Abstract
It is increasingly clear that an extraordinarily diverse range of clinically important conditions—including infections, vaccinations, autoimmune diseases, transplants, transfusion reactions, aging, and cancers—leave telltale signatures in the millions of V(D)J-rearranged antibody and T cell receptor [TR per the Human Genome Organization (HUGO) nomenclature but more commonly known as TCR] genes collectively expressed by a person’s B cells (antibodies) and T cells. We refer to these as the immunome. Because of its diversity and complexity, the immunome provides singular opportunities for advancing personalized medicine by serving as the substrate for a highly multiplexed, near-universal blood test. Here we discuss some of these opportunities, the current state of immunome-based diagnostics, and highlight some of the challenges involved. We conclude with a call to clinicians, researchers, and others to join efforts with the Adaptive Immune Receptor Repertoire Community (AIRR-C) to realize the diagnostic potential of the immunome.
Keywords: adaptive immune receptor repertoire (AIRR), diagnostic test, T-cell receptor repertoire, antibody repertoire, analyses, immunome, immunomics, clinical laboratory testing
Introduction
The convergence of high-throughput sequencing technologies with advances in computation and data science has given sequencing a growing role in clinical diagnosis. Examples of high-throughput sequencing applications that have begun to enter the clinic in recent years include cancer-gene sequencing to identify clinically actionable mutations and whole-genome and metagenomic sequencing to resolve medical mysteries (1–4). The main appeal of sequencing as a diagnostic modality is its potential to detect all of the possible variants of a given gene or genes in a single test. In situations where there are many variants that may be diagnostically or prognostically useful, as is the case for cancer and in genetic disorders such as cystic fibrosis, sequencing has been shown to be more sensitive than tests that target a limited set of variants, for example using PCR (5, 6). NGS-based testing has also advanced the field of immunogenomics, providing a more streamlined means of identifying and cataloguing novel human leukocyte antigen (HLA) genes and associating allelic variants and haplotypes with diseases and immune perturbations (see below). Because of their intimate association with many different diseases, B- and T-cell immunomes will feature prominently in the future of clinical lab testing (7) (Figure 1).
Figure 1.
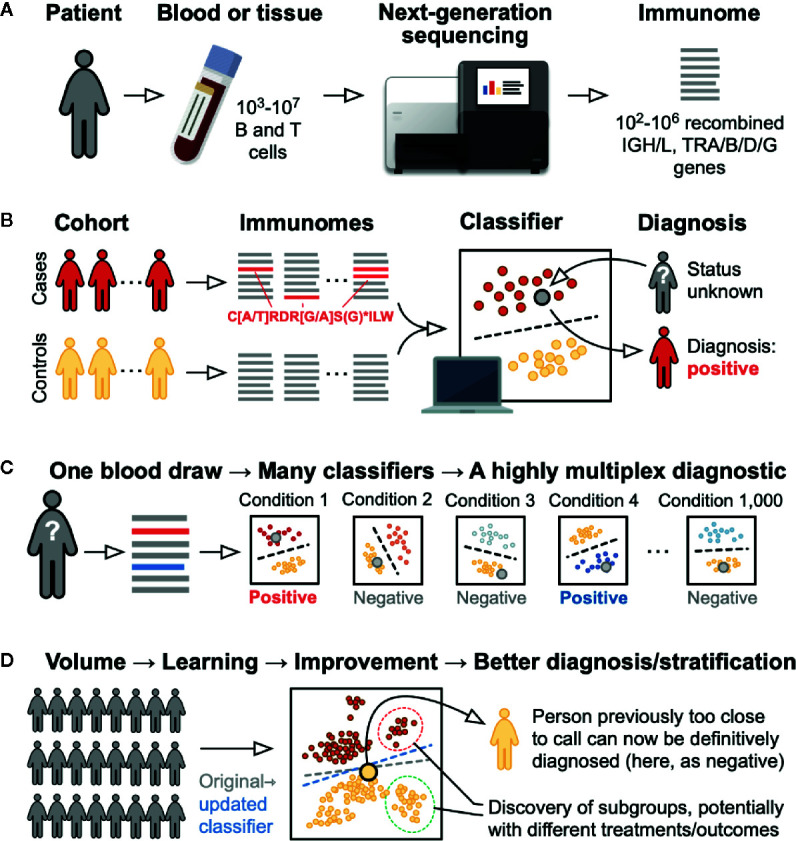
Immunome-based diagnostic testing. (A) Testing begins with a standard clinical blood draw. The recombined immunoglobulin (B cell receptor) and TR (T cell receptor) rearranged genes are sequenced, leading to a list of the hundreds of thousands of different sequences present in the sample: i.e., the patient’s immunome. (B) To develop a test for a specific condition, immunomes are sequenced from a set of cases positive for the condition and an appropriately matched set of controls. Robust statistical and mathematical techniques are used to identify patterns in the form of specific sequences, motifs (e.g., the IGH CDR3 shown in red), and clusters, as well as changes in overall sequence diversity, that are characteristic of the cases but not the controls. Based on these and other sequence features, and with the help of computational techniques, a classifier is developed that reliably separates the two groups. Using this classifier, a patient of unknown status (large gray circle) can be diagnosed by sequencing that patient’s immunome and looking