Abstract
Gliomas account for 50% of primary brain tumours in the central nervous system. Small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3), a newly identified long non-coding RNA (lncRNA), serves an oncogenic role in various types of cancer. The aim of the present study was to investigate the effect of SUMO1P3 on glioma progression. The results demonstrated that SUMO1P3 expression was upregulated in glioma tissues and cell lines. Furthermore, SUMO1P3 was associated with a poor overall survival of patients with glioma. The results of the in vitro cell proliferation and flow cytometry assays demonstrated that SUMO1P3-knockdown suppressed cell proliferation and cell cycle. The results of the wound healing and Transwell assays demonstrated that SUMO1P3-knockdown significantly repressed cell migration and invasion. In addition, SUMO1P3 promoted glioma by regulating the expression levels of β-catenin, cyclin-D1, N-cadherin and E-cadherin. Overall, the results of the present study suggested that SUMO1P3 may act as an oncogene by regulating cell proliferation, cell cycle, cell migration and invasion in glioma, and may represent a novel diagnostic biomarker and therapeutic target for glioma.
Keywords: small ubiquitin-like modifier 1 pseudogene 3, glioma, proliferation, cell cycle, migration
Introduction
Gliomas account for 50% of primary brain tumours in the central nervous system (1). According to the malignancy of the tumour, the World Health Organization (WHO) categorizes glioma into four grades, I-IV (2). Patients incipiently remain asymptomatic until obvious clinical symptoms appear, such as headaches, seizures, nausea, sensory loss and aphasia (3). Standard treatment of glioma includes surgical excision followed by adjuvant chemotherapy and/or radiotherapy; however, the overall survival of patients diagnosed with malignant glioma is poor due to a high risk of relapse (4,5). Thus, an improved understanding of the precise molecular mechanisms underlying glioma pathogenesis is crucial to help provide novel therapeutic candidates for the treatment of glioma.
Long non-coding RNAs (lncRNAs), which are transcripts of >200 nucleotides without protein-coding ability, have been demonstrated to serve an essential role in tumour initiation and progression (6). For example, small nucleolar RNA host gene 12 acts as a tumour promoter by regulating cell proliferation, apoptosis, migration and multidrug resistance in gastric, colorectal, non-small cell lung and triple-negative breast cancer (7-10). Metastasis associated lung adenocarcinoma transcript 1 acts as an oncogene in different types of cancer, such as kidney carcinoma, ovarian cancer and colorectal cancer, by regulating cell viability, apoptosis, invasion, migration and epithelial-to-mesenchymal transition (EMT) (11-13). Recently, several studies have revealed that lncRNAs can affect the initiation and progression of glioma (10-12). For example, lncRNA H19 decreases chemoresistance to temozolomide by suppressing EMT via regulation of the Wnt/β-catenin signalling pathway in glioma cell lines (14). lncRNA X-inactive specific transcript promotes tumorigenesis and angiogenesis by sponging microRNA (miR)-429 in glioma cells (15). P73 antisense RNA 1T acts as a competing endogenous RNA to promote high mobility group proteins B1 (HMGB1) expression via sponging the miR-142 promoter, and thus functions as an oncogenic lncRNA by promoting proliferation and invasion in glioma cells (16).
Small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3), a newly identified lncRNA, serves an oncogenic role in bladder, breast and colon cancer, where it may be used as a potential prognostic biomarker (17-19). However, the effect of SUMO1P3 on glioma progression remains unknown. Thus, the present study investigated the expression pattern and function of SUMO1P3 in glioma and further analysed the downstream molecular signalling pathway. The present study demonstrated that SUMO1P3 may act as a tumour promoter in glioma, and may be used as a novel diagnostic biomarker and therapeutic target for glioma.
Materials and methods
Human tissues
Human brain tissue samples were collected from the Department of Neurosurgery at The Third Affiliated Hospital of Soochow University (Changzhou, China) during excision surgeries, between June 2015 and September 2017. A total of 12 glioma tissues (grade II, n=4; grade III, n=4 and grade IV, n=4) were obtained from patients with glioma (age range, 24-64 years; median age, 47 years) and histologically confirmed by three independent pathologists according to the 2016 WHO Classification of Tumors of the Central Nervous System (20). These patients were followed up by telephone every month for 12 months. A total of 10 tissue samples (n=10) with no tumour complication were obtained from patients with cranial trauma (age range, 21-65 years; median age, 45 years), who served as the control. The tissues were immediately snap-frozen and stored in liquid nitrogen until subsequent experimentation. The present study was approved by the Research Ethics Board of the Third Affiliated Hospital of Soochow University and written informed consent was provided by patients or their guardians prior to the start of the study.
