0381 and (g) *p = 0.0303. (h) Scatter plot showing individual and mean values ± SEM of total hydroxyproline (collagen content) in tumors from IgG1 (n = 6) and anti-CSF1 (n = 6) treated mice. (i-n) Scatter plots showing individual and mean values ± SEM of the levels of total collagen crosslinks (i), LNL (j), HLNL (k), DHLNL (l), Pyr (m), and total HLCCS (n) in 8-week-old IgG1 treated (n = 9 i,k,l,n or 10 j,m) and anti-CSF1 (n = 11) treated PyMT tumors. Quantity of crosslinks per tissue was calculated normalizing crosslinks to dry tissue weight. Values were plotted as normalized peak areas as quantified from LC-MS data. Statistical analyses (h-n) were performed using two-tailed unpaired t-test (h) *p = 0.0447 (i) *p = 0.0174 (j) *p = 0.0359 (k) *p = 0.0235 (l) *p = 0.0311 (n) *p = 0.0366. (o) Scatter plot with individual values and mean ± SEM of Tgfb1 gene expression by RT-qPCR in tumor cells, cancer-associated fibroblasts, and macrophages sorted from PyMT tumors (n = 4). Statistical analysis was performed using Kruskal-Wallis one-way ANOVA for overall comparison and two-tailed Mann-Whitney U test for individual comparisons. Overall ***p = 0.0002, Tumor-CAF *p = 0.0286, Tumor-TAM *p = 0.0286, CAF-TAM *p = 0.0286. (p) Representative images of PyMT tumors from mice treated with IgG1 (n = 6) and anti-CSF1 (n = 5) stained for pan-cytokeratin (green), pS465/467SMAD2 (red), and DAPI (blue). Scale bar is 100 μm. (q) Scatter plot showing individual and mean values ± SEM of the mean nuclear intensity of stromal pS465/467SMAD2 of IgG1 treated (n = 6) and anti-CSF1 treated (n = 5) PyMT mice. Statistical analysis was performed using two-tailed unpaired t-test (p = 0.06). For (a-q) all n values represent biologically independent mouse tissue specimens. (r) Representative IHC images of serial human breast tumor sections stained for pS465/467SMAD2 and CD68 and counterstained with hematoxylin. Scale bar is 100 μm. (s) Scatter plot with linear regression correlation of stromal pS465/467SMAD2 IHC staining with stromal CD68 IHC staining in human breast tumors (n = 10, *p = 0.0248). (t) Scatter plot depicting the linear regression of CD14+ CD11b+ HLA-DR+ tumor-associated macrophage (as %CD45+ by flow cytometry) with mean elastic modulus as measured by AFM microindentation in human breast tumors (n = 15, **p = 0.0025). For (r-t) all n values represent biologically independent human tissue specimens.
Characterization of the TAMs infiltrating PyMT tumors from 8-week-old mice identified a population enriched for CD163 and Relmα, which secrete high quantities of TGFβ and have been implicated in tissue fibrosis (Extended Data Figure 7 b-c)43-45. TAM secreted TGFβ stimulates fibroblast transdifferentiation (cancer-associated fibroblasts; CAFs) that express collagen crosslinking enzymes including LOX14. PCR analysis of flow activated cell sorted (FACS) cells from PyMT tumors (11-week-old) showed that TAMs expressed by far the highest levels of TGFβ, as compared to the tumor epithelial cells and the CAFs (Figure 5o). While the stromal cells in the PyMT tumors from IgG control antibody-treated mice showed enhanced SMAD2 phosphorylation (pS465/467SMAD2), levels in the stromal cells in tumors from mice treated with anti-CSF1 antibody were low (Figure 5p-q). Consistently, analysis of a publicly available human breast tumor gene expression data set showed a positive correlation between the TAM marker, CD163, and LOX and PLOD2 but not LOXL2 (n = 1904) (Extended Data Figure 7k-m). Co-staining of resected human breast tumors with the macrophage/monocyte marker CD68 and pS465/467SMAD2, which reflects TGFβ response signaling, revealed a positive correlation between pS465/467SMAD2 and the infiltrating TAMs at the invasive front of human breast tumors (Figure 5r-s). FACS analysis established a correlation between the infiltrating TAMs, as demonstrated by CD14hiCD11b+ HLA-DR+ cell surface markers normalized to total CD45 infiltrating cells, and the elastic modulus of the invasive front of human invasive breast cancers (Figure 5t). The findings implicate factors such as TGFβ secreted by CD163 and Relmα positive infiltrating TAMs in the LOX/LH2-dependent collagen crosslinking, stromal stiffening, and fibrosis associated with aggressive breast tumors.
LH2 predicts poor prognosis in breast cancer patients
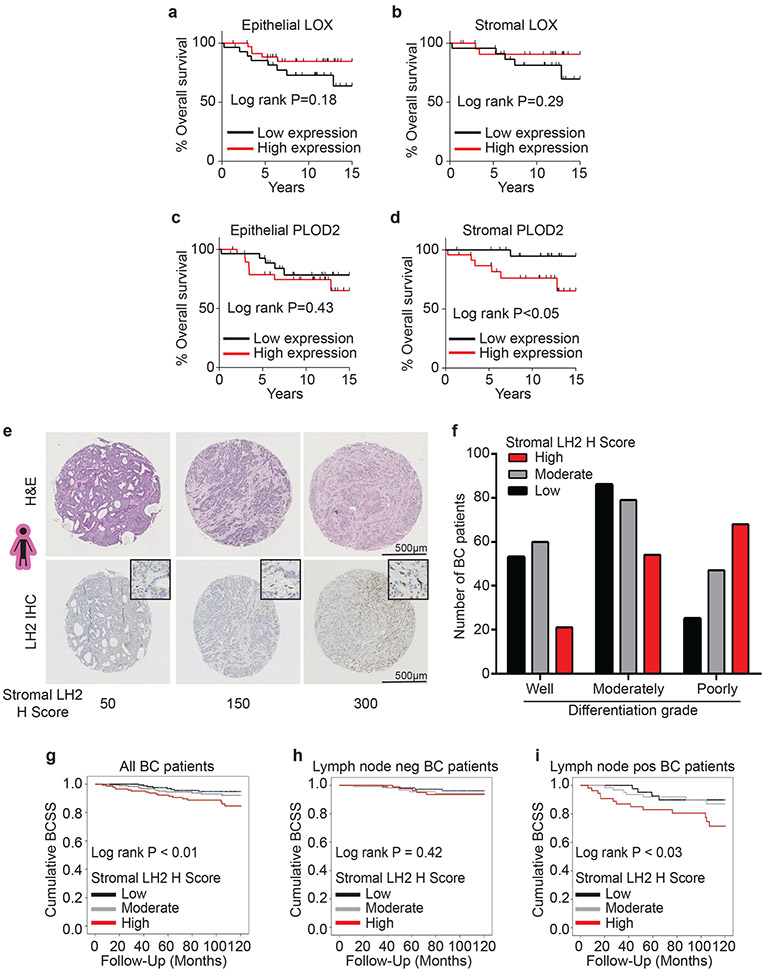
We next assessed the contribution of stromal LOX and PLOD2 expression to overall survival in breast cancer patients. Surprisingly, neither stromal cell nor epithelial LOX predicted overall patient survival at least in this cohort (Figure 6a-b). Instead, overexpression of stromal cell PLOD2, but not epithelial PLOD2, correlated with poor breast cancer patient prognosis (Figure 6c-d).
Figure 6: Stromal LH2 predicts poor patient outcomes.
(a-b) Kaplan-Meier plots showing overall survival for patients based on LOX expression in epithelial cells (low n = 28, high n = 36) (a) or stromal cells (low n = 23, high n = 24) (b). (c-d) Kaplan-Meier plots showing overall survival for patients based on PLOD2 expression in epithelial cells (low n = 28, high n = 29) (c) or stromal cells (low n = 23, high n = 24) (d). For (a-d) the median expression was defined as the cutoff for low and high expression. (e) Representative phase contrast images from tissue microarrays (TMAs) of human breast cancers. Sections were stained with H&E and lysyl hydroxylase two (LH2) via IHC. (f) Bar graphs showing clinical correlation between stromal LH2 H score and tumor grade (see Supplementary Table 3 for number of patients). LH2 IHC staining was assessed by a pathologist with the semi-quantitative stromal specific H-score from 0 to 300. The lowest tertile of LH2 H-scores was defined as H-scores between 0 and less or equal to 120, the moderate H-score to above 120 and equal or less than 230, and the highest stromal LH2 score as above 230. For tumor grade and LH2 H score, statistical analysis was performed using a linear-by-linear association (****p<0.0001). (g) Kaplan-Meier curves indicating cumulative breast cancer specific survival (BCSS) based on stromal LH2 H score assessed in breast cancer patients up to 10 years after diagnosis (LH2 low n = 175, moderate n = 188, high n = 146). (h) BCSS curves by stromal LH2 H score including only axillary lymph node negative patients (LH2 low n = 116, moderate n = 116, high n = 90). (i) BCSS curves by stromal LH2 H score including only axillary lymph node positive patients (LH2 low n = 44, moderate n =63, high n = 54). For Kaplan-Meier curves, statistical analyses were performed by LogRank test (g) **p = 0.008, (i) *p = 0.026. All n values represent biologically independent human tissue specimens.
