Abstract
The development of sequencing technology has expanded our knowledge of the human gastric microbiome, which is now known to play a critical role in the maintenance of homeostasis, while alterations in microbial community composition can promote the development of gastric diseases. Recently, carcinogenic effects of gastric microbiome have received increased attention. Gastric cancer (GC) is one of the most common malignancies worldwide with a high mortality rate. Helicobacter pylori is a well-recognized risk factor for GC. More than half of the global population is infected with H. pylori, which can modulate the acidity of the stomach to alter the gastric microbiome profile, leading to H. pylori-associated diseases. Moreover, there is increasing evidence that bacteria other than H. pylori and their metabolites also contribute to gastric carcinogenesis. Therefore, clarifying the contribution of the gastric microbiome to the development and progression of GC can lead to improvements in prevention, diagnosis, and treatment. In this review, we discuss the current state of knowledge regarding changes in the microbial composition of the stomach caused by H. pylori infection, the carcinogenic effects of H. pylori and non-H. pylori bacteria in GC, as well as the potential therapeutic role of gastric microbiome in H. pylori infection and GC.
Keywords: dysbiosis, gastric microbiome, gastric cancer, gastritis, Helicobacter pylori, peptic ulcers
Introduction
Gastric cancer (GC) is the third leading cause of cancer death after lung and colorectum cancers, and accounts for 782,685 deaths worldwide each year (Bray et al., 2018). Risk factors for GC include Helicobacter pylori infection, age, high salt intake, low fruit and vegetables intake, alcohol consumption, and smoking (Parsonnet et al., 1991; Jakszyn and Gonzalez, 2006; Anderson et al., 2010; Butt et al., 2019; Kumar et al., 2020; Li et al., 2021). The early stage of disease is asymptomatic or has non-specific symptoms (Luan et al., 2020); therefore, most patients are not diagnosed until an advanced stage.
Helicobacter pylori infection is common, affecting >50% of the global population with a higher incidence in developing countries (Hooi et al., 2017). The prevalence of H. pylori infection varies by age, ethnicity and living conditions (Seyda et al., 2007; Laszewicz et al., 2014; Alberts et al., 2020), and most cases occur in childhood (Banatvala et al., 1993). Only a small percentage of people develop pathological conditions related to H. pylori infection such as chronic gastritis, peptic ulcers, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (Wang et al., 2014). Chronic gastritis is the early presentation of persistent inflammation caused by H. pylori infection. As the condition progresses, injury to gastric epithelial cells can lead to the development of GC (Kidane, 2018). H. pylori has been listed as a Type I carcinogen by the World Health Organization (WHO) (International Agency for Research on Cancer, 1994). Therefore, detecting and eradicating H. pylori in the early phase of infection can prevent GC and other gastrointestinal diseases.
Microbiomes are complex microbial communities composed of bacteria, fungi, and viruses that reside in distinct habitats in the human body (Human Microbiome Project Consortium, 2012). The colon is among the most widely studied human microbial ecosystems as it contains the largest population of microorganisms (Villéger et al., 2019). The human microbiome and its metabolites have both physiologic and pathologic functions in homeostasis maintenance and disease development (Gilbert et al., 2018). In recent decades, there has been increasing interest in the relationship between the human microbiome and diseases. Despite the evidence that disruption the balance between microbiome and host in the stomach can promote the development of GC, the mechanism is not clearly understood.
To gain a better understanding of the relationship between the gastric microbiome and gastric carcinogenesis, here we provide an update on the gastric microbiome in healthy state and H. pylori-associated pathological conditions, including gastritis, peptic ulcer disease, and GC. Especially, we focus on the possible mechanisms of gastric microbiome in the development of GC. The potential therapeutic role of gastric microbiome in H. pylori infection and GC is also discussed.
The Human Gastric Microbiome
The Healthy Gastric Microbiome
The stomach was previously thought to be a sterile organ because of its strongly acidic environment. However, the discovery of H. pylori in the stomach of patients with gastritis and peptic ulcers by Marshall and Warren in 1982 refuted this notion (Marshall and Warren, 1984). Classical methods for studying the human gastric microbiome relied on microbiologic techniques including culture, isolation, and identification. However, as only a small number of gastric microorganisms can be grown under standard culture conditions, most microorganisms cannot be identified by this approach. The microorganisms most frequently isolated from the human stomach by culture-dependent methods were Veillonella, Lactobacillus, and Clostridium spp. (Zilberstein et al., 2007). Additionally, a large number of taxa have since been detected with newer technologies such as random shotgun sequencing, microarrays, and next-generation sequencing. The microbial load of the stomach is approximately 102–104 colony−forming units (CFU)/ml, which is much lower than that of the intestine (1010–1012 CFU/ml) (Delgado et al., 2013). Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria are the most highly represented phyla in gastric mucosa under normal conditions (Bik et al., 2006; Delgado et al., 2013; Liu et al., 2019a). Human gastric juice also has a diverse microbial community that is distinct from that of the gastric mucosa (Sung et al., 2016): the former is dominated by Firmicutes, Actinobacteria and Bacteroidetes, while the latter mainly includes Proteobacteria and Firmicutes (Bik et al., 2006; Nardone and Compare, 2015; Sung et al., 2016). Additionally, bacteria present in the oral cavity and duodenum such as Veillonella, Lactobacillus, and Clostridium can transiently colonize the stomach (Zilberstein et al., 2007; Nardone and Compare, 2015). Thus, the microbial community in gastric juice may not be representative of the gastric microbiome as a whole.
The specific mechanisms contributing to inter-individual variations in gastric microbiome composition are not well understood. Microbiome composition is affected by childbirth delivery mode (in infants), age, sex, ethnicity, diet, lifestyle, geography, use of antibiotics, use of proton pump inhibitors (PPI) or histamine H2 receptor antagonists, and the presence of H. pylori (Tsuda et al., 2015; Bokulich et al., 2016; Haro et al., 2016; Lloyd-Price et al., 2016; Yang et al., 2016; Nardone et al., 2017). The acidic environment of the healthy stomach prevents the overproliferation of bacteria and reduces the risk of infection (Howden and Hunt, 1987). Long-term treatment with PPI or H2 antagonists reduces gastric acid secretion, leading to bacterial overgrowth (Alarcón et al., 2017). Antibiotic usage, immunosuppression, and gastric fluid pH > 4 were found to be associated with reduced bacterial diversity in the stomach (Von Rosenvinge et al., 2013). Interestingly, a study of the gastric microbiome in twins showed that genetic background had no influence on gastric microbial community structure (Dong et al., 2017); similar findings were also reported for different niches of the human body (Lee et al., 2011; Lee J.E. et al., 2013).
Effects of H. pylori Infection on the Gastric Microbiome
The interactions between H. pylori and stomach-resident bacteria are not fully known. H. pylori is the predominant bacterium in the stomach of H. pylori-infected patients (Yu et al., 2017). However, low numbers of H. pylori have been detected by broad-range polymerase chain reaction and 16S rDNA sequence analysis in patients who were found to be negative for H. pylori infection by traditional methods such as histopathology, rapid urease test, serologic analysis, and culture (Bik et al., 2006). Thus, pyrosequencing has been used to define a cutoff value for H. pylori infection in human gastric samples (Kim et al., 2015).
Most H. pylori strains can modulate the gastric environment, thus altering the habitat of resident microorganisms (Mitchell et al., 2017). Alterations in gastric microbiome profile can increase the risk for developing GC (Liu et al., 2019a). A barcoded pyrosequencing analysis of 6 Swedish patients without or with H. pylori infection found that those who were H. pylori-negative had a more diverse gastric microbiome than patients testing positive (Andersson et al., 2008). In another study, children infected with H. pylori had a lower alpha diversity than H. pylori-negative children (Llorca et al., 2017). It was also reported that eradication of H. pylori increased microbial diversity in the stomach (Li et al., 2017). An examination of patients at different histologic stages of gastric carcinogenesis (gastritis, gastric intestinal metaplasia, and GC) revealed an inverse relationship between H. pylori abundance and microbial diversity in non-cancer gastric biopsies, but GC was associated with a lower diversity compared to other samples with similar H. pylori abundance; the difference was abrogated by antibiotic treatment (Li et al., 2017). A pyrosequencing analysis of the gastric microbiome demonstrated that Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria were the major phyla in both H. pylori-negative and -positive patients (Jo et al., 2016). Similarly, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were the most abundant phyla in a pediatric population (51 pediatric patients; 18 positive and 33 negative for H. pylori), but their relative proportions differed between H. pylori-positive vs. -negative patients (Llorca et al., 2017). Another study found that Proteobacteria, Spirochetes, and Acidobacteria were highly represented in H. pylori-positive patients whereas Actinobacteria, Bacteroidetes, and Firmicutes were detected at low levels (Maldonado-Contreras et al., 2011).
Helicobacter pylori Infection and Gastric Microbiome in Gastric Diseases
Gastritis
Chronic atrophic gastritis is a gastric premalignant condition (Sugano et al., 2015). Most cases of chronic atrophic gastritis are caused by persistent H. pylori infection (Sipponen and Maaroos, 2015), which triggers an inflammatory response that has various effects on gastric epithelial cells such as disruption of the gastric barrier (Gajewski et al., 2016), induction of apoptosis (Wan et al., 2016), and stimulation of proinflammatory cytokine secretion (Peek et al., 1995). Microbial composition in the stomach was shown to be altered in patients with chronic atrophic gastritis, with Helicobacteraceae, Streptococcaceae, Fusobacteriaceae, and Prevotellaceae as the major taxa (Parsons et al., 2017). Moreover, the abundance of Tannerella, Treponema, and Prevotella spp. was shown to be reduced in atrophic gastritis patients compared to healthy controls (Zhang et al., 2019), and a recent study showed that several pathways were significantly altered in patients with H. pylori-induced atrophic gastritis, including underrepresented (e.g., succinate dehydrogenase and tagaturonate reductase) and overrepresented (e.g., fumarate reductase, ketol-acid reductor isomerase, glycolate oxidase, and alanine dehydrogenase) pathways (Parsons et al., 2017). H. pylori infection stimulates the production and release of proinflammatory factors (Outlioua et al., 2020): gastric mucosal interleukin (IL)-8 level was correlated with the severity of the H. pylori-induced atrophic gastritis (Xuan et al., 2005; Lee K.E. et al., 2013); serum tumor necrosis factor (TNF)-α level was found to be related to the degree of chronic inflammation and neutrophil infiltration (Siregar et al., 2015); and serum vascular endothelial growth factor (VEGF) level was linked to the severity of gastric lesions in patients with gastritis (Siregar et al., 2015).
Peptic Ulcer Disease
Peptic ulcer disease is defined as acid-induced peptic injury of the mucosa reaching the submucosa (Lanas and Chan, 2017). Most cases of peptic ulcer disease are caused by H. pylori infection and use of non-steroidal anti-inflammatory drugs (NSAIDs), and other risk factors include gastrinoma, smoking, and use of other medications (Bernersen et al., 1996; Kavitt et al., 2019). H. pylori infection and the use of NSAIDs were shown to synergistically promote the development of peptic ulcer disease (Huang et al., 2002). However, only a small proportion of patients with H. pylori infection or who use NSAIDs progress to peptic ulcer disease, suggesting that decreased mucosal resistance to bacterial virulence factors and drug toxicity contribute to its pathogenesis (Lanas and Chan, 2017). H. pylori eradication therapy was shown to reduce the rate of recurrence (Tomita et al., 2002; Feder et al., 2018). A matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis showed that non-H. pylori bacteria were more abundant in patients with non-ulcer dyspepsia than in those with gastric ulcers, and the predominant non-H. pylori bacteria in H. pylori-positive patients were Streptococcus, Neisseria, Rothia, and Staphylococcus (Hu et al., 2012). Another study in Malaysian patients showed similar microbiome profiles in H. pylori-positive and -negative patients as well as an association between the isolation of Streptococci and peptic ulcer disease, suggesting that Streptococci colonize the stomach and that their interactions with H. pylori contribute to the development of peptic ulcer disease (Khosravi et al., 2014).
