Abstract
Angiostrongylus cantonensis is a well-known pathogen causing eosinophilic meningitis associated with angiostrongyliasis. Humans, as accidental hosts, are infected by consuming undercooked snails containing third-stage larvae. A. malaysiensis is closely related to A. cantonensis and has been described as a potential human pathogen. The two species distribution was recently reported to overlap in the same endemic area, particularly in the Indochina Peninsula. Similar morphological characteristics of the third-stage larva in the snail-intermediate host often lead to misidentification of the two species. Thus, we aimed to develop a sensitive and specific method to detect and discriminate Angiostrongylus third-stage larva by designing species-specific primers based on the mitochondrial cytochrome b gene. We developed the SYBR Green quantitative real-time PCR (qPCR) method for two species-specific detection assays, which could be conducted simultaneously. The method was subsequently employed to detect and identify third-stage larvae of Angiostrongylus isolated from infected Achatina fulica collected from six public parks in Bangkok Metropolitan, Thailand. The method was also a preliminary applied to detect parasite tissue debris in the patients' cerebrospinal fluid (CSF). SYBR Green qPCRs quantitatively detected approximately 10−4 ng of genomic DNA from one larva, facilitating species-specific detection. Based on the pools of third-stage larvae isolated individually from the tissue of each infected A. fulica collected from the public parks, the qPCR results revealed that A. malaysiensis was the predominant species infecting 5.26% of the collected snails. In comparison, coinfection between A. malaysiensis and A. cantonensis was 5.97%, and no single infection of A. cantonensis was detected in A. fulica. Our SYBR Green qPCR method is a useful and inexpensive technique for A. cantonensis and A. malaysiensis discrimination, and the method has sufficient sensitivity to detect isolated larvae from a snail-intermediate host. The ratio of A. cantonensis and A. malaysiensis larvae infecting the snails can also be estimated simultaneously. Our qPCRs can be employed in a molecular survey of A. cantonensis and A. malaysiensis within intermediate hosts and for clinical diagnosis of angiostrongyliasis with CSF specimens in future studies.
Keywords: SYBR Green-based quantitative real-time PCR, Angiostrongylus cantonensis, Angiostrongylus malaysiensis, Infective stage larva, Achatina fulica, Mitochondrial cytochrome b
Abbreviations: SYBR, Green qPCR; SYBR, Green quantitative real-time polymerase chain reaction; Cytb, cytochrome b; CSF, cerebrospinal fluid; CV, coefficients of variation; E, efficiencies of reactions; Cq, quantification cycle value
Graphical abstract
Highlights
-
•
The SYBR Green qPCRs were developed to detect and discriminate Angiostrongylus third-stage larvae by designing species-specific primers based on the mitochondrial cytochrome b gene.
-
•
A. malaysiensis is the predominant species in Bangkok Metropolitan, Thailand.
-
•
Coinfection between A. cantonensis and A. malaysiensis has occurred in the Achatina fulica population in Bangkok
1. Introduction
The genus Angiostrongylus Kamensky, 1905, contains the zoonotic nematode parasites of vertebrates, commonly known as “lungworms” (Spratt, 2015). Several species in this genus have been reported to be human pathogens, including A. cantonensis and A. costaricensis, while A. mackerrasae and A. malaysiensis are considered potential human pathogenic parasites (Bhaibulaya and Cross, 1971; Sawabe and Makiya, 1994; Aghazadeh et al., 2016; Ansdell and Wattanagoon, 2018). Among them, A. cantonensis is a well-known pathogen that causes eosinophilic meningitis associated with angiostrongyliasis in humans (Barratt et al., 2016), particularly in Thailand (Eamsobhana, 2013). Humans are accidental hosts and are infected by eating undercooked snails, slugs, paratenic hosts, and contaminated fresh vegetables containing the infective third-stage larvae (Wang et al., 2012). The clinical symptoms can range from headaches to coma, depending on the number of parasitic larvae (Wang et al., 2012; Pozio, 2015; Ansdell and Wattanagoon, 2018; Johnston et al., 2019). Many terrestrial and freshwater snails, including, Achatina fulica, Cryptozona spp., Pomacea spp., and Pila spp., are reported to be important vectors of A. cantonensis (Thiengo et al., 2010; Hu et al., 2018; Vitta et al., 2016). Moreover, the widespread distribution of some invasive vector species (A. fulica and Pomacea spp.) has promoted parasite transmission (Thiengo et al., 2010; Fontanilla and Wade, 2012; Yang et al., 2013). Rodents, which are the natural-definitive host of Angiostrongylus, also facilitated parasite transmission through inhabiting urban areas close to humans (Alicata, 1965; Spratt, 2015; Barratt et al., 2016).
Adults of A. cantonensis and A. malaysiensis can be morphologically discriminated by the bursal rays of males and the minute protrusion at the posterior end of females (Bhaibulaya, 1979). However, morphological variations between the two species can confound identification. Furthermore, differences between the morphological characteristics of the larval stages have not yet been described. A. cantonensis was known as the predominant species widely distributed throughout many countries in Indochina (Rodpai et al., 2016). However, a cryptic species of the Malaysian strain of A. cantonensis was recently correctly described as A. malaysiensis (Eamsobhana et al., 2015; Dusitsittipon et al., 2017), due to differences in its pathogenicity. It was recently revealed that A. cantonensis and A. malaysiensis overlap in their distribution, and misidentifications mostly in the specimens from Thailand were discovered (Rodpai et al., 2016; Dusitsittipon et al., 2017, Dusitsittipon et al., 2018). Thus, an under-estimation of the prevalence of A. malaysiensis could result, especially in the Indochina Peninsula, due to the difficulties in the morphological identification of adult and third-stage larva.
To overcome the challenges in the morphological identification of Angiostrongylus, several polymerase chain reaction (PCR)-based identification techniques were developed. However, these techniques were aimed at increasing the sensitivity and specificity of detecting A. cantonensis in blood, the peripheral tissues of wild Hawaiian rats, and the cerebrospinal fluid (CSF) of patients (Eamsobhana et al., 2013; Jarvi et al., 2015; Qvarnstrom et al., 2016). To date, no technique with both high sensitivity and specificity have been developed for detecting and discriminating between A. cantonensis and A. malaysiensis.