for presence or absence of the pattern. (C) By applying classifiers for many different conditions to the sequence from a single blood draw, many different conditions can be diagnosed simultaneously, yielding a highly multiplexed diagnostic assay. (D) As more individuals are tested for a specific condition, the classifier for that condition will be refined—in AI terms, it “learns”—allowing individuals who were previously unclassifiable to be diagnosed and potentially allowing stratification of patients who might benefit from different treatments or who might have a different prognosis or risk of disease development.
The term “immunome” refers to the repertoire of a person’s antibodies and TRs, most often measured from the blood, which contains roughly 50,000–440,000 B cells and 600,000–3,500,000 T cells per ml in a healthy adult (8–10). Antibodies and TRs are encoded by genes of extraordinary diversity: each person’s immunome contains millions of distinct rearranged antibody and TR genes (henceforth simply “genes”) (11–14). This diversity is what makes it possible for an individual’s immune system to recognize and respond to different antigens in vaccination, infection, autoimmunity, cancer, and other conditions. The binding of an antibody expressed on the B-cell surface to one of its specific antigens—for example, influenza hemagglutinin or the spike protein of SARS-CoV-2—can promote B cell activation, division, and differentiation, resulting in the production of antibodies. For alpha-beta TRs on the surface of T cells, the antigen is typically a peptide that is presented to the TR in the context of the major histocompatibility complex (gamma-delta T cells do not necessarily require MHC).
The Immunome as Diagnostic
For diagnostic purposes, the expansion of antigen-specific B or T cell clones acts as a signal amplifier, indicating a response to a specific antigen or antigens in the form of an increased frequency of cells expressing antigen-specific antibody and/or TR genes in the immunome. Such increases can now be measured quantitatively through high-throughput sequencing in an application known as adaptive immune receptor repertoire sequencing, or AIRR-seq. In principle, repeated cycles of antigen encounter, clonal expansion, and repertoire diversification result in a personalized record of a patient’s immune status across vaccination, infection, autoimmunity, transplant rejection, transfusion reactions, and cancer. AIRR-seq makes it possible to read this record. The past few years have seen an explosion of proofs of principle in the research literature. For example, in patients who have had influenza or received an influenza vaccine, AIRR-seq has demonstrated an increase in influenza-specific antibody and TR genes in the blood and in tissues (15–20). Similar results have been demonstrated in viral infections as diverse as dengue and SARS-CoV-2 (21–28). Noteworthy in this regard is the current effort to discern a T cell fingerprint for SARS-CoV-2 exposure, immune status and possibly even immunopathology in the ImmuneCODE project, a collaboration between Adaptive Biotechnologies and Microsoft, which leverages a rapidly growing and publicly accessible dataset of over 1,400 TR immunomes from individuals who were exposed to SARS-CoV-2 (28, 29). Patterns have been reported in a host of autoimmune diseases such as lupus (30–33), and antibodies and TRs against neoantigens have been reported across solid tumors and in specific cancers such as melanoma (34). These examples and many others increasingly support the view that disease-specific patterns in immunomes are widespread and are likely to be clinically useful.