Gastric Cancer
Helicobacter pylori infection leads to persistent inflammation of gastric mucosa that causes changes in the cell cycle of gastric epithelial cells, which eventually result in atrophy of the glands, intestinal metaplasia, and GC (Correa, 1992). In addition to H. pylori, other microorganisms in the stomach have been implicated in gastric carcinogenesis (Table 1). Therefore, clarifying their distribution and functions can serve as a basis for the development of new therapeutic strategies.
TABLE 1.
Studies on dysbiosis of gastric microbiome in gastric cancer.
| Authors | Year | Sample size | Result |
| Aviles-Jimenez et al. | 2014 |
|
|
| Wang et al. | 2016 |
|
|
| Jo et al. | 2016 |
|
|
| Yu et al. | 2017 |
|
|
| Castaño-Rodríguez et al. | 2017 |
|
|
| Hsieh et al. | 2018 |
|
|
| Ferreira et al. | 2018 |
|
|
| Coker et al. | 2018 |
|
|
| Liu et al. | 2019a |
|
|
| Ling et al. | 2019 |
|
|
| Chen et al. | 2019 |
|
|
| Gunathilake et al. | 2019 |
|
|
| Gantuya et al. | 2020 |
|
|
| Wang Z. et al. | 2020 |
|
|
| Wang L. et al. | 2020 |
|
|
| Ravegnini et al. | 2020 |
|
|
Dysbiosis is defined as the compositional and functional alterations of the microbiome (Levy et al., 2017). In spite of the growing number of studies exploring the microbial dysbiosis during gastric carcinogenesis, there is still no consensus in terms of the alteration pattern of the gastric microbiome. There are some studies suggesting that the microbial diversity is markedly reduced in inflammatory diseases and cancer (Ahn et al., 2013; Aviles-Jimenez et al., 2014; Gong et al., 2016). Certainly, GC is no exception (Ferreira et al., 2018). However, some studies indicated that the richness and diversity of gastric microbiome in GC tissues were increased compared to control tissues (Castaño-Rodríguez et al., 2017; Chen et al., 2019). It has been reported that the bacterial richness and diversity decreased gradually from healthy control, through non-atrophic chronic gastritis, intestinal metaplasia, IN to GC (Wang Z. et al., 2020). However, another study showed that the bacterial diversity reduced gradually from normal, intestinal metaplasia, GC, gastritis to atrophy (Gantuya et al., 2020). Moreover, microbial diversity was found to be increased in advanced-stage GC compared to the early stage, which did not differ significantly from that in chronic gastritis (Wang L. et al., 2020). Novosphingobium, Ralstonia, Ochrobactrum, Anoxybacillus, and Pseudoxanthomonas were enriched in early GC whereas Burkholderia, Tsukamurella, Uruburuella, and Salinivibrio were more abundant in advanced as compared to early GC (Wang L. et al., 2020). However, another study reported that there was no significant difference in microbial community composition between early- and late-stage GC, while microbial richness decreased from normal to peritumoral to tumoral tissues (Liu et al., 2019a). Additionally, Prevotella copri and Bacteroides uniformis were reduced whereas Prevotella melaninogenica, Streptococcus anginosus, and Propionibacterium acnes were enriched in tumor tissue compared to normal and peritumoral tissues (Liu et al., 2019a). A recent study gave insight into the microbial composition in different subtypes of GC. Fusobacteria, Bacteroidetes, Patescibacteria were enriched in signet-ring cell carcinoma, whereas Proteobacteria and Acidobacteria were enriched in adenocarcinoma (Ravegnini et al., 2020).
Dysbiosis of the oral microbiome has been linked to inflammatory bowel disease, colorectal cancer, and pancreatic cancer (Nakatsu et al., 2015; Atarashi et al., 2017; Gaiser et al., 2019). The abundance of oral microbiota including Peptostreptococcus, Streptococcus, and Fusobacterium was shown to be higher in GC samples than in adjacent non-tumor samples (Chen et al., 2019). Similarly, an investigation of the gastric microbiome in superficial gastritis, atrophic gastritis, intestinal metaplasia, and GC by 16S rRNA gene sequencing revealed that oral bacteria such as Peptostreptococcus stomatis, S. anginosus, Parvimonas micra, Slackia exigua, and Dialister pneumosintes were enriched in GC compared to tissue samples from precancerous stages (Coker et al., 2018). It is possible that alterations in the acidic environment of the stomach in GC enable colonization by oral bacteria (Chen et al., 2019). However, further studies are needed to clarify the role of the oral microbiome in gastric carcinogenesis.
Lactobacillus is a major genus in the gut microbiome and is used to relieve various gastrointestinal conditions (Salazar-Lindo et al., 2007). Lactic acid production has several biologically important functions including immunomodulation and anti-inflammatory and anti-cancer effects (Han et al., 2015; Ghosh et al., 2019). However, lactic acid-producing bacteria also play a role in gastric carcinogenesis (Vinasco et al., 2019). A study conducted in Taiwan found that the abundance of Lactobacillus was increased in patients with GC compared to those with gastritis or intestinal metaplasia (Hsieh et al., 2018), which is partly consistent with other reports (Castaño-Rodríguez et al., 2017; Ferreira et al., 2018). Another study examining the abundance of lactic acid-producing bacteria in the tumor microenvironment reported that lactic acid-producing bacteria such as Lactococcus lactis and Lactobacillus brevis were enriched in adjacent non-tumor tissue (Chen et al., 2019). Animal experiment also revealed the potential carcinogenic role of Lactobacillales in GC (Bali et al., 2021).
Microbial interactions determine the microbiome homeostasis and influence the disease-associated microenvironment. The reduced complexity of the microbial interaction network in GC was attributed to the lower abundance of H. pylori (Liu et al., 2019a; Wang L. et al., 2020). Other studies have suggested that the interaction network is more complex in GC, which may be the result of reduced H. pylori abundance accompanied by an increased abundance of other microorganisms (Ferreira et al., 2018; Chen et al., 2019).
Mechanisms of Carcinogenesis
Mechanisms employed by gastric microbiota to promote gastric carcinogenesis include activation of inflammation, modulation of the host immune response, regulation of tumor growth and angiogenesis, production of microbial metabolites, and induction of DNA damage (Ou et al., 2019; Lee et al., 2020; Leite et al., 2020; Sierra et al., 2020).
Helicobacter pylori and Gastric Carcinogenesis
Helicobacter pylori is a Gram-negative spiral-shaped bacterium with urease, catalase, and oxidase activities (Kusters et al., 2006). It is thought that H. pylori colonized modern humans over 50,000 years ago and has evolved to resist the harsh acidic environment of the human stomach (Linz et al., 2007). The flagellum and spiral shape of H. pylori allow it to achieve primary infection and colonize the gastric mucosa (Sycuro et al., 2012; Gu, 2017). H. pylori urease transforms urea into ammonia to neutralize stomach acid (Scott et al., 2010). The functional and structural features of H. pylori allow it to traverse the gastric mucus, form a protected niche adjacent to the surface of the gastric epithelium, and deliver its products to host cells, which is critical for its survival in the human stomach and evasion of the host immune response, and for promoting disease development (Olofsson et al., 2014; Zhang et al., 2017; Pachathundikandi et al., 2019). Genetic diversity – a prominent feature of H. pylori strains that arises from point mutations and intra-/intergenomic recombination (Kraft and Suerbaum, 2005) – was shown to be correlated with the pathogenicity of H. pylori strains and influence the risk of malignant transformation (Yadegar et al., 2019).
Helicobacter pylori Virulence Factors
Helicobacter pylori strains have multiple virulence factors that directly or indirectly influence the risk of GC development (Ansari and Yamaoka, 2019). One of the most important of these virulence factors is VacA, which was initially identified by its ability to induce vacuolation in epithelial cells (Cover and Blaser, 1992). VacA is a multifunctional toxin that exhibits effects in different host cell types (e.g., gastric epithelial cells, antigen-presenting cells, phagocytic cells, mast cells, and T cells) (Supajatura et al., 2002; Torres et al., 2007; Manente et al., 2008; Altobelli et al., 2019). Aside from vacuolation, VacA impairs host gastric epithelial cells in a variety of ways – for example, by increasing mitochondrial membrane permeability, disrupting endocytic trafficking, and inducting apoptosis (Willhite and Blanke, 2004; Gauthier et al., 2005; Matsumoto et al., 2011). Additionally, VacA modulates the host immune response by inhibiting the activation and proliferation of immune cells and stimulating the production of proinflammatory cytokines (e.g., TNF-α and IL-6) by mast cells to promote the development of H. pylori-associated gastritis, peptic ulcer disease and GC (Supajatura et al., 2002; Torres et al., 2007).
Another virulence factor associated with the development of GC is CagA, which is encoded by the cagA gene located at one end of the cag pathogenicity island (cag PAI) (Hatakeyama, 2017). The cag PAI also encodes the type IV bacterial secretion system (T4SS) (Knorr et al., 2019), which forms a complex that delivers CagA from adherent H. pylori to host cells across the outer and inner bacterial membranes (Chung et al., 2019). After translocation, CagA can act on gastric epithelial cells to promote carcinogenesis by promoting inflammation, inducing proliferation, inhibiting apoptosis, disrupting cell–cell junctions, and causing the loss of cell polarity (Bagnoli et al., 2005; Buti et al., 2011, 2020; Yang et al., 2018).
Helicobacter pylori and Immune Response
Helicobacter pylori infection can stimulate both innate and adaptive immune responses (Bimczok et al., 2010; Freire de Melo et al., 2014). H. pylori violence factors activate the host immune response (Karkhah et al., 2019). The pattern recognition receptors (PRRs) of host cells recognize H. pylori pathogen-associated molecular patterns (PAMPs), triggering the initial stage of the innate immune response (Uno et al., 2007). Toll-like receptors (TLRs) are major components of PRRs that can bind to lipopolysaccharide, lipoproteins, lipoteichoic acid, double-stranded RNA, flagellin, unmethylated nucleic acids, and CpG repeats of H. pylori (Satoh and Akira, 2016). After recognizing PAMPs, TLRs activate NF-κB, interferon regulatory factor (IRF), and activator protein (AP)-1 to stimulate the expression of inflammatory mediators such as interferon (IFN)-γ, IL-1, IL-2, IL-6, IL-8, IL-12, and TNF-α (Kawasaki and Kawai, 2014; Nejati et al., 2018). Interestingly, H. pylori can evade the recognition by host PRRs in the innate immune response, thereby ensuring its long-term survival (Sun et al., 2013; Devi et al., 2015). In adaptive immunity, cluster of differentiation (CD)4 + T cells are the main mediators of the host immune response to H. pylori infection (Karkhah et al., 2019). CD4 + T cells were more abundant in GC samples compared to peritumoral and normal tissue samples, whereas CD8 + T cells showed the opposite distribution pattern (Huang et al., 2014). The release of inflammatory mediators induced by H. pylori virulence factors during the innate immune response activates T helper (Th)1/Th17 cell responses and stimulates the production of IFN-γ, IL-17, and TNF-α (Bimczok et al., 2010; Beigier-Bompadre et al., 2011). Thus, Th1/Th17 cells mediate the inflammatory response in H. pylori-infected patients (Bimczok et al., 2010; Beigier-Bompadre et al., 2011). Inflammation causes the loss of acid-secreting parietal cells, leading to an increase in pH in the stomach, which results in the decreased abundance of H. pylori and increased colonization by other bacteria (Pereira-Marques et al., 2019). H. pylori and the chronic inflammation that it induces enhance the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), which cause DNA damage such as point mutations and double-strand DNA breaks, dysregulate signal transduction pathways, and induce apoptosis or autophagy of gastric epithelial cells (Handa et al., 2010; Toller et al., 2011; Shimizu et al., 2017). The DNA repair system was found to be impaired in H. pylori-positive gastric epithelial cells (Han et al., 2020). Thus, H. pylori may promote gastric carcinogenesis by causing genetic instability in host cells. Additionally, ROS induce DNA mutations in H. pylori that allow it to adapt to the host environment (Gobert and Wilson, 2017).
