Abstract
Gardner's syndrome is a rare autosomal dominant hereditary disease that is characterized by multiple colorectal polyps combined with extra-colonic presentation (such as osteoma or desmoid tumors) of familial adenomatous polyposis syndrome. Gardner's syndrome is caused by the mutation of the adenomatous polyposis coli (APC) gene, which is located at 5q21. The aim of the current study was to investigate the APC gene mutations present in a Han Chinese family diagnosed with Gardner's syndrome. The 38-year-old proband presented with clinical symptoms, and was later diagnosed with Gardner's syndrome. Genomic DNA was extracted from the peripheral venous blood of 150 normal controls as well as the family members of the proband. Analysis of the respective APC gene sequences was performed using PCR amplification and Sanger sequencing. Pathogenesis associated with the APC mutation was investigated using reverse-transcription quantitative PCR and determined through bioinformatics approaches. Haplotype analysis was performed to identify the genetic source of the mutation(s). In the initial screening for APC variants, the APC c.4621C>T variant was detected in the proband and his son, but was not detected in the proband's affected mother. The mRNA expression changed significantly according to age and the presence of the mutation in the blood of the patients. Haplotype analysis suggested the presence of maternal mosaicism for this mutation. Haplotype analysis revealed that the APC c.4621C>T variant in a patient with Gardner's syndrome was most likely derived from his mother through mosaicism. These results indicate the necessity to verify the possibility of gonadal mosaicism when a proband diagnosed with Gardner's syndrome appears to exhibit a de novo mutation.
Keywords: Gardner's syndrome, maternal mosaicism, adenomatous polyposis coli, variant, haplotype analysis
Introduction
Gardner's syndrome is also known as familial multiple colon polyposis-osteoma-soft tissue tumor syndrome. In addition to the development of intestinal polyposis and colorectal adenocarcinoma, which are key features of Gardner's syndrome, Gardner's syndrome also exhibits extra-colonic presentation of the familial adenomatous polyposis syndrome, which include dental abnormalities, osteomas, soft-tissue tumors (including desmoid tumors) and epidermoid cysts (1,2). Dental abnormalities include impacted and additional teeth, and osteomas typically occur in the mandible, but can also present in the skull and long bones (3). Osteoma and dental abnormalities may be early sensitive indicators for the diagnosis of Gardner's syndrome (4,5). Affected individuals also have an increased risk for extra-colonic malignancies, including gastric (6) and papillary tumors of the duodenum (7).
Gardner's syndrome is an autosomal-dominant disorder that is caused by germline mutations in the adenomatous polyposis coli (APC) gene (3). There is no significant difference in the incidence of Gardner's syndrome in various regions of the world, and its worldwide incidence ranges between 1 in 4,000 and 1 in 12,000(8). Mutational analysis of the APC gene indicates that the majority of germline variants include nonsense mutations, which leads to the formation of a truncated protein (9). According to genotypic and phenotypic correlation studies involving APC mutations, the mutation site associated with Gardner's syndrome mainly occurs in the 3' end of the APC gene (10-14). For example, in patients with Gardner's syndrome, truncating mutations between codons 1403 and 1578 are associated with an increase in mandibular lesions (10), while mutations beyond codon 1444 are associated with a 2-fold increased risk in osteomas (11).
The majority of patients carrying APC mutations have a family history of colorectal polyps and cancers (9). However, 25-30% of APC mutations are de novo and lack clinical or genetic indications in unaffected family members (15,16). It is now recognized that this can be partially explained as a result of mutational mosaicism (17). The incidence of Gardner's syndrome in China may be low, since it is currently very rare in the Han Chinese population. The majority of current studies about Chinese Gardner's syndrome are sporadic case reports or family studies. In the current study, mutation analysis of the APC gene in a Chinese patient with Gardner syndrome was performed. The mutational analysis included determining whether the variant was de novo or chimeric.
Materials and methods
Research subjects
The current study was approved by the Southern Medical University Institutional Review Board. Informed consent was obtained from all research participants. The proband, a 38-year-old male, was from the Chaoshan area of Guangdong Province. A period of 5 years prior to diagnosis, pus was repeatedly present in the individuals left lower posterior tooth. In 2016, after diagnosis of fully impacted extraneous teeth and following treatment, the left lower posterior tooth was extracted. The tooth extraction wound underwent a prolonged period of healing, with postoperative gum restoration being absent. Upon further examination, the mandibular CT plain scan indicated that multiple osteomas were present in the upper and lower jaw, part of the skull, paranasal sinus and cervical vertebra. Multiple embedded teeth and a partially lost tooth were observed. The left lower jaw indicated the presence of an odontogenic infection. These results suggested that the patient may have Gardner's syndrome. In 2018, a tumor was identified in the right forearm of the proband and was surgically removed. During consultation on the familial history of disease, it was revealed that the patient's 62 year old mother had developed mandibular hyperplasia when middle-aged. The patient's mother died from colorectal cancer 3 months after the collection of a peripheral blood sample.
Clinical examination
An X-ray dental panorama was performed on the oral cavity and an X-ray examination was performed on the right forearm of the proband. Pathological sections were taken from hyperosteogeny tissue of the mandibular bone, and the right tibial. Hyperosteogeny tissues were harvested and stored at formalin for further analyses. Biochemical tests were performed on the 3 ml peripheral blood obtained from the proband and his mother. After collecting peripheral blood, the serum was separated via centrifugation at 1,610 x g for 5 min at room temperature. The electrochemiluminescence immunoassay and the kits for the instrument was performed using Elecsys Cobas e 601 (Roche Diagnostics) to detect the serum levels of carcinoembryonic antigen (CEA; Elecsys CEA kit) and carbohydrate antigens including CA15-3 (Elecsys CA15-3 kit), CA19-9 (Elecsys CA19-9 kit) and CA72-4 (Elecsys CA72-4 kit). All Elecsys kits were all obtained from Roche Diagnostics GmbH.