Immune repertoire profiling is already in clinical use for the diagnosis and monitoring of hematologic malignancies, most frequently of the B cell lineage. The hypervariable third complementarity determining region of the antibody heavy chain or TR beta-chain genes can be used as a clonal fingerprint. Identifying expansions of the same CDR3 sequence and tracking these expansions over time can be used to monitor the frequency of a specific B cell or T cell clone and thereby test for minimal residual disease in these conditions (35). Furthermore, the presence of somatic hypermutations in the antibody variable gene sequence can be used as a prognostic marker in chronic lymphocytic leukemia (36). In addition, patterns within the immunome have been shown to correlate with likelihood of disease or treatment response in several clinical contexts. For example, limited diversity of the repertoire has been associated with frailty during aging (37, 38) and provides evidence of immunodeficient states that are caused by disease or therapies such as bone marrow transplantation. Skewing of immunomes towards overrepresentation of particular variable region genes, CDR3 sequences, or other more complex motifs may serve as fingerprints for disease or disease susceptibility, and studies of immunologic exposure and motifs may be useful for monitoring of immune responses to pathogens as well as therapeutic vaccines (32, 33, 39–41). Immunome profiling thus provides an opportunity to mine immune systems at the individual and population levels to identify signatures that, combined, would represent a “universal” diagnostic laboratory test. Such a clinical test would represent a natural next step for investigations into the role of adaptive immunity in clinical laboratories.
Advantages of AIRR-SEQ
AIRR-seq provides a global view of the immune system in a single snapshot that can be interpreted with different computational methods. Recent progress has already brought the cost and turnaround time of AIRR-seq tests within range of other clinical NGS tests, especially those that must be performed at reference laboratories, also known as “send-out tests” (as opposed to tests that can be performed on site at the hospital). Most critically, sequencing has the fundamental advantage of being an unbiased method, which can in principle reveal atypical immune responses, or responses to new or emerging pathogens such as SARS-CoV-2 without the cost and delay required to develop new pathogen-specific reagents (42–44). Regardless of the condition, the sequencing procedure is the same; what differs is the computational query for each test result: one for influenza, another for lupus, and so on. Each query parses the data for immunoglobulin or TR sequences, motifs, or clusters specific for the given condition. In this manner, AIRR-seq testing can reveal unusual or unexpected patterns in a series of patients with a shared condition. Immunomes and specific sequences can be made easily available through existing resources that have been developed in part in anticipation of diagnostic purposes; these resources include the AIRR Data Commons, which can be accessed via the iReceptor Plus portal (45) and using the AIRR-C API (46). Reference sequences for specific variable genes, alleles and haplotypes can be found at IMGT (47), the Open Germline Reference Database (OGRDB) (48), and the VDJdb database (49) and antigen-specific antibodies and TRs can be queried in the Immune Epitope Database (IEDB) using a suite of tools (50).
Immunome-based diagnostics also align well with the vision of so-called learning health systems, in which the experience of each patient and physician contribute to an improved understanding of disease prevention and management for the benefit of all (51–57). With enhanced data collection in the electronic medical record, secure data sharing, and advances in machine learning, this vision is gradually moving closer to becoming a reality. Just as the growth of repositories such as the UK Biobank increases the power for finding patterns across many conditions, each newly sequenced immunome adds statistical power for finding immunome patterns across the broader population. Data from AIRR-seq based tests could be used for continuing improvement of their diagnostic power. Indeed, efforts by the growing Adaptive Immune Receptor Repertoire Community (AIRR-C; www.antibodysociety.org/the-airr-community/) (58) and by EuroClonality (www.euroclonality.org/) have already led to the development of several high-quality curated public, searchable databases of sequences and their specificities, as well as standards for repertoire data (58–60).