In addition to its impact on effector T cells, H. pylori activates immunosuppressive responses in the host. Regulatory T cells (Tregs) inhibit aberrant or excessive immune responses that can be damaging to the host (Liu et al., 2015). Forkhead box protein (Foxp)3 + Tregs are the most important regulators of immune suppression (Deng et al., 2010). Mutation of Foxp3 is associated with severe autoimmune disease in both humans and animal models (Colobran et al., 2016; Ujiie and Shevach, 2016). An increased number of Foxp3 + Tregs was observed in tumor and peritumoral samples (Ling et al., 2019), and increased expression of Foxp3 in tumor-infiltrating Tregs suppressed T cell proliferation and was associated with tumor-node-metastasis stage in GC patients (Yuan et al., 2010). In H. pylori-positive individuals, the impaired immune response of CD4 + memory T cells to H. pylori antigens was rescued by depletion of Foxp3 + Tregs (Lundgren et al., 2003, 2005). Meanwhile, in mice infected with H. pylori, Treg deficiency resulted in severe gastric inflammation and reduced colonization by H. pylori (Rad et al., 2006). Foxp3 + Tregs can be subdivided into inducible costimulator (ICOS) + and ICOS-cells (Ito et al., 2008). ICOS + Foxp3 + Tregs secrete IL-10 and transforming growth factor (TGF)-β to suppress the function of dendritic cells (DCs) and T cells, respectively (Ito et al., 2008), and have been linked to poor clinical outcome in GC (Liu et al., 2019b). Thus, the current evidence suggests that Tregs enhance H. pylori-induced inflammation and promote the development of GC by suppressing host immune responses.
DCs function as a bridge between innate and adaptive immunity. The maturation status of DCs determines their immune function (enhancing immunity or promoting immunologic tolerance) and clinical outcomes in cancer (Karthaus et al., 2012). As one DCs subtype, plasmacytoid (p)DCs promote immunologic tolerance and tumor development (Hartmann et al., 2003; Perrot et al., 2007). The number of blood DC antigen (BDCA)2 + pDCs was found to be increased in tumor and peritumoral samples (Ling et al., 2019), which predicted a poor prognosis in patients with GC (Liu et al., 2019b). It has been suggested that tumor-infiltrating pDCs induce the activation and expansion of ICOS + Foxp3 + Treg cells to achieve immune suppression (Conrad et al., 2012).
Other Gastric Bacteria and Gastric Carcinogenesis
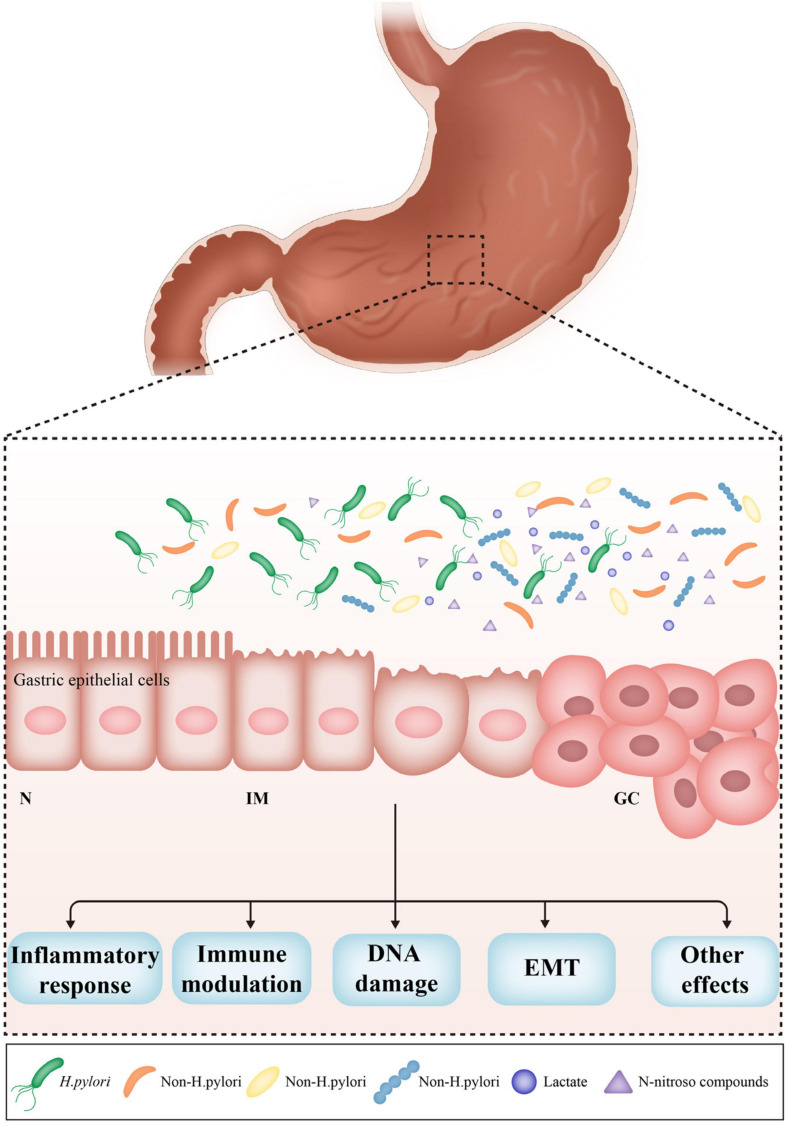
Non-H. pylori bacteria also contribute to gastric carcinogenesis. A prospective randomized controlled trial demonstrated that the incidence of GC was similar in patients receiving H. pylori eradication therapy vs. a placebo over a period of 7.5 years in a high-risk region of China (Wong et al., 2004). H. pylori monoassociation accelerated the progression of atrophic gastritis and gastrointestinal intraepithelial neoplasia (GIN) in germ-free insulin-gastrin mice, but induced less severe gastric lesions and delayed the onset of GIN compared to mice harboring a complex microbiome. These results indicate that some microorganisms in the stomach play a critical role in the development of GC (Figure 1). However, it is unclear which non-H. pylori bacteria dominate this process, and the pathogenic mechanisms have yet to be established.
FIGURE 1.
The effects of non-Helicobacter pylori bacteria on gastric carcinogenesis. H. pylori infection triggers the inflammatory response, which causes the loss of acid-secreting parietal cells, leading to the increased pH in the stomach. The alteration of acidic environment of the stomach allows colonization of other bacteria, subsequently results in dysbiosis of gastric microbiome. Non-H. pylori bacteria promote gastric carcinogenesis through their own characteristics and microbial metabolites, such as N-nitroso compounds and lactate. The main possible mechanisms include induction of inflammatory response, modulation of immune response, induction of DNA damage, and promotion of EMT. N, normal; IM, intestinal metaplasia; GC, gastric cancer; EMT, epithelial–mesenchymal transition.
Non-H. pylori Bacteria That Promote Gastric Carcinogenesis
A recent case-control study conducted in Korean investigating gastric microbiome profiles found that patients with GC had a higher abundance of P. acnes and P. copri than control subjects, suggesting that the presence of these species increases the risk of GC (Gunathilake et al., 2019). P. acnes and its products may trigger corpus-dominant lymphocytic gastritis via activation of the natural killer group 2 member D (NKG2D) system and secretion of the proinflammatory cytokine IL−15 (Montalban-Arques et al., 2016). In fact, NKG2D ligand (NKG2DL) was shown to be upregulated in tumor cells (Guerra et al., 2008), and the NKG2D–NKG2DL system and IL−15 have been implicated in carcinogenesis (Oppenheim et al., 2005). However, H. pylori is unable to activate NKG2D–NKG2DL system and IL−15 expression (Montalban-Arques et al., 2016). Prevotella is the predominant bacterial genus in the human gut microbiome (Gálvez et al., 2020). P. copri has a proinflammatory function in several diseases that involves enhancing resistance to host-derived ROS and producing the redox protein thioredoxin (Hofer, 2014). However, in a cohort of 276 Chinese patients, P. acnes abundance was increased whereas that of P. copri was decreased in GC tissue (Liu et al., 2019a). Given these incongruent findings, additional studies are needed to elucidate the role of P. copri in gastric carcinogenesis.
The gut microbiome can modulate the human immune system (Schluter et al., 2020), and the same is likely true for the gastric microbiome. A study involving 64 patients with GC found that BDCA2 + pDCs and Foxp3 + Tregs were more abundant in tumoral and peritumoral tissues than in normal tissues (Ling et al., 2019). Moreover, Stenotrophomonas and Selenomonas abundance was positively correlated with the number of BDCA2 + pDCs and Foxp3 + Tregs, respectively, whereas Comamonas and Gaiella abundance was negatively correlated with BDCA2 + pDCs and Foxp3 + Tregs numbers, respectively (Ling et al., 2019). Additionally, a serum microbiome analysis found that Comamonas was enriched in healthy controls relative to GC patients (Dong et al., 2019). This suggests that alterations in the gastric microbiome profile can modulate immune cell populations that contribute to the establishment of an immunosuppressive microenvironment (Ling et al., 2019). pDCs and Tregs have been reported to suppress antitumor immunity, enabling tumor cell evasion of immune surveillance mechanisms (Huang et al., 2014). There is little known about the interaction between Stenotrophomonas and human DCs, although Stenotrophomonas achieved immune escape by circumventing phagocytosis and stimulated the expression of TNF-α and IL-12 by DCs to promote inflammation (Roscetto et al., 2015). More research is required to clarify the mechanism of microbiome-driven immune modulation.
The increased abundance of Fusobacterium in patients with GC was shown to have diagnostic value (Hsieh et al., 2018). Fusobacterium nucleatum predicted worse prognosis in Lauren’s diffuse-type GC but not in intestinal-type GC (Boehm et al., 2020). Moreover, Fusobacterium sp. infection was positively correlated with the tumor-infiltrating lymphocytes and p53 expression in GC tissues (Nie et al., 2021). F. nucleatum has been detected in various diseases including appendicitis, inflammatory bowel disease, pancreatic cancer, and colorectal cancer (Swidsinski et al., 2011; Castellarin et al., 2012; Shaw et al., 2016; Del Castillo et al., 2019). However, the pathogenic mechanisms of F. nucleatum in GC are unknown. In colorectal cancer, interaction between the F. nucleatum adhesin FadA and E-cadherin of epithelial cells activated β-catenin and the Wnt signaling pathway (Rubinstein et al., 2013). F. nucleatum also drives the activation of NF-κB signaling and increased the expression of proinflammatory cytokines such as IL-1β, IL-6, IL-8, and TNF (Kostic et al., 2013; Brennan and Garrett, 2019). Outer membrane vesicles of F. nucleatum can interact with host epithelial cells to induce inflammatory responses and epithelial-mesenchymal transition (EMT) (Hashemi Goradel et al., 2019). Another F. nucleatum adhesin, fibroblast activation protein (Fap)2, can bind to T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT) receptor expressed by natural killer cells to block antitumor immune responses (Gur et al., 2015). A similar mechanism may be employed by F. nucleatum to promote the development of GC.