Studies have used the mitochondrial cytochrome b (Cytb) gene for species discrimination of A. cantonensis and A. malaysiensis due to high genetic variation between both species and its ability to provide robust phylogenetic relationships (Dusitsittipon et al., 2017, Dusitsittipon et al., 2018). The SYBR Green quantitative real-time PCR (qPCR) method was considered suitable for quantitative detection and species discrimination and has been used successfully in many studies to detect, quantify, and discriminate species. For example, Opisthorchis viverrini and Haplorchis taichui were detected from human stool samples, Leishmania was quantified in human samples, and Salmonella subspecies were all successfully discriminated (Weirather et al., 2011; Barbau-Piednoir et al., 2013; Lamaningao et al., 2017).
Through the design of species-specific primers to the partial Cytb gene sequences of A. cantonensis and A. malaysiensis, we, therefore, aimed to develop a sensitive and specific method using SYBR Green qPCRs for the detection and species discrimination of Angiostrongylus third-stage larvae. We also determined the efficacy of the technique by detecting Angiostrongylus genomic material within the CSF of infected patients. The developed qPCRs were subsequently implemented to detect, discriminate, and estimate the ratio between A. cantonensis and A. malaysiensis third-stage larvae isolated from A. fulica collected from Bangkok public parks to explore the prevalence of A. cantonensis and A. malaysiensis distributing in Bangkok Metropolitan.
2. Materials and methods
2.1. Specimens used
2.1.1. Reference specimens for evaluating sensitivity and specificity
The third-stage larvae of A. cantonensis, adults of A. cantonensis and A. malaysiensis, and CSF of patients with diseases related to eosinophilic meningitis, including neurocysticercosis, gnathostomiasis, and angiostrongyliasis, were used as the reference specimens. Disease diagnoses of the CSF from patients were performed using the immunoblot assay by the immunological diagnostic service unit of the Department of Helminthology, Faculty of Tropical Medicine, Mahidol University. The positive CSF specimens were kept as the archived research specimen stock of the Department. The active larvae and adult worms of Angiostrongylus were preserved in 70% ethanol at −20 °C, while the CSF specimens were kept at −80 °C. The Angiostrongylus specimens were used to evaluate the sensitivity, specificity, and reproducibility of the species-specific primers designed for the SYBR Green qPCRs. One sample per disease was used to determine the specificity of the qPCRs.
2.1.2. Third-stage larvae isolated from wild A. fulica to implement with the developed SYBR Green qPCRs
285 A. fulica snails were collected by hand from six public parks around Bangkok, Thailand (see Fig. 1). After collection, the snails were transported to the laboratory within an hour and subsequently euthanized at 0 °C for 10 min. The shell of each snail was removed, the foot and mantel parts were dissected and incubated with a digestion solution (1% HCl and 1% pepsin) at 37 °C for 1 h, following the protocol of Vitta et al. (Vitta et al., 2011). According to Ash's criteria, the larvae isolated from each snail were morphologically identified (Ash, 1970). From each Angiostrongylus-positive snail, larvae were counted under a stereomicroscope and preserved in 70% ethanol at −20 °C before being used for DNA extraction. The number of Angiostrongylus-positive snails and the number of larvae infecting each snail are listed in Table S1. All the larva was taken into the total count, regardless of whether it was active, weak, or had a degraded body. The collection of A. fulica was performed with permission from the Animal Care and Use Committee, Faculty of Tropical Medicine, Mahidol University, Bangkok (No. FTM-ACUC 024/2018).
Fig. 1.
Map of the six public parks in the Bangkok Metropolitan where wild A. fulica were collected.
2.2. Genomic DNA extraction
2.2.1. Preparation of reference gDNA and CSF samples
The individual adult A. cantonensis and A. malaysiensis, and the third-stage larvae of A. cantonensis of the described reference specimens were transferred into a 1.7-ml centrifuge tube and washed thoroughly with sterile distilled water to remove the ethanol. The third-stage larvae of A. cantonensis were grouped into pools of 1, 5, 10, 50, 100, and 200 larvae. Before DNA extraction, the larval cuticles were homogenized using TissueLyser LT at 50 Hz for 30 s (Qiagen, Hilden, Germany) with 20 mg of 0.1 mm silica beads in 200 μl lysis buffer. For the CSF samples, we centrifuged them separately at 2000 rpm to obtain tissue debris before DNA extraction. Total genomic DNA (gDNA) was extracted from the adult, pooled larval specimens, and CSF samples using the Genomic DNA mini kit (Geneaid Biotech Ltd., Taipei, Taiwan) following the manufacturer's instructions. The eluted gDNA concentration was measured using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, MA, USA). The gDNA was stored at 4 °C until use.
2.2.2. DNA extraction of the isolated third-stage larvae from wild A. fulica
The gDNA of the isolated Angiostrongylus larvae from each positive snail was extracted following the third-stage larvae method described above. In instances where the pooled larvae did not exceed 100 per snail, all of the larvae were pooled and extracted together. In instances where the pooled larvae exceeded 100 per snail, we randomly picked up only 100 larvae. When the pooled larvae were less than 10, we did not perform gDNA extraction and excluded them from the qPCRs to avoid a false negative, resulting from either the degraded or weakened larvae present in the sample pool.
2.3. Development of SYBR Green qPCRs
2.3.1. Design of species-specific primers
Species-specific primers were designed manually based on the partial Cytb gene sequences of A. cantonensis (GenBank accession numbers KC995188, KC995190, KC995193, KC995268, KC995262, KC995265, KC995248, KC995214, KC995211, KC995226, and KC995223, KX147425) and A. malaysiensis (GenBank accession numbers KX147395, KX147380, KX147409, KX147398, KX147455, KX147406, KX147442, KX147445, KX147405, NC_030332, and KT947979). The oligonucleotide primer properties, including GC content, amplicon size, melting temperature, and hairpin formation, were predicted by OligoCalc version 3.27 and Primer3 (Kibbe, 2007; Untergasser et al., 2007). To determine the specificity of the designed primers, in-silico PCR was performed using FastPCR 6.6 (Kalender et al., 2014) by testing cross-amplification with the Cytb gene sequences of A. costaricensis (AP017675), A. vasorum (NC_018602) and A. mackerrasae (MN793157). The nucleotide sequences of the designed species-specific primers are provided in Table 1.
