Abstract
Antisense technology is beginning to deliver on the broad promise of the technology. Ten RNA-targeted drugs including eight single-strand antisense drugs (ASOs) and two double-strand ASOs (siRNAs) have now been approved for commercial use, and the ASOs in phase 2/3 trials are innovative, delivered by multiple routes of administration and focused on both rare and common diseases. In fact, two ASOs are used in cardiovascular outcome studies and several others in very large trials. Interest in the technology continues to grow, and the field has been subject to a significant number of reviews. In this review, we focus on the molecular events that result in the effects observed and use recent clinical results involving several different ASOs to exemplify specific molecular mechanisms and specific issues. We conclude with the prospective on the technology.
Keywords: antisense, RNase H1, splicing, molecular mechanisms, clinical results
Abbreviations: ds, double-stranded; FCS, familial chylomicronemia; GalNAc, N-acetyl galactosamine; GFR, glomerular filtration rate; PD, pharmacodynamics; PK, pharmacokinetic; RTD, RNA-targeted drug; SGLT2, sodium-glucose cotransmitter 2; SMN1, survival motor neuron; ss ASO, single-strand antisense oligonucleotide; TIE, translation inhibitory element
Recent progress and the performance of RNA-targeted drugs (RTDs) in well-controlled clinical trials argue that RTD discovery technology is beginning to deliver the potential value that encouraged investment beginning 3 decades ago. To date, ten RTDs have been approved for commercial use (Table 1), including eight single-strand antisense oligonucleotide (ss ASO) drugs (ASOs) and two double-stranded (ds) antisense drugs that are referred to as siRNAs (1, 2, 3, 4). Nusinersen, a PS ASO that corrects the splicing of the SMN2 pre-mRNA to treat spinal muscular atrophy (SMA) is the first “blockbuster” RTD (3). Though all of the approved RTDs are designed to treat patients with rare diseases, Table 1 shows that while the rare disease pipeline is robust, there are numerous RTDs in advanced clinical trials that are focused on diseases with very high incidences. In fact, Pelacarsen, formerly AKCEA-APO(a)-LRx and ION-TTR- LRx, is enrolling patients into cardiovascular outcome studies and in advanced phase 2 studies, there are PS ASOs such as IONIS-FXI-LRx, IONIS APOCIII-LRx, Vupanorsen (formerly AKCEA-ANGPTL3-LRx), and a number of ASOs designed to treat high-incidence neurological diseases including treatments for Alzheimer’s and Parkinson’s diseases (Table 1) (4). Table 1 also shows that ss PS ASOs can be administered by multiple routes for both systemic and local therapeutic effects. The versatility of the technology is further demonstrated by the fact that post-RNA-binding mechanisms of action include both ASOs that cause RNA reduction via RNase H1 and those that increase protein production by correcting RNA splicing defects. Preclinical data suggest that in the coming years, a range of new post-RNA-binding mechanisms will be used to bring clinical benefit (5, 6, 7, 8). That ASOs in the clinic are being used to treat diseases caused in full or partly by toxic RNAs, as well as more traditional protein-caused diseases argue that more targets and opportunities will emerge as the roles of RNAs are better understood. Finally, several chemical classes and the critical importance of advances in medicinal chemistry are apparent. The significance of advances in oligonucleotide medicinal chemistry and in ASO designs is best shown by comparing the performances of phosphorothioate (PS) oligodeoxynucleotides (first generation) to those PS ASOs containing 2ʹ methoxyethyl (2ʹ-MOE) substitutions (second generation) to PS ASOs containing 2ʹ constrained ethyl (2ʹcEt) modifications (generation 2.5) to 2ʹ-MOE and 2ʹcEt PS ASOs conjugated with N-acetyl galactosamine (GalNAc) (3, 9).
Table 1.
Clinical development activities of antisense medicines
| A - approved RNA targeted drugs | |||||
|---|---|---|---|---|---|
| Drug | Indication | Target RNA | Mechanism | Chemistry | Year approved |
| Fomivirsen | Cytomegalovirus retinitis | HCMV UL122 | RNase H1 | PS, DNA | 1998 |
| Mipomersen | Homozygous familial hypercholesterolemia | APOB | RNase H1 | PS, MOE | 2013 |
| Nusinersen | Spinal muscular atrophy | SMN2 | Splicing, intron 7 | PS, MOE | 2016 |
| Eteplirsen | Duchenne muscular dystrophy | DMD | Splicing, exon 51 | PMO | 2016 |
| Inotersen | Hereditary transthyretin- mediated amyloidosis | TTR | RNase H1 | PS, MOE | 2018 |
| Volanesorsen | Familial chylomicronemia syndrome | ApoC-III | RNase H1 | PS, MOE | 2019 |
| Patisiran | Hereditary transthyretin- mediated amyloidosis | TTR | Ago2 | PO-siRNA Cationic lipid formulation |
2018 |
| Golodirsen | Duchenne muscular dystrophy | DMD | Splicing, exon 53 | PMO | 2019 |
| Givosiran | Acute hepatic porphyria | ALAS1 | Ago2 | Ome/F- siRNA, GalNAc | 2019 |
| Viltolarsen | Duchenne muscular dystrophy | DMD | Splicing, exon 53 | PMO | 2020 |
| B. ASO drugs under development | ||||||
|---|---|---|---|---|---|---|
| Chemistrya | Drug | Target | Organ | Dose | Route | Indication | Key observations | References |
| Phase 3 | ||||||
| 2ʹ-MOE | Tofersen (BIIB067/ISIS 666853) | SOD1, CNS | ALS (Ionis/Biogen) | Phase 2 findings of dose-dependent reduction in SOD1 CSF concentration; slowing decline in clinical function, respiratory function, and muscle strength, versus placebo; well tolerated with multiple dose administrations at doses of 20–100 mg | NCT02623699 (185) | |
| 2ʹ-MOE | Tominersen (RG6042/ISIS 443139) | HTT, CNS | 10−120 mg once every 4 weeks, IT | Huntington’s Disease (Ionis/Roche) | Phase ½a findings of dose-dependent reduction of mutant huntingtin CSF concentration; well tolerated with multiple dose administrations at monthly doses of 10–120 mg | NCT03761849 NCT02519036 (186) |
| undisclosed | Sepofarsen (QR-110) | LCA10, p.Cys998X | ITV | Leber’s Congenital Amaurosis Type 10 (ProQR) | Phase ½ findings included improvement in visual acuity | NCT03913143 |
| 2ʹ-MOE, GalNAc | Pelacarsen (TQJ230/AKCEA- APO(a)-LRx) | Apo(a), Liver | 80 mg once monthly, SC | CVD (Ionis/Akcea/Novartis) | Phase 2 findings of dose-dependent reduction in serum lipoprotein (a) levels; no difference compared with placebo-treated patients in platelet counts, liver and renal function tests | NCT04023552 NCT03070782 (129) |
| 2ʹ-MOE, GalNAc | AKCEA- TTR-LRx (ION 682884) | TTR, Liver | 45 mg once monthly, SC | ATTR (Ionis/Akcea) | Phase 1 findings of dose-dependent reduction in plasma TTR; well tolerated with multiple dose administrations; no clinically relevant effect on platelet counts, or liver and renal function tests | NCT04136184 NCT04136171 NCT03728634 (187) |
| PMO | Casimersen (SRP-4045) | Dystrophin Exon 45, Muscle | 30 mg/kg once weekly, IV | DMD (Sarepta) | Statistically significant increase from baseline in dystrophin protein (versus placebo) at week 48 interim end point Study ongoing (double-blind, placebo- controlled to 96 weeks) | NCT02500381 (188) |
| 2ʹ-H | Alicaforsen | ICAM-1, Colon | 240 mg once daily, Enema | Chronic Pouchitis and Ulcerative Colitis (Ionis) | Phase 2 findings included reduction in Pouchitis Disease Activity Index and endoscopy subscore | NCT02525523 (171, 189) |
| Phase 2 | ||||||
| 2ʹ-MOE | BIIB080 (IONIS- MAPTRx) | TAU, CNS | Once monthly, IT | Alzheimer’s Disease, FTD (Ionis/Biogen) | NCT03186989 | |
| 2ʹ-MOE | IONIS-FXIRx/BAY 2306001 | Factor XI, Liver | 100–300 mg once weekly, SC | Clotting Disorders (Ionis/Bayer) | Phase 2 findings included reduction of Factor XI protein, and reduction of thrombotic events without increase in bleeding. | NCT02553889 NCT01713361 (190) |
| 2ʹ-MOE | IONIS- GCGRRx | GCGR, Liver | 50−200 mg once weekly, SC | T2D (Ionis/Suzhou-Ribo) | Phase 2 findings included attenuation of glucagon-induced increase in blood glucose levels, dose-dependent reduction of HbA1c, no cases of severe or symptomatic hypoglycemia, dose- dependent increase in liver transaminase levels consistent with pharmacology, and no increase in hepatic glycogen content | NCT01885260 NCT02583919 NCT02824003 (191) |
| 2ʹ-MOE | IONIS- DGAT2Rx | DGAT2, Liver | 250 mg once weekly, SC | NASH (Ionis) | Phase 2 findings included significant absolute reduction in liver fat, versus placebo, and 50% of patients treated had at least a 30% relative reduction in liver fat; no changes in liver or renal function, no cases of thrombocytopenia | NCT03334214 (192) |
| 2ʹ-MOE | Apatorsen | HSP27, Tumor Cells | 200−1000 mg once weekly, IV | Cancer (Ionis/OncoGenex) | Phase 1 findings included decrease in tumor markers and decline in CTCs | NCT01454089 (193) |
| 2ʹ-MOE | ATL1102 | CD49d, Immune Cells | DMD: 25 mg weekly, SC MS: 200 mg twice weekly, SC | DMD, MS (Ionis/ATL) | Phase 2 findings in nonambulatory DMD patients included positive effects on modulating CD49d+ T cells in blood Phase 2 findings in MS patients included reduction in new active lesions, and moderate thrombocytopenia | ACTRN1261800 0970246 (194) |
| 2ʹ-MOE | Atesidorsen/ATL1103 | GHR, Liver | 200 mg once or twice weekly, SC | Acromegaly (Ionis/ATL) | Phase 2 findings included significant reduction in IGF-1 in patients with acromegaly who received ATL1103 200 mg twice weekly, versus once weekly | ACTRN1261500 0289516 (195) |
| 2ʹ-MOE | IONIS- HBVRx | HBV surface Ag, Liver | 150–300 mg once weekly, SC | HBV, chronic atypical (Ionis/GSK) | Phase 2 findings included dose dependent reductions of HbsAg and HBV DNA, and an acceptable safety profile to proceed to longer treatment durations | NCT02981602 (196) |
| 2ʹ-Ome Stereo-pure PS |
WVE-120101 | mHTT (rs362307), CNS | IT | Huntington’s Disease (Wave) | In progress | NCT03225833 |
| 2ʹ-Ome Stereo-pure PS |
WVE-120102 | mHTT (rs362331), CNS | 2–32 mg, IT | Huntington’s Disease (Wave) | In progress | NCT03225846 |
| cEt | DYN101 | DNM2, MTM1 | 1.5–9.0 mg/kg, IM | Centronuclear Myopathy (Ionis/Dynacure) | In progress | NCT04033159 |
| cET | IONIS- ENAC-2.5Rx | ENAC, Lung | Inhaled/Nebulized | Cystic Fibrosis (Ionis) | In progress | NCT03647228 |
| cET | AZD9150/IONIS- STAT3-2.5Rx | STAT3, Cancer and Stromal Cells | 2−4 mg/kg once weekly, IV | Cancer (Ionis/Astrazeneca) | Phase 1b findings included (1) reduction of STAT3, (2) reduction in serum IL6, and (3) reduction in tumor burden. | NCT02549651 NCT01563302 (197, 198) |
| cET | AZD5312/IONIS-AR- 2.5Rx | AR, Cancer Cells | 150−1150 mg once weekly, IV | Prostate cancer (Ionis/Suzhou- Ribo) | Phase 1 findings included declines in PSA and circulating tumor cells in some patients. | NCT03300505 NCT02144051 (199) |
| LNA | Cobomarsen (MRG-106) | miR-155, Cancer Cells | 75−900 mg once weekly, ITM/SC/IV | Hematological malignancies (miRagen) | Phase 1 findings included improvements in cutaneous lesions, and transcriptional changes consistent with target activity | NCT03713320 NCT02580552 (200) |
| LNA | Civi 007 | PCSK9, Liver | SC | CVD (Civi) | In progress | NCT04164888 NCT03427710 |
| PO, 2ʹ-Ome, ENA | DS-5141b | Dystrophin Exon 45, Muscle | 0.1−6.0 mg/kg once weekly, SC | DMD (Daiichi) | In progress | NCT02667483 |
| PMO Peptide | SRP-5051 | Dystrophin Exon 51, Muscle | Multiple ascending dose, IV | DMD (Sarepta) | In progress | NCT04004065 NCT03375255 |
| 2ʹ-MOE, GalNAc | Vupanorsen (AKCEA- ANGPTL3-LRx/ION 702803) | ANGPTL3, Liver | 40–80 mg total monthly dose, SC | Dyslipidemias (Ionis/Akcea/Pfizer) | Phase 2 findings in patients with HTG, T2D and NAFLD included dose-dependent reductions in ANGPTL3, TGs, ApoC-III, VLDL and non-HDL cholesterol, and total cholesterol with no reductions in liver fat or HbA1c; favorable safety and tolerability profile | NCT03371355 NCT02709850 (97, 201) |
| 2ʹ-MOE, GalNAc | AKCEA- APOCIII-LRx | ApoC-III, Liver | 10–50 mg total monthly dose, SC | CVD (Ionis/Akcea) | Phase 2 findings in patients with HTG and CVD, or high-risk of CVD, included >90% patients at 50 mg monthly dose achieved TG ≤ 150 mg/dl compared to 5% placebo, significant reduction in multiple risk factors, and no safety signals, including those related to platelet counts, liver or renal function | NCT03385239 NCT02900027 (98, 202) |
| 2ʹ-MOE, GalNAc | IONIS-AGT-LRx | AGT, Liver | Once weekly, SC | Treatment-resistant Hypertension (Ionis) | In progress | NCT04083222 NCT03714776 NCT03101878 |
| 2ʹ-MOE, GalNAc | GSK3389404/IONIS- HBV-LRx | HBV surface Ag, Liver | 30−120 mg single dose/once weekly, SC | Chronic HBV (Ionis/GSK) | In progress | NCT03020745 (203) |
| 2ʹ-MOE, GalNAc | IONIS-FB-LRx | Factor B, Liver | 10−40 mg once every 2 weeks, SC | Primary IgA Nephropathy Ocular Disease (Ionis/Roche) | Phase 1 findings included (1) dose- dependent reduction in factor B levels accompanied by similar reduction in factor B function and complement split factor Bb, and (2) no drug-related adverse events | NCT04014335 NCT03815825 ACTRN1261600 0335493 (204) |
| 2ʹ-MOE, GalNAc | IONIS-PKK-LRx | Kallikrein B1, Liver | 20–80 mg, once monthly, SC | HAE (Ionis) | Phase 1 findings included dose- dependent reduction of plasma prekallikrein levels with target reduction maintained during dosing intervals as predicted by PK properties | NCT04307381 NCT03263507 (177, 205) |
| 2ʹ-MOE, GalNAc | IONIS-GHR-LRx | GHR, Liver | Monthly, SC | Acromegaly (Ionis) | In progress | NCT03548415 |
| 2ʹ-MOE, GalNAc | IONIS-FXI-LRx | FXI, Liver | Monthly, SC | Clotting Disorders (Ionis/Bayer) | In progress | NCT03582462 |
| PO, 2ʹ-H Liposome | Prexigebersen | GRB2, Tumor cells | Twice weekly, IV | AML, CML, solid tumors (Bio-Path) | In progress | NCT04196257 NCT02923986 NCT02781883 |
| Undisclosed | QR-1123 (ION357) | RHO, P23H mutation, Eye | ITV | Autosomal Dominant Retinitis Pigmentosa (Ionis/ProQR) | In progress | NCT04123626 |
| Undisclosed | QR-421a | USH2A, exon 13 mutation, Eye | 50–200 μg, ITV | Retinitis pigmentosa (ProQR) | In progress | NCT03780257 |
| Undisclosed | QR-313 | COL7A1, exon 73 mutation, Skin | Once daily, topical cream | Recessive dystrophic epidermolysis bullosa (ProQR/Wings) | In progress | NCT03605069 |
| Undisclosed | RG-012 | miR-21, Kidney | 110−220 mg once weekly, SC | Alport Syndrome (Regulus/Genzyme) | In progress | NCT02855268 |
| Phase 1 | ||||||
| 2ʹ-MOE | BIIB078 (IONIS- C9Rx) | C9orf72, CNS | Multiple ascending doses, IT | ALS (Ionis/Biogen) | In progress | NCT04288856 NCT03626012 |
| 2ʹ-MOE | BIIB094 (ION859) | LRRK2, CNS | Single and multiple ascending doses, IT | Parkinson’s Disease (Ionis/Biogen) | In progress | NCT03976349 |
| 2ʹ-MOE | BIIB101 (ION464) | SNCA, CNS | IT | Multiple System Atrophy (Ionis/Biogen) | NCT04165486 | |
| LNA | RG6127 | Host target | SC | HBV (Roche) | NCT03762681 | |
| LNA | ISTH0036 | TGF-β2, Eye | 6.75−225 μg single dose, IVT | Glaucoma (Isarna) | Phase 1 findings included (1) dose–response trend observed in postoperative introcular pressure, and (2) no adverse events | NCT02406833 (206) |
| SNA Lipid nanoparticle | XCUR17 | IL17RA, Skin | Daily, Topical gel | Psoriasis (Exicure) | Phase 1 finding included decrease in the levels of psoriasis and inflammation markers downstream of target, with a significant reduction in keratin 16 expression and clinical improvement in epidermal thickness | (207) |
| Undisclosed | JNJ- 64991524 | Undisclosed | Oral | Undisclosed (Janssen) | NCT03346122 | |
| Nonactive | ||||||
| 2ʹ-MOE | IONIS- PTP1BRx | PTP1B, Liver | 200 mg once weekly, SC | T2D (Ionis) | Phase 2 findings included (1) reduction of HbA1c, (2) improved leptin and adiponectin levels, and (3) decreased body weight. | NCT00455598 (208) |
| 2ʹ-MOE | IONIS- GCCRRx | GCCR, Liver | 60−420 mg once weekly, SC | T2D (Ionis) | Phase 1 findings included (1) improvement in lipid profile, and (2) attenuation of dexamethasone-induced hepatic insulin resistance. | NCT01968265 (209) |
| 2ʹ-MOE | IONIS- FGFR4Rx | FGFR4, Liver | 100−200 mg once weekly, SC | Obesity (Ionis) | NCT02463240 | |
| LNA | Miravirsen | miR-122, Liver | 3−7 mg/kg once weekly, SC | HCV (Santaris/Roche) | Phase 2 findings included inhibition of miR-122 function | NCT01200420 (90, 91) |
| Discontinued | ||||||
| Stereo-pure PS 2ʹ-F, 2ʹ-Ome | Suvodirsen | Dystrophin Exon 51, Muscle | 3.5–5.0 mg/kg weekly, IV infusion | DMD (Wave) | Failure to demonstrate efficacy in multidose phase 1 open-label extension | NCT03907072 NCT03508947 |
| 2ʹ-MOE | IONIS- PKKRx | Kallikrein B1, Liver | 200 mg once weekly, SC | HAE Chronic Migraine (Ionis) |
Replaced with GalNAc conjugate | NCT03108469 (205, 210) |
| 2ʹ-H | Mongersen | SMAD7, Intestine | 160 mg once daily, Oral | Crohn’s Disease (Nogra Pharma/Celgene) | Phase 3 failed to demonstrate benefit in patients with active Crohn’s Disease with clinical remission (CD Activity Index score <150) attained in 22.8% of patients on GED-0301 versus 25% on placebo (p = 0.6210). Adverse events | NCT02596893 NCT02601300 (211, 212) |
| 2ʹ-Ome | Drisapersen | Dystrophin Exon 51, Muscle | 6 mg/kg once weekly, SC | DMD (Prosensa/BioMarin) | Rejected by FDA | (213, 214, 215) |
| 2ʹ-MOE | Custirsen | CLU, Tumor Cells | 640 mg once weekly, IV | Prostate Cancer and NSCLC (Ionis/OncoGenex) | Failure to meet primary endpoints in phase 3 trials | NCT01578655 NCT01630733 (216, 217) |
| 2ʹ-MOE | IONIS- APO(a)Rx | Apo(a), Liver | 300 mg once weekly, SC | CVD (Ionis) | Replaced with GalNAc conjugate | (218) |
| 2ʹ-MOE | ISIS 388626 | SGLT2, Kidney | 50−200 mg once weekly, SC | T2D (Ionis) | Availability of small-molecule inhibitors of SGLT2 | (63) |
| 2ʹ-MOE | ISIS 333611 | SOD1, CNS | 0.15−3.0 mg single dose, IT | Familial ALS (Ionis) | Replaced by more potent compound | (219) |
| 2ʹ-MOE | ISIS 104838 | TNFα, Immune Cells | 0.1−6 mg/kg IV, 200 mg once weekly, SC | Inflammatory Disease (Ionis) | Inadequate activity | (220) |
| 2ʹ-MOE | ISIS 113715 | PTP1B, Liver | 100−600 mg once weekly, SC | T2D (Ionis) | Replaced by more potent compound | |
| cET, 2ʹ-MOE | IONIS- DMPK2.5Rx | DMPK, Muscle | 100−600 mg once weekly, SC | MD type 1 (Ionis) | Inadequate activity | NCT02312011 (221) |
| LNA | EZN-4176 | AR, Cancer Cells | 0.5−10 mg/kg once weekly, IV | Prostate Cancer | ALT elevations | NCT01337518 (222) |
| Undisclosed, GalNAc | AZD4076/RG-125 | miR-103/107, Liver | SC | Diabetic NASH (Regulus/Astra Zeneca) | Failed in development | NCT02826525 |
| Undisclosed, GalNAc | RG-101 | miR-122, Liver | SC | HCV (Regulus) | Cases of hyperbilirubinemia | EudraCT 2016–002069–77 (223) |
All drugs are modified with PS linkages, except for the PMOs. All 2’-MOE chemistries include 2’-deoxy sugar residues to support Rnase H1 activity, unless specified as fully modified. Abbreviations are provided by column heading. Chemistry: 2’-H, 2’-deoxy; 2’-F, 2’-fluoro; 2’-Ome, 2’-methoxy; 2’- MOE, 2’-O-methoxy ethyl; cET, (S)-constrained ethyl; ENA, 2’-O,4’-C-ethylene-bridged nucleic acid; GalNAc, triantennary N-acetylgalactosamine; LNA, locked nucleic acid; PMO, phosphorodiamidate morpholino oligomer; PO, phosphodiester linkage; PS, phosphorothioate linkage; SNA, spherical nucleic acid nanoparticle. Drugs: 2.5, generation 2.5 ASO drugs containing cEt modification; L, ligand conjugate. Target/Organ: AGT, angiotensinogen; ANGPTL3, angiopoietin like 3; Apo(a), apolipoprotein A1; ApoB-100, apolipoprotein B-100; ApoC-III, apolipoprotein C-III; AR, androgen receptor; C9orf72, chromosome 9 open reading frame 72; CD49d, integrin subunit alpha 4; CEP290, centrosomal protein 290; CLU, clusterin; CMV IE2, cytomegalovirus immediate early gene 2; COL7A1, collagen type VII alpha 1 chain; DGAT2, diacylglycerol O-acyltransferase 2; DMD, dystrophin; DMPK, DM1 protein kinase; FGFR4, fibroblast growth factor receptor 4; GCCR, glucocorticoid receptor (nuclear receptor subfamily 3, group C, member 1); GCGR, glucagon receptor; GHR, growth hormone receptor; GRB2, growth factor receptor bound protein 2; HBV, hepatitis B virus; HCV, hepatitis C virus; HSP27, heat shock protein 27; HTT, huntingtin; ICAM1, intercellular adhesion molecule 1; IL17RA, interleukin 17 receptor alpha; KLKB1, kallikrein B1; LRRK2, leucine rich repeat kinase 2; MAPT (TAU), microtubule-associated protein tau; mHTT, mutant huntingtin; miR, microRNA; PCSK9, proprotein convertase subtilisin/kexin type 9; PTP1B, protein tyrosine phosphatase, nonreceptor type 1; RHO, Rhodopsin; SCNN1A (EnaC), sodium channel epithelial 1 alpha subunit; SGLT2, sodium/glucose cotransporter 2 (solute carrier family 5 member 2); SMAD7, SMAD family member 7; SMN2, survival of motor neuron 2; SNCA, alpha-synuclein; SOD1, superoxide dismutase 1; STAT3, signal transducer and activator of transcription 3; TGF-β2, transforming growth factor beta 2; TNFα, tumor necrosis factor alpha; TTR, transthyretin; USH2A, usherin. Dose/Route: IM, intramuscular; IT, intrathecal; ITM, intratumoral; IV, intravenous; IVT, intravitreal; SC, subcutaneous. Indication: ALS, amyotrophic lateral sclerosis; AML, acute myeloid leukemia; ATTR, transthyretin amyloidosis; CML, chronic myelogenous leukemia; CMV, cytomegalovirus; CVD, cardiovascular disease; DMD, Duchenne muscular dystrophy; FCS, familial chylomicronemia syndrome; HAE, hereditary angioedema; HBV, hepatitis B virus; HCV, hepatitis C virus; HoFH, homozygous familial hypercholesterolemia; MD, myotonic dystrophy; MS, multiple sclerosis; NASH, nonalcoholic steatohepatitis; NSCLC, non-small-cell lung cancer; SMA, spinal muscular atrophy; T2D, type 2 diabetes. Key Observations: AIDS, acquired immune deficiency syndrome; ALT, alanine aminotransferase; CTC, circulating tumor cells; FDA, Food and Drug Administration; HbA1c, hemoglobin A1c; IL6, interleukin 6; LDL, low-density lipoprotein; Lp(a), lipoprotein (a); PSA, prostate antigen. References: NCT, national clinical trial (registry, clinicaltrials.gov).
