Abstract
The introduction of SARS-CoV-2 containing human stool and sewage into water bodies may raise public health concerns. However, assessment of public health risks by faecally contaminated water is limited by a lack of knowledge regarding the persistence of infectious SARS-CoV-2 in water. In the present study the decay rates of viable infectious SARS-CoV-2 and SARS-CoV-2 RNA were determined in river and seawater at 4 and 20°C. These decay rates were compared to S. typhimurium bacteriophage MS2 and pepper mild mottle virus (PMMoV). Persistence of viable SARS-CoV-2 was temperature dependent, remaining infectious for significantly longer periods of time in both freshwater and seawater at 4°C than at 20°C. T90 for infectious SARS-CoV-2 in river water was 2.3 days and 3.8 days at 20°C and 4°C, respectively. The T90 values were 1.1 days and 2.2 days in seawater at 20°C and 4°C, respectively. In contrast to the rapid inactivation of infectious SARS-CoV-2 in river and sea water, viral RNA was relatively stable. The RNA decay rates were increased in non-sterilised river and seawater, presumably due to the presence of microbiota. The decay rates of infectious MS2, MS2 RNA and PMMoV RNA differed significantly from the decay rate of SARS-CoV-2 RNA, suggesting that their use as surrogate markers for the persistence of SARS-CoV-2 in the environment is limited.
Keywords: SARS-CoV-2, MS2, PMMoV, River water, Sseawater, Persistence
Graphical Abstract
1. Introduction
Sewage may contain a variety of human viruses belonging to at least 41 viral families, including Adenoviridae, Polyomaviridae and Papillomaviridae, which may cause gastrointestinal and respiratory disease among other pathologies (Cantalupo et al., 2011; Fernandez-Cassi et al., 2018; Martínez-Puchol et al., 2020). Pollution of coastal waters and rivers by sewage or by faecal contamination from non-point animal sources therefore poses a serious public health risk due to ingestion of contaminated water during recreational activities or contamination for of shellfish for human consumption (Ballesté et al., 2020; Bellou et al., 2013; Soller et al., 2010; Yamahara et al., 2007). Human faecal contamination originating from sewerage misconnections, combined sewage overflows or discharges from wastewater treatment plants may occur frequently in urban areas (Ahmed et al., 2019; Kay et al., 2008; Panasiuk et al., 2015; Reynolds et al., 2020).
SARS-CoV-2, a positive sense single stranded RNA coronavirus, causes respiratory disease that varies in severity and may ultimately prove to be fatal, although the majority of infected individuals exhibit mild symptoms or are asymptomatic (Gorbalenya et al, 2020; Wang, Horby et al., 2020). To date, there are over 119 million confirmed cases globally, resulting in 2.6 million deaths (WHO, 2020). Although SARS-CoV-2 initially causes infection of the upper respiratory tract, it subsequently disseminates to other parts of the body. Recent studies have shown that 27 to 89% of infected individuals shed SARS-CoV-2 RNA in their stool, with 107 RNA/g faeces one week after symptoms develop, dropping to 103 g/faeces after three weeks (Wölfel et al., 2020; Zhang et al., 2020). Viral RNA remains present in faeces after respiratory tract samples are PCR negative, suggesting gastrointestinal replication (Chen et al., 2020; Lin et al., 2020; Y. Wu et al., 2020). Viable virus particles have been detected in faeces in a recent study the first week after symptoms develop (Wang, Xu et al., 2020).
The presence of SARS-CoV-2 RNA in stool samples of both symptomatic and asymptomatic infected individuals implies that wastewater surveillance is a viable means to monitor the circulation of SARS-CoV-2 in the population (Bivins et al., 2020b; Kitajima et al., 2020). Indeed, following initial reports of detection of viral RNA in the influent of wastewater treatment plants, it now has been firmly established that the levels of SARS-CoV-2 RNA are a valuable and robust indicator of the prevalence of the virus in the community (Haramoto et al., 2020; Medema et al., 2020; Patel et al., 2020; Randazzo et al., 2020). SARS-CoV-2 RNA is relatively stable in sewage and non-chlorinated drinking water (Ahmed et al., 2020). A subsequent study showed that viable SARS-CoV-2 decays in dechlorinated tap water and in sewage with T90 ranging between 1.6 and 2.1 days (Bivins et al., 2020a). However, the persistence of the infectious SARS-CoV-2 particle in the natural aquatic environment remains unknown. It therefore not clear to what extent environmental contamination with untreated human sewage poses a risk to public health due to transmission of SARS-CoV-2 to individuals coming into contact with sewage contaminated water (La Rosa et al., 2020).
Pepper mild mottle virus (PMMoV) has been recently proposed as human pollution indicator since it is the most abundant RNA virus in human faeces and is present in high relative concentrations in sewage (Kitajima et al., 2018). Furthermore, it has been evaluated as a virus process indicator for drinking water and sewage treatments (Symonds et al., 2019). F-specific coliphages have been widely used as an indicator of water quality, as well as an indicator for the of enteric viruses in water disinfection treatments (Jofre et al., 2016; Lucena and Jofre, 2014; McMinn et al., 2017). Bacteriophages have been included in water quality guidelines and rapid tests have been developed to detect infectious particles (Blanch et al., 2020). PMMoV and S. typhimurium bacteriophage MS2 (F-specific phage) are non-enveloped single-strand RNA viruses (Jofre et al., 2016; Rosario et al., 2009) but are both abundant in human faeces and used as a process control. This study evaluates their persistence and their usefulness in aquatic environments as compared to infectious SARS-CoV-2.
In this study we determined the decay rates of viable infectious SARS-CoV-2 and SARS-CoV-2 RNA in river and seawater. These decay rates are compared to S. typhimurium bacteriophage MS2 and PMMoV showing that the latter behave differently than SARS-CoV-2 and may therefore not be potential surrogates. We show that the presence of microbiota increases the decay rates of SARS-CoV-2 RNA at 20°C.
