Abstract
Objective:
To provide a large-scale assessment of serum protein dysregulations in diffuse cutaneous systemic sclerosis (dcSSc) and to investigate serum protein correlates of SSc fibrotic features.
Methods:
Baseline serum protein profile of 66 participants with dcSSc enrolled in the Scleroderma: Cyclophosphamide Or Transplant trial and 66 healthy, age- and gender-matched controls was investigated. A panel of 230 proteins, including several cytokines and chemokines was determined. Whole blood gene expression profiling in concomitantly collected samples was performed.
Results:
Mean disease duration was 2.3 years, all had interstitial lung disease (ILD), and none were treated with immunosuppressive agents at baseline visit. Ninety proteins were differentially expressed compared to controls. Similar to previous global skin transcript results, hepatic fibrosis, granulocyte and agranulocyte adhesion and diapedesis were the top over-represented pathways. Eighteen proteins correlated with modified Rodnan Skin Score (mRSS). Soluble EGFR was significantly down-regulated in dcSSc and showed the strongest negative correlation with mRSS and predicted its course, whereas Alpha 1 Antichymotrypsin was significantly up-regulated in dcSSc and showed the strongest positive correlation with mRSS. Furthermore, higher CA15.3 correlated with more severe ILD, based on lower forced vital capacity and higher high-resolution CT scores. Only 14 genes showed significant differential expression in the same direction in serum protein and whole blood RNA gene expression analyses.
Conclusions:
dcSSc has a distinct serum protein profile with prominent dysregulation of proteins related to fibrosis and immune cell adhesion/diapedesis. The differential expression for most serum proteins in SSc is likely to originate outside the peripheral blood cells.
Introduction
Systemic Sclerosis (SSc - scleroderma) is a complex autoimmune disorder in which vascular involvement, immune dysregulation with autoantibody production and fibrosis are the main pathological processes (1). As evident by a standardized mortality ratio of 3.5 (2), SSc is associated with a substantial mortality and disease burden. Development of effective treatment strategies for SSc has been hampered by an incomplete understanding of disease pathogenesis and the underlying molecular heterogeneity. Genome wide association studies (3,4) and whole genome microarray studies (5,6) have provided new insights into disease pathogenesis at the DNA and RNA levels. An interferon signature is the most prominent gene expression signature in SSc peripheral blood cells (5) and interferon inducible chemokines correlate with disease severity (7).
Although proteins are in closer proximity to disease pathogenesis than findings at the DNA/RNA level, data on large-scale examination of serum proteins are still scarce in SSc. Investigations performed on small panels of proteins, showed that chemokines, vascular growth factors and adhesion molecules such as interleukin 8 (IL-8), vascular endothelial growth factor (VEGF), angiopoietin 2 (ANGPT2), platelet endothelial cellular adhesion molecule-1 (PECAM-1) are markers of early disease and reflect the endothelial dysregulation in SSc (8–10). Interleukin 6 (IL-6) is related to lung fibrosis and might be predictive of disease progression in early SSc-interstitial lung disease (ILD) (11). Moreover, adipokines such as leptin and adiponectin correlate negatively with changes in SSc skin fibrosis (12–14).
Recently, a study performed with SOMAscan aptamer technology in 34 participants with diffuse cutaneous SSc (dcSSc) and 15 controls identified tumor necrosis factor-α, interferon-γ, transforming growth factor-β (TGF-β), and interleukin 13 (IL-13) as potential upstream regulators in SSc (15). Few other studies have recently investigated larger panels of serum proteins in SSc (16,17).
In the present study, we investigated an extended panel of 230 serum proteins in the baseline serum samples of individuals with dcSSc (18), enrolled in the Scleroderma: Cyclophosphamide Or Transplantation (SCOT) Trial in comparison to 1:1 matched unaffected controls in order to provide a hypothesis generating assessment of serum protein dysregulation and its clinical correlates in SSc (19). Moreover, availability of concomitantly collected whole blood RNA samples enabled direct comparison between SSc serum protein and whole blood gene expression profiles, showing the differential expression for most serum proteins in SSc is likely to originate outside the peripheral blood cells..
Materials and Methods
Selection of study population
Sixty-six of 75 individuals with dsSSc who participated in the SCOT trial had a baseline serum sample available for analysis. Baseline samples were examined in the present study. None of the participants were on immunosuppressive agents except for prednisone or its equivalent ≤ 10 mg per day during baseline blood sample collection. However, 27 participants had received immunosuppressive agents in the prior two months before baseline sample collection.
Briefly, the inclusion criteria included diffuse cutaneous involvement, lung or kidney involvement, a disease duration of less than 5 years (calculated from the onset of the first non-Raynaud symptom). Exclusion criteria included: significant prior treatment with cyclophosphamide (either oral or intravenous); presence of clinically significant rheumatic diseases other than SSc; any active uncontrolled infection or HIV, HCV, HBV infections. All SCOT participants provided informed consent and the SCOT protocol was approved by the Institutional Review Board of all participating institutions. Detailed information on the selection of participants and study design have been published previously (19).
In addition, serum from healthy controls 1:1 matched based on age- (+/- 10 years) and gender was investigated.
The severity of lung involvement was evaluated by forced vital capacity (FVC) % predicted (19). Standardized volumetric high-resolution computed tomography (HRCT) scan was performed and quantitative interstitial lung disease (QILD) score was measured, using an established algorithm, as another surrogate for ILD severity. The QILD score (expressed as a percentage) represents the sum of quantitative lung fibrosis, quantitative ground glass, and quantitative honeycomb for all lung lobes (20,21).
The modified Rodnan skin score (mRSS) was used to assess severity of skin involvement (22). The antibody profile was determined using commercial laboratories at each site.
Serum protein determination
Serum protein assays were performed by Myriad Rules-Based Medicine (Austin, TX, USA) using Human Discovery Multi-Analyte Profiling (MAP) (https://myriadrbm.com/scientific-media/multiplex-assay-development-white-paper/) V.2 multiplexed immune assay, which was the most comprehensive panel of proteins available with this technology at the time of study. This panel includes an extensive list of cytokines, chemokines, metabolic markers, hormones, growth factors, tissue remodelling proteins, angiogenesis markers, acute phase reactants and cancer markers that can reliably and reproducibly be measured with this technology. The levels of 228 serum proteins were determined with this assay.
All samples were stored at less than −70°C and had not been previously thawed. An aliquot of each sample was added to individual microsphere multiplexes of the selected MAP and blocker. After incubation, multiplexed cocktails of biotinylated reporter antibodies were added. Multiplexes were labelled using an excess of streptavidin-phycoerythrin solution. The resulting data were interpreted using proprietary software developed by Myriad Rule Based Medicine.
In addition, two cytokines considered to be pertinent to SSc pathogenesis, interleukin 10 (IL-10) and IL-6 were determined by ultra-sensitive Simoa Assays (Quanterix, Massachusetts, USA) (23).
For each assay, lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ) were determined by Myriad Rules-Based Medicine, representing the concentration at the lower and upper limit of the linear range of the standard curve, respectively (ie the lowest and highest amount of protein that can be detected accurately).