Though traditional drug discovery modalities have advanced incrementally, to a very large extent, these technologies have remained static, dependent on cumbersome, costly, and time-consuming screening processes and are highly inefficient. Rather remarkably, given how much the technology has already advanced and of vital importance is the fact that advances in antisense technology are continuing. Deeper understanding of the molecular mechanisms responsible for the pharmacokinetic (PK) behaviors of PS ASOs in animals and cells, the molecular mechanisms of pharmacodynamic (PD) effects of PS ASOs and the toxicities are yielding ever better performing PS ASOs. Most exciting is that antisense medicinal chemistry, which for most of the 3 decades of research on antisense technology focused on enhancing ASO interactions with target RNAs, now adds a major focus on PS ASO–protein interactions and targeted delivery to specific tissues such as what has been achieved with GalNAc conjugation (10, 11). The progress in the technology and its current status have been the subject of several recent reviews (1, 2, 3, 4).
Nevertheless, whether the technology has the potential to be truly broadly enabling remains to be answered in full. In this review, we will first address this question by describing the known properties of PS ASOs that lead us to believe that the technology has the potential to be even more broadly enabling than small-molecule drug discovery. This will be followed by a more detailed discussion than has been provided in other reviews of the theoretical framework that was the basis on which progress was made and on which the future of the technology will be written. Then, the current understanding of the molecular mechanisms responsible for the effects of ASOs that have been observed will be comprehensively discussed. Finally, we will get a glimpse of the new advances and insights that are defining the future.
Why antisense technology?
The idea of designing oligonucleotides to bind to specific sequences in target RNAs via Watson–Crick hydrogen bonding and the term “antisense” were introduced in 1978 (12). As proposed, the idea was quite simple and the word antisense was nonspecific. The authors were agnostic as to the post-RNA-binding mechanisms that would ensue to alter the behavior and performance of a target RNA, the structure of drug administered (single or double strand) or the chemical modifications that might be required to introduce acceptable properties for therapeutic administration. “Antisense” included an oligonucleotide of any structure or chemistry designed to bind target RNA via Watson–Crick hybridization. In practice, antisense is used to refer to ss ASOs, and siRNA refers to ds ASOs that act via AGO2 to cause degradation of target RNAs (for review see (13, 14)).
Not until the late 1980s, however, did any meaningful effort to convert the concept to reality begin when several companies were founded to pursue the concept. The delay is easily explained because most considered the concept impossible to reduce to practice. And for good reasons. It was impossible to cost-effectively synthesize even gram quantities of an oligonucleotide. It was clear that unmodified oligonucleotides could not be effective therapeutic agents because they are rapidly degraded by nucleases present in all in vitro and in vivo biological systems. Nor had any oligonucleotide medicinal chemistry had been performed, and it was unclear what the scope of the effort might be. Because of their size and negative charge, theoretical concepts of the time argued that these agents could not enter cells and without entering cells, ASOs could not function via an antisense mechanism. Moreover, experience with polynucleotides such as poly(I): poly(C) suggested that oligonucleotides would be unacceptably toxic (15). Given the daunting challenges, why invest the decades, careers, and billions of dollars required to learn whether the concept could be reduced to practice? The simple answer was the limitations of traditional approaches to drug discovery.
Limitations of traditional drug discovery approaches
The intellectual basis of the drug discovery industry was defined around the end of the 19th century independently by Langley, Erlich, and Bernard (16). As the theories of the time were inadequate, all were wrestling with the same question: how could one explain the potency of certain toxins such as curare and arsenicals? Ultimately, the term “receptive substance” was coined to suggest a specific interaction between these toxins and some substance in the body that bound with higher affinity than other sites (for review, see (17)). It is fascinating that an entire industry and thousands of products, to say nothing of the benefits of the products, were created prior to the 1980s when the concept was validated with the purification of the nicotinic receptor (for review, see (18)) and the cloning of the first receptors (for review, see (19)). Though the term receptor or receptive substance was entirely generic, in practice a receptor has been considered a protein and, even today, despite progress in RNA-targeted drug discovery, many pharmacologists assume that receptors are proteins.
Contemporaneously, chemical manufacturers, seeking new applications for their products, found that small molecules such as acetylsalicylic acid, or aspirin, could be effective therapeutics. Thus, traditional small-molecule drug discovery was birthed. The effects of various small-molecule agents were rationalized in the context of receptor theory, leading to the evolution of small-molecule drug discovery. Over time, the so-called 500 Da rule, which suggests that ideal therapeutic agents should be no greater than 500 Da, was established with a focus on designing small molecules designed to bind to specific proteins, either cell surface proteins coupled to signal transduction processes, i.e., receptors, or enzymes involved in cellular processes such as intermediary metabolism (20). Though crude plant extracts had been used for thousands of years as therapeutics, the idea of purifying the active component from biological sources for therapeutic purposes developed later. Arguably the first example of a “purified” natural product was insulin as replacement therapy for type 1 diabetes (21), and protein replacement therapies have added meaningful value for a relatively limited number of diseases. It was not until the use of penicillin during the Second World War that large-scale fermentation and purification of natural products to be used as antiinfectives were proven feasible. The virtual eradication of bacterial infections as a cause of significant mortality in the developed world remains perhaps the greatest achievement of the drug discovery industry (22). Interestingly, natural products are typically chemically more diverse and higher molecular weight than the “500 Da rule.” A single novel drug discovery technology was reasonably advanced at the time, but still not fully proven and of course that was monoclonal antibody technology. Over the last 3 decades, this technology has advanced and has proven to be an important dug discovery platform, used relatively broadly, but certainly not as broadly enabling as small-molecule drug discovery (23).
Clearly, the great strength of small-molecule drug discovery is that there are small-molecule drugs that are pharmacologically active after all routes of administration, in all the organs and most cells in the body. However, the limitations are well known and those limits have shaped the industry: inadequate selectivity, numerous “undruggable” targets, inefficient drug discovery and development, the inability to learn general principles from past experience. (The phrase “change a methyl, change the drug” is sadly true.)
The major reason to invest in the creation and advancement of antisense technology was that it had the potential to address all the limitations of small-molecule drug discovery. Evolutionary processes chose pattern recognition in nucleic acids polymers, as the means to store, retrieve, and use critical genetic information because of the specificity afforded by the genetic code. Since ASOs were to be designed to use the same code, the rules that govern binding of ASOs to RNA were well understood and could be directly applied to the drug discovery process. Moreover, the basic rules governing the specificity of binding to nucleic acid sequences were simple enough to calculate and then prove experimentally that 16 to 20 nucleotides were optimal (24). Consequently, in principle, drug discovery should be significantly more efficient and the drugs should be more specific than small molecules. It was further hoped that essentially all targets should be druggable since the biosynthesis of proteins depends on an RNA that should be targetable. The efficiency of drug development might also be greatly enhanced because all ASOs within the same chemical class differ only in sequence, thus should share many properties in common and the failure rate of drugs in development should be reduced because the lessons learned with an ASO representative of a specific chemical class could be applied (25). These were powerful incentives to invest, and the advantages have been to a large extent realized. The evidence supporting these conclusions and the questions that remain will be discussed. As several thorough reviews of the technology have recently been published that reviewed recent clinical results in detail (1, 2, 3, 4), we will not reiterate the summary of recent clinical results. Rather, we will discuss the results of selected clinical studies to exemplify novel specific mechanisms and the impacts of those advances.
History
Prerequisite advances
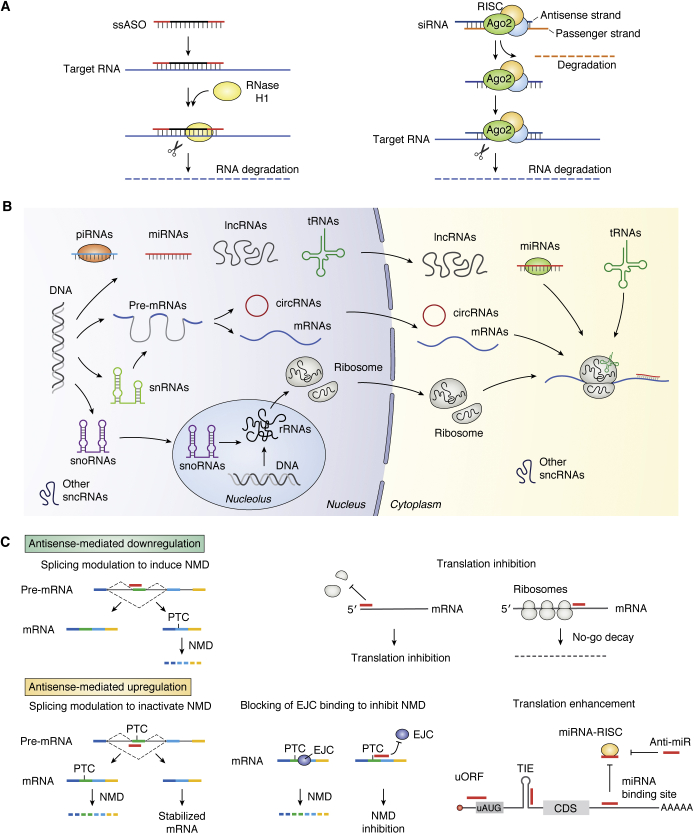
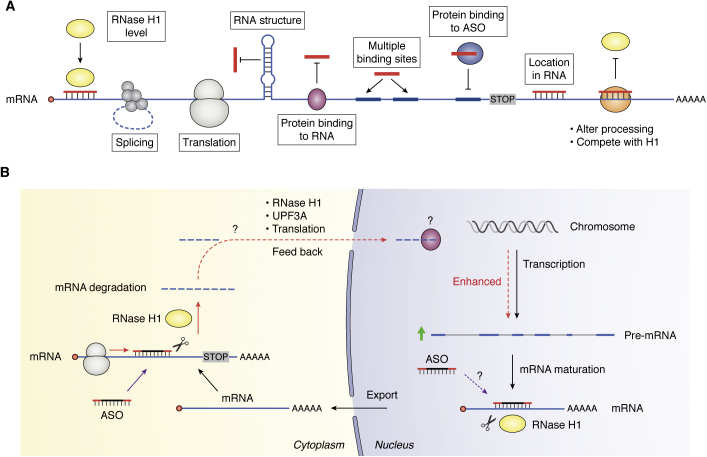
Without the evolution in understanding the RNA world (Fig. 1), antisense technology would not be possible. Because of continuing advances in understanding the structures and functions of RNAs, new opportunities for antisense technology continue to be identified. In fact, a simple and correct way to think of antisense drugs is that they alter the intermediary metabolism of RNAs. ASOs can be designed to alter the anabolism of precursor RNAs to mature RNAs, including, pre-mRNAs (26, 27), pre-rRNA (28), long noncoding RNAs (29), snRNAs and snoRNAs (30), antisense transcripts (25), and toxic RNAs such as those containing triplet repeats (31). ASOs can also be designed to alter the catabolism of precursor and mature RNAs via several mechanisms (3, 4, 5, 6) and to alter translation (7, 8, 25).
Figure 1.
Antisense technology can modulate gene expression through different mechanisms.A, commonly used antisense mechanisms to degrade target RNAs, including RNase H1-dependent and RISC-dependent mechanisms. B, various types of RNAs. In addition to the three traditional types of RNAs (rRNAs, tRNAs, and mRNAs) that are directly involved in protein synthesis, many types of small noncoding RNAs (<200 nt) have been identified to participate in various biological processes: this includes snRNAs in pre-mRNA splicing, snoRNAs in rRNA processing and modification, miRNAs in translation modulation and mRNA stability, piRNAs in RNA silencing and epigenetics, and other small ncRNAs (e.g., RNase P and MRP RNA) in tRNA and rRNA maturation. Moreover, a large number of long ncRNAs (>200 nt) have also been identified in recent years to play important roles in multiple biological processes, from chromatin remodeling (e.g., Xist, HOTAIR), transcription (e.g., asRNA, eRNA), and RNA processing (e.g., NATS) in the nucleus to translation (e.g., as-Uchl1) and protein modification (e.g., NKILA) in the cytoplasm. In addition, circular RNAs have also been identified that may modulate gene expression by acting as sponges for mRNAs or RNA-binding proteins. C, antisense oligonucleotides can modulate gene expression through additional mechanisms, including downregulation by induced NMD through altering splicing (upper left), inhibit translation initiation by binding to cap region or triggering mRNA no-go decay by binding to coding region of mRNAs (upper right panels). In addition, antisense oligonucleotides can also upregulate gene expression by altering splicing to skip PTC containing exons (lower left panel) or by inhibiting EJC binding to inhibit NMD (lower middle panel) or by enhancing translation through masking translation inhibitory elements, including uORF, TIE, miRNA-binding sites or even miRNAs (lower right panel). The circles of different colors indicate proteins. CDS, coding region sequence; EJC, exon–exon junction complex; NMD, non-sense-mediated decay; PTC, premature termination codon; RISC, RNA-induced silencing complex; TIE, translation inhibit element; uORF, upstream open reading frame. The red bars indicate antisense oligonucleotides or miRNAs.
Similarly, the sequences of target RNAs must be available to support the design of ASOs. Thus, advances in DNA sequencing that ultimately led to the sequencing of the human and other genomes (32) are a prerequisite for antisense drug discovery. Not only are modern genomic sequencing and transcriptomic methods essential for target selection, but they also support approaches to prove mechanisms of action and the selectivity of RNA target reduction (5).
Intellectual framework
An essential first step in the creation of antisense technology was the elaboration of an intellectual framework in which ASOs could be considered. Though chemically modified oligonucleotides had never been considered as possible therapeutic agents prior to the advent of antisense technology and specific sequences in targeted RNAs, it made sense to rationalize behaviors of ASOs in the context of traditional receptor theory (33). As trivial as this may seem, framing the questions that needed to be answered in this context has been critical.
Broad-based medicinal chemistry efforts
ASOs are chemically modified oligonucleotides. This is because phosphodiester oligonucleotides are rapidly degraded by exo- and endonucleases and lack the necessary affinity for receptor sequences in target RNAs. Though a great deal of medicinal chemistry had been performed on nucleosides, no focused effort to create the medicinal chemistry of oligonucleotides had been performed in the early years. Thus, a broad-based program that modified essentially every position in an oligonucleotide, except sites required for Watson–Crick hydrogen bonding, was developed and many thousands of modifications have synthesized and tested in vitro and in vivo. These modifications include those involving the nucleobase, the sugar, and the phosphate. Additionally, numerous groups of diverse structures have been conjugated at the 5ʹand 3ʹ termini and various positions on the nucleobase and the 2ʹ position of the sugar (1, 9). However, based on first principles, the major focus was on the 2ʹ position of the sugar and the phosphate, and indeed, the major advances in performance derived from modifications at these sites. Since ds ASOs or siRNAs are comprised of the same building blocks, the medicinal chemistry created to modify ASOs served as the basic chemical tool kit for siRNAs as well (10, 13, 14, 34, 35). Because these advances have been stochastic, they have been dubbed second-generation and generation 2.5 chemistries (3, 4).
More recently with the success of GalNAc conjugation in targeted delivery of both ss PS ASOs and ds PS containing siRNAs (1, 3, 4, 5, 10, 35), focus has shifted to targeted delivery to other tissues. Additionally, because of breakthroughs in understanding the molecular mechanisms by which PS ASOs induce toxicities, site-specific modifications have evolved (3, 5, 36).
The molecular mechanisms by which ASOs produce their effects
The major advances here are focused primarily on PS ASOs and have included insights into the mechanisms of cellular uptake and distribution, pharmacodynamics, and cellular and organ toxicity, and these in turn have resulted in new approaches to enhance the performance of PS ASOs (1, 3, 4, 37, 38).
Process, analytical and manufacturing chemistries
In 1989, the synthesis of even gram quantities of PS ASOs was impossible and prohibitively costly. Similarly, analytical methods to determine the purity of PS ASOs were primitive as were methods to measure PS ASOs and metabolites in biological systems. Advances in these areas were critical to the development first of PS ASOs for rare diseases, then as potency increased, the synthesis of the ton quantities of PS ASOs necessary for very large indications. Similarly, analytical methods have improved dramatically in precision and sensitivity (1, 39).
In fact, considered from a process chemical perspective, scale-up was comparatively simple as there were no new chemistries that needed to be invented, no exotherms, or high-pressure reactions, and PS ASOs are quite stable when stored as dry powders. Today, at ton scale, cost of goods is sufficiently low to support commercialization for very large indications and even oral administration.
Structural classes of RTDs
Depending on the length, ss PS (or PO)-modified ASOs range from approximately 6.5 to 7.5 kDa. Single-stranded molecules behave as highly flexible rods and can adopt a wide range of conformations in solution. Waters of hydration and counterions are present at each PS (or PO) linkage (40, 41). The diffusion coefficient and behavior on filtration confirm the claim that ss PS ASOs behave like flexible rods (42). Molecular modeling demonstrates several interesting characteristics of these molecules, including substantial flexibility and partial helical structures. However, no modeling studies have been reported for PS containing ASOs or the chimeric PS ASOs that are broadly used today, but it is highly likely that PS ASOs behave similarly to ss oligonucleotides in solution. ASOs have a hydrophilic surface, the PS or PO linkages and a hydrophobic surface, the nucleobases, and thus are relatively amphipathic, an important property that affects protein binding and consequently distribution and cell uptake (5). The nucleobases in an ss ASO are free to engage in Watson–Crick hybridization, hydrophobic, and base stacking interactions with nucleic acids and amino acids.
A ds ASO (siRNA) is, of course, at least double the molecular weight of a comparable ss ASO. In an siRNA, the sense strand meets the formal definition of a drug delivery device; it is noncovalently bound, enhances the stability of the antisense strand, and must be removed by the AGO2 loading complex before the pharmacophore, the antisense strand, is active (10, 14, 34, 43). To date, very little information is available on waters of hydration, counterions, or diffusion coefficients for chemically modified ds ASOs, though a 2ʹ F ds RNA was shown to have limited hydration (44), but it is reasonable to conclude that these agents are also rod-like, but with wider transverse dimensions and less flexibility than an ss ASO. More importantly, ds ASOs present two hydrophilic faces and the nucleobases are not free to engage in the stacking, hydrogen bonding, or hydrophobic interactions that are available to ss ASOs. These fundamental physicochemical properties are reflected in quite significant differences in the pharmacokinetic behavior of ss PS ASOs compared with ds PS containing siRNAs, which will be discussed in the pharmacokinetics section.
Oligonucleotide medicinal chemistry
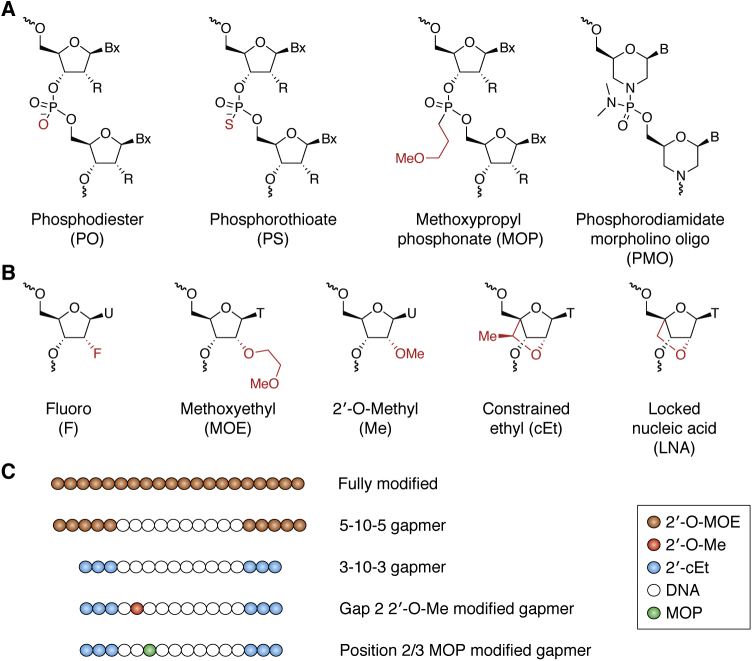
As summarized above, the evolution of the medicinal chemistry of oligonucleotides has been critical to the steadily improving performance of ASOs in the clinic. More recently, based on understanding the importance of protein binding to the in vivo distribution, cell uptake, and subcellular distribution, focus has shifted to targeted delivery to specific cell types and organ systems and point modifications that alter interactions with specific cellular proteins (1, 3, 5). Two phosphate modifications have proven to be most valuable, PMO (phosphorodiamidate morpholino oligomer) and PS. The PMO modification is neutral and is used in ASOs that have been shown to be very modestly active at high doses only in the treatment of patients with Duchenne’s muscular dystrophy (DMD), a disease of skeletal muscles (3, 4, 45, 46). The PS modification has proven far more important and is broadly used in all major classes of ASOs and all chemically modified siRNAs (Fig. 2A).
Figure 2.
Oligonucleotide chemical modifications.A, commonly used backbone modifications. MOP, methoxypropyl phosphonate; PMO, phosphorodiamidate morpholino oligomer; PO, phosphodiester; PS, phosphorothioate. B, commonly used 2’ modifications. cEt, constrained ethyl; F, fluoro; LNA, locked nucleic acid; Me, 2’-O-methyl; MOE, methoxyethyl. C, ASO designs. Fully modified ASOs that do not activate RNase H1 are commonly used to modulate splicing and translation. Gapmer ASOs that contain DNA gap and activate RNase H1 are normally modified at both ends of ASOs with different 2’ modifications. To further improve ASO performance, 2’Me or MOP (methoxypropyl phosphonate) modification is introduced into certain positions of the gap region.
The replacement of one nonbridging oxygen with a sulfur alters the physicochemical characteristics of the phosphate in important ways. This modification results in the creation of a chiral center at each internucleotide link, so not surprisingly, the effects of the chiral centers have been extensively studied, as have the effects of pure R or S isomers (47, 48, 49, 50, 51). Suffice it to say that no reproducible systemic advantages for the use of chirally pure PS ASOs have been identified. The issue of chirality will be further developed as the results of recent clinical studies with chirally pure PS ASOs are discussed a bit later in this review. Because the sulfur atom is twice as large as the oxygen atom, the charge distribution, bond angles, and stretching of PS links differ substantially from PO linkages (41, 52). Put simply, the sulfur substitution spreads the charge and makes the phosphate more “lipophilic,” thereby facilitating binding to proteins. As a general rule, for proteins that require PS moieties to bind ASOs, the minimum number of this modification needed to support meaningful protein interactions is 10. In ASOs that contain both PS and PO moieties, the placement of PS units plays an important role, but, to date, no general rules have emerged, although PS placed at 5ʹ end of ASOs tends to bind more proteins and have higher affinities for key proteins (53, 54). The enhanced protein binding enabled by PS substitutions is critical as protein binding of ss PS ASOs plays crucial roles in the absorption, distribution, cellular uptake, intracellular distribution, activity, and toxicity of PS ASOs. This topic will be discussed in detail in a later section.
Appropriate modifications of the 2ʹ-position in the ribose result in “pre-organization” of the RNA-PS ASO heteroduplex and thus enhance the affinity for cognate sequences in target RNAs. Commonly used 2ʹ modifications are listed in Figure 2B. This in turn increases the potency of PS ASOs, reducing the doses required to produce pharmacological effects (3, 9). Some 2ʹ modifications, e.g., 2ʹ-MOE, also substantially reduce proinflammatory effects (9, 55).
More recently, focus has shifted to target delivery to specific tissues and cells. To date, the major success is the conjugation of GalNAc moieties to PS ASOs and siRNAs to enhance productive delivery to hepatocytes (1, 3, 4, 5, 10, 35). On average, this modification increases the potency of PS ASO with 2ʹ-MOE or cEt modifications 15- to 30-fold in humans (56). More recently, encouraging data have been published showing that a GLP-1 peptide conjugated to PS ASOs effectively delivered PS ASOs for the first time to beta cells of the pancreas (57). Work continues to evaluate other ligands to enhance productive delivery of PS ASOs and siRNAs to other tissues and cells (58, 59, 60).
Architectural features of ASOs
ASOs are oligomeric and comprised of nucleotide analogs. Because ASOs can be designed to work through a variety of post-RNA-binding mechanisms, numerous designs have been evaluated. As new molecular mechanisms of action are identified and new insights into the molecular mechanisms of distribution, cellular uptake and subcellular distributions, and various toxicities are reported, the designs are becoming progressively more complex. For example, ASOs of various motifs and compositions including various 2ʹ modifications, pendant groups, 5ʹ and 3ʹ termini and ever, more frequently, select modifications of specific nucleotides are being incorporated (36, 61).
Length
A question that was answered early in the development of antisense technology was: what is the optimal length of an ASO? Theoretical calculations suggested that approximately 16 nucleotides should result in sufficient affinity to bind a cognate sequence and sufficient specificity for an ASO to affect a single transcript (33). The theoretical calculations were confirmed experimentally (24). Affinity and specificity vary, as a function of not only the length of the ASO but also the affinity per nucleotide for the cognate sequence. Thus, as new higher-affinity modifications were introduced, the length required to achieve binding to a target RNA and induce pharmacological effects was reduced. In fact, a 2ʹ-MOE chimeric PS ASO of 12 nucleotides was shown to effectively reduce a target expressed in renal proximal convolutes tubules, sodium-glucose cotransmitter 2 (SGLT2), in multiple species and to induce changes in glomerular filtration rate (GFR) in man (62, 63). However, the specificity for the cognate sequence versus all other RNAs varies inversely with length to at least 18 to 20 nucleotides. Theoretical analyses and experimental results have shown that higher-affinity PS ASOs have optimal affinity and specificity between 16 and 18 nucleotides (64). The propensity of PS ASOs to form self-structures that inhibit binding to cognate sequences and other effects increases when ASO length exceeds 20 to 22 nucleotides, resulting in current approaches being generally limited to 16 to 20 nucleotides (1, 33).