Cell lines and culture
A total of three glioma cell lines, including U87, LN229 and U251, and the human glial cell line, HEB, were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The U87 cell line used in the present study was authenticated by Shanghai VivaCell Biosciences Ltd., with STR profiling (http://www.vivacell.com.cn/Helps/STRAuthentication.html) as the U87 MG American Type Culture Collection (ATCC) version, which was a glioblastoma of unknown origin. Cells were cultured in Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2.
Cell transfection
Synthetic SUMO1P3 small interfering (si)RNA and a scrambled non-targeting siRNA as negative control siRNA (si-NC) were purchased from Shanghai GenePharma Co., Ltd. Cells were transfected using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Briefly, 10 µl siRNA (20 µmol/l) was mixed with 150 µl Opti-Mem medium (Gibco; Thermo Fisher Scientific, Inc.) in one tube. 5 µl Lipofectamine® 2000 reagent was mixed with 150 µl Opti-Mem medium in another tube. The contents of the two tubes were mixed and incubated at room temperature for 5 min. The mixture was subsequently added to the cells in 6-well plates and the plates were incubated at 37˚C. After 24 h, the medium was replaced with DMEM with 10% FBS. The cells were collected for subsequent experimentation following 24 h of further culture. The sequences of these oligonucleotides were as follows: siSUMO1P3-302, 5'-GGCGUUCCAAUGAAUUCAUTT-3'; siSUMO1P3-877, 5'-CUUAAUUCAAGCUACUCUTT-3'; siSUMO1P3-946, 5'-GAUAACUGAUAAGGAGAGATT-3'; and si-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'. Cells were transfected using Lipofectamine®™ 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from cells and human tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The purity and concentration of the RNA was evaluated with NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Subsequently, 1 µg RNA was used to synthesize cDNA using the PrimeScript™ RT Reagent kit (Takara Bio, Inc.) according to the manufacturer's instructions. qPCR was subsequently performed using SYBR Green PCR Master mix (Takara Bio, Inc.) on the ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 10 min at 95˚C, followed by 40 cycles for 10 sec at 95˚C and 40 sec at 60˚C. GAPDH served as the endogenous control for SUMO1P3 and the relative gene expression was calculated using the 2-ΔΔCq method (21). The following primer sequences were used for qPCR: SUMO1P3 forward, 5'-ACTGGGAATGGAGGAAGA-3' and reverse, 5'-TGAGAAAGGATTGAGGGAAAAG-3'; GAPDH forward, 5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse, 5'-GGCTGTTGTCATACTTCTCATGG-3'.
Western blotting
Proteins were extracted from cells using RIPA buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined using BCA assays. Subsequently, proteins (30 µg/lane) were separated on 10% SDS-PAGE gels and then transferred onto PVDF membranes. After blocking with 10% non-fat milk for 1 h at room temperature, the membranes were incubated with antibodies against N-cadherin (1:1,500; cat. no. 13116), E-cadherin (1:1,000; cat. no. 14472), β-catenin (1:1,000; cat. no. 8480), cyclin D1 (1:1,000; cat. no. 2978) and β-actin (1:2,500; cat. no. 3700) overnight at 4˚C. Subsequently, the membranes were incubated with a secondary horseradish peroxidase (HRP)-labelled goat anti-mouse IgG (1:3,000; cat. no. A0216) or HRP-labelled goat anti-rabbit IgG (1:3,000; cat. no. A0208) for 1 h at room temperature. All the primary antibodies were purchased from Cell Signalling Technology, Inc., and the secondary antibodies were purchased from Beyotime Institute of Biotechnology. Protein bands were visualized using the ECL Advanced Western Blot Detection kit (Thermo Fisher Scientific, Inc.) on the Bio-Rad ChemiDoc™ Touch (Bio-Rad Laboratories, Inc.) and subsequently quantified using Image Lab software (version 5.2; Bio-Rad Laboratories, Inc.).