2. Materials and methods
2.1. Virus stock preparation
A clinical strain of SARS-CoV-2 (nCoV-Italy-INMI1), obtained from the European Virus Archive Global (EVAg), was propagated in VeroE6 cells (ATCC CRL-1586). A master stock was passaged to P2 in VeroE6 cells at a multiplicity of infection (MOI) of 0.01 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 2% heat-inactivated foetal bovine serum (FBS), following two freeze-thaw cycles at -80°C. Virus stocks were titred on VeroE6 cells and infectivity quantified by TCID50 assay, in which cytopathic effect (CPE) was scored 48 hours post-infection and calculated according to the method of Reed and Muench (Reed and Muench, 1938).
Pepper mild mottle virus was purchased from the DSMZ under catalogue number PV-0093 and was resuspended according to the strain collection protocol.
A high titre stock of MS2 bacteriophages (ATCC 15597-B1) was obtained using Salmonella typhimurium strain WG49 (NCTC 12484). Phage lysates were prepared by plating plaque-purified phage according to (ISO, 1995). The plaques were resuspended in tryptone-yeast extract-glucose (TYGB) medium broth, centrifuged at 3000 g for 10 min and the phage lysate was filtered through a 0.22 μm filter (Dennehy and Turner, 2004). MS2 bacteriophage and S. typhimurium WG49 were kindly provided by Dr. Anicet Blanch, University of Barcelona.
2.2. Experimental microcosm design
Microcosm experiments were carried out in river water and seawater obtained from the Grand Canal, Dublin and from Dublin Bay Water samples (150 ml), respectively, were filter sterilized (0.22 μm) and spiked with infectious SARS-CoV-2 (final concentration 3.16 × 104 TCID50/ml), pepper mild mottle virus (final concentration 3.2 × 1010 GC/ml) and MS2 (final concentration 6.5 × 106 pfu/ml). Spiked water (500 μl) was aliquoted into sterile 2 ml screw cap tubes. Microcosms were maintained in duplicate in the dark at either 4°C±2°C or at 20°C±2°C under BSL-3 laboratory conditions. Samples were taken after the microcosms were prepared, to determine the starting concentration, and subsequently at 4 and 8 hours. For the first week, samples were taken daily and after this point sampling took place every two days until day 20. Microcosm samples were stored at -80°C prior to analysis.
In parallel, microcosms with non-filter sterilised fresh and sea water were established in duplicate to assess the effect of filtration via the difference in MS2 persistence under BSL-2 conditions. Water samples (64 ml) were spiked with heat-inactivated SARS-CoV-2 (final concentration 8.1 × 106 GC/ml), pepper mild mottle virus (final concentration 3.2 × 1010 GC/ml) and MS2 (final concentration 6.5 × 107 pfu/ml). SARS-CoV-2 was heat inactivated at 60°C for 30 minutes prior to spiking. To ensure that there was no difference in RNA quality and stability following heat inactivation, we compared SARS-CoV-2 RNA from matched non-heat inactivated and heat inactivated virus, and did not observe any difference between these in RT-qPCR assays as was previously reported (Wang, Lien et al. 2020). The spiked water samples were subsequently aliquoted (1 ml) into sterile 2 ml screw cap tubes. Microcosms were maintained at 4°C±2°C and at 20°C±2°C in the dark. Microcosms were sampled immediately and then at day 1, 2, 3, 6, 8, 14 and 20. Only time points on day 0, 3, 6, 8, 14 and 20 were selected for RNA viral decay analysis. Samples were stored at -80°C until further analysis.
2.3. Enumeration of infectious SARS-CoV-2 and MS2
At each time point, water samples were titred on VeroE6 cells and infectious titre determined by TCID50 assay. Cytopathic effect (CPE) was scored 48 hours post-infection and TCID50 calculated as described (Reed and Muench, 1938).
Infectious MS2 was enumerated using the double agar layer plaque assay method adapted from EPA Method 1602 (USEPA, 2001). Briefly, 100 μl microcosm subsample was mixed with 1 ml of Escherichia coli strain HS culture and 2.5 ml of 0.7% (w/v) tryptic soy agar (TSA) before pouring onto 1.5% TSA plates. Plates were incubated at 37°C±2°C for 16-24 hours. The bacterial host strain Escherichia coli HS was prepared according to the manufacturer's instructions (Bluephage S.L., Spain). A positive control for the detection of F-specific coliphages (Bluephage S.L., Spain) was included in each assay.
2.4. Viral RNA extraction
Viral RNA was extracted from 140 µl of each microcosm subsample using the QIAamp Viral RNA Mini Kit (Qiagen, Germany). In addition, RNA extraction controls (positive: SARS-CoV-2, negative: PCR grade water) were added. RT-qPCR assays were performed within 24 hours of RNA extraction.
2.5. RT-qPCR analysis
RT-qPCR assays were performed on the Roche Lightcycler 96 platform (Roche Diagnostics, Germany) in a total volume of 20 µl, containing 5 µl of template (1 µl for pepper mild mottle virus assays) using LightCycler Multiplex RNA Virus Master (Roche Diagnostics, Germany). Primer sequences and concentrations and thermal cycling conditions for each gene are given in Table 1 .
Table 1.
Primers, probes, cycling conditions, Limit of Detection (LoD) and Quantification (LoQ) used in this study.