Designs of ASOs
PS ASOs designed to bind to sites involved in the processing, translation, or degradation of target RNAs are typically fully 2ʹ modified (Fig. 2C). The 2ʹ modifications are used to enhance affinity for the target RNA and increase stability against nucleases, the enzymes that degrade PS ASOs (PMOs are used to alter splicing as well), and to avoid creating a heteroduplex that is a substrate for enzymes that degrade the RNA in a DNA/RNA-like duplex (RNase H1) or enzymes that degrade the RNA in an RNA/RNA-like duplex (AGO2) (25, 26, 27). Any 2ʹ modification or neutral backbone at the nucleotides involved the enzymatic mechanism for either RNase H1 or AGO2 inhibits those enzymes (13, 65). Though, at first blush, designing ASOs for occupancy-only mediated mechanisms of action is straightforward, as new occupancy-only mechanisms are discovered, PS ASO designs have become more complex. For example, the affinity of an ASO designed to enhance the translation of a protein by binding to an upstream reading frame (uORF) translation initiation codon or a translation inhibitory element (TIE) must be “fine-tuned” so that it prevents the translation initiation complex from binding effectively to the upstream translation initiation site or to bind to and disrupt the structure of a TIE, yet be removed by helicases such that translation read through can occur (7, 8). Therefore, these ASOs may include fewer PS moieties, lower-affinity 2ʹ modifications such as 2ʹ methoxy and at the 3ʹ terminus, higher-affinity nucleotides such as cEts. In contrast to PS ASOs that are designed to bind to uORFs and TIEs, which are located in the 5ʹ UTR of mRNAs, to exploit the No-Go Decay pathway, PS ASOs must be fully 2ʹ modified and of high affinity, but must bind in the coding region of the RNA (6).
In contrast, PS ASOs designed to cause degradation of target RNAs by creating an ASO-RNA duplex that is a substrate for a nuclease (as opposed to a complex pathway such as the No-Go Decay pathway) must meet the specific requirements imposed by the enzyme. RNase H1 requires a minimum of deoxynucleotides and charged phosphate analogs in the region of the ASO that participates in the enzymatic mechanism. Since the enzyme behaves essentially as a pair of calipers, the entire enzyme that measures the number of nucleotides per helical turn (this is defined by whether the duplex is DNA/DNA, RNA/RNA, or DNA/RNA-like) and the catalytic domain measures the width of the minor groove and senses whether the phosphate and 2ʹ OH contributed by the ASO are present (65). PS ASO must be designed to meet the enzymes requirements, and this has led to the “gapmer” design in which various 2ʹ and phosphate modifications can be used in the “wings,” but only 2ʹ OH and PS or PO moieties are tolerated in the portion of the ASO (the central gap) that participates in the catalytic mechanism of RNase H1. Thus “gapmers” can tolerate a wide range of modifications in the wing and modifications at specific nucleotides in the gap can be employed to make the cleavage selective for an even more specific site or mutation and to alter interactions with proteins to enhance the therapeutic index of PS ASOs (36, 61, 66). Designing ss PS ASOs to create heteroduplexes that are substrates for AGO2 is much more complex because of the structural requirements of AGO2 and components of the loading complex, specifically Dicer. In the end, by alternating 2ʹF and 2ʹOMe nucleosides, placing PS modifications only at sites tolerated by AGO2 and employing a 3ʹ adenosine dinucleotide to optimize interactions with Dicer and a stable phosphate analog, 5ʹ vinyl (E) phosphonamidite to bind to the phosphate binding pocket of AGO2 were ss ASOs active in vivo (67). Once again, as new insights accumulate, the designs of ASOs to exploit various mechanisms of action become more complex and contain ever more site-specific modifications and cell and tissue targeting ligands (1).
Properties of ASOs currently being evaluated
PMO ASOs
Pharmacokinetics
PMOs are readily absorbed after subcutaneous (SQ) administration, but to demonstrate even a modest pharmacological effect in the clinic, these two agents are dosed weekly at 30 to 80 mg/kg, making SQ dosing unattractive. All three approved drugs are administered as IV infusions. PMOs are readily absorbed after SQ dosing (t1/2 absorption: 30–120 min). Though they do not bind plasma proteins (or other proteins), they have a plasma elimination half-life of approximately 2 to 15 h (68, 69). PMOs distribute to peripheral tissues, notably skeletal muscle, but detailed analyses of tissue concentrations have not been published. They have been reported to have tissue elimination half-lives of 7 to 14 days, and the main route of elimination is as intact molecules in urine (45, 69).
Pharmacodynamics
Because the internucleotide linkage in PMO ASOs is neutral and RNase H1 requires a phosphate or charged phosphate analog at internucleotide linkages near the site of cleavage of RNA and because RNase H1 requires an RNA/DNA-like helical conformation and minor groove dimensions, PMOs do not support RNase H1 cleavage (45). Though to date no studies of PMO ASOs or PMO-containing siRNAs have been published, given the structural requirements to support AGO2 cleavage, it is highly unlikely that PMOs would support AGO2 cleavage (13). Thus, in principle, PMO ASOs can induce pharmacological effects only via occupancy-only mechanisms such as a steric block of translation or alterations in splicing (45). To date, the only mechanism to display consistent activity in mammalian cells has been the alteration of splicing (45). In fact, the only transcript to be shown to be amenable to shifts in splicing due to PMOs is dystrophin. The sole tissue in which consistent pharmacological effects have been demonstrated is skeletal muscle, and the only disease to show even modest consistent benefit is DMD. Thus, in mammalian systems, PMO ASOs have demonstrated very limited applicability.
Toxicology
In both animals and humans, PMO ASOs have been shown to have excellent safety profiles.
Eteplirsen
To date, three PMO ASOs have been approved for commercial use in a major market (Table 1). All three of these PMO ASOs target the dystrophin transcript, work via the same mechanism, have similar pharmacokinetics and potencies, and are used to treat the same disease, DMD (Table 1). Consequently, we will focus on eteplirsen, perhaps the best characterized of the three provides a solid perspective on the performance of PMOs in the clinic.
DMD is an X-linked genetic disease caused by mutations in the dystrophin gene, a large gene with multiple introns and in which a wide range of mutations have been observed. Dystrophin is an essential component of the sarcolemma. Thus, all skeletal muscles are adversely affected by loss-of-function mutations in dystrophin. The disease typically is manifested around ages 3 to 5 and progresses where patients are confined to wheelchairs as teenagers often leading to death in the 20s. As all skeletal muscles are affected, pulmonary function also declines with age.
Given the observations that some truncation mutations in the dystrophin gene result in partially functional truncated protein, the PMO strategy is to cause skipping of an exon that results in a coding sequence that can be translated into a partially functional partial protein. About 14% of DMD patients have mutations that are amenable to exon 51 skipping resulting in a functional partial coding sequence and partial protein. Eteplirsen is designed to cause exon 51 skipping. It has been evaluated in several clinical trials in ambulatory DMD patients typically at 30 to 50 mg/kg weekly as an IV infusion. In shorter-term trials, evidence of increased dystrophin levels was achieved, but little evidence of clinical benefit was observed. However, longer-term studies have confirmed the increase in dystrophin protein and modest clinical benefit. In ambulatory patients, a 6-min walk test is usually performed. Upper limb strength is also evaluated and is the main efficacy measure in nonambulatory patients. Though eteplirsen resulted in a modest reduction in the rate of loss in skeletal muscle performance in a 6-min walk test, the differences were statistically significant and even in patients who became nonambulatory, upper limb strength and cardiac performance were stabilized (70). In other studies, eteplirsen also reduced the progression in loss of lung function (71). Thus, for a select group of DMD patients, this drug appears to be safe and effective therapy. The approval of eteplirsen was controversial because the phase 3 trial failed to provide statistically compelling evidence of efficacy. Furthermore, the FDA required that the manufacturer note that production of functional protein has not been proven in the label. Only a small fraction of DMD patients have this specific mutation.
Impact of the PS modification on antisense technology
With the single exception of PMO drugs, all currently employed ASOs and all chemically modified siRNAs contain at least some PS linkages and, in many cases, PS moieties at all internucleotide links (1, 5). The importance of PS modifications to RTDs cannot be overstated for without this modification, it is unlikely that RTD technology would exist today. Fortunately, as a result of progress reported in the past few years, we now have detailed molecular mechanisms to explain the effects of PS modified RTDs on their interactions in biological systems (3, 5). These will be discussed in subsequent sections of this review. In this section, we will describe the properties of the various chemical classes of PS ASOs.
Chiral PS ASOs
The substitution of sulfur for a nonbridging oxygen in a phosphate group creates a chiral center at every PS linkage (referred to as Sp and Rp), so in a fully phosphorylated 20 mer compound, there are 219 Rp or Sp stereoisomers. Quite naturally, significant efforts have been invested in developing stereochemical synthetic schemes and in the evaluation of the effect of chirality on the behavior of PS ASOs in biological systems (72, 73, 74). It is thought that the increased nuclease resistance of chirally impure PS ASOs is due to the sulfur displacing the divalent cation that most nucleases use as a part of the enzymatic mechanism from the catalytic center of the enzymes. Alternatively, the sulfur may be less able to take on a negative charge during the reaction (72). In either case, it is sensible to assume that the Rp and Sp stereoisomers might differ in their sensitivity to nucleases, and indeed, that is the case. The Sp isomer is more nuclease-resistant than the Rp isomer (47). So, an Sp chirally pure PS ASO is more stable to nucleases than the Rp chirally pure ASO. Similarly, it is logical to assume that the stereoisomers might differ in their effects on hybridization of the PS ASO and the cognate RNA sequence, and once again the assumption is correct. Unfortunately, and not surprisingly, the Rp stereoisomer results in improved affinity per Rp linkage compared with the Sp or a diastereomeric chiral mixture. Thus, what is “seen” as the more “natural” nucleic acid hybridizes to the cognate sequence with higher affinity and is readily degraded by nucleases. This means that a chirally impure diastereomeric PS ASO might bind to the target RNA with somewhat lower affinity than natural DNA and be more nuclease-resistant, and that is what is observed.
Many PS ASOs are designed to activate RNase H1 and therefore are chimeric with wings comprised of 2ʹ modified sugars and a PS oligodeoxynucleotide gap (25). Since PS ASOs interact with RNase H1 at both the hybrid binding domain and in the catalytic center and the enzyme displays sequence preferences for cleavage, one would expect that the overall effects of chiral purity on activity and toxicity might be complex, and indeed they are. But the bottom line is this: thorough studies to evaluate the overall impact of chiral purity on activity and toxicity have shown that chiral purity or chiral purity at individual PS units provides no meaningful improvement in therapeutic index and overall performance of PS ASOs (48). Clinical results to date appear to reflect that theoretical analysis.
Huntington’s disease (HD)
WVE-120101 and WVE-120102 are chirally pure PS 2ʹ methoxy gapmer ASOs that target the mutant forms of Huntingtin (HTT), rs362307 and rs362331, respectively, which are currently under evaluation in two separate phase 1 b/2a studies (75).
WVE-120102 was administered intrathecally at doses of 2 to 16 mg every 2 months to patients with HD. An interim analysis was presented. No safety or tolerability data were shown, but the drug was reported to be safe and well tolerated. A comparison of all patients treated at any dose level showed a 12.4% reduction in mutant HTT protein in the CSF, achieving a p-value <0.05, and an analysis of the effects on HTT protein at each dose level suggested a dose–response relationship. The study is now enrolling HD patients in the 32 mg every 2-week cohort (76).
Duchenne’s muscular dystrophy
Suvodirsen (WVE-210201), a fully 2ʹ methoxy, 2ʹ-F modified PS ASO with chirally pure PS moieties at selected sites designed to increase skipping of exon 51, was evaluated at doses of 0.5 to 10 mg/kg in a multidose 12-week study. While safe and well tolerated, the drug did not show increases in dystrophin protein in muscle in the phase 1 open-label extension with weekly IV doses of 3.5 or 5.0 mg/kg. Nor was there evidence of effects at any dose of any parameter tested (77). Development of this PS ASO was subsequently terminated (78).
2ʹ-O-methoxyethyl (2ʹ-MOE) PS ASOs
2ʹ-MOE is one of several thousand 2ʹ modifications of the furanose evaluated (9). Obviously 2ʹ-MOE is substantially bulkier than hydrogen and is more hydrophilic and thus more hydrophilic. Because it fixes the furanose in a favorable conformation in effect “pre-organizing the duplex,” 2ʹ-MOE increases the Tm per nucleotide by more than 2 degrees. In a typical chimeric ASO designed to serve as a substrate for RNase H1, there is a central PS deoxynucleotide center of 8 to 10 nucleotides flanked by PS 2ʹ modified nucleotides. Thus, a typical “gapmer” ASO has 8 to 10 2ʹ-MOE nucleotides resulting an increase in affinity for cognate RNA sequences of several orders of magnitude (79). This, in turn translates into a 10- to 20-fold increase in potency compared with PS oligodeoxynucleotides in animal models. Because it is not clear that PS oligonucleotides administered systemically in humans resulted in antisense-mediated pharmacological effects, direct comparison with PS 2ʹ-MOE ASOs is difficult, but a 50- to 100-fold increase in potency in humans is a reasonable estimate. That the increase in affinity for the target RNA is not fully reflected in the increase in potency compared with PS oligodeoxynucleotides is explained by several factors. In a “gapmer” ASO, the potential substrate for RNase H1 is reduced to 8 to 10 nucleotides. Human RNase H1 requires at least four contiguous deoxynucleotides to cleave RNA in an ASO–RNA duplex, and enzyme activity is not optimal until about eight contiguous deoxynucleotides are present. Moreover, human RNase H1 (and all mammalian RNase H1 enzymes) displays sequence preferences. Therefore, the efficiency of the enzyme is substantially reduced (65).
The 2ʹ-MOE moiety provides additional significant advantages. It substantially increases resistance to nucleases, the enzymes that degrade PS ASOs. This is a result of the steric bulk of the substitution and its hydrophilic character (79). This translates into an increase in tissue elimination half-life from about 48 h to 2 to 4 weeks facilitating weekly to monthly systemic administration (79, 80). Additionally, 2ʹ-MOE substitution significantly reduces proinflammatory effects (55, 72). The molecular mechanisms responsible for the reduced inflammatory effects are not fully understood, but probably related to changes in interactions with toll-like receptors (TLRs), particularly TLR 9 (72, 79).
PS 2ʹ-MOE ASOs are, by far, the best characterized of any RTD chemical or structural class. More than 10,000 patients in clinical trials have been treated at doses that range from 50 mg to 1200 mg/weekly and treatment durations ranging from 6 weeks to >5 years. In the integrated safety database for 2ʹ-MOE ASOs, there are data from more than 8000 patients treated in randomized, placebo-controlled trials (56, 81, 82). Systemic delivery of PS 2ʹ-MOE ASOs is achieved after IV, SQ, and IM administration. Mipomersen, a PS 2ʹ-MOE gapmer ASO designed to reduce apoB-100, the key structural protein of LDL cholesterol, has also been administered orally to several species in enteric-coated (to prevent acid-dependent precipitation and depurination in the stomach) formulations containing decanoic acid (C10) as a penetration enhancer. In humans, approximately 6% oral bioavailability and statistically significant reductions of plasma apoB-100 and LDL cholesterol were observed in normal volunteers (83). The total dose and number of capsules required meant that this formulation was not appropriate for commercial use. However, the study provided important proof of principle and suggested that with more potent ASOs, commercially attractive oral dosing might be achievable. Administration for local therapy has been achieved after intravitreal (IVT), aerosol, intradermal, intrathecal (IT), and rectal administration (3).
The pharmacokinetics of PS MOE ASOs have been well studied in multiple species, including humans. After SQ or IM administration, these agents are rapidly and completely absorbed (t1/2 absorption approximately 60 min). They are protein bound in plasma (>90% at doses of 300 mg/week in man), and the main proteins in plasma to which they bind have been identified (5, 84). Albumin is the major protein that binds to the ASOs with a kd of approximately 13 μM, but they do not bind to the major drug-binding sites in that protein. They are then rapidly distributed to peripheral tissues with the liver, kidney, fat cells, and spleen accumulating the most drug at low doses. At higher doses, those tissues saturate and distribution to secondary tissues occurs. The suborgan distribution has also been characterized in the liver and kidney after systemic administration and the CNS and lung after local administration. They are cleared slowly from peripheral tissues with elimination half-lives of 2 to 4 weeks (3, 4, 72, 80). Elimination after IT administration is limited by the flow of CSF and supports quarterly or less frequent dosing. Clearance from the lung after aerosol delivery is similar to other tissues after systemic administration and therefore, weekly aerosol dosing is typical. Systemic bioavailability after either IT or aerosol administration is limited and since the doses by either route are quite low, little systemic exposure results from ASOs delivered by either route. Elimination is due to endo- and exonuclease digestion with clearance of metabolites (oligonucleotide fragments) in urine (3, 4, 72, 80)
After SQ administration, the ED50 for targets expressed in hepatocytes is about 150 mg/week in man. Somewhat higher doses are required for other tissues and doses correlate well with the distribution of PS 2ʹ-MOE ASOs. Dose–response curves are log-linear (79). Thus, these agents are straightforward to administer, and different PS ASOs of class behave quite similarly, which facilitates dose selection and development (79, 80). The safety profile is very well understood after administration by all routes of delivery (37, 81, 82).
All PS ASOs result in peak plasma-related transient inhibition of clotting, which has proven of no clinical significance. In nonhuman primates (NHP), they also result in peak plasma-related activation of the alternative complement cascade, but in humans this, once again, has not been an issue. All PS ASOs result in transient clinically insignificant reduction in platelet count of about 20 to 30%, which is also peak plasma concentration dependent. The most common adverse event in humans is injection site reactions (ISRs) after SQ administration that are dose-dependent and typically mild, but can be associated with discontinuation of treatment. At a dose of 300 mg/week, two PS 2ʹ-MOE ASOs, volanesorsen and inotersen, are associated with clinically significant thrombocytopenia in a few patients suffering from familial chylomicronemia (FCS) (85) and transthyretin amyloidosis (TTR amyloidosis). In TTR patients, the mechanism of thrombocytopenia appears to be related to ASO-dependent antiplatelet antibodies (86).
PS 2ʹ S-constrained ethyl (cEt)
PS 2ʹ S-constrained ethyl (cEts), also referred to as generation 2.5 PS ASOs, contain a bicyclic sugar that constrains the furanose surrogate to adopt a conformation favorable to hybridization to RNA. Therefore, the affinity per nucleotide for RNA is increased by approximately 2º in thermal melting experiments compared with PS 2ʹ-MOE ASOs. cEts are members of the bicyclic sugar modification class, several of which have been evaluated preclinically and in the clinic. Another 2ʹ bicyclic sugar, locked nucleic acid (LNA) results in even greater affinity per nucleotide than the cEt modification (9, 87). The increase in affinity per nucleotide of the cEt modification leads to an approximately tenfold increase in potency. In addition to the obvious benefit of being able to employ lower doses in tissues that accumulate PS ASOs at low doses, in principle an increase in potency might enhance the pharmacological activity in tissues that accumulate less PS ASOs, such as various types of cancer or skeletal muscle. The likelihood that an increased activity in lower accumulating tissues is enhanced because the pharmacokinetic behaviors of 2ʹ PS MOE, PS 2ʹ-cEt, and PS 2ʹ LNA ASOs are quite similar. Indeed, this supposition has been proven to be true.
Arguably, the best example of using an increase in potency to achieve better pharmacological activity in a tissue in which lower concentrations accumulate is various types of cancer. PS 2ʹ- MOE ASOs have been thoroughly evaluated in multiple cancer types against a variety of targets. At very high doses (>900 mg/week IV), limited pharmacological activity was observed is a few malignant cells, e.g., prostate or lymphoid tissues, but certainly insufficient to justify commercial approval (79). STAT3 2.5 LRx is a PS 2ʹ cEt targeting STAT3, a well-understood target necessary for survival of cancers. Broad phase 2 studies of this drug demonstrated meaningful antitumor activity in several cancers including head and neck (88).
However, the enhanced potency of PS cEt ASOs was associated with a significant increase in both the incidence and severity of cytotoxicity compared with PS 2ʹ-MOE ASOs in multiple cell lines in vitro and hepatotoxicity in mouse, rats, and NHP. PS LNA ASOs are another example of constrained carbohydrate containing ASOs. As mentioned previously, the LNA substitution results in approximately double the increase in Tm per nucleotide and thus was considered quite promising. Screening of PS LNA ASOs demonstrated an increase in the incidence and severity of cytotoxicity and hepatoxicity that was even greater than observed with PS cEt ASOs (9, 87, 89, 90, 91, 92). In humans, PS LNA ASOs caused severe hepatotoxicity, thrombocytopenia, and nephrotoxicity (90, 93). The apparent correlation between affinity increases beyond that provided by the 2ʹ-MOE substitution resulted in speculation that the toxicities of very high affinity PS ASOs is due to off-target hybridization and RNase H1 cleavage. Cleavage of very large transcripts was thought to be a principle driver of the toxicities because in very long transcripts, there is a higher probability of sequences that are near or perfect matches to the cognate sequence for which the ASO was designed (89). When RNase H1 knockout mice were created and studied, the hypothesis was apparently confirmed because the hepatotoxicity of the very high affinity PS ASOs was substantially ameliorated (89, 94). However, additional work that will be discussed defined a step-by-step mechanism to explain the behavior of most toxic PS ASOs that is entirely unrelated to off-target hybridization (36).
PS 2ʹ-MOE GalNAc ASOs
Targeted delivery to hepatocytes has had an important positive impact of PS ss ASOs and an even more profound influence on modified PS containing DS ASOs, siRNAs (38). The conjugation of a GalNAc to PS 2ʹ-MOE ASOs increases the potency of these agents in humans by 15- to 30-fold compared with the parent molecules by enhancing productive delivery of PS MOEASOs to hepatocytes without altering the overall distribution of these drugs to peripheral tissues (1, 11, 56, 95, 96, 97, 98). Fortunately, it has been possible to compare the behavior of GalNAc conjugated PS 2ʹ-MOE ASOs to the behavior of the unconjugated ASO in several clinical trials, including large phase 2 studies (56).
To highlight this increase in potency observed in the clinic, the dose–response curves for target reduction of three liver-derived proteins that are present in plasma are presented and adding a GalNAc targeting ligand consistently increases potency by 15- to 30-fold compared with the parent PS 2ʹ-MOE PS ASO. The increased potency supports 15- to 30-fold lower dosing, which results in reduction to nearly zero events that could affect compliance such as ISRs and fever and chills that occur in a few patients with 200 to 300 mg/week doses of the parent drugs. Moreover, the GalNAc-conjugated PS 2ʹ-MOE ASOs can be administered monthly to quarterly with essentially no safety or tolerability issues.
A comparison of the performance of Factor XIRx, a well-behaved PS MOE ASO that targets a critical hepatocyte-derived clotting factor in the intrinsic coagulation pathway to the behavior of Factor XI-LRx, the same PS MOE ASO, is another example of the power of GalNAc conjugation. The parent PS 2ʹ-MOE ASO has completed several phase 2 studies and is poised to enter phase 3 trials. In a study of 300 patients who underwent voluntary knee replacement, three dose levels of the ASO were studied and compared with the standard of care, heparin and the 300 mg/week dose showed a sevenfold lower incidence of deep vein thrombosis with no significant increase in bleeding (99, 100). A similar profile has been observed in patients with end-stage renal disease. The GalNAc version of the Factor XI PS MOE ASO has advanced to phase 2 studies. These agents represent a substantial advance in the management of thromboembolic disorders, and the discovery of these agents is representative of the power of antisense technology. In mice, multiple liver-derived clotting factors were reduced simultaneously with PS MOE ASOs without evidence of nonspecificity and no adverse events. All of the ASOs were then evaluated in various models of thrombosis, and the Factor XI ASO displayed a clearly more attractive profile than other factors (101).
PS cEt GalNAc ASOs
Recall that PS cEt and PS MOE ASOs share common pharmacokinetic properties, but PS cEt ASOs are approximately ten times more potent. Not surprisingly, conjugating a GalNAc moiety increases the potency for hepatocyte targets by 15- to 30-fold resulting in the most potent chemical class of PS ASOs identified to date. The ED50 of PS cEt ASOs is 1 to 3 mg/week, meaning the total annual dose for a patient is 50 to 150 mg. Obviously then, given that the elimination half-life from tissues is 3 to 4 weeks as discussed previously and the excellent safety profile of the current cET PS Gal-nac ASOs, monthly, quarterly, or semiannual dosing SQ is feasible (1, 3, 4). Though the total clinical experience is less than with PS 2ʹ-MOE GalNAc ASOs, several hundred humans have been treated systemically (SQ) and by aerosol administration. To date, no ISRs and no drug-related adverse events have been observed.
Given the oral bioavailability and oral activity demonstrated by 2ʹ-MOE mipomersen (a PS 2ʹ- MOE ASO) in several species including humans (102, 103), the enormous increase in potency achieved by combining the cEt and GalNAc modifications suggests that a PS 2ʹ cEt GalNAc ASO might be bioavailable at doses and formulations that are commercially attractive. Indeed, that appears to be the case. Formulated essentially as mipomersen, PS cEt GalNAc ASOs to several different targets expressed in hepatocytes were approximately 7 to 10% bioavailable and reduced their specific mRNAs in several animal species (3, 83, 104, 105). One of these ASOs is currently undergoing clinical testing of both the SQ and oral forms of the drug (103, 106).
That the intact PS cEt GalNAc ASO was found in the liver in all species studied is an interesting observation because the GalNAc moiety is coupled to the PS ASO via a cleavable linker suggesting that a clinically meaningful fraction of the intact drug is absorbed from the gut. The observation that significant pharmacological activity was observed in humans at a dose that can be administered in a single capsule or tablet and at this dose suggests that commercially attractive oral administration of this chemical class of PS ASOs is feasible, which would be a major advance in the technology that could expand the breadth of utility of antisense drugs yet again (3, 83, 103).