Hematoxylin and eosin staining
Bone tissues were pre-fixed in 10% neutral formalin solution for 24 h at room temperature and rinsed with running tap water for 24 h. After washing, decalcification was performed at 4˚C under continuous shaking. The decalcifying solution (3% nitric acid) was changed daily. When the bone was easily penetrated through by a needle without any force, the decalcification process was concluded. Decalcification lasted for a total of 8 days. Finally, samples were neutralized with 0.1% aqueous ammonia solution for 30 min at room temperature. After decalcification, samples were washed in running tap water for 24 h and dehydrated with an ascending ethanol gradient (70, 80, 90, 95 and 100%). The fixed samples were embedded in paraffin and sliced to a thickness of 3 µm. H&E staining was conducted according to routine protocols (18). The section was stained at room temperature with hematoxylin (cat. no. G4070; Beijing Solarbio Science & Technology, Co., Ltd.) for 5 min and eosin Y (cat. no. E8080; Beijing Solarbio Science & Technology, Co., Ltd.) for 1 min.
Molecular analysis
All genomic DNA was extracted from peripheral blood using the phenol-chloroform isoamyl alcohol (PCI) method (19). DNA samples were stored at -20˚C until subsequent use. DNA integrity was evaluated by 1% agarose gel electrophoresis and NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Based on the nucleotide sequence of the APC gene (RefSeq: NM_000038, OMIM: 611731), 39 pairs of primers for 15 exons and untranslated regions were designed (Table I). There were seven members of the family that were available for DNA screening, except for II-1 and II-4 (Fig. 2A). The APC gene was amplified using reverse-transcription quantitative PCT (RT-qPCR) and sequenced using APC primers. RT-qPCR was performed using GoTaq qPCR Master Mix (Promega Corporation) and the CFX96™ Real-Time System (Bio-Rad Laboratories, Inc.). The thermocycling conditions were as follows: 5 min at 95˚C, followed by 40 cycles of denaturation for 20 sec at 95˚C, annealing for 20 sec at 60˚C and extension for 20 sec at 72˚C. Final melting curve analysis was used to monitor the purity of the PCR product. The 2-∆∆Cq method was used to quantify mRNA abundance (20). Relative gene expression levels were normalized to GAPDH. SeqMan software (Lasergene, Version 7.1.0; DNASTAR, Inc.) was used to view and align the sequencing results. Total cellular RNA was isolated using TRIzol™ reagent (Gibco; Thermo Fisher Scientific Inc.), according to the manufacturer's protocol. Complementary DNA (cDNA) synthesis was performed by using the First-Strand cDNA Synthesis Kit (Toyobo Life Science). Based on the complete mRNA sequence of APC gene (GenBank: M74088.1; http://www.ncbi.nlm.nih.gov/nuccore/M74088.1), primers forward, 5'-AGGGTCCAGGTTCTTCCAGA-3' and reverse, 5'-AGGCTGCTCTGATTCTGTTTCA-3' were designed to amplify the extensive APC mRNA, and primers forward, 5'-GTGAAGGTCGGAGTCAACG-3' and reverse, 5'-TGAGGTCAATGAAGGGGTC-3' to amplify the extensive GAPDH mRNA as a control. Each study sample was analyzed in triplicate, and this assay was performed repeatedly four times. The results were expressed as the average (mean ± SD) of four independent experiments. A panel of 25 heterozygous short tandem repeat (STR) chromosomal markers were used to confirm maternity and paternity (21) (Huaxia Platinum PCR Amplification kit; Applied Biosystems; Thermo Fisher Scientific, Inc.). As presented in Table II, these included D3S1358, vWA, D16S539, CSF1PO and TPOX. Linkage equilibrium was assumed between alleles on the same chromosome. At each locus, non-maternal and non-paternal probabilities for each offspring were calculated as the likelihood that unknown individuals could have been the parents. The prevalence of each detected STR allele, for which published allele frequencies are available online (http://www.ncbi.nlm.nih.gov/projects/SNP/). For each child, the likelihood of parentage was calculated to be (1-likelihood of non-parentage) and was 99.999%. Genetic marker PCR products were determined using an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Table I.