Wound healing assay
Transfected cells were seeded into 6-well plates at a density of 50,000 cells/well. When the cell confluence reached ~85%, scratches were performed and the cells were incubated in serum-free medium. The plates were observed under a IX71 light microscope (magnification, x100; Olympus Corporation) at 0 and 48 h after the scratch, and the distance was measured using ImageJ software (National Institutes of Health).
Cell proliferation assay
Cell proliferation was assessed via the Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. Transfected cells were seeded into 96-well plates at a density of 3,000 cells/well. At the indicated time points (0, 24, 48 and 72 h), 10 µl CCK-8 regent was added into each well, and the plates were incubated at 37˚C for 2 h. The absorbance was measured at a wavelength of 450 nm using BioTek Elx800 (BioTek Instruments, Inc.).
Migration and invasion assays
As previously described, Transwell plates were used to assess cell migration and invasion in vitro (22,23). For cell invasion assays, the chambers (Corning, Inc.) were precoated with Matrigel (BD Biosciences) at 37˚C for 5 h. The chambers were observed in six randomly selected fields under a IX71 light microscope (magnification, x200; Olympus Corporation). The number of migrated cells in every field was counted.
Cell cycle analysis
Transfected cells were collected, fixed in 70% ethanol solution for 6 h at 4˚C. Cells were then washed with PBS and subsequently incubated with PI and RNase from a cell cycle and apoptosis analysis kit (cat. no. C1052; Beyotime Institute of Biotechnology) for 30 min at 37˚C according to the manufacturer's instructions. The cell cycle was analysed on a Guava EasyCyte 6HT-2L flow cytometer (EMD Millipore), and assessed using the ModFit LT software (Version 3.1; Verity Software House, Inc.).
Statistical analysis
All experiments were performed independently in triplicates, and data were presented as the mean ± standard error of the mean. Statistical analysis was performed using GraphPad Prism (version 8.3.0; GraphPad Software, Inc.) and the SPSS statistical software package (version 22.0; IBM Corp.). Unpaired Student's t-test was performed for comparisons between two groups. One-way ANOVA followed by Tukey's post-hoc test was used for multiple comparisons. Kaplan-Meier analysis and log-rank test were used for survival analysis of patients with glioma with low and high expression levels of SUMO1P3. P<0.05 was considered to indicate a statistically significant difference.
Results
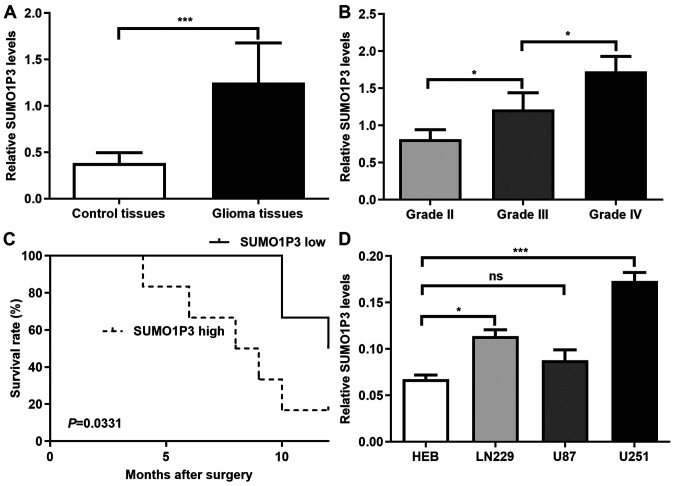
SUMO1P3 expression is upregulated in glioma and high SUMO1P3 expression is associated with a poor prognosis
SUMO1P3 expression was assessed in 12 glioma and 10 control tissues. The clinical parameters of the tissue donors are described in Table I. As presented in Fig. 1A, SUMO1P3 expression was significantly higher in glioma tissues compared with in control tissues. The association between SUMO1P3 expression and glioma malignancy was also analysed. As presented in Fig. 1B, SUMO1P3 expression increased with increasing glioma grade. The present study further investigated whether SUMO1P3 was a prognostic factor for glioma. The median expression value of SUMO1P3 expression was used as a cut-off value to divide all 12 glioma patients into two groups. Patients with SUMO1P3 levels lower than the cut-off value were placed in the low expression group and patients with higher SUMO1P3 expression were assigned to the high expression group. Kaplan-Meier survival analysis indicated that high SUMO1P3 expression was associated with a poorer overall survival in patients with glioma compared with low SUMO1P3 expression (Fig. 1C). Furthermore, SUMO1P3 expression was assessed in 3 glioma cell lines. The results demonstrated that SUMO1P3 expression was upregulated in U251 and LN229 glioma cells compared with in HEB cells, however there was no significant difference between U87 and HEB cells (Fig. 1D). Thus, U251 and LN229 glioma cells were selected for subsequent experiments. Overall, the present results suggested that SUMO1P3 may be a prognostic biomarker for glioma and may act as a tumour promoter in glioma.
