Abstract
Under consecutive monoculture, the abundance of pathogenic fungi, such as Fusarium oxysporum in the rhizosphere of Radix pseudostellariae, negatively affects the yield and quality of the plant. Therefore, it is pertinent to explore the role of antagonistic fungi for the management of fungal pathogens such as F. oxysporum. Our PCR-denatured gradient gel electrophoresis (DGGE) results revealed that the diversity of Trichoderma spp. was significantly declined due to extended monoculture. Similarly, quantitative PCR analysis showed a decline in Trichoderma spp., whereas a significant increase was observed in F. oxysporum. Furthermore, seven Trichoderma isolates from the R. pseudostellariae rhizosphere were identified and evaluated in vitro for their potentiality to antagonize F. oxysporum. The highest and lowest percentage of inhibition (PI) observed among these isolates were 47.91 and 16.67%, respectively. In in vivo assays, the R. pseudostellariae treated with four Trichoderma isolates, having PI > 30%, was used to evaluate the biocontrol efficiency against F. oxysporum in which T. harzianum ZC51 enhanced the growth of the plant without displaying any disease symptoms. Furthermore, the expression of eight defense-related genes of R. pseudostellariae in response to a combination of F. oxysporum and T. harzianum ZC51 treatment was checked, and most of these defense genes were found to be upregulated. In conclusion, this study reveals that the extended monoculture of R. pseudostellariae could alter the Trichoderma communities in the plant rhizosphere leading to relatively low level of antagonistic microorganisms. However, T. harzianum ZC51 could inhibit the pathogenic F. oxysporum and induce the expression of R. pseudostellariae defense genes. Hence, T. harzianum ZC51 improves the plant resistance and reduces the growth inhibitory effect of consecutive monoculture problem.
Keywords: defense genes, Trichoderma, PCR-DGGE, monoculture, Radix pseudostellariae
Introduction
Due to the allelopathy and the dysbiosis of microorganisms, continuous planting of many Chinese medicinal herbs in the same land results in a significant decrease in yield and quality, which is known as continuous monoculture problem or soil sickness (Zhang and Lin, 2009; Zhao et al., 2015; Wu et al., 2016). Radix pseudostellariae is a perennial herb of the Caryophllaceae family, and its tuberous roots are used for medicinal purposes, which has very high economic value (Zhao et al., 2015). However, successive cultivation of R. pseudostellariae on the same piece of land leads to a decline in both the quality and yield owing to poor plant performance and insufficient biotic stress resistance (Lin et al., 2015). In consecutive monocultures, previous studies have reported the imbalance in the rhizosphere microbial community of R. pseudostellariae, especially the abundance of the pathogenic fungi (Fusarium oxysporum) increased significantly under consecutive monoculture (Zhao et al., 2014; Wu et al., 2016a, b, c; Chen et al., 2017). Moreover, most of researches in continuous monoculture problem of R. pseudostellariae and their potential biological microorganisms are focused on prokaryotes (i.e., Pseudomonas spp. and Burkholderia spp.) (Wu et al., 2016a; Chen et al., 2017). Therefore, to develop a reliable system of biological control against plant pathogens, we need to explore the antagonizing role of potentially important eukaryotic microorganisms like fungi as well.
The importance of the beneficial microbes in improving nutrient availability and promoting plant growth, antagonizing soil-borne pathogens, and priming the plant’s immune system is well established and abundantly used in biocontrol strategies (Cotxarrera et al., 2002; Howell, 2006; Lorito et al., 2010; Mendes et al., 2011; Matarese et al., 2012; Walters et al., 2013). Trichoderma spp. is a fungal genus in the family Hypocreaceae, which is found in the soil, rotting wood, plants, and the ocean. Many species are characterized as opportunistic avirulent, symbiotic and can be used as biological control agents against important plant pathogenic fungi (Harman et al., 2004). For example, T. harzianum (SQR-T307) and T. asperellum (T-34) are effective biological control agents against F. oxysporum (Corrales et al., 2010; Yang et al., 2011). T. asperellum isolates could significantly reduce the incidence of tomato wilt when used to suppress Fusarium wilt of tomato (Cotxarrera et al., 2002). T. gamsii 6085 was used in a competitive test against F. subtilis and F. graminearum, which confirmed that T. gamsii has the ability to antagonize the pathogens of rice (Matarese et al., 2012).
These root-associated mutualistic microbes, besides impacting on plant nutrition and growth, can further boost plant defenses, rendering the entire plant more resistant to pathogens (Romera et al., 2019). To cope with biotic stresses incited by biological agents, like insects and pathogens, plants develop responses, and some of these responses systemically spread far from the infected tissue into the whole plant. These responses include the systemic acquired resistance (SAR) and the induced systemic resistance (ISR) (Shoresh et al., 2010; Pieterse et al., 2014). SAR is induced by insects and pathogens, while ISR is mediated by beneficial microbes present in the rhizosphere, like bacteria and fungi (Mukherjee et al., 2013). Studies have shown that many species of Trichoderma could colonize on the root surface that interacts with the first cell layer of the root bark and the epidermis. This symbiotic relationship can effectively protect the plant from pathogens (Mukherjee et al., 2018; Galletti et al., 2020). When Trichoderma interacts with plants, it induces the expression of genes involved in the defense responses of plants (Brotman et al., 2013; Mayo et al., 2016; Manganiello et al., 2018; De Palma et al., 2019; Pimentel et al., 2020) and promotes plant growth and root development (Hermosa et al., 2012). However, the role of root-associated mutualistic plant symbiont, Trichoderma spp., in activation of R. pseudostellariae immunity by triggering the expression of defense-related genes is never explored.
The objectives of this present study are as follows: (1) to analyze the changes of Trichoderma communities in rhizosphere soil under R. pseudostellariae monoculture using denatured gradient gel electrophoresis (DGGE) combined with quantitative PCR (qPCR) technique and (2) further, to study the effect of different Trichoderma strains on the growth and defense response of R. pseudostellariae against the F. oxysporum and also to assess the expression level of defense-related genes in R. pseudostellariae treated with the selected Trichoderma isolate.
Materials and Methods
Site Overview and Experimental Design
The study was conducted at Ningde City, Fujian Province (27°26′ N, 120°04′ E). This station has a subtropical monsoon climate with an annual mean air temperature of 18.4°C and precipitation of 1,668.3 mm. The root tuber propagation materials of Radix pseudostellariae variety “Zhenshen 2” were used as the experimental plant, which was planted on 20th November and harvested on 10th July of the following year. A loam soil was used in the experiments. Physical–chemical characterization of soil used for the experiments was performed, using the protocol described by Jackson (1958) and Watanabe and Olsen (1965). To keep the soil and climatic conditions during the experimental period uniform and subjected to the same field and fertilization management, four types of plots were established within a single experimental field: (1) unplanted soil (CK), (2) containing R. pseudostellariae cultivated in fresh soil (FP), (3) plot under cultivation of R. pseudostellariae for two consecutive years (SP), and (4) plot under cultivation of R. pseudostellariae for three consecutive years (TP). Each type carried three replicate plots with a completely randomized design. The samples were taken in three biological replicates.
Soil Sampling and DNA Extraction
According to our previous study (Chen et al., 2017), after 5 months of planting R. pseudostellariae, its above-ground and underground biomass was significantly different (expanding period of root tubers). Therefore, we randomly collected soil samples from five different locations within each plot on 25th April, 2018 (Figure 1A). Moreover, for yield determination, we harvested the plants on 10th July, 2018 (Figure 1B). While taking soil samples, the rhizosphere soil clung to the root system of R. pseudostellariae was collected. DNA of soil (0.5 g) was extracted with BioFast Soil Genomic DNA Extraction Kit (BioFlux, Hangzhou, China) following the instructions. Furthermore, DNA concentration was measured using NanoDrop 2000C Spectrophotometer (Thermo Scientific, United States) and diluted to 20 ngμl–1.
