Abstract
Intracellular organelles do not, as thought for a long time, act in isolation but are dynamically tethered together by entire machines responsible for interorganelle trafficking and positioning. Among the proteins responsible for tethering is the family of VAMP-associated proteins (VAPs) that appear in all eukaryotes and are localized primarily in the endoplasmic reticulum. The major functional role of VAPs is to tether the endoplasmic reticulum with different organelles and regulate lipid metabolism and transport. VAPs have gained increasing attention because of their role in human pathology where they contribute to infections by viruses and bacteria and participate in neurodegeneration. In this review, we discuss the structure, evolution, and functions of VAPs, focusing more specifically on VAP-B for its relationship with amyotrophic lateral sclerosis and other neurodegenerative diseases.
Keywords: ALS, endoplasmic reticulum, tethering proteins, VAMP
Abbreviations: AD, Alzheimer's disease; ALS, amyotrophic lateral sclerosis; ER, endoplasmic reticulum; FUS, fused in sarcoma/translocated in liposarcoma protein; HCV, hepatitis C virus; Ig, immunoglobulin; MSP, major sperm protein; nsps, nonstructural proteins; PD, Parkinson's disease; PDB, Protein Data Bank; TDP-43, transactive response DNA binding protein 43 kDa; VAP, VAMP-associated protein
The eukaryotic cell has been thought of for a long time as a sort of container of several other smaller containers, the organelles, separated from each other by organellar membranes and without direct physical links. This view has been completely revolutionized by electron microscopy evidence that has first raised and then proven that organelles might be talking to each other through connectors mostly constituted by proteins or protein complexes that form fixed or transient molecular tethers (for comprehensive reviews, refer for instance the study by Eisenberg-Bord et al., 2016 (1), and Henne et al., 2016 (2), and references within).
The discovery of tethers has offered a new perspective on the functioning of a cell and proposed a new route for regulation and trafficking between organelles. The list of proteins reported to be involved in tethering has increasingly been expanding. To date, proteins that act as tethers between the endoplasmic reticulum (ER) and the plasma membrane, mitochondria, and endosomes/lysosomes have been identified in several different organisms (1). It is thus well accepted that many, if not all organelles, are physically tethered, providing a description of the cell interior in which organelles are spatially organized and functionally interconnected in a static and/or dynamic fashion.
Tethering can involve proteins that physically bridge two organelles attaching them to each other, correctly positioning organelles for division or inheritance (1, 3). Other tethers also have specific functions, such as playing a role in the transfer of small molecules (e.g., lipids, calcium) in a nonvesicular manner. Recent studies have started to uncover the molecular mechanisms of tethering to understand their spatial, temporal, and functional organization. We are beginning to understand that each contact site is a functional microenvironment based on the concerted activity of distinct types of tether and nontether components.
In this review, we want to zoom in on a specific family of tethering proteins, the VAMP-associated proteins (VAPs), where VAMP stands for vesicle-associated membrane protein. Discovered in the last decade of last century, only recently has enough interest been paid to these proteins both because they constitute one of the best described tethering systems and because of their association with the disease. We focus on these proteins because of their important role in neurodegeneration. They constitute a family of integral membrane proteins that tether the ER to plasma membrane, mitochondria, endosomes, and the Golgi apparatus. We will discuss here the architecture, evolution, interactions, and the multiple functional roles of VAPs. We relate these features to the importance of VAPs in disease, both in viral and bacterial infections, and in neurodegeneration including amyotrophic lateral sclerosis (ALS), frontotemporal dementia, Alzheimer's disease (AD), and Parkinson's disease (PD). We have chosen not to include links of VAPs to cancer not for the lack of interest but to focus the review on two aspects, infectious and neurodegenerative diseases, which have only relatively recently been linked together (4, 5, 6, 7, 8). The role of VAPs in cancer is in its infancy and possibly still indirect (9).
The VAP family
The first VAP was identified in Aplysia californica by a yeast two-hybrid screen using as a bait the v-SNARE VAMP (also known as synaptobrevin), a protein required for synaptic-vesicle fusion. From this work, the protein was named “VAP” for its association with vesicles or “VAP33”, as the Aplysia protein migrated as a 33-kDa protein on SDS-PAGE (10, 11, 12, 13, 14). VAPs are found in all eukaryotic organisms where they play important functions in membrane trafficking (10), neurotransmitter release, and lipid transport (15). They also participate in the unfolded protein response machine, an ER reaction that suppresses accumulation of misfolded proteins to maintain cell viability and function (16).
VAPs are ubiquitously expressed, but their expression levels depend on cell and tissue types (17, 18, 19). In addition to their main localization in the ER (19, 20, 21, 22), they have also been found in other intracellular membranes, including the Golgi, the ER–Golgi intermediate compartment (11, 12), recycling endosomes (14, 23), tight and neuromuscular junctions (16), and plasma membrane (15).
Amino acid sequences of VAPs are highly conserved from yeast to mammals with the human protein sharing 99% identity with mouse and 35% with yeast (17, 18, 19, 24, 25). Vertebrates have two closely related VAP genes, VAP-A and VAP-B. VAP-A and VAP-B share high (typically, ca., 63%) amino acid sequence identity. An alternative spliced isoform of VAP-B comprised of the N-terminal 70 with additional 29 amino acids was also identified and named VAP-C (18).
The three-dimensional structure of VAPs
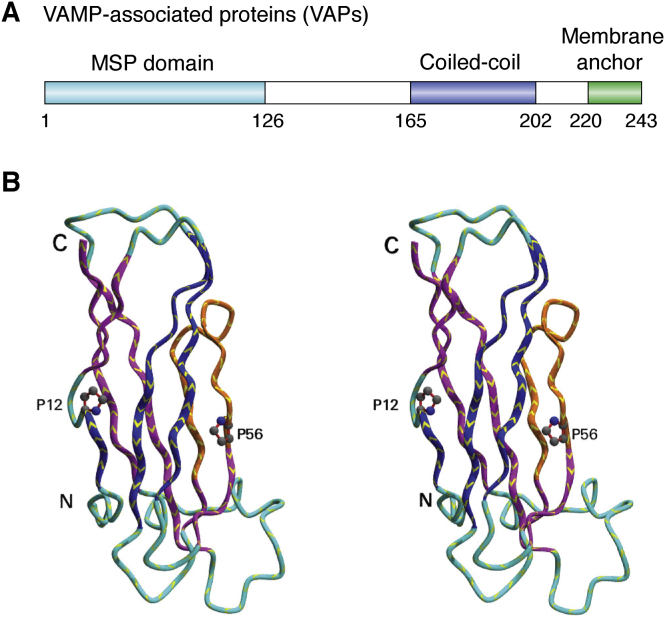
The architecture of VAPs is modular and composed of three conserved domains (Fig. 1A): an N-terminal major sperm protein (MSP) domain (residues 1–126 of human VAP-B), followed by a central amphipathic domain that is predicted to comprise a coiled-coil domain (residues 165–202), and a C-terminal hydrophobic transmembrane domain (residues 220–243). The MSP is a nematode protein homologous to VAPs that shares with these the same domain architecture and evinces a high degree of amino acid sequence similarity in the N-terminal domain (48% identity). The MSP domain has an immunoglobulin (Ig) fold (26), a structure found in very diverse protein families throughout the living systems. Ig-like domains are involved in a variety of functions, including cell–cell recognition, cell–surface receptors, muscle structure, and, of course, the immune system. As in all Ig folds, the domain consists of a β-sandwich assembled by two β-sheets packed face-to-face and enclosing the hydrophobic core of the protein (Fig. 1B, Supplemental Data S1). The nearly ellipsoidal shape of the domain measures approximately 15 Å × 20 Å × 45 Å. A stretch of 16 amino acids in the MSP domain, the so-called VAP consensus sequence, is particularly conserved (21, 27). The structures of the MSP domains of VAP-A and VAP-B proteins are available in the Protein Data Bank (PDB) (Table 1).