| Target Gene | Primer/Probe Sequence (5’ -3’) | Primer/Probe Concentration | LoD gc per μl of reaction | LoQ gc per μl of reaction | Cycling Condition | Reference |
|---|---|---|---|---|---|---|
| N1 SARS-CoV-2 Nucleocapsid | 2019-nCoV_N1-F GACCCCAAAAT CAGCGAAAT |
500nM | 4.5 gc/µl | 4.5 gc/µl | RT (50°C - 600s), 95°C - 30s, 45 cycles (95°C - 5s, 60°C -30s) | (Centers for Disease Control and Prevention, 2020) |
| 2019-nCoV_N1-R TCTGGTTACTGC CAGTTGAATCTG |
500nM | |||||
| 2019-nCoV_N1-P FAM-ACCCCGCATTAC GTTTGGTGGACC-IBFQ |
125nM | |||||
| Pepper Mild Mottle Virus | PMMV-FP1-rev GAGTGGTTTGACC TTAACGTTTGA PMMV-RP1 |
900nM | 160 gc/ µl | 160 gc/ µl | RT (50°C - 600s), 95°C - 30s, 45 cycles (95°C - 15s, 60°C -60s) | (Haramoto et al., 2013; Zhang et al., 2006) |
| TTGTCGGTTGCA ATGCAAGT |
900nM | |||||
| PMMV-Probe1 FAM-CCTACCGAAG CAAATG-BHQ1 |
200nM | |||||
| MS2 | Pecson-2F AAGGTGCCTACAA GCGAAGT |
1000nM | 17.6 gc/ µl | 17.6 gc/ µl | RT (50°C - 600s), 95°C - 30s, 45 cycles (95°C - 5s, 60°C -30s) | (Carratalà et al., 2013; Pecson et al., 2009) |
| Pecson-2R TTCGTTTAGGGCAA GGTAGC |
1000nM | |||||
| PecP-2 FAM-ATCGTGGGGTCGC CCGTACG-BHQ1 |
250nM |
MS2 and pepper mild mottle virus were quantified using 10-fold dilutions of gBlock Gene Fragments (Integrated DNA Technologies, USA). An N1 amplicon was used as standard for SARS-CoV-2 quantification. The 2019-nCoV_N_Positive Control (Integrated DNA Technologies, USA) was amplified by end-point PCR following the Centers for Disease Control and Prevention protocol (Centers for Disease Control and Prevention, 2020). The amplicon was purified using QIAquick PCR purification kit (Qiagen, Germany) and quantified with Qubit (Invitrogen, USA)
All samples, negative controls and extractions blanks were analyzed in duplicate and four triplicate quantification standards were included in each 96-well plate. Results were expressed in GC/ml. The efficiency of each reaction was determined using the E = 10(1/slope)-1 equation (Rutledge 2003). The limit of detection was determined as the lowest concentration of DNA detected in 95% or more of replicates and the limit of quantification was determined as the lowest concentration of DNA quantified within 0.5 standard deviations of the log10 concentration (Table 1) (Blanchard et al., 2012; Rutledge and Stewart, 2008).
2.6. Data analysis
The decay rates were determined with a first order decay model (Chick, 1908) and a biphasic decay model.
The first-order decay rate constant was calculated using the following formulas:
Where Nt and N0 are the concentrations of TCID50, pfu or GC in the microcosm at time t and time 0 and k is the decay rate.
The biphasic decay model was modeled using the following formula:
Where Nt is the concentration at time t, Nf0 is the initial concentration of the fast decay period and Ns0, the initial concentration of the slow decay period. kf and ks are the fast and slow decay rates.
The fit of the model was evaluated by r 2, RMSE and Wald-Wolfowitz runs test. The models were compared with the extra sum-of-squares F test, the best fitting model was chosen. Significant differences were determined at an alpha level of 0.05. The days needed to achieve a 90% reduction in initial concentration was calculated as follows:
Prism8 (GraphPad Software, USA) was used to perform the statistical analysis and to prepare data plots.
3. Results
3.1. Decay rates of infectious SARS-CoV-2 are higher in seawater than in river water
To determine the persistence of SARS-CoV-2 in the environment and to assess public health risks of sewage spills it is critically important to know the decay rates of infectious viral particles under environmental conditions. We therefore set up microcosm experiments in which filter-sterilised river water and seawater was spiked with infectious SARS-CoV-2 and incubated at 4 and 20°C. In addition, bacteriophage MS2 and PMMoV were added at the start of the experiment to evaluate their usefulness as a surrogate for SARS-CoV-2.
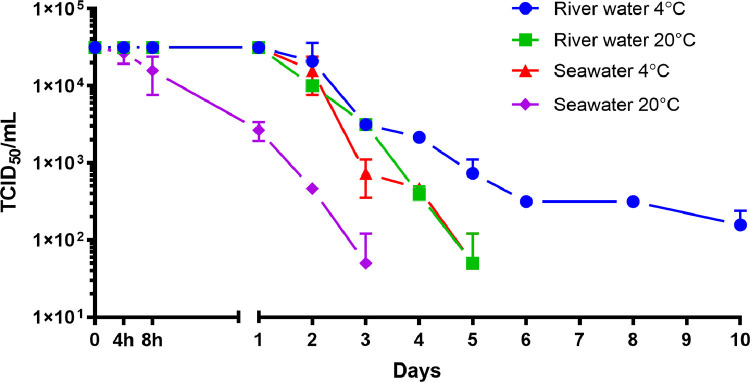
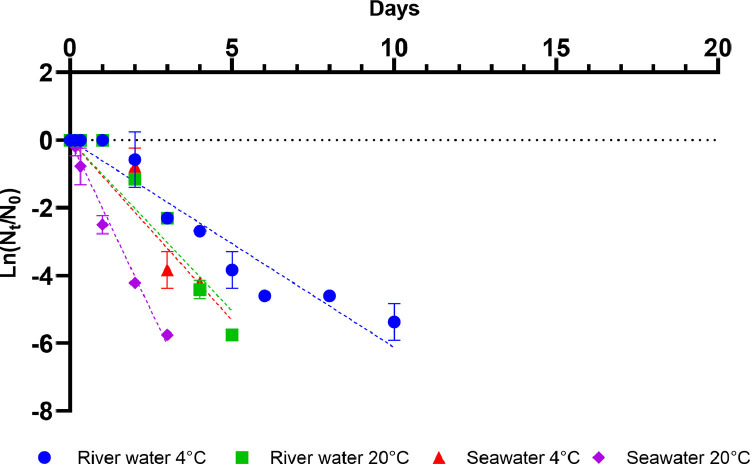
Following inoculation at a titre of 3.16 × 104 TCID50/ml, infectious SARS-CoV-2 titres remained stable for 24 hours in river water at both 4 and 20°C and in seawater at 4°C. In contrast, SARS-CoV-2 infectivity in seawater at 20°C started to decline after 4 hours (Fig 1 ). Infectious SARS-CoV-2 was more stable in river water than in seawater, and more stable at 4 than at 20°C (Fig. 2 ). The first-order decay rates ranged from 0.61 to 2.02 day−1 (Table 2 ). Interestingly, the decay rates for infectious SARS-CoV-2 in seawater at both temperatures were higher than those for river water at the either temperature. The estimated decay rate in river water at 4°C was 0.61 day −1 (T90= 3.8 days) and 1.01 day−1 (T90= 2.3 days) at 20°C, whereas in seawater the decay rates were 1.07 day−1 (T90= 2.2 days) and 2.02 day−1 (T90= 1.1 days) (Table 3 ).