APC primers.
| Primer | Exon position | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|---|
| APC-5'UTR | 5'UTR | GGAAGCGGAGAGAGAAGCAG | TGACACTGGTATCTGTTTGCCAC |
| APC-1 | Exon 1 | AGGCAAATGTATTCAGACAC | GTACTTGCCAAATAAGACAAC |
| APC-2 | Exon 2 | GTTTCTAATACCTTGCACAG | AGATATCCTTTTAAAACTGCT |
| APC-3 | Exon 3 | TGATAATAATTGAAGCCAGAC | ATAGGTCTTCAGAATCCCAG |
| APC-4 | Exon 4 | AGCCTTTGGTGAAGTGTAAG | CCTATAGAGTATGTTTGGCAATTC |
| APC-5 | Exon 5 | TTTAAGTGAAATAGGCCAATC | TAGTCATTAGCTACCGGAAG |
| APC-6 | Exon 6 | CAATTTGTTATTAAAGGGTG | GTTATCACTTGTTTACCTGCT |
| APC-7 | Exon 7 | TGCAGCTCCATTAAATGTCA | CATAATCAAAATGCAGCTTAGAGT |
| APC-8 | Exon 8 | TTCATACTTAGTTTTCTGGCAAT | TCCAACTATTCTCATGCCTC |
| APC-9 | Exon 9 | GGCCACTCATACTATTTACTCAC | CTTTGAAACATGCACTACGAT |
| APC-10 | Exon 10 | TGATCCACTAAAATTCCGTG | ATAATAATTCCCTCTGATGCTC |
| APC-11 | Exon 11 | AAGAGTGTTTATAAAGCCCTA | ACAAATGAGTAAAGATAAGCG |
| APC-12 | Exon 12 | ATCATTTCTCACCACTTATTC | ACTAAATACTGAGCAACAATC |
| APC-13 | Exon 13 | TTATGGCTCACAGTAACCTCA | AAAATACAAATAGCCGGGAG |
| APC-14 | Exon 14 | TTGTGTTCTGCTTGTTTTATAGAG | TCTCTGGATCACGCTCATAG |
| APC-15-1 | Exon 15 | AGAGTGGCACCCAACCATAG | TCCCATAATGCTTCCTGGTC |
| APC-15-2 | Exon 15 | CAGGCAAATCCTAAGAGAGAACA | CTTGATGAAGAGGAGCTGGG |
| APC-15-3 | Exon 15 | GCTCAAGCTTGCCATCTCTT | TATGGGCAGCAGAGCTTCTT |
| APC-15-4 | Exon 15 | CCAGGAACTTCTTCAAAGCG | GTGAAGGACTTTGCCTTCCA |
| APC-15-5 | Exon 15 | GTCAATACCCAGCCGACCTA | AGGCTGATCCACATGACGTT |
| APC-15-6 | Exon 15 | TTCCAACCACATTTTGGACA | GAGCTGATTCTGCCTCTTGG |
| APC-15-7 | Exon 15 | AACGTCATGTGGATCAGCCT | TGCTGGATTTGGTTCTAGGG |
| APC-15-8 | Exon 15 | CAGACGACACAGGAAGCAGA | GCAGCTTGCTTAGGTCCACT |
| APC-15-9 | Exon 15 | GTGAACCATGCAGTGGAATG | TGTTGGCATGGCAGAAATAA |
| APC-15-10 | Exon 15 | TTTGCCACGGAAAGTACTCC | TATCATCCCCCGGTGTAAAA |
| APC-15-11 | Exon 15 | CTGTGGCAAGGAAACCAAGT | TGATTTTTGTTGGGTGCAGA |
| APC-15-12 | Exon 15 | CCCAAAGGGAAAAGTCACAA | GCTGATTGTTGGTTGGAGGT |
| APC-15-13 | Exon 15 | TCACCTCATCATTACACGCC | GGTTCTCCCTGTGAGTCAGG |
| APC-15-14 | Exon 15 | ACTCCGGTTTGCTTTTCTCA | AGCAGCAGCAGCTTGATGTA |
| APC-15-15 | Exon 15 | GCCTTCAAGACTCAAGGGTG | TTGTCCTGCCTCGAGAGATT |
| APC-15-16 | Exon 15 | GCTGCTGCTGCATGTTTATC | TGGCAACAGGGCTTAATTCT |
| APC-15-17 | Exon 15 | AATCTCTCGAGGCAGGACAA | TCCTTTGGAGGCAGACTCAC |
| APC-15-18 | Exon 15 | CAGGTTTATCCAAGAATGCCA | TTCAGAATGAGACCGTGCAA |
| APC-15-19 | Exon 15 | CCCACCTAATCTCAGTCCCA | CAATCACCGGGGGAGTATTA |
| APC-15-20 | Exon 15 | AAATGGCACCTGCTGTTTCT | TTCCACTGGATTCTGTGCTG |
| APC-15-21 | Exon 15 | TTGGAAAATCGCCTGAACTC | TGGCTTCCAGAACAAAAACC |
| APC-3’UTR-1 | 3'UTR | ACAAAGAAGCGAGATTCCAA | CCACTGTAGCTATCTCTATGCAC |
| APC-3’UTR-2 | 3'UTR | CAGTAATATGGTTCCCGATG | CCAATGCTTAGTCTGTGCTAG |
| APC-3’UTR-3 | 3'UTR | GAAGACTGTTGCCACTTAACC | TATTTGGCCTGCTATCGATT |
APC, Adenomatous polyposis coli; UTR, untranslated region.
Figure 2.
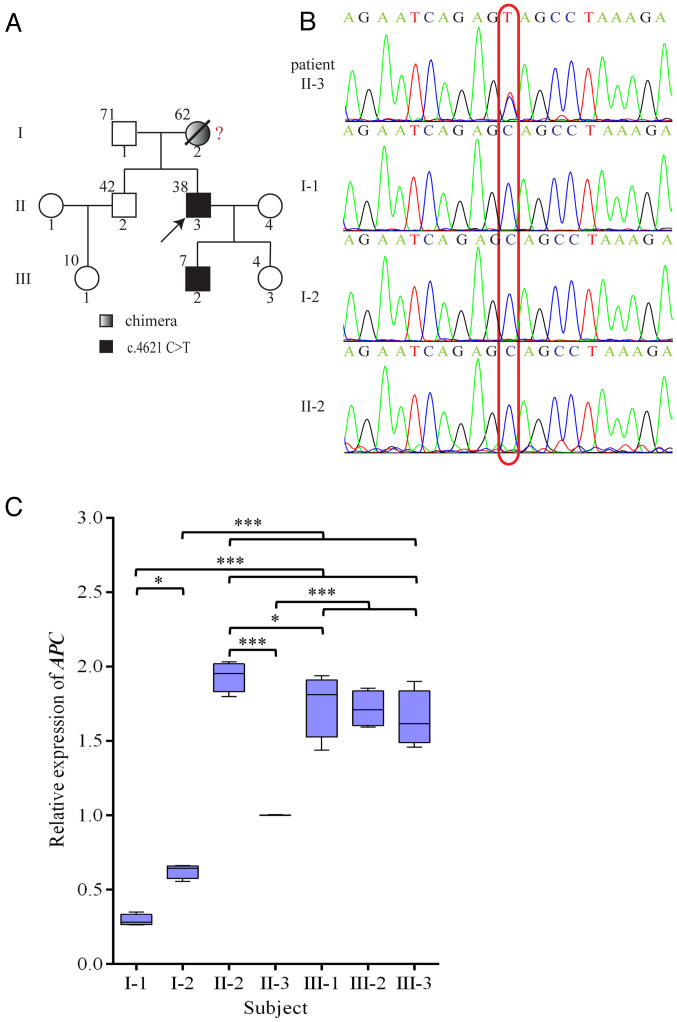
(A) Family diagram. The upper left corner of the family members indicates the age. Red question mark indicates a possible chimera. (B) Sequencing map. (C) Expression of the APC gene. Box-and-whisker plots of pairwise comparisons for relative expression of APC gene. *p<0.05, and ***p<0.001 vs. II-3. APC, adenomatous polyposis coli.