Challenges and Opportunities
There are of course several challenges to address for immunome-based blood testing to become a practical and useful clinical reality, just as there were (and are) for incorporating cancer sequencing, whole-genome sequencing, and metagenomics into patient care. First, the timing of sample collection is important to consider, particularly in investigations of infections or vaccine responses. Recall responses can arise within days of an exposure, but initial responses may take a week or longer to become detectable; durations of responses vary from months or less to lifelong. These considerations mean that immunome-based testing may eventually play multiple different roles for screening, diagnosis, and rule-out testing, depending on the condition and durability of the immune response. Second, protocols for sample preparation will have to be standardized and validated. These will need to cover cases in which sequencing will be performed from whole blood or peripheral blood mononuclear cells (PBMCs) vs. select subsets (e.g. memory B cells, effector T cells); whether single IGH, IGK/L, TRA, TRB, TRG, TRD chains or paired chains will be sequenced (the latter is currently lower throughput and more expensive and requires intact cells); which regions of each chain are sequenced; whether the sequencing is from genomic DNA (agnostic to the cell subset or activation state), mRNA (potentially including isotype information but also may over-represent activated cells), or both; and so on. Third, computational pipelines will likewise need to be clearly described and validated, from sequence assembly (where necessary), to sequence annotation, to correction for sampling and sequencing errors, to the statistical and/or machine learning methods used. All of the stages of this process will need to be rigorously validated to achieve regulatory approval. Fourth, sequencing and computational resources will have to be in place to guarantee clinically viable price points and turnaround times. Fifth, samples will have to be clinically annotated, and this and indeed every process will have to be carefully vetted for adherence to patient privacy directives and related legislation. Sixth, systems for storing and sharing the new clinical data must be set up, to avoid silos and instead maximize the statistical power of widespread testing (as in the vision for learning health systems). And seventh, the signatures, signals, and patterns on which the tests are based must be robust and interpretable, which will likely require the development of “reference ranges” that are relatable to age, gender, genetic background (e.g. ethnicity, gene-segment variants, MHC dependency), and geography (for environmental or infectious exposures).
Fortunately, none of these issues is unique to the immunome; all are or have been regularly encountered in clinical pathology/laboratory medicine, and especially in the development of existing high-throughput sequence-based tests. Moreover, these issues have long been at the center of discussions by the AIRR-C and others to develop guidelines to facilitate eventual clinical adoption (58, 59, 61). For the first time, AIRR-seq offers the possibility of a universal laboratory test that potentially addresses many day-to-day issues in clinical practice: diagnosis and disease prognosis, resistance or response to drugs, immunodeficiency, anti-tumor and immunotherapy responses, and monitoring for progression vs. recovery. In addition, it introduces bioinformatics as the key testing component, suitable for the current era of personalized immuno-medicine, data science, and data-driven patient care. For AIRR-seq based clinical testing to reach its full potential, broad implementation will be required. Similar excitement and partnership have already led to the development of sequencing-based tests in cancer and inherited disease, and is also occurring for the microbiome. We hope that clinicians, laboratorians, researchers, and funding organizations will join efforts to further realize the diagnostic potential of the immunome to help patients with a wide variety of important health conditions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
RA, FR, NS, ELP, and the AIRR-C conceived of the work. RA wrote the initial drafts and prepared the figure. RA, FR, NS, ELP, and the AIRR-C edited the final draft. All authors contributed to the article and approved the submitted version.
Conflict of Interest
Author FR was employed by Roche Sequencing Solutions.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are members of the Diagnostics Working Group of the AIRR Community of The Antibody Society (TAbS). The authors gratefully acknowledge support from TAbS. RA was supported by NIH R01AI148747.