Fig. 1.
Decay of infectious SARS-CoV-2 in filter sterilised river water and seawater at 4°C and 20°C. Data points represent mean values obtained from duplicate experiments. Error bars represent standard deviation.TCID50: fifty percent tissue culture infectious dose.
Fig. 2.
Mean decay curves of infectious SARS-CoV-2 in filter sterilised river water and seawater at 4°C and 20°C. Data points represent mean values obtained from duplicate experiments. Error bars represent standard deviation
Table 2.
Inactivation parameters of infectious SARS-CoV-2 and MS2 in river water and seawater, first-order decay rates were estimated by linear regression. 95% confidence interval (CI) have been included. ns: non significant deviation from the model. sv: significant variation. s: stable for the duration (20 days) of the experiment.
| kmean (day −1) | 95% CI | r2 | RMSE | Runs test | |||
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 filter sterilised water | River water | 4°C | 0.61 | 0.69 to 0.53 | 0.93 | 0.57 | p=0.05 sv |
| 20°C | 1.01 | 1.22 to 0.80 | 0.92 | 0.66 | p=0.10 ns | ||
| Seawater | 4°C | 1.07 | 1.30 to 0.83 | 0.91 | 0.73 | p=0.71 ns | |
| 20°C | 2.02 | 2.21 to 1.83 | 0.99 | 0.28 | p=0.50 ns | ||
| MS2 filter sterilised water | River water | 4°C | s | ||||
| 20°C | 0.71 | 1.08 to 0.34 | 0.73 | 1.22 | p=0.67 ns | ||
| Seawater | 4°C | 0.23 | 0.36 to 0.09 | 0.31 | 1.39 | p=0.50 ns | |
| 20°C | 0.30 | 0.49 to 0.09 | 0.04 | 2.04 | p=0.20 ns | ||
| MS2 non-filter sterilised water | River water | 4°C | s | ||||
| 20°C | 1.17 | 1.24 to 1.09 | 0.99 | 0.32 | p=0.20 ns | ||
| Seawater | 4°C | 0.34 | 0.47 to 0.22 | 0.69 | 1.46 | p=0.54 ns | |
| 20°C | 1.23 | 1.71 to 0.75 | 0.63 | 1.99 | p=0.20 ns |
Table 3.
T90 (days) with 95% confidence intervals (CI) for infectious SARS-CoV-2 and MS2 in river water and seawater. s: stable for the duration (20 days) of the experiment.
| T90 (days) | River water 4°C | River water 20°C | Seawater 4°C | Seawater 20°C |
|---|---|---|---|---|
| SARS-CoV-2 filter sterilised water | 3.8 (3.3-4.3) | 2.3 (1.9-2.9) | 2.2 (1.8-2.8) | 1.1 (1.0-1.3) |
| MS2 filter sterilised water | s | 3.2 (2.6-6.8) | 10.2 (6.4-25.3) | 7.8 (4.7-23.5) |
| MS2 non-filter sterilised water | s | 2.0 (1.9-2.1) | 6.7 (4.9-10.7) | 1.9 (1.3-3.1) |
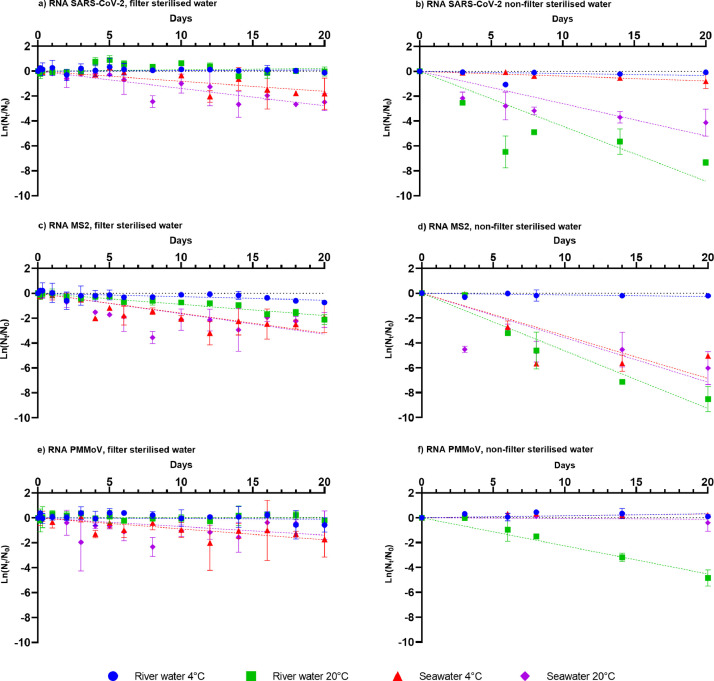
In contrast to the rapid inactivation of infectious SARS-CoV-2 in river and sea water, the N1 RNA marker was relatively stable, showing no decline in river water for the duration (20 days) of the experiment at either 4 or 20°C (Fig 4a). At 4 °C in seawater, a decline was observed, however, it did not achieve 1 log10 reduction. Although the levels of the N1 marker did decline in sea water at 20°C, the decay rate (0.14 day−1, T90=16.6 days) was lower than those of the infectious particle (Fig 4a, Table 4 ).