Microbial Metabolites of Non-H. pylori Bacteria That Promote Gastric Carcinogenesis
Nitrosating agents play an important role in gastric carcinogenesis (Correa, 1992). Humans are exposed to N-nitroso compounds (NOCs) from exogenous sources such as processed meat, smoked fish, and certain vegetables as well as from endogenous synthesis (Tsugane and Sasazuki, 2007). It is well-established that NOCs have carcinogenic effects. Epidemiologic studies have shown that patients with GC have higher NOC levels than healthy subjects (Walters et al., 1982; Xu et al., 2015). However, food contains only small amounts of nitrite, which is mainly derived from the reduction of nitrate to nitrite by oral bacteria in saliva (Hyde et al., 2014; Burleigh et al., 2018). After entering stomach, nitrite is converted to NOCs, which could be inhibited by ascorbic acid (Kobayashi, 2018). Several bacteria such as Veillonella, Clostridium, Haemophilus, Staphylococcus, Neisseria, Lactobacillus, and Nitrospirae contribute to gastric carcinogenesis by stimulating the production of NOCs (Wang et al., 2016; Zhang et al., 2019). Nitrosating or nitrate−reducing bacteria were found to be more abundant in GC patients than control subjects, although the difference between the 2 groups was not statistically significant (Jo et al., 2016). Higher nitrate and nitrite reductase activities associated with the microbiome were observed in GC than in chronic gastritis (Ferreira et al., 2018). Subtotal gastrectomy significantly altered the gastric microbiome profile and decreased the level of nitrate and nitrite reductases and the expression of genes involved in nitrosation (Tseng et al., 2016). These results indicate that changes in the gastric environment that reduce acid secretion leading to the growth of NOC-producing bacteria, thereby increasing the risk of gastric carcinogenesis.
The abundance of lactic acid bacteria was shown to be increased in patients with GC (Yu et al., 2017). Although a large number of literatures have discussed the protective effects of lactic acid bacteria against GC, it may increase the risk of GC through multiple mechanisms including increased production of ROS, NOCs, and lactate as well as induction of EMT and immune tolerance. In vitro and in vivo experiments have demonstrated that lactic acid bacteria stimulate the generation of ROS that cause DNA damage and enhance the formation of NOCs that induce mutagenesis, angiogenesis, and protooncogene expression and inhibit apoptosis (Jones et al., 2012; Vinasco et al., 2019). Lactic acid bacteria also increase the production of lactate; the levels of L- and D-lactate and lactate dehydrogenase were higher in patients with gastric carcinoma than in those with gastric ulcers and healthy controls (Armstrong et al., 1984). Lactate is a source of energy for cancer cells (Faubert et al., 2017) and plays a regulatory role in various aspects of carcinogenesis including tumor angiogenesis, immune escape, tumor cell migration, and metastasis (San-Millán and Brooks, 2017). Lactic acid bacteria can promote EMT – a process characterized by the loss of adherence junctions and polarity in epithelial cells and emergence of mesenchymal phenotypes that contribute to tumor invasion, migration, and metastasis (Yeung and Yang, 2017) – by inducing a state of multipotency (Vinasco et al., 2019). Finally, lactic acid bacteria promote the establishment of immune tolerance, allowing colonization by other carcinogenic bacteria (van Baarlen et al., 2009; Vinasco et al., 2019).
The role of the extragastric microbiome in gastric carcinogenesis has also been investigated using animal models. Colonization of mice with enterohepatic Helicobacter species prior to H. pylori infection influenced the severity of H. pylori-induced gastric lesions, suggesting that the interaction between H. pylori and the extragastric microbiome is an important mechanism underlying gastric carcinogenesis (Lemke et al., 2009; Ge et al., 2011).
Role of Bacteria in the Treatment of H. pylori Infection
Eradication of H. pylori infection is an effective strategy for reducing the risk of GC. The Maastricht V/Florence Consensus Report recommends PPI-clarithromycin-containing triple therapy as a first-line treatment for this purpose (Malfertheiner et al., 2017). Several studies have demonstrated that antibiotic treatment of H. pylori infection altered the composition of the gastric microbiome (Li et al., 2017; Zhang et al., 2019). However, given the increased rates of antibiotic resistance in H. pylori, there is an urgent need for novel H. pylori eradication strategies.
The Food and Agriculture Organization of the United Nations and the WHO define probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (Hill et al., 2014). Some probiotics prevent antibiotic-induced adverse effects, increase H. pylori eradication rates, and reduce fluctuations in the gut microbiome profile (Oh et al., 2016). One of the most widely studied probiotics is Lactobacillus, which is widely used in food production and clinical practice to balance the microbial ecosystem in the human gastrointestinal tract (Seddik et al., 2017). Some strains of Lactobacillus mitigate H. pylori infection by preventing its adhesion to epithelial cells, producing bacteriocins or organic acids, and suppressing mucosal inflammation (Gotteland et al., 2006; Yang and Sheu, 2012; Sakarya and Gunay, 2014). Lactobacillus acidophilus and Lactobacillus bulgaricus were shown to decrease the adhesion of H. pylori to gastric mucosal cells (Song et al., 2019), and L. bulgaricus inhibited IL-8 production by mucosal cells by modulating the TLR4/IκBα/NF-κB pathway (Song et al., 2019). Lactobacillus supplementation was effective in eradicating H. pylori infection in patients with chronic gastritis (Francavilla et al., 2008; Fang et al., 2019; Yu et al., 2019). Multi-strain probiotics can also increase H. pylori eradication rates and prevent adverse events (McFarland et al., 2016): a Lactobacillus- and Bifidobacterium-containing probiotic mixture was shown to exert beneficial effects against H. pylori, with a low incidence of side effects (Wang et al., 2013); and a combination of Bifidobacterium infantis, L. acidophilus, Enterococcus faecalis, and Bacillus cereus enhanced host immunity and reduced inflammation in GC patients who underwent gastrectomy (Zheng et al., 2019).
Role of Bacteria in the Treatment of GC
Conventional treatments for GC including surgery, chemotherapy, and radiation therapy are not very effective. Therefore, novel treatment strategies are needed. Although bacteria were traditionally regarded as carcinogenic, there is now available evidence for their anticancer properties. The role of the microbiome in cancer treatment was proposed as early as 1867, when it was reported that infection with Streptococcus pyogenes caused cancer remission in a patient (Sawant et al., 2020). Bacteria exert anticancer effects via multiple mechanisms including (i) colonizing tumors, (ii) releasing substances, (iii) suppressing nutrients required for tumor metabolism and proliferation, (iv) serving as a vehicle for anticancer drugs delivery, (v) forming biofilms, and (vi) enhancing host immunity (Soleimanpour et al., 2020; Yaghoubi et al., 2020).
Helicobacter pylori ribosomal protein (HPRP)-A1 and its enantiomer HPRP-A2 are 15-mer cationic peptides that are derived from the N terminus of the ribosomal protein L1 of H. pylori (Mai et al., 2015). HPRP-A1 and HPRP-A2 show strong antimicrobial and anticancer activities. HPRP-A1 is a membrane-active peptide that can disrupt the tumor cell membrane, and is thus often used to aid the delivery of other drugs to cancer cells (Zhao et al., 2015b). The KLA peptide exerts anticancer effects by inducing apoptosis via disruption of mitochondrial membranes, but has low membrane penetration (Hu et al., 2018). HPRP-A1 facilitates the entry of KLA peptide into cancer cells, which localizes to mitochondrial membranes to promote tumor cell death (Hao et al., 2019). HPRP-A2 induces the apoptosis of GC cells by enhancing ROS production; activating caspase-3, caspase-8 and caspase-9; reducing mitochondrial membrane potential; and causing cell cycle arrest in the G1 phase (Zhao et al., 2015a).
Besides HPRP-A1 and HPRP-A2, ieodoglucomides B – a glycolipopeptide isolated from the marine bacterium Bacillus licheniformis – has been demonstrated to have cytotoxic activity against stomach cancer cell lines (Tareq et al., 2012). Additionally, FW523-3 – a lipopeptide compound isolated from the culture broth of the marine bacterium Micromonospora chalcea – was shown to inhibit the proliferation of multiple cancer cells types including GC cells (Xie et al., 2011).
Conclusion
Growing evidence suggests the relationship between gastric microbiome and the development of GC. Changes of the gastric microbiome across different disease stages have been described. This is probably owed to the interactions between the gastric microbiota, environment, and host immune response. Although numerous studies investigating the carcinogenic mechanisms of H. pylori have been performed, limited progress has been made regarding the definite role of non-H. pylori in the development of GC over the past decades. Currently, only few studies have focused on the possible carcinogenic roles of non-H. pylori and their metabolites, including induction of inflammatory response, modulation of immune response, induction of DNA damage, and promotion of EMT. Therefore, further investigations are required to elucidate the detailed carcinogenic mechanisms of gastric microbiome to provide novel insights for the diagnosis, prevention, and treatment of GC.
Author Contributions
ZL and FJ designed the review and revised the manuscript. JY performed the literature search and wrote the manuscript. XZ performed the literature search and analyzed the literature. XL prepared the manuscript figure and revised the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This present work was funded by the grants of the National Natural Science Foundation of China (81972671, 81771724, 31700800, and 81790631) and the National S&T Major Project of China (2018YFC2000500).