Table 1.
Species-specific primers designed for A. cantonensis and A. malaysiensis using the CytB gene.
| Species | Primer | Sequencea | Amplicon size (bp) |
|---|---|---|---|
| A. cantonensis | AC4_cytb_F | 5′AAT GTT TGT TGA GGC AGA TC 3′ | 117 |
| AC5_cytb_R | 5′ GCT ACA ACA CCC ATA ACC T 3′ | ||
| A. malaysiensis | AM3_cytb_F | 5′ CGA GAT ATT TAT TGA GGC TG 3′ | 141 |
| AM4_cytb_R | 5′GAC AAA ACC CTC ATC AAT AA 3′ |
2.3.2. SYBR Green quantitative real-time PCRs
The SYBR Green qPCRs were performed separately for A. cantonensis and A. malaysiensis using the designed species-specific primers. Following the protocol of the Luna® Universal qPCR master mix (New England Biolabs, MA, USA), each 20 μl reaction contained 10 μl of the master mix, 1.0 μM of each pair of species-specific primers, 1 μl of gDNA, and RNAase-free water. The qPCRs of both species were performed simultaneously on a CFX96 TouchTM Real-Time PCR machine (Bio-Rad Laboratories, CA, USA), and the thermal cycling profile included an initial denaturation at 95 °C for 60 s, followed by 45 cycles of denaturation at 95 °C for 15 s, and an extension at 60 °C for 30 s. A final melting analysis program was applied at 60–95 °C and each cycle duration was 5 s with a 0.5 °C increment per cycle. The SYBR Green qPCRs described above were used for standard curve preparation, specificity and sensitivity assays, and reproducibility assay. These qPCRs were also implemented to detect and discriminate species of the third-stage larvae of Angiostrongylus isolated from wild A. fulica.
2.3.3. Standard curve construction for SYBR Green qPCRs
gDNA of the A. cantonensis and A. malaysiensis adults were serially diluted in ten-folds (ranging from 10−4 to 1 ng/μl) and used as templates for the species-specific SYBR Green qPCRs to construct the standard curve of each species. Three replicates of each gDNA concentration were performed. The precision of the standard curve and the qPCR robustness for both A. cantonensis and A. malaysiensis were verified by considering the slope values, correlation coefficient, and qPCR efficiency.
2.4. Assessment of the developed SYBR Green qPCRs
2.4.1. Specificity assay
With the thermocycling profile describe above, the species-specific primers developed for A. cantonensis and A. malaysiensis were evaluated for their specificity to the various target gDNA templates. Triplicates were performed for each gDNA template. The qPCR specificity was also tested with the CSF of patients with gnathostomiasis, cysticercosis, and angiostrongyliasis. PCR products were visualized on 2% agarose gel stained with SYBR™ Safe (Life Technologies, CA, USA) to determine the species-specific band sizes together with positive controls. The purified DNA samples were sequenced by Macrogen (Seoul, South Korea), an external biotechnology company, using the Sanger sequencing method with PCR primers. The obtained nucleotide sequences (query sequences) were confirmed as the target sequences by comparing them with annotated sequences in the NCBI databases using the standard nucleotide BLAST (Johnson et al., 2008). The specificity of qPCR reactions was considered by the melting curve analysis to determine the qPCR amplicons of A. malaysiensis- and A. cantonensis.
2.4.2. Sensitivity assay
To test the sensitivity of the SYBR Green qPCR method, we used gDNA from groups of 1, 5, 10, 50, 100, and 200 third-stage A. cantonensis larvae. Triplicates were performed for each group, and the quantitation cycle (Cq) values obtained for each group were then estimated with the standard curve to determine the amount of larval gDNA.
2.4.3. Reproducibility assay
To evaluate the reproducibility of the developed SYBR Green qPCRs, inter-and intra- reproducibility experiments were conducted using the same serially diluted gDNA concentrations used previously to construct the standard curve. The intra-reproducibility assay was performed by amplifying three replicates of each gDNA concentration. The inter-reproducibility assay was conducted through three separate sets of qPCR runs, with each set conducted on a different day. The mean, standard deviation (SD), and coefficient of variation (CV) were calculated separately using the Cq values for evaluation. The percentage of the CV of inter-and intra-reproducibility assays were then calculated to determine the precision within and between assays, respectively.
2.5. Implementation of SYBR Green qPCRs to estimate the ratio of A. cantonensis and A. malaysiensis third-stage larvae from wild A. fulica
Using the gDNA of the pooled larvae isolated from each A. fulica as a template, the qPCRs were conducted separately with each Angiostrongylus species-specific primers. From the Cq values of each species-specific qPCR assay, the approximate number for each species of larva were determined based on the qPCR standard curve of the sensitivity assay. The prevalence of A. cantonensis, A. malaysiensis, and the coinfection evidence of both Angiostrongylus species in A. fulica were then determined by estimating the ratio of A. cantonensis to A. malaysiensis infected in each A. fulica.
3. Results
3.1. Standard curve for SYBR-green quantitative real-time PCRs
The effectiveness of the SYBR Green qPCRs was determined using the Cytb species-specific primers for A. cantonensis and A. malaysiensis. The standard curves for A. cantonensis (Slope = −3.379, R2 = 0.999) and A. malaysiensis (Slope = −3.412, R2 = 0.999) are shown in Figs. 2A and B. The qPCR efficiency values for both species-specific primers of A. cantonensis and A. malaysiensis were 97.78% and 96.4%, respectively.
Fig. 2.
Amplification plots of the SYBR Green qPCR standard curves for A. cantonensis (A) and A. malaysiensis (B). The SYBR Green qPCRs were performed separately using the species-specific Cytb primers with the 10-fold serial dilutions of gDNA from 10−4 to 1 ng/μl as a template. The standard curve was plotted based on the Cq values of each gDNA concentration. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Sensitivity and specificity of SYBR Green quantitative real-time PCRs
The sensitivity assay results revealed that approximately 1 ng of gDNA was extracted from 100 larvae, while 10−4 ng of gDNA was extracted from 1 larva (Fig. 3). The number of the third-stage larvae was then estimated using the standard curve. From the results, we estimated that the gDNA of a single larva is approximately 10−4 ng (average Cq value of 31), while the gDNA of 100 pooled larvae is approximately 1 ng (average Cq value of 16). The Cq value of 35 was defined as the cut-off for no larva detected.