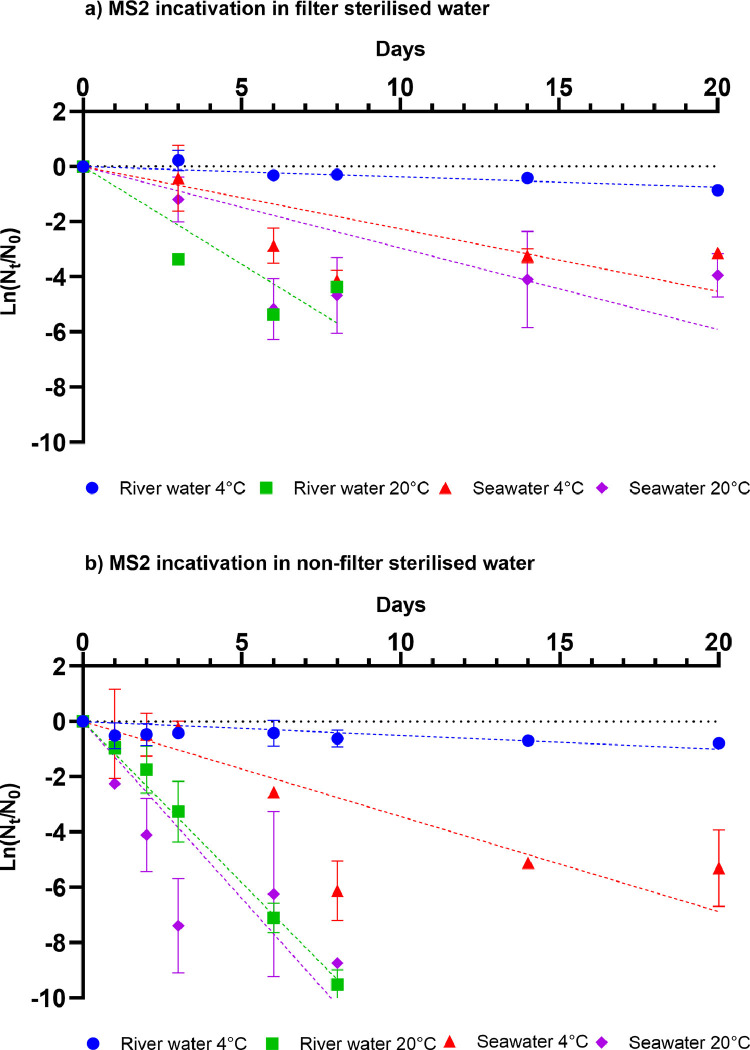
Fig. 4.
Mean decay curves of SARS-CoV-2, MS2 and PMMoV in filter sterilised river water and seawater and non-filter sterilised river water and seawater at 4°C and 20°C. Data points represent mean values obtained from duplicate experiments. Error bars represent standard deviation.
Table 4.
Inactivation parameters of SARS-CoV-2, MSA and PMMoV RNA in river water and seawater, first-order decay rates were estimated by linear regression. 95% confidence intervals (CI) have been included. ns: non-significant deviation from the model. s: stable for the duration (20 days) of the experiment.
| Filter sterilised water |
Non-filter sterilised water |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kmean (day −1) | 95% CI | r2 | RMSE | Runs test | kmean (day −1) | 95% CI | r2 | RMSE | Runs test | |||
| SARS-CoV-2 RNA | River water | 4°C | s | s | ||||||||
| 20°C | s | 0.44 | 0.64 to 0.25 | 0.45 | 2.03 | p=0.20 ns | ||||||
| Seawater | 4°C | s | s | |||||||||
| 20°C | 0.14 | 0.16 to 0.11 | 0.80 | 0.30 | p=0.76 ns | 0.26 | 0.36 to 0.16 | 0.46 | 1.08 | p=0.40 ns | ||
| MS2 RNA | River water | 4°C | s | p=0.15 ns | s | |||||||
| 20°C | s | p=0.76 ns | 0.46 | 0.55 to 0.38 | 0.94 | 0.84 | p=0.50 ns | |||||
| Seawater | 4°C | 0.16 | 0.20 to 0.13 | 0.62 | 0.65 | p=0.17 ns | 0.34 | 0.50 to 0.18 | 0.61 | 1.67 | p=0.50 ns | |
| 20°C | 0.17 | 0.21 to 0.12 | 0.46 | 0.84 | p=0.06 ns | 0.36 | 0.54 to 0.18 | 0.22 | 1.86 | p=0.20 ns | ||
| PMMoV RNA | River water | 4°C | s | s | ||||||||
| 20°C | s | 0.23 | 0.27 to 0.19 | 0.96 | 0.39 | p=0.20 ns | ||||||
| Seawater | 4°C | s | s | |||||||||
| 20°C | s | s | ||||||||||
3.2. Decay of potential SARS-CoV-2 surrogates MS2 and PMMoV in filter-sterilised sea and river water
Bacteriophages or viruses abundant in human faeces, e.g., MS2 or PMMoV are used to monitor the water quality and may be used as surrogates to evaluate the environmental persistence of human pathogenic viruses, which is why these two potential surrogates were included in SARS-CoV-2 microcosms using filter sterilised sea and river water. Water was filter sterilised to avoid any risk of bacterial contamination or non-specific cytopathic effects in the VeroE6 cells used in the infectivity assays of SARS-CoV-2.
Viable MS2 particles were most stable at 4°C in river and sea water (Fig 3 a). The highest inactivation rate was observed in river water at 20°C (T90=3.2 day−1). The T90 values in seawater were 10.2 and 7.8 days at 4 and 20°C, respectively (Table 2, 3). Under all microcosm conditions tested here, viable MS2 particles were more stable than infectious SARS-2-CoV-2 (Fig 2, 3a, Table 2, 3). The largest difference in decay rates was observed in river water at 4°C, where the decay rate for viable SARS-CoV-2 was 0.61 day−1, and MS2 particles did not exceed 1 log10 reduction during the course of the experiment (20 days). The decay rates of infectious SARS-CoV-2 and MS2 were most similar in river water microcosms at 20°C, with only a 1.4-fold difference between SARS-CoV-2 and MS2 (Table 2). In sharp contrast, SARS-CoV-2 RNA was more stable than MS2 RNA under all conditions tested. SARS-CoV-2 and MS2 RNA decay rates were determined with a first order decay model so that these could be compared to the decay rates of the infectious particle.
Fig. 3.
Mean decay curves of infectious MS2 in (a) filter sterilised river water and seawater and (b) non-filter sterilised river water and seawater at 4°C and 20°C. Data points represent mean values obtained from duplicate experiments Error bars represent standard deviation.
The difference was most notable in river water in which SARS-CoV-2 RNA, but not MS2 RNA, was completely stable for the duration (20 days) of the experiment at 20 °C (Fig 4 a,c, Table 4). The differences were less pronounced in sea water, where the decay rate for MS-2 RNA was 1.2-fold larger than for SARS-CoV-2 RNA at 20°C (Table 4).