FIGURE 1.
(A) Photographs of R. pseudostellariae under FP, SP, and TP treatments. (B) Yield of R. pseudostellariae under FP, SP, and TP treatments. FP, plot with R. pseudostellariae cultivated in fresh soil; SP, plot with R. pseudostellariae, monocultured for two consecutive years; TP, plot with cultivation of R. pseudostellariae for three consecutive years. Different letters show significant differences according to least significant difference (LSD) (P ≤ 0.05), n = 3.
PCR-DGGE Analysis
To evaluate the changes of Trichoderma community in the rhizosphere, the nested PCR strategy was designed and applied. In the first round of PCR, we used ITS1F and ITS4, taxon-selective ITS primers. PCR amplification protocol is described in Supplementary Table 1. The 50-μl PCR reaction contains 1 μl of each primer (10 mM), 2 μl template DNA (20 ngμl–1), and 25 μl 2X EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China). The products of PCR were subsequently diluted (1:5) for the second PCR reaction via DG-GC (a 40-bp GC-clamp at its 5′ end) and DT primers (Supplementary Table 1). PCR reaction followed for the second round was similar to the first round.
We performed DGGE using the Junyi JY-TD331A system (JUNYI, Beijing, China) using an 8% (w/v) polyacrylamide gel with a denaturation gradient of 30–60%. The gels were electrophoresed in 1X TAE buffer at 80 V and 60°C for 12 h. After electrophoresis, gels were immediately stained with silver stain.
Quantitative PCR for F. oxysporum and Trichoderma Spp.
We performed real-time PCR quantifications of F. oxysporum (ITS1F and AFP308R) and Trichoderma spp. (DG and DT, as mentioned above) in four soil samples (CK, FP, SP, and TP), and amplification protocol is described in Supplementary Table 1. PCR reaction of 15 μl contains 7.5 μl 2X TransStart Green qPCR SuperMix (TransGen Biotech, Beijing, China), 1 μl of each primer (10 μM), 2 μl of template DNA (20 ngμl–1), and 3.5 μl H2O. Meanwhile, serial dilutions of plasmid DNA were set as standard curves. The standard curve was generated by log10 value against the threshold cycle (Ct) value. Four independent quantitative PCR assays were performed for each treatment.
Isolation of Trichoderma spp. With Antagonistic Activity Toward F. oxysporum
To isolate Trichoderma spp., soil suspensions were made by adding 10 g of fresh soil into a flask containing 90 ml of sterile water (10–1 gl–1). After dilution (10–2 gl–1), a total of 100 μl suspensions were plated onto Thayer-Martin agar medium (containing 0.25 gl–1 pentachloronitrobenzene and 30 mgl–1 streptomycin sulfate). The plates were placed in an incubator at 28°C for 4 days and then each single colony was separated and purified.
After purification, we used the CTAB-based method (Rogers and Bendich, 1985) for DNA extraction from different isolates. For sequencing, three primer sets (ITS1F and ITS4, EF1 and EF2, and fRPB2-5f and fRPB2-7cr) were used for amplification (protocol as described in Supplementary Table 1). PCR products were cloned into the pEASY-T1 Cloning vector and sent to BoShang (Fuzhou, China) for sequencing. We further used the BlastN search method to screen for similar sequences in the NCBI and Tricho-BLAST at the website of the International Subcommission on Trichoderma and Hypocrea Taxonomy. For phylogram, published ITS1, rpb2, and tef1 sequences for biocontrol isolates were obtained from GenBank. ClustalX2 programs were used for sequence alignment. Gaps/missing data treatment was set to complete deletion. Phylogenetic analysis was carried out with the MEGA6 software. Maximum likelihood was used for statistical method. Neighbor-joining (NJ) trees were constructed for each data set (ITS1, rpb2, and tef1) using the Tamura–Nei distance measure. The robustness of the internal branches was assessed with 1,000 bootstrap replications (Saitou and Nei, 1987).
Evaluation of the Biocontrol Effects of Trichoderma spp.
For in vitro antagonism assays, Trichoderma isolates were used to evaluate the biocontrol effects against F. oxysporum (pathogenic fungi of R. pseudostellariae), which was part of the microbial collection of our lab (Chen et al., 2017). The strains (F. oxysporum and Trichoderma isolates) were inoculated in potato dextrose agar (PDA) slant culture medium at 4°C. We inoculated each Trichoderma isolates in dual culture with F. oxysporum. Two different isolates were placed 5.5 cm apart on the same PDA plate with three replicates. After incubation at 30°C for 5 days, the parameters of the antagonistic activity of Trichoderma isolates against F. oxysporum were recorded. Thus, the percentage of inhibition (PI) was calculated by the following formula:%PI = [(r1 - r2)/r1] × 100, where r1 is the distance between the furthest point and sowing point of the F. oxysporum and r2 represents the distance between the sowing point and the edge of the F. oxysporum from where F. oxysporum and Trichoderma mycelia came into contact (Supplementary Figure 1).
Unplanted soil (CK) was used for pot assays. R. pseudostellariae were planted in plastic pots and placed in a greenhouse on December 15, 2017. After 5 months of planting, a 2-ml spore suspension (a spore suspension of F. oxysporum with a concentration of 106 spores · ml–1 was made by rinsing mycelia with sterile water) of isolated F. oxysporum was added to the soil for observing the effects of Fusarium wilt on R. pseudostellariae. In addition, 7 days after inoculation with F. oxysporum, spore suspensions of four isolated strains (ZC4, ZC5, ZC51, and CC2-7) were added to pots to evaluate the biocontrol potential of Trichoderma spp. Each treatment was three replicates. After 16 days, rhizospheric soil was collected from each treatment (FOX, ZC4, ZC5, ZC51, and CC2-7), and then, soil DNA was immediately extracted for qPCR assays (as mentioned above) of F. oxysporum and Trichoderma spp.
Expression Analysis of Defense-Related Genes in R. pseudostellariae
To further determine the effect of Trichoderma treatment and/or F. oxysporum infection on the expression of defense-related genes in R. pseudostellariae, we prepared MS medium for in vitro culture of R. pseudostellariae. Seedlings of R. pseudostellariae were transferred in the medium with tweezer and placed in a culture room at 26°C. After 60 days of incubation in the culture room, four treatments were set up: (1) inoculated with F. oxysporum into tissue-cultured seedlings of R. pseudostellariae (F); (2) inoculated with T. harzianum ZC51 into tissue-cultured seedlings of R. pseudostellariae (T); (3) simultaneously inoculated with F. oxysporum and T. harzianum ZC51 into the tissue-cultured seedlings of R. pseudostellariae (TF); and (4) tissue-cultured seedlings of R. pseudostellariae without any treatment (NTF). Seven days after the inoculation, the tissue culture seedlings of R. pseudostellariae were taken out; the plants were washed with sterile water, quickly treated with liquid nitrogen, and frozen in a refrigerator at -80°C for later extraction of RNA.
Plant RNA Isolation and Real-Time PCR Analysis
Plants were ground into powder with liquid nitrogen, and plant RNA was extracted with TransZol Up Plus RNA Kit (TransGen Biotech, Beijing, China) in accordance with the instructions. Furthermore, RNA concentration was measured using NanoDrop 2000C Spectrophotometer (Thermo Scientific, United States). According to the kit’s instructions, the first-strand cDNA was synthesized using TransScript®, miRNA First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Each sample used 1 μg of total RNA, and the products were immediately diluted to 80 μl with DEPC water as a template.