Table II.
Genotyping.
| Individual | ||||||
|---|---|---|---|---|---|---|
| Short tandem repeat | I-1 | I-2 | II-3 (proband) | |||
| D3S1358 | 15 | 16 | 15 | 16 | 15 | 16 |
| vWA | 15 | 17 | 17 | 18 | 17 | 17 |
| D16S539 | 11 | 13 | 9 | 11 | 11 | 13 |
| CSF1PO | 12 | 12 | 10 | 10 | 10 | 12 |
| TPOX | 8 | 9 | 8 | 11 | 8 | 9 |
| Yindel | 1 | 0 | 0 | 0 | 1 | 0 |
| AMEL | X | Y | X | X | X | Y |
| D8S1179 | 10 | 15 | 14 | 16 | 14 | 15 |
| D21S11 | 29 | 30 | 30 | 31 | 29 | 31 |
| D18S51 | 13 | 16 | 17 | 19 | 16 | 17 |
| Penta E | 11 | 14 | 16 | 21 | 11 | 21 |
| D2S441 | 10 | 11 | 11 | 11.3 | 11 | 11 |
| D19S433 | 14 | 15.2 | 14.2 | 15.2 | 15.2 | 15.2 |
| TH01 | 9 | 9 | 9 | 9 | 9 | 9 |
| FGA | 22 | 24 | 21 | 22 | 22 | 22 |
| D22S1045 | 16 | 17 | 15 | 16 | 16 | 16 |
| D5S818 | 12 | 12 | 11 | 12 | 11 | 12 |
| D13S317 | 11 | 12 | 11 | 12 | 11 | 11 |
| D7S820 | 8 | 12 | 11 | 11 | 8 | 11 |
| D6S1043 | 13 | 14 | 10 | 17 | 14 | 17 |
| D10S1248 | 14 | 16 | 15 | 16 | 14 | 15 |
| D1S1656 | 11 | 14 | 15 | 15 | 14 | 15 |
| D12S391 | 19 | 23 | 17 | 23 | 17 | 19 |
| D2S1338 | 17 | 19 | 24 | 24 | 19 | 24 |
| Penta D | 9 | 9 | 9 | 9 | 9 | 9 |
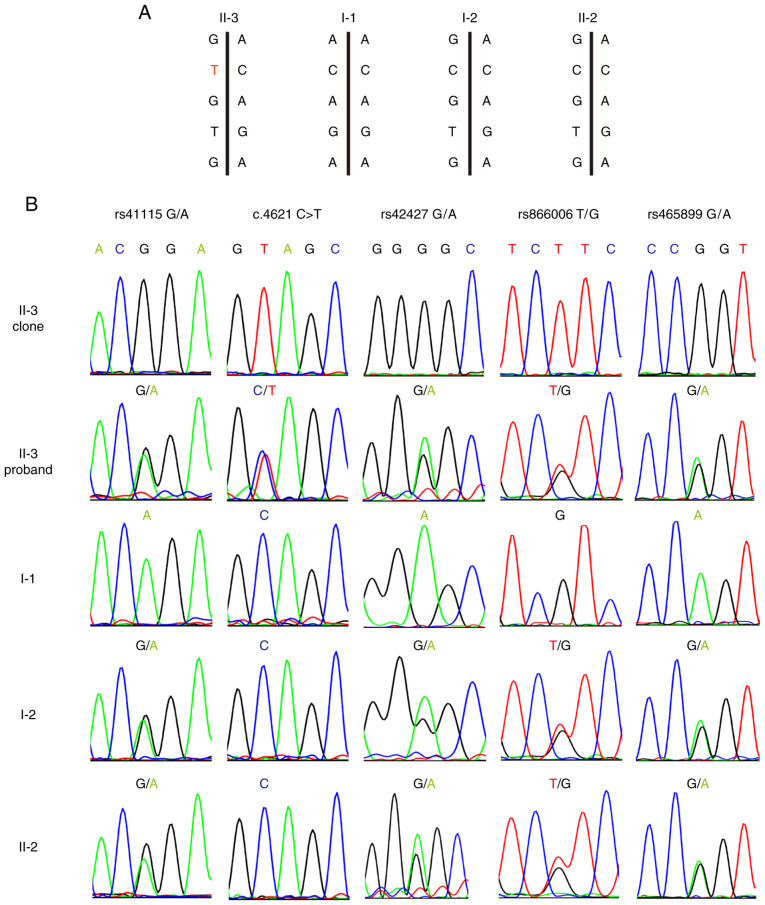
In order to avoid the interference of homologous recombination exchange, primers forward, 5'-TCAAACAGCTCAAAC CAAG-3' and reverse, 5'-AAATGATTTAGGAGCATAGCC-3' were designed for an APC fragment that included the c.4621C>T variant and four single nucleotide polymorphisms (SNPs). The SNPs included rs41115 G>A, rs42427 G>A, rs866008 T>G and rs465899 G>A, which are 142 base-pairs upstream, 413 base-pairs downstream, 647 base-pairs downstream and 1259 base-pairs downstream of the mutation site, respectively.
The APC PCR product from lymphoblast DNA was cloned into the pMD19-T vector (Takara Bio, Inc.) and transformed into chemically competent E. coli strain DH5α (Vazyme Biotech Co., Ltd.). PCR amplification of the cloned APC fragment amplicon was performed on isolated colonies, which were selected using 100 µg/ml ampicillin (Sigma-Aldrich; Merck KGaA). The products were then sequenced to determine which SNPs alleles were in cis with the APC c.4621C>T variant (22). Three bacterial colonies were sequenced per subject in order to obtain a consensus sequence.