Table I.
Clinical parameters of tissue donors.
| Clinicopathological parameters | Glioma (n=12) | Control (n=10) |
|---|---|---|
| Sex | ||
| Male | 6 | 5 |
| Female | 6 | 5 |
| Age, years | ||
| <50 | 6 | 5 |
| ≥50 | 6 | 5 |
| WHO grade | ||
| II | 4 | NA |
| III | 4 | NA |
| IV | 4 | NA |
| Follow-up | ||
| Alive | 4 | NA |
| Dead | 8 | NA |
| Mean survival time, months | 9.8±5.9 | NA |
The data of mean survival time are presented as mean survival time ± standard error of the mean. WHO, World Health Organization; NA, not applicable.
Figure 1.
SUMO1P3 expression is upregulated in glioma. (A) Relative SUMO1P3 expression in control and glioma tissues. (B) Relative SUMO1P3 expression in glioma tissues with different grades. (C) Kaplan-Meier survival curves for patients with glioma with low and high expression levels of SUMO1P3. (D) Reverse transcription-quantitative PCR analysis was performed to detect relative SUMO1P3 expression in different cell lines. Data are presented as the mean ± standard error of the mean. *P<0.05; ***P<0.001. ns, not significant; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3.
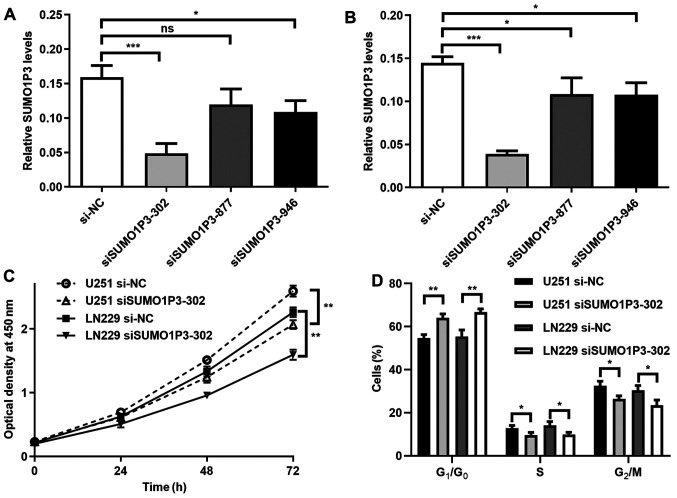
SUMO1P3-knockdown suppresses cell proliferation and cell cycle in vitro
To investigate the function of SUMO1P3 in glioma, its expression was suppressed via transfection with siRNA in glioma cell lines. As presented in Fig. 2A and B, SUMO1P3 expression was significantly decreased following transfection with siSUMO1P3-302 in U251 and LN229 cells. The relative expression level in the siSUMO1P3-302 group was 30% of the si-NC group, thus siSUMO1P3-302 was selected for subsequent experimentation. The results of the cell proliferation assay demonstrated that SUMO1P3-knockdown significantly supressed U251 and LN229 cell proliferation (Fig. 2C). The influence of SUMO1P3 on cell cycle distribution was subsequently assessed. As presented in Figs. 2D and S1, SUMO1P3-knockdown resulted in the accumulation of cells in G1 phase, with 11.24 and 13.46% more cells in G1/G0 phase in the U251 and LN229 glioma cell lines compared with the si-NC groups. Meanwhile, there was a significant decrease in cells in S and G2/M phases in the siSUMO1P3-302 groups compared with cells in the si-NC groups. Collectively, the current results suggested that SUMO1P3-knockdown suppressed cell proliferation in glioma via G0/G1 cell cycle arrest.