Based on the previous transcriptome data of R. pseudostellariae in our laboratory (Qin et al., 2017), nine primer pairs were used (Supplementary Table 2) to analyze the expression of defense-related genes in R. pseudostellariae as a result of Trichoderma and/or F. oxysporum infection. The actin gene (Supplementary Table 2) was used as an internal reference gene. The 15 μl of the PCR reaction contains 7.5 μl of 2 × SYBR Green qPCR Master Mix (TransGen Biotech, Beijing, China), 1 μl of each primer (10 μM), 0.6 μl of cDNA template, and 5.9 μl H2O. The PCR program was as follows: 94°C for 30 s, followed by 40 cycles of 94 C for 5 s and 60°C for 30 s. After RT-PCR, the 2–Δ Δ CT method (Livak and Schmittgen, 2001) was used to calculate the relative gene expression levels.
Statistical Analysis
Grayscale of DGGE bands was performed with the Quantity One v4.6.2 software to detect the band of gel. Principal component analysis (PCA) of DGGE was performed by SPSS 20.0 software. Diversity analysis of DGGE was performed by DPS 7.05. For RT-PCR, comparison between two groups was done with independent sample T-test by Excel 2013 software. Multiple comparison was carried out by one-way analysis of variance (ANOVA) followed by least significant difference (LSD) test (P ≤ 0.05) using DPS 7.05 software for all parameters.
Results
The Yield of R. pseudostellariae Under Consecutive Monoculture
The yield of R. pseudostellariae in the FP was significantly higher (P < 0.05) than SP and TP (Figure 1B). The fresh weights of roots in FP were 459.8 kg⋅per 666.7 m2, while it was 153.4 kg and 20.9 kg⋅per 666.7 m2 in SP and TP, respectively (Figure 1B).
Soil Nutritional Status
The chemical composition of the soil evaluated was as follows: total nitrogen 1.65 g⋅kg–1, available nitrogen 36.42 mg⋅kg–1, total phosphorus 0.51 g⋅kg–1, effective phosphorus 100.31 mg⋅kg–1, total K 7.66 g⋅kg–1, and effective potassium 322.52 mg⋅kg–1.
Trichoderma-Specific DGGE
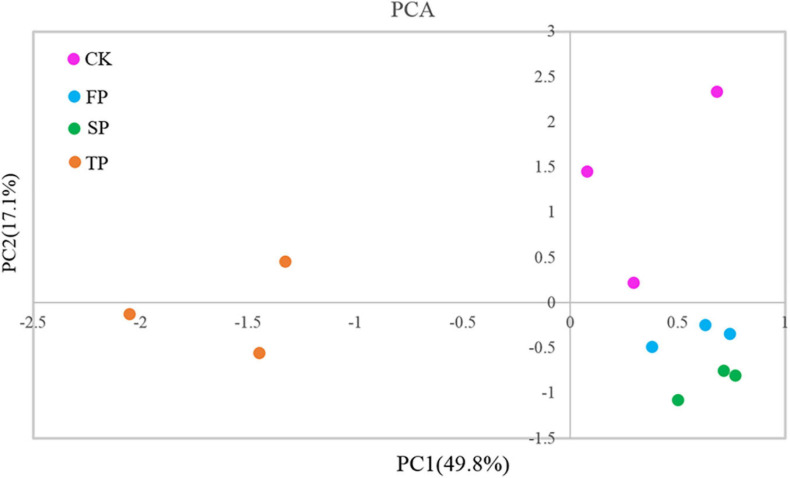
Trichoderma-specific DGGE analysis indicated that the shifts of the Trichoderma community in the rhizosphere changed with increasing period of monoculture (Supplementary Figure 2). Based on DGGE profiles, we performed principal component analysis (PCA) to explore Trichoderma rhizosphere community structure between the four different soil conditions. Among them, the first principal component revealed 49.80% of the total variance, and the second principal component indicated 17.10% of the total variance. The results of PCA also showed that the Trichoderma community in TP was separated from CK, FP, and SP by the first principal component, and CK was separated from FP and SP by the second principal component (Figure 2).
FIGURE 2.
Principal component analysis of Trichoderma DGGE. CK, unplanted soil; FP, plot with R. pseudostellariae cultivated in fresh soil; SP, plot with R. pseudostellariae, monocultured for two consecutive years; TP, plot with cultivation of R. pseudostellariae for three consecutive years; DGGE, denatured gradient gel electrophoresis.
We also analyzed the diversity of Trichoderma-specific DGGE. The results showed that the Simpson, Shannon, and Brillouin’s indices decreased significantly with increasing period of monoculture (P ≤ 0.05). However, there was no significant difference in evenness index among the four samples (Table 1).
TABLE 1.
Estimated Simpson, Shannon, evenness, and Brillouin’s indices for all the samples using Trichoderma-specific DGGE.
| Treatments | Simpson | Shannon | Evenness | Brillouin |
| CK | 0.9094 + 0.0082a | 3.5867 + 0.0571a | 0.9693 + 0.0154a | 3.5603 + 0.054a |
| FP | 0.9147 + 0.0015a | 3.6275 + 0.011a | 0.9803 + 0.003a | 3.6006 + 0.0094a |
| SP | 0.907 + 0.0016a | 3.5073 + 0.0093b | 0.9783 + 0.0026a | 3.4811 + 0.0084b |
| TP | 0.8835 + 0.0025b | 3.1317 + 0.0144c | 0.9879 + 0.0046a | 3.104 + 0.0136c |
Different letters within a column show significant differences according to LSD (P ≤ 0.05), n = 3. CK, unplanted soil; FP, planting of R. pseudostellariae in the newly planted soil; SP, planting of R. pseudostellariae in two consecutive years; TP, planting of R. pseudostellariae in three consecutive years.
Abundance of Trichoderma spp. and F. oxysporum by Quantitative PCR Under Different Continuous Years
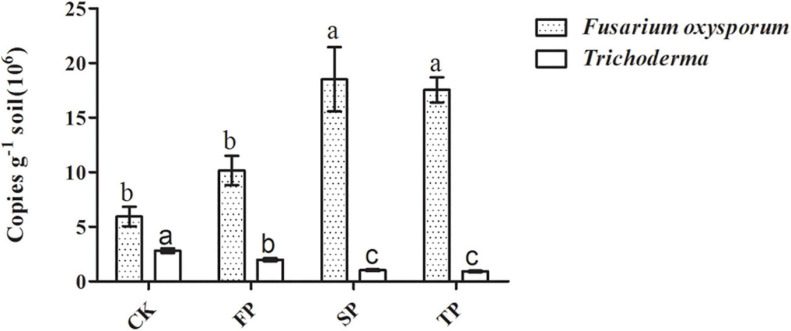
Quantitative PCR was used to analyze changes in Trichoderma spp. and F. oxysporum abundance in four soil samples (Figure 3). For Trichoderma spp. and F. oxysporum qPCR analyses, standard curves of y = −0.2271x + 10.763 (R2 = 0.9944) and y = −0.2673x + 10.607 (R2 = 0.9986), respectively, were developed. Abundance of Trichoderma spp. was significantly decreased with prolonged monoculture (Figure 3). These results were consistent with the Trichoderma-specific DGGE analysis (Supplementary Figure 2). However, the quantitative PCR results for F. oxysporum were the opposite (Figure 3).