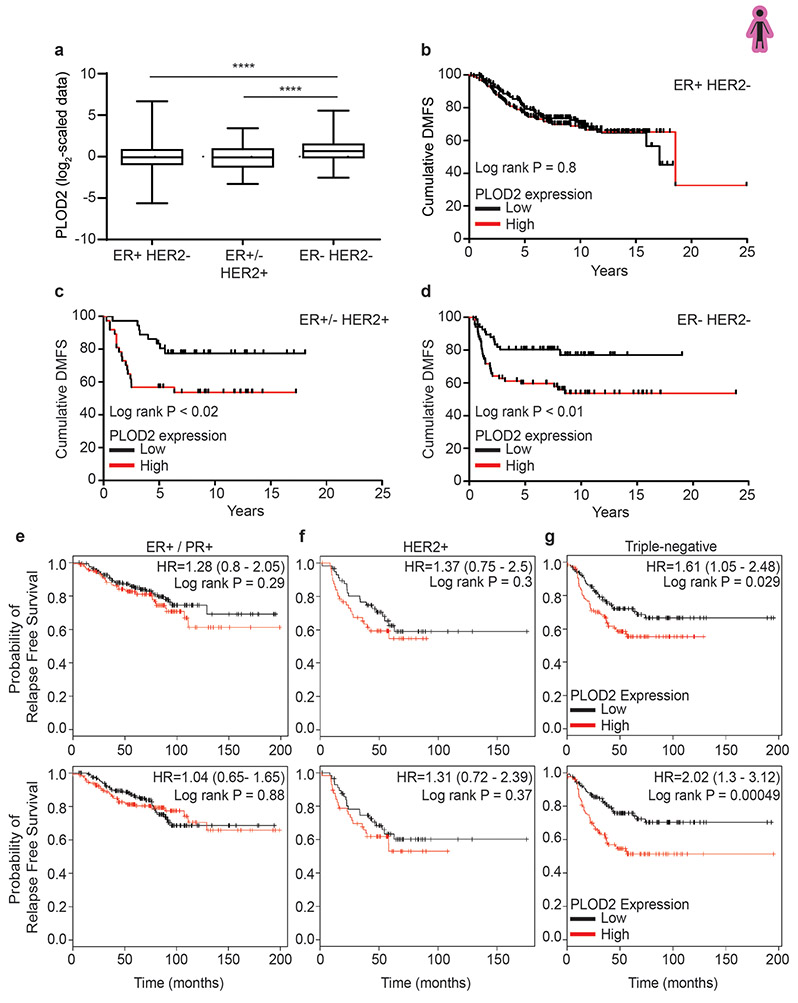
LH2 (the protein encoded by PLOD2) is a key enzyme that regulates the level of HLCC crosslinking of fibrillar collagen to increase the tensile properties of the stroma18,22,46. We quantified the highest level of HLCCs and the stiffest invasive front in the breast tissue from women with the more aggressive breast cancers (Figure 1c,j). When we analyzed the gene expression data from a published cohort of breast cancer patients we observed that ER−/HER2− (TN; n=133) breast cancers express the highest level of the PLOD2 gene transcript as compared to the levels expressed in HER2+ (n=73) and ER+/HER2− (luminal, n=314) breast tumors (Extended Data Figure 9a)47. High PLOD2 expression in breast cancer patients with HER2+ and TN tumors also predicted reduced distant metastasis-free survival (DMFS), as well as increased risk of relapse in TN tumors (Extended Data Figure 9b-g)47,48. These findings implicate PLOD2/LH2 in breast tumor aggression possibly because it induces HLCC in fibrillar collagens that contribute more significantly to stromal stiffening.
We next assessed the contribution of stromal versus neoplastic epithelial LH2 protein expression to human breast tumor aggression. We developed an LH2 immunostaining protocol (IHC), and then analyzed neoplastic epithelial and stromal expression of LH2 enzyme in tissue biopsies from a large cohort of histopathologically classified breast cancer patients (N=505 for stromal and N=468 for epithelial) with accompanying clinical information and follow up data (Supplementary Table 2 and Supplementary Table 3). LH2 IHC staining in the epithelium and stromal cells was scored as low, moderate, and high, and the relationship between epithelial versus stromal expression and breast cancer patient outcome was calculated (Figure 6e; Extended Data Figure 10c). IHC scoring analysis revealed that only a modest number of patients with poorly differentiated breast tumors expressed moderate to high levels of epithelial LH2 (Extended Data Figure 10d); a finding that accords with prior links between tumor size, hypoxia and LH2-dependent tumor aggression22,23. A disproportionately high number of breast cancer patients with moderately and poorly differentiated breast tumors had high stromal cell LH2 (indicated by an H score of above 230; Figure 6f). The data and our observation that even well differentiated breast tumors express moderate levels of stromal cell LH2 (H score above 120 and equal or less than 230) imply that stromal cell LH2, rather than epithelial LH2, may be a more robust indicator of breast tumor aggression. Stratification of breast tumor patient analysis into ER+/HER2− (N=296); HER2+ (N=36) and TN (N=32) also showed an enrichment for moderate and high stromal LH2 H-score in the more aggressive cancer subtypes (Supplementary Table 2and Supplementary Table 3). We uncovered a correlation between high stromal cell LH2 H-score and shorter breast cancer patient-specific survival when adjusted for age at diagnosis (Figure 6g). We also established an association between stromal—not epithelial—LH2 levels and survival in lymph node–positive breast cancer patients but not lymph node–negative breast cancers (Figure 6h-i; Extended Data Figure 10e-g). The clinical data identify stromal cell LH2 as a novel biomarker with potential to predict metastatic disease and poor patient survival among breast cancers overall, as well as within the highest risk TN breast cancer subtype.
Discussion:
We implicated a subset of CD163/Relmα positive TAMs in stromal cell-mediated collagen crosslinking, stromal stiffening and tumor aggression. Clinical findings corroborated our experimental results, revealing significant associations between TAMs and collagen crosslinking enzymes and evidence that inflammatory stromal cell-mediated collagen crosslinking and stromal stiffening promote tumor aggression and compromise patient outcome. Diverse subsets of myeloid cells account for poor patient outcomes because they differentially promote angiogenesis, tumor cell intravasation, and immunosuppression49. Our results expand this repertoire to include a role for CD163/Relmα TAMs in collagen crosslinking and stiffening. We found that TN breast cancers contain more LH2− and LOX-derived collagen crosslinks that could explain their higher stromal stiffness and aggression2. Collagen analysis revealed that each breast tumor subtype exhibits a distinct collagen organization, stiffness and crosslinking profile, suggesting distinct collagen architectures and crosslinking signatures may reflect differences in tissue pathobiology. Consistently, TN breast cancers are characterized by abundant macrophages, consistent with our AFM measurements, second harmonic imaging and collagen crosslinking analysis2. Given that the immune cell infiltrate modifies tumor treatment, it is possible distinct collagen architectures, collagen crosslinking signatures and stromal rigidity contribute to this impact on therapeutic efficacy.
Using genetically engineered mouse models of spontaneous mammary cancer in which LOX was targeted to either the stromal cells or the mammary epithelium, we implicated fibroblasts—and not the epithelium—in tumor fibrosis, collagen crosslinking and stromal stiffening50,51. The data are consistent with earlier work in which alterations in fibrosis, collagen organization or stromal stiffness were not detected when LOXL2 was genetically-ablated, or ectopically-increased in the mammary epithelium of PyMT tumors, despite documenting changes in metastasis38. The observed discrepancies could be due to lack of collagen accessibility by the bulk of the tumor epithelium due to anatomic compartmentalization, or intrinsic biochemical differences in crosslinking enzyme secretion or activity between epithelial cells versus stromal cells. While in vitro cultured and orthotopically injected immortalized tumor epithelial cells engineered to overexpress LOX or LOXL2 appear to modify collagen, few reports exist that quantify changes in collagen crosslinking21,51,52. Furthermore, it remains unclear whether collagen modifications induced upon orthotopic tumor cell injections are truly mediated by crosslinking enzymes secreted by the injected tumor epithelium or whether these modifications result from indirect activation of the surrounding stromal fibroblasts. Clearly further studies will be needed to address this issue including a rigorous assessment of the relative efficiency of stromal fibroblasts versus epithelial cells on collagen crosslinking. Regardless, our results highlight the necessity of choosing an appropriate model to study ECM remodeling and suggest orthotopic models may fail to recapitulate the natural evolution of ECM remodeling in tumor progression38.
Our studies indicate that stromal LH2 is a robust predictor of breast cancer survival, especially in those patients that are lymph node–positive, thereby linking collagen crosslinking and HLCC abundance to patient outcome. The data revealed that increased expression of collagen crosslinking enzymes and collagen crosslinks occur early during malignancy, concurrent with tumor cell invasion and the accumulation of infiltrating macrophages53. This could explain why therapeutics targeting LOX and LOXL2 in late stage tumors have thus far failed to prolong cancer patient survival. The stroma in established invasive tumors likely harbor high levels of crosslinked collagens, so that while these agents may prevent further collagen crosslinking activity, they are not capable of reversing crosslinks or collagen modifications already present in the tissue. Instead, a detectable increase in stromal LH2 may provide an early prognostic marker of disease aggression that could inform interventions and treatment strategies. Indeed, our results show for the first time that LH2-derived collagen crosslinks are not only a distinct feature of stromal stiffness in TN breast cancer but can predict distant metastasis-free survival. Future studies using experimental models will be needed to confirm whether there is a causal relationship between stromal LH2, tumor aggression and metastasis. Regardless, the findings imply that the profile of collagen crosslinks and not merely crosslink abundance, may critically regulate tumor aggression.
Methods:
Human breast specimen acquisition and processing
Fresh human breast specimens from breast reduction, prophylaxis, or breast tumor mastectomy were either embedded in an optimum cutting temperature (OCT) aqueous embedding compound (Tissue-Plus, Scigen, Cat# 4583) within a disposable plastic base mold (Fisher, Cat# 22-363-554) and were snap frozen by direct immersion into liquid nitrogen and kept at −80°C freezer until cryo-sectioning for analysis, or specimens were formalin fixed and paraffin embedded (FFPE). All human breast specimens were collected from prospective patients undergoing surgical resection at UCSF or Duke University Medical Center between 2010 and 2014. The selected specimens were de-identified, stored, and analyzed according to the procedures described in Institutional Review Board (IRB) Protocol #10-03832 and #10-05046, approved by the UCSF Committee of Human Resources and the Duke’s IRB (Pro00034242)2.
Mouse studies
Macrophages were depleted in MMTV-PyMT mice by i.p. injections of 1mg of anti-CSF1 antibody (Bio X Cell, Cat# BE0204-A025, Clone 5A1) or an IgG1 control (Bio X Cell, Cat# BP0088-A025) every 7 days starting at 4 weeks of age. Mice were sacrificed at 8 and 11 weeks of age for tissue analysis.
Generation of mice
All mouse studies were maintained under pathogen-free conditions and performed in accordance with the Institutional Animal Care and Use Committee and the Laboratory Animal Research Center at the University of California, San Francisco.