Fig. 3.
The Mean Cq values for the pooled larvae samples of A. cantonensis. Error bars represent standard deviation. The unfilled-green circles are the Cq values for each gDNA concentration. The numbers of A. cantonensis larvae used for estimating gDNA concentration based on the mtCytb target gene are (A) 200 larvae, (B) 100 larvae, (C) 50 larvae, (D) 10 larvae, (E) 5 larvae, and (F) 1 larva. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
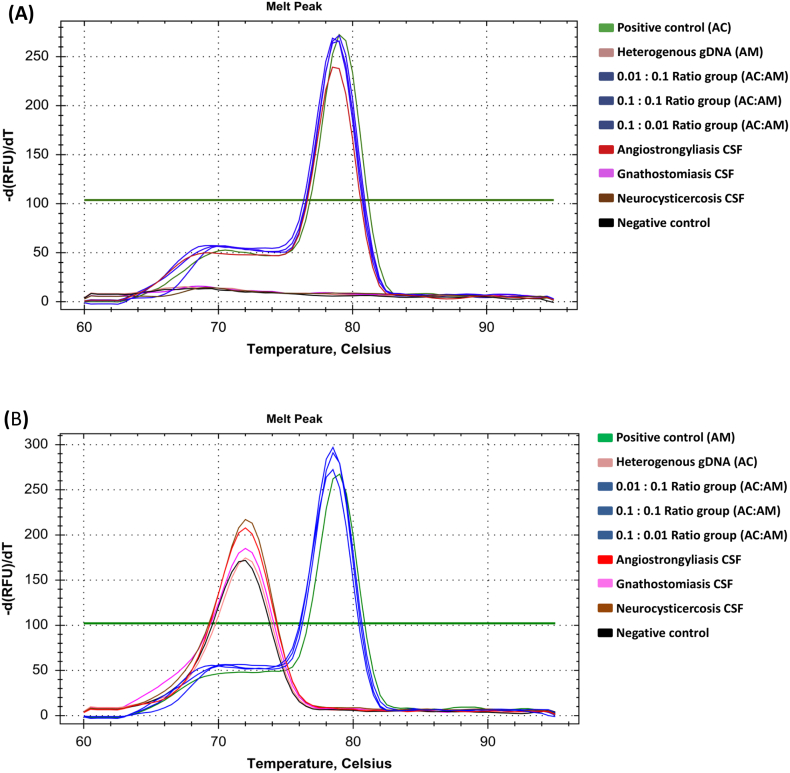
For the specificity assay, both species-specific primers did not amplify the gDNA from the different species (i.e., A. cantonensis primers only amplified A. cantonensis gDNA and not A. malaysiensis gDNA). As shown in Fig. 4A, the high specificity of AC4_cytb_F and AC5_cytb_R primers for A. cantonensis was also demonstrated in the artificially mixed A. cantonensis and A. malaysiensis gDNA, in equal concentration for each species, and when there was a lower concentration of A. cantonensis than A. malaysiensis gDNA. These primers also did not amplify the gnathostomiasis and neurocysticercosis samples, and the heterogeneous sample containing 1 ng of A. malaysiensis gDNA. Using the A. malaysiensis primers, the specificity was like the A. cantonensis primers, where only A. malaysiensis was amplified in the artificially mixed gDNA (Fig. 4B). The gnathostomiasis, neurocysticercosis, and angiostrongyliasis samples from the CSF of patients were not amplified. The primer-dimers present for the qPCR reactions of the A. malaysiensis-specific primers can be distinguished from the qPCR target by the melting curve analysis. The melting curve of the primer-dimers was around 72 °C and whereas the amplicons for A. malaysisensis is 78 °C.
Fig. 4.
The SYBR Green qPCR melting curve of A. cantonensis (AC) and A. malaysiensis (AM) at 79 °C and 78 °C, respectively, showing primer specificity for A. cantonensis (A) and A. malaysiensis (B) to the CSF of angiostrongyliasis, gnathostomiasis, and neurocysticercosis, the various artificially mixed gDNA containing AC and AM and heterogeneous and homogenous gDNA as the control for each qPCR assay. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Intra- and inter-reproducibility assays of SYBR Green qPCRs
The inter-reproducibility results showed consistent amplification between the replicated series of gDNA concentrations in the same assay. Moreover, the low percentage of the CV values of the inter-and intra-reproducibility assays indicated high precision, both within and between amplification (Table S2).
3.4. Detection of the third-stage Angiostrongylus larvae from naturally infected A. fulica
From the 285 A. fulica collected from the six public parks, 32 snails were positive with the third-stage larvae of Angiostrongylus (11.22%), as shown in Table S1. Of the 32 pools of larvae isolated from each positive A. fulica, 17 pools had more than 10 larvae, and the species-specific SYBR Green qPCRs were performed on these 17 pools. The qPCR results showed that all positive A. fulica was infected with A. malaysiensis (Table S3). Five out of 17 A. fulica (29.14%) revealed the coinfection of both A. malaysiensis and A. cantonensis. One A. fulica showed relatively high numbers of infections of A. cantonensis together with A. malaysiensis (Table S3).
4. Discussion
We developed species-specific SYBR Green qPCRs that are simple, economical, highly sensitive, and specific, with the ability to detect, discriminate, and estimate the ratio of the third-stage larvae of A. cantonensis and A. malaysiensis that were isolated from A. fulica. Recent studies reported overlap in the distributions of A. cantonensis and A. malaysiensis within the terrestrial snail host, particularly in Indochina (Rodpai et al., 2016). Although the gold standard for Angiostrongylus identification is based on morphological characteristics, the third-stage larvae of these two species are morphologically similar (Ash, 1970; Bhaibulaya and Cross, 1971; Bhaibulaya, 1979), resulting in misidentification. The SYBR Green qPCR method was then used to explore the distribution of Angiostrongylus in Bangkok Metropolitan to reflect the risk of human contact to angiostrongyliasis.