References
- Ahn J., Sinha R., Pei Z., Dominianni C., Wu J., Shi J., et al. (2013). Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 105 1907–1911. 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón T., Llorca L., Perez-Perez G. (2017). Impact of the microbiota and gastric disease development by Helicobacter pylori. Curr. Top. Microbiol. Immunol. 400 253–275. 10.1007/978-3-319-50520-6_11 [DOI] [PubMed] [Google Scholar]
- Alberts C. J., Jeske R., de Martel C., den Hollander W. J., Michel A., Prins M., et al. (2020). Helicobacter pylori seroprevalence in six different ethnic groups living in Amsterdam: the HELIUS study. Helicobacter 25:e12687. 10.1111/hel.12687 [DOI] [PubMed] [Google Scholar]
- Altobelli A., Bauer M., Velez K., Cover T. L., Müller A. (2019). Helicobacter pylori VacA targets myeloid cells in the gastric lamina propria to promote peripherally induced regulatory T-cell differentiation and persistent infection. mBio 10:e0261-19. 10.1128/mBio.00261-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W. F., Camargo M. C., Fraumeni J. F. J., Correa P., Rosenberg P. S., Rabkin C. S. (2010). Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA 303 1723–1728. 10.1001/jama.2010.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A. F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S., Yamaoka Y. (2019). Helicobacter pylori virulence factors exploiting gastric colonization and its pathogenicity. Toxins 11:677. 10.3390/toxins11110677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. P., Dent D. M., Berman P., Aitken R. J. (1984). The relationship between gastric carcinoma and gastric juice lactate (L + D) and lactate dehydrogenase. Am. J. Gastroenterol. 79 675–678. [PubMed] [Google Scholar]
- Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., Narushima S., et al. (2017). Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science 358 359–365. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviles-Jimenez F., Vazquez-Jimenez F., Medrano-Guzman R., Mantilla A., Torres J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4:4202. 10.1038/srep04202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoli F., Buti L., Tompkins L., Covacci A., Amieva M. R. (2005). Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. U.S.A. 102 16339–16344. 10.1073/pnas.0502598102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali P., Coker J., Lozano-Pope I., Zengler K., Obonyo M. (2021). Microbiome signatures in a fast- and slow-progressing gastric cancer murine model and their contribution to gastric carcinogenesis. Microorganisms 9:189. 10.3390/microorganisms9010189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banatvala N., Mayo K., Megraud F., Jennings R., Deeks J. J., Feldman R. A. (1993). The cohort effect and Helicobacter pylori. J. Infect. Dis. 168 219–221. 10.1093/infdis/168.1.219 [DOI] [PubMed] [Google Scholar]
- Beigier-Bompadre M., Moos V., Belogolova E., Allers K., Schneider T., Churin Y., et al. (2011). Modulation of the CD4+ T-cell response by Helicobacter pylori depends on known virulence factors and bacterial cholesterol and cholesterol α-glucoside content. J. Infect. Dis. 204 1339–1348. 10.1093/infdis/jir547 [DOI] [PubMed] [Google Scholar]
- Bernersen B., Johnsen R., Straume B. (1996). Non-ulcer dyspepsia and peptic ulcer: the distribution in a population and their relation to risk factors. Gut 38 822–825. 10.1136/gut.38.6.822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik E. M., Eckburg P. B., Gill S. R., Nelson K. E., Purdom E. A., Francois F., et al. (2006). Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U.S.A. 103 732–737. 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimczok D., Clements R. H., Waites K. B., Novak L., Eckhoff D. E., Mannon P. J., et al. (2010). Human primary gastric dendritic cells induce a Th1 response to H. pylori. Mucosal Immunol. 3 260–269. 10.1038/mi.2010.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm E. T., Thon C., Kupcinskas J., Steponaitiene R., Skieceviciene J., Canbay A., et al. (2020). Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci. Rep. 10:16240. 10.1038/s41598-020-73448-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Chung J., Battaglia T., Henderson N., Jay M., Li H., et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8:343ra82. 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Garrett W. S. (2019). Fusobacterium nucleatum – symbiont, opportunist and Oncobacterium. Nat. Rev. Microbiol. 17 156–166. 10.1038/s41579-018-0129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh M. C., Liddle L., Monaghan C., Muggeridge D. J., Sculthorpe N., Butcher J. P., et al. (2018). Salivary nitrite production is elevated in individuals with a higher abundance of oral nitrate-reducing bacteria. Free Radic. Biol. Med. 120 80–88. 10.1016/j.freeradbiomed.2018.03.023 [DOI] [PubMed] [Google Scholar]
- Buti L., Ruiz-Puig C., Sangberg D., Leissing T. M., Brewer R. C., Owen R. P., et al. (2020). CagA-ASPP2 complex mediates loss of cell polarity and favors H. pylori colonization of human gastric organoids. Proc. Natl. Acad. Sci. U.S.A. 117 2645–2655. 10.1073/pnas.1908787117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti L., Spooner E., Van der Veen A. G., Rappuoli R., Covacci A., Ploegh H. L. (2011). Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc. Natl. Acad. Sci. U.S.A. 108 9238–9243. 10.1073/pnas.1106200108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt J., Varga M. G., Wang T., Tsugane S., Shimazu T., Zheng W., et al. (2019). Smoking, Helicobacter Pylori serology, and gastric cancer risk in prospective studies from China, Japan, and Korea. Cancer Prev. Res. 12 667–674. 10.1158/1940-6207.CAPR-19-0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodríguez N., Goh K.-L., Fock K. M., Mitchell H. M., Kaakoush N. O. (2017). Dysbiosis of the microbiome in gastric carcinogenesis. Sci. Rep. 7:15957. 10.1038/s41598-017-16289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M., Warren R. L., Freeman J. D., Dreolini L., Krzywinski M., Strauss J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22 299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-H., Wang A., Chu A.-N., Gong Y.-H., Yuan Y. (2019). Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front. Microbiol. 10:1261. 10.3389/fmicb.2019.01261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J. M., Sheedlo M. J., Campbell A. M., Sawhney N., Frick-Cheng A. E., Lacy D. B., et al. (2019). Structure of the Helicobacter pylori Cag type IV secretion system. eLife 8:e47644. 10.7554/eLife.47644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker O. O., Dai Z., Nie Y., Zhao G., Cao L., Nakatsu G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67 1024–1032. 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colobran R., Álvarez de la Campa E., Soler-Palacín P., Martín-Nalda A., Pujol-Borrell R., de la Cruz X., et al. (2016). Clinical and structural impact of mutations affecting the residue Phe367 of FOXP3 in patients with IPEX syndrome. Clin. Immunol. 163 60–65. 10.1016/j.clim.2015.12.014 [DOI] [PubMed] [Google Scholar]
- Conrad C., Gregorio J., Wang Y.-H., Ito T., Meller S., Hanabuchi S., et al. (2012). Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells. Cancer Res. 72 5240–5249. 10.1158/0008-5472.CAN-12-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P. (1992). Human gastric carcinogenesis: a multistep and multifactorial process–First American cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 52 6735–6740. [PubMed] [Google Scholar]
- Cover T. L., Blaser M. J. (1992). Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267 10570–10575. [PubMed] [Google Scholar]
- Del Castillo E., Meier R., Chung M., Koestler D. C., Chen T., Paster B. J., et al. (2019). The microbiomes of pancreatic and duodenum tissue overlap and are highly subject specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol. Biomarkers Prev. 28 370–383. 10.1158/1055-9965.EPI-18-0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., Cabrera-Rubio R., Mira A., Suárez A., Mayo B. (2013). Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb. Ecol. 65 763–772. 10.1007/s00248-013-0192-5 [DOI] [PubMed] [Google Scholar]
- Deng L., Zhang H., Luan Y., Zhang J., Xing Q., Dong S., et al. (2010). Accumulation of foxp3+ T regulatory cells in draining lymph nodes correlates with disease progression and immune suppression in colorectal cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 16 4105–4112. 10.1158/1078-0432.CCR-10-1073 [DOI] [PubMed] [Google Scholar]
- Devi S., Rajakumara E., Ahmed N. (2015). Induction of Mincle by Helicobacter pylori and consequent anti-inflammatory signaling denote a bacterial survival strategy. Sci. Rep. 5:15049. 10.1038/srep15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Xin Y., Wang L., Meng X., Yu X., Lu L., et al. (2017). Characterization of gastric microbiota in twins. Curr. Microbiol. 74 224–229. 10.1007/s00284-016-1176-8 [DOI] [PubMed] [Google Scholar]
- Dong Z., Chen B., Pan H., Wang D., Liu M., Yang Y., et al. (2019). Detection of microbial 16S rRNA Gene in the serum of patients with gastric cancer. Front. Oncol. 9:608. 10.3389/fonc.2019.00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H.-R., Zhang G.-Q., Cheng J.-Y., Li Z.-Y. (2019). Efficacy of Lactobacillus-supplemented triple therapy for Helicobacter pylori infection in children: a meta-analysis of randomized controlled trials. Eur. J. Pediatr. 178 7–16. 10.1007/s00431-018-3282-z [DOI] [PubMed] [Google Scholar]
- Faubert B., Li K. Y., Cai L., Hensley C. T., Kim J., Zacharias L. G., et al. (2017). Lactate metabolism in human lung tumors. Cell 171 358–371.e9. 10.1016/j.cell.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder R., Posner S., Qin Y., Zheng J., Chow S.-C., Garman K. S. (2018). Helicobacter pylori-associated peptic ulcer disease: a retrospective analysis of post-treatment testing practices. Helicobacter 23:e12540. 10.1111/hel.12540 [DOI] [PubMed] [Google Scholar]
- Ferreira R. M., Pereira-Marques J., Pinto-Ribeiro I., Costa J. L., Carneiro F., MacHado J. C., et al. (2018). Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67 226–236. 10.1136/gutjnl-2017-314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla R., Lionetti E., Castellaneta S. P., Magistà A. M., Maurogiovanni G., Bucci N., et al. (2008). Inhibition of Helicobacter pylori infection in humans by Lactobacillus reuteri ATCC 55730 and effect on eradication therapy: a pilot study. Helicobacter 13 127–134. 10.1111/j.1523-5378.2008.00593.x [DOI] [PubMed] [Google Scholar]
- Freire de Melo F., Rocha G. A., Rocha A. M. C., Teixeira K. N., Pedroso S. H. S. P., Pereira Junior J. B., et al. (2014). Th1 immune response to H. pylori infection varies according to the age of the patients and influences the gastric inflammatory patterns. Int. J. Med. Microbiol. 304 300–306. 10.1016/j.ijmm.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Gaiser R. A., Halimi A., Alkharaan H., Lu L., Davanian H., Healy K., et al. (2019). Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 68 2186–2194. 10.1136/gutjnl-2018-317458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski A., Mnich E., Szymañski K., Hinc K., Obuchowski M., Moran A. P., et al. (2016). Helicobacter pylori antigens, acetylsalicylic acid, LDL and 7-ketocholesterol – their potential role in destabilizing the gastric epithelial cell barrier. An in vitro model of Kato III cells. Acta Biochim. Pol. 63 145–152. 10.18388/abp.2015_1122 [DOI] [PubMed] [Google Scholar]
- Gálvez E. J. C., Iljazovic A., Amend L., Lesker T. R., Renault T., Thiemann S., et al. (2020). Distinct polysaccharide utilization determines interspecies competition between intestinal Prevotella spp. Cell Host Microb. 28 838–852.e6 10.1016/j.chom.2020.09.012 [DOI] [PubMed] [Google Scholar]