Statistical analysis
Proteins with levels below the LLOQ in more than 50% of baseline SSc samples were excluded from the analysis. A total of 182 proteins (79.1%) had detectable level in more than 50% of SSc samples. Out of these 182 proteins, 128 proteins (70.2%) had measurements above LLOQ in all samples. For the remainder of proteins, levels below the LLOQ were replaced by the LLOQ. Moreover, protein measurements above the ULOQ were replaced with ULOQ. As shown in Supplementary Figure 1, the raw values of majority of the serum proteins showed a right-skewed distribution, protein expression data were natural log-transformed to approximately conform to normality. Principal component analysis (PCA) was performed to identify outliers. T-tests were used to estimate differential expression between SSc and controls for each protein. P-values were corrected for multiple testing using the Benjamini-Hochberg method (24). Proteins with a false discovery rate (FDR) of <0.05 were considered to be differentially expressed in SSc vs. control comparison. Subsequently, the differentially expressed proteins were modelled using Ingenuity Pathway Analysis software (QIAGEN Inc., https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/) to identify the over-represented canonical pathways and to predict the activated upstream cytokines/growth factors. The goal of Upstream Regulator Analysis is to identify upstream regulators of a molecular profile and predict whether they are activated or inhibited. This analysis is based on expected causal effects between upstream cytokines/growth factors and targets; the expected causal effects are derived from the literature compiled in Ingenuity Knowledge Base (25). A Z score algorithm is used to make predictions. The primary purpose of the activation z‐score is to infer the activation states of predicted expression regulators. Given the observed differential regulation of a molecule (“up” or “down”) in the dataset, the activation state of an upstream regulator is determined by the regulation direction associated with the relationship from the regulator to the molecule. In practice, z‐scores greater than 2 or smaller than ‐2 can be considered significant.
For correlation with clinical variables (i.e. mRSS, FVC, HRCT QILD score), Pearson’s correlation was calculated and proteins that reached nominal significance level (p<0.05) in the univariable analysis and Pearson’s correlation ≥ 0.3 or ≤-0.3 were considered as significantly correlated. In this part, we did not account for multiple comparisons because this was a hypothesis generating analysis. Multivariable analyses after adjustment for age and gender were also performed. Partial correlation coefficients after adjustment for these demographic factors were also provided.
In an exploratory analysis, the predictive significance of serum proteins found to correlate with mRSS and FVC was examined for the serial measurement of mRSS and FVC obtained 3–14 months after randomization in the CYC (representing the active treatment period) and transplant arms, separately. For this analysis, mixed effect linear regression modelling was utilized after controlling for baseline disease severity (ie baseline mRSS or FVC) and time variable. The fixed effects were baseline serum protein, baseline mRSS or FVC, and timepoint (all as continuous). The random effects were intercept and timepoint. An unstructured correlation matrix was used.
Analyses were performed using RStudio Version 0.99.489 (RStudio Consortium, Boston, MA, USA) and SAS 9.4 (SAS Institute Inc., Cary, NC, USA).
Analysis of Gene expression and its relationship to serum proteins
Global gene expression studies on whole blood RNA samples from baseline for SCOT participants (stored in Tempus tubes) and from matched controls were examined using Illumina Human HT-12 bead arrays. All microarray experiments were performed in a single batch (for further details, please see (26)). Data were normalized according to the quantile method. A corresponding transcript was present on the microarray platform for the majority of investigated proteins. We focused on the transcripts that corresponded to the investigated proteins. Specifically, 282 transcripts corresponding to 172 examined proteins were identified. Only 10 (5.5%) of the 182 serum proteins did not have a matching probe on the microarray platform. For the transcript analysis, the data set was filtered by the list of corresponding 282 transcripts. Differentially expressed transcripts in the comparison of SSc to controls with an FDR < 0.1 were identified using two-sample t-test analysis. We used a less stringent FDR cut-off for the transcript than protein analysis (FDR <0.1 vs. <0.05) because the effect size (i.e. fold change) tends to be lower at the RNA level than protein level. For example, the fold changes in SSc to control comparison in the investigated 282 transcripts ranged from 0.61 to 1.41 while the fold change in the corresponding proteins ranged from 0.4 to 3.68. Subsequently, the list of differentially expressed transcripts was intersected with the list of differentially expressed proteins. The microarray analysis was performed with BRB-ArrayTools (National Cancer Institute, National Institutes of Health, USA) (27).
Interferome database search
We examined whether the differentially expressed serum proteins in SSc vs. control comparison were interferon inducible based on Interferome v 2.01 database (http://interferome.its.monash.edu.au/interferome/) (28). For the Interferome database, homo sapiens was chosen as species, lung, skin and blood were selected as organs. A list of Type I IFN inducible proteins was generated.
Results
Demographic data
Baseline demographic and clinical characteristics of investigated samples are presented in Table 1. As expected in diffuse disease, anti-topoisomerase (Scl-70) was the most common disease-specific autoantibody (39.4%), followed by anti-RNP antibodies (15.2%). All SCOT participants had signs of alveolitis on HRCT as evidenced by visual confirmation of ground glass opacity. The mean disease duration was 2.3 years.
Table 1:
Demographic and clinical characteristics of SCOT participants and controls
| Features | dcSSc (66) | Controls (66) | ||
|---|---|---|---|---|
| Age, mean years (SD) | 45.3 (10.3) | 46.3 (9.5) | ||
| Gender, Female n (%) | 40 (60.6%) | 40 (60.6%) | ||
| Race, n (%) | ||||
| Black | 6 (9.1%) | 9 (13.6%) | ||
| Asian | 3 (4.5%) | 0 (0 %) | ||
| Other | 6 (9.1%) | 0 (0 %) | ||
| White | 51 (77.3%) | 57 (86.4%) | ||
| Autoantibody, n (%) | Pos | Neg | Not Done | |
| ANA | 57(86.4%) | 7(10.6%) | 2(3%) | N.A. |
| ACA | 4(6.1%) | 56(84.8) | 6(9.1%) | N.A. |
| ATA | 26(39.4%) | 39(59.1%) | 1(1.5%) | N.A. |
| RNP | 10(15.2%) | 55(83.3%) | 1(1.5%) | N.A. |
| Disease duration mean (SD) | 2.31 (1.25) | N.A. | ||
| mRSS mean (SD) | 29.22 (9.35) | N.A. | ||
| FVC (%) mean (SD) | 74.62 (15.61) | N.A. | ||
| QILD score mean (SD) | 22.92 (11.63) | N.A. | ||
| QILD score >0, n (%) | 66 (100%) | N.A. | ||
Abbreviations: ANA: anti-nuclear antibodies; ACA: anti-centromere antibodies; ATA: anti-topoisomerase antibodies; RNP: anti-Ul ribonucleoprotein antibodies; mRSS: modified Rodnan skin score; FVC: forced vital capacity; QILD score: quantitative interstitial lung disease score; N: number; NA: not applicable; SD: Standard deviation
Serum proteins data
Ninety of 182 proteins were differentially expressed in the SSc versus control comparison with a FDR<0.05. Supplementary Figure 2 shows a heatmap of the 90 differentially expressed proteins in the baseline SCOT participant and healthy control samples. The ten most up- and down-regulated proteins based on the fold change are presented in Table 2. The complete list of ninety differential expressed proteins in SSc is available in Supplementary Table 1. The complete analysis results for all examined 182 serum proteins can be found Additional Data File at https://www.uth.tmc.edu/scleroderma/. The individual level protein data are available at ImmPort (https://www.immport.org).