Figure 1.
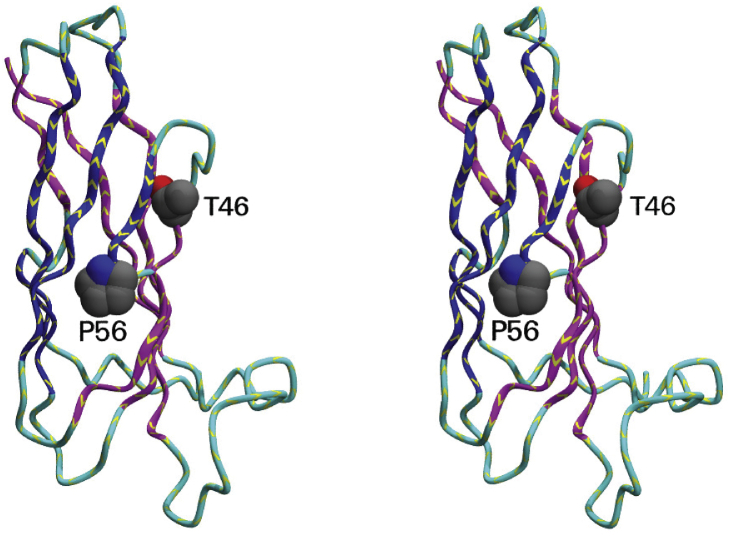
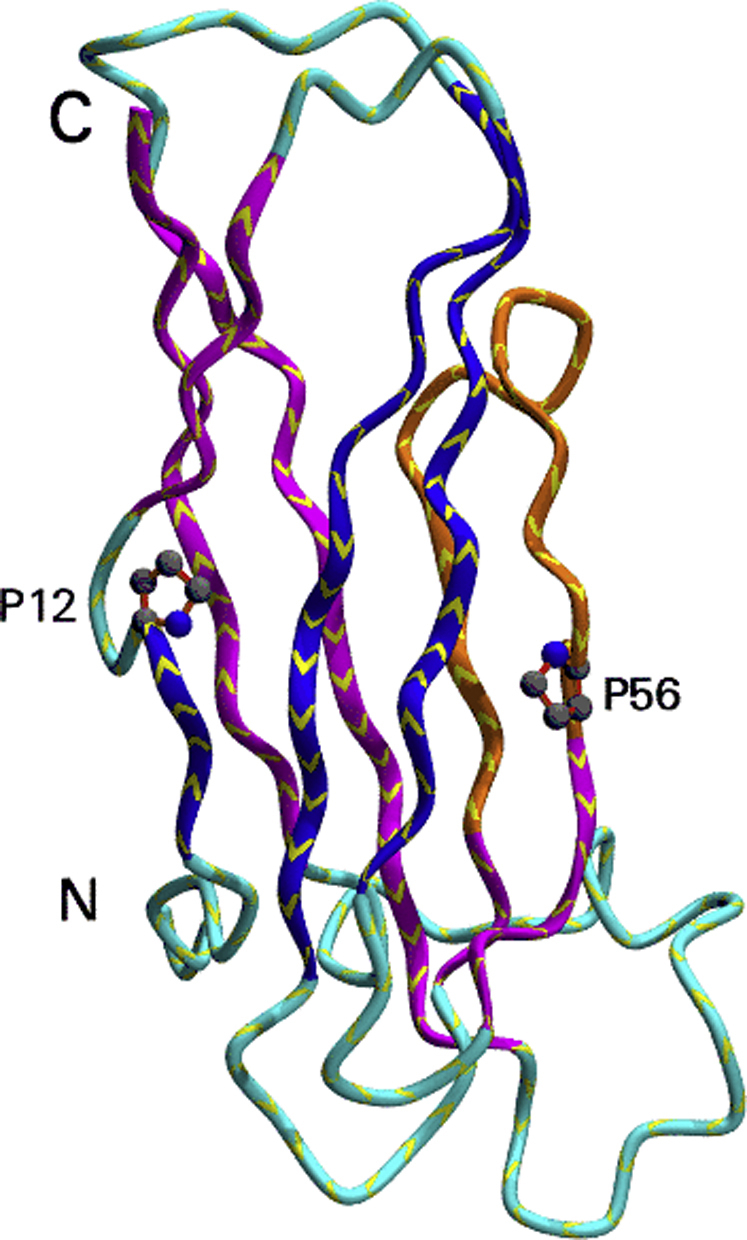
The structure of VAPs.A, schematic representation of the domain architecture along the sequence. B, three-dimensional structure of VAP-B (PDB code: 3ikk) as a stereo view. The two prolines indicated (P12 and P56) are the points at which the strands switch from one sheet to the other as described in section “The peculiarities of the Ig-fold of the MSP family.” The region shown in orange (residues 42–57) shows the VAP consensus sequence. PDB, Protein Data Bank.
Table 1.
Summary of the available structures of VAPs
| PDB code | Protein description | Method | Resolution |
|---|---|---|---|
| 1z9o | Rat VAP-A MSP homology domain in complex with the rat ORP1 FFAT motif | X-ray | 1.9 Å |
| 1z9l | Rat VAP-A MSP homology domain | X-ray | 1.7 Å |
| 2cri | Mouse MSP domain of VAP-A | NMR | Not applicable |
| 2rr3 | Complex of human VAP-A MSP domain and the OSBP FFAT motif | NMR | Not applicable |
| 2mdk | MSP-P56S domain VAP-B in dodecylphosphocholine | NMR | Not applicable |
| 3ikk | Human MSP domain | X-ray | 2.5 Å |
MSP, major sperm protein; PDB, Protein Data Bank.
The recombinant isolated MSP domain is a monomer in the solution, but it may contribute to dimer formation of the full-length protein together with other regions of the protein (28, 29). The coiled-coil domain that follows the MSP is a motif often found in VAMP and SNARE proteins and in several other proteins implicated in vesicular transport (18). These proteins share clear sequence homology that goes beyond the presence of leucine-zipper heptads (30). The evolutionary conservation of the SNARE coiled-coil homology domain has thus suggested that the domain fulfills a similar function in different membrane fusion proteins. The C-terminal hydrophobic transmembrane domain contains a putative dimerization motif GxxxG that could favor both homodimerization and heterodimerization (18, 31).
The peculiarities of the Ig fold of the MSP family
It is worth spending a few words about the MSP domain because it has some interesting peculiarities. The Ig domain of VAPs is at the sequence level homologous to the nematode MSP, and similar, at a structural but not at the sequence level, to the bacterial pilus PapD chaperone. In nematodes, the MSP is a small (∼14 kDa) basic protein encoded by a multigene family of up to 28 members and prone to dimerization (32). Locomotion in amoeboid nematode sperm cells (which instead of swimming like typical sperm crawl like amoebae) is produced by bundling of filaments formed by MSP, constructed from two helical subfilaments that coil around each other and are arranged into long, branching, fiber complexes. PapD is a periplasmic chaperone that assists the assembly of adhesive pili in gram-negative bacteria. Its function is to bind, stabilize, and cap the different filamental subunits until they are assembled into the pilus. The chaperone provides the seventh missing strand necessary to complete the Ig fold of the pilus subunits.