- Gantuya B., El Serag H. B., Matsumoto T., Ajami N. J., Uchida T., Oyuntsetseg K., et al. (2020). Gastric mucosal microbiota in a mongolian population with gastric cancer and precursor conditions. Aliment. Pharmacol. Ther. 51 770–780. 10.1111/apt.15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier N. C., Monzo P., Kaddai V., Doye A., Ricci V., Boquet P. (2005). Helicobacter pylori VacA cytotoxin: a probe for a clathrin-independent and Cdc42-dependent pinocytic pathway routed to late endosomes. Mol. Biol. Cell 16 4852–4866. 10.1091/mbc.e05-05-0398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z., Feng Y., Muthupalani S., Eurell L. L., Taylor N. S., Whary M. T., et al. (2011). Coinfection with Enterohepatic Helicobacter species can ameliorate or promote Helicobacter pylori-induced gastric pathology in C57BL/6 mice. Infect. Immun. 79 3861–3871. 10.1128/IAI.05357-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T., Beniwal A., Semwal A., Navani N. K. (2019). Mechanistic insights into probiotic properties of lactic acid bacteria associated with ethnic fermented dairy products. Front. Microbiol. 10:502. 10.3389/fmicb.2019.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. (2018). Current understanding of the human microbiome. Nat. Med. 24 392–400. 10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A. P., Wilson K. T. (2017). Polyamine- and NADPH-dependent generation of ROS during Helicobacter pylori infection: a blessing in disguise. Free Radic. Biol. Med. 105 16–27. 10.1016/j.freeradbiomed.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D., Gong X., Wang L., Yu X., Dong Q. (2016). Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol. Res. Pract. 2016:6951091. 10.1155/2016/6951091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotteland M., Brunser O., Cruchet S. (2006). Systematic review: are probiotics useful in controlling gastric colonization by Helicobacter pylori? Aliment. Pharmacol. Ther. 23 1077–1086. 10.1111/j.1365-2036.2006.02868.x [DOI] [PubMed] [Google Scholar]
- Gu H. (2017). Role of flagella in the pathogenesis of Helicobacter pylori. Curr. Microbiol. 74 863–869. 10.1007/s00284-017-1256-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra N., Tan Y. X., Joncker N. T., Choy A., Gallardo F., Xiong N., et al. (2008). NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 28 571–580. 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunathilake M. N., Lee J., Choi I. J., Kim Y.-I., Ahn Y., Park C., et al. (2019). Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci. Rep. 9:13589. 10.1038/s41598-019-50054-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M., et al. (2015). Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42 344–355. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. J., Lee N.-K., Park H., Paik H.-D. (2015). Anticancer and anti-inflammatory activity of probiotic Lactococcus lactis NK34. J. Microbiol. Biotechnol. 25 1697–1701. 10.4014/jmb.1503.03033 [DOI] [PubMed] [Google Scholar]
- Han T., Jing X., Bao J., Zhao L., Zhang A., Miao R., et al. (2020). H. pylori infection alters repair of DNA double-strand breaks via SNHG17. J. Clin. Invest. 130 3901–3918. 10.1172/JCI125581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa O., Naito Y., Yoshikawa T. (2010). Helicobacter pylori: a ROS-inducing bacterial species in the stomach. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 59 997–1003. 10.1007/s00011-010-0245-x [DOI] [PubMed] [Google Scholar]
- Hao W., Hu C., Huang Y., Chen Y. (2019). Coadministration of kla peptide with HPRP-A1 to enhance anticancer activity. PLoS One 14:e0223738. 10.1371/journal.pone.0223738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro C., Rangel-Zúñiga O. A., Alcalá-Díaz J. F., Gómez-Delgado F., Pérez-Martínez P., Delgado-Lista J., et al. (2016). Intestinal microbiota is influenced by gender and body mass index. PLoS One 11:e0154090. 10.1371/journal.pone.0154090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Wollenberg B., Rothenfusser S., Wagner M., Wellisch D., Mack B., et al. (2003). Identification and functional analysis of tumor-infiltrating plasmacytoid dendritic cells in head and neck cancer. Cancer Res. 63 6478–6487. [PubMed] [Google Scholar]
- Hashemi Goradel N., Heidarzadeh S., Jahangiri S., Farhood B., Mortezaee K., Khanlarkhani N., et al. (2019). Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell. Physiol. 234 2337–2344. 10.1002/jcp.27250 [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. (2017). Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 93 196–219. 10.2183/pjab.93.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). Expert consensus document. The international scientific association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Hofer U. (2014). Pro-inflammatory prevotella? Nat. Rev. Microbiol. 12:5. 10.1038/nrmicro3180 [DOI] [PubMed] [Google Scholar]
- Hooi J. K. Y., Lai W. Y., Ng W. K., Suen M. M. Y., Underwood F. E., Tanyingoh D., et al. (2017). Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153 420–429. 10.1053/j.gastro.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Howden C. W., Hunt R. H. (1987). Relationship between gastric secretion and infection. Gut 28 96–107. 10.1136/gut.28.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.-Y., Tung S.-Y., Pan H.-Y., Yen C.-W., Xu H.-W., Lin Y.-J., et al. (2018). Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 8:158. 10.1038/s41598-017-18596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Chen X., Huang Y., Chen Y. (2018). Synergistic effect of the pro-apoptosis peptide kla-TAT and the cationic anticancer peptide HPRP-A1. Apoptosis 23 132–142. 10.1007/s10495-018-1443-1 [DOI] [PubMed] [Google Scholar]
- Hu Y., He L.-H., Xiao D., Liu G.-D., Gu Y.-X., Tao X.-X., et al. (2012). Bacterial flora concurrent with Helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J. Gastroenterol. 18 1257–1261. 10.3748/wjg.v18.i11.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Q., Sridhar S., Hunt R. H. (2002). Role of Helicobacter pylori infection and non-steroidal anti-inflammatory drugs in peptic-ulcer disease: a meta-analysis. Lancet 359 14–22. 10.1016/S0140-6736(02)07273-2 [DOI] [PubMed] [Google Scholar]
- Huang X. M., Liu X. S., Lin X. K., Yu H., Sun J. Y., Liu X. K., et al. (2014). Role of plasmacytoid dendritic cells and inducible costimulator-positive regulatory T cells in the immunosuppression microenvironment of gastric cancer. Cancer Sci. 105 150–158. 10.1111/cas.12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde E. R., Luk B., Cron S., Kusic L., McCue T., Bauch T., et al. (2014). Characterization of the rat oral microbiome and the effects of dietary nitrate. Free Radic. Biol. Med. 77 249–257. 10.1016/j.freeradbiomed.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Ito T., Hanabuchi S., Wang Y.-H., Park W. R., Arima K., Bover L., et al. (2008). Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity 28 870–880. 10.1016/j.immuni.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakszyn P., Gonzalez C.-A. (2006). Nitrosamine and related food intake and gastric and oesophageal cancer risk: a systematic review of the epidemiological evidence. World J. Gastroenterol. 12 4296–4303. 10.3748/wjg.v12.i27.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H. J., Kim J., Kim N., Park J. H., Nam R. H., Seok Y.-J., et al. (2016). Analysis of gastric Microbiota by Pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 21 364–374. 10.1111/hel.12293 [DOI] [PubMed] [Google Scholar]
- Jones R. M., Mercante J. W., Neish A. S. (2012). Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr. Med. Chem. 19 1519–1529. 10.2174/092986712799828283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhah A., Ebrahimpour S., Rostamtabar M., Koppolu V., Darvish S., Vasigala V. K. R., et al. (2019). Helicobacter pylori evasion strategies of the host innate and adaptive immune responses to survive and develop gastrointestinal diseases. Microbiol. Res. 218 49–57. [DOI] [PubMed] [Google Scholar]
- Karthaus N., Torensma R., Tel J. (2012). Deciphering the message broadcast by tumor-infiltrating dendritic cells. Am. J. Pathol. 181 733–742. 10.1016/j.ajpath.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Kavitt R. T., Lipowska A. M., Anyane-Yeboa A., Gralnek I. M. (2019). Diagnosis and treatment of peptic ulcer disease. Am. J. Med. 132 447–456. 10.1016/j.amjmed.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Kawai T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5:461. 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi Y., Dieye Y., Poh B. H., Ng C. G., Loke M. F., Goh K. L., et al. (2014). Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci. World J. 2014:610421. 10.1155/2014/610421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane D. (2018). Molecular mechanisms of H. pylori-induced DNA double-strand breaks. Int. J. Mol. Sci. 19:2891. 10.3390/ijms19102891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim N., Jo H. J., Park J. H., Nam R. H., Seok Y.-J., et al. (2015). An appropriate cutoff value for determining the colonization of Helicobacter pylori by the pyrosequencing method: comparison with conventional methods. Helicobacter 20 370–380. 10.1111/hel.12214 [DOI] [PubMed] [Google Scholar]
- Knorr J., Ricci V., Hatakeyama M., Backert S. (2019). Classification of Helicobacter pylori virulence factors: is CagA a toxin or not? Trends Microbiol. 27 731–738. 10.1016/j.tim.2019.04.010 [DOI] [PubMed] [Google Scholar]
- Kobayashi J. (2018). Effect of diet and gut environment on the gastrointestinal formation of N-nitroso compounds: a review. Nitric oxide Biol. Chem. 73 66–73. 10.1016/j.niox.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Kostic A. D., Chun E., Robertson L., Glickman J. N., Gallini C. A., Michaud M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microb. 14 207–215. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Suerbaum S. (2005). Mutation and recombination in Helicobacter pylori: mechanisms and role in generating strain diversity. Int. J. Med. Microbiol. 295 299–305. [DOI] [PubMed] [Google Scholar]
- Kumar S., Metz D. C., Ellenberg S., Kaplan D. E., Goldberg D. S. (2020). Risk factors and incidence of gastric cancer after detection of Helicobacter pylori infection: a large cohort study. Gastroenterology 158 527–536.e7. 10.1053/j.gastro.2019.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J. G., van Vliet A. H. M., Kuipers E. J. (2006). Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19 449–490. 10.1128/CMR.00054-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanas A., Chan F. K. L. (2017). Peptic ulcer disease. Lancet 390 613–624. 10.1016/S0140-6736(16)32404-7 [DOI] [PubMed] [Google Scholar]
- Laszewicz W., Iwańczak F., Iwańczak B. (2014). Seroprevalence of Helicobacter pylori infection in Polish children and adults depending on socioeconomic status and living conditions. Adv. Med. Sci. 59 147–150. 10.1016/j.advms.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Lee J. E., Lee S., Lee H., Song Y.-M., Lee K., Han M. J., et al. (2013). Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. 10.1371/journal.pone.0063514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. E., Khoi P. N., Xia Y., Park J. S., Joo Y. E., Kim K. K., et al. (2013). Helicobacter pylori and interleukin-8 in gastric cancer. World J. Gastroenterol. 19 8192–8202. 10.3748/wjg.v19.i45.8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Sung J., Lee J., Ko G. (2011). Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl. Environ. Microbiol. 77 7433–7437. 10.1128/AEM.05490-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Um J., Hwang B., Lee Y. C., Chung B. C., Hong J. (2020). Assessing the progression of gastric cancer via profiling of histamine, histidine, and bile acids in gastric juice using LC-MS/MS. J. Steroid Biochem. Mol. Biol. 197:105539. 10.1016/j.jsbmb.2019.105539 [DOI] [PubMed] [Google Scholar]
- Leite M., Marques M. S., Melo J., Pinto M. T., Cavadas B., Aroso M., et al. (2020). Helicobacter Pylori targets the EPHA2 receptor tyrosine kinase in gastric cells modulating key cellular functions. Cells 9:513. 10.3390/cells9020513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke L. B., Ge Z., Whary M. T., Feng Y., Rogers A. B., Muthupalani S., et al. (2009). Concurrent Helicobacter bilis infection in C57BL/6 mice attenuates proinflammatory H. pylori-induced gastric pathology. Infect. Immun. 77 2147–2158. 10.1128/IAI.01395-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Kolodziejczyk A. A., Thaiss C. A., Elinav E. (2017). Dysbiosis and the immune system. Nat. Rev. Immunol. 17 219–232. 10.1038/nri.2017.7 [DOI] [PubMed] [Google Scholar]