PS 2ʹ F ASOs and siRNAs
A fluoro (F) substitution at the 2ʹ deoxy position is another modification that has been studied. It is rarely used in PS ASOs, but is still frequently substituted at a number of nucleotide positions in both the sense and antisense strands of siRNAs and is present at some level in almost all GalNAc-conjugated siRNAs (1, 10, 35, 43, 107). It is, of course, electron withdrawing and hydrophobic. It increases the Tm per nucleotide by approximately 2º, but does not impart any resistance to degradation by nucleases (9). Several properties of 2ʹ F nucleosides and 2ʹ F containing oligonucleotides have generated concern. The ultimate metabolic product of any oligonucleotide is a nucleotide, which may be further metabolized by phosphatases. 2ʹ F nucleotides and nucleosides are substrates for the enzymes of the nucleotide salvage pathway and therefore are incorporated into DNA and RNA and inhibitory of both DNA and RNA polymerases (5, 108). 2ʹ F nucleoside analogs have been studied extensively as anticancer and antiviral agents and were found to be quite toxic. Fialuridine (FIAU), a fluoro-containing nucleoside analog that showed promise initially, but in humans resulted in severe hepatotoxicity, mitochondrial dysfunction, metabolic acidosis, and death (104, 109). More recently, PS and 2ʹ F containing ASOs were shown to be hepatotoxic due to binding avidly to and causing the cellular degradation of several paraspeckle proteins including P54nrb (Non-POU domain-containing octamer-binding protein) and PSF (splicing factor proline and glutamine rich), which resulted in P53 activation, necrosis, and apoptosis in vitro and in mouse liver (110, 111). This issue is an important concern because most currently used siRNAs contain 2ʹ F nucleotides, and several have been terminated from development, including revusiran. Revusiran is GalNAc-conjugated siRNA that contains 2ʹ F, 2ʹ methoxy and PS moieties. It is designed to treat patients with TTR amyloidosis. A phase 3 study of patients with TTR cardiomyopathy was discontinued because treated patients experienced severe hepatotoxicity, metabolic acidosis, and death significantly greater than the placebo group. The concern is exacerbated by the fact that details of all the adverse events including time courses for affected patients, as well as mechanistic studies in those patients have never been published (3, 106, 112). Rather, a superficial report of the study that suggested a minor difference in the age of patients in the revusiran arm of the study might be accountable. However, in a later published mechanistic study in animals, the authors concluded that most likely cause of the adverse events was off-target effects due to the promiscuity of AGO2 (106, 112).
PS 2ʹ LNA ASOs
As discussed earlier, PS 2ʹ LNA ASOs are more toxic in in vitro assays and mouse and NHPs than other 2ʹ modified PS drugs. In the clinic, several different PS 2ʹ LNA ASOs were shown to be toxic with severe thrombocytopenia, severe liver toxicity, and nephrotoxicity being observed. Thus, no clinical trials with ASOs of this chemical class are currently underway. However, efforts to select safe LNA-modified ASOs and to ameliorate the toxicities continue.
PS ASO interactions with proteins
Several years ago, the conceptual basis and practice of antisense technology underwent a tectonic shift occasioned by cumulative data that supported the conclusion that the fate of PS ASOs in biological systems is defined by proteins (5). This means that if a PS ASO is found at a site in a biological system, a protein or proteins are responsible for the PS ASO being there. It also means that all known post-RNA-binding mechanisms are mediated by proteins and often can be influenced by other proteins. Finally, it means that the major mechanism that explains the toxicities of some PS ASOs, irrespective of 2ʹ modification, is driven by unique interactions with paraspeckle proteins and RNase H1 that cause the formation of a unique PS ASO ribonucleoproteins (RNPs) that relocalize to the nucleolus causing nucleolar toxicity and apoptosis (3, 5, 36, 65, 113). Two lines of research, targeted delivery and studies on molecular mechanisms, drove the advances in understanding PS ASO behavior and the shift in focus. The data supporting this important conclusion is substantial, continues to accumulate, and will be discussed as part of the discussions of pharmacokinetics, cell uptake and distribution, mechanisms of pharmacodynamics, and toxicities.
For more than 2 decades, the primary focus was to understand the specifics of PS ASO interactions with RNA. In effect, the field was focused on the nucleic acid language and gratifyingly, today we have a solid working knowledge of the factors that determine the ability of a specific PS ASO of a specific chemical class to bind specifically to its cognate “receptor” sequence (Table 2).We also have a large chemical toolbox (1, 3, 4, 5, 9, 25) such that the primary focus today is to understand the effects that posttranscriptional modifications may have on PS ASO–RNA interactions. However, given the obvious importance of PS ASO interactions with proteins, during the last several years substantial efforts to understand the chemistry of PS ASO–protein interactions have rapidly yielded important and useful insights (5).
Table 2.
Structure activity relationships of PS ASO-RNA and PS-ASO protein interactions
| PS ASO | Protein |
|---|---|
| ✓ Phosphorothioates | ✓ Domains (for key proteins) |
| o Number | ✓ Structures |
| o Placement | ✓ Charge |
| ✓ Charge | ✓ Hydrophobicity |
| ✓ 2′ modifications | ▪ Modifications |
| o Hydrophobicity | ▪ Acylation |
| o Number | ▪ Mesylation |
| o Orientation (5′ or 3′) | ▪ Phosphorylation |
| ▪ Sequence | ▪ Glycosylation |
| ✓ Base modifications | ▪ Lipidation |
| ▪ Pendant groups (conjugates) | ▪ Ubiquitinylation |
✓ More solid knowledge.
▪ Less knowledge.
Several high-yield efficient assays contributed to the rapid progress. A biotin pull-down assay with PS ASOs as bait and a variety of PS ASOs used as competitors has supported rapid identification of the total proteins that bind a specific PS ASO, identifying how different cellular conditions affect PS ASO–protein interactions and preliminary structure–activity relationships (5, 53, 114). A modification of the NanoBRET assay that was originally designed to measure protein interactions that supports the evaluation of PS ASO–protein interactions has been particularly instrumental because it is rapid, works well with isolated proteins or mixtures, does not require a protein-specific antibody (antibodies to nano-luciferase can be used), supports direct or competitive binding assays, is rapid, reproducible, and consistent with other binding assays. This procedure even supports estimating the region in a protein that binds and the orientation of the oligonucleotide (5, 115, 116). A modified NanoBiT assay has allowed the evaluation of the binding of PS ASOs in live cells, the characterization of the RNP complexes that some PS ASOs induce, and the effects of PS ASOs on the proteins with which they interact (36, 117). The Bio-ID assay has played an important role in understanding the compositions of various PS ASO-induced protein–protein and RNP complexes (L. Zhang et al., unpublished).
Interactions with proteins in blood
PS ASOs of all chemical classes interact with plasma proteins and proteins on the surface of cells resident in plasma, including platelets (72, 80). The interactions with plasma proteins play critical roles in the pharmacokinetics of PS ASOs as well as toxicities. The plasma proteins that bind PS 2ʹ-MOE ASOs have been well characterized (Table 3) (84), and it is likely that other PS chemical classes experience similar interactions, but to date, these have not been evaluated. Substantial binding to plasma proteins requires a minimum of 10 to 12 PS moieties. Since PS ASOs are water-soluble, 6 to 7 kDa molecular weight molecules, were they not to bind to plasma proteins, they would be cleared by glomerular filtration. At therapeutic doses, >90% of circulating PS ASO is bound to plasma proteins. Albumin is the main repository for these compounds in blood, but PS ASOs also interact with higher affinity with many other plasma proteins. Although PS ASOs do bind to sites bound by small-molecule drugs on albumin, no albumin binding related drug–drug interactions have been observed (72, 80).
Table 3.
Plasma proteins that interact with PS ASOs
| Protein | pI | Plasma Conc. (μM) | Kd (μM) | Function |
|---|---|---|---|---|
| Serum Albumin | 4.8 | 600 | 12.7 | Carrier of acidic/neutral/lipophilic molecules, control of osmolality |
| IgG | 6.6–7.2 | 75 | 1.6 | Antibody, humoral immunity |
| Apolipoprotein A-I | 5.6 | 40 | 5.3 | Lipoparticle, heparan binding protein (HBP) |
| Apolipoprotein A-II | 5.7 | 24 | >500 | Lipoparticle, HBP |
| Complement Factor C3 | 6.0 | 20 | 0.5 | Innate immunity, HBP |
| Transferrin | 5.6 | 12 | 7.0 | Iron transport |
| α-1 Antitrypsin | 4 | 11 | >100 | Protease inhibitor |
| Haptoglobin | 5.6 | 11 | 54.7 | Free plasma Hb binding |
| Hemopexin | 8.5 | 9.9 | 13.9 | Heme scavenger |
| Fibrinogen | 5.5 | 9 | 0.87 | Blood clotting, HBP |
| α-2-Macroglobulin | 6.5 | 6 | 0.05 | Protease inhibitor |
| Prealbumin/TTR | 4.6 | 6 | 132 | Transport of thyroxin and retinol |
| Antithrombin III | 4.9–5.3 | 3.5 | 8.7 | Inactivates coagulation enzymes, HBP |
| α-1-Antichymotrypsin | 6.0 | 3.3 | 21.3 | Serine protease inhibitor |
| β-2-Glycoprotein | 9.1 | 2.7 | 57.1 | Binds cardiolipin, anticoagulant |
| Ceruloplasmin | 5.4 | 2 | 22.6 | Copper carrier protein |
| α-1 Acid Glycoprotein | 2.7 | 1.7 | >500 | Carrier of basic/neutral lipophilic molecules, acute-phase protein (APP) |
| Complement Component C1q | NA | 1.6 | 3.4 | Immunoglobulin binding protein |
| Complement Factor C4 | 6.6 | 1.4 | 0.43 | Innate immunity, HBP |
| Histidine-rich glycoprotein | 6.5 | 1.3 | 0.009 | Immunity, vascularization, coagulation, HBP, APP |
| Plasminogen | 6.2 | 1.2 | 2.1 | Dissolves fibrin blood clots |
| Fibronectin | 5.5–6.0 | 0.9 | 0.54 | Cell adhesion, hemostasis, HBP |
| ApoB100 | 6.6 | 0.7 | >10 | Lipoparticle, HBP |
| Factor H | 6.2 | 0.6 | 0.5 | Regulates alternative complement pathway |
| Apolipoprotein E | 5.4 | 0.5 | 0.027 | Chylomicrons, lipoparticle, HBP |
| Factor V | 5.7 | 0.02 | 0.032 | Coagulation, cofactor |
Adapted from ref (84).
Model proteins
Table 4 provides a full list of cellular proteins with which PS ASOs interact. Table 4A lists the membrane-localized proteins while Table 4B summarizes the intercellular proteins with which PS ASOs interact. As expected, many proteins that have nucleic acid binding domains that bind PS ASOs have been identified, but others that do not have canonical nucleic acid binding domains bind PS ASOs as well. This table also demonstrates that interactions with proteins can affect essential every facet of PS ASO behavior. Given the number and diversity of the proteins that bind PS ASOs, a few were selected for detailed characterization (Table 5 & Fig. 3A). All proteins selected were known to bind to PS ASOs. Other considerations were important, including the characteristics of the domains expressed, the number of sites for PS ASO binding, the effects of the protein on the pharmacological activity, the subcellular localization or PS ASO toxicity, and the subcellular localization of the protein. PC4 was of particular interest because a crystal structure of the protein complexed with a PO oligodeoxynucleotide has been published (118, 119, 120). While the other model proteins are important, substantial advances in characterizing the biochemistry of PS ASOs have been reported, and the overall progress has been summarized recently (5). Consequently, in this review, we will simply discuss the general principles that have been identified.
-
•
PS ASOs bind to many more proteins with greater affinity than PO oligonucleotides.
-
•
The principal reasons for the enhanced interactions of PS ASOs with proteins are the relatively greater lipophilicity of the PS moiety compared with PO and the size of the sulfur, which increase the area over which the negative charge is spread.
-
•
Though PS ASOs interact extensively with nucleic acid domains, they also bind to proteins that have no canonical nucleic acid domains. They also bind to non–nucleic acid binding domains in proteins with nucleic acid binding domains.
-
•
The number and placement of PS moieties are influential in PS ASO–protein binding.
-
•
Ionic interactions with cationic amino acid side chains play an important role, but hydrophobic and base-stacking interactions with aromatic amino acids are also prominent and important.
-
•
Many proteins bind preferentially to the 5ʹ pole of both “gapmer ASOs” and fully 2ʹ modified PS ASOs with binding footprints that typically involve 7 to 9 nucleotides. The most likely reason for this is the partial helical character of ss oligonucleotides.
-
•
Though most proteins prefer the 5ʹ pole of PS ASOs, there are a few proteins that are indifferent to the poles and others that prefer the 3ʹ pole.
-
•
The nature of the 2ʹ substitution plays a critical role in total proteins bound and the interactions of many specific proteins and PS ASOs.
-
•
The binding of PS ASOs to many proteins is significantly affected by the sequence of the PS ASO.
-
•
Nucleolin (NCL) is an interesting exception to many of the general observations. It has four nucleic acid binding domains to which PS ASOs bind, but each domain appears to be promiscuous regarding the orientation, 2ʹ modification, or sequence of PS ASOs.
-
•
Though waters of hydration and counterions are present at each internucleotide link, no obvious effects on binding have been identified to date.
-
•
For membrane-localized proteins, the phospholipid composition and stability of the membrane influence PS ASO–protein binding and the cellular uptake and endosomal release of PS ASOs, there is no evidence of direct interactions of PS ASO with membrane phospholipids (121, 122).
Table 4.
Cellular proteins that bind PS ASOs
| A. PS ASO binding proteins localized to plasma membranes | ||
|---|---|---|
| Protein | Effects on ASO activity | Ref. |
| ASGPR | Enhance | (224) |
| EGFR | Enhance | (121) |
| LDLR | N/D | S. Wanget al., unpublished |
| M6PR | Enhance | (164) |
| TLRs | (mediate inflammatory effects) | (225) |
| Stabilin | Enhance | (226) |
| SRB | N/D | S. Wang et al., unpublished |
| Nucleolin | N/D | (227) |
| B. Cellular PS-ASO binding proteins | ||||||
|---|---|---|---|---|---|---|
| Protein | Feature | ASO activity upon protein knockdown | ASO localization | Impact on ASO toxicity | ASO-protein colocalization | References |
| Nucleic acid binding proteins (33) | ||||||
| Ago2 | RNA Binding | Reduced (free uptake) | Not characterized | Yes (P-bodies) | (228) | |
| CArG binding factor | RNA binding | Not characterized | Not characterized | (53) | ||
| DDX21 | RNA binding | Yes | Yes | Yes (toxic ASO) | L. Zhang et al., unpublished | |
| DHX30 | RNA binding | No change | No change | (53) | ||
| EIF2S2 | RNA binding | Not characterized | Not characterized | |||
| eIF4H | RNA binding | No change | No change | |||
| GRSF | RNA binding | No change | No change | |||
| HMGB1 | DNA binding | No change | No change | No | ||
| hnRNP D1Like | RNA binding | Not characterized | Not characterized | |||
| hnRNPA1 | RNA binding | Not characterized | Not characterized | (229) | ||
| hnRNPA2 | RNA binding | Not characterized | Not characterized | (53) | ||
| hnRNPF | RNA binding | Not characterized | Not characterized | |||
| hnRNPH1 | RNA binding | Not characterized | Not characterized | |||
| hnRNPK | RNA binding | Increase | Yes | Yes | (123) | |
| hnRNPQ | RNA binding | No change | No change | (53) | ||
| hnRNPU | RNA binding | Not characterized | Not characterized | |||
| hnRNPUL | RNA binding | Not characterized | Not characterized | |||
| ILF2 | RNA binding | Not characterized | Not characterized | |||
| ILF3 | RNA binding | No change | No change | |||
| KHSRP | RNA binding | No change | No change | |||
| Ku70 | DNA binding | Increase | No change | No | ||
| Ku80 | DNA binding | Increase | No change | No | ||
| La/SSB | RNA binding | Reduce | Yes | No | ||
| NCL | RNA binding | No change | No change | Yes (toxic ASO) | (230) | |
| NPM1 | RNA binding | Reduce | Yes | Yes (toxic ASO) | (53) | |
| P54nrb | RNA/DNA binding | Increase | Yes | Yes | Yes | (123) |
| PC4/Sub1 | DNA binding | Reduce | No change | No | (53) | |
| PSF | RNA/DNA binding | Increase | Yes | Yes | Yes | (123) |
| PSPC1 | RNA binding | Increase | Yes | Yes | (53) | |
| RHA | RNA binding | No change | No change | |||
| RNase H1 | DNA/RNA duplex binding | Reduce | No change | Yes | ||
| RNF163/ZNF9 | DNA binding | No change | No change | |||
| YBX1 protein | RNA binding | No change | Not characterized | Yes | ||
| Chaperone proteins (11) | ||||||
| GRP78/Bip | Chaperone protein | No change | No change | (53) | ||
| HSC70 | Chaperone protein | Reduce | No change | No | X.H. Liang et al., unpublished | |
| HSP90-AA1 | Chaperone protein | Reduce | No change | Yes | No | (54) |
| Hsp90-AB | Chaperone protein | Reduce | No change | Yes | ||
| HSPA1L | Chaperone protein | Not characterized | Not characterized | (53) | ||
| TCP1-alpha | Chaperone protein | Reduce | Not characterized | Yes (LE) | (114) | |
| TCP1-beta | Chaperone protein | Reduce | Not characterized | Yes (PS-body, LE) | ||
| TCP1-delta | Chaperone protein | Not characterized | Not characterized | Yes (LE) | ||
| TCP1-episilon | Chaperone protein | Reduce | Not characterized | Yes (LE) | ||
| TCP1-gamma | Chaperone protein | Not characterized | Not characterized | Yes (LE) | ||
| TCP1-Theta | Chaperone protein | Not characterized | Not characterized | Yes (LE) | ||
| Other proteins (17) | ||||||
| ACLY | Enzyme | No change | No change | (53) | ||
| Albumin | Secreted | Not characterized | Not characterized | (84) | ||
| Annexin A2 | Membrane binding | Reduce | Yes | Yes (LE) | (231) | |
| ATAD3A | Membrane | No change | No change | |||
| FTCD/58K | Enzyme | Reduce | Yes | Yes (LE) | X.H. Liang et al., unpublished | |
| IMP9 | Transport | Reduce | Yes | |||
| JKTPB1 delta 6 | hnNRP-like | Not characterized | Not characterized | (53) | ||
| KCTD12 | Membrane receptor | No change | No change | |||
| LRPPRC | Transport/transcription | No change | No change | |||
| NARS | tRNA synthase | No change | No change | |||
| NDKA | Enzyme | Not characterized | Not characterized | |||
| RAN | Transport | Reduce | Yes | |||
| SHMT2 | Enzyme | Not characterized | Not characterized | |||
| Thymidylate kinase | Enzyme | Not characterized | Not characterized | |||
| VARS | tRNA synthase | Reduce | No change | |||
| β-actin (ACTB) | Structure | Reduce | Not characterized | |||
| β-tubulin (TUBB2C) | Structure | No change | Not characterized | |||
Adapted from ref (38).
Bolded proteins demonstrated to influence ASO activity or localization.
Table 5.
Characteristics of model PS-ASO-binding proteins
| Model proteins | DNA/RNA binding | RBD/DBD | ASO binding | ASO activity | Toxic ASO response | ASO/protein localization |
|---|---|---|---|---|---|---|
| NCL | RNA | 4 | 4 | No | Mislocalize | No |
| FUS | RNA | 1 | 2 | No | Mislocalize | Colocalization |
| SSBP1 | DNA | 1 | 1 | No | N/A | No |
| La | RNA | 2 | 2 | Enhance | No | ASO Nuclear retention |
| PC4 | DNA | 1 | 1 | Enhance | N/A | No |
| P54 | RNA/DNA | 2 | 2 | Inhibit | Mislocalize | Colocalization |
| PSF | RNA | 2 | 2 | Inhibit | Mislocalize | Colocalization |
| H1 | Hybrid | 2 | 2 | Essential | Mislocalize | No (adjacent) |
Figure 3.
Schematic prediction of the domains of model PS-ASO binding proteins.A, protein domains involved in PS-ASO binding. PS-ASO binding domains are induced with red bars. Tighter binding is marked with thicker bars. LaM, La motif; NLS, nuclear localization signal; NOPS, Nona/Paraspeckle domain; pQ, proline and glutamine-rich domain; RGG, arginine/glycine–glycine-rich domain; RRM, RNA recognition motif; ZFD, zinc-finger domain. B, schematic illustration of the relationship of HBD and catalytic domains on a heteroduplex. The HBD and Cat domains act as a caliper to measure approximately one helical turn. This enables the enzyme to bind to any duplex, but discriminate RNA/DNA heteroduplex from RNA/RNA and DNA/DNA duplexes based on slight differences in helical geometry. The catalytic domain measures the width of the minor groove and detects the presence or absence of a 2’OH and phosphate positioned properly. The enzyme is minimally processive, cleaving at most one or two nucleotides 3’ to its initial cleavage site. CAT, catalytic domain; HBD, hybrid binding domain.
PS ASOs affect the fates of many of the proteins with which they interact
The corollary of the statement that proteins determine the fate of PS ASO, that PS ASOs affect the fate of many of the proteins with which they interact, is also true (3, 5). For example, PS 2ʹ F ASOs have been shown to cause rapid cellular degradation of paraspeckle proteins (111), induce the formation of several protein–PS ASO–protein or PS ASO–RNP aggregates in both the cytoplasm and nucleus, and even replace long noncoding RNAs as architectural organizing elements (3, 5, 29, 36, 61, 117, 123). PS ASOs can also cause cellular mislocalization of the proteins and formation of the protein–PS ASO–RNA aggregates (3, 5, 36, 114, 117, 124, 125). The earliest aggregate observed microscopically was called a PS body. These structures were shown to be nuclear and innocuous. Subsequently, the compositions of PS bodies have shown to be quite simple and include the PS ASO and a single protein, TCP1β (53, 114). Under conditions of stress, PS bodies can be observed in the cytoplasm and contain several TCP isoforms. PS ASOs not only localize in P-bodies, cytoplasmic RNP granules, but also in cell lines incapable of forming P-bodies, and can induce P-body-like aggregates (124). PS ASOs can be found in stress granules as well under certain conditions (126). Numerous additional aggregates of paraspeckle proteins, RNAs, PS ASOs, and nonparaspeckle proteins such as RNase H1 have been shown to form in the nucleus and the nucleolus (3, 5, 36, 117). Each aggregate forms at relatively specific concentrations after transfection or electroporation and all display unique kinetics of formation, migration, and dissolution. Remarkably, most of these aggregates are benign, with nucleolar accumulation of toxic PS ASOs, paraspeckle proteins, RNase H1, and other proteins being the exception (3, 4, 5).
Molecular mechanisms of pharmacodynamics
Identification of the optimal sites within a target RNA for PS ASOs to bind
Target RNAs, which are highly structured, are all elements of complex RNPs. The structure of the target RNAs is one of the major factors that defines the activity of an ASO. Because these structures must be invaded by the ASO to allow Watson–Crick hydrogen bonding between the ASO and the cognate sequence (the factors that influence PA ASO activity are discussed in a later section), screening multiple sites in a target RNA is a critical component of antisense drug discovery (64). Of course, target RNAs vary in length and structure, as well as the types of repeated sequences that are present. The longer a transcript, the more likely there will be sequences that are similar enough to the cognate sequence to support ASO binding and RNase H1 cleavage. Therefore, numerous sites throughout the transcript are screened. Thanks to 30 years of RNA site screening experience and insights in the molecular mechanisms resulting in the effects observed, in silico analyses assure that PS ASOs with motifs that may be problematic, such as clusters of guanosines, are not even tested. As a rule, at least several hundred sites are screened, where activity will range from 0 to 90%.
Typically, 20 to 50 PS ASOs will be selected for full dose–response evaluations and assessment of specificity for the target RNA. At Ionis, this process is fully automated and very rapidly results in multiple candidate PS ASOs ready to be tested in animal models (64, 105).
A typical example is the identification of Apo(a)Rx and the GalNAc conjugated form of Apo(a)Rx, pelacarsen. The apo(a) gene is relatively large (13.3 Kb) and more than 2200 PS 2ʹ-MOE gapmer ASOs were screened. Since there are ten kringle domains, and these structures contain repetitive sequences and previous work had shown that PS ASOs that target repetitive cognate sites in transcripts have significantly enhanced potency (127, 128), particular attention was focused on these repeat sequences, and indeed, the most potent PS ASO was targeted to one of those sites that binds to the exon 24 to 25 splice site of the mature human apo(a) transcript (127). The GalNAc form of the PS 2ʹ-MOE targets the same site. The selection of this site demonstrates how facile and automated the basic screening process is as the process consumed a few weeks and very limited resources because of the automation. It also reflects the importance of basic research into the molecular mechanisms of PS ASOs and the interplay between new knowledge and screening processes. Both the parent PS 2ʹ-MOE ASO and the GalNAc version selectively reduced apo(a) mRNA and protein in multiple species, and the GalNAc version was approximately 30-fold more potent in humans. The GalNAc PS 2ʹ-MOE ASO, pelacarsen, has completed a placebo-controlled study in 286 patients who had experienced a prior cardiovascular event, had Lp(a) levels >60 mg/dl with all other cardiovascular risk factors that were well controlled. Several dose levels and schedules were compared after 6 months of treatment and many patients continued to be treated for 1 year. Administration of the ASO resulted in significant, dose-dependent, and prolonged reductions in Lp(a) and oxidized phospholipids. At a monthly dose of 60 mg, 81% of treated patients achieved Lp(a) levels below 50 <mg/dl and at a dose of 20 mg/week (80 mg/month), 98% of treated patients had Lp(a) levels <50 mg/dl, an approximately 80% reduction. These reductions were maintained for multiple weeks. The safety and tolerability were excellent, with approximately 95% compliance and no serious drug-related adverse events. ISRs were highly infrequent (129). Based on these results, pelacarsen is now being evaluated in a cardiovascular outcomes trial in the same type of patients that were evaluated in the phase 2 study.