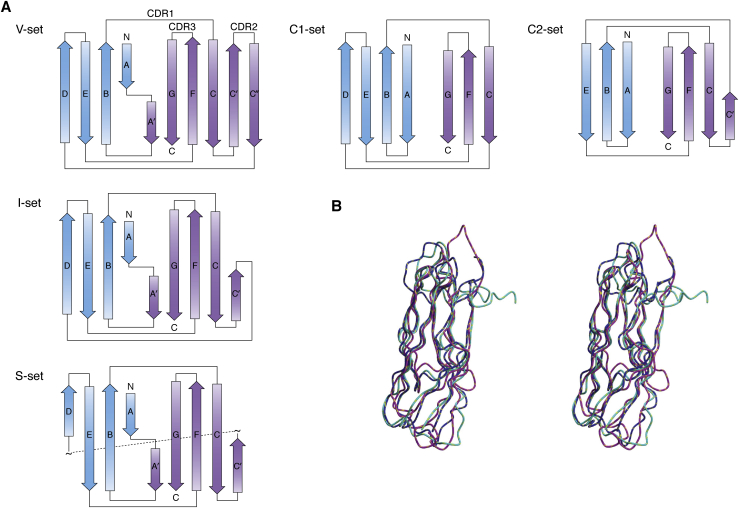
Despite their completely different origin and functions, the Ig folds of MSP, VAP, and PapD are remarkably similar and can be classified as S-type (for switching type) as compared with the C-, V-, and I-types (Fig. 2, Supplemental Data S2). The S-subfamily has several features that make it distinct from other Ig folds despite a general closeness to the I-type subfamily (33, 34). The β-sheets in S-type Ig domains have two switching points. Strand 1 begins in one of the sheets but transfers to the other, through an S-shaped bulge centered, in MSP and VAP on a proline (P12 in VAP-B numbering) in an unusual cis-conformation. In PapD, the switching is shifted and different. Switching within the first strand occurs also in the I and V subfamily but does not involve a cis-proline. A second switching between strands occurs at the C’ strand that forms another S-shaped bulge centered on P56 (VAP-B numbering). Another noticeable difference between MSP and more classical Ig domains is the absence of the buried tryptophan strongly conserved in the V-, C-, and I-Ig subfamilies. At this position, VAP-B has V44 that packs tightly against the rest of the hydrophobic core (Fig. S1).
Figure 2.
Similarities and differences within the Ig superfamily.A, schematic representation of the Ig C-, V-, I- and S-fold types. The two distinct sheets of the β-sandwich are indicated in blue and purple. B, comparison in stereo of the three-dimensional structures of VAP-B (PDB code: 3ikk), MSP (PDB code: 1grw), and PapD (PDB code: 3dpa). Ig, immunoglobulin; PDB, Protein Data Bank.
Interesting questions raised by comparison of VAP-B MSP domain, MSP, and PapD are whether the S-type fold arose from convergent or divergent evolution from the rest of the Ig superfamily and whether its specific features are dictated by function. Although involved in different contexts, all three proteins have functions that require specific mechanical properties that might be achieved by this specific variant of the Ig fold. It may thus be interesting, in the future, to probe the resistance against unfolding and resilience of the S-type fold and compare its properties with those of other members of the Ig family.
Molecular evolution of VAPs
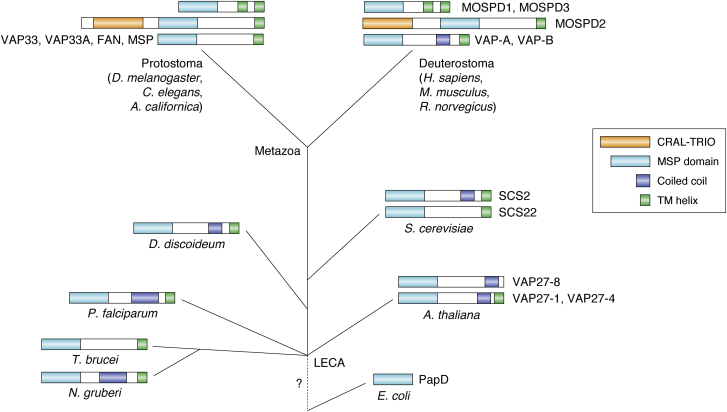
VAP homologs that contain the MSP domain (PFAM number PF00635) and the C-terminal transmembrane helix appear to be widespread among eukaryotes, with experimentally characterized representatives in A. californica (10), Drosophila melanogaster (35), Saccharomyces cerevisiae (27), Caenorhabditis elegans (32), Arabidopsis thaliana (36), and with predicted proteins in species such as Plasmodium falciparum (PF3D7_1439800), Trypanosoma brucei (TbgDal_XI14770), Naegleria gruberi (NAEGRDRAFT_80113), and Phytophthora infestans (PITG_02627) (Fig. 3). These examples cover the major branches of the eukaryotic tree (16). Most, but not all, of the homologs also contain a coiled-coil region implicated in dimerization. Homology between VAPs and MSPs appears clear from sequence comparison (37) and includes, for example, the 16-aa VAP consensus sequence (27). In contrast, a possible homology with PapD is not at all clear, as the E-values are insignificant even using sensitive homology detection (38). PapD proteins do not contain the conserved sequence motif or the C-terminal transmembrane helix. The origin of the VAP family therefore appears to be at the root of the eukaryotes, consistent with the origin of the endomembrane system in which they play a major role.
Figure 3.
Schematic overview of the evolution of proteins containing the VAP domain based on sequence analysis, with emphasis on the three domain architectures observed in the metazoa. Homology with the prokaryotic PapD cannot be established and is indicated with a question mark. The VAP-A architecture with the MSP domain, a coiled-coil, and a transmembrane helix can be found throughout eukaryotes and therefore probably dates back to the last eukaryotic common ancestor (LECA). Many variations on this architecture can be found, including the loss of the coiled-coil region or the C-terminal transmembrane helix. There are also several gene duplications in the family in the various taxa, such as in plants. Species for which structures have been determined are named in figure. MSP, major sperm protein.
In humans, VAP-B has, besides VAP-A, three other homologs, MOSPD1, MOSPD2, and MOSPD3. These proteins have been identified independently from VAP-A/B but belong to the same MSP domain containing family. MOSPD1 and MOSPD3 play a pivotal role in developmental regulation (39), and MOSPD2 has been linked to membrane tethering (37). As compared with VAP-A and VAP-B, MOSPD2 has an extra CRAL–TRIO domain (40), N-terminal to the MSP domain, whereas MOSPD1 and MOSPD3 have an extra C-terminal transmembrane helix. The CRAL–TRIO domain is a structural module with an α/β fold that binds small lipophilic molecules. These are hosted in a large hydrophobic cavity formed between a six-strand β-sheet and a cluster of α-helices. Examination of the SMART database showed that the combination of the MSP domain with the CRAL–TRIO domain as present in one nematode MSP and in MOSPD2 is only observed in metazoa and therefore appears to have evolved later than the simpler architecture present in VAP-A and VAP-B.
VAPs as molecular tethers and lipid metabolism proteins
Having discussed the structure and evolution of VAPs, it is now to consider their functions. A major role of VAPs is that of tethering the ER with other organelles and/or to regulate lipid transport and metabolism. The VAP interactome is thus rich because it reflects the different localizations and the associated specific functions of these proteins. Overall, the endogenous VAP-interacting proteins can be divided into two broad groups: SNARE and FFAT motif–containing proteins (Table 2). VAP-A, for instance, binds promiscuously several different SNAREs. This suggests that, rather than being associated with the regulation of a specific protein, VAP-A could have a role in overall SNARE-mediated fusion events (17).
Table 2.
