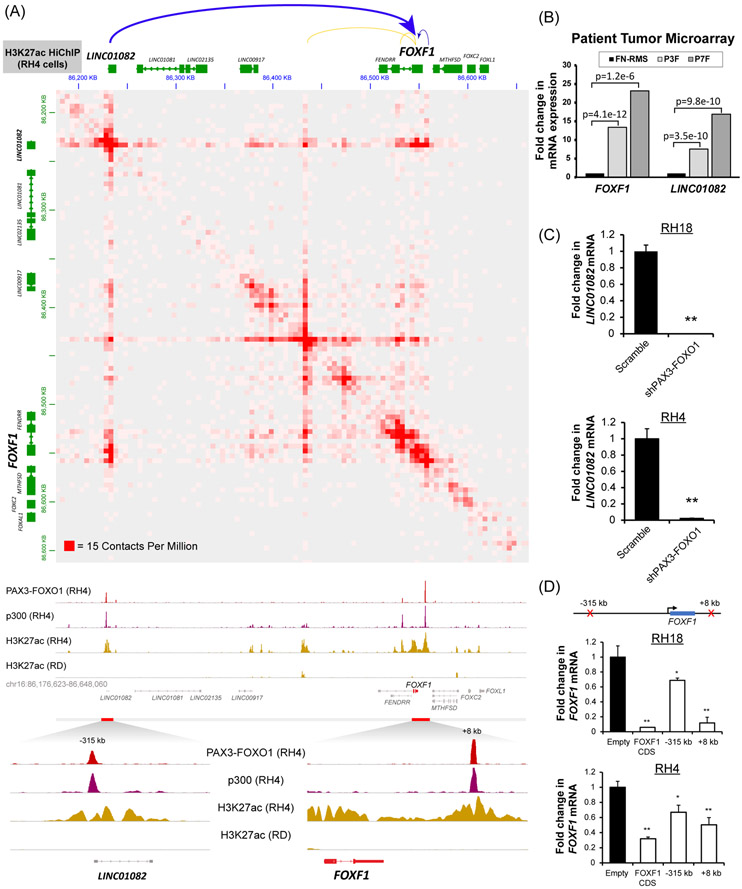
Figure 2. FOXF1 is a direct transcriptional target of the PAX3-FOXO1 protein.
(A) (top) HiChIP analysis shows long range chromatin interactions at the FOXF1 gene locus. Both the −315kb and the +8kb enhancers loop to the FOXF1 gene locus (top panel). H3K27ac HiChIP was performed using RH4 cells and is shown at 5kb resolution (GSE120770). ChIP-Seq shows binding of PAX3-FOXO1 fusion protein and p300 to DNA near the FOXF1 gene and LINC01082 in RH4 cells, but not in RD cells (bottom panel). Both enhancers are positive for H3K27ac. Data from GSE19063 and GSE83726. (B) Both the FOXF1 and the LINC01082 mRNA are increased in PAX3-FOXO1-positive (n=26) and PAX7-FOXO1-positive (n=7) RMS but not in fusion-negative FN-RMS (n=25). Data from microarray data GEO series accession number GSE66533 (80). (C) Depletion of PAX3-FOXO1 using shRNA decreased LINC01082 mRNA, shown by qRT-PCR using RH4 and RH18 cells. Values were normalized to β-actin (n=3). Data reported as mean ± SEM. (D) Repression of either −315kb upstream or +8kb downstream enhancers using CRISPRi decreased FOXF1 mRNA. RH4 and RH18 cells were transduced with a lentivirus containing a dCas9:KRAB and a gRNA targeting either −315kb upstream binding site or +8kb downstream binding site. The gRNA targeting the FOXF1 coding sequence (CDS) was used as a positive control. Values were normalized to β-actin (n=3). Data reported as mean ± SEM. * p<0.05; ** p<0.01.