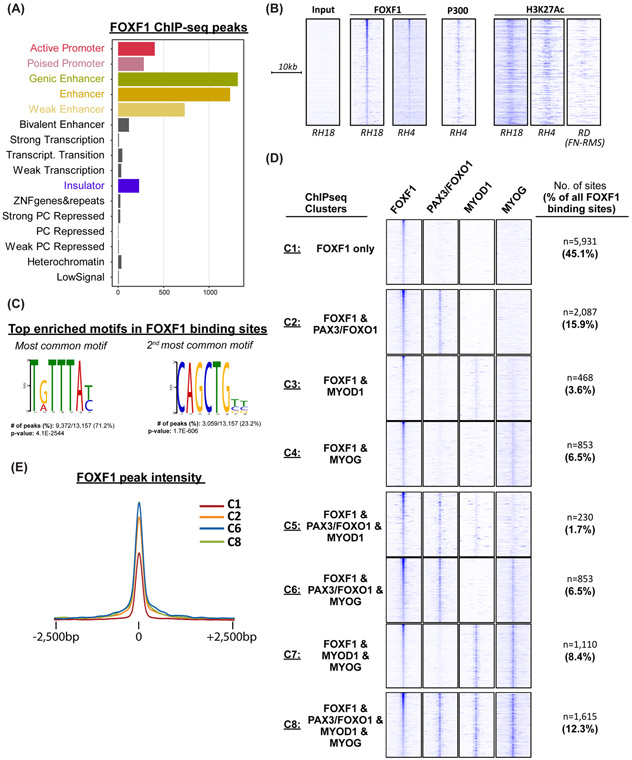
Figure 6. FOXF1 DNA-binding is associated with active enhancers and with binding of PAX3-FOXO1 and myogenic transcription factors.
(A) ChromHMM analysis of FOXF1 binding sites in fusion-positive RH18 cells. (B) FOXF1 binds to the enhancers that are active in FP-RMS but not in FN-RMS. Heatmaps of FOXF1, P300 and H3K27ac ChIPseq data in FP-RMS cell lines RH4, RH18 and FN-RMS cell line RD. (C) FOXF1 DNA-binding sites are enriched in Forkhead and E-box motifs. De novo motif analysis was performed using MEME-ChIP. (D) FOXF1 binding sites are shared with PAX3-FOXO1, MYOD1, and/or MYOG. The most common binding partner is PAX3-FOXO1. Out of all FOXF1 binding sites, 36% are also occupied by PAX3-FOXO1 (Cluster C2+C5+C6+C8). Each of the 13,157 FOXF1 binding sites were assigned to a single cluster based on the presence or absence of PAX3-FOXO1, MYOD1, and/or MYOG. Heatmaps are normalized for each transcription factor and the number of enhancers in each cluster are shown. (E) Tag density plot showing that FoxF1 signal is higher when co-bound with PAX3-FOXO1. Pairwise analysis of FOXF1 peak quality across clusters was performed. Enrichment of FOXF1 is highest in clusters C2, 6 and 8.