TetO-mLOX-eGFP construct and transgenic mouse generation
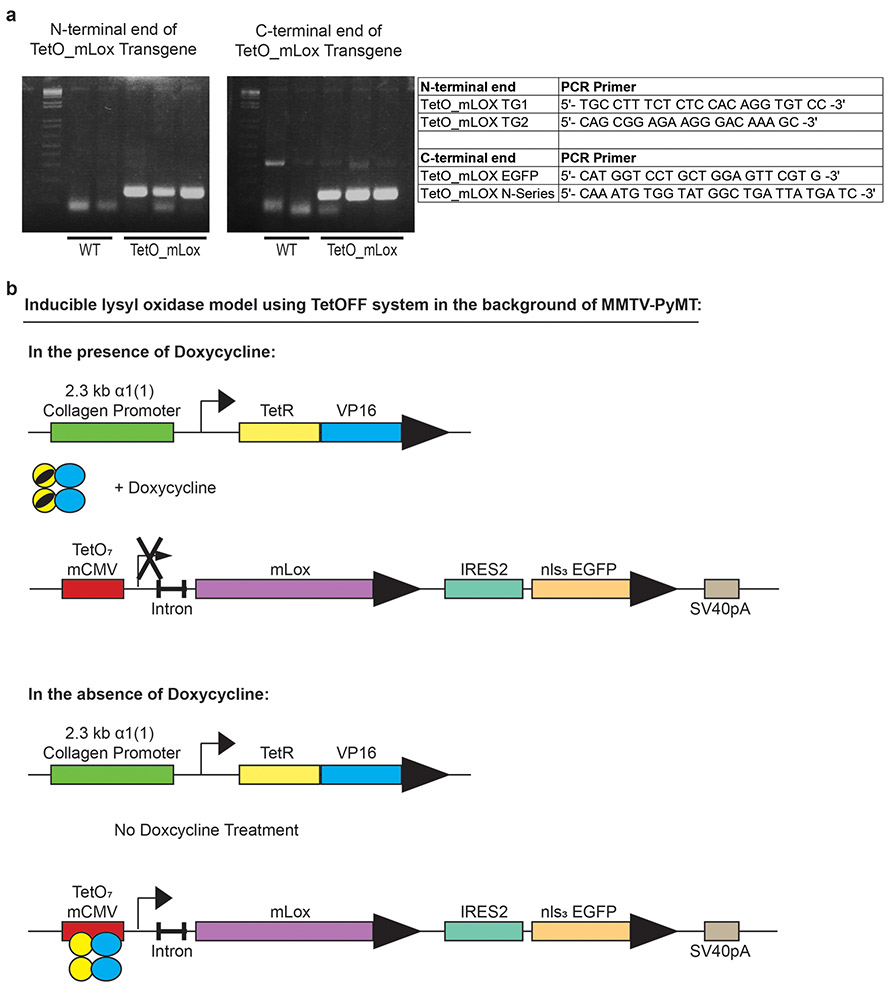
Full length mouse Lox (mLox) cDNA was purchased from OriGene. The full length ORF was amplified by PCR using forward and reverse primers respectively: GCAGGGATCCGCCACCATGCGTTTCGCCTGGGCTG and GGCGTCTAGAGCACCATGCGTTTCGCCTGGGCTGTGC. Following digestion with BamHI and XbaI, the PCR product was inserted into pSK TetO IRES 3xnlsEGFP54 downstream of the Tet regulated minimal CMV promoter and a 5’ UTR containing a chimeric intron of human β-globin and immunoglobulin heavy chain genes, which was expressed as a bicistronic mRNA via an internal ribosome entry site (IRES2) with eGFP targeted to the nucleus by a N terminal in frame fusion of 3 tandem repeats of the SV40 nuclear localization sequence (nls). The fragment containing the expression cassette from Tet regulated promoter to SV40 polyadenylation signal (SV40pA) was agarose gel purified from XhoI– EagI digested donor plasmid and was used to generate TetO-mLox-eGFP transgenic mice by pronuclear injection into FVB/n oocyte (Mouse Biology Program at UC Davis; project number MBP-834; colony number PN663).
Generation of MMTV-PyMT/Col1a1(2.3)-tTA/TetO-mLox-eGFP mice
The MMTV-PyMT/Col1a1(2.3)-tTA/TetO-Rs1 triple transgenic mice were generated by heterozygote or homozygote crosses of mice carrying the TetO-mLox-eGFP transgene with mice carrying heterozygote of the Col1a1(2.3)-tTA transgene (line 139)55 or MMTV-rtTA and MMTV-PyMT transgene56 to generate the experimental triple-transgenic genotype. In all breeding thereafter, MMTV-PyMT/Col11a1-tTA or MMTV-rtTA/TetO-mLox-eGFP male mice were crossed with TetO-mLox-eGFP female mice. Two mg/mL doxycycline hyclate (Alfa Aesar; Cat# J60579) was added to 5% sucrose water to modulate TetO-mLox-eGFP transgene expression.
Preparation of human breast specimens for hydrolysis
OCT was removed from tissue blocks by first transferring biospecimens to a conical tube and then performing 5X washes with 70% ethanol followed by 5X washes with 18 MΩ H2O48. Each wash consisted of vortexing the sample for 15 minutes at 4°C and then centrifuging at 18,000 x g for 15 minutes at 4°C. Between 1 and 3 milligrams of tissue was washed with 1X PBS buffer by vortexing for 15 minutes at 4°C and then sonicated on ice for 20 seconds using a Sonic Dismembrator M100 (ThermoFisher, San Jose, CA, USA). The homogenate was then centrifuged at 18,000 x g for 20 minutes at 4°C. The supernatant was removed, and the pellet was re-suspended in 1mg/mL NaBH4 (prepared in 0.1N NaOH) in 1X PBS for 1 hour at 4°C with vortexing. The reaction was neutralized by adding glacial acetic acid to a final concentration of 0.1% (pH ~ 3 −4)57. The sample was then centrifuged at 18,000 x g for 20 minutes at 4°C. The supernatant was removed, and the pellet was washed three times with 18 MΩ H2O to remove residual salt that could interfere with downstream LC-MS/MS analysis. The remaining pellet was dried under vacuum for further analysis.
Protein hydrolysis
The dried sample was placed in a glass hydrolysis vessel and hydrolyzed in 6N HCl, 0.1% phenol. The hydrolysis vessel is flushed with N2 gas, sealed and placed in a 110°C oven for 24 hours. After hydrolysis, the sample was cooled to room temperature and then placed at −80°C for 30 minutes prior to lyophilization. The dried sample was re-hydrated in 100μL of 18 MΩ H2O for 5 minutes, then 100 μL of glacial acetic acid for 5 minutes and finally 400 μL of butan-1-ol for 5 minutes. Importantly, 10 μL of sample is removed after re-hydration in water and saved for determination of hydroxyproline content.
Preparation of crosslink enrichment column
CF-11 cellulose powder is loaded in a slurry of butan-1-ol: glacial acetic acid, water (4:1:1) solution onto a Nanosep MF GHP 0.45μm spin columns until a settled resin bed volume of approximately 5mm is achieved. The resin is washed with 1.5 mL 4:1:1 organic mixture using an in-house vacuum manifold set up. Re-hydrated samples are then loaded onto individual columns, the vacuum is turned on and the sample is pulled through the resin into glass collection vials. The flow through is again passed over the resin to ensure maximal binding of crosslinked amino acids and set aside. The column is then washed with 1.5 mL of fresh 4:1:1 organic mixture. A fresh collection vessel is placed under the column and 750 μL of 18 MΩ H2O is used to elute crosslinked amino acids off of the CF-11 resin. The eluent is then placed in a speed vac and run until complete dryness. Dried eluent is then reconstituted in a buffer appropriate for downstream MS analysis on amide HILIC UHPLC columns.
UHPLC analysis
Up to 20 μL of tissue hydrolysates were analyzed on a Vanquish UPHLC system (ThermoFisher, San Jose, CA, USA) using an Acquity UHPLC BEH Amide column (2.1 × 100mm, 1.7 μm particle size – Waters, Milford, MA, USA). Samples were separated using a 5 minute gradient elution (55% - 40% Mobile phase B) at 250 μL/min (mobile phase: (A) 10mM ammonium acetate adjusted to pH 10.2 with NH4OH (B) 95% acetonitrile, 5% Mobile Phase A, pH 10.2, column temperature: 35°C.
MS data acquisition
The Vanquish UPHLC system (ThermoFisher, San Jose, CA, USA) was coupled online with a QExactive mass spectrometer (Thermo, San Jose, CA, USA), and operated in two different modes – 1. Full MS mode (2 μscans) at 70,000 resolution from 75 to 600 m/z operated in positive ion mode and 2. PRM mode at 17,500 resolution with an inclusion list of in-tact crosslinked amino acid masses (Supplementary Table 2), and an isolation window of 4 m/z. Both modes were operated with 4 kV spray voltage, 15 sheath gas and 5 auxiliary gas. Calibration was performed before each analysis using a positive calibration mix (Piercenet – Thermo Fisher, Rockford, IL, USA). Limits of detection (LOD) were characterized by determining the smallest injected crosslinked amino acids (LNL, DHLNL, d-Pyr,) amount required to provide a signal to noise (S/N) ratio greater than three using < 5 ppm error on the accurate intact mass. Based on a conservative definition for Limit of Quantification (LOQ), these values were calculated to be threefold higher than determined LODs.
MS data analysis
MS Data acquired from the QExactive were converted from a raw file format to .mzXML format using MassMatrix (Cleveland, OH, USA). Assignment of crosslinked amino acids was performed using MAVEN (Princeton, NJ, USA)58. The MAVEN software platform provides the means to evaluate data acquired in Full MS and PRM modes and the import of in-house curated peak lists for rapid validation of features. Normalization of crosslinked amino acid peak areas was performed using two parameters, 1. Hydroxyproline content and 2. Tissue dry weight pre-hydrolysis (in milligrams)57. Hydroxyproline content is determined by running a 1:10 dilution of the pre-enrichment sample through the Full MS mode (only) described above and exporting peak areas for each run.
Quantification of crosslinked amino acids
Relative quantification of crosslinked amino acids was performed by exporting peak areas from MAVEN into GraphPad (La Jolla, CA, USA) and normalizing based on the two parameters described above. All crosslink values are normalized to total collagen content (i.e. hydroxyproline abundance) and dry tissue weight and are plotted as log2 transformed normalized peak areas from LC-MS data unless otherwise noted in the figure legend. Statistical analysis, including T test and ANOVA (significance threshold for P values <0.05) were performed on normalized peak areas. Total crosslink plots were generated by summing normalized peak areas for all crosslinks in a given sample. Total HLCC plots were generated by summing normalized peak areas for all HLCC (DHLNL, Pyr, dPyr) crosslinks in a given sample.
Picrosirius red staining and quantification
FFPE tissue sections were stained using 0.1% Picrosirius red (Direct Red 80, Sigma-Aldrich, Cat# 365548 and picric acid solution, Sigma-Aldrich, Cat# P6744) and counterstained with Weigert’s hematoxylin (Cancer Diagnostics, Cat# CM3951), as previously described2. Polarized light images were acquired using an Olympus IX81 microscope fitted with an analyzer (U-ANT) and a polarizer (U-POT, Olympus) oriented parallel and orthogonal to each other. Images were quantified using an ImageJ macro to determine percentage area coverage per field of view using three to five fields of view per tissue. The ImageJ macro is available at https://github.com/northcottj/picrosirius-red.