FIGURE 3.
Quantification of Trichoderma spp. and F. oxysporum in the different plots. CK, control with unplanted soil; FP, plot with R. pseudostellariae cultivated in fresh soil; SP, plot with R. pseudostellariae, monocultured for two consecutive years; TP, plot with cultivation of R. pseudostellariae for three consecutive years. Different letters in the same color show significant differences according to least significant difference (LSD) (P ≤ 0.05); data are means ± standard errors (one-way analysis of variance, n = 4).
Screening for Trichoderma Isolates With Antagonistic Activity Toward F. oxysporum
For in vitro antagonism assays, we screened seven isolates of Trichoderma from four different soils. The results of sequencing showed that the seven isolates belonged to three species of Trichoderma (Table 2). The accession number (ITS, tef1, and rpb2) of seven isolates are in Table 2. Among these, ZC5 isolate showed the highest antagonistic activity (72.77%) (Table 2) toward F. oxysporum, whereas ZC13 showed the lowest antagonistic activity.
TABLE 2.
In vitro antifungal activity of Trichoderma strains against F. oxysporum.
| Lab. Code | Inhibition in growth assay | Identify | Accession number (ITS) | Accession number (rpb2) | Accession number (tef1) |
| ZC5 | 47.91 ± 3.41a | T. harzianum | MW376900.1 | MW407164 | MW415424 |
| ZC51 | 47.66 ± 3.21a | T. harzianum | MW376903.1 | MW407167 | MW415425 |
| ZC4 | 30.66 ± 1.36b | T. asperelloides | MW376899.1 | MW407163 | WM588808 |
| CC2-7 | 30.16 ± 11.31b | T. rugulosum | MW376897.1 | MW407161 | MW588806 |
| ESK2 | 24.41 ± 4.44bc | T. asperelloides | MW376898.1 | MW407162 | MW588807 |
| ZC11 | 23.62 ± 1.67bc | T. asperelloides | MW376901.1 | MW407165 | MW588809 |
| ZC13 | 16.67 ± 2.67c | T. asperelloides | MW376902.1 | MW588811 | MW588810 |
Different letters within a column show significant differences according to LSD (P ≤ 0.05), n = 3.
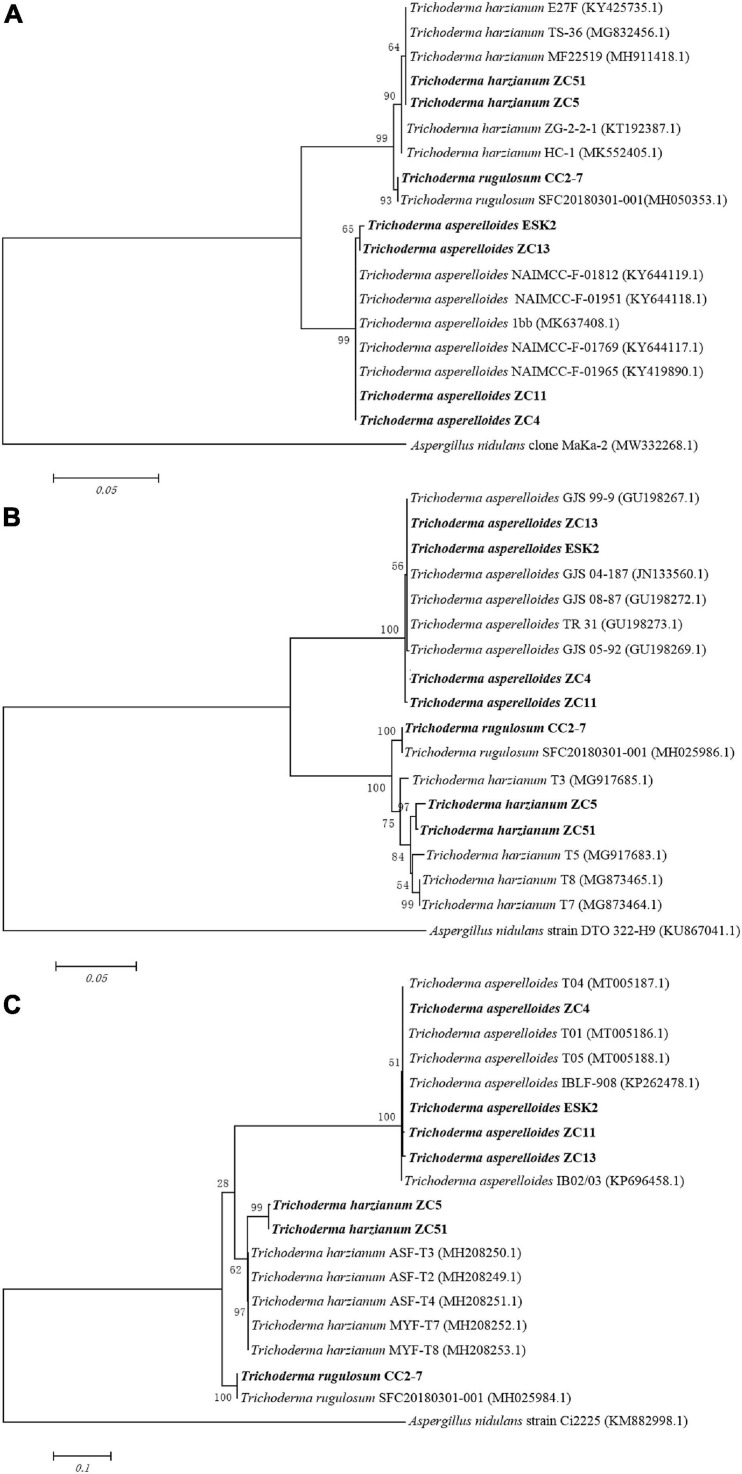
The phylogenetic analysis (ITS) using neighbor-joining method generated a dendrogram with three main branches, where the first branch included T. harzianum ZC5 and ZC51; the second branch comprised T. rugulosum CC2-7; and the third branch comprised T. asperelloides ESK2, ZC13, ZC11, and ZC4 (Figure 4A). For rpb2, a dendrogram contained three main branches, where the first branch included T. asperelloides ZC13, ESK2, ZC14, and ZC11; the second branch comprised T. rugulosum CC2-7; and the third branch comprised T. harzianum ZC5 and ZC51 (Figure 4B). For tef1, a dendrogram contained three main branches, where the first branch included T. asperelloides ZC4, ESK2, ZC11, and ZC13; the second branch comprised T. harzianum ZC5 and ZC51; and the third branch comprised T. rugulosum CC2-7 (Figure 4C).
FIGURE 4.
Neighbor-joining tree based on sequence analysis of Trichoderma isolates used in this study. Values of the bootstrap analysis (1,000 repetitions) are given at the nodes. (A) ITS; (B) rpb2; (C) tef1. Aspergillus nidulans was the outgroup. Sequences of biocontrol isolates used for this comparison were obtained from GenBank.
Biocontrol Effects of Trichoderma spp.
Based on the in vitro antagonism assays, four isolates of Trichoderma (ZC4, ZC5, ZC51, and CC2-7) that showed inhibition activity against F. oxysporum higher than 30% were selected for further in vivo biocontrol assay (Figure 5). In the pot experiment, compared with the control (FOX), we found that T. harzianum ZC51 significantly inhibited the growth of F. oxysporum and enhanced the growth of R. pseudostellariae, whereas no disease symptoms developed during the period of the experiment (Figure 5D).
FIGURE 5.