Statistical analysis
Statistical analysis was performed with software package of SPSS 20.0 (IBM Corp.). Figures were produced with Adobe Photoshop/Illustrator CS6 imaging processing and drawing system (Adobe, Inc.) and GraphPad Prism 6.0 software (GraphPad Software, Inc.). Relative expression of APC gene among individuals in the research family were compared using a one-way ANOVA and Least-significant Difference (LSD) test. Bonferroni correction was performed for further pairwise comparisons. Data were presented as the mean ± SD from four independent experiments with a two-sided P-value <0.05 for the difference was considered to be statistically significant.
Results
Clinical outcomes
The postoperative X-ray dental panorama (Fig. 1A) of the proband showed that there were impacted teeth in the right lower jaw, and that most of the left mandibular teeth were absent. Regions of dense radiopacity were observed around the teeth. Radiological reporting of these scans confirmed the dense radio opacities to be consistent with the presence of osteomas.
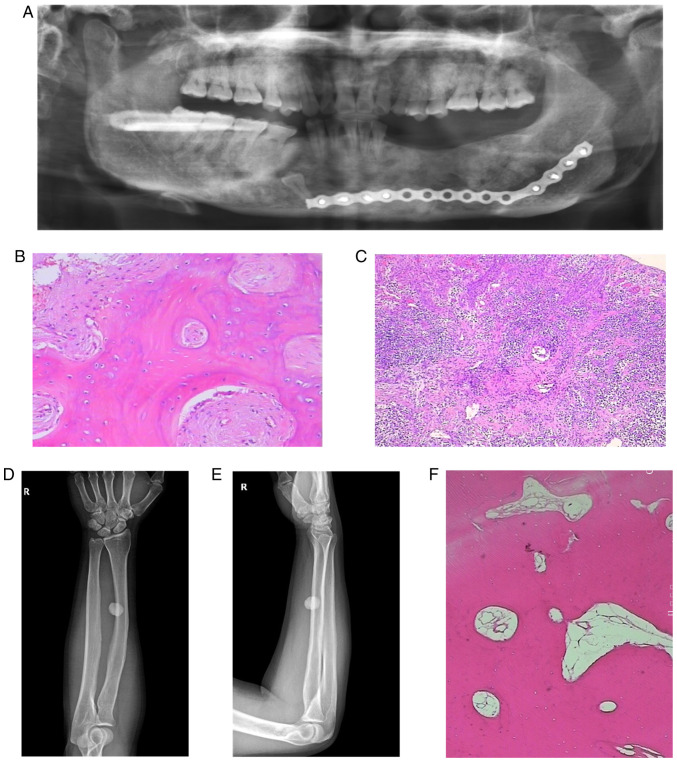
Figure 1.
(A) Postoperative oral X-ray, panoramic film. (B and C) Pathological sections on hyperosteogeny tissues of the left and right mandibular bone (magnification, x400 and x100, respectively). (D) The radius and ulna of right forearm on anteroposterior radiograph. (E) The radius and ulna of right forearm on lateral radiograph. (F) Pathological sections on hyperosteogeny tissue of the right tibial (magnification, x400).
Pathological sections were obtained from gray-brown bone tissue of the left and right mandible (Fig. 1B and C). The results confirmed that the mandibular osteoma tissue consisted of a well-differentiated lamellar bone. The trabeculae were connected with each other with scattered, loose connective tissue was indicated between them. There was no obvious abnormal shape in the bone cells with osteoblasts being visible. A large number of acute and chronic inflammatory cells had localized to the site of injury. The left mandibular alveolar had lesioned with hyperplastic spindle cells growing with a spiral shape in this area. The trabeculae in this region were curved, and osteoblasts were visible around them. Osteoblasts showed no obvious abnormalities.
The radius and ulna of the right forearm on anteroposterior and lateral radiographs (Fig. 1D and E) revealed the presence of a circular high density shadow. The boundary of the shadow was clear with an approximate size of 1.7 by 1.5 cm. The shape and density of the ulna and tibia were normal. No definite hyperosteogeny or destruction was indicated. No fracture line was observed and the soft tissue was normal.
Pathological sections were obtained from hyperosteogeny tissue of the right tibial (Fig. 1F). The results confirmed that the tissue consisted of a well-differentiated lamellar bone, and that the cells were not abnormal. These results were consistent with a diagnosis of osteoma.
Serum biochemical test demonstrated that the proband (Fig. 2A; II-3) was normal (Table III), but that his mother's (I-2) serum CEA was 30.060 ng/ml (reference value range: 0.000-3.400), while her carbohydrate antigen 19-9 (CA19-9) was >1,000.000 U/ml (reference value range: 0.000-27.000).
Table III.
Results of the biochemical tests of the proband and his mother.
| Parameter | Proband | Proband's mother | Reference value range |
|---|---|---|---|
| CEA (ng/ml) | 0.389 | 30.060 | 0.000-3.400 |
| CA15-3 (U/ml) | 15.670 | 15.380 | 0.000-25.000 |
| CA19-9 (U/ml) | 7.910 | >1,000.000 | 0.000-27.000 |
| CA72-4 (U/ml) | 3.860 | 2.860 | 0.000-6.900 |
Laboratory outcomes
Genetic testing was conducted on each of the family members. A nonsense variant, c.4621C>T (23), was indicated in the proband (II-3) and his son (III-2) by sequencing of the APC gene from peripheral blood. None of the other members of this family had this mutation (Fig. 2A and B). The mutation causes the codon at position 1541 to change from a CAG of the encoded glutamine to a stop codon (UAG). This resulted in early termination of translation.
The amplification level of APC cDNA in the proband (II-3) was 1.0, while the relative amplification from the proband's unaffected brother (II-2), who is 4 years older than the proband, was ~2.0 (Fig. 2C). The relative amplification from the proband's father and mother was ~0.3 and 0.6, respectively. The relative amplification from the third generation was approximated between 1.65 and 1.75. Paternity testing was used to confirm the genetic contributions of the mother and father to their children. This was conducted using 25 chromosomal markers. Based on known frequencies of these alleles in the general population, non-paternity can be ruled out with 99.999% certainty (Table II).