Summary of some of the best described interactions involving VAP-B
| Region involved | Protein partner | Disease associated | References |
|---|---|---|---|
| N.D.a | CERT | Chlamydia infections | (63, 64) |
| MSP domain | NS5A | Hepatitis C | (56) |
| MSP domain | NS5B | Hepatitis C | (57) |
| N.D. | NS3/4A (NS4A) | Hepatitis C | (59) |
| MSP domain | Eph receptors (EphA2, EphA4, EphA5, EphA6, EphA7, EphA8) | Hepatitis C | (51, 56, 57) |
| MSP domain | ATF6 | Hepatitis C, ALS8 | (76) |
| MSP domain | NS1/2 | Diarrheab | (55) |
| N.D. | OSBP1 | Common coldc | (60) |
| N.D. | PTPIP51 | AD, PD, FTD, ALSd | (46) |
| MSP domain | α-Synuclein | PDd | (47, 71) |
| MSP domain | p97–FAF1 complex | ALS8 | (88) |
| MSP domain | ASNA1–TRC complex | ALS8 | (88) |
AD, Alzheimer's disease; ALS, amyotrophic lateral sclerosis; FTD, frontotemporal dementia; MSP, major sperm protein; PD, Parkinson's disease.
Not determined.
Caused by norovirus infection.
Caused by rhinovirus infection.
These pathologies are associated with tethering disruption.
FFAT motif–containing proteins are characterized by a short linear motif with the consensus sequence EFFDAxE (i.e., two phenylalanines in an acidic tract) (41). These proteins are implicated in lipid transfer between the ER and other organelles, such as the Golgi apparatus, endosomes, and plasma membrane (42, 43, 44). Interactions with FFAT-containing proteins map to the MSP domain. Accordingly, mutations in the MSP domain that disrupt FFAT motif binding inhibit VAP-dependent ER–plasma membrane association (45).
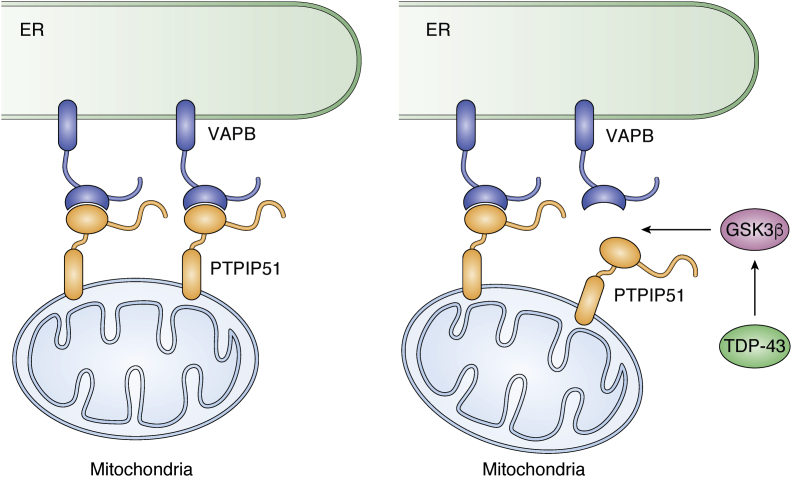
Another FFAT motif–containing protein that interacts with VAP-B is the tyrosine phosphatase-interacting protein 51 (PTPIP51). Several neuronal functions are regulated by signaling between mitochondria and the ER. This signaling involves direct interaction between the two organelles that is mediated by binding of VAP-B to the outer mitochondrial membrane protein PTPIP51, as confirmed by independent biochemical assays (46). VAP-B and PTPIP51 act as interorganellar bridges between ER and the mitochondria (46, 47, 48). Both proteins are involved in the regulation of intracellular Ca2+ homeostasis (48). Accordingly, modulation of VAP-B and PTPIP51 expression results in variations in ER–mitochondrion contacts and in Ca2+ exchange between the two organelles. VAP-B–PTPIP51 tethers also regulate autophagy through mediating Ca2+ delivery from the ER to mitochondria (49).
Interestingly, proteins of the MOSPD family (MOSPD1, 2, and 3), which share with VAP-A/B a similar architecture, are also intraorganelle tethers but with high partner specificity. MOSPD1 and MOSPD3 prefer unconventional FFAT-related FFNT (two phenylalanines [FF] in a neutral tract) (50) motifs. VAPA/VAPB/MOSPD2 and MOSPD1/MOSPD3 assemble into two different ER-attached complexes in control of interactions between intracellular compartments.
Although quite different among each other, these interactions confirm an important role of VAPs in organelle tethering and lipid metabolism. In addition to these interactions, the MSP domain of VAP-B was found as a cleavage product to serve as a ligand of Eph receptors (51). Secreted MSP domains usually bind to the extracellular signal domains of these receptor tyrosine kinases and sit on the cell surface.
VAPs in infectious diseases
Interactions of VAPs with various viral nonstructural proteins (nsps) have been conclusively demonstrated (52, 53, 54). Both VAP-A and VAP-B enhance the replication of various RNA viruses, including murine norovirus, hepatitis C virus (HCV), rhinovirus, and tombusvirus (Table 2). In many cases, the viral proteins mimic cellular proteins that normally interact with VAPs. VAP-A, for instance, binds through conserved residues in the MSP domain, a conserved motif in the NS1 domain of the alternative nsp1/2 protein from murine norovirus, a leading cause of gastroenteritis (55). The residues of nsp1/2 interacting with VAP-A include the acidic residues E40, E42, D43, E44, E52, and E54 and the bulky aromatic Y47. This stretch faithfully mimics the classical FFAT motif in which aromatic residues are flanked by acidic ones.
Both VAPs bind the nsps NS5A and NS5B of HCV playing an essential role as host factors for viral replication. NS5A/B are RNA-dependent RNA polymerase phosphoenzymes that appear to possess multiple and diverse functions in viral replication, interferon resistance, and pathogenesis. They have an N-terminal catalytic domain connected to a membrane-binding region by an autoregulatory C-linker. It is this linker that binds the VAP MSP domain. Although the details of how the interaction works are still unknown, it was shown that overexpression of VAP-B enhances viral RNA replication (56, 57). Conversely, disruption through mutation or downregulation of VAP-A and VAP-B, but not VAP-C, inhibits viral replication (58).
By a combined proteomics/genomics analysis, it was also found that the VAPs have strong interactions with the viral nsp3/4A protease through nsp4A, which may interfere with the unfolded protein response machine, preventing host cells from detecting and degrading viral proteins at the ER (59). Other interactions with proteins involved in viral replication comprise proteins implicated in the metabolism of sterol because this is a major component of the cellular membranes where viruses assemble viral replicase complexes. Examples of these interacting proteins are oxysterol-binding protein 1 that is a component of the cycle fueled by cholesterol-driving human rhinovirus replication (60, 61) and various oxysterol-binding protein–related proteins involved in the replication of the RNA tombusviruses in plants (62).
VAPs play a role also in bacterial infection as, for instance, Chlamydia trachomatis, an obligate intracellular human pathogen responsible for ocular and genital infections. The mechanism involves lipid metabolism and interaction with a vesicular compartment in the host cell membrane known as an inclusion, required for the reproductive cycle of the bacterium. Inclusion of C. trachomatis relies on a set of effector proteins that are injected into the host cells or inserted into the inclusion membrane. Interactions of the bacterial proteins with host VAP-B enhance lipid import into the bacterial inclusion. It was proposed that insertion of the C. trachomatis effector protein IncD in the inclusion membrane contributes to the recruitment of VAP-B, the lipid transfer ceramide transfer protein CERT, and the sphingomyelin synthases, SMS1 and SMS2 (63, 64, 65). Together, the complex of these proteins establishes an on-site sphingomyelin biosynthetic factory critical for replication in which CERT converts ceramide into sphingomyelin (65).