Biocontrol potential of Trichoderma against F. oxysporum. (A) R. pseudostellariae was treated with F. oxysporum (FOX); (B) R. pseudostellariae was treated with F. oxysporum and T. asperellum ZC4 (ZC4); (C) R. pseudostellariae was treated with F. oxysporum and T. harzianum ZC5 (ZC5); (D) R. pseudostellariae was treated with F. oxysporum and T. harzianum ZC51 (ZC51); (E) R. pseudostellariae was treated with F. oxysporum and T. hamatum CC2-7 (CC2-7).
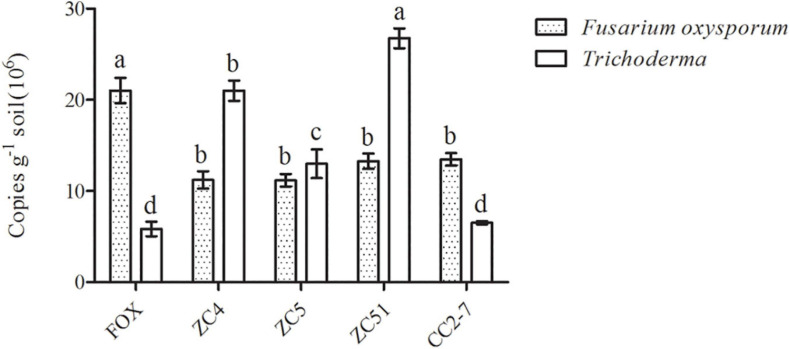
Moreover, quantitative PCR was used to analyze changes in Trichoderma spp. and F. oxysporum abundance in the pot experiments. Compared with the control (FOX), the abundance of Trichoderma spp. in the soil treated with Trichoderma strains ZC4, ZC5, and ZC51 increased significantly, with T. harzianum ZC51 showing the highest abundance (Figure 6). Finally, the quantitative PCR results for F. oxysporum in the pot experiments showed that all treatments significantly decreased as compared to control (Figure 6). The results showed that T. harzianum ZC51 strain, potentially, could be used as a biological control agent against F. oxysporum.
FIGURE 6.
Quantification of Trichoderma spp. and F. oxysporum in the pot experiment. FOX, R. pseudostellariae was treated with F. oxysporum; ZC4, R. pseudostellariae was treated with F. oxysporum and T. asperellum ZC4; ZC5, R. pseudostellariae was treated with F. oxysporum and T. harzianum ZC5; ZC51, R. pseudostellariae was treated with F. oxysporum and T. harzianum ZC51; CC2-7, R. pseudostellariae was treated with F. oxysporum and T. hamatum CC2-7. Different letters in the same color show significant differences according to least significant difference (LSD) (P ≤ 0.05); data are means ± standard errors (one-way analysis of variance, n = 4).
Effect of Trichoderma Treatment and/or F. oxysporum in the Expression of R. pseudostellariae Defense-Related Genes
The T. harzianum ZC51 strain was selected, based on its positive effect on R. pseudostellariae phenotype without infection (Figure 5D) and the highest abundance among all treatments (Figure 6). The expression of defense-related genes was examined in these plants, i.e., Trichoderma ZC51-non-inoculated and F. oxysporum-infected plants (F), Trichoderma ZC51-inoculated and F. oxysporum-non-infected plants (T), Trichoderma ZC51-inoculated and F. oxysporum-infected (TF) plants, or Trichoderma ZC51-non-inoculated and F. oxysporum-non-infected plants (NTF).
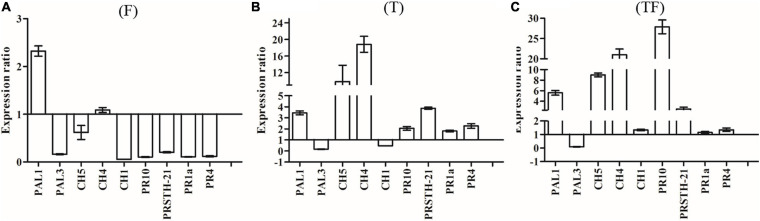
To analyze the expression of defense-related genes, we used actin as a housekeeping gene to determine the relative expression levels of other genes. Expressions of PAL1 and PAL3 were studied to determine the involvement of phenylalanine ammonia lyase in R. pseudostellariae response to T. harzianum ZC51 treatment and/or F. oxysporum. There was no significant difference of the PAL1 and PAL3 expression in the three treatments (Figures 7A–C). We also examined the expression of CH1, CH4, and CH5 of chitinase by treated plants. Expression of CH4 and CH5 increased in plants inoculated with Trichoderma ZC51 (T) (Figure 7B) and Trichoderma ZC51-inoculated and F. oxysporum-infected (TF) (Figure 7C). In contrast, the expression of CH1 and CH5 decreased in plants only infected with F. oxysporum (F) (Figure 7A). In addition to these genes, we further analyzed the expression of PRSTH-21, PR1a, PR4, and PR10 involved in the process of plant disease resistance. The expression of PRSTH-21, PR1a, PR4, and PR10 increased in Trichoderma ZC51-inoculated (T) (Figure 7B) and F. oxysporum-infected (TF) (Figure 7C) plants. However, the opposite was true for the plants only infected with F. oxysporum (F) (Figure 7A).
FIGURE 7.
Expression of PAL1, PAL3, CH5, CH4, CH1, PR10, PRSTH-21, PR1a, and PR4 genes in comparison with α-actin reference genes. (A) Inoculated with F. oxysporum into tissue-cultured seedlings of R. pseudostellariae; (B) inoculated with T. harzianum ZC51 into tissue-cultured seedlings of R. pseudostellariae; (C) simultaneously inoculated with F. oxysporum and T. harzianum ZC51 into the tissue-cultured seedlings of R. pseudostellariae. Data are means ± standard errors (one-way analysis of variance, n = 4).
Also, in order to understand the effect of Trichoderma ZC51 (T) on gene expression of F. oxysporum (F)-infected R. pseudostellariae, we compared the expression of the same gene in different treatments (Table 3). Among these genes, the expression of PAL1, CH5, CH4, CH1, PR10, PRSTH-21, PR1a, and PR4 significantly increased in plants which were treated with T and TF treatments (Table 3).
TABLE 3.
Expression of PAL1, PAL3, CH5, CH4, CH1, PR10, PRSTH-21, PR1a, and PR4 genes in comparison with α-actin reference genes.
| Treatments | F | T | TF |
| PAL1 | 2.3231 ± 0.1857c | 3.4471 ± 0.3011b | 5.5851 ± 0.7776a |
| PAL3 | 0.1622 ± 0.0157a | 0.1699 ± 0.0257a | 0.0929 ± 0.026b |
| CH5 | 0.6188 ± 0.2544b | 9.8098 ± 6.8382a | 8.9884 ± 0.6374a |
| CH4 | 1.0905 ± 0.0894b | 18.8246 ± 3.3248a | 21.0104 ± 2.4285a |
| CH1 | 0.0565 ± 0.0014c | 0.471 ± 0.0095b | 1.3374 ± 0.1003a |
| PR10 | 0.1049 ± 0.0136b | 2.0501 ± 0.2914b | 27.8775 ± 2.9425a |
| PRSTH-21 | 0.2035 ± 0.0206c | 3.872 ± 0.1593a | 2.5438 ± 0.5835b |
| PR1a | 0.1075 ± 0.005c | 1.8026 ± 0.1541a | 1.1476 ± 0.1243b |
| PR4 | 0.1188 ± 0.021c | 2.2681 ± 0.3655a | 1.3469 ± 0.2162b |
Different letters within a row show significant differences according to LSD (P ≤ 0.05), n = 4. F, inoculated with F. oxysporum into tissue-cultured seedlings of R. pseudostellariae; T, inoculated with T. harzianum ZC51 into tissue-cultured seedlings of R. pseudostellariae; TF, simultaneously inoculated with F. oxysporum and T. harzianum ZC51 into the tissue-cultured seedlings of R. pseudostellariae.