Second harmonic generation image acquisition
Second harmonic generation (SHG) imaging was performed using a custom-built two-photon microscope setup equipped resonant-scanning instruments based on published designs containing a five-PMT array (Hamamatsu, C7950), as previously published2. The setup was used with two channel simultaneous video rate acquisition via two PMT detectors and an excitation laser (2W MaiTai Ti-Sapphire laser, 710–920 nm excitation range). SHG imaging was performed on a Prairie Technology Ultima System attached to an Olympus BX-51 fixed stage microscope equipped with a 25X (NA 1.05) water immersion objective. Paraformaldehyde-fixed or FFPE tissue sections were exposed to polarized laser light at a wavelength of 830 nm and emitted light was separated using a filter set (short pass filter, 720 nm; dichroic mirror, 495 nm; band pass filter, 475/40 nm). Images of x–y planes at a resolution of 0.656 mm per pixel were captured using at open-source Micro-Magellan software suite.
Immunofluorescence/Immunohistochemistry
Immunofluorescence staining was performed as previously described2. Briefly, mouse tissues were harvested and fixed with 10% buffered formalin phosphate (Fisher, Cat# 100-20) for 16-24 hours at room temperature and then further processed and paraffin-embedded. Five-μm sections dried for 30 minutes in 60°C, follow by deparaffinization and rehydration. Antigen retrieval was perform using DAKO Target Retrieval Solution (DAKO, Cat# S1699) for five minutes in a pressure cooker set to high pressure. Tissue sections were incubated with anti-pY397FAK antibody (Abcam, Cat# Ab39967, dilution 1:25) overnight at 4°C and with anti-rabbit IgG Alex Fluor 633 (ThermoFisher, Cat# A-21070, dilution 1:2000) for one hour at room temperature. Antigen retrieval for immunofluorescent staining of pS465/467SMAD2, cytokeratin 8+18, cytokeratin 5, F4/80, vimentin, and PDGFRα was performed using Diva Decloaker (BioCare, Cat# DV2004MX) for five minutes in a pressure cooker set to high pressure. Tissue sections were incubated with anti-pS465/467SMAD2 antibody (Millipore, Cat# AB3849-I, dilution 1:100), anti-F4/80 antibody (AbD Serotec, clone CI:A3-1, Cat# MCA497GA, dilution 1:400), anti-F4/80 biotin (Biolegend, clone BM8, Cat# 123105, dilution 1:400), anti-cytokeratin 8+18 antibody (Fitzgerald, Cat# 20R-CP004, dilution 1:400), anti-cytokeratin 5 antibody (Fitzgerald, Cat# 20R-CP003, dilution 1:400), anti-vimentin antibody (Cell Signaling, Cat# 5741, dilution 1:100), anti-PDGFRα (CD140a) antibody (Biolegend, Cat# 135901, dilution 1:100), anti-mouse I-A/I-E antibody (Biolegend, Cat# 107601, dilution 1:100), anti-CD86 antibody (Biolegend, Cat# 105001, dilution 1:100), anti-CD163 antibody (ThermoFisher, Cat# 14-1631-80, dilution 1:100), anti-CD11b antibody (Abcam, Cat# ab133357, 1:800), anti-CD4 antibody (ThermoFisher, Cat# 14-9766-82, dilution 1:100), anti-CD8 antibody (Abcam, Cat# ab203035, dilution 1:100), anti-Relmα (Abcam, Cat# ab39626, dilution 1:200), and anti-CD45 (BD Biosciences, Cat# 550539, dilution 1:100) overnight at 4°C and with anti-rat IgG Alexa Fluor 488 (ThermoFisher, Cat# A-11006, dilution 1:1000), anti-guinea pig IgG Alexa Fluor 568 (ThermoFisher, Cat# A-11075, dilution 1:1000), anti-rabbit IgG Alexa Fluor 633 (ThermoFisher, Cat# A-21070, dilution 1:2000), anti-rabbit IgG Alexa Fluor 633 (ThermoFisher, Cat# A-21070, dilution 1:1000) for one hour at room temperature or streptavidin conjugated with Alexa Fluor 555 (ThermoFisher, Cat# S32355, dilution (1:2000) for 30 minutes at room temperature.
Quantification of stromal nuclear pS465/467SMAD2 was performed using Imaris 9. Surfaces were created around each nucleus and epithelial nuclei were manually excluded based on cytokeratin signal and cell morphology. The means of the mean nuclear signal intensity for all stromal nuclei were calculated for each field of view and averaged for every animal.
Lungs from 11-week-old IgG1 control and anti-CSF1 treated PyMT mice were cut into 5-micron sections from 5 layers with 100 microns between the first three layers and 50 microns between the last two layers. Sections were analyzed for metastases by PyMT staining. Antigen retrieval was performed in Tris-EDTA buffer at pH 9 for four minutes in a pressure cooker set to low pressure. Tissue sections were incubated with anti-PyMT antibody (Novus Biologicals, Cat# NB-100-2749, dilution 1:250) overnight at 4°C and with a biotinylated anti-rat antibody for 1 hour at room temperature. Vectastain Elite ABC (Vector, Cat# PK6100) and ImmPACT DAB Peroxidase (Vector, Cat# SK-4105) were used for signal detection and nuclei were counterstained with methyl green.
mRNA In Situ Hybridization
Fresh, RNase-free FFPE sections were stained with RNAscope multiplex fluorescent reagent kit V2 according to standard manufacturer protocol. Target retrieval was performed using an eight-minute incubation in a pressure cooker set to low pressure. Opal 520 and Opal 570 (PerkinElmer) were used at 1:1500 for target visualization. Five to six images per tissue were used for Lox mRNA quantification and four to five images per tissue were used for Plod2 mRNA quantification.
Gene expression by RT-qPCR
Total RNA was reverse transcribed using random primers (Amersham Bioscienes) and results were normalized to 18S RNA to control for varying cDNA concentration between samples. The primer sequences used are 18s forward 5’-GGATGCGTGCATTTATCAGA-3’ and reverse 5’-GGCGACTACCATCGAAAGTT-3’, Lox forward 5’-CGGGAGACCGTACTGGAAGT-3’ and reverse 5’-CCCAGCCACATAGATCGCAT-3’, Loxl2 forward 5’-CACAGGCACTACCACAGCAT-3’ and reverse 5’-CCAAAGTTGGCACACTCGTA-3’, and Tgfb1 forward 5’-TCATGTCATGGATGGTGCCC-3’ and reverse 5’-GTCACTGGAGTTGTACGGCA-3’.
Atomic force microscopy data acquisition
Atomic force microscopy (AFM) measurements were performed as previously described2. Briefly, 20μm OCT-embedded frozen human breast tissue or 30μm mouse mammary gland sections were fast thawed by immersion in PBS at room temperature. Next, these sections were immersed in PBS containing phosphatase inhibitors (Roche, Cat# 04906845001), protease inhibitor (Roche, Cat# 04693124001), and propidium iodide (ACROS, Cat# 440300250) and placed on the stage for AFM measurements. AFM indentations were performed using an MFP3D-BIO inverted optical AFM (Asylum Research) mounted on a Nikon TE2000-U inverted fluorescent microscope. Silicon nitride cantilevers were used with a spring constant of 0.06 N m−1 and a borosilicate glass spherical tip with 5 μm diameter (Novascan Tech). The cantilever was calibrated using the thermal oscillation method prior to each experiment. The indentation rate was held constant within each study but varied between 2-20 μms−1 with a maximum force of 2 nN between studies. Force maps were obtained as a raster series of indentations utilizing the FMAP function of the IGOR PRO build supplied by Asylum Research. Three to five maps were taken per tissue sample. Elastic properties of ECM were reckoned using the Hertz model. A Poisson’s ratio of 0.5 was used in the calculation of the Young’s elastic modulus.
Western blotting
Snap frozen tissues were ground while frozen and lysed in 2% SDS containing protease and phosphatase inhibitor. Samples were boiled for 5 minutes (95°C) and loaded onto the SDS-polyacrylamide gel, and protein was separated at 110 constant volts.
The protein was transferred onto a pre-wet polyvinylidene difluoride (PVDF) membrane (100% methanol, 1 minute) at 250 mA for 2 hours. The PVDF membrane was rinsed with TBST and non-specific binding was blocked with 5% nonfat dry milk dissolved in TBST. The membrane was then incubated with the primary antibody overnight at 4°C, washed with TBST, incubated with horseradish-peroxidase conjugated secondary antibody (1 hour, room temperature; dilution 1:5000), washed with TBST, and detected with the chemiluminescence system Quantum HRP substrate (Advansta #K-12042). Quantification was performed using gel densitometry in ImageJ. Primary antibodies used are anti-LOX (1:1000, Abcam Cat# ab174316) and anti-E-cadherin (1:1000, Cell Signaling Cat#3195).