The PMMoV RNA decay profile was similar to that of SARS-CoV-2 (Fig 4a,e). In both cases viral RNA was stable in river water at 4 and 20°C for the duration (20 days) of the experiment. Both SARS-CoV-2 RNA and PMMoV RNA levels decayed slightly in seawater at both temperatures, however, they did not exceed 1 log10 reduction for the duration of the experiment (20 days).
3.3. Water microbiota increases viral and phage decay
We studied the decay of infectious SARS-CoV-2 in filter sterilised sea and river water to avoid any risk of bacterial contamination or non-specific cytopathic effects in the VeroE6 cells used in the infectivity assays. However, this removes the microbial microbiota from the microcosm experiment. We therefore repeated the microcosm experiments using non-sterilised river and sea water to examine the impact of the microbiota on viral and phage decay.
The effect of non-filtered sea and river water was pronounced for infectious MS2. The decay rates of infectious MS2 in non-filter sterilised water were 1.4 to 4.1- fold higher than in filter sterilised water (Table 2). The decay rates of infectious MS2 in non-filter sterilised river (1.17 day−1,T90=2.0 days) and non-filter sterilised seawater (1.23 day−1, T90=1.9 days) were similar at 20°C. At 4°C, MS2 T90 was 6.7 days (0.34 day−1) in seawater, in contrast, MS2 in non-filter sterilised river water was stable for the duration of the experiment (20 days
Similar observations were made for SARS-CoV-2, MS2 and PMMoV (river water 20°C) RNA where greater decay rates were observed in non-filter sterilised than in filter sterilised water (Table 4). SARS-CoV-2 RNA was more stable in non-filter sterilised water at 4°C, whereas rapid inactivation occurred at 20°C (Table 4, 5 ). As was the case for non-filter sterilised water, SARS-CoV-2 RNA is more stable in sea water (T90 = 8.9 days) than in river water (T90 = 5.2 days). Similar results were obtained for MS2, while PMMoV RNA (at 4°C and seawater at 20°C) was stable. Only between SARS-CoV-2 RNA and MS2 RNA in non-filter sterilised freshwater at 20°C were decay rates similar. T90 values were 5.2 and 5.0 days for SARS-CoV-2 RNA and MS2 RNA, respectively.
Table 5.
T90 (days) with 95% confidence intervals (CI) for SARS-CoV-2, MSA and PMMoV RNA in river water and seawater. s: stable for the duration (20 days) of the experiment.
| Filter sterilised water |
Non-filter sterilised water |
|||||||
|---|---|---|---|---|---|---|---|---|
| River water | River water | Seawater | Seawater | River water | River water | Seawater | Seawater | |
| T90 (days) | 4°C | 20°C | 4°C | 20°C | 4°C | 20°C | 4°C | 20°C |
| SARS-CoV-2 RNA | s | s | s |
16.6 (14.0-20.2) |
s | 5.2 (3.6-9.4) |
s |
8.9 (6.3-14.9) |
| MS2 RNA | s | s | 14.4 (11.8-18.4) |
14.0 (11.0-19.3) |
s | 5.0 (4.2-6.0) |
6.7 (4.6-12.7) |
6.4 (4.3-12.9) |
| PMMoV RNA | s | s | s | s | s | 10.2 (8.7-12.2) |
s | s |
4. Discussion
In this paper we show that although SARS-CoV-2 RNA is relatively stable in both sea and river water, infectious SARS-CoV-2 decays rapidly in both water matrices, with faster decay rates in sea than in river water, and at 20°C than at 4°C. This is in agreement with other studies investigating persistence of coronaviruses in water matrices; SARS-CoV-1 infectivity has been reported to persist for 3-5 days in river water (Duan et al., 2003; Wang et al., 2005), and SARS-CoV-2 infectivity in dechlorinated tap water had a T90 of 1.7 days (Bivins et al., 2020a). In contrast, mouse hepatitis virus, a β-coronavirus, persisted for more than 14 days in lake water incubated at 4°C and for 10 days at 25°C (Casanova et al., 2009; Sizun et al., 2000). SARS-CoV-2 and other coronaviruses are enveloped viruses, and are unable to maintain infectivity in the environment including in water matrices as long as non-enveloped viruses such as norovirus, an enteric virus that is also shed in faeces and can persist for >61 days in water at room temperature (Seitz et al., 2011). Salinity has previously been demonstrated to have a negative effect on influenza virus stability, which is an enveloped RNA virus (Dublineau et al., 2011), in contrast to non-enveloped enteric viruses which maintain infectivity in salinated water (Lo et al., 1976).
Shedding of infectious SARS-CoV-2 in the stool and urine of infected COVID-19 patients is extremely low or undetectable, despite detection of viral RNA in these samples and in sewage (Ahmed et al., 2020; Bivins et al., 2020b; Wang, Xu et al., 2020; Wölfel et al., 2020). This, together with the rapid inactivation of SARS-CoV-2 in both river and seawater, strongly indicates that the likelihood of viral transmission through contact with sewage contaminated waters is low.
In contrast to SARS-CoV-2, MS2 phage infectivity persisted for longer periods of time in all matrices and at both temperatures evaluated. Our data are similar to previous reports investigating MS2 infectivity in river water and seawater (Governal and Gerba, 1997; Muniesa et al., 2009; Ogorzaly et al., 2010), and the decay rates for SARS-CoV-2 and MS2 were significantly different in all conditions tested. This indicates that MS2 does not represent a relevant surrogate for persistence of coronaviruses and other enveloped viruses in water. Interestingly, MS2 infectivity decayed more rapidly in non-filter sterilised than in filter sterilised water. This has been reported previously, where the presence of a microbial population in water has a negative effect on virus survival (Rzezutka and Cook, 2004; Yang and Griffiths, 2013) and filtration of lake water significantly prolonged infectivity of duck plague virus (Wolf and Burke, 1982). This may be due to the production of enzymes such as lipases and proteases as well as predation by for example protozoa. Heat treatment of water resulted in similar removal of antiviral factors (Hawley and Garver, 2008; Yang and Griffiths, 2013). It therefore seems likely that infectious SARS-CoV-2 decays more rapidly in the presence of microbiota than reported here for filter sterilised water.