Figure 2.
SUMO1P3-knockdown represses cell proliferation and cell cycle in vitro. Reverse transcription-quantitative PCR analysis was performed to detect relative SUMO1P3 expression in (A) U251 and (B) LN229 cells after siRNA transfection. Role of SUMO1P3-knockdown in (C) cell proliferation and (D) cell cycle. Data are presented as the mean ± standard error of the mean. *P<0.05; **P<0.01; ***P<0.001. ns, not significant; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; siRNA, small interfering RNA; NC, negative control.
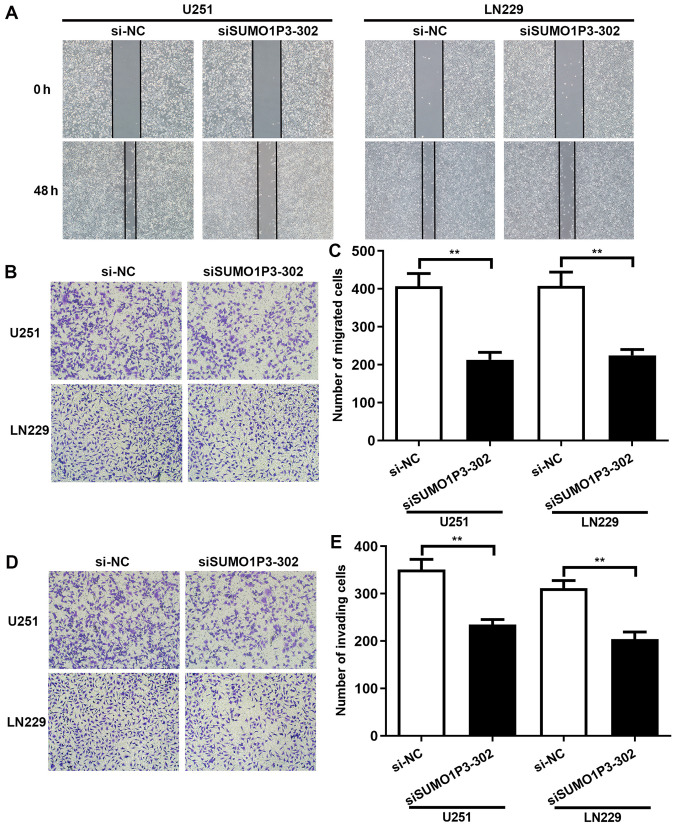
SUMO1P3-knockdown represses cell migration and invasion in vitro
To assess the role of SUMO1P3 in cell migration and invasion, wound healing and Transwell assays were performed. The results of the wound healing assay demonstrated that SUMO1P3-knockdown significantly decreased the speed of wound closure in U251 and LN229 cells. Statistical analysis has been performed and the results showed that the relative wound closure percentage of U251 and LN229 cells at 48 h was 23.38 and 27.79% in the siSUMO1P3-302 group, respectively, and 44.87 and 39.21% in the si-NC group, respectively (Fig. 3A). The role of SUMO1P3 on cell migration and invasion was also investigated. The results of the Transwell assay demonstrated that the number of migrated U251 and LN229 cells in the siSUMO1P3-302 group was 213 and 225 at 48 h, significantly lower than the 407 and 408 cells in the si-NC group, respectively (Fig. 3B and C). In accordance with the results of cell migration, SUMO1P3-knockdown significantly supressed the invasive ability of U251 and LN229 cells (Fig. 3D and E). Overall, the present results suggested that SUMO1P3-knockdown supressed the migratory and invasive abilities of U251 and LN229 cells.
Figure 3.
SUMO1P3-knockdown represses cell migration and invasion in vitro. (A) Representative results of the wound healing assay in SUMO1P3-knockdown cells (magnification, x100). (B) Representative results of the cell migration assay in SUMO1P3-knockdown cells (magnification, x200). (C) Statistical analysis for the results of the cell migration assay. (D) Representative results of the cell invasion assay in SUMO1P3-knockdown cells (magnification, x200). (E) Statistical analysis for the results of the cell invasion assay. Data are presented as the mean ± standard error of the mean. **P<0.01. SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; si, small interfering RNA; NC, negative control.