This evidence confirms an active involvement of VAPs in bacterial and viral infectivity.
VAPs in neurodegenerative diseases
VAP-B has been connected to various neurodegenerative diseases, including ALS, frontotemporal dementia, AD, and PD through disruption of mechanisms regulating the ER–mitochondria tethering (46, 66, 67). In particular, association with ALS was observed and closely studied in animal models (68). The RNA-binding proteins fused in sarcoma/translocated in liposarcoma protein (FUS) (69) and transactive response DNA binding protein 43 kDa (TDP-43) (67, 70), and both WT and mutant α-synuclein (47, 71) are for instance known to disrupt the VAP-B-PTPIP51 interaction with consequent loosening of the ER–mitochondria interaction, which induces autophagosome formation. As a consequence, Ca2+ uptake by mitochondria is perturbed and mitochondrial ATP production is impaired. Results of coimmunoprecipitation and pull-down assays argue against a direct interaction between VAP-B and TDP-43 or FUS. The link seems mediated by GSK3β: several groups have reported that, in disease states, both TDP-43 and FUS may activate GSK3β to inhibit binding of VAP-B to PTPIP51 and reduce the ER–mitochondria connections (69, 70) (Fig. 4). GSK3β is strongly implicated also in AD and PD as a regulator of the VAP-B–PTPIP51 interaction (70). Regulation might require phosphorylation of PTPIP51 and/or VAP-B, resulting in inhibition of the GSK3β interaction (70, 71). α-Synuclein binds instead to the VAP-B MSP domain directly and disrupts the VAP-B–PTPIP51 interaction (71). Accordingly, overexpression of WT α-synuclein and its PD-related mutant disrupts the VAP-B–PTPIP51 tether.
Figure 4.
VAP-B tether between the ER and mitochondria and its link to neurodegeneration. VAP-B and PTPIP51 are normally linked together with interactions between VAP-B and a complementary region of PTPIP51. TDP-43 and fused in sarcoma/translocated in liposarcoma protein may activate GSK3β that, in turn, inhibits binding of VAP-B to PTPIP51 and reduce the ER–mitochondria connections. This is thought to cause dysregulation and disease. ER, endoplasmic reticulum.
Overall, these studies strongly support a direct link between the VAPs, the VAP-B–PTPIP51 tether, and neurodegeneration.
The effect of ALS-related clinical mutations on the VAP fold
Several mutations of VAP-B have been observed in patients with ALS: P56S, T46I, del160, D130E, A145V, S160Δ, and V234I (72, 73, 74, 75). Among them, the first two are in the MSP domain, two involve truncations or deletions of the protein, affecting function, two are in the coiled-coil region, and one more in the membrane-binding domain. Only the effects of P26S and T46I mutations have been studied in sufficient detail.
The effects of P56S and T46I on the VAP-B structure were more closely monitored (Fig. 5, Supplemental Data S3). The P56S variant is by far the most prevalent form that cosegregates with disease in patients with ALS (76). Overexpression of the P56S mutant of VAP-B leads to misregulation of ER stress regulatory pathways, inducing aggregation and mislocalization of the protein in non-ER regions (51, 77). It was demonstrated that mislocalization of the protein resulted in potentiation of TDP-43–induced motor neuronal cell death, at variance with WT VAP-B that inhibited it (78).
Figure 5.
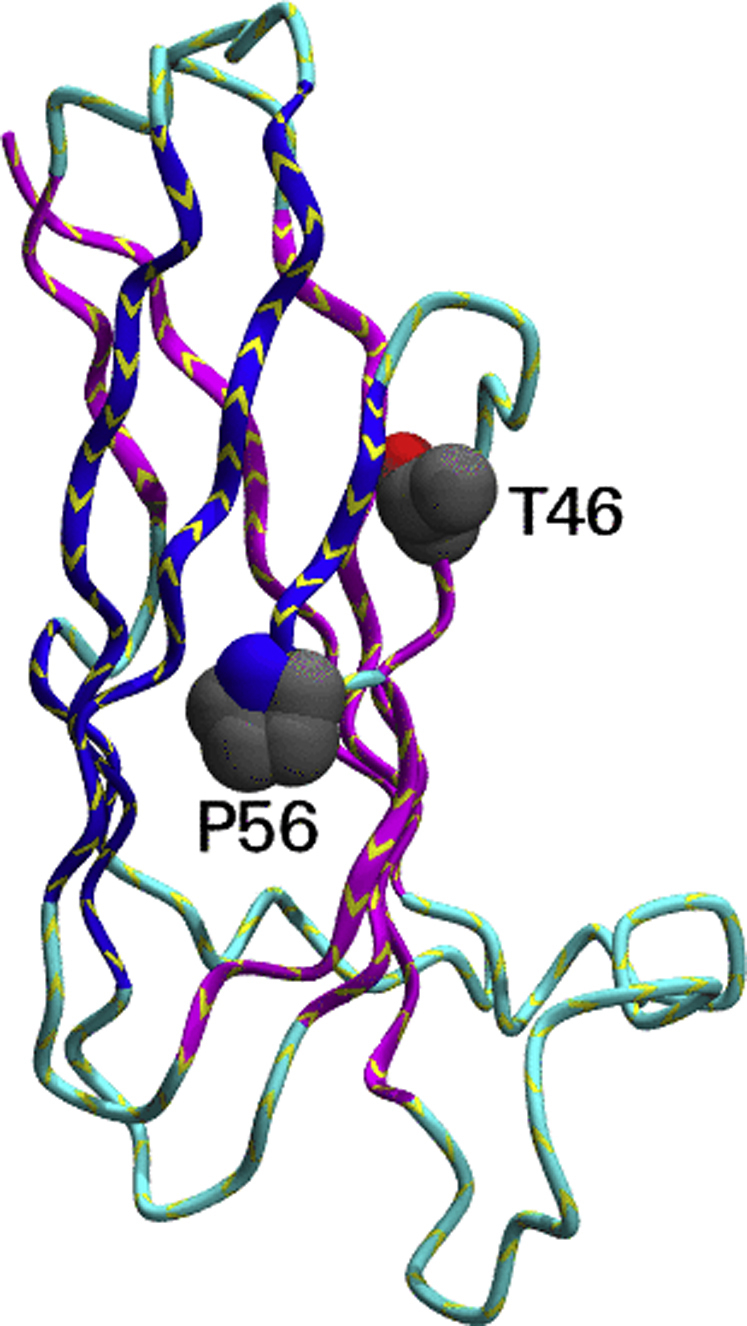
Mapping ALS-related mutations onto the structure of the MSP domain of VAP-B (PDB code: 3ikk). The backbone of the protein is shown as a ribbon in stereo. The adopted view differs from that of Figure 1 by a rotation of 90 degrees around an axis on the page. The side chains of T46 and P56 are shown. T46 has an accessible surface area of 31 Å2 and is thus semiexposed. P56 is also semiexposed (48 Å2) and is key in defining the switching point of strand C’. ALS, amyotrophic lateral sclerosis; MSP, major sperm protein; PDB, Protein Data Bank.
The P56S mutation was subjected to a detailed structural investigation and a close comparison with the WT using recombinantly produced proteins expressed in bacteria (29). Isolated WT VAP-B MSP domain was demonstrated to be well-folded and able to bind to FFAT motifs at a neutral pH. Acidification of the solution induced reversible unfolding as it is common to several other well-behaved proteins. Conversely, the recombinant P56S mutant was proven to be unfolded already at neutral pH. This behavior can be rationalized by bearing in mind that P56 is crucially involved in the formation of the S-shaped conformation of the bulge that allows switching of the C’ strand between the two sheets. By analogy, independent mutation of P12 located in the other S-shaped loop of the MSP domain produced a highly insoluble and unfolded protein. This study thus strongly suggested that the P56S mutation causes a crucial destabilization of the MSP native structure, with consequent aggregation of the protein and loss of function.