Flow cytometry
Mouse tissue was harvested and chopped with a razor blade. Chopped tissue was digested in 100 U/mL Collagenase Type 1 (Worthington Biochemical Corporation, Cat# LS004196), 500 U/mL Collagenase Type 4 (Worthington Biochemical Corporation, Cat# LS004188), and 200 μg/mL DNase I (Roche, Cat# 10104159001) while shaking at 37°C. Digested tissue was filtered using a 100 μm filter to remove remaining pieces. Red blood cells were lysed in ACK buffer (ThermoFisher Scientific, Cat# A1049201) and remaining cells were counted. Cells were stained with fluorophore-conjugated primary antibodies for 30 minutes on ice and subsequently stained with a viability marker. Antibodies used for staining were anti-CD3e BV421 antibody (Biolegend, Cat# 145-2C11, dilution 1:100, Monoclonal, Clone 145-2C11), anti-CD11b PerCP-Cy5.5 antibody (Biolegend, Cat# 101227, dilution 1:500, Clone M1/70), anti-CD11c BV605 antibody (Biolegend, dilution 1:200, Monoclonal, Clone N418), anti-CD14 BV510 antibody (Biolegend, Cat# 123323, dilution 1:200, Monoclonal, Clone Sa 14-2), anti-CD24 PE antibody (BD Biosciences, dilution 1:400, Monoclonal, Clone M1/69), anti-CD24 BV650 antibody (Biolegend, dilution 1:200, Monoclonal, Clone M1/69), anti-CD29 AF488 antibody (Biolegend, Cat# 102211, dilution 1:100, Monoclonal, Clone HMβ1-1), anti-CD31 APC antibody (Biolegend, Cat# 10240, dilution 1:40, Monoclonal MEC13.3),anti-CD45 APC antibody (Biolegend, Cat# 103112, dilution 1:160, Monoclonal, Clone 30-F11), anti-CD45 AF700 antibody (Biolegend, Cat# 103127, dilution 1:100, Monoclonal, Clone 30-F11), anti-CD140a PE antibody (Biolegend, Cat# 135905, dilution 1:50, Monoclonal, Clone APA5), anti-F4-80 BV510 antibody (Biolegend, Cat# 123135, dilution 1:200, Monoclonal, Clone BM8), anti-Ly6C BV421 antibody (Biolegend, Cat# 128031, dilution 1:200, Monoclonal, Clone HK1.4), anti-Ly6C BV711 antibody (Biolegend, Cat# 128037, dilution 1:200, Monoclonal, Clone HK1.4), and anti-MHCII PE-Cy7 antibody (Biolegend, Cat# 107629, dilution 1:300, Monoclonal, Clone M5/114.15.2), and anti-TER-119 APC (Biolegend, Cat# 116212, dilution 1:80, Monoclonal, Clone TER-119). Zombie NIR Fixable Viability Dye (Biolegend, Cat# 423105) was also used. Primary human TAMs were analyzed as described in Ruffell et al. PNAS 201259.
Patient gene expression analysis
For the stroma and epithelium specific gene expression analysis, the breast cancer datasets from Finak et al. 2008 and Gruosso et al. 2019 have been used (GSE9014, GSE58644, GSE88715, and GSE88847)36,37. Briefly, whole Human Genome 44 K arrays (Agilent Technologies, product G4112A) were used for stroma and epithelial expression profiles. Details of laser capture microdissection, RNA extraction, labeling, hybridization, scanning and quality filters are described in Finak et al., 2006 and 200860. Briefly, the dataset was normalized using loess (within-array) and quantile (between-array) normalization. Probes were ranked by Inter-quartile range (IQR) values, and the most variable probe per gene across expression data were selected for further analysis. Replicate arrays with a concordance above 0.944 were averaged before assessing differential expression.
An association between PLOD2 and distant metastasis-free survival (DMFS) has been determined using an online tool (http://xena.ucsc.edu) to download GEO data (GSE2034, GSE5327, and GSE7390) from 683 patients analyzed on Affymetrix U133A platform as described in Yau et al.47. Patients have been excluded from analyses if their molecular subtyping of ER/HER2 status and PAM50 did not align: ER+/HER2− must always be luminal, ER+ or−/HER2+ must be HER2+, and ER−/HER2− must always be basal-like. PLOD2 expression levels have been divided based on the median for each tumor subtype: ER+/HER2− (low n=157; high=157), ER− or +/HER2+ (low n=36; high=37), and ER−/HER2− (low n=66; high=67). All statistical analyses were done using GraphPad Prism Version 6.01: Kruskal-Wallis one-way ANOVA test was applied to assess the relationship in PLOD2 expression levels among tumor subtypes and log rank P value (Mantel-Cox) tests for DMFS curves.
An association between PLOD2 gene expression and relapse-free survival (RFS) has been determined using an online tool (http://kmplot.com/analysis/) from 1,809 patients analyzed on Affymetrix platform (HGU133A and HGU133+2 microarrays)48. Affymetrix ID 202619 or 202620 were used for PLOD2 probes (2014 version) in these analyses. All breast cancer patients in this database were included regardless to lymph node status, TP53 status, or grade. No restrictions were placed in term of patient treatment. PLOD2 expression levels have been divided based on the median for each tumor subtype: ER+/PR+/HER2− (low n=170; high=169), ER−/PR−/HER2+ (low n=58; high=57), and ER−/PR−/HER− (low n=128; high=127). Hazard ratio (and 95% confidence intervals) and log rank P values were calculated and displayed once the data were plotted using the online tool.
The cBioPortal for Cancer Genomics was used to determine the levels of LOX, PLOD2, and LOXL2 gene expression in breast cancer patients segregated by ER and HER2 status61,62. ER, PR, and HER2 status were determined by gene expression levels. Samples positive for both ER/PR and HER2 overexpression were excluded from subtype analysis. The cBioPortal was also used to assess gene expression associations of LOX, PLOD2, and LOXL2 with CD163. All data accessed via cBioPortal are from the 1904 patients in the METABRIC dataset analyzed for gene expression by Illumina human v3 microarray63. All 1904 samples were included in correlation analyses.
Statistical analysis
GraphPad Prism Version 6.01 was used to perform all statistical analyses with the exception of LH2 IHC and RFS correlations with PLOD2. Statistical significance was determined using the appropriate tests as noted in the corresponding figure legends or method section.
Tissue Microarray (TMA) LOX IHC:
LOX IHC was performed on Biomax BC081116c human breast tissue microarray. Citrate buffer (pH = 6.0) was used for antigen retrieval and tissue was stained using monoclonal recombinant anti-LOX (Abcam Cat# ab174316; dilution 1:800). Neoplastic epithelial and stromal staining intensity was evaluated by a researcher on a scale from 0-3 with zero as no signal and three as strong signal. HER2+ subtype included all HER2+ patients regardless of ER status.
LH2 IHC and prognostic analyses:
Study population
The female Malmö Diet and Cancer Study (MDCS) cohort consists of women born 1923–1950)64,65. Information on incident breast cancer is annually retrieved from the Swedish Cancer Registry and the South Swedish Regional Tumor Registry. Follow-up until December 31, 2010, identified a total of 910 women with incident breast cancer, the following conditions excluded patients: 1) with in situ only cancers (n=68), 2) who received neo-adjuvant treatments (n=4), 3) with distant metastasis at diagnosis (n=14), 4) those who died from breast cancer-related causes ≤ 0.3 years from diagnosis (n=2), and finally 5) patients with bilateral cancers (n=17). In addition, one patient who declined treatment for four years before accepting surgery was excluded. Patient characteristics at diagnosis and pathological tumor data were obtained from medical records. Of note, HER2+ subtype included all HER2+ patients regardless of ER status. Information on cause of death and vital status was retrieved from the Swedish Causes of Death Registry, with last follow-up December 31st, 2014. Ethical permission was obtained from the Ethical Committee at Lund University (Dnr 472/2007). All participants originally signed a written informed consent form.
Tumor evaluation
Tumor samples from incident breast cancer cases in MDCS were collected, and a tissue microarray (TMA) including two 1-mm cores from each tumor was constructed (Beecher, WI, USA). Within the study population (N=910), tumor tissue cores were accessible from 718 patients. Four-μm sections dried for one hour in 60°C were automatically pretreated using the Autostainer plus, DAKO staining equipment with Dako kit K8010 (Dako, DK). A primary mouse monoclonal Lysyl Hydroxylase 2 (LH2) antibody (Origene; Cat# TA803224, dilution 1:150) was used for the immunohistochemical staining.
TMA cores were analyzed by a cohort of 4 anatomic pathologists (ACN, AC, JG, AN) using the PathXL digital pathology system (http://www.pathxl.com, PathXL Ltd., UK) blinded to all other clinical and pathologic variables. Immunohistochemistry for LH2 was assessed separately for stromal and neoplastic epithelial components of the tumors. Stromal LH2 staining was assessed with the semi-quantitative H-score which combines intensity and proportion positive assessments into a continuous variable from 0-30066. Cellular stromal components were assessed (including fibroblasts, macrophages, endothelial cells, adipocytes, and other stromal cell types) while areas of significant lymphocytic infiltrate were specifically excluded from the percent positive estimation. Neoplastic epithelial LH2 staining intensity was scored 0-3+ based on the predominant intensity pattern in the tumor — invasive tumor cells did not display significant intra-tumoral heterogeneity of LH2 staining within each core. Verification of inter-observer reproducibility for the H-score was established in a training series of 16 cases evaluated by all study pathologists to harmonize scoring. Inter-observer agreement in the training set was very high, evaluating the IHC scores both as continuous variables (Pearson correlation coefficients ranging from 0.912-0.9566, all p values < 0.0001), and after transformation into categorical data (negative, low, and moderate/high; weighted kappa coefficients ranging from 0.673-0.786). In addition, 50 cases of the study cohort were evaluated blindly by two pathologists to confirm data fidelity; the Pearson correlation coefficient = 0.7507 (p = 5.7 E-05), considered a strong level of agreement.
After exclusion of cases for which LH2 was not evaluable on the TMA, H-scores for 505 patients were included for statistical associations with clinicopathologic features and patient outcome. Each patient was represented by two cores, and TMA core 1 and core 2 were merged into a joint variable favoring the highest stromal LH2 H-score or epithelial LH2 intensity because we predict that higher H-score would drive patient outcome in accordance with our gene expression data demonstrating high LH2 expression correlated with poor outcome. The Pearson correlation coefficient between cores = 0.647, demonstrating moderate agreement among the stromal LH2 H-scores for the two cores. In cases with only one TMA core providing a LH2 score, the expression of this core was used. Further, the joint stromal LH2 variable was categorized into tertiles based on the study population with valid LH2 annotation (N=505). The lowest tertile of LH2 H-scores were defined as scores between 0 and less or equal to 120 (N=171), the moderate H-score as above 120 and equal or less than 230 (N=188), and the highest stromal LH2 score as above 230 (N=146).
Statistical analyses for LH2 IHC analyses
Patient and tumor characteristics at diagnosis in relation to stromal LH2 expression were categorized and presented as percentages. Continuous variables are presented as the mean and min/max. The associations between LH2 expression and grade or tumor size, respectively, were analyzed through linear-by-linear association. The association between LH2 expression and prognosis was examined using breast cancer-specific mortality as endpoint, which was defined as the incidence of breast cancer-related death. Follow-up was calculated from the date of breast cancer diagnosis to the date of breast cancer-related death, date of death from another cause, date of emigration or the end of follow-up as of December 31st, 2014. Main analyses included the overall population; additional analyses were performed in subgroup analyses stratified by estrogen receptor (ER) or axillary lymph node involvement (ALNI) status. The prognostic impact of stromal LH2 expression was analyzed through Cox proportional hazards analyses, which yielded hazard ratios (HR) and 95% confidence intervals (CI) for crude models, and multivariate models adjusted for age at diagnosis (model 1) and tumor characteristics ER (dichotomized, cut-off 10% stained nuclei), ALNI (none or any positive lymph node involvement), histological grade (Nottingham grade I-III), and tumor size (dichotomized using cut-off 20 mm). Kaplan-Meier curves including the LogRank test indicated LH2 status to particularly impact the first 10 years after diagnosis and survival variables constructed to capture these effects were used in Cox regression models investigating the effects during the first post-diagnostic decade. All statistical analyses were performed in SPSS version 22.0 (IBM).