4.1. Development of SYBR Green qPCRs
Using the species-specific primers, the SYBR Green qPCRs developed could detect a small amount of DNA (<10−4 ng) for both Angiostrongylus species. We also established the standard curve and demonstrated high qPCR efficiency (see Figs. 2A and B). The CV percentage of reproducibility confirmed that the standard curve construction had low system variation in the manipulation assay. However, the Cq values were slightly fluctuated, especially the values estimated from the small larval pool. We postulated that DNA damage and loss during the larvae preservation and DNA extraction processes might have resulted in fluctuating Cq values, particularly for a small amount of DNA. In contrast, no fluctuation effect was observed when the larvae quantity exceeded 10 (Fig. 3).
We also confirmed the high specificity and the absence of cross-amplification of the developed method using heterogeneous gDNA in the species-specific SYBR Green qPCRs for both primer sets (Figs. 4A and B). Furthermore, the developed method showed the preliminary result to detect low amounts of Angiostrongylus genetic material in the CSF specimens. We demonstrated the potential of the method for diagnosing angiostrongyliasis by no cross-reaction with other diseases that also cause eosinophilic meningitis. Although the species-specific primers sometimes could amplify non-specific amplicons, the melting curve analysis can distinguish the target amplicon from the non-specific ones. As the diagnostic capability of the qPCRs was tested only will a small number of CSF specimens, further investigation is required with larger sample size.
Cost-effectiveness in terms of time, specificity, and the cost was also achieved through the developed species-specific SYBR Green qPCR method. Although two separate PCR mixtures had to be conducted for each species-specific primer, the reactions can be conducted simultaneously using the same qPCR thermocycling conditions, saving time for the user. This method does not require the design of a separate probe for saving the cost of the qPCRs (Maeda et al., 2003; Tajadini et al., 2014; Zhang et al., 2016).
The TaqMan probe real-time PCR was previously developed using the ribosomal internal transcribed spacer 1 (ITS1) as the genetic marker to detect third-stage A. cantonensis larvae in mollusks, and it was more sensitive as compared to the 18S rDNA-based conventional PCR (Qvarnstrom et al., 2007, Qvarnstrom et al., 2010). The TaqMan assay targeting ITS1 region was also applied for the diagnostics of patients with eosinophilic meningitis by detecting A. cantonensis DNA (Qvarnstrom et al., 2016). However, it was designed to detect only A. cantonensis detection (Qvarnstrom et al., 2010). Our newly developed SYBR Green qPCRs serve as an alternative to the TaqMan assays that have already been developed and can detect both A. cantonensis and A. malaysiensis. Although the newly developed SYBR Green qPCRs using the mitochondrial Cytb gene as a marker has not been proven suitable for molecular diagnosis yet, it may be an alternative method in the future. The new method also has the potential to confirm the occurrence of A. malaysiensis infection in patients living in areas where the distributions of A. cantonensis and A. malaysiensis overlap.
4.2. Distribution of Angiostrongylus in Bangkok metropolitan
We implemented the developed species-specific qPCRs for the molecular discrimination of A. cantonensis and A. malaysiensis third-stage larvae isolated from A. fulica collected from Bangkok's public parks. The qPCR results are indicative that A. malaysiensis is the predominant population, and no single infection of A. cantonensis was detected from the pooled larvae (Table S3). The coinfection of A. cantonensis and A. malaysiensis in A. fulica was relatively substantial (5.29%) in the representative population. However, the numbers of larvae determined by the qPCRs were smaller than the number counted under the stereomicroscope, possibly due to inactive and dead larvae and the DNA extraction process starting with a TissueLyser homogenizing step for breaking down the larval cuticle, resulting in discrepancies. Thus, we suggest using a ratio instead of larval numbers for coinfection evidence and parasite intensity.
Until now, A. cantonensis is the species of human pathogen distributing worldwide. However, in the Indochina Peninsula, co-existence between A. cantonensis and its closely related A. malaysiensis have been reported (Barratt et al., 2016; Rodpai et al., 2016; Dusitsittipon et al., 2017). A. malaysiensis is a potential human pathogenic species in Thailand and Southeast Asia, causing neurological disease in experimental monkeys (Cross, 1979). However, A. malaysiensis infection in humans has not been reported, even though the parasite (Liat, 1974) has been reported in snail-intermediate hosts in the Indochina region (Rodpai et al., 2016; Dusitsittipon et al., 2017). Recently, the coinfection evidence in the same snail intermediate hosts and overlapping distribution of A. cantonensis and A. malaysiensis lead to the question of the parasitic ability of A. malaysiensis to be a human pathogen. Comparative studies on the pathogenicity between A. cantonensis and A. malaysiensis in a mice-nonpermissive host of A. cantonensis (OuYang et al., 2012) are required to predict human pathogenicity differences between these closely related species. Our species-specific qPCRs developed may support further research on pathogenicity study by detecting and isolating the A. malaysiensis from A. cantonensis in the wild snail populations before in vivo experiments with the mice.
A fulica is one of the harmful-invasive species originating from East Africa and is highly predacious and can reproduce quickly. The presence of A. fulica in the ecosystem resulted in the extinction of native fauna and decreased biodiversity (Fisher et al., 2006). Modification of environments due to increasing human activity could have aided in the deployment and dissemination of A. fulica. Therefore, this snail is abundant in areas with high population density in urban areas (Ohlweiler et al., 2010; Albuquerque et al., 2008). Not only have A. cantonensis and A. malaysiensis been reported in A. fulica, but it is also a crucial vector known to harbor A. cantonensis and A. costaricensis (Carvalho et al., 2003; Caldeira et al., 2007). Notably, the distribution of A. fulica may be useful to predict the host-parasite association of parasite transmission (Pavanelli et al., 2017).
In this study, we revealed that A. malaysiensis, instead of A. cantonensis, is the predominant species in Bangkok Metropolitan. Although A. cantonensis was previously recognized as the main Angiostrongylus species distributed throughout Thailand (Dusitsittipon et al., 2015), the underestimation of A. malaysiensis in Thailand could have occurred because of morphological similarity. The coinfection of both A. cantonensis and A. malaysiensis presented in the A. fulica population in Bangkok. The coinfection evidence may support the investigation of the F1 hybrid in Bangkok city, which was revealed previously by the microsatellite genotyping (Dusitsittipon et al., 2017).