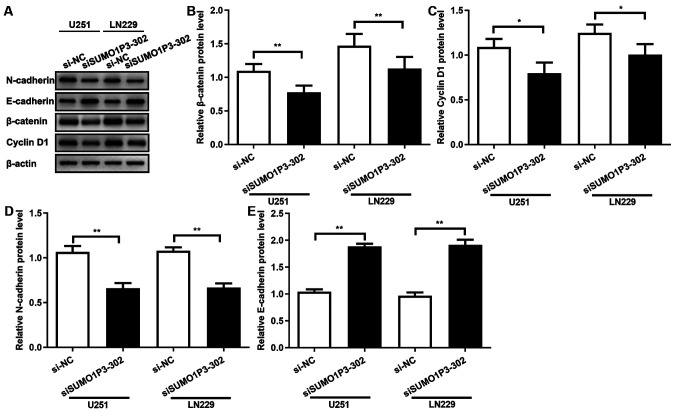
SUMO1P3 acts as a tumour promoter via regulating the expression levels of β-catenin, cyclin-D1, N-cadherin and E-cadherin
The results of the present study demonstrated that SUMO1P3 acted as a tumour promoter in glioma cells by regulating cell proliferation, migration and invasion, whereas the underlying molecular mechanism remains unknown. β-catenin protein expression was assessed following transfection with siRNA, and the results demonstrated that SUMO1P3-knockdown significantly supressed β-catenin expression in both cell lines (Fig. 4A and B). Cyclin-D1 expression was also assessed, which is associated with the cell cycle and a downstream signalling protein of β-catenin (24). As presented in Fig. 4A and C, SUMO1P3-knockdown significantly supressed cyclin-D1 expression. The expression levels of proteins associated with migration and invasion were subsequently assessed. As presented in Fig. 4A, D and E, SUMO1P3-knockdown significantly supressed N-cadherin expression and significantly increased E-cadherin expression. Overall, the current results suggested that SUMO1P3 regulated cell proliferation, cell cycle, migration and invasion by regulating the expression levels of β-catenin, cyclin-D1, N-cadherin and E-cadherin.
Figure 4.
SUMO1P3 acts as a tumour promoter by regulating the expression levels of β-catenin, cyclin-D1, N-cadherin and E-cadherin. (A) Western blot analysis was performed to detect relative protein expression levels of N-cadherin, E-cadherin, β-catenin and cyclin-D1 in U251 and LN229 siRNA-transfected glioma cells. (B-E) Statistical analysis for the western blotting results of (B) β-catenin, (C) cyclin-D1, (D) N-cadherin and (E) E-cadherin. Data are presented as the mean ± standard error of the mean. *P<0.05; **P<0.01. SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; siRNA, small interfering RNA; NC, negative control.
Discussion
Previous studies have reported that lncRNAs are aberrantly expressed in cancer and may serve a crucial role during tumour progression (6,13,16,25). To the best of our knowledge, Mei et al (26) has been the first to report that SUMO1P3 expression is upregulated in gastric cancer tissues and may serve as a potential biomarker for the diagnosis of gastric cancer. Since then, multiples studies have demonstrated the expression pattern and function of SUMO1P3 in different types of tumour. SUMO1P3 expression is upregulated in bladder cancer tissues, and SUMO1P3-knockdown inhibits cell proliferation and migration, while inducing apoptosis in bladder cancer cells (17). Additionally, SUMO1P3 expedites the malignant behaviours of colon cancer in vivo, such as growth, metastasis and angiogenesis, by regulating the expression levels of cyclin D1, vimentin, vascular endothelial growth factor A and E-cadherin (19). A recent study on breast cancer has suggested that SUMO1P3 acts as an oncogenic lncRNA by targeting miR-320a (18), identified as a tumour suppressor in different types of cancer, such as gastric cancer, non-small cell lung cancer and colorectal cancer (27-29). miR-320a supresses β-catenin expression by directly targeting the 3'-untranslated region of β-catenin mRNA in prostate cancer cells (30). In addition, SUMO1P3 promotes hepatocellular carcinoma progression through enhancing the Wnt/β-catenin signalling pathway by sponging miR-320a (31). Thus, it is possible that SUMO1P3-knockdown represses β-catenin expression by targeting miR-320a in glioma.