Data Availability
All gene expression array and RNA-Seq data have been deposited in GEO and are publicly available. For the stroma and epithelium specific gene expression analysis, the breast cancer datasets from Finak et al. 2008 and Gruosso et al. 2019 have been used (GSE9014, GSE58644, GSE88715, and GSE88847)36,37. The association between PLOD2 and distant metastasis-free survival (DMFS) was determined using an online tool (http://xena.ucsc.edu) to download GEO data (GSE2034, GSE5327, and GSE7390) from 683 patients analyzed on Affymetrix U133A platform as described in Yau et al.47. The association between PLOD2 gene expression and relapse-free survival (RFS) was determined using an online tool (http://kmplot.com/analysis/) from 1,809 patients analyzed on Affymetrix platform (HGU133A and HGU133+2 microarrays)48. Affymetrix ID 202619 or 202620 were used for PLOD2 probes (2014 version) in these analyses. The cBioPortal for Cancer Genomics was used to access the publicly available METABRIC dataset to determine the levels of LOX, PLOD2, and LOXL2 gene expression in breast cancer patients segregated by ER and HER2 status and to assess gene expression associations of LOX, PLOD2, and LOXL2 with CD16363. Macrophage RNA-Seq data has been deposited into GEO (GSE157290).
Extended Data
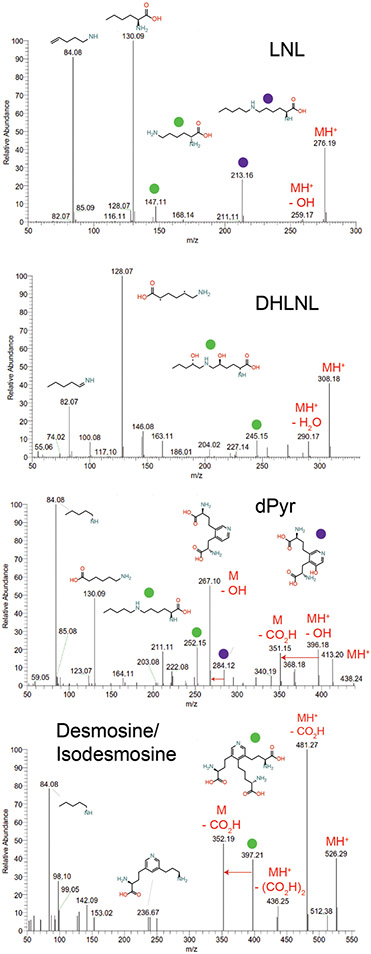
Extended Data Figure 1: Partial assignment of MS2 fragmentation spectra of crosslinked amino acid standards.
MS2 fragmentation spectra of commercially available crosslinked amino acid standards with partial assignment of MS2 fragmentation spectra (LNL, DHLNL, dPyr). Protonated forms of precursor ions are denoted by MH+ labels. Colored circles above fragment ions are matched to their suggested fragment ion structure above the full spectra or are listed above the ion if space permits.
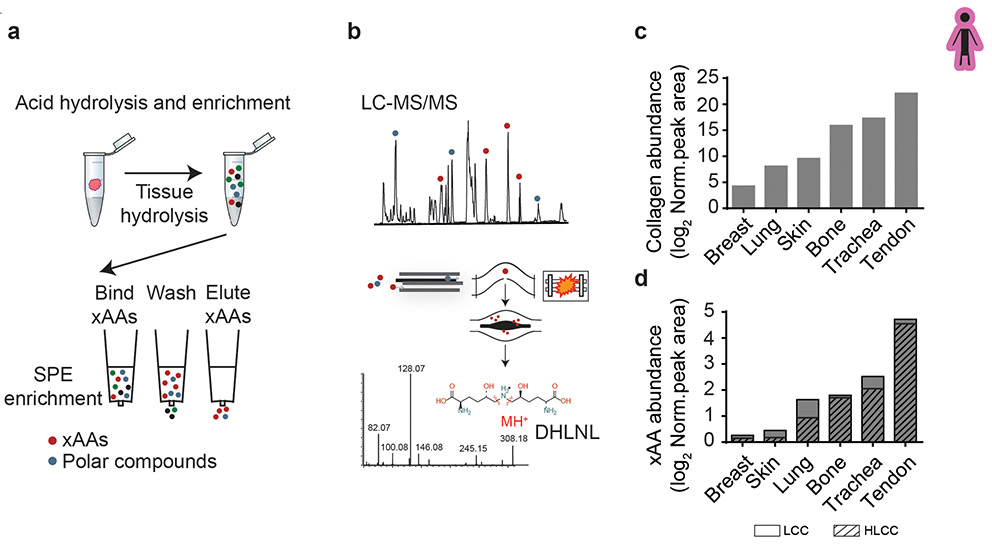
Extended Data Figure 2: Optimized crosslinking amino acid analysis (xAAA) enables accurate measurement of collagen state across a diverse range of tissue types.
(a) xAAA workflow schematic. Clinical specimens are hydrolyzed and enriched by solid phase extraction (SPE). (b) The enriched hydrolysate is analyzed by LC-SRM on a hybrid quadrupole orbitrap instrument. MS2 spectra is used to accurately identify crosslinked amino acids (xAAs) such as dihydroxy lysinonorleucine (DHLNL) (c) Bar graphs showing quantification of tissue collagen and (d) total xAA measured in human breast, lung, skin, bone, trachea and tendon (pooled n = 3 each tissue). The calculated amino acid crosslink abundancies are normalized to total tissue collagen content (i.e. hydroxyproline abundance) which is calculated based on dry tissue weight. The final values have been plotted as relative abundance based on peak area. Bar graphs depict relative abundance of Lysine derived-collagen crosslink (LCC) and hydroxylysine-derived collagen crosslink (HLCC) species.
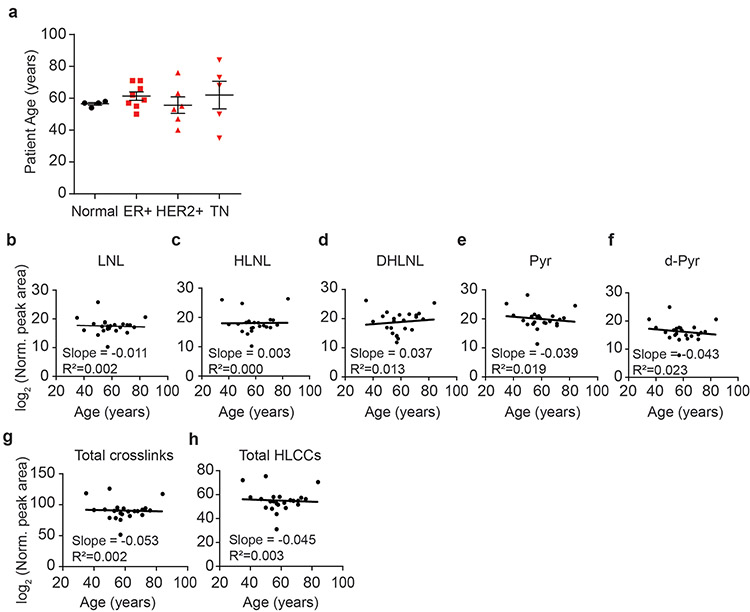
Extended Data Figure 3: Abundance of LOX-mediated crosslinks does not correlate with patient age.
(a) Scatter plot showing the mean and SEM of patient age for normal (n = 4) and tumor tissue (ER+ n = 8, HER2+ n = 6, TN n = 5) samples by subtype. (b-f) Scatter plots showing the levels of each LCC and HLCC crosslink measured in normal and tumor breast tissues versus patient age. The total abundance of crosslinks (g) was calculated by summing all individual crosslinks and the total tissue HLCC abundance (h) was calculated by summing DHLNL, Pyr, and d-Pyr. All crosslink values are normalized to total collagen content (i.e. hydroxyproline abundance) and dry tissue weight and are plotted as log2 transformed normalized peak areas from LC-MS data. The best fit line and its slope and r2 value are displayed on each plot. All n values represent biologically independent human tissue specimens.
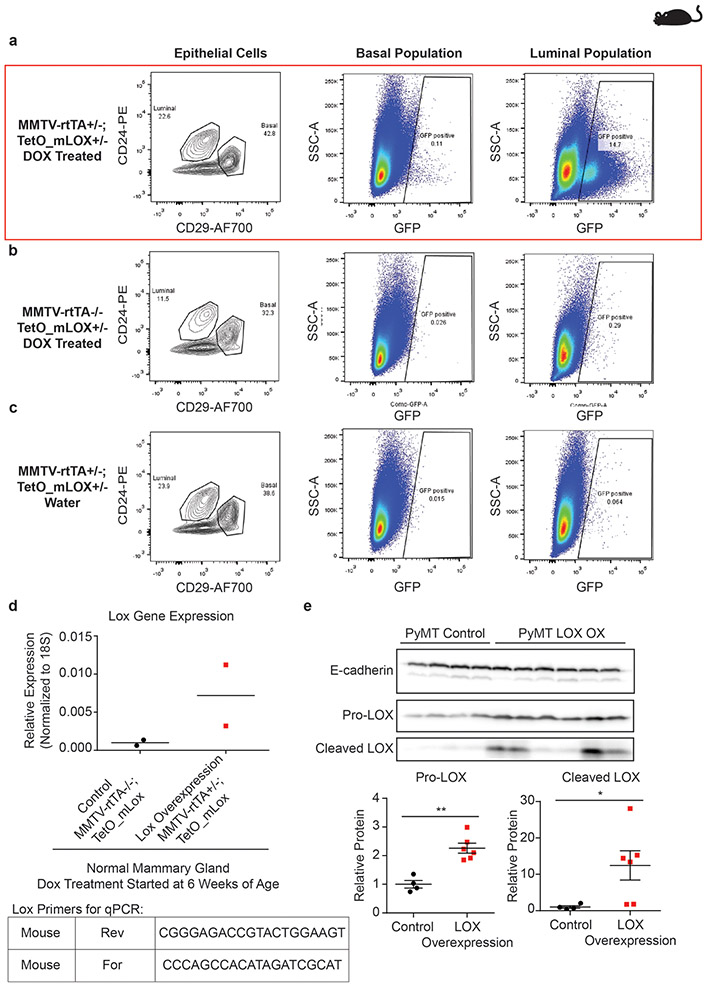
Extended Data Figure 4: Validation of LOX overexpression in doxycycline (DOX) treated MMTV-rtTA+/−; TetO_mLox+/− mice.