4.3. Limitations
We could not design probes from the available Cytb sequences in GenBank for both Angiostrongylus species due to high intraspecific variation for A. cantonensis and A-T rich region for A. malaysiensis. Also, we could not develop a duplex SYBR Green qPCR due to the closeness of the melting curve between A. cantonensis (79 °C) and A. malaysiensis (78 °C). However, we overcame this limitation by separating the PCR reaction between the two species. Moreover, the specificity of the two species-specific primer sets should be further evaluated with real specimens of other Angiostrongylus such as A. costaricensis, A. vasorum, and A. mackerrasae to confirm the predicted in silico results.
5. Conclusions
The developed species-specific SYBR Green qPCRs are a useful and alternative technique with high sensitivity and specificity. They can detect a single larva of Angiostrongylus and discriminate between A. cantonensis and A. malaysiensis simultaneously, rendering the method comparable to the probe-based approach. The developed method can also be used to estimate the ratio of larvae infecting the snail-intermediate host. A. malaysiensis was found to be the predominant species infecting A. fulica in the Bangkok Metropolitan. The coinfection evidence of both A. malaysiensis and A. cantonensis larvae in the same A. fulica host was detected through our method. Our newly developed qPCRs can be used to perform a molecular survey of A. cantonensis and A. malaysiensis in the natural intermediate host, particularly in Indochina Peninsula, to investigate parasite prevalence and to increase our understanding of host-parasite relationships. The newly developed method will be require further testing with various environmental samples to evaluate the specificity of the method to render it valuable for field applications. Moreover, the suitability of the method for clinical diagnosis using CSF from patients will also require further evaluation to assess the usefulness of the method for implementation.
Ethical approval statement
Ethical clearance was provided by the Animal Care and Use Committee, Faculty of Tropical Medicine, Mahidol University, Bangkok (No. FTM-ACUC 024/2018 and No. FTM-ACUC 025/2020).
Funding
The fieldwork for this research was supported by the Office of the Higher Education Commission and the Thailand Research Fund (MRG6180023), Thailand.
Availability of data and materials
All data generated during this study are included in the published article.
Declaration of Competing Interest
None.
Acknowledgments
We wish to acknowledge the Department of Helminthology, Faculty of Tropical Medicine, Mahidol University, for technical support and specimen collection. We thank Suzanne Leech Ph.D., from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fawpar.2021.e00119.
Contributor Information
Kittipong Chaisiri, Email: kittipong.cha@mahidol.ac.th.
Naowarat Saralamba, Email: naowarat.sar@mahidol.ac.th.
Yanin Limpanont, Email: yanin.lim@mahidol.ac.th.
Sirilak Dusitsittipon, Email: sirilak.dus@mahidol.ac.th.
Vachirapong Charoennitiwat, Email: vachirapong.cha@mahidol.edu.
Abigail Hui En Chan, Email: abigailhuien.cha@student.mahidol.ac.th.
Urusa Thaenkham, Email: urusa.tha@mahidol.edu.
Appendix A. Supplementary data
The following are the supplementary data related to this article.
The number of Angiostrongylus larvae infection in the collected Achatina fulica by digestion method
Intra- and inter-reproducibility assays of the qPCRs for the species-specific primers of A. cantonensis and A. malaysiensis
Number of Angiostrongylus-positive snails, number of larvae infecting in each snail based on morphological and molecular identifications and the number of larvae estimated based on the qPCR method
References
- Aghazadeh M., Harvie M.C., Owen H.C., Verissimo C., Aland K.V., Reid S.A., Traub R.J., McManus D.P., McCarthy J.S., Jones M.K. Comparative pathogenesis of eosinophilic meningitis caused by Angiostrongylus mackerrasae and Angiostrongylus cantonensis in murine and Guinea pig models of human infection. Parasitology. 2016;143:1243–1251. doi: 10.1017/S003118201600069X. [DOI] [PubMed] [Google Scholar]
- Albuquerque F.S., Peso-Aguiar M.C., Assunção-Albuquerque M.J. Distribution, feeding behavior and control strategies of the exotic land snail Achatina fulica (Gastropoda: Pulmonata) in the northeast of Brazil. Braz. J. Biol. Sci. 2008;68:837–842. doi: 10.1590/s1519-69842008000400020. [DOI] [PubMed] [Google Scholar]
- Alicata J.E. Advance Parasitology. Academic Press; London: 1965. Biology and distribution of the rat lungworm, Angiostrongylus cantonensis, and its relationship to eosinophilic meningoencephalitis and other neurological disorders of man and animals; pp. 223–248. [DOI] [PubMed] [Google Scholar]
- Ansdell V., Wattanagoon Y. Angiostrongylus cantonensis in travelers: clinical manifestations, diagnosis, and treatment. Curr. Opin. Infect. Dis. 2018;31:399–408. doi: 10.1097/QCO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- Ash L.R. Diagnostic morphology of the third-stage larvae of Angiostrongylus cantonensis, Angiostrongylus vasorum, Aelurostrongylus abstrusus, and Anafilaroides rostratus (Nematoda: Metastrongyloidea) J. Parasitol. 1970;56:249–253. [PubMed] [Google Scholar]
- Barbau-Piednoir E., Bertrand S., Mahillon J., Roosens N.H., Botteldoorn N. SYBR® Green qPCR Salmonella detection system allowing discrimination at the genus, species and subspecies levels. Appl. Microbiol. Biotechnol. 2013;97:9811–9824. doi: 10.1007/s00253-013-5234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J., Chan D., Sandaradura I., Malik R., Spielman D., Lee R., Marriot D., Harkness J., Ellis J., Stark D. Angiostrongylus cantonensis: a review of its distribution, molecular biology and clinical significance as a human pathogen. Parasitology. 2016;143:1087–1118. doi: 10.1017/S0031182016000652. [DOI] [PubMed] [Google Scholar]