Contributor Information
the Adaptive Immune Receptor Repertoire Community:
Ramy A. Arnaout, Rohit Arora, Rachael Bashford-Rogers, Felix Breden, Syed Ahmad Chan Bukhari, Brian Corrie, Lindsay G. Cowell, Sol Efroni, Christopher Gooley, Victor Greiff, Jason Vander Heiden, Yoshinobu Koguchi, Ton Langerak, Theam Soon Lim, Eline Luning Prak, Encarnita Mariotti-Ferrandiz, Susanna Marquez, Pieter Meysman, Enkelejda Miho, Keshav Motwani, Nima Nouri, Milena Pavlović, Florian Rubelt, Geir Kjetil Sandve, Nicholas Schwab, Igor Snapkov, Cinque Soto, Ulrik Stervbo, Johannes Trück, Henk-Jan van den Ham, Corey Watson, and Cédric R. Weber
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.626793/full#supplementary-material
References
- 1. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet (2019) 20:341–55. 10.1038/s41576-019-0113-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morganti S, Tarantino P, Ferraro E, D’Amico P, Duso BA, Curigliano G. Next Generation Sequencing (NGS): A Revolutionary Technology in Pharmacogenomics and Personalized Medicine in Cancer. Adv Exp Med Biol (2019) 1168:9–30. 10.1007/978-3-030-24100-1_2 [DOI] [PubMed] [Google Scholar]
- 3. Prokop JW, May T, Strong K, Bilinovich SM, Bupp C, Rajasekaran S, et al. Genome sequencing in the clinic: the past, present, and future of genomic medicine. Physiol Genomics (2018) 50:563–79. 10.1152/physiolgenomics.00046.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabbagh R, Van den Veyver IB. The current and future impact of genome-wide sequencing on fetal precision medicine. Hum Genet (2020) 139:1121–30. 10.1007/s00439-019-02088-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beauchamp KA, Johansen Taber KA, Grauman PV, Spurka L, Lim-Harashima J, Svenson A, et al. Sequencing as a first-line methodology for cystic fibrosis carrier screening. Genet Med Off J Am Coll Med Genet (2019) 21:2569–76. 10.1038/s41436-019-0525-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao EY, Jones M, Jones SJM. Whole-Genome Sequencing in Cancer. Cold Spring Harb Perspect Med (2019) 9:1–13. 10.1101/cshperspect.a034579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson WH. Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery. Nat Rev Rheumatol (2015) 11:171–82. 10.1038/nrrheum.2014.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanco E, Pérez-Andrés M, Arriba-Méndez S, Contreras-Sanfeliciano T, Criado I, Pelak O, et al. Age-associated distribution of normal B-cell and plasma cell subsets in peripheral blood. J Allergy Clin Immunol (2018) 141:2208–19.e16. 10.1016/j.jaci.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 9. Kokuina E, Breff-Fonseca MC, Villegas-Valverde CA, Mora-Díaz I. Normal Values of T, B and NK Lymphocyte Subpopulations in Peripheral Blood of Healthy Cuban Adults. MEDICC Rev (2019) 21:16–21. [DOI] [PubMed] [Google Scholar]
- 10. Valiathan R, Deeb K, Diamante M, Ashman M, Sachdeva N, Asthana D. Reference ranges of lymphocyte subsets in healthy adults and adolescents with special mention of T cell maturation subsets in adults of South Florida. Immunobiology (2014) 219:487–96. 10.1016/j.imbio.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 11. Briney BS, Willis JR, McKinney BA, Crowe JE, Jr. High-throughput antibody sequencing reveals genetic evidence of global regulation of the naïve and memory repertoires that extends across individuals. Genes Immun (2012) 13:469–73. 10.1038/gene.2012.20 [DOI] [PubMed] [Google Scholar]
- 12. Britanova OV, Putintseva EV, Shugay M, Merzlyak EM, Turchaninova MA, Staroverov DB, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol Baltim Md 1950 (2014) 192:2689–98. 10.4049/jimmunol.1302064 [DOI] [PubMed] [Google Scholar]
- 13. Kaplinsky J, Arnaout R. Robust estimates of overall immune-repertoire diversity from high-throughput measurements on samples. Nat Commun (2016) 7:11881. 10.1038/ncomms11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soto C, Bombardi RG, Branchizio A, Kose N, Matta P, Sevy AM, et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature (2019) 566:398. 10.1038/s41586-019-0934-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen G, Yang X, Ko A, Sun X, Gao M, Zhang Y, et al. Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8+ TCR Repertoires to Immunodominant Viral Antigens. Cell Rep (2017) 19:569–83. 10.1016/j.celrep.2017.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crowe JE. Influenza Virus-Specific Human Antibody Repertoire Studies. J Immunol Baltim Md 1950 (2019) 202:368–73. 