Table 2:
Top up-/ down-regulated serum proteins in SCOT participants vs. controls comparison
| Protein name | Gene name | Fold Change | P raw | P FDR | Direction |
|---|---|---|---|---|---|
| Growth Hormone | GH1* | 3.69 | <0.001 | <0.001 | Upregulated |
| Ferritin | FTH1 | 3.04 | <0.001 | <0.001 | Upregulated |
| C-Reactive Protein | CRP | 2.98 | <0.001 | <0.001 | Upregulated |
| Chromogranin-A | CHGA | 2.77 | <0.001 | <0.001 | Upregulated |
| Macrophage inflammatory protein 3 beta | CCL19* | 2.48 | <0.001 | <0.001 | Upregulated |
| Monocyte Chemotactic Protein 1 | CCL2* | 2.48 | <0.001 | <0.001 | Upregulated |
| Myoglobin | MB | 2.38 | <0.001 | <0.001 | Upregulated |
| Monokine Induced by Gamma Interferon | CXCL9* | 2.30 | <0.001 | <0.001 | Upregulated |
| B Lymphocyte Chemoattractant | CXCL13* | 2.19 | <0.001 | <0.001 | Upregulated |
| Prolactin | PRL | 2.08 | <0.001 | <0.001 | Upregulated |
| Lactoylglutathione lyase | GLO1* | 0.49 | <0.001 | <0.001 | Down regulated |
| Neuron-Specific Enolase | ENO2* | 0.56 | 0.002 | 0.007 | Down regulated |
| Vitamin K-Dependent Protein S | PROS1* | 0.56 | 0.005 | 0.013 | Down regulated |
| Superoxide Dismutase 1 | SOD1 | 0.65 | <0.001 | <0.001 | Down regulated |
| Protein S100-A6 | S100A6* | 0.69 | 0.002 | 0.006 | Down regulated |
| Macrophage Migration Inhibitory Factor | MIF | 0.71 | 0.023 | 0.046 | Down regulated |
| Adiponectin | ADIPOQ | 0.72 | <0.001 | <0.001 | Down regulated |
| Kallikrein-7 | KLK7* | 0.73 | <0.001 | <0.001 | Down regulated |
| Insulin like Growth Factor Binding Protein 6 | IGFBP6 | 0.73 | <0.001 | <0.001 | Down regulated |
| Tetranectin | CLEC3B* | 0.75 | <0.001 | <0.001 | Down regulated |
Abbreviations: FDR: false discovery rate, FC: Fold Change. Values>1 refer to up-regulated expression of proteins and values<1 refer to down-regulated expression of proteins in relation to the value observed in controls. For example: FC=2.30 is equivalent to an increase of 130% from the reference value. FC=0.65 is equivalent to a decrease of 35% from the reference value.
Indicates interferon inducible proteins.
As shown in the Supplementary Figure 3, principal component analysis (PCA) identified only one outlier. The PCA showed that the majority of SCOT participants had a different serum protein profile than controls. Furthermore, the 27 individuals who received immunosuppressive therapy two months prior to sample collection did not group separately from other SCOT participants. Supplementary Tables 2 and 3 show the demographic/clinical characteristic, as well as list and duration of prior immunosuppressive treatment in SCOT participants dichotomized based on whether they were treated with immunosuppressive agents two months prior to sample collection.
Prominent role of profibrotic and granulocyte/agranulocyte extravasation pathways in SSc serum profile
An Ingenuity Canonical Pathway Analysis of differentially expressed serum proteins in SSc vs. control revealed hepatic fibrosis, granulocyte adhesion and diapedesis, and agranulocyte adhesion and diapedesis as the top three over-represented pathways. Of note, the same three pathways were found to be the top dysregulated pathways in our previously published global gene expression study of SSc skin (29). Interestingly, the top over-represented Canonical pathways in the concomitantly collected whole blood RNA samples were antigen presentation, interferon, and natural killer cell pathways. The complete list of over-represented canonical pathways in both datasets is shown in the Additional Data File at https://www.uth.tmc.edu/scleroderma/.
As shown in Figure 1, the top predicted activated upstream cytokines/growth factors (based on an activation Z-score >2) for the observed SSc serum protein profile included prominent pro-fibrotic proteins such as oncostatin (OSM), IL-6, interleukin-18 (IL-18), interleukin-4 (IL-4), interleukin-33 (IL-33), B cell activating factor (gene name: TNFSF13B) and Monocyte Chemotactic Protein 1 (gene name: CCL2).
Figure 1.
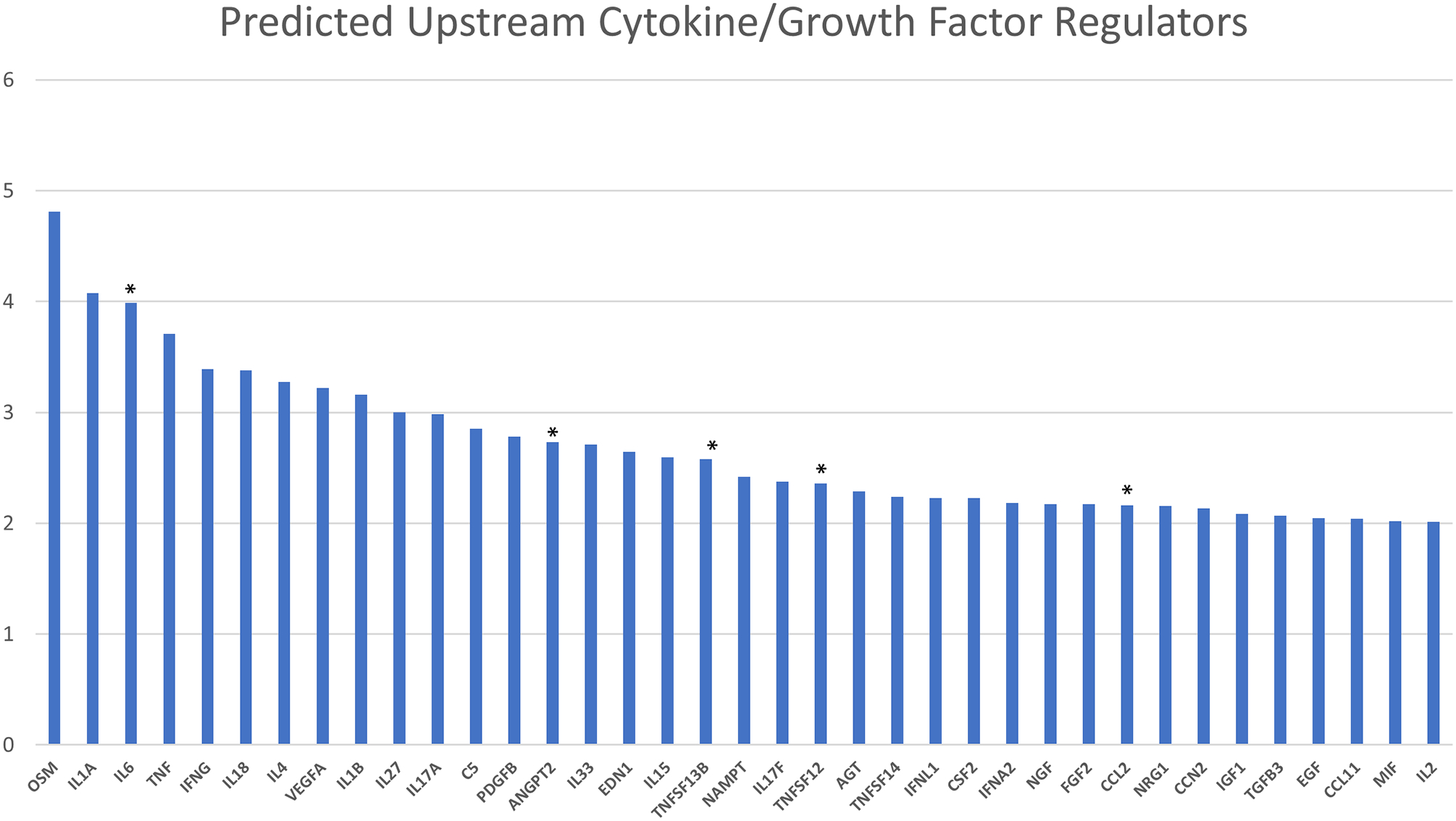
Top predicted upstream cytokine and growth factor regulators based on Ingenuity Knowledge Base.