- Bhaibulaya M. Studies on Angiostrongylus in Eastern Asia and Australia Taipei, Taiwan. Maryland: US Naval Medical Research Unit. Vol. 2. 1979. Morphology and taxonomy of major Angiostrongylus species of Eastern Asia and Australia; pp. 4–13. [Google Scholar]
- Bhaibulaya M., Cross J. Angiostrongylus malaysiensis (Nematoda: meta-strongylidae), a new species of rat lung-worm from Malaysia. Southeast Asian J. Trop. Med. Public Health. 1971;2:527–533. [PubMed] [Google Scholar]
- Caldeira R.L., Mendonça C.L., Goveia C.O., Lenzi H.L., Graeff-Teixeira C., Lima W.S., Mota E.M., Pecora I.L., Medeiros A.M.Z., Carvalho O.S. First record of molluscs naturally infected with Angiostrongylus cantonensis (Chen, 1935) (Nematoda: Metastrongylidae) in Brazil. Mem. Inst. Oswaldo Cruz. 2007;102:887–889. doi: 10.1590/S0074-02762007000700018. [DOI] [PubMed] [Google Scholar]
- Carvalho O.D., Teles H., Mota E.M., Mendonça C.L., Lenzi H.L. Potentiality of Achatina fulica Bowdich, 1822 (Mollusca: Gastropoda) as intermediate host of the Angiostrongylus costaricensis Morera & Céspedes 1971. Rev. Soc. Bras. Med. Trop. 2003;36:743–745. doi: 10.1590/s0037-86822003000600017. [DOI] [PubMed] [Google Scholar]
- Cross J.H. Experimental studies on Angiostrongylus species and strains in monkeys and laboratory animals. In: Cross J.H., editor. Studies on Angiostrongyliasis in Eastern Asia and Australia. US Naval Medical Research Unit; Taipei, Taiwan: 1979. pp. 118–137. (NAMRU-2 Special Publication No. 44). [Google Scholar]
- Dusitsittipon S., Thaenkham U., Watthanakulpanich D., Adisakwattana P., Komalamisra C. Genetic differences in the rat lungworm, Angiostrongylus cantonensis (Nematoda: Angiostrongylidae), in Thailand. J. Helminthol. 2015;89:545–551. doi: 10.1017/S0022149X14000388. [DOI] [PubMed] [Google Scholar]
- Dusitsittipon S., Criscione C.D., Morand S., Komalamisra C., Thaenkham U. Cryptic lineage diversity in the zoonotic pathogen Angiostrongylus cantonensis. Mol. Phylogenet. Evol. 2017;107:404–414. doi: 10.1016/j.ympev.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Dusitsittipon S., Criscione C.D., Morand S., Komalamisra C., Thaenkham U. Hurdles in the evolutionary epidemiology of Angiostrongylus cantonensis: pseudogenes, incongruence between taxonomy and DNA sequence variants, and cryptic lineages. Evol. Appl. 2018;11:1257–1269. doi: 10.1111/eva/12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamsobhana P. Angiostrongyliasis in Thailand: epidemiology and laboratory investigations. Hawaii J. M. Public Health. 2013;72(Suppl. 2):28. [PMC free article] [PubMed] [Google Scholar]
- Eamsobhana P., Wanachiwanawin D., Dechkum N., Parsartvit A., Yong H.S. Molecular diagnosis of eosinophilic meningitis due to Angiostrongylus cantonensis (Nematoda: Metastrongyloidea) by polymerase chain reaction-DNA sequencing of cerebrospinal fluids of patients. Mem. Inst. Oswaldo Cruz. 2013;108:116–118. doi: 10.1590/s0074-027620130001000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamsobhana P., Lim P., Yong H. Phylogenetics and systematics of Angiostrongylus lungworms and related taxa (Nematoda: Metastrongyloidea) inferred from the nuclear small subunit (SSU) ribosomal DNA sequences. J. Helminthol. 2015;89:317–325. doi: 10.1017/S0022149X14000108. [DOI] [PubMed] [Google Scholar]
- Fisher M.L., Simião M., Colley E., Zenni R.D., Silva D.A.T., Latoski N. O caramujo exótico invasorna vegetação nativa em Morretes, PR: diagnóstico da população de Achatina fulica Bowdich,1822 em um fragmento de floresta ombrófila densa aluvial. Biota Neotropica. 2006;6:1–5. [Google Scholar]
- Fontanilla I., Wade C. Research Note: first report of Angiostrongylus cantonensis in the giant African land snail Achatina fulica in French Polynesia detected using the SSU rRNA gene. Trop. Biomed. 2012;29:642–645. [PubMed] [Google Scholar]
- Hu Q.-A., Zhang Y., Guo Y.-H., Lv S., Xia S., Liu H.-X., Fang Y., Liu Q., Zhang Q.-M., Yang C.-L., Lin G.-Y. Small-scale spatial analysis of intermediate and definitive hosts of Angiostrongylus cantonensis. Infect. Dis. Poverty. 2018;7:100. doi: 10.1186/s40249-018-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvi S.I., Pitt W.C., Farias M.E., Shiels L., Severino M.G., Howe K.M., Jacquier S.H., Shiels A.B., Amano K.K., Luiz B.C., Maher D.E., Allison M.L., Holtquist Z.C., Scheibelhut N.T. Detection of Angiostrongylus cantonensis in the blood and peripheral tissues of wild Hawaiian rats (Rattus rattus) by a quantitative PCR (qPCR) assay. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(suppl_2):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D.I., Dixon M.C., Elm J.L., Jr., Calimlim P.S., Sciulli R.H., Park S.Y. Review of cases of angiostrongyliasis in Hawaii, 2007–2017. Am. J. Trop. Med. Hyg. 2019;101:608–616. doi: 10.4269/ajtmh.19-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender R., Lee D., Schulman A.H. FastPCR software in PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol. Biol. 2014;1116:271–302. doi: 10.1007/978-1-62703-764-8_18. [DOI] [PubMed] [Google Scholar]
- Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35(suppl_2):W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaningao P., Kanda S., Laimanivong S., Shimono T., Darcy A.W., Phyaluanglath A., Mishima N., Nishiyama T. Development of a PCR assay for diagnosing trematode (Opisthorchis and Haplorchis) infections in human stools. Am. J. Trop. Med. Hyg. 2017;96:221–228. doi: 10.4269/ajtmh.16-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liat L.B. New hosts of Angiostrongylus malaysiensis Bhaibulaya and Cross, 1971 in Malaysia. Southeast Asian J. Trop. Med. Public Health. 1974;5:379. [PubMed] [Google Scholar]