Cyclin D1 is a downstream molecule of β-catenin (24). Cyclin D1 mediates the progression from G1 to S phase, thus slowing the proliferation of cancer cells (32). In the present study, SUMO1P3-knockdown supressed proliferation in glioma cells probably via cell cycle arrest caused by suppression of cyclin D1. β-catenin and E-cadherin are epithelial cell markers, and N-cadherin is a mesenchymal phenotype marker during the EMT process (33). Increasing evidence suggests that an E-cadherin to N-cadherin shift is mainly involved in EMT and serves a key role in glioma progression and invasion (34). According to the present results, it may be speculated that SUMO1P3-knockdown may supress cell migration and invasion by regulating the expression levels of β-catenin, N-cadherin and E-cadherin. However, the current study presents some limitations. Firstly, only 12 glioma tissues and 10 control tissues were used in the present study. The number of patients was relatively small, therefore further studies using more human tissues should be performed to confirm the relationship between SUMO1P3 expression and glioma malignancy. Furthermore, control tissues were obtained from patients with severe acute traumatic brain injury during a needed surgery, thus theses tissues were not healthy tissues. Despite consent for the collection of tissues was provided by the patients or their guardians, it would have been unethical for healthy tissue to be used. A recent study has reported that SUMO1P3 promotes glioma cell proliferation, migration, and invasion via the Wnt/β-catenin pathway (35). The results and conclusions are consistent with the present study, except for the expression levels of SUMO1P3 in U87 glioma cell line. Lou et al (35) demonstrated high SUMO1P3 expression in U87 cells, whereas the present study identified there was no significant difference between U87 and HEB cells. Since there are two versions of U87 cell line, one is the original glioblastoma cell line established in the University of Uppsala (https://web.expasy.org/cellosaurus/CVCL_GP63) and the other is the U87 MG ATCC version, which is most probably a glioblastoma but whose origin is unknown (https://web.expasy.org/cellosaurus/CVCL_0022), there are differences in the behaviours of these cell lines. The present study confirmed the U87 cell line used was the ATCC version by STR profiling. Regarding Lou et al have not clarified the version of U87 cell line used, it is possible that they used the original glioblastoma cell line established in the University of Uppsala.
In conclusion, the present study revealed the expression pattern of SUMO1P3 and its tumorigenic function in glioma tissues and cells. SUMO1P3-knockdown was demonstrated to suppress cell proliferation, cell cycle, migration and invasion by regulating the expression levels of β-catenin, cyclin-D1, N-cadherin and E-cadherin in U251 and LN229 glioma cells. Overall, the current results suggest that SUMO1P3 may act as a novel diagnostic biomarker and therapeutic target for glioma.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was funded by Changzhou Municipal Commissions of Health and Family Planning Major Scientific and Technological Project (grant no. ZD201620), Changzhou Municipal Commission of Health and Family Planning Youth Talent Scientific and Technological Project (grant no. QN201807) and Funding from Young Talent Development Plan of Changzhou Health Commission (grant no. CZQM2020042).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DD and JC collaborated to design the study, performed the cell cycle assay, analysed the data, drafted the initial manuscript and confirmed the authenticity of the raw data. YM performed RT-qPCR, western blotting and the wound healing assays. LX performed cell proliferation, migration and invasion assays. NS acquired and analysed the data of patients, collected the human tissues and helped in revising the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Board of the Third Affiliated Hospital of Soochow University (grant no. 2020031; Changzhou, China). Written informed consent was provided by all patients or their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Millaer KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Bielle F. Building diagnoses with four layers: WHO 2016 classification of CNS tumors. Rev Neurol (Paris) 2016;172:253–255. doi: 10.1016/j.neurol.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Boussiotis VA, Charest A. Immunotherapies for malignant glioma. Oncogene. 2018;37:1121–1141. doi: 10.1038/s41388-017-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 2017;318:2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50(e6079) doi: 10.1590/1414-431X20176079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, Wang J, Tan Q, Cheng Y, Xia E, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9:533–545. [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8:84086–84101. doi: 10.18632/oncotarget.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. 2018;17:2743–2749. doi: 10.3892/mmr.2017.8143. [DOI] [PubMed] [Google Scholar]