* Proteins differentially expressed in dcSSc versus Controls.
Y axis: activation Z score calculated based on the Ingenuity Pathway Analysis for identifying upstream regulators (see Method section for further details).
Serum protein correlates with mRSS
Alpha 1 antichymotrypsin, NT proBNP, endostatin, osteopontin, tenascin and insulin growth factor binding protein 4 (IGFBP-4) were upregulated in SSc versus controls (p<0.05) (see Supplementary Table 1) and showed a positive correlation with mRSS (Table 3). Epidermal growth factor receptor (EGFR), Macrophage derived chemokine, kallikrein-5 and tetranectin were down-regulated in SSc vs. control comparison (p <0.05) (see Supplementary Table 1) and were negatively correlated with mRSS. Please see Table 3 for all serum proteins correlating with mRSS. Furthermore, the complete analysis results for all proteins can be found at https://www.uth.tmc.edu/scleroderma/.
Table 3:
Serum proteins correlating significantly with mRSS at baseline
| Protein name | Gene name | Correlation | Adjusted for age, gender | ||
|---|---|---|---|---|---|
| r | Pᵘ | Partial correlation | P | ||
| Alpha 1 Antichymotrypsin ↑ | SERPINA3 | 0.42 | 0.001 | 0.42 | 0.001 |
| NT proBNP ↑ | NPPB | 0.38 | 0.002 | 0.38 | 0.002 |
| Endostatin ↑ | COL18A1 | 0.37 | 0.002 | 0.38 | 0.002 |
| Osteopontin ↑ | SPP1 | 0.34 | 0.006 | 0.38 | 0.002 |
| Angiopoietin 2 ↑ | ANGPT2 | 0.33 | 0.005 | 0.33 | 0.007 |
| Serum Amyloid P Component | APCS | 0.32 | 0.008 | 0.32 | 0.010 |
| Tenascin C ↑ | TNC | 0.31 | 0.011 | 0.32 | 0.010 |
| Alpha 1 Microglobulin | AMBP | 0.31 | 0.011 | 0.32 | 0.012 |
| Insulin like Growth Factor Binding Protein 4 ↑ | IGFBP4 | 0.30 | 0.018 | 0.34 | 0.009 |
| Interleukin 22 | IL22 | −0.30 | 0.016 | −0.29 | 0.020 |
| Hepatocyte Growth Factor receptor | MET | −0.30 | 0.016 | −0.31 | 0.012 |
| Tetranectin ↓ | CLEC3B | −0.31 | 0.011 | −0.34 | 0.007 |
| Kallikrein 5 ↓ | KLK5 | −0.33 | 0.007 | −0.33 | 0.007 |
| Urokinase type Plasminogen Activator | PLAU | −0.34 | 0.005 | −0.35 | 0.005 |
| Thymus and activation regulated chemokine | CCL17 | −0.35 | 0.003 | −0.36 | 0.003 |
| Tamm Horsfall Urinary Glycoprotein | UMOD | −0.40 | 0.001 | −0.42 | 0.001 |
| Macrophage Derived Chemokine ↓ | CCL22 | −0.43 | <0.001 | −0.44 | <0.001 |
| Epidermal Growth Factor Receptor ↓ | EGFR | −0.43 | <0.001 | −0.43 | 0.001 |
Abbreviations: r: Pearson correlation coefficient; Pᵘ: p value from univariable model; P: p value from multivariable model after adjustment for age and gender
Proteins differentially expressed and upregulated in dcSSc vs. controls;
Proteins differentially expressed down regulated in dcSSc vs. controls
In an exploratory analysis, the predictive significance of serum proteins listed in Table 3 for the course of mRSS 3 to 14 months after randomization in CYC (representing active treatment period, sample size=32) and transplants arms (sample size=30) was investigated. As shown in Supplementary Table 4, baseline serum NT proBNP and Angiopoietin 2 levels predicted higher subsequent mRSS (p=0.013 and p=0.038, respectively) while baseline serum EGFR and Kallikrein-5 levels predicted lower subsequent mRSS (p=0.034 and 0.003, respectively) in CYC arm after adjustment for baseline mRSS values. Similarly, EGFR and Kallikrein5 predicted lower subsequent mRSS in the transplant arm (p=0.019 and 0.004, respectively).
Serum protein correlates of ILD severity
Supplementary Tables 5 and 6 show the serum protein correlates of FVC and HRCT QILD score. Noteworthy, Carcinoma Antigen 15.3 (CA 15.3) and Growth Regulated alpha protein were associated with more severe involvement (ie lower FVC and higher HRCT QILD scores) in both analyses (Figure 2). As shown in Supplementary Table 7, none of the proteins correlating with FVC at the baseline visit predicted the course of FVC 3 to 14 months after randomization in both treatment arms (sample size= 32 for CYC and for transplant).
Figure 2.