Because maternal and paternal inheritance was not an issue, haplotype analysis of the APC locus was performed to determine if the mutant allele originated from the mother. The rs41115 SNP was homozygous (AA) in the father (I-1), but heterozygous (GA) in the mother (I-2). Sequence analysis confirmed that each of the two children were heterozygous and carried the GA alleles. DNA cloning and sequencing of individual alleles demonstrated that the c.4621C>T variant was co-inherited with the rs41115 G allele at position 4479. This showed that the mutation must occur in the C allele of the maternal line at nucleotide position 4621, thus indicating the maternal origin of the chromosome (Fig. 3A and B). In addition, rs42427 G>A, rs866008 T>G and rs465899 G>A confirmed this result. Furthermore, both affected children (II-3) and an unaffected sibling (II-2) inherited the identical chromosome from the mother (Fig. 3A and B).
Figure 3.
Haplotype analysis in the family with a confirmed mutation in the APC gene. (A) Schematic diagram of SNPs in two homologous chromosomes. (B) PCR-amplified mutation-specific clone and sequence analysis of genomic DNA at the variant-associated SNP position in the APC gene. APC, adenomatous polyposis coli; SNP, single nucleotide polymorphisms.
Discussion
As an extra-colonic presentation of the familial adenomatous polyposis syndrome, Gardner's syndrome has prominent characteristics. These include dental abnormalities, soft-tissue tumors (including desmoid tumors), osteomas and epidermoid cysts. Clinically, the proband in the current study exhibited dental abnormalities and osteoma, which was consistent with the early signs of Gardner's syndrome. Through mutation screening of the APC gene, the proband was indicated to carry a rare c.4621C>T variant. The genotype and phenotype of this mutation correlated with Gardner's syndrome. Consistent with a mechanism of nonsense-mediated mRNA decay (NMD) (24), APC gene expression in the blood of the proband was half that of his wild-type brother. However, the APC gene expression of the proband's father and mother were less than the proband, which may be due to the fact the expression of mRNA decreases with age. In the third generation, the APC gene expression of the child with the mutation was similar to those who lacked the mutation, indicating that the pathogenic mutation did not affect APC expression in childhood. This was also consistent with the characteristic of Gardner's syndrome in that lesions only present after middle age. The specific reason for this requires further investigation. To the best of our knowledge, no study on the decrease of expression level with increasing age or the antagonistic assumption that during childhood the level of APC gene expression was not affected by NMD has been performed. Therefore, to verify this speculation, further follow-up observations on the research family should be performed to study whether the expression dose of APC gene changes with age. The location of the conserved sites is important as mutations in such regions usually result in a genetic disorder (25). In the present study, the results demonstrated that mutations at the conserved c.4621 position in the APC gene also caused Gardner syndrome. Combining the clinical phenotype of Gardner's syndrome and genetic analysis, the proband was confirmed to be a patient suffering from Gardner's syndrome. Concerning the development of clinical characteristics of Gardner’s syndrome, dental malformations and mandibular tumors preceded the development of other forms of the syndrome, including intestinal polyps. Therefore, dentists should be aware that dental malformations and mandibular tumors serve an important role in the development of Gardner's syndrome.
Interestingly, mutation screening in other members of the family revealed that the parents of the proband did not carry the c.4621C>T variant. As this mutation was only detected in the peripheral blood of the proband and his son, the c.4621C>T variant appeared to be a de novo variant. However, the proband's mother exhibited similar oral symptoms and clinical history to the proband. Having died from cancer, it was therefore necessary to determine if the transmission of the mutation to the progeny was through maternal mosaicism. Serum biochemical tests indicated that the CEA and CA19-9 levels of the proband's mother were greater than the reference range. Among numerous tumor markers, CEA and CA19-9 are the most common markers to perform clinicopathological investigations on within colorectal carcinoma (26,27). The results of the current study indicated that the mother suffered from colorectal cancer. The high levels of CEA and CA19-9 markers were associated with advanced tumors which was consistent with the mother's disease course (28). Unfortunately, the mother of the proband died before the collection and analysis of the tissue specimen, which included screening intestinal polyps for cell mosaicism (29). Consequently, a paternity test and haplotype analysis was designed to obtain more information on the mosaic nature associated with the identified variant. The results of paternity testing supported the parental relationship between the proband and his parents. Haplotype analysis demonstrated that the chromosome with the c.4621C>T variant originated from the mother. As reported in the literature, ~20% of apparently sporadic cases of familial adenomatous polyposis in which APC mutations can be identified are known to display mosaicism (30). Therefore, combined with the clinical phenotype of the mother, haplotype analysis and literary references, this mutation was more likely to have arisen from maternal mosaicism than due to a de novo event in the proband.