In contrast to infectious SARS-CoV-2 and MS2, viral RNA from SARS-CoV-2, MS2 and PMMoV remained detectable for the duration of the experiment (20 days). In non-filter sterilised water, MS2 and SARS-CoV-2 RNA decayed at a faster rate than in filter sterilised water. This was particularly evident in seawater and river water incubated at room temperature. Similar to our observations that MS2 infectivity decreased at a faster rate in non-sterile compared to sterile water, indigenous microbial populations in water have been shown to have a negative effect on viral RNA stability (Rzezutka and Cook, 2004). The persistence of viral RNA in water in the present study supports the use of nucleic acid detection methods, including PCR, in the detection and quantification of SARS-CoV-2 RNA in environmental water samples. PMMoV RNA remained stable for the 20 day period measured in the current study, and MS2 RNA decay rates differed significantly from those of SARS-CoV-2 RNA, with the exception of non-filtered water at 20°C. PMMoV is stable and persist longer times in aquatic environments, in previous studies, no significant decline has been observed, thus PMMoV could be considered as a conservative marker with respect to virus reduction (Kitajima et al., 2018; Rachmadi et al., 2016). In sewage, a recent study demonstrated that mouse hepatitis virus (MHV) RNA decay rates were similar to that of SARS-CoV-2, indicating that this β-coronavirus may represent a suitable surrogate for studies investigating the persistence of SARS-CoV-2 RNA in sewage and other water matrices (Ahmed et al., 2020). In contrast, MS2 and PMMoV RNA persistence did not correlate with SARS-CoV-2 RNA in the present study.
5. Conclusion
We demonstrated that infectivity of SARS-CoV-2 decreases rapidly in filter-sterilised river and seawater water, particularly at higher temperatures. The data therefore suggest that river water and seawater contaminated with sewage containing faecal matter from SARS-CoV-2 infected individuals is unlikely to contain high levels of infectious virus due to the rapid inactivation of the virus in these matrices. Infectious MS2 persists longer than infectious SARS-CoV-2 in both river and seawater and is therefore not an appropriate surrogate for the persistence of infectious SARS-CoV-2 under these conditions. The decay rates of infectious SARS-CoV-2 in river and seawater at 4 and 20°C far exceeds those of SARS-CoV-2 RNA, even when the decay rates of SARS-CoV-2 RNA were determined in the presence of microbiota. Although measurement of SARS-CoV-2 RNA therefore demonstrates that sea or river water has been contaminated with the virus at some point in time in the past, it is not a valid methodology to assess the presence of infectious SARS-CoV-2 in the environment. The decay rates of SARS-CoV-2 RNA, MS2 RNA and PMMoV RNA differed, indicating that their use as surrogates for SARS-CoV-2 is limited.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was part funded by the European Regional Development Fund through the Ireland Wales Cooperation programme (Acclimatize) and by Science Foundation Ireland (20-CoV-0159).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117090.
Appendix. Supplementary materials
References
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Besley C. Enhanced insights from human and animal host-associated molecular marker genes in a freshwater lake receiving wet weather overflows. Sci. Rep. 2019;9:12503. doi: 10.1038/s41598-019-48682-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesté E., Demeter K., Masterson B., Timoneda N., Sala-Comorera L., Meijer W.G. Implementation and integration of microbial source tracking in a river watershed monitoring plan. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139573. [DOI] [PubMed] [Google Scholar]
- Bellou M., Kokkinos P., Vantarakis A. Shellfish-Borne Viral Outbreaks: A Systematic Review. Food Environ. Virol. 2013 doi: 10.1007/s12560-012-9097-6. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00730. acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., De Los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., Van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., Van Der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight against COVID-19. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Blanch A.R., Lucena F., Muniesa M., Jofre J. Fast and easy methods for the detection of coliphages. J. Microbiol. Methods. 2020;173 doi: 10.1016/j.mimet.2020.105940. [DOI] [PubMed] [Google Scholar]
- Blanchard P., Regnault J., Schurr F., Dubois E., Ribière M. Intra-laboratory validation of chronic bee paralysis virus quantitation using an accredited standardised real-time quantitative RT-PCR method. J. Virol. Methods. 2012;180:26–31. doi: 10.1016/j.jviromet.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Cantalupo P.G., Calgua B., Zhao G., Hundesa A., Wier A.D., Katz J.P., Grabe M., Hendrix R.W., Girones R., Wang D., Pipas J.M. Raw sewage harbors diverse viral populations. MBio. 2011;2 doi: 10.1128/mBio.00180-11. e00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carratalà A., Rodriguez-Manzano J., Hundesa A., Rusiñol M., Fresno S., Cook N., Girones R. Effect of temperature and sunlight on the stability of human adenoviruses and MS2 as fecal contaminants on fresh produce surfaces. Int. J. Food Microbiol. 2013;164:128–134. doi: 10.1016/j.ijfoodmicro.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. 2019-novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes.https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf URL. (accessed 10.15.20) [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chick H. An investigation of the laws of disinfection. J. Hyg. (Lond). 1908;8:92–158. doi: 10.1017/S0022172400006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. Gorbalenya A.E., Baker S.C., et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J.J., Turner P.E. Reduced fecundity is the cost of cheating in RNA virus ϕ 6. Proc. R. Soc. London. Ser. B Biol. Sci. 2004;271:2275–2282. doi: 10.1098/rspb.2004.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S.M., Zhao X.S., Wen R.F., Huang J.J., Pi G.H., Zhang S.X., Han J., Bi S.L., Ruan L., Dong X.P. Stability of SARS Coronavirus in Human Specimens and Environment and Its Sensitivity to Heating and UV Irradiation. Biomed. Environ. Sci. 2003;16:246–255. [PubMed] [Google Scholar]
- Dublineau A., Batéjat C., Pinon A., Burguière A.M., Leclercq I., Manuguerra J.-C. Persistence of the 2009 Pandemic Influenza A (H1N1) Virus in Water and on Non-Porous Surface. PLoS One. 2011;6:e28043. doi: 10.1371/journal.pone.0028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Timoneda N., Martínez-Puchol S., Rusiñol M., Rodriguez-Manzano J., Figuerola N., Bofill-Mas S., Abril J.F., Girones R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018;618:870–880. doi: 10.1016/j.scitotenv.2017.08.249. [DOI] [PubMed] [Google Scholar]
- Governal R.A., Gerba C.P. Persistence of MS-2 and PRD-1 bacteriophages in an ultrapure water system. J. Ind. Microbiol. Biotechnol. 1997;18:297–301. doi: 10.1038/sj.jim.2900388. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley L.M., Garver K.A. Stability of viral hemorrhagic septicemia virus (VHSV) in freshwater and seawater at various temperatures. Dis. Aquat. Organ. 2008;82:171–178. doi: 10.3354/dao01998. [DOI] [PubMed] [Google Scholar]
- ISO, 1995. ISO 10705-1:1995 - Water quality — Detection and enumeration of bacteriophages — Part 1: Enumeration of F-specific RNA bacteriophages.