Unlike P56S, T46I was shown not to lead to complete disruption of the native fold of the protein but to reduce appreciably its thermodynamic stability and unfolding cooperativity (79). The key to understanding the causes of this destabilization is to consider that T46 is a partially buried residue in the available structure of Vap-B (3ikk) and creates a pocket in the MSP fold. Substitution of a small buried semihydrophilic residue by a bulky hydrophobic isoleucine is more than likely to destabilize the fold causing misfolding and aggregation. Accordingly, studies of the interactome of the mutant have resulted in loss or loosening of some interactions (79).
Altogether, these results provide models to explain the mechanism for the role of ALS-related mutations in the pathology.
A relationship between ALS and viral infection
We have described the role of VAPs both in infective and neurodegenerative diseases. Do these events occur through independent mechanisms? Probably not: increasing evidence supports a relationship between disregulation of the gut microbiota with the risk of neurodegeneration through alteration of circulating levels of inflammatory cytokines and/or production of neurotoxins that would affect the central nervous system, as observed in PD, AD, and other disorders (80). Independent studies have, for instance, detected differences in the bacterial taxa of patients with PD as compared with healthy controls (81). First linked to herpes simplex virus in early pioneering studies (82), it has been more recently suggested that AD may be linked to an imbalance of the gut microbiota through what is called the gut–brain axis (83).
A pathogenic connection between ALS and retroviruses was suggested by the observation of HIV cases associated with ALS or ALS-like diseases (4, 5, 6, 7). Accordingly, a recent report disclosed that a patient with chronic HCV developed an ALS-like syndrome (8). Reverse transcriptase, the enzyme utilized by retroviruses to replicate, has been detected in a significantly higher percent of patients with sporadic ALS than healthy controls (84, 85). Postmortem analysis of brain tissues of patients with ALS has also revealed a significantly higher expression of human endogenous retrovirus K (86). Interestingly, antiviral therapies seem to improve ALS-like symptoms in some individuals (87). It is thus not inconceivable to explore whether this link could involve VAP-mediated processes, given the role that these proteins play in several essential cellular functions.
The ways this link could be realized are multifold. Misregulation of the ER stress regulatory pathways seems to be at the root of both the pathological mechanism for ALS and of HCV infection (76). Viral NS5A, FFAT-containing proteins, and the EphA4 receptor bind, for instance, overlapping surfaces on the MSP domain and have been shown to compete for binding (56). In some patients with HCV, the NS5A protein has been found to accumulate abnormally with consequent potential perturbation of the physiological function of the VAPs in a manner similar to that induced by the T46I mutation. The hypothesis is supported by the fact that the two residues affected by the ALS-causing mutations P56 and T46 are both located at the interaction surface of MSP with NS5A (56). Some authors have suggested that HCV infection might not only induce an ALS-like syndrome but also trigger pathogenesis associated with signaling pathways mediated by EphA2 and EphA5 (57). Eph receptors and their ephrin ligands are polymorphic families of cell–surface signaling molecules important in many aspects of development, including cell differentiation and spatial organization of growing tissues. Dysfunction of ephrin signaling is implicated in several pathological conditions including but not limited to cancer.
The evidence presented could thus explain a VAP-mediated link between infectious and neurodegenerative diseases.
Conclusions
In this review, we have analyzed the structure and functions of proteins from the VAP family, an important tethering component of the eukaryotic cell. We have discussed how the Ig fold of the conserved globular MSP domain differs from more canonical Ig fold types and shares similarity with two other protein families, nematode MSP and bacterial PapD. Although in different ways, all three protein families seem to be implicated in functions that would require specific mechanical properties which might be encoded in the S-type fold. Detailed sequence and structural comparison would suggest that only the MSP family is evolutionarily related to VAPs.
We have then discussed the wide interactome of VAPs and how they interact both with endogenous and viral proteins often with overlapping regions, suggesting competition among these interactions. Clear links seem also to associate VAPs and specifically VAP-B with neurodegenerative diseases, such as ALS. Altogether, our review demonstrates how the VAP family has an important role in the healthy metabolism and how destabilization of the structure and/or interactions leads to pathology. Much more work remains to be done to solve the structures at atomic resolution of the several complexes involving the VAPs, which will constitute an essential step to understand their functions.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The research was supported by UK Dementia Research Institute (RE1 3556), which is funded by the Medical Research Council, Alzheimer’s Society, and Alzheimer’s Research UK, and by Alzheimer's Research UK (grant ARUK-PG2019B-020).
Author contributions
E. F. D. wrote the initial overall draft and drew Figure 4; M. A. H. traced the evolution of the VAP family and contributed to Figure 3; A. M. L. prepared the structural figures; and A. P. put together the various contributions. All authors helped to improve and finalize the text and the figures.
Edited by Phyllis Hanson
Supporting information
Supplemental Data S1.
Supplemental Data S2.

Supplemental Data S3.
References
- 1.Eisenberg-Bord M., Shai N., Schuldiner M., Bohnert M. A tether is a tether is a tether: Tethering at membrane contact sites. Dev. Cell. 2016;39:395–409. doi: 10.1016/j.devcel.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Henne W.M. Organelle remodeling at membrane contact sites. J. Struct. Biol. 2016;196:15–19. doi: 10.1016/j.jsb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnert M. Tether me, tether me not-dynamic organelle contact sites in metabolic rewiring. Dev. Cell. 2020;54:212–225. doi: 10.1016/j.devcel.2020.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Verma A., Berger J.R. ALS syndrome in patients with HIV-1 infection. J. Neurol. Sci. 2006;240:59–64. doi: 10.1016/j.jns.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Lyons J., Venna N., Cho T.A. Atypical nervous system manifestations of HIV. Semin. Neurol. 2011;31:254–265. doi: 10.1055/s-0031-1287653. [DOI] [PubMed] [Google Scholar]
- 6.Farhadian S., Patel P., Spudich S. Neurological complications of HIV infection. Curr. Infect. Dis. Rep. 2017;19:50. doi: 10.1007/s11908-017-0606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfahad T., Nath A. Retroviruses and amyotrophic lateral sclerosis. Antiviral Res. 2013;99:180–187. doi: 10.1016/j.antiviral.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastos A.F., Orsini M., Machado D., Mello M.P., Nader S., Silva J.G., da Silva Catharino A.M., de Freitas M.R., Pereira A., Pessoa L.L., Sztajnbok F.R., Leite M.A., Nascimento O.J., Bastos V.H. Amyotrophic lateral sclerosis: One or multiple causes? Neurol. Int. 2011;3 doi: 10.4081/ni.2011.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia H., Liang K., Liu G., Zhang Z., Shi Y., Liang H., Liu P. lncRNA DANCR promotes proliferation and metastasis of breast cancer cells through sponging miR-4319 and upregulating VAPB. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2020.3675. [DOI] [PubMed] [Google Scholar]
- 10.Skehel P.A., Martin K.C., Kandel E.R., Bartsch D. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 1995;269:1580–1583. doi: 10.1126/science.7667638. [DOI] [PubMed] [Google Scholar]