A number of literature reports have indicated that mosaicism of APC mutations can be identified through sequence analysis of peripheral blood samples and/or adenomas (17,29,31-34). However, if only peripheral blood is inspected for the presence of mutation, mosaicism may be missed due to very low, or even null presentation of the mutated alleles (34). The present study presents an example of this. If only the presence or absence of a mutation in the blood is examined without considering the abnormality of the clinical phenotype and further verification through haplotype analysis, such a variant may be considered to be de novo in origin. Therefore, it can be suggested that if de novo mutations are detected in cases with Gardner's syndrome, the presence of mutational mosaicism should be verified. Researchers should carefully consider the clinical symptoms of the parents, and collect colorectal polyps, or preferably, tumor tissue cells for investigating the presence of mutational mosaicism (35). This strategy should be incorporated in suspected familial adenomatous polyposis cases for which APC mutation have been ruled out. Familial adenomatous polyposis is an autosomal dominant genetic disorder that is typically characterized by the development of hundreds to thousands of adenomas in the colon and rectum during youth (36). If no APC mutations are indicated in the DNA from peripheral blood of suspected familial adenomatous polyposis cases, samples from tissues (such as colon polyps and osteomas) should be collected to detect APC mutations. If different members of the proband's family or different specimen types have inconsistent APC mutation, the possibility of mosaicism should be considered. According to the human geneticist's point of view (37), there are three main types of mosaicism. Mosaicism may exist in various parts of the body with different mosaic proportions or only exist in certain parts of the body. Detecting this will aid future in-depth research and prevent missed diagnosis of familial adenomatous polyposis. If no tissue is collected, haplotype analysis could be used as indirect evidence for mutational mosaicism. The current study had certain limitations that should be acknowledged. Firstly, the possibility of mosaicism was not considered prior to experimentation, as the colon tissue samples of the mother of the proband could not be collected in time. Additionally, there was no direct evidence that could verify the existence of mosaicism. Secondly, the detection of chimera is challenging, which mainly depends on the type of chimera, the location and the proportion of the chimera.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the Natural Science Foundation of Guangdong Province (grant no. 2018B030311033), the Science and Technology Program of Guangzhou (grant no. 201707010301) and the National Natural Science Foundation of China (grant no. 31970558).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DC designed and performed research, analyzed data, and was a major contributor in writing the manuscript; DC and FH performed experiments; XX analyzed the data and revised the manuscript; FX and LZ designed and supervised the research, and wrote the manuscript. All authors reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Southern Medical University Institutional Review Board. Informed consent was obtained from all research participants.
Patient consent for publication
Informed consent for the publication of any associated data and accompanying images was obtained from all research participants.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kamel SG, Kau CH, Wong ME, Kennedy JW, English JD. The role of Cone beam CT in the evaluation and management of a family with Gardner's syndrome. J Craniomaxillofac Surg. 2009;37:461–468. doi: 10.1016/j.jcms.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Dinarvand P, Davaro EP, Doan JV, Ising ME, Evans NR, Phillips NJ, Lai J, Guzman MA. Familial adenomatous polyposis syndrome: An update and review of extraintestinal manifestations. Arch Pathol Lab Med. 2019;143:1382–1398. doi: 10.5858/arpa.2018-0570-RA. [DOI] [PubMed] [Google Scholar]
- 3.Gómez García EB, Knoers NV. Gardner's syndrome (familial adenomatous polyposis): A cilia-related disorder. Lancet Oncol. 2009;10:727–735. doi: 10.1016/S1470-2045(09)70167-6. [DOI] [PubMed] [Google Scholar]
- 4.Yu D, Ng Cw B, Zhu H, Liu J, Lin Y. Bone and dental abnormalities as first signs of familial Gardner's syndrome in a Chinese family: A literature review and a case report. Med Sci (Paris) null: 2018;34:20–25. doi: 10.1051/medsci/201834f104. [DOI] [PubMed] [Google Scholar]
- 5.Adisen MZ, Okkesim A, Misirlioglu M. The importance of early diagnosis of Gardner's syndrome in dental examination. Niger J Clin Pract. 2018;21:114–116. doi: 10.4103/njcp.njcp_381_16. [DOI] [PubMed] [Google Scholar]
- 6.de Oliveira JC, Viana DV, Zanardo C, Santos EMM, de Paula AE, Palmero EI, Rossi BM. Genotype-phenotype correlation in 99 familial adenomatous polyposis patients: A prospective prevention protocol. Cancer Med. 2019;8:2114–2122. doi: 10.1002/cam4.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yachida T, Nakajima T, Nonaka S, Nakamura K, Suzuki H, Yoshinaga S, Oda I, Moriya Y, Masaki T, Saito Y. Characteristics and clinical outcomes of duodenal neoplasia in Japanese patients with familial adenomatous polyposis. J Clin Gastroenterol. 2017;51:407–411. doi: 10.1097/MCG.0000000000000555. [DOI] [PubMed] [Google Scholar]
- 8.Basaran G, Erkan M. One of the rarest syndromes in dentistry: Gardner syndrome. Eur J Dent. 2008;2:208–212. [PMC free article] [PubMed] [Google Scholar]
- 9.Half E, Bercovich D, Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis. 2009;4(22) doi: 10.1186/1750-1172-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juhn E, Khachemoune A. Gardner syndrome: Skin manifestations, differential diagnosis and management. Am J Clin Dermatol. 2010;11:117–122. doi: 10.2165/11311180-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 11.Galiatsatos P, Foulkes WD. Familial adenomatous polyposis. Am J Gastroenterol. 2006;101:385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 12.Wallis YL, Macdonald F, Hultén M, Morton JE, McKeown CM, Neoptolemos JP, Keighley M, Morton DG. Genotype-phenotype correlation between position of constitutional APC gene mutation and CHRPE expression in familial adenomatous polyposis. Hum Genet. 1994;94:543–548. doi: 10.1007/BF00211023. [DOI] [PubMed] [Google Scholar]