- Jofre J., Lucena F., Blanch A., Muniesa M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water. 2016;8:199. doi: 10.3390/w8050199. [DOI] [Google Scholar]
- Kay D., Crowther J., Stapleton C.M., Wyer M.D., Fewtrell L., Edwards A., Francis C.A., McDonald A.T., Watkins J., Wilkinson J. Faecal indicator organism concentrations in sewage and treated effluents. Water Res. 2008;42:442–454. doi: 10.1016/j.watres.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018 doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: Occurrence, persistence and concentration methods - A scoping review. Water Res. 2020 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Zhenling, Huang S., Zhang Zhenyi, Fang Z., Gu Z., Gao L., Shi H., Mai L., Liu Y., Lin X., Lai R., Yan Z., Li X., Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Lo S., Gilbert J., Hetrick F. Stability of human enteroviruses in estuarine and marine waters. Appl. Environ. Microbiol. 1976;32:245–249. doi: 10.1128/aem.32.2.245-249.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena F., Jofre J. Bacteriophages in the Control of Food- and Waterborne Pathogens. ASM Press; Washington, DC, USA: 2014. Potential Use of Bacteriophages as Indicators of Water Quality and Wastewater Treatment Processes; pp. 103–118. [DOI] [Google Scholar]
- Martínez-Puchol S., Rusiñol M., Fernández-Cassi X., Timoneda N., Itarte M., Andrés C., Antón A., Abril J.F., Girones R., Bofill-Mas S. Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens. Sci. Total Environ. 2020;713 doi: 10.1016/j.scitotenv.2020.136604. [DOI] [PubMed] [Google Scholar]
- McMinn B.R., Ashbolt N.J., Korajkic A. Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett. Appl. Microbiol. 2017;65:11–26. doi: 10.1111/lam.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Muniesa M., Payan A., Moce-Llivina L., Blanch A.R., Jofre J. Differential persistence of F-specific RNA phage subgroups hinders their use as single tracers for faecal source tracking in surface water. Water Res. 2009;43:1559–1564. doi: 10.1016/j.watres.2008.12.038. [DOI] [PubMed] [Google Scholar]
- Ogorzaly L., Bertrand I., Paris M., Maul A., Gantzer C. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl. Environ. Microbiol. 2010;76:8019–8025. doi: 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasiuk O., Hedström A., Marsalek J., Ashley R.M., Viklander M. Contamination of stormwater by wastewater: A review of detection methods. J. Environ. Manage. 2015 doi: 10.1016/j.jenvman.2015.01.050. [DOI] [PubMed] [Google Scholar]
- Patel M., Chaubey A.K., Pittman C.U., Mlsna T., Mohan D. Coronavirus (SARS-CoV-2) in the environment: Occurrence, persistence, analysis in aquatic systems and possible management. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.142698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Martin L.V., Kohn T. Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: Advantages and limitations of an enzymatic treatment to reduce false-positive results. Appl. Environ. Microbiol. 2009;75:5544–5554. doi: 10.1128/AEM.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmadi A.T., Kitajima M., Pepper I.L., Gerba C.P. Enteric and indicator virus removal by surface flow wetlands. Sci. Total Environ. 2016;542:976–982. doi: 10.1016/j.scitotenv.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Reynolds L.J., Sala-Comorera L., Martin N.A., Nolan T.M., Stephens J.H., Gitto A., O'Hare G.M.P., O'Sullivan J.J., Meijer W.G. Correlation between antimicrobial resistance and faecal contamination in small urban streams and bathing waters. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.140242. [DOI] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge R.G. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge R.G., Stewart D. Critical evaluation of methods used to determine amplification efficiency refutes the exponential character of real-time PCR. BMC Mol. Biol. 2008;9:96. doi: 10.1186/1471-2199-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzezutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004 doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Seitz S.R., Leon J.S., Schwab K.J., Lyon G.M., Dowd M., McDaniels M., Abdulhafid G., Fernandez M.L., Lindesmith L.C., Baric R.S., Moe C.L. Norovirus infectivity in humans and persistence in water. Appl. Environ. Microbiol. 2011;77:6884–6888. doi: 10.1128/AEM.05806-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizun J., Yu M.W.N., Talbot P.J. Survival of human coronaviruses 229E and OC43 in suspension and after drying on surfaces: A possible source of hospital-acquired infections. J. Hosp. Infect. 2000;46:55–60. doi: 10.1053/jhin.2000.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller J.A., Schoen M.E., Bartrand T., Ravenscroft J.E., Ashbolt N.J. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 2010;44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- Symonds E.M., Rosario K., Breitbart M. 2019. Pepper mild mottle virus: Agricultural menace turned effective tool for microbial water quality monitoring and assessing (waste) water treatment technologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA, 2001. USEPA Method 1602: Male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure.
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020 doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Lien C., Liu S., Selveraj P. Effective Heat Inactivation of SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.04.29.20085498. 2020.04.29.20085498. [DOI] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y., Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. URL https://www.who.int/emergencies/diseases/novel-coronavirus-2019. (accessed 16.03.21).
- Wolf K., Burke C.N. Survival of duck plague virus in water from Lake Andes National Wildlife Refuge, South Dakota. J. Wildl. Dis. 1982;18:437–440. doi: 10.7589/0090-3558-18.4.437. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahara K.M., Layton B.A., Santoro A.E., Boehm A.B. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environ. Sci. Technol. 2007;41:4515–4521. doi: 10.1021/es062822n. [DOI] [PubMed] [Google Scholar]
- Yang Y., Griffiths M.W. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl. Environ. Microbiol. 2013;79:4564–4567. doi: 10.1128/AEM.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:0108–0118. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.Lou, Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.