Phases of antisense drug action
The pharmacodynamic effects of PS ASOs can be divided into three distinct steps; prehybridization or receptor binding, a hybridization, and a posthybridization phase. The kinetics of these processes have been characterized for PS ASOs that activate RNase H1, and it is likely that the kinetics of PS ASOs that use other post-RNA-binding mechanisms are similar (130). As described above, during the prehybridization phase, PS ASOs bind to cell surface proteins, enter early endosomes, and are shuttled to late endosomes, interact with the Golgi system to induce interactions of late endosomes with COP II vesicles, and are released into the cytosol by late endosomes (1, 3, 38). Release of PS ASOs from late endosomes is the rate-limiting step in the cell uptake and distribution process. On average it takes about 60 min to accumulate enough PS ASO in the cytosol and nucleus to hybridize to the target RNA sufficiently to result in pharmacological activity. Every step in this process depends on interactions with specific proteins. Though the process between release from late endosomes and hybridization is not defined, we believe that they are mediated by shuttling of PS ASOs between proteins and that nuclear uptake and exit are active processes (T.A. Vickers et al., unpublished data). Hybridization is slow, taking about 20 min for sufficient target RNA to be bound to recruit RNase H1 and that process takes about 40 min before sufficient target RNA is cleaved to generate a measurable onset of action. Whether PS ASO hybridization is facilitated by proteins or lipid is unknown, but this seems likely. The impact of the PS ASO and RNase H1 cleavage is to double the normal cellular rate of RNA cleavage (130).
Types of post-RNA-binding mechanisms
PS ASOs can be designed to work via an occupancy-only or occupancy-dependent RNA degradation after binding to target RNAs (25) (Table 6). As is apparent in Table 6, it is possible today to design PS ASOs to work via several post-RNA mechanisms that reduce or increase the production of a target protein or reduce a noncoding target RNA. Though translation arrest and other approaches can be used to reduce the production of a target protein, RNase H1, AGO2, or no-go decay-mediated degradation of the pre- or mRNA is more effective (3). However, occupancy-only approaches are required to increase the translation of target proteins. Certainly, the most advanced approach used to increase protein production is altering the splicing of a pre-mRNA. In fact, four ASOs, including nusinersen, that alter splicing have been approved in a major market (Table 1). The pioneering work demonstrating that ASOs can alter splicing was performed using PMO ASOs by Kole and colleagues (26) and studies that suggested that pre-mRNAs that are amenable to alteration of splicing typically are slowly spliced because of weak splice signals (27). Certainly, the best understood, most broadly used ASO that alters splicing is nusinersen.
Table 6.
Post-RNA-binding mechanisms of action of PS ASOs
| Occupancy only |
Occupancy induced degradation | |
|---|---|---|
| Loss of function | Gain of function | |
| 5’ cap inhibition | Altered processing | RNase H1 |
| Translational arrest | uORF utilization | AGO2 |
| Alternate polyadenylation signal utilization | TIE utilization | NMD |
| NMD inhibition | No-Go decay | |
| PTC readthrough | ||
Occupancy-only
The design, discovery, and development of a PS ASO that alters RNA splicing: nusinersen is exemplary of an occupancy only mechanism. For centuries, patients who suffered from SMA simply progressed and died, never having a diagnosis or an incorrect diagnosis, but all that changed when the gene responsible for the disease, survival motor neuron (SMN1), and the mutations responsible for SMA were discovered (131). The mutation is a homozygous loss of segment of the gene that results in full loss of function. The identification of the genetic cause of the disease enabled far better diagnosis of the disease, the development of natural history studies, and better characterization of the various forms of the disease (132). The subsequent discovery of SMN2, a duplicate gene present in only humans, both provided an improved understanding of the molecular basis for variations in the severity of the disease and a potential direct solution to the cause of the disease (133). In this gene, a mutation in exon 7 results in the exclusion of exon 7 in 90% of the transcripts, which leads to a partial SMN protein that is unstable (134). Nusinersen is a PS fully 2ʹ-MOE-modified 18 nucleotide ASO. It binds to a site in the transcript that in the absence of nusinersen binds hnRNP A1/A2, a splicing receptor. Nusinersen prevents the binding of hnRNP A1/A2 and results in efficient inclusion of exon 7 (135). Given the precision of the site targeted by nusinersen, screening activities were obviously limited to a small region of the transcript and various types of 2ʹ modifications or backbones. Nusinersen provides an excellent example of the interplay between genomics, RNA biology, and antisense technology as well as the precision with which PS ASOs can be targeted to sites in RNAs.
Nusinersen was shown to be highly effective in mouse models of SMA when administered intraventricularly (136) and was then shown to be well tolerated in animal toxicity studies that paved the way to clinical evaluations. Clinical trials have demonstrated remarkable efficacy in symptomatic patients with all forms of SMA and excellent safety and tolerability for as long as 7 years of treatment. The treatment of presymptomatic infants has demonstrated even more substantial benefit and, in many patients, growth and development consistent with normal healthy children (137, 138, 139, 140).
Other occupancy-only mechanisms
The number of occupancy-only mechanisms available to PS ASOs continues to increase. Certainly, the first approach studied was translation arrest, which was shown to work and is available (25). Both mechanisms that enhance the translation of specific transcripts and those that result in the reduction of a specific protein are shown in Table 6. For example, PS ASOs designed to bind to sites involved in splicing of introns with weaker splice signals can bind, block the binding of critical splicing factors including hnRNPs H and F, and this leads to reduced splicing and loss of the pre- and mRNA. In fact, using PS gapmer ASOs that form heteroduplexes with target RNAs that are substrates for RNase H1, competition between the two mechanisms was observed (113). Similarly, PS fully 2ʹ-MOE-modified ASOs that bind to polyadenylation signals in the 3ʹ-UTR have been shown to cause a shift in the site of polyadenylation that leads to a less stable pre-mRNA that ultimately reduces expression of the protein (141). More recently, PS fully modified 2ʹ-MOE ASOs that bind to the coding region near the 3’terminus of the open reading frame were shown to reduce target RNAs via no-go decay. Reduction of proteins necessary for no-go decay, PELO and HBS1L, reduced or ablated the reduction of the target RNA (6).
In contrast, it has been shown that it is possible to design PS ASOs that can selectively increase translation of specific proteins. This is a potentially important advance because it provides new opportunities other than altering splicing to increase the levels of specific proteins, in effect using ASOs to be “agonists” rather than the reduction of targets or “antagonist activity” often considered the major use of PS ASOs. These mechanisms once again derive from advances in understanding the biology of RNA. Relatively recent studies suggest that translation of approximately 50% or more of mammalian mRNAs is suppressed by either sequence or structural elements in the 5 ʹ UTRs. The sequence elements are referred to as uORFs and the structural elements are stem-loop structures referred to translation inhibitory elements (TIEs) (7, 8). uORFs are sequence elements that approximate a translation initiation signal and though not all uORFs are actively used, the characteristics, such as the number of uORFs, the position of the uORF in the 5ʹ UTR, and strength of the Kozak sequence can be used to identify uORFs that are likely to be active (8). To block a uORF such that the proper translation initiation signal is used by the polysome, a PS ASO must be fully 2ʹ modified to assure that RNase H1 does not cleave the RNA, have affinity for the uORF sufficient to prevent the initiation complex from forming at that site, and yet the ribosome and associated proteins must be able to remove the PS ASO to “read through” the uORF sequence. Consequently, the affinity of the PS ASO for the target sequence must be “fine-tuned,” and modifications that inhibit helicases are avoided. These factors resulted in the optimal design being an 18-nucleotide PS 2ʹ methoxy with three cEts at the 3ʹ terminus. An interesting example of the importance of uORFs and the therapeutic potential of uORF ASOs is RNase H1. The translation of mammalian RNase H1 is carefully regulated by a uORF (142, 143). Given the vital roles that RNase H1 plays in removing R loops and maintaining mitochondrial health, regulation of translation to control RNase H1 protein level might be important for normal biological processes (94). To identify the optimal site at the uORF the correct length, Tm, and chemistry of the PS ASO, a position and structure activity study was conducted and processes similar to this are required to fine-tune the activity at any functional uORF. The optimally designed PS ASO consistently increases RNase H1 translation such that there was a 2- to 4-fold increase in the total amount of the enzyme in several cell lines and mouse liver. This level of increase was sufficient to enhance the potency of PS gapmer ASOs (7, 8).
TIEs are stem-loop structures present in the 5ʹ UTR of many mRNAs. To create PS ASOs that can interact with TIEs to enhance translation is a more formidable challenge than designing uORF PS ASOs because, by definition, a stem loop has two sides and directionality as well as a loop of varying sequence and size (7). For the TIEs tested, PS fully modified ASOs 16 to 18 nucleotides long that bind to the 3ʹ stem appeared to work most effectively. While these two approaches to enhancing the levels of targeted proteins represent important additions to the opportunities to use PS ASOs, as drug discovery approaches, a key challenge is that most uORFs and TIEs are poorly conserved across species making studies in animal models of disease difficult. Nevertheless, progress in developing PS ASOs that use the mechanisms is occurring.
Occupancy mediated RNA degradation
RNase H1
The RNase H1 post-RNA-binding mechanism of action is certainly the most frequently and most broadly used mechanism of action of PS ASOs. It is also the best understood and is exemplary of the sophistication in understanding the molecular events that underlie the effects observed (25). RNase H1 is a double-strand nuclease that cleaves RNA only in RNA/DNA-like duplexes (65, 144). In humans, two canonical RNase H enzymes are expressed, RNase H1 and H2. The principle function of RNase H2 is to participate in degrading Okazaki fragments, and it is not involved in cleaving RNAs in PS ASO/RNA duplexes because it is extensively chromatin-bound. In cell homogenates, since RNase H2 is at least 20 times as abundant as RNase H1, it is responsible for most of the cleavage of RNA in RNA/ASO duplexes (145).
Mammalian RNase H1 is comprised of a hybrid-RNA-binding domain, a spacer domain, and a catalytic domain (146). The enzyme also has an N-terminal mitochondria leader sequence that is either not translated or proteolytically removed from most RNase H1 molecules in the nucleus and cytosol. The catalytic domain is homologous to E. coli RNase H1. The catalytic site includes a glutamic acid, several aspartic acids, and a lysine located in the highly basic alpha helical region (147). Enzymatic activity is Mg+2-dependent, the pH optimum is 7 to 8, and the enzyme is sensitive to ionic strength, pH, and Mn+2 concentrations. A pair of vicinal cysteines at positions 147 and 148 results in the enzyme being highly sensitive to oxidation and sulfhydryl reagents. The enzyme exhibits multiple pIs because there are multiple phosphorylation sites (148, 149). The hybrid binding domain will bind to any double-stranded nucleic acid duplex but binds 10 to 20 times more avidly to DNA/RNA duplexes. Using two lysine residues and a tryptophan, the hybrid binding domain positions the enzyme precisely on the heteroduplex (147). The hybrid binding domain enhances the affinity for the heteroduplex by reducing the off-rate and thus significantly reduces the Kcat of the enzyme compared with the catalytic domain only or E. coli RNase H1 (65). The enzyme can be considered two calipers. The entire enzyme measures one DNA/RNA helical turn while the catalytic domain assesses the minor grove dimensions and the presence of a 2ʹ OH on the RNA strand (Fig. 3B) (65). Thus, the hybrid binding domain acts in concert to assure that human RNase H1 is highly specific for DNA/RNA duplexes.
Human RNase H1 can discriminate between subtle changes in the helical geometry of a duplex at the catalytic center of the enzyme. Consequently, it displays sequence sensitivity (150, 151) and quite variable cleavage patterns. In some sequences, a single cleavage site is predominant, while in others multiple sites may contribute to the overall cleavage activity (65). No consensus sequence for cleavage has been identified, but from a pharmacological perspective, this property is important in enhancing the specificity of the enzyme, limiting off-target cleavages, and has been used to discriminate between sequences that differ at a single nucleotide (66). The structure of human RNase H1 in toto is the principal factor that limits the value of chiral PS substitutions because the S and R forms of PS have opposite effects on the interactions of substrates with the hybrid binding domain and the catalytic domain (48). The enzyme displays minimal processivity, cleaving only one or two nucleotides beyond the initial cleavage site and can cleave one or two nucleotides in a single-stranded RNA overhang (65). After initial RNase H1 cleavages, RNAs are further degraded via the normal cellular RNA clearance pathways (152) (Fig. 4).
Figure 4.
RNA cleavage fragments by RNase H1 or siRNA are degraded by 5’-3’ exonucleases and 3’-5’ exonucleases. In the nucleus, the 5’ cleaved fragments are further degraded from 3’ end by exosomes and Dis3 and from 5’ end by XRN2 after decapping. The 3’ fragment is further degraded by 5’-3’ exonuclease XRN2. In the cytoplasm, the 5’ fragment is degraded by XRN1 from 5’ end after decapping and by cytoplasmic exosome and Dis3L1 from 3’ end. The 3’ fragment is degraded from 5’ end by XRN1. Adapted from ref (152).
Potential for hybridization-based cleavage of nontargeted RNAs
An important consideration for any process that leads to cleavage of target RNAs is the potential for cleavage of nontarget RNAs. The potential of RNase H1 to cleave off-targets has been thoroughly investigated using the SOD1 minigene system that supports precise control of the main variables that influence the propensity to cleave off-targets in both a cell-free and a cellular assay and compared with the parent human transcript for SOD1 (113, 130, 153). In short, near-perfect complementarity is required, but if there are multiple sites in an RNA with sequences that are near matches for the cognate site, the extent of off-target cleavage can be greater. This is an important asset for the pharmacological use of PS ASOs, but the use of rapid sequencing methods has suggested that some off-target cleavages can occur. Unfortunately, in no study were sufficient dose responses and time courses performed with adequate controls to discriminate between true hybridization-based off-target cleavages and secondary or compensatory changes due to target reduction, low-level cytotoxic effects, or effects secondary to binding to cellular proteins. Such thorough studies are important because the steps to take to reduce such effects are dependent on the mechanism(s) underlying the observations.
Factors that influence the activities of PS ASOs designed to activate RNase H1
The factors that affect the activities of RNase H1-dependent PS ASOs have been determined, as have the rates of the steps in the process after PS ASO entry into cells (Fig. 5A) (113, 128, 130). Except for very rapidly transcribed RNAs, transcription rate does not affect RNase H1-dependent PS ASO activities. Nor does the transcript number up to several thousand copies per cell (154). On the other hand, many factors can affect the activities of these PS ASOs. Certainly, the secondary structures of target RNAs have a significant effect. The binding of the exon junction complex can block access to the target RNA, but the other RNA-binding proteins that have been tested have affinities for RNAs that are insufficient to block access to currently used chemical classes of PS ASOs However, proteins that compete with RNase H1 for the PS ASO/RNA heteroduplex certainly can inhibit access to the heteroduplex by RNase H1 and reduce the activities of these ASOs (28, 53, 113, 155). The level of RNase H1 is another important component. Additionally, since PS ASOs alter the intermediary metabolism of RNAs and the rates of each step in PS ASO-directed RNase H1 activity are slow (minutes to hours), the rates of steps in the processing and translation of RNAs can have significant effects as can the number and positioning of cognate sequences (27, 130, 156).
Figure 5.
Many factors affect the activity of PS-ASOs.A, schematic representation of factors that affect PS-ASO activity. Though the level of RNase H1 and the number of PS-ASO binding sites in an RNA positively correlate with ASO activity, other factors, such as RNA structure, RNA-binding protein, or PS-ASO-binding protein, can inhibit PS-ASO activity. In addition, both splicing and translation can affect the activity of PS ASOs. On the other hand, RNA copy number and RNA half-life do not normally affect ASO activity. B, a feedback transcriptional upregulation can cause tolerance effect of gapmer PS ASOs. Some gapmer ASOs targeting coding region of mRNAs can trigger RNase H1 cleavage of the mRNA, leading to enhanced transcription and increased pre-mRNA levels, in an RNase H1, translation, and UPF3A-dependent manner. This in turn can reduce the activity of PS ASOs in decreasing the mRNA levels. Adapted from ref (157).
Very recently, the first mechanism of tolerance to RNase H1-dependent PS ASOs has been characterized (Fig. 5B). RNase H1-dependent PS ASOs that bind in the coding region of target RNAs can initiate a process that is dependent on RNase H1 cleavage of the target RNA on the polysome and triggers increases in the transcription rates of targeted RNAs that are dependent on the protein, regulator of nonsense transcripts 3A (UPF3A) (157). This mechanism is consistent with the observations that the number of transcripts is immaterial, but that the rates of transcription, processing, nuclear export, translation, and degradation of target RNAs are determinative. It is also consistent with earlier studies that demonstrated that for RNase H1-dependent PS ASOs that bind in introns, the rate at which the intron is spliced is a critical determinant of activity (27). Studies using the SOD1 minigene system demonstrated that splicing rate was a critical factor in defining the activity of intronic RNase H1-dependent PS ASOs and that the presence of multiple cognate sites in a target RNA resulted in increased activity because the rate of RNase H1 cleavage increased (130).
Comparison of AGO2 versus RNase H1-mediated RNA cleavage
The other broadly used RNA cleavage mechanism is AGO2. This is the sole mechanism shown to be available for ds siRNA agents, and it is a mechanism that can be used by properly designed PS ASOs (10, 13, 14, 35, 43, 67, 158). The structural requirements for activity of an ss siRNA ASO are consistent with the properties of AGO2, the most challenging of which was the identification of a stable charged phosphate analog that can bind to the phosphate-binding pocket in AGO2 (67). Thus, it is important to compare the properties of the two mechanisms that are accessible by PS ASOs.
Activities in the nucleus and cytosol
The distribution of RNase H1 and AGO2 differs. RNase H1 is present in the nucleus, cytosol, and mitochondria (65). AGO2, on the other hand, is localized to the cytoplasm (13). Thorough studies have demonstrated that RNase H1-dependent PS ASOs are active in both the cytosol and nucleus, and if one corrects for volume, the activity of these agents in the cytosol is roughly comparable with the activity observed in the nucleus (159). Certainly, there are many examples of PS ASOs that affect RNA processing events that occur in the nucleus, PS ASOs that simultaneously cause degradation of target pre- and mRNA in both the nucleus and cytosol, RNase H1-dependent degradation on the polysome, and enhanced translation induced by PS ASOs and reduction of nuclear retained RNAs (7, 8, 28, 30, 130, 152, 156, 159). Though there have been sporadic reports of AGO2 activity in the nucleus, in our hands we have never confirmed activity for AGO2 ASOs or siRNAs in the subcellular compartment (160). Competing or alternative mechanisms in the nucleus may explain some of the discrepant results. For example, siRNAs targeting some polyadenylation sites resulted in reduction of the target RNA because they blocked the primary polyadenylation site, shifting polyadenylation to an area that resulted in a less stable form of the RNAs (160).
Potency and duration of effect
In principle, the potency of the two mechanisms might differ generically or there might be RNAs that are more sensitive to one mechanism versus the other. In direct head-to-head comparisons of matched PS 2ʹ-MOE gapmers and siRNAs, no general differences in potency were observed and none the PS ASOs and siRNAs showed greater potency than the other for any of the targets studied (160). Obviously, a much larger series of targets in various cell lines and animals would be required to identify target RNAs that may be more sensitive to one of the mechanisms. Equally obviously, the more potent PS ASOs such as cEts and LNAs need to be compared with the optimized designs of siRNAs used today before meaningful conclusions can be drawn.
To study the duration of effect, equi-effective doses must be compared, ideally at the concentrations of each agent that reduce the target RNA by 50% (IC50 doses) as this is the site in a dose–response curve that is most accurate. Theoretically, one might expect significant differences in the duration of effect of RNase H1 and AGO2 agents because, once an antisense strand is loaded into AGO2, the residence time in AGO2 is quite long (13, 35, 43). However, once again head-to-head studies failed to find a significant difference between the mechanisms (160). Frequently claims are made that suppression of a target in the clinic is prolonged using siRNAs, but if one compares equi-effective doses, no obvious differences are observed. Obviously, what is needed is more thorough studies in animals and humans before definitive conclusions can be reached.
Specificity and potential for hybridization-dependent RNase H1 or AGO2-mediated off-target degradation
Though both enzymes have RNase H motifs, they are enzymatically and biologically quite different, and this results in disparate pharmacological properties and behaviors. RNase H1 is remarkably specific for RNA/DNA like duplexes and for specific sequences. It uses the entirety of the information contained in a 20mer PS ASO, is sensitive to the sequence and helical structure of the heteroduplex at the catalytic site (65). These properties result in a very limited propensity for true hybridization- and -RNase H1-dependent off-target cleavage. In contrast, AGO2 is designed to be somewhat promiscuous as it uses just the seven-nucleotide seed region to discriminate target from nontarget RNAs. The propensity for off-target effects due to AGO2 has been confirmed in cells and animals (160, 161). Given this risk, efforts to identify chemical modifications that reduce the potential for off-target cleavage have taken place, but to date no systemic solution has been proven with thorough systematic studies. The question of whether off-target cleavage of RNAs by RNase H1 or AGO2 contributes to clinical toxicities remains poorly understood. Certainly, there have been several siRNAs withdrawn for clinical trials because of side effects, and revusiran is a particularly concerning example. Revusiran is a GalNAc-conjugated siRNA that contains PS, 2ʹ methoxy and 2ʹ F nucleotides. It targets transthyretin (TTR), a protein produced in hepatocytes and exported to peripheral tissue in which it functions. WT or mutants of the protein can precipitate causing TTR amyloidosis (3). In patients with TTR amyloidosis caused cardiomyopathy, revusiran was administered weekly at a dose of 500 mg/week. The phase 3 study was discontinued after 7 months because of a significant imbalance in deaths, metabolic acidosis, and liver toxicity in the treated group. Because revusiran contained 2ʹ F nucleotide and the toxicities were reminiscent of toxicities observed with fialuridine (FIAU), 2ʹ F nucleoside and 2ʹ F siRNAs have been shown to cause toxicities by enhancing the degradation of paraspeckle proteins (109, 111), there has been controversy about the mechanism of toxicity (106, 112). Most recently, the authors concluded that the toxicities were likely due to AGO2-mediated off-target cleavage.
From a pharmacological and therapeutic perspective, irrespective of the causal mechanism, the concern is untoward effects of which true off-target effects are one possible source. SiRNAs (and probably ss siRNAs) can also cause adverse events in other ways. For example, a portion of off-target events have been shown to be mediated by AGO1 (162) or altered polyadenylation (155). Even less predictable and therefore more worrisome are effects secondary to siRNA competition for binding to AGO2 with microRNAs (miRNAs) since AGO2 molecules are typically fully loaded with miRNAs. In fact, alpha tubulin was reduced secondary to dislodging miRNAs from AGO2 that regulate the level of a protease that degrades alpha tubulin (163). Additionally, adverse effects can be mediated by the chemical composition of an siRNA such as 2ʹ F or PS moieties (106, 112).
Taking a similar perspective with PS ASOs off-target cleavage caused adverse events are clearly less of a concern than AGO2-based approaches. However, chemical class toxicities certainly are observed. For example, PS LNA ASOs have resulted in severe thrombocytopenia, nephrotoxicity, hepatotoxicity, and injection site reactions (ISRs) in clinical trials (3), and at doses> 200 mg/week all tested PS ASOs cause moderate self-limited platelet reductions. PS-modified ASOs (and siRNAs) also interact extensively with proteins and as will be discussed later, the major mechanism of toxicity for all tested chemical classes of PS ASOs is nucleolar toxicity mediated by protein interactions (36). Finally, with PS 2ʹ- MOE ASOs dosed at 300 mg/week, evidence of drug–disease interaction results in severe thrombocytopenia in patients with FCS and TTR amyloidosis (3).
In conclusion, it is fair to say that true off-target degradation-mediated effects are clearly of greater concern for ASOs that act via AGO2. Potential competing mechanisms that may result in unexpected effects exist for both RNase H1 and AGO2, chemical class-mediated events exist for both PS ASOs and siRNAs. However, the major mechanisms of toxicity for PS ASOs are now understood and straightforward medicinal chemical solutions to reduce toxicity and maintain potency have been identified (36, 61). On balance, for ss ASOs, RNase H1 is today the preferred mechanism because of its robustness, predictability, and the depth of mechanistic understanding. Nevertheless, work continues to improve the performance of ASOs that work via both mechanisms (3, 4, 5).
No-Go Decay
More recently, PS fully modified 2ʹ-MOE ASOs that bind to the coding region near the 3ʹ terminus of the open reading frame were shown to reduce target RNAs via No-Go Decay. Reduction of proteins necessary for No-Go Decay, PELO, and HBS1L, reduced or ablated the reduction of the target RNA (6) Though the mechanism does not involve the creation of a heteroduplex that is a substrate for specific enzyme, the net pharmacological effect is the same. This mechanism is of interest because fully 2ʹ modified PS ASOs are used and there may be some mRNAs that are more amenable to this mechanism than RNase H1 or AGO2.
Pharmacokinetics of PS ASOs
Molecular mechanisms of cell uptake and intracellular distribution
The mechanisms by which PS ASOs enter and distribute within cells are now reasonably well understood and have been reviewed extensively (1, 4, 5, 38). Consequently, here we will provide only a high-level summary. The first step is simple adsorption to extracellular domains of proteins located in plasma membranes. The process is temperature and energy-independent, and many cell surface proteins that bind PS ASOs or GalNAc-conjugated PS ASOs have been identified, but we suspect that there are others that are involved as well (Fig. 6). The decision about whether the cellular uptake is productive (supports antisense activities) or nonproductive is made at the cell surface and determined by the proteins to which the PS ASO binds. Micropinocytosis has been identified as a major nonproductive pathway. As mentioned, membrane lipid composition and turnover can affect cell uptake and distribution, but to date no specific interactions between PS ASO and the lipid components of membranes have been identified (122). All productive pathways identified to date converge on late endosomes where release of PS ASOs appears to occur. The fraction of PS ASO that is taken up by cells and released is very low (<0.1%) (3, 38, 125, 164). Accumulation of PS ASOs in the nucleus is rapid if the PS ASOs are transfected or electroporated, but requires hours by free uptake. Activity in either the cytoplasm or nucleus correlates with the kinetics of cell uptake and release of PS ASOs (3).
Figure 6.
Mechanisms of PS ASO cellular uptake and release.A, cell surface PS ASO adsorption. Different cell surface proteins, including receptors, can interact with PS ASOs either directly or via ligands conjugated to PS ASOs. Cell surface proteins can direct the internationalization of PS ASOs through clathrin- or caveolin-dependent endocytic pathways or via nonconventional endocytic pathways. B, PS ASOs can enter cells via different endocytic pathways including macropinocytosis. The macropinocytosis pathway may represent a nonproductive pathway for unformulated ASOs. PS ASO internalization via nonconventional endocytic pathways is only conjecture. Internalized PS ASOs normally traffic from early endosomes (EE) to late endosomes (LE) and to lysosomes. PS ASOs must escape from endocytic organelles to be effective. LEs appear to be the major productive release site. Several release pathways may exist, including back-fusion-mediated, membrane leakage, and vesicle-mediated PS ASO release. Released PS ASOs can interact with cellular proteins and enter nucleus.