- Li T. H., Qin Y., Sham P. C., Lau K. S., Chu K.-M., Leung W. K. (2017). Alterations in gastric microbiota after H. Pylori eradication and in different histological stages of gastric carcinogenesis. Sci. Rep. 7:44935. 10.1038/srep44935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Eshak E. S., Shirai K., Liu K., Dong J. Y., Iso H., et al. (2021). Alcohol Consumption and Risk of Gastric Cancer: the Japan collaborative cohort study. J. Epidemiol. 31 30–36. 10.2188/jea.JE20190304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z., Shao L., Liu X., Cheng Y., Yan C., Mei Y., et al. (2019). Regulatory T cells and plasmacytoid dendritic cells within the tumor microenvironment in gastric cancer are correlated with gastric microbiota dysbiosis: a preliminary study. Front. Immunol. 10:533. 10.3389/fimmu.2019.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B., Balloux F., Moodley Y., Manica A., Liu H., Roumagnac P., et al. (2007). An African origin for the intimate association between humans and Helicobacter pylori. Nature 445 915–918. 10.1038/nature05562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Shao L., Liu X., Ji F., Mei Y., Cheng Y., et al. (2019a). Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBiomedicine 40 336–348. 10.1016/j.ebiom.2018.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu H., Yan C., Mei Y., Lin C., Hong Y., et al. (2019b). Plasmacytoid dendritic cells and ICOS+ regulatory T cells predict poor prognosis in gastric cancer: a pilot study. J. Cancer 10 6711–6715. 10.7150/jca.34826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gerner M. Y., Van Panhuys N., Levine A. G., Rudensky A. Y., Germain R. N. (2015). Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528 225–230. 10.1038/nature16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca L., Pérez-Pérez G., Urruzuno P., Martinez M. J., Iizumi T., Gao Z., et al. (2017). Characterization of the gastric microbiota in a pediatric population according to Helicobacter pylori status. Pediatr. Infect. Dis. J. 36 173–178. 10.1097/INF.0000000000001383 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Abu-Ali G., Huttenhower C. (2016). The healthy human microbiome. Genome Med. 8 1–11. 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan F., Li X., Cheng X., Huangfu L., Han J., Guo T., et al. (2020). TNFRSF11B activates Wnt/β-catenin signaling and promotes gastric cancer progression. Int. J. Biol. Sci. 16 1956–1971. 10.7150/ijbs.43630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren A., Strömberg E., Sjöling A., Lindholm C., Enarsson K., Edebo A., et al. (2005). Mucosal FOXP3-expressing CD4+ CD25high regulatory T cells in Helicobacter pylori-infected patients. Infect. Immun. 73 523–531. 10.1128/IAI.73.1.523-531.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren A., Suri-Payer E., Enarsson K., Svennerholm A.-M., Lundin B. S. (2003). Helicobacter pylori-specific CD4+ CD25high regulatory T cells suppress memory T-cell responses to H. pylori in infected individuals. Infect. Immun. 71 1755–1762. 10.1128/iai.71.4.1755-1762.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai X. T., Huang J., Tan J., Huang Y., Chen Y. (2015). Effects and mechanisms of the secondary structure on the antimicrobial activity and specificity of antimicrobial peptides. J. Pept. Sci. 21 561–568. 10.1002/psc.2767 [DOI] [PubMed] [Google Scholar]
- Maldonado-Contreras A., Goldfarb K. C., Godoy-Vitorino F., Karaoz U., Contreras M., Blaser M. J., et al. (2011). Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 5 574–579. 10.1038/ismej.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfertheiner P., Megraud F., O’Morain C. A., Gisbert J. P., Kuipers E. J., Axon A. T., et al. (2017). Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut 66 6–30. 10.1136/gutjnl-2016-312288 [DOI] [PubMed] [Google Scholar]
- Manente L., Perna A., Buommino E., Altucci L., Lucariello A., Citro G., et al. (2008). The Helicobacter pylori’s protein VacA has direct effects on the regulation of cell cycle and apoptosis in gastric epithelial cells. J. Cell. Physiol. 214 582–587. 10.1002/jcp.21242 [DOI] [PubMed] [Google Scholar]
- Marshall B. J., Warren J. R. (1984). Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 323 1311–1315. 10.1016/S0140-6736(84)91816-6 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Isomoto H., Nakayama M., Hisatsune J., Nishi Y., Nakashima Y., et al. (2011). Helicobacter pylori VacA reduces the cellular expression of STAT3 and pro-survival Bcl-2 family proteins, Bcl-2 and Bcl-XL, leading to apoptosis in gastric epithelial cells. Dig. Dis. Sci. 56 999–1006. 10.1007/s10620-010-1420-1 [DOI] [PubMed] [Google Scholar]
- McFarland L. V., Huang Y., Wang L., Malfertheiner P. (2016). Systematic review and meta-analysis: multi-strain probiotics as adjunct therapy for Helicobacter pylori eradication and prevention of adverse events. United Eur. Gastroenterol. J. 4 546–561. 10.1177/2050640615617358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. R., Derakhshan M. H., Wirz A. A., Orange C., Ballantyne S. A., Going J. J., et al. (2017). The gastric acid pocket is attenuated in H. pylori infected subjects. Gut 66 1555–1562. 10.1136/gutjnl-2016-312638 [DOI] [PubMed] [Google Scholar]
- Montalban-Arques A., Wurm P., Trajanoski S., Schauer S., Kienesberger S., Halwachs B., et al. (2016). Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 240 425–436. 10.1002/path.4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G., Li X., Zhou H., Sheng J., Wong S. H., Wu W. K. K., et al. (2015). Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 6:8727. 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone G., Compare D. (2015). The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 3 255–260. 10.1177/2050640614566846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone G., Compare D., Rocco A. (2017). A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet. Gastroenterol. Hepatol. 2 298–312. 10.1016/S2468-1253(16)30108-X [DOI] [PubMed] [Google Scholar]
- Nejati S., Karkhah A., Darvish H., Validi M., Ebarhimpour S., Nouri H. R. (2018). Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders. Microb. Pathog. 117 43–48. 10.1016/j.micpath.2018.02.016 [DOI] [PubMed] [Google Scholar]
- Nie S., Wang A., Yuan Y. (2021). Comparison of clinicopathological parameters, prognosis, micro-ecological environment and metabolic function of Gastric Cancer with or without Fusobacterium sp. infection. J. Cancer 12 1023–1032. 10.7150/jca.50918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (1994). Schistosomes, liver flukes and Helicobacter pylori. IARC working group on the evaluation of carcinogenic risks to Humans. Lyon, 7-14 June 1994. IARC Monogr. Eval. Carcinog. Risks Hum. 61 1–241. [PMC free article] [PubMed] [Google Scholar]
- Oh B., Kim B.-S., Kim J. W., Kim J. S., Koh S.-J., Kim B. G., et al. (2016). The Effect of Probiotics on gut microbiota during the Helicobacter pylori eradication: randomized controlled trial. Helicobacter 21 165–174. 10.1111/hel.12270 [DOI] [PubMed] [Google Scholar]
- Olofsson A., Nygård Skalman L., Obi I., Lundmark R., Arnqvist A. (2014). Uptake of Helicobacter pylori vesicles is facilitated by clathrin-dependent and clathrin-independent endocytic pathways. mBio 5:e00979-14. 10.1128/mBio.00979-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim D. E., Roberts S. J., Clarke S. L., Filler R., Lewis J. M., Tigelaar R. E., et al. (2005). Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat. Immunol. 6 928–937. 10.1038/ni1239 [DOI] [PubMed] [Google Scholar]
- Ou Y., Ren H., Zhao R., Song L., Liu Z., Xu W., et al. (2019). Helicobacter pylori CagA promotes the malignant transformation of gastric mucosal epithelial cells through the dysregulation of the miR-155/KLF4 signaling pathway. Mol. Carcinog. 58 1427–1437. 10.1002/mc.23025 [DOI] [PubMed] [Google Scholar]
- Outlioua A., Badre W., Desterke C., Echarki Z., El Hammani N., Rabhi M., et al. (2020). Gastric IL-1β, IL-8, and IL-17A expression in Moroccan patients infected with Helicobacter pylori may be a predictive signature of severe pathological stages. Cytokine 126:154893. 10.1016/j.cyto.2019.154893 [DOI] [PubMed] [Google Scholar]
- Pachathundikandi S. K., Tegtmeyer N., Arnold I. C., Lind J., Neddermann M., Falkeis-Veits C., et al. (2019). T4SS-dependent TLR5 activation by Helicobacter pylori infection. Nat. Commun. 10:5717. 10.1038/s41467-019-13506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J., Friedman G. D., Vandersteen D. P., Chang Y., Vogelman J. H., Orentreich N., et al. (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325 1127–1131. 10.1056/NEJM199110173251603 [DOI] [PubMed] [Google Scholar]
- Parsons B. N., Ijaz U. Z., D’Amore R., Burkitt M. D., Eccles R., Lenzi L., et al. (2017). Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 13:e1006653. 10.1371/journal.ppat.1006653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek R. M. J., Miller G. G., Tham K. T., Perez-Perez G. I., Zhao X., Atherton J. C., et al. (1995). Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab. Invest. 73 760–770. [PubMed] [Google Scholar]
- Pereira-Marques J., Ferreira R. M., Pinto-Ribeiro I., Figueiredo C. (2019). Helicobacter pylori infection, the gastric microbiome and gastric cancer. Adv. Exp. Med. Biol. 1149 195–210. 10.1007/5584_2019_366 [DOI] [PubMed] [Google Scholar]
- Perrot I., Blanchard D., Freymond N., Isaac S., Guibert B., Pachéco Y., et al. (2007). Dendritic cells infiltrating human non-small cell lung cancer are blocked at immature stage. J. Immunol. 178 2763–2769. 10.4049/jimmunol.178.5.2763 [DOI] [PubMed] [Google Scholar]
- Rad R., Brenner L., Bauer S., Schwendy S., Layland L., da Costa C. P., et al. (2006). CD25+/Foxp3+ T cells regulate gastric inflammation and Helicobacter pylori colonization in vivo. Gastroenterology 131 525–537. 10.1053/j.gastro.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Ravegnini G., Fosso B., Saverio V., Di Sammarini G., Zanotti F., Rossi G., et al. (2020). Gastric adenocarcinomas and signet-ring cell carcinoma: unraveling gastric cancer complexity through microbiome analysis-deepening heterogeneity for a personalized therapy. Int. J. Mol. Sci. 21:9735. 10.3390/ijms21249735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscetto E., Vitiello L., Muoio R., Soriano A. A., Iula V. D., Vollaro A., et al. (2015). In vitro interaction of Stenotrophomonas maltophilia with human monocyte-derived dendritic cells. Front. Microbiol. 6:723. 10.3389/fmicb.2015.00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microb. 14 195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarya S., Gunay N. (2014). Saccharomyces boulardii expresses neuraminidase activity selective for α2,3-linked sialic acid that decreases Helicobacter pylori adhesion to host cells. APMIS 122 941–950. 10.1111/apm.12237 [DOI] [PubMed] [Google Scholar]
- Salazar-Lindo E., Figueroa-Quintanilla D., Caciano M. I., Reto-Valiente V., Chauviere G., Colin P. (2007). Effectiveness and safety of Lactobacillus LB in the treatment of mild acute diarrhea in children. J. Pediatr. Gastroenterol. Nutr. 44 571–576. 10.1097/MPG.0b013e3180375594 [DOI] [PubMed] [Google Scholar]
- San-Millán I., Brooks G. A. (2017). Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 38 119–133. 10.1093/carcin/bgw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Akira S. (2016). Toll-Like receptor signaling and its inducible proteins. Microbiol. Spectr. 4:microbiolsec.MCHD-0040-2016. 10.1128/microbiolspec.MCHD-0040-2016 [DOI] [PubMed] [Google Scholar]
- Sawant S. S., Patil S. M., Gupta V., Kunda N. K. (2020). Microbes as medicines: harnessing the power of bacteria in advancing cancer treatment. Int. J. Mol. Sci. 21:7575. 10.3390/ijms21207575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter J., Peled J. U., Taylor B. P., Markey K. A., Smith M., Taur Y., et al. (2020). The gut microbiota is associated with immune cell dynamics in humans. Nature 588 303–307. 10.1038/s41586-020-2971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. R., Marcus E. A., Wen Y., Singh S., Feng J., Sachs G. (2010). Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4(+), is necessary for acid survival of Helicobacter pylori. J. Bacteriol. 192 94–103. 10.1128/JB.00848-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddik H. A., Bendali F., Gancel F., Fliss I., Spano G., Drider D. (2017). Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 9 111–122. 10.1007/s12602-017-9264-z [DOI] [PubMed] [Google Scholar]
- Seyda T., Derya C., Füsun A., Meliha K. (2007). The relationship of Helicobacter pylori positivity with age, sex, and ABO/Rhesus blood groups in patients with gastrointestinal complaints in Turkey. Helicobacter 12 244–250. 10.1111/j.1523-5378.2007.00500.x [DOI] [PubMed] [Google Scholar]
- Shaw K. A., Bertha M., Hofmekler T., Chopra P., Vatanen T., Srivatsa A., et al. (2016). Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 8:75. 10.1186/s13073-016-0331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Chiba T., Marusawa H. (2017). Helicobacter pylori-mediated genetic instability and gastric carcinogenesis. Curr. Top. Microbiol. Immunol. 400 305–323. 10.1007/978-3-319-50520-6_13 [DOI] [PubMed] [Google Scholar]