- Maeda H., Fujimoto C., Haruki Y., Maeda T., Kokeguchi S., Petelin M., Arai H., Tanimoto I., Nishimra F., Takashiba S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 2003;39:81–86. doi: 10.1016/S0928-9244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- Ohlweiler F.P., Guimarães M.C., Takahashi F.Y., Eduardo J.M. Current distribution of Achatina fulica, in the State of São Paulo including records of Aelurostrongylus abstrusus (Nematoda) larvae infestation. Rev. Inst. Med. Trop. Sao Paulo. 2010;52:211–214. doi: 10.1590/s0036-46652010000400009. [DOI] [PubMed] [Google Scholar]
- OuYang L., Wei J., Wu Z., Zeng X., Li Y., Jia Y., Ma Y., Zhan M., Lei W. Differences of larval development and pathological changes in permissive and nonpermissive rodent hosts for Angiostrongylus cantonensis infection. Parasitol. Res. 2012;111:1547–1557. doi: 10.1007/s00436-012-2995-6. [DOI] [PubMed] [Google Scholar]
- Pavanelli G.C., Yamaguchi M.U., Calaça E.A., Oda F.H. Scientometrics of zoonoses transmitted by the giant African snail Achatina fulica Bowdich, 1822. Rev. Soc. Bras. Med. Trop. 2017:59. doi: 10.1590/s1678-9946201759015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozio E. Foodborne nematodes. In: Gajadhar A.A., editor. Foodborne Parasites in the Food Supply Web. Woodhead Publishing; Cambridge: 2015. pp. 165–199. [Google Scholar]
- Qvarnstrom Y., Sullivan J.J., Bishop H.S., Hollingsworth R., da Silva A.J. PCR-based detection of Angiostrongylus cantonensis in tissue and mucus secretions from molluscan hosts. Appl. Environ. Microbiol. 2007;73:1415–1419. doi: 10.1128/AEM.01968-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y., da Silva A.C.A., Teem J.L., Hollingsworth R., Bishop H., Graeff-Teixeira C., da Silva A.J. Improved molecular detection of Angiostrongylus cantonensis in mollusks and other environmental samples with a species-specific internal transcribed spacer 1-based TaqMan assay. Appl. Environ. Microbiol. 2010;76:5287–5289. doi: 10.1128/AEM.00546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y., Xayavong M., da Silva A.C.A., Park S.Y., Whelen A.C., Calimlim P.S., Sciulli R.H., Honda A.S.S., Higa K., Kitsutani P., Chea N., Heng S., Johnson S., Graeff-Teixeira C., Fox L.M., da Silva A.J. Real-time polymerase chain reaction detection of Angiostrongylus cantonensis DNA in cerebrospinal fluid from patients with eosinophilic meningitis. Am. J. Trop. Med. Hyg. 2016;94:176–181. doi: 10.4269/ajtmh.15-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodpai R., Intapan P.M., Thanchomnang T., Sanpool O., Sadaow L., Laymanivong S., Aung W.P., Phosuk I., Laummaunwai P., Maleewong W. Angiostrongylus cantonensis and A. malaysiensis broadly overlap in Thailand, Lao PDR, Cambodia and Myanmar: a molecular survey of larvae in land snails. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawabe K., Makiya K. Genetic variability in isozymes of Angiostrongylus malaysiensis. Southeast Asian J. Trop. Med. Public Health. 1994;25:728–736. [PubMed] [Google Scholar]
- Spratt D.M. Species of Angiostrongylus (Nematoda: Metastrongyloidea) in wildlife: a review. Int. J. Parasitol. Parasites Wildl. 2015;4:178–189. doi: 10.1016/j.ijppaw.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadini M., Panjehpour M., Javanmard S.H. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014;3:85. doi: 10.4103/2277-9175.127998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiengo S., Maldonado A., Mota E.M., Torres E.J.L., Caldeira R., Carvalho O.S., Oliveira A.P.M., Simoes R.O., Fernandez M.A., Lanfredi R.M. The giant African snail Achatina fulica as natural intermediate host of Angiostrongylus cantonensis in Pernambuco, Northeast Brazil. Acta Trop. 2010;115:194–199. doi: 10.1016/j.actatropica.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(suppl_2):W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitta A., Nateeworanart S., Tattiyapong M. Survey of Angiostrongylus cantonensis in rats and giant African land snails in Phitsanulok province, Thailand. Asian Pac J Trop Med. 2011;4:597–599. doi: 10.1016/S1995-7645(11)60154-5. [DOI] [PubMed] [Google Scholar]
- Vitta A., Polsut W., Fukruksa C., Yimthin T., Thanwisai A., Dekumyoy P. Levels of infection with the lungworm Angiostrongylus cantonensis in terrestrial snails from Thailand, with Cryptozona siamensis as a new intermediate host. J. Helminthol. 2016;90:737–741. doi: 10.1017/S0022149X15001042. [DOI] [PubMed] [Google Scholar]
- Wang Q.-P., Wu Z.-D., Wei J., Owen R., Lun Z.-R. Human Angiostrongylus cantonensis: an update. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:389–395. doi: 10.1007/s10096-011-1328-5. [DOI] [PubMed] [Google Scholar]
- Weirather J.L., Jeronimo S.M., Gautam S., Sundar S., Kang M., Kurtz M.A., Haque R., Schriefer A., Talhari S., Carvalho E.M., Donelson J.E., Wilson M.E. Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J. Clin. Microbiol. 2011;49:3892–3904. doi: 10.1128/JCM.r00764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.-B., Wu Z.-D., Lun Z.-R. The apple snail Pomacea canaliculata, a novel vector of the rat lungworm, Angiostrongylus cantonensis: its introduction, spread, and control in China. Hawaii J. Med. Public Health. 2013;72(Suppl. 2):23. [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Sambono J.L., Morgan J.A., Venus B., Rolls P., Lew-Tabor A.E. An evaluation of quantitative PCR assays (TaqMan® and SYBR Green) for the detection of Babesia bigemina and Babesia bovis, and a novel fluorescent-ITS1-PCR capillary electrophoresis method for genotyping B. bovis isolates. Vet. Sci. 2016;3:23. doi: 10.3390/vetsci3030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of Angiostrongylus larvae infection in the collected Achatina fulica by digestion method
Intra- and inter-reproducibility assays of the qPCRs for the species-specific primers of A. cantonensis and A. malaysiensis
Number of Angiostrongylus-positive snails, number of larvae infecting in each snail based on morphological and molecular identifications and the number of larvae estimated based on the qPCR method
Data Availability Statement
All data generated during this study are included in the published article.