Routes of administration of PS MOE and PS cEt ASOs and GalNAc conjugates
Ss PS ASOs can be given by all routes of administration, including oral (Table 7) (1, 3). However, the oral delivery reported was not commercially attractive. Recent advances in medicinal chemical modifications including the incorporation of 2’cEts and GalNAc conjugation suggest that commercially attractive oral administration may be possible (103). Ss PS ASOs have also been shown to be effective when administered intrathecally to treat neurological diseases, and aerosol administration is effective for pulmonary delivery (165, 166). SiRNAs may be administered intravenously in cationic lipid delivery systems and when conjugated to GalNAc, subcutaneously (10, 11).
Table 7.
Routes of administration validated for PS ASOs
| A. Local administration | ||
|---|---|---|
| Route | Example | Indication |
| Intradermal | Ionis CTGF ASO | Scarring |
| Intravitreal | Vitravene | CMV retinitis |
| Pulmonary | ENAC-2.5 | Cystic fibrosis (CF) |
| Intrathecal | Nusinersen | Spinal Muscular Atrophy |
| Rectal | Alicaforsen | Pouchitis |
| B. Systemic | ||
|---|---|---|
| Route | Example | Indication |
| Subcutaneous | Inotersen | TTR Amyloidosis |
| Subcutaneous, intramuscular | IONIS HCV ASO | Hepatitis C |
| Subcutaneous, oral | Mipomersen | Homozygous Familial Hypercholesterolemia |
Systemic administration
An important advantage of ss PS MOE and PS cEt ASOs that is often overlooked is that they can be systemically administered and distributed to various organs and cells by all parenteral routes, as well as orally. The only effect of GalNAc conjugation on the pharmacokinetics of ss PS ASOs is that they display a greater productive distribution to hepatocytes. Though the pharmacokinetic properties of PS 2ʹ-MOE ASOs also apply to PS cEt ASOs, it is not yet proven that PS 2ʹ cEt-GalNAc ASOs can be given to humans at doses and in formulations that are attractive and can be administered by multiple routes to achieve local pharmacological effects (3, 4). Therefore, we will briefly describe the routes of administration.
Oral administration
Ss PS ASOs can be administered orally with formulation. The formulations must be enteric coated to avoid acid-based depurination and precipitation in the stomach and must include a penetration enhancer. Penetration enhancers are thought to transiently open the tight junctions in gut epithelium to increase bioavailability (102). In humans, mipomersen (Kynamro) was administered orally daily at a dose of 500 mg and resulted in >6% bioavailability and reduction of apoB-100 and atherogenic lipids (83). Recently, an oral formulation of a PS cEt ASO conjugated with GalNAc showed about 10% oral bioavailability and target reduction in the liver of rats (103).
Targeted delivery of PS ASOs
GalNAc
The conjugation of a triantennary GalNAc moiety to a PS ASO with either 2ʹ-MOE or 2ʹ cEt modifications has been shown to increase the affinity for asialoglycoprotein receptors expressed on hepatocytes (3, 4, 11, 57, 58, 96, 97, 98). We assume that PS ASOs with other 2ʹ modifications behave similarly, but this has not been thoroughly evaluated. Asialoglycoprotein receptors are high-capacity scavenger receptors that are highly expressed in the plasma membranes of hepatocytes (167). The conjugation of GalNAc results in a significant increase in productive delivery of conjugated PS ASOs to hepatocytes by shifting the suborgan distribution of these compounds to the hepatocytes from the nonparenchymal cells of the liver without altering the overall tissue distribution of PS ASOs (95). Since the GalNAc moiety is cleaved very rapidly in the early endosome, there is no detectable change in subcellular distribution (95). As previously indicated, this results in a 15- to 30-fold increase in the potency of PS 2ʹ-MOE and PS 2ʹ cEt ASOs in the clinic (96). Moreover, as previously mentioned, in several species, oral bioavailability of PS 2ʹ cEt ASOs has been reported (103).
The effects of GalNAc conjugation on siRNAs are quite different. In the absence of cationic lipids or GalNAc conjugation, siRNAs do not distribute to a meaningful extent to any peripheral tissue (10, 35, 43). Therefore, the concentration of a GalNAc-conjugated siRNA achieves a significantly higher concentration in the liver than unconjugated siRNA (112).
Progress in identifying ligands for targeted delivery of PS ASOs to cells other than hepatocytes
Arguably, the most interesting advance published to date on ligand-conjugated ASOs for targeted delivery involves conjugation of a glucagon-like peptide 1 (GLP-1) peptide to PS ASOs. Unconjugated PS ASOs do not distribute at meaningful levels to the beta cells of the pancreas and are inactive. In cells in vitro and in mice, GLP-1 effectively delivers PS ASOs to the beta cells of the pancreas and results in excellent pharmacological activity. These receptors are G-protein-coupled receptors and signaling appears to be required for PS ASO delivery (57), thus scavenger receptors, tyrosine kinase-type receptors, and other G-protein-coupled receptors can serve as binding sites for targeted delivery of PS ASOs.
Another approach that is mechanistically more interesting involves the conjugation of epidermal growth factor (EGF) to PS ASOs. This enhances affinity for EGF receptors (EGFR) and increases productive delivery to cells that express EGFR. Interestingly EGFR are tyrosine kinase-coupled receptors, and these receptors are not considered high-capacity as GalNAc receptors and other scavenger receptors are. It is also interesting that cellular uptake of EGF-conjugated PS ASOs requires receptor internalization, but not EGFR signaling (121).
Local administration to treat diseases of specific organs
PS ASOs can be administered by several routes to achieve therapeutic concentrations in a specific organ (102). This will be discussed in greater detail below.
Intravitreal administration
PS ASOs can be administered intravitreally in saline. After intraocular dosing, absorption of PS ASOs by the retina and slow elimination from the vitreous via vitreous flow have been shown. Little to no degradation of PS ASOs in the eye was observed, supporting infrequent administration. Target reduction in retinal cells has been demonstrated in several species (168). Fomivirsen, a PS oligodeoxynucleotide administered intravitreally every 3 months to treat cytomegalovirus (CMV) caused retinitis in AIDS patients (169), was the first RNA targeted drug to be approved for commercial use. Currently, a PS 2ʹ-MOE ASO is in phase 1/2 studies in patients with retinitis pigmentosa (168).
Intradermal administration
PS ASOs can be administered in saline formulations intradermally to treat local cutaneous diseases and various dermatological conditions. An interesting example of PS ASOs administered intradermally is work that was performed on wound healing. Scarring after plastic surgery is a meaningful issue for patients and plastic surgeons that often requires resection of the scar and, unfortunately, that is relatively ineffective. A PS 2ʹ-MOE ASO targeting connective tissue growth factor (CTGF) was evaluated in animal models of wound healing and in several clinical trials. The drug was well tolerated in the clinic and produced a statistically significant improvement in scarring (170).
Rectal administration
Alicaforsen, a PS oligodeoxynucleotide that targets intercellular adhesion molecule 1 (ICAM- 1), has been administered rectally to patients with left-sided ulcerative colitis and pouchitis. In both patient groups, alicaforsen was safe and well tolerated with no significant drug-related side effects and resulted in statistically significant improvement in diseases severity scores (171, 172, 173).
Aerosol delivery for pulmonary diseases
PS ASOs can be administered by nebulizers or other devices to treat pulmonary diseases. A number of studies have shown that PS 2ʹ-MOE and PS cEt ASOs can be administered safely and distributed to both large and small airways and pulmonary cells (174). A 2ʹ-MOE PS ASO targeted to interleukin 4 alpha receptor (IL4Rα) administered by aerosol delivery in a randomized double-blind clinical trial demonstrated excellent safety and tolerability, target reduction in pulmonary gavage-obtained cells, and evidence of clinical benefit. More recently a PS 2ʹ methoxy ASO that targets the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) was studied in patients with phenylamine 508 mutations. The drug was well tolerated and produced dose-dependent evidence of benefit, but oddly the dose–response relationship was bell-shaped with the top dose providing no apparent benefit. Finally, the first of the more potent class of PS cEt ASOs, targeting the epithelial sodium channel (ENaC), is in phase 2 trials in patients with CF and in patients suffering from chronic bronchitis (175).
Intrathecal administration
The IT administration of PS ASOs has opened a new chapter in the neurosciences and in the treatment of neurological diseases. Based on a substantial and growing database derived from controlled clinical trials involving multiple ASOs, we know that repeated doses of PS 2ʹ-MOE ASOs for as long as 7 years are safe and well tolerated in humans (105). At therapeutic doses, these agents distribute broadly throughout the spinal column and CNS and display pharmacological activities (P. Jafar-Nejad et al., unpublished). Nusinersen, a fully 2ʹ-MOE modified PS ASO that revolutionized the treatment of spinal muscular atrophy (SMA) (138), is commercialized worldwide and is the first RTD blockbuster (3, 176). Furthermore, advances in the design and chemistries used intrathecally continues and hopefully will result in even better-performing PS ASOs.
Toxicological effects of PS ASOs
Integrated safety databases
Because PS ASOs within a specific chemical class differ only in sequence, they behave similarly with regard to many of their behaviors. Consequently, databases have been assembled that integrate all safety findings in toxicological studies in NHP and all placebo-controlled clinical trials for PS 2ʹ-MOE, PS cEt, PS MOE –GalNAc, and PS cEt-GalNAc ASOs, and the results of analyses from these extensive date have been published (56, 81, 82, 177) These databases are an invaluable asset as they display the potential adverse events of each drug and the doses at which they are observed. PS MOE ASOs have been studied in more than 10,000 patients and that database is comprised of data from more than 7000 to 8000 patients involving many different PS MOE ASOs. Though the experience with other chemical classes is less, experience is accumulating rapidly. Unfortunately, safety databases for other chemical classes of ASOs and siRNAs have not been published.
Systemic toxicities
Toxicities caused by interactions of PS ASOs with proteins in blood
As PS ASOs interact extensively with plasma proteins and proteins on the surface of cells in blood, a wide range of adverse events due to these interactions have been identified. As a general rule, PS ASOs of all chemical classes are associated with these events, but there are significant variations in the incidence, severity, and dose at which a number of the events are a function of the type of 2ʹ substituents. These events are peak plasma-related and therefore dose-related, transient, and self-limiting. Some occur acutely while others are observed only with repeated chronic administration (3, 25, 55, 56, 82, 177)
Effects on the complement system
PS ASOs interact with complement Factor H, a protein that inhibits the alternate complement pathway. PS oligodeoxynucleotides (PS ODNs) bind to and inhibit complement Factor H as do other chemical classes, but PS MOE ASOs appear to bind Factor H differently from PS ODNs and are less inhibitory. In NHP, at high doses, PS ODNs and other chemical classes can activate the alternative complement cascade and cause cardiovascular collapse, platelet count reductions, and inflammation in various tissues. The effects are dose-dependent and less common for the more potent PS MOE and cEt ASOs. Fortunately, in humans, these effects have not been observed (55).
Effects on the clotting cascade
All chemical classes of PS ASOs studied produce a peak plasma concentration-dependent (and therefore dose-dependent) transient prolongation of clotting measured by an increase in the activated prothrombin time (APPT) in all species studied. The maximal increase in APPT is about 30% and is not associated with physiological sequelae. Since the distribution half-life of PS ASOs is 60 to 90 min, the effects typically reverse within 60 min. The mechanism responsible has been shown to be the reversible inhibition of the intrinsic tenase complex (55, 81).
Effects on platelets
All chemical classes of PS ASOs tested result in a dose-dependent, peak plasma concentration-dependent reduction in platelet counts with no effect on platelet activation in NHP and humans. The reduction in platelet number is reproducible, predictable, and requires no treatment. Though the precise mechanism is not defined, the reduction in platelets is most likely due to PS ASOs binding to platelets and causing increased clearance by the reticuloendothelial system (RES) (55). A more problematic type of effect on platelet number has also been observed. In both NHP and humans, with chronic administration, sporadic unpredictable platelet reductions that can be severe are observed in a few NHP in toxicological study. The mechanisms to explain these events are not fully defined, but in humans treated with PS 2ʹ-MOE and PS 2ʹ-MOE GalNAc ASOs, the integrated safety databases show that severe thrombocytopenia has not been encountered in humans treated with a wide range of doses for prolonged periods (56, 81, 82, 177). However, importantly in patients with TTR amyloidosis and FCS treated with 300 mg/week of inotersen or volanesorsen respectively, severe thrombocytopenia was experienced by a few patients (3, 85, 178), demonstrating the potential for drug–disease interactions. For inotersen, the mechanism appears to be dependent on the underlying inflammatory processes secondary to the disease resulting in increases in a number of inflammatory markers that lead to inotersen-dependent antiplatelet IgG antibodies in a few patients (86). Since FCS patients also experience similar immune activation, a similar mechanism is suspected for volanesorsen.
Experience with PS cEts and PS cEt-GalNAc ASOs is more limited, but since PS MOE GalNAc and the cEt ASOs are more potent and administered at lower doses, severe thrombocytopenia is unlikely. However, at doses of PS cEt ASOs in excess of 600 mg/week, in cancer patients on multiple chemotherapeutic agents, thrombocytopenia was observed, so additional experience is necessary (88). Severe thrombocytopenia was also seen in patients administered PS LNAs, but no data on the mechanism to explain the severe thrombocytopenia have been published (3).
Immunotoxicity
PS ASOs are immunostimulatory. In addition to substantially increasing potency compared with PS ODNs, PS MOE ASOs are also significantly less proinflammatory. PS cEts appear to behave similarly to PS MOE cEts with regard to immune stimulation and approximately tenfold more potent than PS MOE ASO. Since the effects on the immune system are dose-dependent, as potency increases, the potential to induce immune stimulation decreases at therapeutic doses (3). Pelacarsen and other PS GalNAc ASOs clearly demonstrate the value of reducing therapeutic doses as the incidence of ISRs was dramatically reduced and compliance improved compared with PS 2ʹ-MOE ASOs (129).
Innate immune activation
PS ASOs can induce the innate immune system. PS CpG motif-containing ASOs are potent stimulators of the innate immune system, but PS ASOs that do not contain CpG motifs can also activate innate immunity, albeit at much higher doses and to a lesser extent (55). Binding to toll-like receptor 9 (TLR9) is a principle factor in the stimulation of the innate immune system for both CpG containing oligonucleotides and non-CpG containing PS ASOs (55), but other TLRs, such as TLR7 may be involved with some PS ASO sequences (176, 179). The stimulation of the innate immune system is myeloid differentiation protein (Myd 88) dependent (180) and the receptor for advanced glycation readout (RAGE) may play a role in some circumstances (180). The nature of the 2ʹ position is important with PS ODNs >PS 2ʹ methoxy > PS MOE ASOs being the rank order of potency for stimulation of innate immunity. Five methyl C substitution reduces this effect (181).
Other immunostimulatory effects
While CpG oligonucleotides can cause increases in IL1, 6, and 12, with PS 2ʹ-MOE ASOs, no increase in IL6 or IL12 was observed suggesting differences with regard to T helper cells Type 1 (TH1) responses (181). In normal human volunteers, mipomersen, a PS MOE ASO, was thoroughly evaluated for immune stimulatory effects. Multiple cytokines, chemokines, and acute-phase reactants were measured in the clinic. Aside from a transient increase in CRP that typically occurred only after the first dose, the other observation was a transient increase in IL-6 (182). Additionally, chronic dosing of mipomersen was associated with the emergence of low-titer nonneutralizing antibodies that did not affect the pharmacokinetics or the pharmacological effects of the drug (183).
Cyto- and organ toxicities
The major mechanism by which PS ASOs produce cytotoxicity, hepatotoxicity, and some nephrotoxicities is now known, and it is thought that this mechanism underlies most adverse events in most organs. The mechanism is secondary to unique interactions of toxic PS ASOs with paraspeckle proteins and RNase H1 that result in mislocalization of paraspeckle proteins and RNase H1 to the nucleolus and the inhibition of nucleolar RNA transcription and processing, leading to nucleolar stress and apoptosis (Fig. 7) (36). Remarkably, the substitution of a 2ʹ methoxy at the second nucleotide from the 5ʹ junction of the oligodeoxynucleotide has been shown to ablate or reduce the toxicity with little effect on potency. Other substitutions at position 2 can effect similar reductions in the toxicity (36, 61).
Figure 7.
A major toxic mechanism of PS ASOs mediated by PS ASO protein interactions. Toxic PS ASOs bind more proteins more tightly. The tight interactions between PS ASOs and proteins can cause paraspeckle protein mislocalization to the nucleolus, in an RNase H1-dependent manner, and can affect pre-rRNA synthesis, leading to nucleolar stress and apoptotic cell death.
Certainly, other mechanisms of toxicity also have been observed. In addition to the various effects related to interactions with plasma proteins enumerated above, PS 2ʹ F-modified PS ASOs have been shown to cause rapid cellular degradation of paraspeckle proteins (111) and toxicities secondary to true off-target degradation have been observed and with specific PS ASOs are risks that must be avoided. Adverse events secondary to effects on the intended target can occur and as has been observed with both inotersen and volanesorsen, adverse events secondary to drug–disease interactions may occur. However, the effort to understand the molecular mechanisms responsible for each type of toxicity is paying enormous dividends and the significant increases in potency achieved greatly enhance the therapeutic index of PS ASOs sufficiently to support the use of these agents in the very common diseases as well as rare diseases.
Conclusions and Prospectus
In conclusion, antisense technology is beginning to deliver on the promise envisioned more than 30 years ago though many questions remain to be answered and the final proof of value awaits the completion of cardiovascular outcome trials and other long-term studies in a wide range of diseases. Moreover, it is reasonable to conclude that antisense technology is broadly enabling. All known classes of RNAs have been shown to be amenable to the effects of PS ASO including highly structured small noncoding nuclear RNAs (30), preribosomal RNA (28), long noncoding RNAs (29, 123), micro-RNAs (1), pre- and m-RNAs (3), and circular RNAs (184). PS ASOs can be designed to take advantage of a widening range of potential post-RNA-binding mechanisms of action. Importantly, in addition to the proven ability of PS ASOs to increase the levels of specific proteins by altering splicing, new approaches that exploit the recently discovered translation regulatory mechanisms that also can be used to increase the levels of specific proteins are now enabled, and it seems likely that other approaches will be identified to do this. Thus “agonist”-like activity may become more broadly accessible. PS ASOs can be administered by all routes of administration for system and local therapeutic purposes (3). They distribute broadly and with the more potent PS ASOs, activity in human cancers has been demonstrated (88). Though more progress is required to have sufficient potency to treat skeletal and cardiac muscle with PS ASOs, current efforts are encouraging and new ligand to target tissues other than the liver may be on the horizon. Obviously, a key final step remains and that is the development of commercially attractive oral formulations, but current clinical trials of new oral formulations suggest that this route of administration may be feasible. Finally, the definition of the mechanisms of toxicities has facilitated solutions to many of the issues encountered in the past.
Antisense technology continues to advance
The current status of the technology demonstrates both the need for and the value of consistent and long-term investment in research to advance a new drug discovery technology. It is gratifying that the major advances have resulted from the commitment to understand not just what PS ASOs do, but also why they behave as observed by understanding molecular mechanisms in ever greater detail. It seems likely that new pharmacodynamic mechanisms, deeper understanding of molecular mechanisms by which PS ASOs enter cells and distribute intracellularly, produce pharmacological effects, and cause adverse events has already shifted focus from simply RNA targets or the nucleic acid language to protein–PS ASO interactions and the language with which PS ASOs communicate with proteins. This will continue to be an ever more important source of progress. New approaches to targeted delivery of PS ASOs to cells other than hepatocytes seem likely to be identified. Since only a tiny fraction of intracellular PS ASOs is available at the sites at which hybridization to target RNAs occurs, efforts to target intracellular delivery and to increase the rate of release from late endosomes may be a fruitful area of research. Finally understanding the molecular mechanisms that explain why some PS ASOs are toxic has already resulted in new medicinal chemical solutions to toxicities. This should support the use of more potent PS ASOs and enhance the efficiency of drug discovery as fewer active PS ASOs will need to be discarded because of toxicities.
Next-generation PS ASOs will be chemically more complex
Based on available data, it seems likely that several new PS ASO designs will emerge and that they will “decorated” at specific sites to target specific tissues, target to specific sites in cells, and reduce the potential to cause toxic effects. This may increase the cost of synthesis a bit, but that increase will be more than offset by the enhanced potency, reduced potential for adverse events, and new mechanisms of action that should once again expand the versatility of the technology.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We wish to thank Kim Butler for excellent assistance in manuscript preparation, and Tracy Reigle for help in figure preparation. This work is supported by an internal funding from Ionis Pharmaceuticals.
Author contributions
X.-H. L. developed many of the figures and contributed numerous edits. B. F. B. prepared Table 1 and provided edits. R. M. C. provided numerous edits. S. T. C. determined the content and outline, wrote the primary text, and contributed to figures and tables and edits.