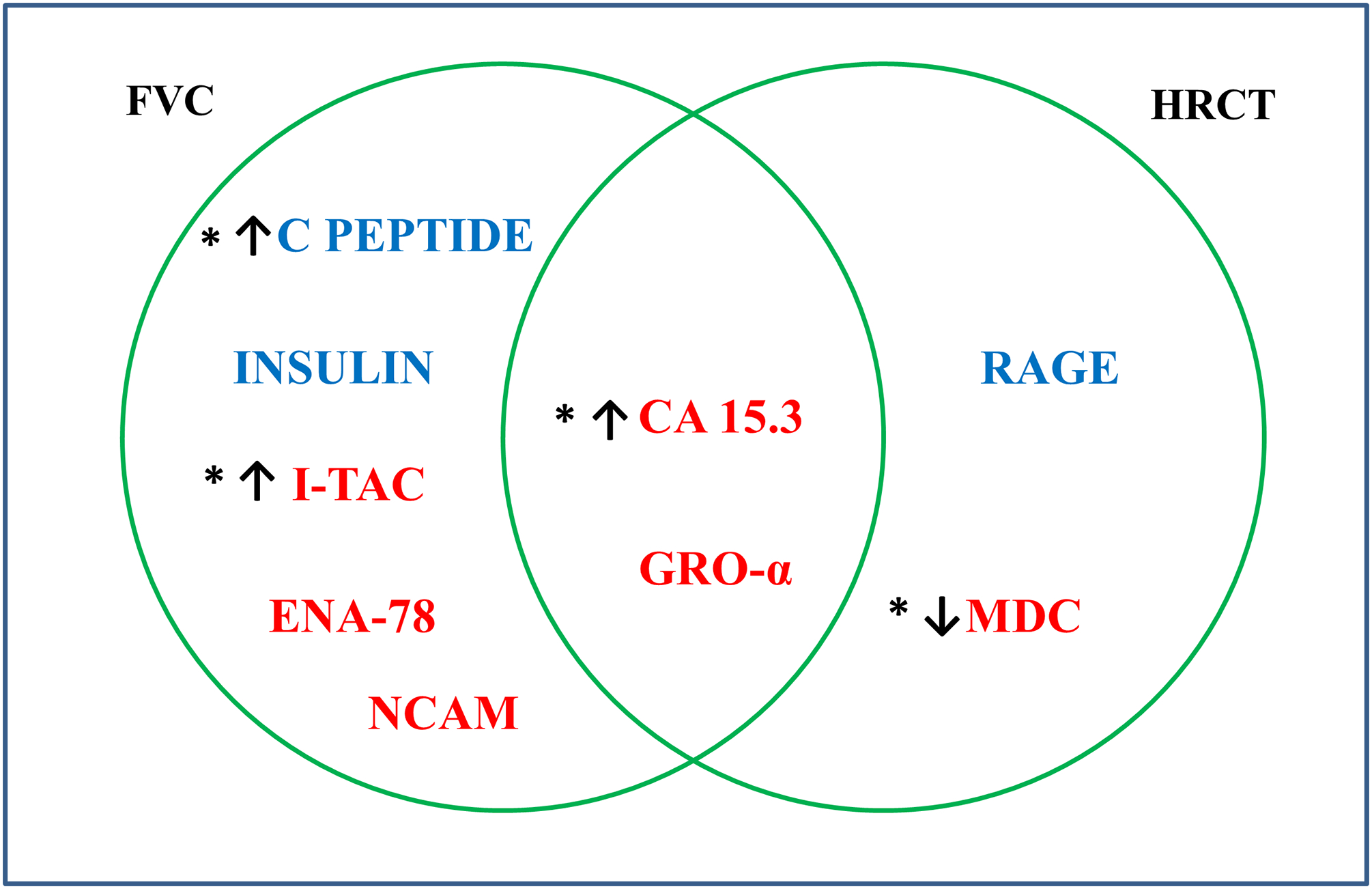
Venn diagram of serum proteins correlating with FVC and HRCT QILD score, blue font indicates association with better FVC and HRCT score and red indicates association with worse FVC and HRCT score. Abbreviations: NCAM: Neuronal Cell Adhesion Molecule; CA 15.3: Cancer Antigen 15.3; GRO-α: Growth Regulated alpha protein; I-TAC: Interferon inducible T cell alpha chemoattractant; ENA-78: Epithelial Derived Neutrophil Activating Protein78; RAGE: Receptor for advanced glycosylation end products; MDC: Macrophage Derived Chemokine.
* Proteins differentially expressed in dcSSc versus Controls. ↑ Proteins upregulated in dcSSc vs. controls. ↓ Proteins down regulated in dcSSc vs. controls
Whole blood Gene expression vs serum proteins
After filtering the whole blood gene expression data set by 282 transcripts corresponding to 172 examined proteins, we performed a comparison between SSc and control transcript profiles. A total of 52 transcripts were differentially expressed (FDR <0.1) in dcSSc versus controls. Among this list, 24 transcripts had a corresponding differentially expressed protein, of which only 17 transcripts (belonging to 14 genes) showed a differential expression in the concordant direction in its corresponding protein (Table 4). Of note,7 transcripts were differentially expressed in the opposite direction, indicating that the observed differential expression in the serum proteins does not stem from peripheral blood cells. Among concordantly expressed transcript-serum protein pairs, Interleukin 1β (IL-1β) was the only one with a decreased level at both whole blood gene expression and serum protein levels while the remainder were up-regulated in SSc at both levels. Moreover, endostatin was upregulated both at RNA transcript and protein levels and positively correlated with mRSS (Table 3). CA 15.3 was upregulated both at transcript and serum protein levels and positively correlated with lung fibrosis (see Supplementary Table 5 and 6).
Table 4:
Whole blood gene expression versus serum protein expression at baseline
| Analyte name | Gene name | Direction* | Whole blood gene expr | Serum proteins | ||||
|---|---|---|---|---|---|---|---|---|
| FC | P raw | P FDR | FC | P raw | P FDR | |||
| Interleukin 1 beta | IL1B | Down regulated | 0.71 | <0.001 | <0.001 | 0.89 | 0.015 | 0.032 |
| Alpha 1 Antitrypsin | SERPINA1 | Upregulated | 1.04 | 0.018 | 0.092 | 1.22 | <0.001 | 0.001 |
| Monokine Induced by Gamma Interferon | CXCL9 | Upregulated | 1.05 | 0.001 | 0.010 | 2.30 | <0.001 | <0.001 |
| Interleukin 2 receptor alpha | IL2RA | Upregulated | 1.05 | 0.017 | 0.091 | 1.97 | <0.001 | <0.001 |
| Cancer Antigen 15.3 | MUC1 | Upregulated | 1.06 | <0.001 | 0.006 | 1.67 | 0.001 | 0.004 |
| Tumor necrosis factor ligand superfamily member 13 | TNFSF13 | Upregulated | 1.08 | 0.001 | 0.008 | 1.23 | 0.017 | 0.037 |
| Tumor necrosis factor ligand superfamily member 13 | TNFSF13 | Upregulated | 1.1 | <0.001 | 0.005 | 1.23 | 0.017 | 0.037 |
| Cancer Antigen 15.3 | MUC1 | Upregulated | 1.11 | <0.001 | <0.001 | 1.67 | 0.001 | 0.004 |
| Monocyte Chemotactic Protein 1 | CCL2 | Upregulated | 1.12 | <0.001 | 0.001 | 2.48 | <0.001 | <0.001 |
| Interleukin 16 | IL16 | Upregulated | 1.15 | <0.001 | 0.005 | 1.08 | 0.019 | 0.040 |
| Lectin Like Oxidized LDL Receptor 1 | OLR1 | Upregulated | 1.15 | 0.006 | 0.040 | 1.38 | <0.001 | <0.001 |
| Endostatin | COL18A1 | Upregulated | 1.16 | <0.001 | 0.001 | 1.28 | <0.001 | <0.001 |
| B cell activating factor | TNFSF13B | Upregulated | 1.17 | 0.015 | 0.081 | 1.83 | <0.001 | <0.001 |
| B cell activating factor | TNFSF13B | Upregulated | 1.23 | 0.001 | 0.009 | 1.83 | <0.001 | <0.001 |
| Interferon gamma Induced Protein 10 | CXCL10 | Upregulated | 1.26 | <0.001 | 0.001 | 2.03 | <0.001 | <0.001 |
| Haptoglobin | HP | Upregulated | 1.29 | 0.001 | 0.008 | 1.71 | <0.001 | <0.001 |
| Myeloperoxidase | MPO | Upregulated | 1.41 | <0.001 | 0.006 | 1.84 | <0.001 | <0.001 |
Concordant direction of whole blood RNA vs serum protein expression
Abbreviations: FC: fold change; FDR: false discovery rate
Type I IFN inducible proteins
Among the ninety differentially expressed proteins in SSc, forty upregulated molecules are known to be Type I IFN inducible (44.4%) while ten downregulated proteins were Type I IFN inducible (11.1%) accordingly to the Interferome database search (Supplementary Table 1). MCP-1, Monokine Induced by Gamma Interferon (MIG), Interferon gamma Induced Protein 10 (IP-10), Interferon inducible T cell alpha chemoattractant (I-TAC) as well as CA 15.3 are among this list (see Supplementary Table 8).