- 11.Soussan L., Burakov D., Daniels M.P., Toister-Achituv M., Porat A., Yarden Y., Elazar Z. ERG30, a VAP-33–related protein, functions in protein transport mediated by COPI vesicles. J. Cell Biol. 1999;146:301–312. doi: 10.1083/jcb.146.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weir M.L., Xie H., Klip A., Trimble W.S. VAP-A binds promiscuously to both v- and tSNAREs. Biochem. Biophys. Res. Commun. 2001;286:616–621. doi: 10.1006/bbrc.2001.5437. [DOI] [PubMed] [Google Scholar]
- 13.Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 glued and late endosome positioning. J. Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuijpers M., van Dis V., Haasdijk E.D., Harterink M., Vocking K., Post J.A., Scheper W., Hoogenraad C.C., Jaarsma D. Amyotrophic lateral sclerosis (ALS)-associated VAPB-P56S inclusions represent an ER quality control compartment. Acta Neuropathol. Commun. 2013;1:24. doi: 10.1186/2051-5960-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy S.E., Levine T.P. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Lev S., Ben Halevy D., Peretti D., Dahan N. The VAP protein family: From cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Weir M.L., Klip A., Trimble W.S. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): A broadly expressed protein that binds to VAMP. Biochem. J. 1998;333(Pt 2):247–251. doi: 10.1042/bj3330247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura Y., Hayashi M., Inada H., Tanaka T. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 1999;254:21–26. doi: 10.1006/bbrc.1998.9876. [DOI] [PubMed] [Google Scholar]
- 19.Skehel P.A., Fabian-Fine R., Kandel E.R. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1101–1106. doi: 10.1073/pnas.97.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawano M., Kumagai K., Nishijima M., Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 21.Kagiwada S., Hosaka K., Murata M., Nikawa J., Takatsuki A. The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol. 1998;180:1700–1708. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyles J.P., McMaster C.R., Ridgway N.D. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 23.Phillips M.J., Voeltz G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikawa J., Murakami A., Esumi E., Hosaka K. Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J. Biochem. 1995;118:39–45. doi: 10.1093/oxfordjournals.jbchem.a124889. [DOI] [PubMed] [Google Scholar]
- 25.Pennetta G., Hiesinger P.R., Fabian-Fine R., Meinertzhagen I.A., Bellen H.J. Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron. 2002;35:291–306. doi: 10.1016/s0896-6273(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 26.Williams A.F., Barclay A.N. The immunoglobulin superfamily--domains for cell surface recognition. Annu. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 27.Loewen C.J., Levine T.P. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 28.Mitne-Neto M., Machado-Costa M., Marchetto M.C., Bengtson M.H., Joazeiro C.A., Tsuda H., Bellen H.J., Silva H.C., Oliveira A.S., Lazar M., Muotri A.R., Zatz M. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum. Mol. Genet. 2011;20:3642–3652. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J., Lua S., Tong J.S., Song J. Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis. Biochemistry. 2010;49:3887–3897. doi: 10.1021/bi902057a. [DOI] [PubMed] [Google Scholar]
- 30.Weimbs T., Low S.H., Chapin S.J., Mostov K.E., Bucher P., Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: A new superfamily. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russ W.P., Engelman D.M. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 32.Roberts T.M., Stewart M. Acting like actin: The dynamics of the nematode major sperm protein (MSP) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility. J. Cell Biol. 2000;149:7–12. doi: 10.1083/jcb.149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bork P., Holm L., Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 34.Harpaz Y., Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 35.Ma Z., Liu Z., Huang X. OSBP- and FAN-mediated sterol requirement for spermatogenesis in Drosophila. Development. 2010;137:3775–3784. doi: 10.1242/dev.049312. [DOI] [PubMed] [Google Scholar]
- 36.Wang P., Richardson C., Hawkins T.J., Sparkes I., Hawes C., Hussey P.J. Plant VAP27 proteins: Domain characterization, intracellular localization and role in plant development. New Phytol. 2016;210:1311–1326. doi: 10.1111/nph.13857. [DOI] [PubMed] [Google Scholar]
- 37.Di Mattia T., Wilhelm L.P., Ikhlef S., Wendling C., Spehner D., Nominé Y., Giordano F., Mathelin C., Drin G., Tomasetto C., Alpy F. Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep. 2018;19 doi: 10.15252/embr.201745453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Thaler R., Rumpler M., Spitzer S., Klaushofer K., Varga F. Mospd1, a new player in mesenchymal versus epidermal cell differentiation. J. Cell Physiol. 2011;226:2505–2515. doi: 10.1002/jcp.22595. [DOI] [PubMed] [Google Scholar]
- 40.Zimmer S., Stocker A., Sarbolouki M.N., Spycher S.E., Sassoon J., Azzi A. A novel human tocopherol-associated protein: Cloning, in vitro expression, and characterization. J. Biol. Chem. 2000;275:25672–25680. doi: 10.1074/jbc.M000851200. [DOI] [PubMed] [Google Scholar]
- 41.Loewen C.J., Roy A., Levine T.P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Angelo G., Vicinanza M., De Matteis M.A. Lipid-transfer proteins in biosynthetic pathways. Curr. Opin. Cell Biol. 2008;20:360–370. doi: 10.1016/j.ceb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 43.De Matteis M.A., Di Campli A., D'Angelo G. Lipid-transfer proteins in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta. 2007;1771:761–768. doi: 10.1016/j.bbalip.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., Emr S.D. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Paillusson S., Stoica R., Gomez-Suaga P., Lau D.H.W., Mueller S., Miller T., Miller C.C.J. There's something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 2016;39:146–157. doi: 10.1016/j.tins.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devine M.J., Kittler J.T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 2018;19:63–80. doi: 10.1038/nrn.2017.170. [DOI] [PubMed] [Google Scholar]
- 48.De Vos K.J., Mórotz G.M., Stoica R., Tudor E.L., Lau K.F., Ackerley S., Warley A., Shaw C.E., Miller C.C. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012;21:1299–1311. doi: 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez-Suaga P., Paillusson S., Stoica R., Noble W., Hanger D.P., Miller C.C.J. The ER-mitochondria tethering complex VAPB-PTPIP51 regulates autophagy. Curr. Biol. 2017;27:371–385. doi: 10.1016/j.cub.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabukusta B., Berlin I., van Elsland D.M., Forkink I., Spits M., de Jong A.W.M., Akkermans J.J.L.L., Wijdeven R.H.M., Janssen G.M.C., van Veelen P.A., Neefjes J. Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-related FFNT motifs. Cell Rep. 2020;33:108475. doi: 10.1016/j.celrep.2020.108475. [DOI] [PubMed] [Google Scholar]