Edited by Karin Musier-Forsyth
References
- 1.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Crooke S.T., Seth P.P., Vickers T.A., Liang X.H. The interaction of phosphorothioate-containing RNA targeted drugs with proteins is a critical determinant of the therapeutic effects of these agents. J. Am. Chem. Soc. 2020;142:14754–14771. doi: 10.1021/jacs.0c04928. [DOI] [PubMed] [Google Scholar]
- 3.Crooke S.T., Liang X.-H., Crooke R.M., Baker B.F., Geary R.S. Antisense drug discovery and development technology considered in a pharmacological context. Biochem. Pharmacol. 2020 doi: 10.1016/j.bcp.2020.114196. [DOI] [PubMed] [Google Scholar]
- 4.Crooke S.T., Baker B.F., Crooke R.M., Liang X.-H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2020 doi: 10.1038/s41573-021-00162-z. In press. [DOI] [PubMed] [Google Scholar]
- 5.Crooke S.T., Vickers T.A., Liang X.H. Phosphorothioate modified oligonucleotide-protein interactions. Nucleic Acids Res. 2020;48:5235–5253. doi: 10.1093/nar/gkaa299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang X.H., Nichols J.G., Hsu C.W., Vickers T.A., Crooke S.T. mRNA levels can be reduced by antisense oligonucleotides via no-go decay pathway. Nucleic Acids Res. 2019;47:6900–6916. doi: 10.1093/nar/gkz500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang X.H., Sun H., Shen W., Wang S., Yao J., Migawa M.T., Bui H.H., Damle S.S., Riney S., Graham M.J., Crooke R.M., Crooke S.T. Antisense oligonucleotides targeting translation inhibitory elements in 5' UTRs can selectively increase protein levels. Nucleic Acids Res. 2017;45:9528–9546. doi: 10.1093/nar/gkx632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X.-h., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 2016;34:875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- 9.Swayze E.E., Bhat B. The medicinal chemistry of oligonucleotides. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. 2nd Ed. CRC Press; Boca Raton, FL: 2008. pp. 143–182. [Google Scholar]
- 10.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 11.Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S., Kretschmer P., Brown C.R., Gupta S., Willoughby J.L.S., Boshar J.A. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017;45:10969–10977. doi: 10.1093/nar/gkx818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. U. S. A. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigova A., Zamore O.D. Small RNA silencing pathways. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 75–88. [Google Scholar]
- 14.Chernikov I.V., Vlassov V.V., Chernolovskaya E.L. Current development of siRNA bioconjugates: From research to the clinic. Front. Pharmacol. 2019;10:444. doi: 10.3389/fphar.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field A.K., Young C.W., Krakoff I.H., Tytell A.A., Lampson G.P., Nemes M.M., Hilleman M.R. Induction of interferon in human subjects by poly I:C. Proc. Soc. Exp. Biol. Med. 1971;136:1180–1186. doi: 10.3181/00379727-136-35454. [DOI] [PubMed] [Google Scholar]
- 16.Maehle A.-H., Prüll C.-R., Halliwell R.F. The emergence of the drug receptor theory. Nat. Rev. Drug Discov. 2002;1:637–641. doi: 10.1038/nrd875. [DOI] [PubMed] [Google Scholar]
- 17.Maehle A.H. “Receptive substances”: John Newport Langley (1852-1925) and his path to a receptor theory of drug action. Med. Hist. 2004;48:153–174. doi: 10.1017/s0025727300000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Changeux J.P. The nicotinic acetylcholine receptor: The founding father of the pentameric ligand-gated ion channel superfamily. J. Biol. Chem. 2012;287:40207–40215. doi: 10.1074/jbc.R112.407668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay D.B., Duffull S.B., Glass M. 100 years of modelling ligand-receptor binding and response: A focus on GPCRs. Br. J. Pharmacol. 2020;177:1472–1484. doi: 10.1111/bph.14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abou-Gharbia M. Discovery of innovative small molecule therapeutics. J. Med. Chem. 2009;52:2–9. doi: 10.1021/jm8012823. [DOI] [PubMed] [Google Scholar]
- 21.Vecchio I., Tornali C., Bragazzi N.L., Martini M. The discovery of insulin: An important milestone in the history of medicine. Front. Endocrinol. 2018;9:613. doi: 10.3389/fendo.2018.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson N., Czaplewski L., Piddock L.J.V. Discovery and development of new antibacterial drugs: Learning from experience? J. Antimicrob. Chemother. 2018;73:1452–1459. doi: 10.1093/jac/dky019. [DOI] [PubMed] [Google Scholar]
- 23.Goding J.W. Introduction to monoclonal antibodies. In: Goding J.W., editor. Monoclonal Antibodies. Principles and Practice. Academic Press; San Diego, CA: 1996. pp. 116–140. [Google Scholar]
- 24.Monia B.P., Johnston J.F., Geiger T., Muller M., Fabbro D. Antitumor activity of a phosphorothioate antisense oligodeoxynucleotide targeted against C-Raf kinase. Nat. Med. 1996;2:668–675. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- 25.Crooke S.T., Vickers T.A., Lima W.F., Wu H.-J. Mechanisms of antisense drug action, an introduction. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. 2nd Ed. CRC Press; Boca Raton, FL: 2008. pp. 3–46. [Google Scholar]
- 26.Sazani P., Graziewicz M.A., Kole R. Splice switching oligonucleotides as potential therapeutics. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 89–114. [Google Scholar]
- 27.Hodges D., Crooke S.T. Inhibition of splicing of wild-type and mutated luciferase-adenovirus pre-mRNAs by antisense oligonucleotides. Mol. Pharmacol. 1995;48:905–918. [PubMed] [Google Scholar]
- 28.Pollak A.J., Liang X.-H., Crooke S.T. Gapmer ASOs targeting 5S rRNA can reduce mature 5S rRNA by two mechanisms. Nucleic Acid Ther. 2020;30:312–324. doi: 10.1089/nat.2020.0864. [DOI] [PubMed] [Google Scholar]
- 29.Shen W., Liang X.H., Sun H., De Hoyos C.L., Crooke S.T. Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang X.H., Vickers T.A., Guo S., Crooke S.T. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011;39:11–17. doi: 10.1093/nar/gkq1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swinnen B., Robberecht W., Van Den Bosch L. RNA toxicity in non- coding repeat expansion disorders. EMBO J. 2020;39 doi: 10.15252/embj.2018101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCombie W.R., McPherson J.D., Mardis E.R. Next-generation sequencing technologies. Cold Spring Harb. Perspect. Med. 2019;9 doi: 10.1101/cshperspect.a036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooke S.T. Basic Principles of Antisense Technology. Marcel Dekker, Inc; New York, NY: 2001. Antisense drug technology: Principles, strategies, and applications; pp. 1–28. [Google Scholar]
- 34.Manoharan M., Rajeev K.G. Utilizing chemistry to harness RNA interference pathways for therapeutics: Chemically modified siRNAs and antagomirs. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 437–464. [Google Scholar]
- 35.Tang X., Zhang S., Fu R., Zhang L., Huang K., Peng H., Dai L., Chen Q. Therapeutic prospects of mRNA-based gene therapy for glioblastoma. Front. Oncol. 2019;9:1208. doi: 10.3389/fonc.2019.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen W., De Hoyos C.L., Migawa M.T., Vickers T.A., Sun H., Low A., Bell T.A., 3rd, Rahdar M., Mukhopadhyay S., Hart C.E., Bell M., Riney S., Murray S.F., Greenlee S., Crooke R.M. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019;37:640–650. doi: 10.1038/s41587-019-0106-2. [DOI] [PubMed] [Google Scholar]
- 37.Crooke S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017;27:70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 39.Capaldi D.C., Scozzari A.N. Manufacturing and analytical processes for 2O-(2-methoxy-ethyl)-modified oligonucleotides. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies and Applications. CRC Press; Boca Raton: 2008. pp. 401–434. [Google Scholar]
- 40.White A.P., Reeves K.K., Snyder E., Farrell J., Powell J.W., Mohan V., Griffey R.H. Hydration of single-stranded phosphodiester and phosphorothioate oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:3261–3266. doi: 10.1093/nar/24.16.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baraniak J., Frey P.A. Effect of ion pairing on bond order and charge localization in alkyl phosphorothioates. J. Am. Chem. Soc. 1988;110:4059–4060. [Google Scholar]
- 42.Plumridge A., Meisburger S.P., Andresen K., Pollack L. The impact of base stacking on the conformations and electrostatics of single-stranded DNA. Nucleic Acids Res. 2017;45:3932–3943. doi: 10.1093/nar/gkx140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fougerolles A.R.D., Maraganore J.M. Discovery and development of RNAi therapeutics. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 465–486. [Google Scholar]
- 44.Manoharan M., Akinc A., Pandey R.K., Qin J., Hadwiger P., John M., Mills K., Charisse K., Maier M.A., Nechev L., Greene E.M., Pallan P.S., Rozners E., Rajeev K.G., Egli M. Unique gene-silencing and structural properties of 2'-fluoro- modified siRNAs. Angew. Chem. Int. Ed. Engl. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iversen P.L. Morpholinos. In: Crooke S.T., editor. Antisense Drug Technology. CRC Press; Boca Raton, FL: 2008. pp. 565–582. [Google Scholar]
- 46.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metab. 2019;29:501. doi: 10.1016/j.cmet.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Wan W.B., Migawa M.T., Vasquez G., Murray H.M., Nichols J.G., Gaus H., Berdeja A., Lee S., Hart C.E., Lima W.F., Swayze E.E., Seth P.P. Synthesis, biophysical properties and biological activity of second generation antisense oligonucleotides containing chiral phosphorothioate linkages. Nucleic Acids Res. 2014;42:13456–13468. doi: 10.1093/nar/gku1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostergaard M.E., De Hoyos C.L., Wan W.B., Shen W., Low A., Berdeja A., Vasquez G., Murray S., Migawa M.T., Liang X.H., Swayze E.E., Crooke S.T., Seth P.P. Understanding the effect of controlling phosphorothioate chirality in the DNA gap on the potency and safety of gapmer antisense oligonucleotides. Nucleic Acids Res. 2020;48:1691–1700. doi: 10.1093/nar/gkaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koziolkiewicz M., Krakowiak A., Kwinkowski M., Boczkowska M., Stec W.J. Stereodifferentiation--the effect of P chirality of oligo(nucleoside phosphorothioates) on the activity of bacterial RNase H. Nucleic Acids Res. 1995;23:5000–5005. doi: 10.1093/nar/23.24.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koziolkiewicz M., Wojcik M., Kobylanska A., Karwowski B., Rebowska B., Guga P., Stec W.J. Stability of stereoregular oligo(nucleoside phosphorothioate)s in human plasma: Diastereoselectivity of plasma 3'-exonuclease. Antisense Nucleic Acid Drug Dev. 1997;7:43–48. doi: 10.1089/oli.1.1997.7.43. [DOI] [PubMed] [Google Scholar]
- 51.Jahns H., Roos M., Imig J., Baumann F., Wang Y., Gilmour R., Hall J. Stereochemical bias introduced during RNA synthesis modulates the activity of phosphorothioate siRNAs. Nat. Commun. 2015;6:6317. doi: 10.1038/ncomms7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinke C.A., Reeves K.K., Powell J.W., Lee S.A., Chen Y.Z., Wyrzykiewicz T., Griffey R.H., Mohan V. Vibrational analysis of phosphorothioate DNA: II. The POS group in the model compound dimethyl phosphorothioate [(CH3O)2(POS)] J. Biomol. Struct. Dyn. 1997;14:509–516. doi: 10.1080/07391102.1997.10508149. [DOI] [PubMed] [Google Scholar]
- 53.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang X.H., Shen W., Sun H., Kinberger G.A., Prakash T.P., Nichols J.G., Crooke S.T. Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2'-modifications and enhances antisense activity. Nucleic Acids Res. 2016;44:3892–3907. doi: 10.1093/nar/gkw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henry S.P., Kim T.-W., Karmer-Stickland K., Zanardi T.A., Fey R.A., Levin A.A. Toxicologic properties of 2'-O-methoxyethyl chimeric antisense inhibitors in animals and man. In: Crooke S.T., editor. Antisense Drug Technology-Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 327–364. [Google Scholar]
- 56.Crooke S.T., Baker B.F., Pham N.C., Hughes S.G., Kwoh T.J., Cai D., Tsimikas S., Geary R.S., Bhanot S. The effects of 2'-O-methoxyethyl oligonucleotides on renal function in humans. Nucleic Acid Ther. 2018;28:10–22. doi: 10.1089/nat.2017.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ammala C., Drury W.J., 3rd, Knerr L., Ahlstedt I., Stillemark-Billton P., Wennberg- Huldt C., Andersson E.M., Valeur E., Jansson-Lofmark R., Janzen D., Sundstrom L., Meuller J., Claesson J., Andersson P., Johansson C. Targeted delivery of antisense oligonucleotides to pancreatic beta-cells. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aat3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang S., Allen N., Prakash T.P., Liang X.H., Crooke S.T. Lipid conjugates enhance endosomal release of antisense oligonucleotides into cells. Nucleic Acid Ther. 2019;29:245–255. doi: 10.1089/nat.2019.0794. [DOI] [PubMed] [Google Scholar]
- 59.Ostergaard M.E., Jackson M., Low A., Chappell A.E., Lee R.G., Peralta R.Q., Yu J., Kinberger G.A., Dan A., Carty R., Tanowitz M., Anderson P., Kim T.W., Fradkin L., Mullick A.E. Conjugation of hydrophobic moieties enhances potency of antisense oligonucleotides in the muscle of rodents and non-human primates. Nucleic Acids Res. 2019;47:6045–6058. doi: 10.1093/nar/gkz360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prakash T.P., Mullick A.E., Lee R.G., Yu J., Yeh S.T., Low A., Chappell A.E., Ostergaard M.E., Murray S., Gaus H.J., Swayze E.E., Seth P.P. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019;47:6029–6044. doi: 10.1093/nar/gkz354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Migawa M.T., Shen W., Wan W.B., Vasquez G., Oestergaard M.E., Low A., De Hoyos C.L., Gupta R., Murray S., Tanowitz M., Bell M., Nichols J.G., Gaus H., Liang X.H., Swayze E.E. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res. 2019;47:5465–5479. doi: 10.1093/nar/gkz247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zanardi T.A., Han S.C., Jeong E.J., Rime S., Yu R.Z., Chakravarty K., Henry S.P. Pharmacodynamics and subchronic toxicity in mice and monkeys of ISIS 388626, a second-generation antisense oligonucleotide that targets human sodium glucose cotransporter 2. J. Pharmacol. Exp. Ther. 2012;343:489–496. doi: 10.1124/jpet.112.197426. [DOI] [PubMed] [Google Scholar]
- 63.van Meer L., Moerland M., van Dongen M., Goulouze B., de Kam M., Klaassen E., Cohen A., Burggraaf J. Renal effects of antisense-mediated inhibition of SGLT2. J. Pharmacol. Exp. Ther. 2016;359:280–289. doi: 10.1124/jpet.116.233809. [DOI] [PubMed] [Google Scholar]
- 64.Freier S.M., Watt A.T. Basic principles of antisense drug discovery. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Application. CRC Press; Boca Raton, FL: 2008. pp. 117–141. [Google Scholar]
- 65.Lima W., Wu H., Crooke S.T. The RNase H mechanism. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. 2nd Ed. CRC Press; Boca Raton, FL: 2008. pp. 47–74. [Google Scholar]
- 66.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F., Hayden M.R., Seth P.P. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P., Swayze E.E., Crooke S.T. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 68.Frank D.E., Schnell F.J., Akana C., El-Husayni S.H., Desjardins C.A., Morgan J., Charleston J.S., Sardone V., Domingos J., Dickson G., Straub V., Guglieri M., Mercuri E., Servais L., Muntoni F. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology. 2020;94:e2270–e2282. doi: 10.1212/WNL.0000000000009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., Dickson G., Wood M.J., Wilton S.D., Straub V., Kole R. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alfano L.N., Charleston J.S., Connolly A.M., Cripe L., Donoghue C., Dracker R., Dworzak J., Eliopoulos H., Frank D.E., Lewis S., Lucas K., Lynch J., Milici A.J., Flynt A., Naughton E. Long-term treatment with eteplirsen in nonambulatory patients with Duchenne muscular dystrophy. Medicine (Baltimore) 2019;98 doi: 10.1097/MD.0000000000015858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan N., Eliopoulos H., Han L., Kinane T.B., Lowes L.P., Mendell J.R., Gordish- Dressman H., Henricson E.K., McDonald C.M., Eteplirsen I., the C.D.I. Eteplirsen treatment attenuates respiratory decline in ambulatory and non- ambulatory patients with Duchenne muscular dystrophy. J. Neuromuscul. Dis. 2019;6:213–225. doi: 10.3233/JND-180351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levin A.A., Yu R.Z., Geary R.S. Basic principles of the pharmaseokinetics of antisense oligonucleotide drugs. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 183–216. [Google Scholar]
- 73.Eckstein F. Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 74.Oka N., Wada T., Saigo K. An oxazaphospholidine approach for the stereocontrolled synthesis of oligonucleoside phosphorothioates. J. Am. Chem. Soc. 2003;125:8307–8317. doi: 10.1021/ja034502z. [DOI] [PubMed] [Google Scholar]
- 75.Panzara M. ASGCT; Washington, DC: 2019. Selective Targeting of Mutant Huntingtin Using Stereopure Oligonucleotides as a Potential Therapeutic Approach for Huntington’s Disease. [Google Scholar]
- 76.Wave Life Sciences . Globe Newswire; Cambridge, MA: 2019. Wave Life Sciences Announces Topline Data and Addition of Higher Dose Cohort in Ongoing Phase 1b/2a PRECISION-HD2 Trial in Huntington’s Disease. [Google Scholar]
- 77.Wagner K., Phan H., Servais L., Gidaro T., Cripe L., Eagle M., Muntoni F., Straub V., Lonkar P., Schmitz S., Lake S., Hu X., Panzara M. O.28Safety and tolerability of suvodirsen (WVE-210201) in patients with Duchenne muscular dystrophy: Results from a phase 1 clinical trial. Neuromuscul. Disord. 2019;29 [Google Scholar]
- 78.Wave Life Sciences . Globe Newswire; Cambridge, MA: 2019. Wave Life Sciences Announces Discontinuation of Suvodirsen Development for Duchenne Muscular Dystrophy. [Google Scholar]
- 79.Bennett C.F. Pharmacological properties of 2'-O-Methoxyethyl-Modified oligonucleotides. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. 2nd Ed. CRC Press; Boca Raton, FL: 2008. pp. 273–304. [Google Scholar]
- 80.Geary R.S., Yu R.Z., Siwkoswki A., Levin A.A. Pharmacokinetic/pharmacodynamic properties of phosphorothioate 2'-O-(2- methoxyethyl)-modified antisense oligonucleotides in animals and man. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 305–326. [Google Scholar]
- 81.Crooke S.T., Baker B.F., Kwoh T.J., Cheng W., Schulz D.J., Xia S., Salgado N., Bui H.H., Hart C.E., Burel S.A., Younis H.S., Geary R.S., Henry S.P., Bhanot S. Integrated safety assessment of 2'-O-methoxyethyl chimeric antisense oligonucleotides in NonHuman primates and healthy human volunteers. Mol. Ther. 2016;24:1771–1782. doi: 10.1038/mt.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crooke S.T., Baker B.F., Witztum J.L., Kwoh T.J., Pham N.C., Salgado N., McEvoy B.W., Cheng W., Hughes S.G., Bhanot S., Geary R.S. The effects of 2'-O-methoxyethyl containing antisense oligonucleotides on platelets in human clinical trials. Nucleic Acid Ther. 2017;27:121–129. doi: 10.1089/nat.2016.0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tillman L.G., Geary R.S., Hardee G.E. Oral delivery of antisense oligonucleotides in man. J. Pharm. Sci. 2008;97:225–236. doi: 10.1002/jps.21084. [DOI] [PubMed] [Google Scholar]
- 84.Gaus H.J., Gupta R., Chappell A.E., Ostergaard M.E., Swayze E.E., Seth P.P. Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay. Nucleic Acids Res. 2019;47:1110–1122. doi: 10.1093/nar/gky1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Witztum J.L., Gaudet D., Freedman S.D., Alexander V.J., Digenio A., Williams K.R., Yang Q., Hughes S.G., Geary R.S., Arca M., Stroes E.S.G., Bergeron J., Soran H., Civeira F., Hemphill L. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N. Engl. J. Med. 2019;381:531–542. doi: 10.1056/NEJMoa1715944. [DOI] [PubMed] [Google Scholar]
- 86.Narayanan P., Curtis B.R., Shen L., Schneider E., Tami J.A., Paz S., Burel S.A., Tai L.J., Machemer T., Kwoh T.J., Xia S., Shattil S.J., Witztum J.L., Engelhardt J.A., Henry S.P. Underlying immune disorder may predispose some transthyretin amyloidosis subjects to inotersen-mediated thrombocytopenia. Nucleic Acid Ther. 2020;30:94–103. doi: 10.1089/nat.2019.0829. [DOI] [PubMed] [Google Scholar]
- 87.Koch T., Orum H. Locked nucleic acid. In: Crooke S.T., editor. Antisense Drug Discovery. CRC Press; Boca Raton, FL: 2008. pp. 519–565. [Google Scholar]
- 88.MacLeod A.R., Crooke S.T. RNA therapeutics in oncology: Advances, challenges, and future directions. J. Clin. Pharmacol. 2017;57 Suppl 10:S43–S59. doi: 10.1002/jcph.957. [DOI] [PubMed] [Google Scholar]
- 89.Burel S.A., Hart C.E., Cauntay P., Hsiao J., Machemer T., Katz M., Watt A., Bui H.H., Younis H., Sabripour M., Freier S.M., Hung G., Dan A., Prakash T.P., Seth P.P. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2016;44:2093–2109. doi: 10.1093/nar/gkv1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 91.van der Ree M.H., van der Meer A.J., de Bruijne J., Maan R., van Vliet A., Welzel T.M., Zeuzem S., Lawitz E.J., Rodriguez-Torres M., Kupcova V., Wiercinska- Drapalo A., Hodges M.R., Janssen H.L., Reesink H.W. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 92.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hagedorn P.H., Persson R., Funder E.D., Albaek N., Diemer S.L., Hansen D.J., Moller M.R., Papargyri N., Christiansen H., Hansen B.R., Hansen H.F., Jensen M.A., Koch T. Locked nucleic acid: Modality, diversity, and drug discovery. Drug Discov. Today. 2018;23:101–114. doi: 10.1016/j.drudis.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 94.Lima W.F., Murray H.M., Damle S.S., Hart C.E., Hung G., De Hoyos C.L., Liang X.H., Crooke S.T. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016;44:5299–5312. doi: 10.1093/nar/gkw350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F., Riney S., Booten S.L., Murray S.F., Gaus H., Crosby J. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N- acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Viney N.J., van Capelleveen J.C., Geary R.S., Xia S., Tami J.A., Yu R.Z., Marcovina S.M., Hughes S.G., Graham M.J., Crooke R.M., Crooke S.T., Witztum J.L., Stroes E.S., Tsimikas S. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): Two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet. 2016;388:2239–2253. doi: 10.1016/S0140-6736(16)31009-1. [DOI] [PubMed] [Google Scholar]
- 97.Graham M.J., Lee R.G., Brandt T.A., Tai L.J., Fu W., Peralta R., Yu R., Hurh E., Paz E., McEvoy B.W., Baker B.F., Pham N.C., Digenio A., Hughes S.G., Geary R.S. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N. Engl. J. Med. 2017;377:222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 98.Alexander V.J., Xia S., Hurh E., Hughes S.G., O'Dea L., Geary R.S., Witztum J.L., Tsimikas S. N-acetyl galactosamine-conjugated antisense drug to APOC3 mRNA, triglycerides and atherogenic lipoprotein levels. Eur. Heart J. 2019;40:2785–2796. doi: 10.1093/eurheartj/ehz209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buller H.R., Gailani D., Weitz J.I. Factor XI antisense oligonucleotide for venous thrombosis. N. Engl. J. Med. 2015;372:1672. doi: 10.1056/NEJMc1503223. [DOI] [PubMed] [Google Scholar]
- 100.Bethune C., Morgan E., Schulz D., Yu R., Jung S.W., Geary R.S., Bhanot S. USADIA Oligonucleotide-Based Therapeutics Conference. Bethesda North Marriott Hotel and Conference Center; Bethesda, MD: 2019. IONIS-FXI-LRx, a FXI GalNAc conjugated antisense drug, produces potent and sustained reductionin FXI activity in normal volunteers. [Google Scholar]
- 101.Zhang H., Lowenberg E.C., Crosby J.R., MacLeod A.R., Zhao C., Gao D., Black C., Revenko A.S., Meijers J.C., Stroes E.S., Levi M., Monia B.P. Inhibition of the intrinsic coagulation pathway factor XI by antisense oligonucleotides: A novel antithrombotic strategy with lowered bleeding risk. Blood. 2010;116:4684–4692. doi: 10.1182/blood-2010-04-277798. [DOI] [PubMed] [Google Scholar]
- 102.Hardee G.E., Tillman L.G., Geary R.S. Routes and formulations for delivery of antisense oligonucleotides. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 217–236. [Google Scholar]
- 103.Yu R.Z., Kulkarni K.H., Tillman L., Prakash T.P., Seth P., Swayze E., Geary R., Monia B.P., Henry S.P., Wang Y. DIA/FDA Oligonucleotide- Based Therapeutics Conference. 2019. Investigation of oral delivery of GalNac3 conjugated antisense oligonucleotides in rats. North Bethesda, MD. [Google Scholar]
- 104.Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., Lamond A.I. Paraspeckles: A novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 105.Bennett C.F. Therapeutic antisense oligonucleotides are coming of age. Annu. Rev. Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 106.Judge D.P., Kristen A.V., Grogan M., Maurer M.S., Falk R.H., Hanna M., Gillmore J., Garg P., Vaishnaw A.K., Harrop J., Powell C., Karsten V., Zhang X., Sweetser M.T., Vest J. Phase 3 multicenter study of revusiran in patients with hereditary transthyretin-mediated (hATTR) amyloidosis with cardiomyopathy (ENDEAVOUR) Cardiovasc. Drugs Ther. 2020;34:357–370. doi: 10.1007/s10557-019-06919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anguela X.M., High K.A. Entering the modern era of gene therapy. Annu. Rev. Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 108.Jordheim L.P., Durantel D., Zoulim F., Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 109.McKenzie R., Fried M.W., Sallie R., Conjeevaram H., Di Bisceglie A.M., Park Y., Savarese B., Kleiner D., Tsokos M., Luciano C., Pruett T., Stotka J.L., Straus S.E., Hoofnagle J.H. Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N. Engl. J. Med. 1995;333:1099–1105. doi: 10.1056/NEJM199510263331702. [DOI] [PubMed] [Google Scholar]
- 110.Shen W., Liang X.H., Sun H., Crooke S.T. 2'-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 2015;43:4569–4578. doi: 10.1093/nar/gkv298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen W., De Hoyos C.L., Sun H., Vickers T.A., Liang X.H., Crooke S.T. Acute hepatotoxicity of 2' fluoro-modified 5-10-5 gapmer phosphorothioate oligonucleotides in mice correlates with intracellular protein binding and the loss of DBHS proteins. Nucleic Acids Res. 2018;46:2204–2217. doi: 10.1093/nar/gky060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Janas M.M., Zlatev I., Liu J., Jiang Y., Barros S.A., Sutherland J.E., Davis W.P., Liu J., Brown C.R., Liu X., Schlegel M.K., Blair L., Zhang X., Das B., Tran C. Safety evaluation of 2'-deoxy-2'-fluoro nucleotides in GalNAc-siRNA conjugates. Nucleic Acids Res. 2019;47:3306–3320. doi: 10.1093/nar/gkz140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vickers T.A., Crooke S.T. Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014;42:7819–7832. doi: 10.1093/nar/gku484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vickers T.A., Crooke S.T. Development of a quantitative BRET affinity assay for nucleic acid-protein interactions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vickers T.A., Migawa M.T., Seth P.P., Crooke S.T. Interaction of ASOs with PC4 is highly influenced by the cellular environment and ASO chemistry. J. Am. Chem. Soc. 2020;142:9661–9674. doi: 10.1021/jacs.0c01808. [DOI] [PubMed] [Google Scholar]