10.4049/jimmunol.1801459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He X-S, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med (2013) 5:171ra19. 10.1126/scitranslmed.3004794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sant S, Grzelak L, Wang Z, Pizzolla A, Koutsakos M, Crowe J, et al. Single-Cell Approach to Influenza-Specific CD8+ T Cell Receptor Repertoires Across Different Age Groups, Tissues, and Following Influenza Virus Infection. Front Immunol (2018) 9:1453. 10.3389/fimmu.2018.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song I, Gil A, Mishra R, Ghersi D, Selin LK, Stern LJ. Broad TCR Repertoire And Diverse Structural Solutions To Recognition Of An Immunodominant CD8 T Cell Epitope. Nat Struct Mol Biol (2017) 24:395–406. 10.1038/nsmb.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strauli NB, Hernandez RD. Statistical inference of a convergent antibody repertoire response to influenza vaccine. Genome Med (2016) 8:60. 10.1186/s13073-016-0314-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell (2020) 182:73–84.e16. 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature (2020) 584:437–42. 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nielsen SCA, Yang F, Jackson KJL, Hoh RA, Röltgen K, Jean GH, et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe (2020) 28:516–25.e5. 10.1016/j.chom.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol (2020) 5:1–15. 10.1126/sciimmunol.abd7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niu X, Li S, Li P, Pan W, Wang Q, Feng Y, et al. Longitudinal Analysis of T and B Cell Receptor Repertoire Transcripts Reveal Dynamic Immune Response in COVID-19 Patients. Front Immunol (2020) 11:582010. 10.3389/fimmu.2020.582010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schultheiß C, Paschold L, Simnica D, Mohme M, Willscher E, von Wenserski L, et al. Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity (2020) 53:442–55.e4. 10.1016/j.immuni.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shomuradova AS, Vagida MS, Sheetikov SA, Zornikova KV, Kiryukhin D, Titov A, et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity (2020) 53:1245–57.e5. 10.1016/j.immuni.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R, et al. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. MedRxiv Prepr Serv Health Sci (2020) 1–33. 10.1101/2020.07.31.20165647 [DOI]
- 29. Gittelman RM, Lavezzo E, Snyder TM, Zahid HJ, Elyanow R, Dalai S, et al. Diagnosis and Tracking of Past SARS-CoV-2 Infection in a Large Study of Vo’, Italy Through T-Cell Receptor Sequencing. medRxiv (2020) 2020:11.09.20228023. 10.1101/2020.11.09.20228023 [DOI] [Google Scholar]
- 30. Bashford-Rogers RJM, Smith KGC, Thomas DC. Antibody repertoire analysis in polygenic autoimmune diseases. Immunology (2018) 155:3–17. 10.1111/imm.12927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hershberg U, Luning Prak ET. The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos Trans R Soc B Biol Sci (2015) 370:20140239. 10.1098/rstb.2014.0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol (2015) 16:755–65. 10.1038/ni.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bashford-Rogers RJM, Bergamaschi L, McKinney EF, Pombal DC, Mescia F, Lee JC, et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature (2019) 574:122–6. 10.1038/s41586-019-1595-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science (2015) 348:803–8. 10.1126/science.aaa3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caccomo S. FDA authorizes first next generation sequencing-based test to detect very low levels of remaining cancer cells in patients with acute lymphoblastic leukemia or multiple myeloma (2020). Available at: https://www.fda.gov/news-events/press-announcements/fda-authorizes-first-next-generation-sequencing-based-test-detect-very-low-levels-remaining-cancer (Accessed December 23, 2020). FDA. [Google Scholar]
- 36. Kröber A, Seiler T, Benner A, Bullinger L, Brückle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood (2002) 100:1410–6. 10.1182/blood.V100.4.1410.h81602001410_1410_1416 [DOI] [PubMed] [Google Scholar]
- 37. Dunn-Walters DK, Ademokun AA. B cell repertoire and ageing. Curr Opin Immunol (2010) 22:514–20. 10.1016/j.coi.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 38. Tabibian-Keissar H, Hazanov L, Schiby G, Rosenthal N, Rakovsky A, Michaeli M, et al. Aging affects B-cell antigen receptor repertoire diversity in primary and secondary lymphoid tissues. Eur J Immunol (2016) 46:480–92. 10.1002/eji.201545586 [DOI] [PubMed] [Google Scholar]