In general, the results demonstrated that Trichoderma ZC51 interaction with R. pseudostellariae affected the expression of plant defense-related genes related to the chitinase and pathogenesis-related proteins, but does not involve phenylalanine ammonia lyase.
Discussion
The low quality and reduced yield of Chinese medicinal herbs are commonly observed due to recurrent cultivation on the same land for many years. This phenomenon of low yield, compromised medicinal quality, poor growth of plants, and high disease susceptibility is owing to consecutive monoculture problems or soil sickness (Zhang and Lin, 2009; Wu et al., 2016c). Our study revealed the facts for typical growth inhibition effects under consecutive monoculture of R. pseudostellariae, with poor plant performance and insufficient resistance to disease. Soil physical and chemical properties, accumulation of root exudates, and shift in the soil microbial community are some factors responsible for the consecutive monoculture problem of R. Pseudostellariae (Zhang and Lin, 2009; Wu et al., 2016a). The biological relationships between plants and microorganisms in the rhizosphere play a crucial role for the health and growth of a plant, which has been paid much attention in recent days (Haney and Ausubel, 2015; Lebeis et al., 2015).
Trichoderma spp. have been studied commonly because of its ability to inhibit soil-borne pathogens and have good plant defense responses (Papavizas, 1985; Hermosa et al., 2012; Pimentel et al., 2020). In this study, PCR-DGGE results showed significant shifts in Trichoderma community in the rhizosphere of R. pseudostellariae after extended monoculture (Supplementary Figure 2 and Figure 2). Based on PCR-DGGE of Trichoderma, results of diversity showed that the extended monoculture of R. pseudostellariae significantly decreased the Trichoderma spp. diversity (Table 1). Quantitative PCR assay confirmed the decrease in Trichoderma with the increasing years of monoculture (Figure 3A), whereas the abundance of F. oxysporum was significantly increased (Figure 3B). A previous study has also reported the changes in the composition and diversity of Fusarium spp. and increase in the abundance of F. oxysporum with the increasing years of monoculture (Chen et al., 2017). This selective change in the microbial community is due to the difference in response of these microorganisms to the root exudates in the rhizosphere (Huang et al., 2014; Zhalnina et al., 2018).
A negative shift in the composition of the soil microbial community is a consequence of the development of soil-borne diseases (Mazzola, 2004). Therefore, maintaining the biodiversity of beneficial soil microbes is crucial to soil health. Biological control with exploitation of the rhizosphere microorganisms that can directly antagonize with plant pathogens is considered to be the most promising method for preventing plant diseases (Qiu et al., 2012; Shen et al., 2014). These species mostly include antagonistic fungi such as Trichoderma spp. and Penicillium spp. (Cotxarrera et al., 2002; Howell, 2002; Siddiqui and Akhtar, 2009). In this study, most of the isolated strains of Trichoderma can inhibit the growth of F. oxysporum (Table 2). The difference in antagonistic abilities may be due to genotype variability (Debbi et al., 2018). The in vivo assays revealed that R. pseudostellariae treated with T. harzianum ZC51 has the best growth phenotype without displaying any disease symptom. Yedidia et al. (2001) reported that T. harzianum T-203 increased the root length, aerial parts, dry weight, and size of the blade by 75, 95, 80, and 45%, respectively, in cucumber plants. Other studies have shown that Trichoderma spp. could promote plant growth, increase nutrient utilization, and improve crop production (Harman et al., 2004).
There is ample evidence that Trichoderma species could induce plant defense responses (Yedidia et al., 1999; Gallou et al., 2009; Singh et al., 2011). However, little is known about the effect of Trichoderma treatment on the expression of defense-related genes in R. Pseudostellariae. PR proteins are well-known proteins that is induced by pathogens and play an important role in the process of plant disease resistance (Linthorst and Van Loon, 1991; Edreva, 2005). Moreover, chitinase proteins a pathogenesis-related proteins that are induced by pathogens; thus, chitinase constitute a crucial part of the plant’s defense against fungal pathogens (Punja and Zhang, 1993; El Hadrami et al., 2010).
As described, the interaction of R. pseudostellariae with F. oxysporum caused the repression of the seven defense-related genes (PAL3, CH5, CH1, PR10, PRSTH-21, PR1a, and PR4), via a mechanism to overcome plant defense responses and thereby promoting the process of infection in plants (Peix et al., 2001). Zhao et al. (2003) also reported similar results in experiments with tomato plants infected with Pseudomonas syringae, where tomatoes showed repression of PR1 and PR4, suggesting that infection with pathogen would reduce the plant self-defense mechanism, hence promoting the development of the disease.
In this study, when the interaction of R. pseudostellariae with T. harzianum ZC51 was analyzed, PR10, PRSTH-21, PR1a, PR4, CH4, and CH5b were up-regulated. Others have shown that T. harzianum T39 reduces the incidence of downy mildew of grapes by directly regulating the expression of defense-related genes (Perazzolli et al., 2011). Similarly, a study has reported the increase in the expression level of several defense-related genes in olive trees only when T. harzianum (Ths97) was applied together with the root rot pathogen F. solani (Amira et al., 2017). Other studies have shown that Trichoderma spp. may also trigger ISR in plants, mainly related to the expression of pathogenesis-related proteins (i.e., PR1, PR2, and PR5) (Hermosa et al., 2012; Mathys et al., 2012). Phenylalanine ammonia lyase (PAL) is one of the most widely studied enzymes involved in the process of plant disease resistance (Kim and Hwang, 2014). In this study, T. harzianum ZC51 did not change the expression of the PAL genes.
Compared with the “simple” two-partner systems (i.e., plant–pathogen or plant–antagonist), the complex three-way interactions involving Trichoderma, plant, and pathogen has received less attention, and this model can better simulate the natural interactions occurring in soil (Vinale et al., 2008). In our study, when T. harzianum ZC51 and F. oxysporum were applied together on R. pseudostellariae, we observed an upregulation of all the analyzed genes with the exception of PAL3. Similar results in the experiments to bean (Phaseolus vulgaris L.) infected with R. solani and/or Trichoderma were observed. The level of expression of defense-related genes (CH5b, CH1, PR1, PR2, PR3, and PR4) were up-regulated. Marra et al. (2006) studied the three-way interaction of Trichoderma with plant and fungal pathogens using proteomics methods, and the results show that antagonistic fungi will reduce the production of some defense proteins but will lead to the accumulation of others (i.e., PR proteins). This suggests that even in the presence of pathogens, several mechanisms are induced in Trichoderma that potentiates its ability to elicit plant defense responses (Mayo et al., 2016). Our results indicate that Trichoderma activate plants’ defense responses and so could be an optimized defense strategy against different plant stress, including plant pathogens and monocropping disease.
Conclusion
To sum up, this study revealed that the continuous monocropping of R. pseudostellariae favored the growth of pathogenic F. oxysporum but decreased the antagonistic fungi (Trichoderma spp.), which resulted in poor yield of R. pseudostellariae. The exogenous application of T. harzianum ZC51 increased the expression levels of genes (PR10, PRSTH-21, PR1a, PR4, CH4, and CH5b) previously involved in plant defense, leading to enhanced defense response and improved growth of the host plant. These findings can be useful to develop locally customized and innovative approaches to address major threats facing medicinal plant cultivation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
WL, JC, and SL conceived the study. JC, LZ, and ID wrote the manuscript. JC, QL, and JW performed the experiments. TW, LW, LZ, and HW performed the statistical analyses. XQ and YA were involved in field management and soil sampling. GP assisted in English correction. All the authors discussed the results and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Professor Irina Druzhinina (the chair of International Commission on Trichoderma Taxonomy) and Dr. Feng Cai (Nanjing Agricultural University) for assistance in the identification of Trichoderma strains.