Discussion
The present study represents a large-scale analysis of serum proteins in individuals with dcSSc in comparison to 1:1 matched healthy controls. Ninety differentially expressed proteins were identified among the 230 assayed by the utilized platform (Supplementary Table 1). Candidate proteins emerged from correlation analysis with mRSS, FVC and QILD score. Ingenuity Pathway Analysis revealed fibrosis and extravasation related pathways as the top over-represented biological processes. Lastly, transcripts and proteins showing differential expression at whole blood RNA and serum protein levels were identified, showing that only a small portion of differentially expressed serum proteins were also differentially expressed in concordant direction in the whole blood RNA samples.
In the present proteomic analysis the majority of early dcSSc participants showed a distinct serum protein profile from control samples and confirmed the presence of a prominent interferon signature in SSc (5,7,30). Among the 65 up-regulated serum proteins, forty were Type I IFN inducible proteins; ten chemokines (MCP-1, Macrophage Inflammatory Protein 1 beta, Monocyte Chemotactic Protein 2, Monocyte Chemotactic Protein 4 (MCP-4), Macrophage inflammatory protein 3 beta, Myeloid Progenitor Inhibitory Factor 1, MIG, IP-10, I-TAC and B Lymphocyte Chemoattractant) and thirty other proteins, including Osteopontin (SPP-1) and Beta 2 Microglobulin. Additionally, several Type I IFN inducible molecules were commonly dysregulated at both RNA and protein level (CXCL9/MIG, CCL2/MCP-1, MUC1/CA 15.3, IL16/Interleukin-16, CXCL10/IP-10, SERPINA1/alpha 1 antitrypsin, TNFSF13/Tumor necrosis factor ligand superfamily, OLR1/ Lectin Like Oxidized LDL Receptor 1 and TNFSF13B/ B cell activating factor). Of note, plasma IP-10 and I-TAC were previously found to be up-regulated in 266 individuals with early SSc enrolled in the GENISOS cohort and their levels correlated with the peripheral blood cell IFN gene expression score (7). Moreover, in a phase 1 open label clinical trial of Anifrolumab (an anti-IFNAR1 monoclonal antibody) conducted in 34 participants with SSc, SPP-1 correlated with IFN activity (whole-blood type I IFN gene signature score) while Beta 2 Microglobulin, IP-10 and MCP-4 were suppressed after treatment with Anifrolumab, supporting that these proteins are regulated by Type I IFN (16).
In our correlation analysis, serum soluble EGFR showed the strongest negative correlation with mRSS and was significantly down-regulated in SSc vs. control comparison. Soluble EGFR can inhibit the activation of its transmembrane receptor by binding EGF or by binding directly the transmembrane receptor itself which can perturb the EGF/EGFR cell signaling (31). Decreased soluble EGFR in SSc might lead to an activation of EGF pathways, as already described for some lung cancers subtypes (32). Indeed, a recent multi-cohort analysis of SSc skin transcriptome data across 7 datasets from 6 centers composed of 515 samples identified 6 positively correlated signaling proteins for the SSc signature, four of which were EGFR ligands (33). Our data provide further support for EGFR signaling as a potential driver of fibrosis in SSc skin. Of note, a correlation between serum soluble EGFR and lung FVC or QILD score was not observed.
Alpha 1 Antichymotrypsin showed the strongest positive correlation with mRSS in our study. This protein is an acute phase reactant produced by liver (34). Its biological function is to inhibit several serine proteases, mainly cathepsin G which is contained in the neutrophil granules and released at the site of inflammation. Notably, an excess of cathepsin G function, is linked to tissue damage (35). Moreover, two proteins mainly associated with SSc vascular manifestation, endostatin and NT-Pro-BNP also showed moderately positive correlation with mRSS. Endostatin is a peptide derived from the C terminus of collagen XVIII produced by fibroblasts with antiangiogenic properties. Previous studies have shown that endostatin was upregulated in SSc serum (36,37). Its antiangiogenic role suggests a feedback loop between endostatin and features of vascular impairment such as digital ulcers, pulmonary arterial hypertension (PAH) and scleroderma renal crisis (38–40). Vascular involvement and extensive skin involvement are not mutually exclusive. In fact, two previous studies have shown an association of serum endostatin levels with more extensive skin involvement (39,41). Of note, endostatin derived peptides showed antifibrotic properties and were able to prevent and reverse dermal TGF-β induced fibrosis in both ex vivo human skin and in vivo mice models (42). Similarly, NT-Pro-BNP has a more established link with vascular abnormalities in SSc, in particular with PAH, cardiac damage and mortality (43) but previous studies have also reported a positive correlation with mRSS (44–46).
In our correlative analyses with SSc-ILD features, the availability of both FVC and QILD score at the baseline visit in all SCOT participants enabled us to identify serum proteins that correlate with functional lung volume, as well as scleroderma related radiographic findings. CA 15.3 and Growth Regulated alpha protein (GRO-α) showed significant correlations with both FVC and QILD score in a clinically concordant direction but only CA 15.3 was also significantly up-regulated in SSc vs control comparison. CA 15.3 significantly correlated with lower FVC and higher QILD score on HRCT. CA 15.3 is a mucin encoded by the gene MUC1, which also encodes Krebs von den Lungen-6 (KL-6) protein. CA 15.3 is produced by epithelial cells, including type II pneumocytes, and is commonly used as a tumor marker in clinical practice for breast and ovarian cancer (47,48). In a previous retrospective study of 221 individuals with SSc, CA15.3 was useful in identifying those with significant ILD and correlated with decreased FVC and higher lung fibrosis scores (49). Of note, CA15.3 did not show a significant positive correlation with mRSS, underscoring its value as a lung specific marker. GRO-α is a neutrophil chemoattractant, consistent with our results, a previous study indicated that this protein was upregulated in SSc sera and was associated with lung impairment in SSc, correlating with decreased DLCO and FVC (50).
Building on the availability of concomitantly collected serum and whole blood RNA samples, we performed a direct comparison between these two sample types, showing that the differential expression for most proteins in SSc serum is most likely to originate outside peripheral blood cells. Our studies focused on serum and peripheral blood RNA which can be readily obtained and are practical sources of biomarker development during routine clinical care. The three over-represented pathways in SSc serum were exactly the same three over-represented pathways previously identified in our global SSc skin gene expression study (hepatic fibrosis, granulocyte adhesion and diapedesis, agranulocyte adhesion and diapedesis) (29). To further investigate this finding, we compared the list of 90 differentially expressed serum proteins with the differentially expressed transcripts in concomitantly collected peripheral blood cell RNA samples. This analysis yielded only 14 molecules that were differentially expressed in both sample types in the concordant direction. There were even 7 molecules that were differentially expressed in the opposite direction. These results support the notion that the source for the majority of differentially expressed serum proteins is likely to be outside of peripheral blood cells. In line with our findings, a recently published SOMAScan proteome analysis in two cohorts of 14 and 20 participants with SSc found that most of the differentially expressed serum proteins overlapped with two previously published SSc skin mRNA expression datasets (15). Prominently affected endorgans in SSc such as skin and lung are potential sources for the SSc serum protein signature, although it is possible that other organs such as liver are also contributing to the SSc protein profile.