(a-c) Representative dot plots (n = 2) of GFP expression in mouse mammary epithelial cells analyzed by flow cytometry. GFP and mLOX are encoded on the same mRNA transcript with independent translation initiation sites. (a) GFP expression is induced in DOX treated MMTV-rtTA+/−; TetO_mLox+/− mice. (b-c) GFP expression is not induced in control mice lacking the MMTV-rtTA promoter (b) or control mice not treated with DOX (c). To select for mammary epithelial cells were gated out cells positive for anti-mouse CD45-APC, anti-mouse CD31-APC, anti-mouse Ter119-APC antibodies. (d) Quantification of Lox gene expression in whole mouse mammary gland by RT-qPCR in control MMTV-rtTA−/−; TetO_mLox+/− mice (n = 2) and Lox overexpressing MMTV-rtTA+/−; TetO_mLox+/− (n = 2) mice. (e) Western blot of whole tumor lysate from control (n = 4) and epithelial LOX overexpressing (n = 6) PyMT tumors. Scatter plots with mean ± SEM quantify optical density of each band normalized to E-cadherin. Western blot for these lysates was run once. Statistical analyses were performed using two-tailed Mann-Whitney U test Pro-LOX **p = 0.0095, Cleaved LOX *p = 0.0381. All n values represent biologically independent mouse tissue specimens.
Extended Data Figure 5: Design of inducible lysyl oxidase expression in vivo using a TetOFF system in the background of MMTV-PyMT model.
(a) PCR results and sequence of PCR primers of N- and C-terminal ends to confirm TetO_mLox transgene incorporation. (b) Diagram of inducible TetOFF system. TetO_mLox transgene is reversibly turned off or on in the presence or absence of doxycycline, respectively.
Extended Data Figure 6: PyMT collagen crosslinking abundance does not increase between 8- and 11-week tumors.
(a-f) Scatter plots showing individual and mean values ± SEM of the levels of total collagen crosslinks (a), total HLCCs (b), LNL (c), HLNL (d), DHLNL (e), and Pyr (f) in 8-week (n = 10) and 11-week-old (n = 9) PyMT tumors. Quantity of crosslinks per tissue was calculated normalizing crosslinks to total collagen content (i.e., hydroxyproline abundance) and dry tissue weight. Values were plotted as log2 transformed normalized peak areas as quantified from LC-MS data. Statistical analysis was performed using two-tailed unpaired t-test (a) p = 0.6149 (b) p = 0.7451 (c) p = 0.7235 (d) p = 0.1766 (e) p = 0.7563 (f) p = 0.4687. All n values represent biologically independent mouse tumor specimens.
Extended Data Figure 7: Early profibrotic macrophages express M2a markers and are lost upon anti-CSF1 treatment.
(a) Scatter plot showing individual values and mean ± SEM of the number of F4-80 positive cells per field of view in IgG1 treated (n = 6) or anti-CSF1 treated (n = 4) PyMT tumors from 8-week-old mice. (b-c) Scatter plots showing individual values and mean ± SEM of the percentage of F4/80 positive cells expressing markers MHCII and CD86 (b) and markers CD163 and Relmα (c) in IgG1 treated (n = 6) or anti-CSF1 treated (n = 4-5) PyMT tumors from 8-week-old mice. (d-g) Scatter plots showing individual values and mean ± SEM of the number of cells per field of view (FOV) of other non-macrophage immune populations CD11b+ F4-80− (d), MHCIIhi F4-80− (e), CD45+ CD4+ (f), and CD45+ CD8+ (g) in IgG1 (n = 4 e,g; n = 5 d,f) and anti-CSF1 (n = 6) treated mice. Statistical analysis (a-g) was performed using two-tailed Mann-Whitney U test (a) *p = 0.019 (b) MHCIIhi p = 0.7190, CD86+ p = 0.4286 (c) CD163+ *p = 0.0303, Relmα+ p = 0.0381 (d) p = 0.9719 (e) p = 0.1524 (f) p = 0.3074 (g) p = 1.000. (h) Heat map of selected differentially expressed genes from macrophages sorted from 8-week and 11-week PyMT tumors. (i) Representative images from 8-week-old IgG1 treated and anti-CSF1 treated PyMT mice stained for cytokeratin 8/18 (green), PDGFRα (magenta), vimentin (yellow), and DAPI (blue). Scale bar is 100 μm. (j) Scatter plot showing mean ± SEM of fibroblasts (vimentin+ and PDGFRα+) per field of view in 8-week-old IgG1 treated (n = 6) and anti-CSF1 (n = 5) treated PyMT mice. All n for (a-j) values represent biologically independent mouse tissue specimens. (k-m) Scatter plot depicting the Spearman correlation of CD163 gene expression with LOX (k), PLOD2 (l), and LOXL2 (*p = 0.0143) (m) in human breast tumors (n = 1904 biologically independent human tissue specimens).
Extended Data Figure 8: ECM Stiffness for individual specimens.
(a) Scatter plot showing mean and individual values of atomic force microscopy (AFM) microindentation measurements of individual human samples categorized as normal breast tissue (black circles) or invasive ductal carcinoma (IDC; red squares). (b) Scatter plot showing mean and individual values of AFM microindentation measurements of individual mouse mammary tumors from doxycycline treated MMTV-PyMT+/−; Col1a1-tTA+/−; TetO-mLox control mice (DOX-ind PyMT Con; black circles) or water treated mice overexpressing stromal Lox (Water PyMT Lox OX; red squares). (c) Scatter plot showing mean and individual values of AFM microindentation measurements of individual mouse mammary gland tumors from 8-week-old IgG1 treated (IgG Control; red squares) and anti-CSF1 treated (aCSF1 Ab; black circles) PyMT tumors.
Extended Data Figure 9: PLOD2 correlations with distant metastasis free survival and probability of relapse free survival.
(a) Log2 scaled data graphs showing relative PLOD2 gene expression levels in patients stratified by estrogen and epidermal growth factor receptor two (ER/HER2) status (ER+ HER2− n=314; ER− or + /HER2+ n=73; ER−/HER2− n=133). Whiskers denote minima and maxima, box denotes 25th and 75th percentiles and median. Statistical analysis was performed to compare PLOD2 expression levels among subtypes using Kruskal-Wallis one-way ANOVA (****p < 0.0001) and two-tailed Mann-Whitney U test without adjustment for multiple comparisons for individual comparisons (****p < 0.0001). (b) Line graphs showing distant metastasis-free survival (DMFS) for patients with estrogen receptor positive and epidermal growth factor receptor two negative breast tumors (ER+/− HER2−; low n=157 & high=157). (c) Line graph of DMFS for patients with estrogen receptor negative or negative and epidermal growth factor receptor two positive breast tumors (ER− or +/HER2+; low n=36 & high=37). (d) Line graph of DMFS for patients with estrogen receptor negative epidermal growth factor receptor negative breast tumors (ER− HER2−; low n=66 & high=67). Statistical analyses were performed to compare PLOD2 and DMFS for each subtype or Log-rank (Mantel-Cox) test (b) p = 0.8051 (c) *p = 0.0144 (d) p = **0.0050. (e-g) Kaplan-Meier curves indicating the probability of relapse-free survival assessed in ER+/PR+ (e), HER2+ (f), and triple negative (g) breast cancer patients up to 16 years after diagnosis. A correlation between PLOD2 (gene encoding LH2) expression and RFS has been determined using an online tool (http://kmplot.com/analysis/) as described in the methods. Top and bottom panels represent two distinct Affymetrix PLOD2 probes (202619 and 202620) from the same database. For Kaplan-Meier curves, statistical analyses were performed using LogRank test.
Extended Data Figure 10: Assessment of LOX and neoplastic epithelial LH2 expression in breast cancer.
(a) Neoplastic epithelial LOX staining of the Biomax BC081116c TMA was assessed by a researcher with an intensity score and stratified as negative/low, moderate, or high. (b) Stromal LOX staining of the Biomax BC081116c TMA was assessed by a researcher with an intensity score and stratified as negative/low, moderate, or high. Scale bar is 500 μm. (c) Tumor samples from incident breast cancer cases were collected, and a tissue microarray (TMA) including two 1-mm cores from each tumor was constructed. Neoplastic epithelial LH2 staining was assessed by a pathologist with semi-quantitative intensity score and stratified as negative, low, or moderate/high. Scale bars are 500 μm. See Supplementary Table 2 for patient information. (d) Clinical correlation of neoplastic epithelial LH2 intensity score with tumor grades. (e) Kaplan-Meier curves indicating cumulative breast cancer specific survival (BCSS) based on epithelial LH2 intensity assessed in breast cancer patients up to 10 years after diagnosis (LH2 negative n = 271, weak n = 112, moderate/high n = 77). (f) BCSS curves by epithelial LH2 intensity including only axillary lymph node negative patients (LH2 negative n = 175, weak n = 67, moderate/high n = 50). (g) BCSS curves by epithelial LH2 intensity including only axillary lymph node positive patients (LH2 negative n = 84, weak n = 42, moderate/high n = 26). For tumor grade and LH2 intensity score, statistical analysis was performed using a linear-by-linear association (***p < 0.001). For Kaplan-Meier curves, statistical analyses were using LogRank test (e) *p = 0.025 (f) *p = 0.038 (g) p = 0.165.
Supplementary Material
Significance.
We identify tumor-associated macrophages that promote fibroblast expression of collagen crosslinking enzymes LOX and LH2 to stiffen the stroma. Stromal LH2 is upregulated in the most fibrotic breast cancers that have the stiffest stroma, the most trivalent HLCCs and the worst prognosis, underscoring its potential as a biomarker and therapeutic target.
Acknowledgements
We thank J. Northcott for writing the ImageJ Macro, L. Korets for mouse husbandry and N. Korets for histology support, as well as K. Lövgren and S. Baker for LH2 immunostaining on patient biopsies. We thank the UCSF Genomics core for their support with the RNA-Seq analysis on mouse TAMs and Connor Stashko for generating the gene expression heat map. The work was supported by investigator grants through the US National Cancer Institute R33 CA183685 (K.C.H & V.M.W) and R01CA192914 and CA174929 to (V.M.W), and R01CA222508-01 to (V.M.W. and E.S.H.), as well as US DOD Breast Cancer Research Program (BCRP) grant BC122990 (V.M.W). Trainee support was provided by US DOD BCRP grant BC130501 (O.M.), US NIH grants TL1 TR001081 & US NIH T32 HL007171 (A.S.B), and US NIH T32 grant CA 108462 (O.M.). Funding from Eastern Star Scholar-Minnesota Masonic Cancer Center (A.C.N.), the Swedish Research Council (S.B.) and US NIH R01 CA057621 (Z.W.) also supported the work.