Footnotes
Funding. This work was supported by the National Key Research and Development Plan 2017YFE0121800 and grants from the National Natural Science Foundation of China (Nos. 81973412 and 81573530), the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (YB2018002), the Project of Key Laboratory of Ministry of Education (GBMUC-2018-006), and the Science and Technology Development Fund of Fujian Agriculture and Forestry University (KF2015043).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.579920/full#supplementary-material
References
- Amira M. B., Lopez D., Mohamed A. T., Khouaja A., Chaar H., Fumanal B., et al. (2017). Beneficial effect of Trichoderma harzianum strain Ths97 in biocontrolling Fusarium solani causal agent of root rot disease in olive trees. Biol. Control 110 70–78. 10.1016/j.biocontrol.2017.04.008 [DOI] [Google Scholar]
- Brotman Y., Landau U., Cuadros-Inostroza A., Tohge T., Fernie A. R., Chet I., et al. (2013). Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:e1003221. 10.1371/journal.ppat.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu L. K., Xiao Z. G., Wu Y. H., Wu H. M., Qin X. J., et al. (2017). Assessment of the diversity of Pseudomonas spp. and Fusarium spp. in Radix pseudostellariae rhizosphere under monoculture by combining DGGE and quantitative PCR. Front. Microbiol. 8:1748. 10.3389/fmicb.2017.01748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales M., Fernandez A., Pinto M. G. V., Butz P., Franz C. M., Schuele E., et al. (2010). Characterization of phenolic content, in vitro biological activity, and pesticide loads of extracts from white grape skins from organic and conventional cultivars. Food Chem. Toxicol. 48 3471–3476. 10.1016/j.fct.2010.09.025 [DOI] [PubMed] [Google Scholar]
- Cotxarrera L., Trillas-Gay M. I., Steinberg C., Alabouvette C. (2002). Use of sewage sludge compost and Trichoderma asperellum isolates to suppress Fusarium wilt of tomato. Soil Biol. Biochem. 34 467–476. 10.1016/S0038-0717(01)00205-X [DOI] [Google Scholar]
- De Palma M., Salzano M., Villano C., Aversano R., Lorito M., Ruocco M., et al. (2019). Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Horticul. Res. 6:5. 10.1038/s41438-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbi A., Boureghda H., Monte E., Hermosa R. (2018). Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in Algeria and a biocontrol strategy with indigenous Trichoderma spp. Front. Microbiol. 9:282. 10.3389/fmicb.2018.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva A. (2005). Pathogenesis-related proteins: research progress in the last 15 years. Gen. Appl. Plant Physiol. 2005 105–124. [Google Scholar]
- El Hadrami A., Adam L. R., El Hadrami I., Daayf F. (2010). Chitosan in plant protection. Mar. Drugs 8 968–987. 10.3390/md8040968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti S., Paris R., Cianchetta S. (2020). Selected isolates of Trichoderma gamsii induce different pathways of systemic resistance in maize upon Fusarium verticillioides challenge. Microbiol. Res. 233:126406. 10.1016/j.micres.2019.126406 [DOI] [PubMed] [Google Scholar]
- Gallou A., Cranenbrouck S., Declerck S. (2009). Trichoderma harzianum elicits defence response genes in roots of potato plantlets challenged by Rhizoctonia solani. Eur. J. Plant Pathol. 124 219–230. 10.1007/s10658-008-9407-x [DOI] [Google Scholar]
- Haney C. H., Ausubel F. M. (2015). Plant microbiome blueprints. Science 349 788–789. 10.1126/science.aad0092 [DOI] [PubMed] [Google Scholar]
- Harman G. E., Howell C. R., Viterbo A., Chel I., Lorito M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2 43–56. 10.1038/nrmicro797 [DOI] [PubMed] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158 17–25. 10.1099/mic.0.052274-0 [DOI] [PubMed] [Google Scholar]
- Howell C. R. (2002). Cotton seedling preemergence damping-off incited by Rhizopus oryzae and Pythium spp. and its biological control with Trichoderma spp. Phytopathology 92 177–180. 10.1094/PHYTO.2002.92.2.177 [DOI] [PubMed] [Google Scholar]
- Howell C. R. (2006). Understanding the mechanisms employed by Trichoderma virens to effect biological control of cotton diseases. Phytopathology 96 178–180. 10.1094/PHYTO-96-0178 [DOI] [PubMed] [Google Scholar]
- Huang X., Chaparro J. M., Reardon K. F., Zhang R., Shen Q., Vivanco J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92 267–275. 10.1139/cjb-2013-0225 [DOI] [Google Scholar]
- Jackson M. L. (1958). Soil Chemical Analysis, Vol. 498. Englewood Cliffs. NJ: Prentice Hall. Inc., 183–204. [Google Scholar]
- Kim D. S., Hwang B. K. (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65 2295–2306. 10.1093/jxb/eru109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis S. L., Paredes S. H., Lundberg D. S., Breakfield N., Gehring J., McDonald M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349 860–864. 10.1126/science.aaa8764 [DOI] [PubMed] [Google Scholar]
- Lin S., Huangpu J. J., Chen T., Wu L. K., Zhang Z. Y., Lin W. X. (2015). Analysis of soil microbial community structure and enzyme activities associated with negative effects of Pseudostellaria heterophylla consecutive monoculture on yield. Pak. J. Bot. 47 761–769. [Google Scholar]
- Linthorst H. J., Van Loon L. C. (1991). Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci. 10 123–150. 10.1080/07352689109382309 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorito M., Woo S. L., Harman G. E., Monte E. (2010). Translational research on Trichoderma: from ’Omics to the field. Annu. Rev. Phytopathol. 48 395–417. 10.1146/annurev-phyto-073009-114314 [DOI] [PubMed] [Google Scholar]
- Manganiello G., Sacco A., Ercolano M. R., Vinale F., Lanzuise S., Pascale A., et al. (2018). Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic Acid. Front. Microbiol. 9:1966. 10.3389/fmicb.2018.01966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra R., Ambrosino P., Carbone V., Vinale F., Woo S. L., Ruocco M., et al. (2006). Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 50 307–321. 10.1007/s00294-006-0091-0 [DOI] [PubMed] [Google Scholar]
- Matarese F., Sarrocco S., Gruber S. (2012). Biocontrol of Fusarium head blight: interactions between Trichoderma and mycotoxigenic Fusarium. Microbiology 158 98–106. 10.1099/mic.0.052639-0 [DOI] [PubMed] [Google Scholar]
- Mathys J., De Cremer K., Timmermans P., Van Kerkhove S., Lievens B., Vanhaecke M., et al. (2012). Genome-wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front. Plant Sci. 3:108. 10.3389/fpls.2012.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo S., Cominelli E., Sparvoli F., González-López O., Rodríguez-González A., Gutiérrez S., et al. (2016). Development of a qPCR strategy to select bean genes involved in plant defense response and regulated by the Trichoderma velutinum–Rhizoctonia solani interaction. Front. Plant Sci. 7:1109. 10.3389/fpls.2016.01109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M. (2004). Assessment and management of soil microbial community structure for disease suppression. Annu. Rev. Phytopathol. 42 35–59. 10.1146/annurev.phyto.42.040803.140408 [DOI] [PubMed] [Google Scholar]
- Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332 1097–1100. 10.1126/science.1203980 [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Horwitz B. A., Herrera-Estrella A., Schmoll M., Kenerley C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51 105–129. 10.1146/annurev-phyto-082712-102353 [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Hurley J. F., Taylor J. T., Puckhaber L., Lehner S., Druzhinina I., et al. (2018). Ferricrocin, the intracellular siderophore of Trichoderma virens, is involved in growth, conidiation, gliotoxin biosynthesis and induction of systemic resistance in maize. Biochem. Bioph. Res. Commun. 505 606–611. 10.1016/j.bbrc.2018.09.170 [DOI] [PubMed] [Google Scholar]
- Papavizas G. C. (1985). Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu. Rev. Phytopathol. 23 23–54. 10.1146/annurev.py.23.090185.000323 [DOI] [Google Scholar]
- Peix A., Mateos P. F., Martinez-Molina E., Rodriguez-Barrueco C., Velazquez E. (2001). Growth promotion of common bean (Phaseolus vulgaris L.) by a strain of Burkholderia cepacia under growth chamber conditions. Soil. Biol. Biochem. 33 1927–1935. 10.1016/s0038-0717(01)00119-5 [DOI] [Google Scholar]
- Perazzolli M., Roatti B., Bozza E., Pertot I. (2011). Trichoderma harzianum T39 induces resistance against downy mildew by priming for defense without costs for grapevine. Biol Control 58 74–82. 10.1016/j.biocontrol.2011.04.006 [DOI] [Google Scholar]
- Pieterse C. M. J., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C. M., Bakker P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52 347–375. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Pimentel M. F., Arnao E., Warner A. J., Subedi A., Rocha L. F., Srour A. Y., et al. (2020). Trichoderma isolates inhibit Fusarium virguliforme growth, reduce root rot, and induce defense-related genes on soybean seedlings. Plant Dis. 104 1949–1959. 10.1094/PDIS-08-19-1676-RE [DOI] [PubMed] [Google Scholar]
- Punja Z. K., Zhang Y. (1993). Plant chitinases and their roles in resistance to fungal diseases. J. Nematol. 25 526–540. 10.1016/0022-1910(93)90133-C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. J., Wu H. M., Chen J., Wu L. K., Lin S., Khan M. U., et al. (2017). Transcriptome analysis of Pseudostellaria heterophylla in response to the infection of pathogenic fusarium oxysporum. BMC Plant Biol. 17:155. 10.1186/s12870-017-1106-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M. H., Zhang R. F., Xue C., Zhang S. S., Li S. Q., Zhang N., et al. (2012). Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fert. Soils 48 807–816. 10.1007/s00374-012-0675-4 [DOI] [Google Scholar]
- Rogers S. O., Bendich A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 5 69–76. 10.1007/BF00020088 [DOI] [PubMed] [Google Scholar]
- Romera F. J., García M. J., Lucena C., Martínez-Medina A., Aparicio M. A., Ramos J., et al. (2019). Induced systemic resistance (ISR) and Fe deficiency responses in dicot plants. Front. Plant Sci. 10:287. 10.3389/fpls.2019.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. N. M., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Shen Z. Z., Wang D. S., Ruan Y., Xue C., Zhang J., Li R., et al. (2014). Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS One 9:e98420. 10.1371/journal.pone.0098420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M., Harman G. E., Mastouri F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48 21–43. 10.1146/annurev-phyto-073009-114450 [DOI] [PubMed] [Google Scholar]
- Siddiqui Z. A., Akhtar M. S. (2009). Effects of antagonistic fungi, plant growth-promoting rhizobacteria, and arbuscular mycorrhizal fungi alone and in combination on the reproduction of Meloidogyne incognita and growth of tomato. J. Gen. Plant Pathol. 75 144–153. 10.1007/s10327-009-0154-4 [DOI] [Google Scholar]
- Singh B. N., Singh A., Singh S. P., Singh H. B. (2011). Trichoderma harzianum-mediated reprogramming of oxidative stress response in root apoplast of sunflower enhances defence against Rhizoctonia solani. Eur. J. Plant Pathol. 131 121–134. 10.1007/s10658-011-9792-4 [DOI] [Google Scholar]
- Vinale F., Sivasithamparam K., Ghisalberti E. L., Marra R., Woo S. L., Lorito M. (2008). Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 40 1–10. 10.1016/j.soilbio.2007.07.002 [DOI] [Google Scholar]
- Walters D., Ratsep J., Havis N. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64 1263–1280. 10.1093/jxb/ert026 [DOI] [PubMed] [Google Scholar]
- Watanabe F. S., Olsen S. R. (1965). Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soil. Soil Sci. Soc. Am. J. 29 677–680. 10.2136/sssaj1965.03615995002900060025x [DOI] [Google Scholar]
- Wu H. M., Wu L. K., Wang J. Y., Zhu Q., Lin S., Xu J. J., et al. (2016). Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front Microbiol. 7:335. 10.3389/fmicb.2016.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. K., Chen J., Wu H. M., Qin X. J., Wang J. Y., Wu Y. H., et al. (2016a). Insights into the regulation of rhizosphere bacterial communities by application of bio-organic fertilizer in Pseudostellaria heterophylla monoculture regime. Front. Microbiol. 7:1788. 10.3389/fmicb.2016.01788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. K., Chen J., Wu H. M., Wang J. Y., Wu Y. H., Lin S., et al. (2016b). Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. U. K. 6:26601. 10.1038/srep26601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. K., Wang J. Y., Huang W. M., Wu H. M., Chen J., Yang Y. Q., et al. (2016c). Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. U. K. 6:19101. 10.1038/srep15871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen L., Yong X., Shen Q. (2011). Formulations can affect rhizosphere colonization and biocontrol efficiency of Trichoderma harzianum SQR-T037 against Fusarium wilt of cucumbers. Biol. Fert. Soils 47 239–248. 10.1007/s00374-010-0527-z [DOI] [Google Scholar]
- Yedidia I., Benhamou N., Chet I. (1999). Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microb. 65 1061–1070. 10.1128/AEM.65.3.1061-1070.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I., Srivastva A. K., Kapulnik Y., Chet I. (2001). Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil 235 235–242. 10.1023/a:1011990013955 [DOI] [Google Scholar]
- Zhalnina K., Louie K. B., Hao Z., Mansoori N., Da Rocha U. N., Shi S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3 470–480. 10.1038/s41564-018-0129-3 [DOI] [PubMed] [Google Scholar]
- Zhang Z. Y., Lin W. X. (2009). Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. Eco Agric. 17 189–196. 10.3724/sp.j.1011.2009.00189 [DOI] [Google Scholar]
- Zhao W. O., Pang L., Dong N., Yang S. (2015). LC-ESI-MS/MS analysis and pharmacokinetics of heterophyllin B, a cyclic octapeptide from Pseudostellaria heterophylla in rat plasma. Biomed. Chromatog. 29 1693–1699. 10.1002/bmc.3481 [DOI] [PubMed] [Google Scholar]
- Zhao Y. F., Thilmony R., Bender C. L., Schaller A., He S. Y., Howe G. A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36 485–499. 10.1046/j.1365-313X.2003.01895.x [DOI] [PubMed] [Google Scholar]
- Zhao Y. P., Wu L. K., Chu L. X., Yang Y. Q., Li Z. F., Azeem S., et al. (2014). Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f.sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. U. K. 5:8197. 10.1038/srep08197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.