The present study has some weaknesses. It is mainly hypothesis generating and does not include mechanistic experiments. Moreover, although we utilized the most comprehensive proteomic MAP panel provided by the RBM Myriad Rule Based Medicine at the time of study, we cannot provide a comprehensive view of proteomic dysregulations similar to genome wide genetic association and genome-wide gene expression studies due to technical limitations of the available proteomic assays. It is likely that a more comprehensive proteomic platform will lead to identification of additional candidate biomarkers. Furthermore, while we had access to concomitantly collected whole blood RNA samples, samples from affected end-organs (skin or lung) were not available in the SCOT trial. Moreover, an independent validation cohort was not included in the present study; future studies are needed to confirm the association of identified serum proteins with SSc fibrotic features.
This study also has several strengths. To our knowledge, the present study represents the largest serum protein study in SSc with validated and robust multiplex protein assays. We analyzed a well-characterized subset of dcSSc with early progressive fibrotic disease. SCOT participants were matched 1:1 for age and gender to controls in order to avoid the potential confounding effect of demographics and to generate sufficient power for identification of differentially expressed proteins. Moreover, the availability of FVC as well as QILD score on HRCT enabled us to identify serum proteins that correlate with lung function as well as the extent of radiographic involvement. Lastly, to our knowledge, the present study is the first study to directly compare large-scale SSc serum protein profile to the concurrently obtained whole blood transcriptome.
In conclusion, four important observations emerged from the present study. Namely, SSc serum samples showed a distinct proteomic profile in comparison to controls which includes an activation of prominent profibrotic cytokines. Moreover, an upregulation of several Type I IFN inducible proteins was observed, confirming previous genetic and gene expression studies, showing a prominent IFN signature in SSc. Furthermore, a direct comparison between the serum protein and peripheral blood gene expression profile indicated that the primary source for SSc serum proteomic profile lies outside peripheral blood cells. Lastly, we were able to identify serum protein correlates of mRSS and ILD severity suggesting EGFR, Alpha 1 Antichymotrypsin, and CA 15.3 as candidate proteins for future mechanistic studies in SSc.
Supplementary Material
Financial support:
The SCOT study was supported by awards from the NIAID, NIH to Duke University, the study contract holder (N01-AI05419 and HHSN 272201100025C).
The study was also supported by grants from Karen Brown scleroderma Foundation, NIH NIAMS (P30-AR061271), NIH R01AR073284, NIH UL1-TR000371 and Department of Defense (W81XWH-16-1-0296). None of the authors have a significant conflict of interest.
References
- 1.Varga J, Trojanowska M, Kuwana M. Pathogenesis of systemic sclerosis: recent insights of molecular and cellular mechanisms and therapeutic opportunities. J Scleroderma Relat Disord. 2017;2:137–52. [Google Scholar]
- 2.Elhai M, Meune C. Original article Trends in mortality in patients with systemic sclerosis over 40 years: a systematic review and meta-analysis of cohort studies. Rheumatology (Oxford) 2012;51:1017–26. [DOI] [PubMed] [Google Scholar]
- 3.Radstake TRDJ, Gorlova O, Rueda B, Martin J-E, Alizadeh BZ, Palomino-Morales R, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allanore Y, Saad M, Dieudè P, Avouac J, Distler JHW, Amouyel P, et al. Genome-Wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7: e1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assassi S, Mayes MD, Arnett FC, Gourh P, Agarwal SK, Mcnearney TA, et al. Systemic Sclerosis and Lupus Points in an Interferon-Mediated Continuum. Arthritis Rheum 2010;62:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitfield ML, Finlay DR, Murray JI, Troyanskaya OG, Chi J-T, Pergamenschikov A, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100:12319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Mayes MD, Tan FK, Wu M, Reveille JD, Harper BE, et al. Correlation of interferon-inducible chemokine plasma levels with disease severity in systemic sclerosis. Arthritis Rheum 2013;65:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandinelli F, Del Rosso A, Gabrielli A, Giacomelli R, Bartoli F, Guiducci S, et al. CCL2, CCL3 and CCL5 chemokines in systemic sclerosis: The correlation with SSc clinical features and the effect of prostaglandin E1 treatment. Clin Exp Rheumatol. 2012;30:S44–S49. [PubMed] [Google Scholar]
- 9.Cossu M, Andracco R, Santaniello A, Marchini M, Severino A, Caronni M, et al. Serum levels of vascular dysfunction markers reflect disease severity and stage in systemic sclerosis patients. Rheumatology (Oxford). 2016; 55:1112–6 [DOI] [PubMed] [Google Scholar]
- 10.Riccieri V, Stefanantoni K, Vasile M, Macrì V, Sciarra I, Iannace N, et al. Abnormal plasma levels of different angiogenic molecules are associated with different clinical manifestations in patients with systemic sclerosis. Clin Exp Rheumatol. 2011;29:S46–52. [PubMed] [Google Scholar]
- 11.De Lauretis A, Sestini P, Pantelidis P, Hoyles R, David M, Goh NSL, et al. Serum interleukin 6 is predictive of early functional decline and mortality in interstitial lung disease associated with systemic sclerosis. J Rheumatol 2013;40:435–446 [DOI] [PubMed] [Google Scholar]
- 12.Masui Y, Asano Y, Shibata S, Noda S, Aozasa N, Akamata K, et al. Serum adiponectin levels inversely correlate with the activity of progressive skin sclerosis in patients with diffuse cutaneous systemic sclerosis. J Eur Acad Dermatology Venereol. 2012;26:354–60. [DOI] [PubMed] [Google Scholar]
- 13.Fantuzzi G. Adiponectin and inflammation: Consensus and controversy. J Allergy Clin Immunol. 2008;121:326–30. [DOI] [PubMed] [Google Scholar]
- 14.Lakota K, Wei J, Carns M, Hinchcliff M, Lee J, Whitfield ML, et al. Levels of adiponectin, a marker for PPAR-gamma activity, correlate with skin fibrosis in systemic sclerosis: potential utility as biomarker? Arthritis Res Ther. BioMed Central Ltd; 2012;14:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice LM, Mantero JC, Stifano G, Ziemek J, Simms RW, Gordon J, et al. A Proteome-Derived Longitudinal Pharmacodynamic Biomarker for Diffuse Systemic Sclerosis Skin. J Invest Dermatol. 2017;137:62–70 [DOI] [PubMed] [Google Scholar]
- 16.Guo X, Higgs BW, Bay-Jensen AC, Karsdal MA, Yao Y, Roskos LK, et al. Suppression of t cell activation and collagen accumulation by an anti-ifnar1 mab, anifrolumab, in adult patients with systemic sclerosis. J Invest Dermatol. 2015;135:2402–9. [DOI] [PubMed] [Google Scholar]
- 17.Beirne P, Pantelidis P, Charles P, Wells AU, Abraham DJ, Denton CP, et al. Multiplex immune serum biomarker profiling in sarcoidosis and systemic sclerosis. Eur Respir J 2009;34:1376–82. [DOI] [PubMed] [Google Scholar]
- 18.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988; 15:202–5. [PubMed] [Google Scholar]