- 11.Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2019;58:196–205. doi: 10.1002/mc.22919. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Zhang X, Hu X, Zhou W, Zhang P, Zhang J, Yang S, Liu Y. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med. 2018;24(52) doi: 10.1186/s10020-018-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Zhang F, Chen Q, Huang Z, Li M. LncRNA MALAT1 modified progression of clear cell kidney carcinoma (KIRC) by regulation of miR-194-5p/ACVR2B signaling. Mol Carcinog. 2019;58:279–292. doi: 10.1002/mc.22926. [DOI] [PubMed] [Google Scholar]
- 14.Jia L, Tian Y, Chen Y, Zhang G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-Catenin pathway. Onco Targets Therapy. 2018;11:313–321. doi: 10.2147/OTT.S154339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, Xiao X, Wang Y. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J Cancer. 2017;8:4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Jin H, Lou F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J Cell Biochem. 2018;119:3007–3016. doi: 10.1002/jcb.26021. [DOI] [PubMed] [Google Scholar]
- 17.Zhan Y, Liu Y, Wang C, Lin J, Chen M, Chen X, Zhuang C, Liu L, Xu W, Zhou Q, et al. Increased expression of SUMO1P3 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Oncotarget. 2016;7:16038–16048. doi: 10.18632/oncotarget.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Song Z, Feng C, Lu Y, Zhou Y, Lin Y, Dong C. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am J Transl Res. 2017;9:5594–5602. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9:5461–5472. [PMC free article] [PubMed] [Google Scholar]
- 20.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Deng D, Luo K, Liu H, Nie X, Xue L, Wang R, Xu Y, Cui J, Shao N, Zhi F. p62 acts as an oncogene and is targeted by miR-124-3p in glioma. Cancer Cell Int. 2019;19(280) doi: 10.1186/s12935-019-1004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng D, Xue L, Shao N, Qu H, Wang Q, Wang S, Xia X, Yang Y, Zhi F. miR-137 acts as a tumor suppressor in astrocytoma by targeting RASGRF1. Tumour Biol. 2016;37:3331–3340. doi: 10.1007/s13277-015-4110-y. [DOI] [PubMed] [Google Scholar]
- 24.Chattopadhyay S, Chaklader M, Law S. Aberrant Wnt signaling pathway in the hematopoietic stem/progenitor compartment in experimental leukemic animal. J Cell Commun Signal. 2019;13:39–52. doi: 10.1007/s12079-018-0470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Zheng ZP, Li H, Zhang HQ, Ma FQ. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Mol Med Rep. 2016;14:1714–1720. doi: 10.3892/mmr.2016.5409. [DOI] [PubMed] [Google Scholar]
- 26.Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30(709) doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 27.Ge X, Cui H, Zhou Y, Yin D, Feng Y, Xin Q, Xu X, Liu W, Liu S, Zhang Q. miR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol Med Rep. 2017;16:9664–9670. doi: 10.3892/mmr.2017.7819. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W, Sun Q, Yu Z, Mao S, Jin Y, Li J, Jiang Z, Zhang Y, Chen M, Chen P, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Dong T, Zhou H, Wang L, Huang A, Feng B, Quan Y, Jin R, Zhang W, Sun J, et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis. 2014;35:886–895. doi: 10.1093/carcin/bgt378. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 31.Wu S, Chen S, Lin N, Yang J. Long non-coding RNA SUMO1P3 promotes hepatocellular carcinoma progression through activating Wnt/β-catenin signalling pathway by targeting miR-320a. J Cell Mol Med. 2020;24:3108–3116. doi: 10.1111/jcmm.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 33.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noh MG, Oh SJ, Ahn EJ, Kim YJ, Jung TY, Jung S, Kim KK, Lee JH, Lee KH, Moon KS. Prognostic significance of E-cadherin and N-cadherin expression in Gliomas. BMC cancer. 2017;17(583) doi: 10.1186/s12885-017-3591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou JY, Luo J, Yang SC, Ding GF, Liao W, Zhou RX, Qiu CZ, Chen JM. Long non-coding RNA SUMO1P3 promotes glioma progression via the Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2020;24:9571–9580. doi: 10.26355/eurrev_202009_23044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.