- 39. Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet (2017) 49:659–65. 10.1038/ng.3822 [DOI] [PubMed] [Google Scholar]
- 40. Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, et al. Identifying specificity groups in the T cell receptor repertoire. Nature (2017) 547:94–8. 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (2017) 357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galson JD, Schaetzle S, Bashford-Rogers RJM, Raybould MIJ, Kovaltsuk A, Kilpatrick GJ, et al. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. Front. Immunol (2020) 11:605170. 10.3389/fimmu.2020.605170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med (2020) 26:842–4. 10.1038/s41591-020-0901-9 [DOI] [PubMed] [Google Scholar]
- 44. Minervina AA, Komech EA, Titov A, Koraichi MB, Rosati E, Mamedov IZ, et al. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T cell memory formation after mild COVID-19 infection. Elife (2021) 10:e63502. 10.7554/eLife.63502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corrie BD, Marthandan N, Zimonja B, Jaglale J, Zhou Y, Barr E, et al. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol Rev (2018) 284:24–41. 10.1111/imr.12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christley S, Aguiar A, Blanck G, Breden F, Bukhari SAC, Busse CE, et al. The ADC API: A Web API for the Programmatic Query of the AIRR Data Commons. Front Big Data (2020) 3:1–12. 10.3389/fdata.2020.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lefranc M-P, Lefranc G. Immunoglobulins or Antibodies: IMGT® Bridging Genes, Structures and Functions. Biomedicines (2020) 8. 10.3390/biomedicines8090319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohlin M, Scheepers C, Corcoran M, Lees WD, Busse CE, Bagnara D, et al. Inferred Allelic Variants of Immunoglobulin Receptor Genes: A System for Their Evaluation, Documentation, and Naming. Front Immunol (2019) 10:1–13. 10.3389/fimmu.2019.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res (2020) 48:D1057–62. 10.1093/nar/gkz874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC, et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res (2019) 47:W502–6. 10.1093/nar/gkz452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ainsworth J, Buchan I. Combining Health Data Uses to Ignite Health System Learning. Methods Inf Med (2015) 54:479–87. 10.3414/ME15-01-0064 [DOI] [PubMed] [Google Scholar]
- 52. Embi PJ, Payne PRO. Evidence generating medicine: redefining the research-practice relationship to complete the evidence cycle. Med Care (2013) 51:S87–91. 10.1097/MLR.0b013e31829b1d66 [DOI] [PubMed] [Google Scholar]
- 53. Etheredge LM. Rapid learning: a breakthrough agenda. Health Aff Proj Hope (2014) 33:1155–62. 10.1377/hlthaff.2014.0043 [DOI] [PubMed] [Google Scholar]
- 54. Etheredge LM. A rapid-learning health system. Health Aff Proj Hope (2007) 26:w107–18. 10.1377/hlthaff.26.2.w107 [DOI] [PubMed] [Google Scholar]
- 55. Friedman CP, Rubin JC, Sullivan KJ. Toward an Information Infrastructure for Global Health Improvement. Yearb Med Inform (2017) 26:16–23. 10.15265/IY-2017-004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Norgeot B, Glicksberg BS, Butte AJ. A call for deep-learning healthcare. Nat Med (2019) 25:14–5. 10.1038/s41591-018-0320-3 [DOI] [PubMed] [Google Scholar]
- 57. Psek WA, Stametz RA, Bailey-Davis LD, Davis D, Darer J, Faucett WA, et al. Operationalizing the learning health care system in an integrated delivery system. EGEMS Wash DC (2015) 3:1122. 10.13063/2327-9214.1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Breden F, Luning Prak ET, Peters B, Rubelt F, Schramm CA, Busse CE, et al. Reproducibility and Reuse of Adaptive Immune Receptor Repertoire Data. Front Immunol (2017) 8:1418. 10.3389/fimmu.2017.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vander Heiden JA, Marquez S, Marthandan N, Bukhari SAC, Busse CE, Corrie B, et al. AIRR Community Standardized Representations for Annotated Immune Repertoires. Front Immunol (2018) 9:2206. 10.3389/fimmu.2018.02206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brüggemann M, Kotrová M, Knecht H, Bartram J, Boudjogrha M, Bystry V, et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia (2019) 33:2241–53. 10.1038/s41375-019-0496-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rubelt F, Busse CE, Bukhari SAC, Burckert JP, Mariotti-Ferrandiz E, Cowell LG, et al. Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat Immunol (2017) 18:1274–8. 10.1038/ni.3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.