- 19.Sullivan KM, Goldmuntz EA, Keyes-Elstein L, McSweeney PA, Pinckney A, Welch B, et al. Myeloablative autologous stem-cell transplantation for severe scleroderma. N Engl J Med. 2018;378:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Li G, Gjertson D, Elashoff R, Shah SK, Ochs R, et al. Classification of parenchymal abnormality in scleroderma lung using a novel approach to denoise images collected via a multicenter study. Acad Radiol. 2008; 15:1004–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna D, Nagaraja V, Tseng C, Abtin F, Suh R, Kim G, et al. Predictors of lung function decline in scleroderma-related interstitial lung disease based on high-resolution computed tomography: implications for cohort enrichment in systemic sclerosis–associated interstitial lung disease trials. Arthritis Res Ther. 2015; 17:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clements P, Lachenbruch P, Siebold J, White B, Weiner S, Martin R, et al. Inter and intraobserver variability of total skin thickness score (modified Rodnan TSS) in systemic sclerosis. J Rheumatol. 1995; 22:1281–5. [PubMed] [Google Scholar]
- 23.Rivnak AJ, Rissin DM, Kan CW, Song L, Fishburn MW, Piech T, et al. A fully-automated, six-plex single molecule immunoassay for measuring cytokines in blood. J Immunol Methods. 2015; 424:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995. p. 289–300. [Google Scholar]
- 25.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assassi S, Wang X, Chen G, Goldmuntz E, Keyes-Elstein L, Ying J, et al. Myeloablation followed by autologous stem cell transplantation normalises systemic sclerosis molecular signatures. Ann Rheum Dis. 2019;78:1371–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 28.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. INTERFEROME v2.0: An updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:1040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol. 2015;67:3016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Isac E, Martin JE, Assassi S, Simeon CP, Carreira P, Ortego-Centeno N, et al. Brief report: IRF4 newly identified as a common susceptibility locus for systemic sclerosis and rheumatoid arthritis in a cross-disease meta-analysis of genome-wide association studies. Arthritis Rheumatol. 2016;68:2338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu A, Raghunath M, Bishayee S, Das1 M. Inhibition of tyrosine kinase activity of the epidermal growth factor (EGF) receptor by a truncated receptor form that binds to EGF: role for interreceptor interaction in kinase regulation. Mol Cell Biol. 1989; 9:671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lococo F, Paci M, Rapicetta C, Rossi T, Sancisi V, Braglia L, et al. Preliminary evidence on the diagnostic and molecular role of circulating soluble EGFR in non-small cell lung cancer. Int J Mol Sci. 2015; 16:19612–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lofgren S, Hinchcliff M, Carns M, Wood T, Aren K, Arroyo E, et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI insight. 2016;1:e89073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bode JG, Albrecht U, Häussinger D, Heinrich PC, Schaper F. Hepatic acute phase proteins – Regulation by IL-6- and IL-1-type cytokines involving STAT3 and its crosstalk with NF-κB-dependent signaling. Eur J Cell Biol. 2012;91:496–505. [DOI] [PubMed] [Google Scholar]
- 35.Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin). Front Biosci. 2007;12:2821–35. [DOI] [PubMed] [Google Scholar]
- 36.Delle Sedie A, Riente L, Maggiorini L, Pratesi F, Tavoni A, Migliorini P, et al. Potential biomarkers in patients with systemic sclerosis. Int J Rheum Dis. 2018;21:261–5. [DOI] [PubMed] [Google Scholar]
- 37.Distler O, Del Rosso A, Giacomelli R, Cipriani P, Conforti ML, Guiducci S, et al. Angiogenic and angiostatic factors in systemic sclerosis: increased levels of vascular endothelial growth factor are a feature of the earliest disease stages and are associated with the absence of fingertip ulcers. Arthritis Res. 2002;4:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiseter S, Molberg Ø, Gunnarsson R, Lund MB, Aalokken TM, Aukrust P, et al. Associations between circulating endostatin levels and vascular organ damage in systemic sclerosis and mixed connective tissue disease: an observational study. Arthritis Res Ther. 2015;17:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebbar M, Peyrat JP, Hornez L, Hatron PY, Hachulla E, Devulder B. Increased concentrations of the circulating angiogenesis inhibitor endostatin in patients with systemic sclerosis. Arthritis Rheum. 2000;43:889–93. [DOI] [PubMed] [Google Scholar]
- 40.Mecoli CA, Shah AA, Boin F, Wigley FM, Hummers LK. The utility of plasma vascular biomarkers in systemic sclerosis: a prospective longitudinal analysis. Arthritis Rheumatol. 2020; 10.1002/art.41265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farouk HM, Hamza SH, El Bakry SA, Youssef SS, Aly IM, Moustafa AA, et al. Dysregulation of angiogenic homeostasis in systemic sclerosis. Int J Rheum Dis. 2013;16:448–54. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi Y, Takihara T, Chambers RA, Veraldi KL, Larregina AT, Feghali-Bostwick CA. A peptide derived from endostatin ameliorates organ fibrosis. Sci Transl Med. 2012;4:136ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allanore Y, Komocsi A, Vettori S, Hachulla E, Hunzelmann N, Distler J, et al. N-terminal pro-brain natriuretic peptide is a strong predictor of mortality in systemic sclerosis. Int J Cardiol. 2016;223:385–9. [DOI] [PubMed] [Google Scholar]
- 44.Carlo-Stella N, Belloli L, Biondi ML, Marasini B. Serum N-terminal pro-brain natriuretic peptide, a marker of skin thickness in systemic sclerosis? Clin Rheumatol. 2009;28:241–2. [DOI] [PubMed] [Google Scholar]
- 45.Choi HJ, Shin YK, Lee HJ, Kee JY, Shin DW, Lee EY, et al. The clinical significance of serum N-terminal pro-brain natriuretic peptide in systemic sclerosis patients. Clin Rheumatol. 2008;27:437–42. [DOI] [PubMed] [Google Scholar]
- 46.Elshamy HA, Ibrahim SE, Farouk HM, Moustafa AA, Aly IM, Osman WM. N-terminal pro-brain natriuretic peptide in systemic sclerosis: new insights. Eur J Dermatol. 2011;21:686–90. [DOI] [PubMed] [Google Scholar]
- 47.Apostolopoulos V, Stojanovska L, Gargosky SE. MUC1 (CD227): a multi-tasked molecule. Cell Mol Life Sci. 2015;72:4475–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lakshmanan I, Ponnusamy MP, Macha MA, Haridas D, Majhi PD, Kaur S, et al. Mucins in lung cancer: diagnostic, prognostic, and therapeutic implications. J Thorac Oncol 2015;10:19–27. [DOI] [PubMed] [Google Scholar]
- 49.Celeste S, Santaniello A, Caronni M, Franchi J, Severino A, Scorza R, et al. Carbohydrate antigen 15.3 as a serum biomarker of interstitial lung disease in systemic sclerosis patients. Eur J Intern Med. European Federation of Internal Medicine; 2013;24:671–6. [DOI] [PubMed] [Google Scholar]
- 50.Furuse S, Fujii H, Kaburagi Y, Fujimoto M, Hasegawa M, Takehara K, et al. Serum concentrations of the CXC chemokines interleukin 8 and growth-regulated oncogene-alpha are elevated in patients with systemic sclerosis. J Rheumatol. 2003;30:1524–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