- 13.Davies DR, Armstrong JG, Thakker N, Horner K, Guy SP, Clancy T, Sloan P, Blair V, Dodd C, Warnes TW, et al. Severe Gardner syndrome in families with mutations restricted to a specific region of the APC gene. Am J Hum Genet. 1995;57:1151–1158. [PMC free article] [PubMed] [Google Scholar]
- 14.Nieuwenhuis MH, Vasen HFA. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): A review of the literature. Crit Rev Oncol Hematol. 2007;61:153–161. doi: 10.1016/j.critrevonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Bisgaard ML, Fenger K, Bülow S, Niebuhr E, Mohr J. Familial adenomatous polyposis (FAP): Frequency, penetrance, and mutation rate. Hum Mutat. 1994;3:121–125. doi: 10.1002/humu.1380030206. [DOI] [PubMed] [Google Scholar]
- 16.Rozen P, Samuel Z, Rabau M, Goldman G, Shomrat R, Legum C, Orr-Urtreger A. Familial adenomatous polyposis at the Aviv Medical Center: Demographic and clinical features. Fam Cancer. 2001;1:75–82. doi: 10.1023/a:1013888226756. [DOI] [PubMed] [Google Scholar]
- 17.Aretz S, Stienen D, Friedrichs N, Stemmler S, Uhlhaas S, Rahner N, Propping P, Friedl W. Somatic APC mosaicism: A frequent cause of familial adenomatous polyposis (FAP) Hum Mutat. 2007;28:985–992. doi: 10.1002/humu.20549. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Wang L, Ma R, Mu Q, Yu N, Zhang Y, Tang Y, Li Y, Jiang G, Zhao D, et al. JiangTang XiaoKe granule attenuates cathepsin K expression and improves IGF-1 expression in the bone of high fat diet induced KK-Ay diabetic mice. Life Sci. 2016;148:24–30. doi: 10.1016/j.lfs.2016.02.056. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor Laboratory Press, Plainview, NY, ppE3-E15, 1989. [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Schwab AL, Tuohy TM, Condie M, Neklason DW, Burt RW. Gonadal mosaicism and familial adenomatous polyposis. Fam Cancer. 2008;7:173–177. doi: 10.1007/s10689-007-9169-1. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann A, Vogt S, Uhlhaas S, Stienen D, Kurth I, Hameister H, Mangold E, Kötting J, Kaminsky E, Propping P, et al. Analysis of rare APC variants at the mRNA level: Six pathogenic mutations and literature review. J Mol Diagn. 2009;11:131–139. doi: 10.2353/jmoldx.2009.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Rosa M, Scarano MI, Panariello L, Morelli G, Riegler G, Rossi GB, Tempesta A, Romano G, Renda A, Pettinato G, et al. The mutation spectrum of the APC gene in FAP patients from southern Italy: Detection of known and four novel mutations. Hum Mutat. 2003;21:655–656. doi: 10.1002/humu.9151. [DOI] [PubMed] [Google Scholar]
- 24.Castellsagué E, González S, Guinó E, Stevens KN, Borràs E, Raymond VM, Lázaro C, Blanco I, Gruber SB, Capellá G. Allele-specific expression of APC in adenomatous polyposis families. Gastroenterology. 2010;139:439–447. doi: 10.1053/j.gastro.2010.04.047. 447.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. ACMG Laboratory Quality Assurance Committee: Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nozoe T, Rikimaru T, Mori E, Okuyama T, Takahashi I. Increase in both CEA and CA19-9 in sera is an independent prognostic indicator in colorectal carcinoma. J Surg Oncol. 2006;94:132–137. doi: 10.1002/jso.20577. [DOI] [PubMed] [Google Scholar]
- 27.Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745–1755. doi: 10.3748/wjg.v22.i5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Wang X, Yu F, Chen J, Zhao S, Zhang D, Yu Y, Liu X, Tang H, Peng Z. Combined detection of preoperative serum CEA, CA19-9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. Int J Clin Exp Pathol. 2015;8:14853–14863. [PMC free article] [PubMed] [Google Scholar]
- 29.Out AA, van Minderhout IJ, van der Stoep N, van Bommel LS, Kluijt I, Aalfs C, Voorendt M, Vossen RH, Nielsen M, Vasen HF, et al. High-resolution melting (HRM) re-analysis of a polyposis patients cohort reveals previously undetected heterozygous and mosaic APC gene mutations. Fam Cancer. 2015;14:247–257. doi: 10.1007/s10689-015-9780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuohy TM, Burt RW. Somatic mosaicism: A cause for unexplained cases of FAP? Gut. 2008;57:10–12. doi: 10.1136/gut.2007.133108. [DOI] [PubMed] [Google Scholar]
- 31.Filipe B, Albuquerque C, Bik E, Lage P, Rodrigues P, Vossen R, Tops C, Nobre Leitão C. APC somatic mosaicism in a patient with Gardner syndrome carrying the E1573X mutation: Report of a case. Dis Colon Rectum. 2009;52:1516–1521. doi: 10.1007/DCR.0b013e3181ab810f. [DOI] [PubMed] [Google Scholar]
- 32.Hes FJ, Nielsen M, Bik EC, Konvalinka D, Wijnen JT, Bakker E, Vasen HF, Breuning MH, Tops CM. Somatic APC mosaicism: an underestimated cause of polyposis coli. Gut. 2008;57:71–76. doi: 10.1136/gut.2006.117796. [DOI] [PubMed] [Google Scholar]
- 33.Necker J, Kovac M, Attenhofer M, Reichlin B, Heinimann K. Detection of APC germ line mosaicism in patients with de novo familial adenomatous polyposis: A plea for the protein truncation test. J Med Genet. 2011;48:526–529. doi: 10.1136/jmg.2011.089474. [DOI] [PubMed] [Google Scholar]
- 34.Urbanova M, Hirschfeldova K, Obeidova L, Janosikova B, Lastuvkova J, Lukas M, Kotlas J, Stekrova J. Two Czech patients with familial adenomatous polyposis presenting mosaicism in APC gene. Neoplasma. 2019;66:294–300. doi: 10.4149/neo_2018_180731N559. [DOI] [PubMed] [Google Scholar]
- 35.Kanter-Smoler G, Fritzell K, Rohlin A, Engwall Y, Hallberg B, Bergman A, Meuller J, Grönberg H, Karlsson P, Björk J, et al. Clinical characterization and the mutation spectrum in Swedish adenomatous polyposis families. BMC Med. 2008;6(10) doi: 10.1186/1741-7015-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short E, Sampson J. The role of inherited genetic variants in colorectal polyposis syndromes. Adv Genet. 2019;103:183–217. doi: 10.1016/bs.adgen.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Spinner NB, Conlin LK. Mosaicism and clinical genetics. Am J Med Genet C Semin Med Genet. 2014;166C:397–405. doi: 10.1002/ajmg.c.31421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.