- Sierra J. C., Piazuelo M. B., Luis P. B., Barry D. P., Allaman M. M., Asim M., et al. (2020). Spermine oxidase mediates Helicobacter pylori-induced gastric inflammation, DNA damage, and carcinogenic signaling. Oncogene 39 4465–4474. 10.1038/s41388-020-1304-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipponen P., Maaroos H.-I. (2015). Chronic gastritis. Scand. J. Gastroenterol. 50 657–667. 10.3109/00365521.2015.1019918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siregar G. A., Halim S., Sitepu V. R. (2015). Serum TNF-a, IL-8, VEGF levels in Helicobacter pylori infection and their association with degree of gastritis. Acta Med. Indones. 47 120–126. [PubMed] [Google Scholar]
- Soleimanpour S., Hasanian S. M., Avan A., Yaghoubi A., Khazaei M. (2020). Bacteriotherapy in gastrointestinal cancer. Life Sci. 254:117754. [DOI] [PubMed] [Google Scholar]
- Song H., Zhou L., Liu D., Ge L., Li Y. (2019). Probiotic effect on Helicobacter pylori attachment and inhibition of inflammation in human gastric epithelial cells. Exp. Ther. Med. 18 1551–1562. 10.3892/etm.2019.7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano K., Tack J., Kuipers E. J., Graham D. Y., El-Omar E. M., Miura S., et al. (2015). Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64 1353–1367. 10.1136/gutjnl-2015-309252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang M., El-Zataari M., Owyang S. Y., Eaton K. A., Liu M., et al. (2013). TLR2 mediates Helicobacter pylori-induced tolerogenic immune response in mice. PLoS One 8:e74595. 10.1371/journal.pone.0074595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J., Kim N., Kim J., Jo H. J., Park J. H., Nam R. H., et al. (2016). Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J. Cancer Prev. 21 60–65. 10.15430/jcp.2016.21.1.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supajatura V., Ushio H., Wada A., Yahiro K., Okumura K., Ogawa H., et al. (2002). Cutting edge: VacA, a vacuolating cytotoxin of Helicobacter pylori, directly activates mast cells for migration and production of proinflammatory cytokines. J. Immunol. 168 2603–2607. 10.4049/jimmunol.168.6.2603 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Dörffel Y., Loening-Baucke V., Theissig F., Rückert J. C., Ismail M., et al. (2011). Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60 34–40. 10.1136/gut.2009.191320 [DOI] [PubMed] [Google Scholar]
- Sycuro L. K., Wyckoff T. J., Biboy J., Born P., Pincus Z., Vollmer W., et al. (2012). Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 8:e1002603. 10.1371/journal.ppat.1002603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareq F. S., Kim J. H., Lee M. A., Lee H.-S., Lee Y.-J., Lee J. S., et al. (2012). Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 14 1464–1467. 10.1021/ol300202z [DOI] [PubMed] [Google Scholar]
- Toller I. M., Neelsen K. J., Steger M., Hartung M. L., Hottiger M. O., Stucki M., et al. (2011). Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. U.S.A. 108 14944–14949. 10.1073/pnas.1100959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T., Fukuda Y., Tamura K., Tanaka J., Hida N., Kosaka T., et al. (2002). Successful eradication of Helicobacter pylori prevents relapse of peptic ulcer disease. Aliment. Pharmacol. Ther. 16 (Suppl. 2), 204–209. 10.1046/j.1365-2036.16.s2.24.x [DOI] [PubMed] [Google Scholar]
- Torres V. J., VanCompernolle S. E., Sundrud M. S., Unutmaz D., Cover T. L. (2007). Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J. Immunol. 179 5433–5440. 10.4049/jimmunol.179.8.5433 [DOI] [PubMed] [Google Scholar]
- Tseng C.-H., Lin J.-T., Ho H. J., Lai Z.-L., Wang C.-B., Tang S.-L., et al. (2016). Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci. Rep. 6:20701. 10.1038/srep20701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda A., Suda W., Morita H., Takanashi K., Takagi A., Koga Y., et al. (2015). Influence of proton-pump inhibitors on the luminal microbiota in the gastrointestinal tract. Clin. Transl. Gastroenterol. 6:e89. 10.1038/ctg.2015.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane S., Sasazuki S. (2007). Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10 75–83. 10.1007/s10120-007-0420-0 [DOI] [PubMed] [Google Scholar]
- Ujiie H., Shevach E. M. (2016). γδ T cells protect the liver and lungs of mice from autoimmunity induced by scurfy lymphocytes. J. Immunol. 196 1517–1528. 10.4049/jimmunol.1501774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno K., Kato K., Atsumi T., Suzuki T., Yoshitake J., Morita H., et al. (2007). Toll-like receptor (TLR) 2 induced through TLR4 signaling initiated by Helicobacter pylori cooperatively amplifies iNOS induction in gastric epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 293 G1004–G1012. 10.1152/ajpgi.00096.2007 [DOI] [PubMed] [Google Scholar]
- van Baarlen P., Troost F. J., van Hemert S., van der Meer C., de Vos W. M., de Groot P. J., et al. (2009). Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. U.S.A. 106 2371–2376. 10.1073/pnas.0809919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villéger R., Lopès A., Carrier G., Veziant J., Billard E., Barnich N., et al. (2019). Intestinal microbiota: a novel target to improve anti-tumor treatment? Int. J. Mol. Sci. 20:584. 10.3390/ijms20184584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinasco K., Mitchell H. M., Kaakoush N. O., Castaño-Rodríguez N. (2019). Microbial carcinogenesis: lactic acid bacteria in gastric cancer. Biochim. Biophys. Acta – Rev. Cancer 1872:188309. [DOI] [PubMed] [Google Scholar]
- Von Rosenvinge E. C., Song Y., White J. R., Maddox C., Blanchard T., Fricke W. F. (2013). Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 7 1354–1366. 10.1038/ismej.2013.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C. L., Smith P. L., Reed P. I., Haines K., House F. R. (1982). N-nitroso compounds in gastric juice and their relationship to gastroduodenal disease. IARC Sci. Publ. 41, 345–355. [PubMed] [Google Scholar]
- Wan X.-K., Yuan S.-L., Tao H.-X., Diao L.-P., Wang Y.-C., Cao C., et al. (2016). The upregulation of TRAF1 Induced by Helicobacter pylori plays an antiapoptotic effect on the infected Cells. Helicobacter 21 554–564. 10.1111/hel.12311 [DOI] [PubMed] [Google Scholar]
- Wang F., Meng W., Wang B., Qiao L. (2014). Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 345 196–202. 10.1016/j.canlet.2013.08.016 [DOI] [PubMed] [Google Scholar]
- Wang L., Xin Y., Zhou J., Tian Z., Liu C., Yu X., et al. (2020). Gastric mucosa-associated microbial signatures of early gastric cancer. Front. Microbiol. 11:1548. 10.3389/fmicb.2020.01548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gao X., Zeng R., Wu Q., Sun H., Wu W., et al. (2020). Changes of the gastric mucosal microbiome associated with histological stages of gastric carcinogenesis. Front. Microbiol. 11:997. 10.3389/fmicb.2020.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou J., Xin Y., Geng C., Tian Z., Yu X., et al. (2016). Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur. J. Gastroenterol. Hepatol. 28 261–266. 10.1097/MEG.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-H., Gao Q.-Y., Fang J.-Y. (2013). Meta-analysis of the efficacy and safety of Lactobacillus-containing and Bifidobacterium-containing probiotic compound preparation in Helicobacter pylori eradication therapy. J. Clin. Gastroenterol. 47 25–32. 10.1097/MCG.0b013e318266f6cf [DOI] [PubMed] [Google Scholar]
- Willhite D. C., Blanke S. R. (2004). Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell. Microbiol. 6 143–154. 10.1046/j.1462-5822.2003.00347.x [DOI] [PubMed] [Google Scholar]
- Wong B. C.-Y., Lam S. K., Wong W. M., Chen J. S., Zheng T. T., Feng R. E., et al. (2004). Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of chinaa randomized controlled trial. JAMA 291 187–194. 10.1001/jama.291.2.187 [DOI] [PubMed] [Google Scholar]
- Xie J.-J., Zhou F., Li E.-M., Jiang H., Du Z.-P., Lin R., et al. (2011). FW523-3, a novel lipopeptide compound, induces apoptosis in cancer cells. Mol. Med. Rep. 4 759–763. 10.3892/mmr.2011.496 [DOI] [PubMed] [Google Scholar]
- Xu L., Qu Y.-H., Chu X.-D., Wang R., Nelson H. H., Gao Y.-T., et al. (2015). Urinary levels of N-nitroso compounds in relation to risk of gastric cancer: findings from the shanghai cohort study. PLoS One 10:e0117326. 10.1371/journal.pone.0117326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J., Deguchi R., Yanagi H., Ozawa H., Urano T., Ogawa Y., et al. (2005). Relationship between gastric mucosal IL-8 levels and histological gastritis in patients with Helicobacter pylori infection. Tokai J. Exp. Clin. Med. 30 83–88. [PubMed] [Google Scholar]
- Yadegar A., Mohabati Mobarez A., Zali M. R. (2019). Genetic diversity and amino acid sequence polymorphism in Helicobacter pylori CagL hypervariable motif and its association with virulence markers and gastroduodenal diseases. Cancer Med. 8 1619–1632. 10.1002/cam4.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi A., Khazaei M., Jalili S., Hasanian S. M., Avan A., Soleimanpour S., et al. (2020). Bacteria as a double-action sword in cancer. Biochim. Biophys. Acta Rev. Cancer 1874:188388. [DOI] [PubMed] [Google Scholar]
- Yang F., Xu Y., Liu C., Ma C., Zou S., Xu X., et al. (2018). NF-κB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis. 9:12. 10.1038/s41419-017-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I., Woltemate S., Piazuelo M. B., Bravo L. E., Yepez M. C., Romero-Gallo J., et al. (2016). Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci. Rep. 6:18594. 10.1038/srep18594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-J., Sheu B.-S. (2012). Probiotics-containing yogurts suppress Helicobacter pylori load and modify immune response and intestinal microbiota in the Helicobacter pylori-infected children. Helicobacter 17 297–304. 10.1111/j.1523-5378.2012.00941.x [DOI] [PubMed] [Google Scholar]
- Yeung K. T., Yang J. (2017). Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 11 28–39. 10.1002/1878-0261.12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Torres J., Hu N., Medrano-Guzman R., Herrera-Goepfert R., Humphrys M. S., et al. (2017). Molecular characterization of the human stomach microbiota in gastric cancer patients. Front. Cell. Infect. Microbiol. 7:302. 10.3389/fcimb.2017.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Zhang R., Ni P., Chen S., Duan G. (2019). Efficacy of Lactobacillus-supplemented triple therapy for H. pylori eradication: a meta-analysis of randomized controlled trials. PLoS One 14:e0223309. 10.1371/journal.pone.0223309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.-L., Chen L., Li M.-X., Dong P., Xue J., Wang J., et al. (2010). Elevated expression of Foxp3 in tumor-infiltrating Treg cells suppresses T-cell proliferation and contributes to gastric cancer progression in a COX-2-dependent manner. Clin. Immunol. 134 277–288. [DOI] [PubMed] [Google Scholar]
- Zhang H., Lam K. H., Lam W. W. L., Wong S. Y. Y., Chan V. S. F., Au S. W. N. (2017). A putative spermidine synthase interacts with flagellar switch protein FliM and regulates motility in Helicobacter pylori. Mol. Microbiol. 106 690–703. 10.1111/mmi.13829 [DOI] [PubMed] [Google Scholar]
- Zhang S., Shi D., Li M., Li Y., Wang X., Li W. (2019). The relationship between gastric microbiota and gastric disease. Scand. J. Gastroenterol. 54 391–396. 10.1080/00365521.2019.1591499 [DOI] [PubMed] [Google Scholar]
- Zhao J., Hao X., Liu D., Huang Y., Chen Y. (2015a). In vitro characterization of the rapid cytotoxicity of anticancer peptide HPRP-A2 through membrane destruction and intracellular mechanism against gastric cancer cell lines. PLoS One 10:e0139578. 10.1371/journal.pone.0139578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang Y., Liu D., Chen Y. (2015b). Two hits are better than one: synergistic anticancer activity of α-helical peptides and doxorubicin/epirubicin. Oncotarget 6 1769–1778. 10.18632/oncotarget.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Chen T., Wang Y., Gao Y., Kong Y., Liu Z., et al. (2019). A randomised trial of probiotics to reduce severity of physiological and microbial disorders induced by partial gastrectomy for patients with gastric cancer. J. Cancer 10 568–576. 10.7150/jca.29072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberstein B., Quintanilha A. G., Santos M. A. A., Pajecki D., Moura E. G., Alves P. R. A., et al. (2007). Digestive tract microbiota in healthy volunteers. Clinics 62 47–54. 10.1590/s1807-59322007000100008 [DOI] [PubMed] [Google Scholar]