- 51.Tsuda H., Han S.M., Yang Y., Tong C., Lin Y.Q., Mohan K., Haueter C., Zoghbi A., Harati Y., Kwan J., Miller M.A., Bellen H.J. The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell. 2008;133:963–977. doi: 10.1016/j.cell.2008.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao L., Aizaki H., He J.-W., Lai M.M. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helbig K.J., Eyre N.S., Yip E., Narayana S., Li K., Fiches G., McCartney E.M., Jangra R.K., Lemon S.M., Beard M.R. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamamoto I., Nishimura Y., Okamoto T., Aizaki H., Liu M., Mori Y., Abe T., Suzuki T., Lai M.M., Miyamura T., Moriishi K., Matsuura Y. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 2005;79:13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCune B.T., Tang W., Lu J., Eaglesham J.B., Thorne L., Mayer A.E., Condiff E., Nice T.J., Goodfellow I., Krezel A.M., Virgin H.W. Noroviruses co-opt the function of host proteins VAPA and VAPB for replication via a phenylalanine-phenylalanine-acidic-tract-motif mimic in nonstructural viral protein NS1/2. mBio. 2017;8 doi: 10.1128/mBio.00668-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta G., Qin H., Song J. Intrinsically unstructured domain 3 of hepatitis C virus NS5A forms a “fuzzy complex” with VAPB-MSP domain which carries ALS-causing mutations. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta G., Song J. C-terminal auto-regulatory motif of hepatitis C virus NS5B interacts with human VAPB-MSP to form a dynamic replication complex. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tu H., Gao L., Shi S.T., Taylor D.R., Yang T., Mircheff A.K., Wen Y., Gorbalenya A.E., Hwang S.B., Lai M.M. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 59.Ramage H.R., Kumar G.R., Verschueren E., Johnson J.R., Von Dollen J., Johnson T., Newton B., Shah P., Horner J., Krogan N.J., Ott M. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol. Cell. 2015;57:329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roulin P.S., Lötzerich M., Torta F., Tanner L.B., van Kuppeveld F.J., Wenk M.R., Greber U.F. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Roulin P.S., Murer L.P., Greber U.F. A single point mutation in the rhinovirus 2B protein reduces the requirement for phosphatidylinositol 4-kinase class III beta in viral replication. J. Virol. 2018;92 doi: 10.1128/JVI.01462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barajas D., Xu K., Sharma M., Wu C.Y., Nagy P.D. Tombusviruses upregulate phospholipid biosynthesis via interaction between p33 replication protein and yeast lipid sensor proteins during virus replication in yeast. Virology. 2014;471-473:72–80. doi: 10.1016/j.virol.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agaisse H., Derré I. Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect. Immun. 2014;82:2037–2047. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derré I., Swiss R., Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elwell C.A., Jiang S., Kim J.H., Lee A., Wittmann T., Hanada K., Melancon P., Engel J.N. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y., Zhu X. Endoplasmic reticulum-mitochondria tethering in neurodegenerative diseases. Transl. Neurodegener. 2017;6:1–8. doi: 10.1186/s40035-017-0092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krols M., van Isterdael G., Asselbergh B., Kremer A., Lippens S., Timmerman V., Janssens S. Mitochondria-associated membranes as hubs for neurodegeneration. Acta Neuropathol. 2016;131:505–523. doi: 10.1007/s00401-015-1528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han S.M., El Oussini H., Scekic-Zahirovic J., Vibbert J., Cottee P., Prasain J.K., Bellen H.J., Dupuis L., Miller M.A. VAPB/ALS8 MSP ligands regulate striated muscle energy metabolism critical for adult survival in caenorhabditis elegans. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoica R., Paillusson S., Gomez-Suaga P., Mitchell J.C., Lau D.H., Gray E.H., Sancho R.M., Vizcay-Barrena G., De Vos K.J., Shaw C.E., Hanger D.P., Noble W., Miller C.C. ALS/FTD-associated FUS activates GSK-3β to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016;17:1326–1342. doi: 10.15252/embr.201541726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoica R., De Vos K.J., Paillusson S., Mueller S., Sancho R.M., Lau K.-F., Vizcay-Barrena G., Lin W.-L., Xu Y.-F., Lewis J. ER–mitochondria associations are regulated by the VAPB–PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paillusson S., Gomez-Suaga P., Stoica R., Little D., Gissen P., Devine M.J., Noble W., Hanger D.P., Miller C.C.J. α-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca. Acta Neuropathol. 2017;134:129–149. doi: 10.1007/s00401-017-1704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H.J., Anagnostou G., Chai A., Withers J., Morris A., Adhikaree J., Pennetta G., de Belleroche J.S. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2010;285:40266–40281. doi: 10.1074/jbc.M110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kabashi E., El Oussini H., Bercier V., Gros-Louis F., Valdmanis P.N., McDearmid J., Mejier I.A., Dion P.A., Dupre N., Hollinger D., Sinniger J., Dirrig-Grosch S., Camu W., Meininger V., Loeffler J.P. Investigating the contribution of VAPB/ALS8 loss of function in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:2350–2360. doi: 10.1093/hmg/ddt080. [DOI] [PubMed] [Google Scholar]
- 74.Nishimura A.L., Mitne-Neto M., Silva H.C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J.R., Gillingwater T., Webb J., Skehel P., Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Blitterswijk M., van Es M.A., Koppers M., van Rheenen W., Medic J., Schelhaas H.J., van der Kooi A.J., de Visser M., Veldink J.H., van den Berg L.H. VAPB and C9orf72 mutations in 1 familial amyotrophic lateral sclerosis patient. Neurobiol. Aging. 2012;33:2950.e1–2950.e4. doi: 10.1016/j.neurobiolaging.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Gkogkas C., Middleton S., Kremer A.M., Wardrope C., Hannah M., Gillingwater T.H., Skehel P. VAPB interacts with and modulates the activity of ATF6. Hum. Mol. Genet. 2008;17:1517–1526. doi: 10.1093/hmg/ddn040. [DOI] [PubMed] [Google Scholar]
- 77.Kanekura K., Nishimoto I., Aiso S., Matsuoka M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8) J. Biol. Chem. 2006;281:30223–30233. doi: 10.1074/jbc.M605049200. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki H., Matsuoka M. Amyotrophic lateral sclerosis-linked mutant VAPB enhances TDP-43-induced motor neuronal toxicity. J. Neurochem. 2011;119:1099–1107. doi: 10.1111/j.1471-4159.2011.07491.x. [DOI] [PubMed] [Google Scholar]
- 79.Lua S., Qin H., Lim L., Shi J., Gupta G., Song J. Structural, stability, dynamic and binding properties of the ALS-causing T46I mutant of the hVAPB MSP domain as revealed by NMR and MD simulations. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giau V.V., Wu S.Y., Jamerlan A., An S.S.A., Kim S.Y., Hulme J. Gut microbiota and their neuroinflammatory implications in Alzheimer's disease. Nutrients. 2018;10:1765. doi: 10.3390/nu10111765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown E.G., Goldman S.M. Modulation of the microbiome in Parkinson's disease: Diet, drug, stool transplant, and beyond. Neurotherapeutics. 2020;17:1406–1417. doi: 10.1007/s13311-020-00942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jamieson G.A., Maitland N.J., Wilcock G.K., Craske J., Itzhaki R.F. Latent herpes simplex virus type 1 in normal and Alzheimer's disease brains. J. Med. Virol. 1991;33:224–227. doi: 10.1002/jmv.1890330403. [DOI] [PubMed] [Google Scholar]
- 83.Borsom E.M., Lee K., Cope E.K. Do the bugs in your gut eat your memories? Relationship between gut microbiota and Alzheimer's disease. Brain Sci. 2020;10:814. doi: 10.3390/brainsci10110814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCormick A.L., Brown R.H., Cudkowicz M.E., Al-Chalabi A., Garson J.A. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology. 2008;70:278–283. doi: 10.1212/01.wnl.0000297552.13219.b4. [DOI] [PubMed] [Google Scholar]
- 85.Steele A.J., Al-Chalabi A., Ferrante K., Cudkowicz M.E., Brown R.H., Garson J.A. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology. 2005;64:454–458. doi: 10.1212/01.WNL.0000150899.76130.71. [DOI] [PubMed] [Google Scholar]
- 86.Douville R., Liu J., Rothstein J., Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moulignier A., Moulonguet A., Pialoux G., Rozenbaum W. Reversible ALS-like disorder in HIV infection. Neurology. 2001;57:995–1001. doi: 10.1212/wnl.57.6.995. [DOI] [PubMed] [Google Scholar]
- 88.Baron Y., Pedrioli P.G., Tyagi K., Johnson C., Wood N.T., Fountaine D., Wightman M., Alexandru G. VAPB/ALS8 interacts with FFAT-like proteins including the p97 cofactor FAF1 and the ASNA1 ATPase. BMC Biol. 2014;12:39. doi: 10.1186/1741-7007-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.