Footnotes
Declaration of Competing Interests
The authors declare the following competing interests: L.M.C. is a paid consultant for Cell Signaling Technologies, Shasqi Inc., and AbbVie Inc., and received reagent and/or research support from Plexxikon Inc., Pharmacyclics, Inc., Acerta Pharma, LLC, Deciphera Pharmaceuticals, LLC, Genentech, Inc., Roche Glycart AG, Syndax Pharmaceuticals Inc., Innate Pharma, and NanoString Technologies, and is a member of the Scientific Advisory Boards of Syndax Pharmaceuticals, Carisma Therapeutics, Zymeworks, Inc, Verseau Therapeutics, Cytomix Therapeutics, Inc., and Kineta Inc. T.M. and K.C.H. are co-founders of Omix Technologies.
References:
- 1.Laklai H et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med 22, 497–505, doi: 10.1038/nm.4082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acerbi I et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) 7, 1120–1134, doi: 10.1039/c5ib00040h (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvorak HF Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 315, 1650–1659, doi: 10.1056/NEJM198612253152606 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Conklin MW et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. The American journal of pathology 178, 1221–1232, doi: 10.1016/j.ajpath.2010.11.076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive KP et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324, 1457–1461, doi: 10.1126/science.1171362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Provenzano PP et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21, 418–429, doi: 10.1016/j.ccr.2012.01.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Netti PA, Berk DA, Swartz MA, Grodzinsky AJ & Jain RK Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res 60, 2497–2503 (2000). [PubMed] [Google Scholar]
- 8.Jain RK, Lee JJ, Hong D & Kurzrock R Reply to Levy A et al. J Clin Oncol 31, 396, doi: 10.1200/JCO.2012.46.4867 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Egeblad M, Rasch MG & Weaver VM Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol 22, 697–706, doi: 10.1016/j.ceb.2010.08.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri O et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater 13, 970–978, doi: 10.1038/nmat4009 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Provenzano PP, Inman DR, Eliceiri KW & Keely PJ Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 28, 4326–4343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levental KR et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekhdjian AH et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol Biol Cell 28, 1467–1488, doi: 10.1091/mbc.E16-09-0654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickup MW et al. Stromally derived lysyl oxidase promotes metastasis of transforming growth factor-beta-deficient mouse mammary carcinomas. Cancer Res 73, 5336–5346, doi: 10.1158/0008-5472.CAN-13-0012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickup MW, Mouw JK & Weaver VM The extracellular matrix modulates the hallmarks of cancer. EMBO Rep 15, 1243–1253, doi: 10.15252/embr.201439246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provenzano PP et al. Collagen density promotes mammary tumor initiation and progression. BMC Med 6, 11, doi:1741-7015-6-11 [pii] 10.1186/1741-7015-6-11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre DR, Weis MA & Wu J-J Advances in collagen cross-link analysis. Methods 45, 65–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamauchi M & Shiiba M Lysine hydroxylation and cross-linking of collagen. Post-translational Modifications of Proteins, 95–108 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Yamauchi M & Sricholpech M Lysine post-translational modifications of collagen. Essays in biochemistry 52, 113–133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller BW et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Mol Med 7, 1063–1076, doi: 10.15252/emmm.201404827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erler JT & Weaver VM Three-dimensional context regulation of metastasis. Clin Exp Metastasis 26, 35–49, doi: 10.1007/s10585-008-9209-8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y et al. Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. The Journal of clinical investigation 125, 1147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilkes DM et al. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Molecular cancer research 11, 456–466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Lampi MC & Reinhart-King CA Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials. Sci Transl Med 10, doi: 10.1126/scitranslmed.aao0475 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Grivennikov SI, Greten FR & Karin M Immunity, inflammation, and cancer. Cell 140, 883–899, doi: 10.1016/j.cell.2010.01.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack M Inflammation and fibrosis. Matrix Biol 68-69, 106–121, doi: 10.1016/j.matbio.2017.11.010 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Wick G et al. The immunology of fibrosis. Annu Rev Immunol 31, 107–135, doi: 10.1146/annurev-immunol-032712-095937 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ueha S, Shand FH & Matsushima K Cellular and molecular mechanisms of chronic inflammation-associated organ fibrosis. Front Immunol 3, 71, doi: 10.3389/fimmu.2012.00071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X et al. CCL2-driven inflammation increases mammary gland stromal density and cancer susceptibility in a transgenic mouse model. Breast Cancer Res 19, 4, doi: 10.1186/s13058-016-0796-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian BZ et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225, doi: 10.1038/nature10138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohela M et al. Intravital imaging reveals distinct responses of depleting dynamic tumor-associated macrophage and dendritic cell subpopulations. Proc Natl Acad Sci U S A 111, E5086–5095, doi: 10.1073/pnas.1419899111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruffell B et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 26, 623–637, doi: 10.1016/j.ccell.2014.09.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeNardo DG et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov 1, 54–67, doi: 10.1158/2159-8274.CD-10-0028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemkov T, D’Alessandro A & Hansen KC Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino acids 47, 2345–2357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oxlund H, Barckman M, Ørtoft G & Andreassen T Reduced concentrations of collagen cross-links are associated with reduced strength of bone. Bone 17, S365–S371 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Gruosso T et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest 129, 1785–1800, doi: 10.1172/JCI96313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finak G et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14, 518–527, doi: 10.1038/nm1764 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Salvador F et al. Lysyl oxidase-like protein LOXL2 promotes lung metastasis of breast cancer. Cancer Res, doi: 10.1158/0008-5472.CAN-16-3152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyre DR, Paz MA & Gallop PM Cross-linking in collagen and elastin. Annual review of biochemistry 53, 717–748 (1984). [DOI] [PubMed] [Google Scholar]
- 40.Lin EY, Nguyen AV, Russell RG & Pollard JW Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. Journal of Experimental Medicine 193, 727–739, doi:DOI 10.1084/jem.193.6.727 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strachan DC et al. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8(+) T cells. Oncoimmunology 2, e26968, doi: 10.4161/onci.26968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin EY et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163, 2113–2126, doi: 10.1016/S0002-9440(10)63568-7 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novak ML & Koh TJ Macrophage phenotypes during tissue repair. J Leukoc Biol 93, 875–881, doi: 10.1189/jlb.1012512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song E et al. Influence of alternatively and classically activated macrophages on fibrogenic activities of human fibroblasts. Cell Immunol 204, 19–28, doi: 10.1006/cimm.2000.1687 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK & Kitani A IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 12, 99–106, doi: 10.1038/nm1332 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Takaluoma K, Lantto J & Myllyharju J Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix biology 26, 396–403 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Yau C et al. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res 12, R85, doi: 10.1186/bcr2753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gyorffy B et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123, 725–731, doi: 10.1007/s10549-009-0674-9 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Noy R & Pollard JW Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61, doi: 10.1016/j.immuni.2014.06.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D & Semenza GL Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem 288, 10819–10829, doi: 10.1074/jbc.M112.442939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Wong CC et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci U S A 108, 16369–16374, doi: 10.1073/pnas.1113483108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natarajan S et al. Collagen Remodeling in the Hypoxic Tumor-Mesothelial Niche Promotes Ovarian Cancer Metastasis. Cancer Res 79, 2271–2284, doi: 10.1158/0008-5472.CAN-18-2616 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin EY et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res 66, 11238–11246, doi: 10.1158/0008-5472.CAN-06-1278 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Ahn Y, Sanderson BW, Klein OD & Krumlauf R Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development 137, 3221–3231, doi: 10.1242/dev.054668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 55.Hsiao EC et al. Osteoblast expression of an engineered Gs-coupled receptor dramatically increases bone mass. Proc Natl Acad Sci U S A 105, 1209–1214, doi: 10.1073/pnas.0707457105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guy CT, Cardiff RD & Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avery NC, Sims TJ & Bailey AJ Quantitative determination of collagen cross-links. Extracellular Matrix Protocols: Second Edition, 103–121 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Clasquin MF, Melamud E & Rabinowitz JD LC‐MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. Current Protocols in Bioinformatics, 14.11. 11–14.11. 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruffell B et al. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A 109, 2796–2801, doi: 10.1073/pnas.1104303108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finak G et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res 8, R58, doi: 10.1186/bcr1608 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Curtis C et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352, doi: 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berglund G, Elmstahl S, Janzon L & Larsson SA The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med 233, 45–51 (1993). [DOI] [PubMed] [Google Scholar]
- 65.Manjer J et al. The Malmo Diet and Cancer Study: representativity, cancer incidence and mortality in participants and non-participants. Eur J Cancer Prev 10, 489–499 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Cohen DA et al. Interobserver agreement among pathologists for semiquantitative hormone receptor scoring in breast carcinoma. Am J Clin Pathol 138, 796–802, doi: 10.1309/AJCP6DKRND5CKVDD (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene expression array and RNA-Seq data have been deposited in GEO and are publicly available. For the stroma and epithelium specific gene expression analysis, the breast cancer datasets from Finak et al. 2008 and Gruosso et al. 2019 have been used (GSE9014, GSE58644, GSE88715, and GSE88847)36,37. The association between PLOD2 and distant metastasis-free survival (DMFS) was determined using an online tool (http://xena.ucsc.edu) to download GEO data (GSE2034, GSE5327, and GSE7390) from 683 patients analyzed on Affymetrix U133A platform as described in Yau et al.47. The association between PLOD2 gene expression and relapse-free survival (RFS) was determined using an online tool (http://kmplot.com/analysis/) from 1,809 patients analyzed on Affymetrix platform (HGU133A and HGU133+2 microarrays)48. Affymetrix ID 202619 or 202620 were used for PLOD2 probes (2014 version) in these analyses. The cBioPortal for Cancer Genomics was used to access the publicly available METABRIC dataset to determine the levels of LOX, PLOD2, and LOXL2 gene expression in breast cancer patients segregated by ER and HER2 status and to assess gene expression associations of LOX, PLOD2, and LOXL2 with CD16363. Macrophage RNA-Seq data has been deposited into GEO (GSE157290).