- 117.Vickers T.A., Rahdar M., Prakash T.P., Crooke S.T. Kinetic and subcellular analysis of PS-ASO/protein interactions with P54nrb and RNase H1. Nucleic Acids Res. 2019;47:10865–10880. doi: 10.1093/nar/gkz771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Werten S., Moras D. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat. Struct. Mol. Biol. 2006;13:181–182. doi: 10.1038/nsmb1044. [DOI] [PubMed] [Google Scholar]
- 119.Huang J., Zhao Y., Liu H., Huang D., Cheng X., Zhao W., Taylor I.A., Liu J., Peng Y.L. Substitution of tryptophan 89 with tyrosine switches the DNA binding mode of PC4. Sci. Rep. 2015;5:8789. doi: 10.1038/srep08789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Werten S., Langen F.W., van Schaik R., Timmers H.T., Meisterernst M., van der Vliet P.C. High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J. Mol. Biol. 1998;276:367–377. doi: 10.1006/jmbi.1997.1534. [DOI] [PubMed] [Google Scholar]
- 121.Wang S., Allen N., Vickers T.A., Revenko A.S., Sun H., Liang X.H., Crooke S.T. Cellular uptake mediated by epidermal growth factor receptor facilitates the intracellular activity of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2018;46:3579–3594. doi: 10.1093/nar/gky145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang S., Allen N., Liang X.H., Crooke S.T. Membrane destabilization induced by lipid species increases activity of phosphorothioate-antisense oligonucleotides. Mol. Ther. Nucleic Acids. 2018;13:686–698. doi: 10.1016/j.omtn.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen W., Liang X.H., Crooke S.T. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42:8648–8662. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y., Shen W., Liang X.H., Crooke S.T. Phosphorothioate antisense oligonucleotides bind P-body proteins and mediate P-body assembly. Nucleic Acid Ther. 2019;29:343–358. doi: 10.1089/nat.2019.0806. [DOI] [PubMed] [Google Scholar]
- 125.Liang X.H., Sun H., Nichols J.G., Allen N., Wang S., Vickers T.A., Shen W., Hsu C.W., Crooke S.T. COPII vesicles can affect the activity of antisense oligonucleotides by facilitating the release of oligonucleotides from endocytic pathways. Nucleic Acids Res. 2018;46:10225–10245. doi: 10.1093/nar/gky841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bailey J.K., Shen W., Liang X.H., Crooke S.T. Nucleic acid binding proteins affect the subcellular distribution of phosphorothioate antisense oligonucleotides. Nucleic Acids Res. 2017;45:10649–10671. doi: 10.1093/nar/gkx709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Graham M.J., Viney N., Crooke R.M., Tsimikas S. Antisense inhibition of apolipoprotein (a) to lower plasma lipoprotein (a) levels in humans. J. Lipid Res. 2016;57:340–351. doi: 10.1194/jlr.R052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vickers T.A., Freier S.M., Bui H.H., Watt A., Crooke S.T. Targeting of repeated sequences unique to a gene results in significant increases in antisense oligonucleotide potency. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsimikas S., Karwatowska-Prokopczuk E., Gouni-Berthold I., Tardif J.C., Baum S.J., Steinhagen-Thiessen E., Shapiro M.D., Stroes E.S., Moriarty P.M., Nordestgaard B.G., Xia S., Guerriero J., Viney N.J., O'Dea L., Witztum J.L. Lipoprotein(a) reduction in persons with cardiovascular disease. N. Engl. J. Med. 2020;382:244–255. doi: 10.1056/NEJMoa1905239. [DOI] [PubMed] [Google Scholar]
- 130.Vickers T.A., Crooke S.T. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015;43:8955–8963. doi: 10.1093/nar/gkv920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Le Paslier D., Frézal J., Cohen D., Weissenbach J., Munnich A. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 132.Kolb S.J., Kissel J.T. Spinal muscular atrophy. Neurol. Clin. 2015;33:831–846. doi: 10.1016/j.ncl.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mailman M.D., Heinz J.W., Papp A.C., Snyder P.J., Sedra M.S., Wirth B., Burghes A.H., Prior T.W. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 134.Wu X., Wang S.H., Sun J., Krainer A.R., Hua Y., Prior T.W. A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum. Mol. Genet. 2017;26:2768–2780. doi: 10.1093/hmg/ddx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:729–744. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E., Norris D.A., Xia S., Bennett C.F., Bishop K.M. Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 138.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., Topaloglu H., Tulinius M., Montes J., Glanzman A.M., Bishop K. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 139.Mercuri E., Darras B.T., Chiriboga C.A., Day J.W., Campbell C., Connolly A.M., Iannaccone S.T., Kirschner J., Kuntz N.L., Saito K., Shieh P.B., Tulinius M., Mazzone E.S., Montes J., Bishop K.M. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 140.De Vivo D.C., Bertini E., Swoboda K.J., Hwu W.L., Crawford T.O., Finkel R.S., Kirschner J., Kuntz N.L., Parsons J.A., Ryan M.M., Butterfield R.J., Topaloglu H., Ben-Omran T., Sansone V.A., Jong Y.J. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: Interim efficacy and safety results from the Phase 2 NURTURE study. Neuromuscul. Disord. 2019;29:842–856. doi: 10.1016/j.nmd.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vickers T.A., Wyatt J.R., Burckin T., Bennett C.F., Freier S.M. Fully modified 2' MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 2001;29:1293–1299. doi: 10.1093/nar/29.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Barbosa C., Peixeiro I., Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suzuki Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., Crouch R.J. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu H., Lima W.F., Crooke S.T. Molecular cloning and expression of cDNA for human RNase H. Antisense Nucleic Acid Drug Dev. 1998;8:53–61. doi: 10.1089/oli.1.1998.8.53. [DOI] [PubMed] [Google Scholar]
- 145.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 146.Wu H., Lima W.F., Crooke S.T. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 147.Lima W.F., Nichols J.G., Wu H., Prakash T.P., Migawa M.T., Wyrzykiewicz T.K., Bhat B., Crooke S.T. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 148.Lima W.F., Wu H., Nichols J.G., Prakash T.P., Ravikumar V., Crooke S.T. Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3'-DNA/5'-RNA terminus of the heteroduplex substrate. J. Biol. Chem. 2003;278:49860–49867. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 149.Wu H., Lima W.F., Crooke S.T. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 150.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Vasquez G., Swayze E.E., Crooke S.T. The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol. Pharmacol. 2007;71:73–82. doi: 10.1124/mol.106.025429. [DOI] [PubMed] [Google Scholar]
- 151.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Siwkowski A.M., Crooke S.T. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 152.Lima W.F., De Hoyos C.L., Liang X.H., Crooke S.T. RNA cleavage products generated by antisense oligonucleotides and siRNAs are processed by the RNA surveillance machinery. Nucleic Acids Res. 2016;44:3351–3363. doi: 10.1093/nar/gkw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lima W.F., Vickers T.A., Nichols J., Li C., Crooke S.T. Defining the factors that contribute to on-target specificity of antisense oligonucleotides. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Miraglia L., Watt A.T., Graham M.J., Crooke S.T. Variations in mRNA content have no effect on the potency of antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 2000;10:453–461. doi: 10.1089/oli.1.2000.10.453. [DOI] [PubMed] [Google Scholar]
- 155.Vickers T.A., Sabripour M., Crooke S.T. U1 adaptors result in reduction of multiple pre-mRNA species principally by sequestering U1snRNP. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liang X.H., Nichols J.G., Sun H., Crooke S.T. Translation can affect the antisense activity of RNase H1-dependent oligonucleotides targeting mRNAs. Nucleic Acids Res. 2018;46:293–313. doi: 10.1093/nar/gkx1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liang X.H., Nichols J.G., De Hoyos C.L., Crooke S.T. Some ASOs that bind in the coding region of mRNAs and induce RNase H1 cleavage can cause increases in the pre-mRNAs that may blunt total activity. Nucleic Acids Res. 2020;48:9840–9858. doi: 10.1093/nar/gkaa715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liang X.H., Sun H., Nichols J.G., Crooke S.T. RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 2017;25:2075–2092. doi: 10.1016/j.ymthe.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H- dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 161.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 162.Vickers T.A., Lima W.F., Wu H., Nichols J.G., Linsley P.S., Crooke S.T. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 'Slicer' independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liang X.H., Hart C.E., Crooke S.T. Transfection of siRNAs can alter miRNA levels and trigger non-specific protein degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:455–468. doi: 10.1016/j.bbagrm.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 164.Liang X.H., Sun H., Hsu C.W., Nichols J.G., Vickers T.A., De Hoyos C.L., Crooke S.T. Golgi-endosome transport mediated by M6PR facilitates release of antisense oligonucleotides from endosomes. Nucleic Acids Res. 2020;48:1372–1391. doi: 10.1093/nar/gkz1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fey R.A., Templin M.V., McDonald J.D., Yu R.Z., Hutt J.A., Gigliotti A.P., Henry S.P., Reed M.D. Local and systemic tolerability of a 2'O- methoxyethyl antisense oligonucleotide targeting interleukin-4 receptor-alpha delivery by inhalation in mouse and monkey. Inhal. Toxicol. 2014;26:452–463. doi: 10.3109/08958378.2014.907587. [DOI] [PubMed] [Google Scholar]
- 166.Karras J.G., Crosby J.R., Guha M., Tung D., Miller D.A., Gaarde W.A., Geary R.S., Monia B.P., Gregory S.A. Anti-inflammatory activity of inhaled IL-4 receptor-alpha antisense oligonucleotide in mice. Am. J. Respir. Cell Mol. Biol. 2007;36:276–285. doi: 10.1165/rcmb.2005-0456OC. [DOI] [PubMed] [Google Scholar]
- 167.Debacker A.J., Voutila J., Catley M., Blakey D., Habib N. Delivery of oligonucleotides to the liver with GalNAc: From research to registered therapeutic drug. Mol. Ther. 2020;28:1759–1771. doi: 10.1016/j.ymthe.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.ProQR Therapeutics . Globe Newswire; Leiden, Netherlands & Cambridge, MA: 2020. ProQR Announces Positive Findings from an Interim Analysis in the Phase 1/2 Trial of QR-421a for Usher Syndrome and Provides Business Update. [Google Scholar]
- 169.Grillone L.R., Lanz R. Fomivirsen. Drugs Today (Barc.) 2001;37:245–255. doi: 10.1358/dot.2001.37.4.620590. [DOI] [PubMed] [Google Scholar]
- 170.Gale J.D., Jensen J., Berman G., Freimuth W., Li G., Pleil A., Kutty M., Rosenthal A., Boswell C.B., Noah V.E.M., Young L. A placebo-controlled study of PF-06473871 (anti-connective tissue growth factor antisense oligonucleotide) in reducing hypertrophic skin scarring. Plast. Reconstr. Surg. Glob. Open. 2018;6 doi: 10.1097/GOX.0000000000001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Miner P.B., Jr., Wedel M.K., Xia S., Baker B.F. Safety and efficacy of two dose formulations of alicaforsen enema compared with mesalazine enema for treatment of mild to moderate left-sided ulcerative colitis: A randomized, double-blind, active-controlled trial. Aliment. Pharmacol. Ther. 2006;23:1403–1413. doi: 10.1111/j.1365-2036.2006.02837.x. [DOI] [PubMed] [Google Scholar]
- 172.Greuter T., Rogler G. Alicaforsen in the treatment of pouchitis. Immunotherapy. 2017;9:1143–1152. doi: 10.2217/imt-2017-0085. [DOI] [PubMed] [Google Scholar]
- 173.Jairath V., Khanna R., Feagan B.G. Alicaforsen for the treatment of inflammatory bowel disease. Expert Opin. Investig. Drugs. 2017;26:991–997. doi: 10.1080/13543784.2017.1349753. [DOI] [PubMed] [Google Scholar]
- 174.Gregory S.A., Karras J.G. Inflammatory diseases. In: Crooke S.T., editor. Antisense Drug Technology - Principles, Strategies, and Applications. CRC Press; Boca Raton, FL: 2008. pp. 665–697. [Google Scholar]
- 175.Zhao C., Crosby J., Lv T., Bai D., Monia B.P., Guo S. Antisense oligonucleotide targeting of mRNAs encoding ENaC subunits alpha, beta, and gamma improves cystic fibrosis-like disease in mice. J. Cyst. Fibros. 2019;18:334–341. doi: 10.1016/j.jcf.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 176.Bennett C.F., Krainer A.R., Cleveland D.W. Antisense oligonucleotide therapies for neurodegenerative diseases. Annu. Rev. Neurosci. 2019;42:385–406. doi: 10.1146/annurev-neuro-070918-050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Crooke S.T., Baker B.F., Xia S., Yu R.Z., Viney N.J., Wang Y., Tsimikas S., Geary R.S. Integrated assessment of the clinical performance of GalNAc3- conjugated 2'-O-methoxyethyl chimeric antisense oligonucleotides: I. Human volunteer experience. Nucleic Acid Ther. 2019;29:16–32. doi: 10.1089/nat.2018.0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Benson M.D., Waddington-Cruz M., Berk J.L., Polydefkis M., Dyck P.J., Wang A.K., Plante-Bordeneuve V., Barroso F.A., Merlini G., Obici L., Scheinberg M., Brannagan T.H., 3rd, Litchy W.J., Whelan C., Drachman B.M. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PubMed] [Google Scholar]
- 179.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 180.Paz S., Hsiao J., Cauntay P., Soriano A., Bai L., Machemer T., Xiao X., Guo S., Hung G., Younis H., Bennett C.F., Henry S., Yun T.J., Burel S. The distinct and cooperative roles of toll-like receptor 9 and receptor for advanced glycation end products in modulating in vivo inflammatory responses to select CpG and non-CpG oligonucleotides. Nucleic Acid Ther. 2017;27:272–284. doi: 10.1089/nat.2017.0668. [DOI] [PubMed] [Google Scholar]
- 181.Senn J.J., Burel S., Henry S.P. Non-CpG-containing antisense 2'- methoxyethyl oligonucleotides activate a proinflammatory response independent of toll- like receptor 9 or myeloid differentiation factor 88. J. Pharmacol. Exp. Ther. 2005;314:972–979. doi: 10.1124/jpet.105.084004. [DOI] [PubMed] [Google Scholar]
- 182.Flaim J.D., Grundy J.S., Baker B.F., McGowan M.P., Kastelein J.J. Changes in mipomersen dosing regimen provide similar exposure with improved tolerability in randomized placebo-controlled study of healthy volunteers. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McGowan M.P., Moriarty P.M., Backes J. The effects of mipomersen, a second generation antisense oligonucleotide, on atherogenic (apoB containing) lipoproteins in the treatment of homozygous familial hypercholesterolemia. Clin. Lipidol. 2014;9:487–503. [Google Scholar]
- 184.Ng W.L., Marinov G.K., Chin Y.-M., Lim Y.-Y., Ea C.-K. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci. Rep. 2017;7:12227. doi: 10.1038/s41598-017-12550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miller T., Cudkowicz M., Shaw P.J., Andersen P.M., Atassi N., Bucelli R.C., Genge A., Glass J., Ladha S., Ludolph A.L., Maragakis N.J., McDermott C.J., Pestronk A., Ravits J., Salachas F. Phase 1-2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 2020;383:109–119. doi: 10.1056/NEJMoa2003715. [DOI] [PubMed] [Google Scholar]
- 186.Tabrizi S.J., Leavitt B.R., Landwehrmeyer G.B., Wild E.J., Saft C., Barker R.A., Blair N.F., Craufurd D., Priller J., Rickards H., Rosser A., Kordasiewicz H.B., Czech C., Swayze E.E., Norris D.A. Targeting huntingtin expression in patients with Huntington's disease. N. Engl. J. Med. 2019;380:2307–2316. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 187.Viney N.J., Tai L., Jung S., Yu R.Z., Guthrie S., Baker B.F., Geary R.S., CSchneider E., Guo S., Monie B.P. Phase 1 investigation of a ligand- conjugated antisense oligonucleotide with increased potency for the treatment of transthyretin amyloidosis. J. Card. Fail. 2019;25:S80–S81. [Google Scholar]
- 188.Therapeutics Sarepta. Globe Newswire; Cambridge, MA: 2019. Sarepta Therapeutics Announces Positive Expression Results from the Casimersen (SRP-4045) Arm of the ESSENCE Study. [Google Scholar]
- 189.Miner P., Wedel M., Bane B., Bradley J. An enema formulation of alicaforsen, an antisense inhibitor of intercellular adhesion molecule-1, in the treatment of chronic, unremitting pouchitis. Aliment. Pharmacol. Ther. 2004;19:281–286. doi: 10.1111/j.1365-2036.2004.01863.x. [DOI] [PubMed] [Google Scholar]
- 190.Buller H.R., Bethune C., Bhanot S., Gailani D., Monia B.P., Raskob G.E., Segers A., Verhamme P., Weitz J.I., Investigators F.-A.T. Factor XI antisense oligonucleotide for prevention of venous thrombosis. N. Engl. J. Med. 2015;372:232–240. doi: 10.1056/NEJMoa1405760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Morgan E.S., Tai L.J., Pham N.C., Overman J.K., Watts L.M., Smith A., Jung S.W., Gajdosik M., Krssak M., Krebs M., Geary R.S., Baker B.F., Bhanot S. Antisense inhibition of glucagon receptor by IONIS-GCGRRx improves type 2 diabetes without increase in hepatic glycogen content in patients with type 2 diabetes on stable metformin therapy. Diabetes Care. 2019;42:585–593. doi: 10.2337/dc18-1343. [DOI] [PubMed] [Google Scholar]
- 192.Loomba R., Morgan E., Watts L., Xia S., Hannan L.A., Geary R.S., Baker B.F., Bhanot S. Novel antisense inhibition of diacylglycerol O-acyltransferase 2 for treatment of non-alcoholic fatty liver disease: A multicentre, double-blind, randomised, placebo-controlled phase 2 trial. Lancet Gastroenterol. Hepatol. 2020;5:829–838. doi: 10.1016/S2468-1253(20)30186-2. [DOI] [PubMed] [Google Scholar]
- 193.Chi K.N., Yu E.Y., Jacobs C., Bazov J., Kollmannsberger C., Higano C.S., Mukherjee S.D., Gleave M.E., Stewart P.S., Hotte S.J. A phase I dose- escalation study of apatorsen (OGX-427), an antisense inhibitor targeting heat shock protein 27 (Hsp27), in patients with castration-resistant prostate cancer and other advanced cancers. Ann. Oncol. 2016;27:1116–1122. doi: 10.1093/annonc/mdw068. [DOI] [PubMed] [Google Scholar]
- 194.Limmroth V., Barkhof F., Desem N., Diamond M.P., Tachas G. CD49d antisense drug ATL1102 reduces disease activity in patients with relapsing-remitting MS. Neurology. 2014;83:1780–1788. doi: 10.1212/WNL.0000000000000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Trainer P.J., Newell-Price J.D.C., Ayuk J., Aylwin S.J.B., Rees A., Drake W., Chanson P., Brue T., Webb S.M., Fajardo C., Aller J., McCormack A.I., Torpy D.J., Tachas G., Atley L. A randomised, open- label, parallel group phase 2 study of antisense oligonucleotide therapy in acromegaly. Eur. J. Endocrinol. 2018;179:97–108. doi: 10.1530/EJE-18-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yuen M.F., Jang J.-W., Jang J.-H., Yoon J.-H., Kweon Y.-O., Park S.-J., Bennett C.F., Kwoh T.J. Phase 2a, randomized, double-blind, placebo controlled study of an antisense inhibitor (ISIS 505358) in treatment-naïve chronic hepatitis B (CHB) patients: Safety and antiviral efficacy. Hepatology. 2019;70 [Google Scholar]
- 197.Reilley M.J., McCoon P., Cook C., Lyne P., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Curran M., Liu Q., Zhou T., Schmidt J., Jo M. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: Results of a phase 1b trial. J. Immunother. Cancer. 2018;6:119. doi: 10.1186/s40425-018-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hong D., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Zhou T., Schmidt J., Jo M., Lee S.J., Yamashita M., Hughes S.G., Fayad L., Piha- Paul S. AZD9150, a next- generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chowdhury S., Burris H.A., Patel M., Infante J.R., Jones S.F., Voskoboynik M., Parry K., Elvin P., Coleman T., Gardner H., Lyne P.D., Arkenau H.T. A phase I dose escalation, safety and pharmacokinetic (PK) study of AZD5312 (IONIS- ARRx), a first-in-class generation 2.5 antisense oligonucleotide targeting the androgen receptor (AR) Eur. J. Cancer. 2016;69 [Google Scholar]
- 200.Querfeld C., Foss F.M., Pinter-Brown L.C., Porus P., William B.M., Pacheco T., Haverkos B.M., Kim Y.H., HGuitart J., Halwani A.S., DeSimone J., Seto A.G., Pestano L.A., Jackson A.L., Williams P.J. Phase 1 study of the safety and efficacy of MRG-106, a synthetic inhibitor of microRNA-155, in CTCL patients. Blood. 2017;130(Supplement 1):820. [Google Scholar]
- 201.Gaudet D., Karwatowska-Prokopczuk E., Baum S.J., Hurh E., Kingsbury J., Bartlett V.J., Figueroa A.L., Piscitelli P., Singleton W., Witztum J.L., Geary R.S., Tsimikas S., St L.O.D.L. Vupanorsen, an N-acetyl galactosamine-conjugated antisense drug to ANGPTL3 mRNA, lowers triglycerides and atherogenic lipoproteins in patients with diabetes, hepatic steatosis, and hypertriglyceridaemia. Eur. Heart J. 2020;41:3936–3945. doi: 10.1093/eurheartj/ehaa689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Akcea Therapeutics. Ionis Pharmaceuticals . PRNewswire; Boston, MA and Carlsbad, CA: 2020. Akcea and Ionis Report Positive Topline Phase 2 Results of AKCEA-APOCIII-L Rx. [Google Scholar]
- 203.Yuen M., Heo J., Kumada H., Suzuki F., Suzuki Y., Xie Q., Jia J., HKarino Y., Hou J., Chayama K., Imamura M., Iao-Tao J.Y., Lim S.G., Tanaka Y., Xie W. AASLD; Boston, MA: 2019. Results After 12 Weeks Treatment of Multiple Doses of GSK3389404 in Chronic Hepatitis B (CHB) Subjects on Stable Nucleos(t)ide Therapy in a Phase 2a Double-Blind, Placebo- Controlled Study. [Google Scholar]
- 204.Grossman T.R., Carrer M., Shen L., Johnson R.B., Hettrick L.A., Henry S.P., Monia B.P., McCaleb M.L. Reduction in ocular complement factor B protein in mice and monkeys by systemic administration of factor B antisense oligonucleotide. Mol. Vis. 2017;23:561–571. [PMC free article] [PubMed] [Google Scholar]
- 205.Cohn D.M., Viney N.J., Fijen L.M., Schneider E., Alexander V.J., Xia S., Kaeser G.E., Nanavati C., Baker B.F., Geary R.S., Levi M., Meijers J.C.M., Stroes E.S.G. Antisense inhibition of Prekallikrein to control hereditary angioedema. N. Engl. J. Med. 2020;383:1242–1247. doi: 10.1056/NEJMoa1915035. [DOI] [PubMed] [Google Scholar]
- 206.Pfeiffer N., Voykov B., Renieri G., Bell K., Richter P., Weigel M., Thieme H., Wilhelm B., Lorenz K., Feindor M., Wosikowski K., Janicot M., Packert D., Rommich R., Mala C. First-in-human phase I study of ISTH0036, an antisense oligonucleotide selectively targeting transforming growth factor beta 2 (TGF-beta2), in subjects with open-angle glaucoma undergoing glaucoma filtration surgery. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Giljohann D. The 15th Annual Meeting of the Oligonucleotide Therapeutics Society. 2019. Clinical results from XCUR17, a topically applied antisense spherical nucleic acid in patients with psoriasis. Munich, Germany. [Google Scholar]
- 208.Digenio A., Pham N.C., Watts L.M., Morgan E.S., Jung S.W., Baker B.F., Geary R.S., Bhanot S. Antisense inhibition of protein tyrosine phosphatase 1B with IONIS-PTP-1BRx improves insulin sensitivity and reduces weight in overweight patients with type 2 diabetes. Diabetes Care. 2018;41:807–814. doi: 10.2337/dc17-2132. [DOI] [PubMed] [Google Scholar]
- 209.Bhanot S., Morgan E., Bethune C., Larouche R., S X., Geary R. ISIS- GCCRRX, a novel glucocorticoid (GC) receptor antisense drug reduces cholesterol and triglycerides and attenuates dexamethasone induced hepatic insulin resistance without systemic GC antagonism in normal subjects. Diabetologia. 2013;56:S282. [Google Scholar]
- 210.Ferrone J.D., Bhattacharjee G., Revenko A.S., Zanardi T.A., Warren M.S., Derosier F.J., Viney N.J., Pham N.C., Kaeser G.E., Baker B.F., Schneider E., Hughes S.G., Monia B.P., MacLeod A.R. IONIS-PKKRx a novel antisense inhibitor of prekallikrein and bradykinin production. Nucleic Acid Ther. 2019;29:82–91. doi: 10.1089/nat.2018.0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sands B.E., Feagan B.G., Sandborn W.J., Schreiber S., Peyrin-Biroulet L., Frederic Colombel J., Rossiter G., Usiskin K., Ather S., Zhan X., D'Haens G. Mongersen (GED-0301) for active Crohn's disease: Results of a phase 3 study. Am. J. Gastroenterol. 2020;115:738–745. doi: 10.14309/ajg.0000000000000493. [DOI] [PubMed] [Google Scholar]
- 212.Monteleone G., Pallone F. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N. Engl. J. Med. 2015;372:2461. doi: 10.1056/NEJMc1504845. [DOI] [PubMed] [Google Scholar]
- 213.Goemans N.M., Tulinius M., van den Hauwe M., Kroksmark A.K., Buyse G., Wilson R.J., van Deutekom J.C., de Kimpe S.J., Lourbakos A., Campion G. Long-term efficacy, safety, and pharmacokinetics of drisapersen in Duchenne muscular dystrophy: Results from an open-label extension study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., De Kimpe S.J., Eagle M., Guglieri M., Hood S., Liefaard L., Lourbakos A., Morgan A., Nakielny J., Quarcoo N. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo- controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 215.McCarty T., Charlesworth D. Globe Newswire; San Rafael, CA: 2016. BioMarin Announces Withdrawal of Market Authorization Application for Kyndrisa™ (Drisapersen) in Europe. [Google Scholar]
- 216.Saad F., Hotte S., North S., Eigl B., Chi K., Czaykowski P., Wood L., Pollak M., Berry S., Lattouf J.B., Mukherjee S.D., Gleave M., Winquist E., Canadian Uro- Oncology G. Randomized phase II trial of Custirsen (OGX-011) in combination with docetaxel or mitoxantrone as second-line therapy in patients with metastatic castrate-resistant prostate cancer progressing after first-line docetaxel: CUOG trial P-06c. Clin. Cancer Res. 2011;17:5765–5773. doi: 10.1158/1078-0432.CCR-11-0859. [DOI] [PubMed] [Google Scholar]
- 217.OncoGenex Pharmaceuticals, Inc. PRNewswire; Bothell, WA and Vancouver, BC: 2016. OncoGenex Announces Results from the Phase 3 AFFINITY Trial of Custirsen in Men with Metastatic Castrate-Resistant Prostate Cancer. [Google Scholar]
- 218.Tsimikas S., Viney N.J., Hughes S.G., Singleton W., Graham M.J., Baker B.F., Burkey J.L., Yang Q., Marcovina S.M., Geary R.S., Crooke R.M., Witztum J.L. Antisense therapy targeting apolipoprotein(a): A randomised, double-blind, placebo-controlled phase 1 study. Lancet. 2015;386:1472–1483. doi: 10.1016/S0140-6736(15)61252-1. [DOI] [PubMed] [Google Scholar]
- 219.Miller T.M., Pestronk A., David W., Rothstein J., Simpson E., Appel S.H., Andres P.L., Mahoney K., Allred P., Alexander K., Ostrow L.W., Schoenfeld D., Macklin E.A., Norris D.A., Manousakis G. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sewell K.L., Geary R.S., Baker B.F., Glover J.M., Mant T.G., Yu R.Z., Tami J.A., Dorr F.A. Phase I trial of ISIS 104838, a 2'-methoxyethyl modified antisense oligonucleotide targeting tumor necrosis factor-alpha. J. Pharmacol. Exp. Ther. 2002;303:1334–1343. doi: 10.1124/jpet.102.036749. [DOI] [PubMed] [Google Scholar]
- 221.Mignon L., Norris D., Bishop K., Derosier F., LANE R., Bennett F. ISIS- DMPKRx in healthy volunteers: A placebo-controlled, randomized, single ascending- dose phase 1 study (P3.166) Neurology. 2016;86 [Google Scholar]
- 222.Bianchini D., Omlin A., Pezaro C., Lorente D., Ferraldeschi R., Mukherji D., Crespo M., Figueiredo I., Miranda S., Riisnaes R., Zivi A., Buchbinder A., Rathkopf D.E., Attard G., Scher H.I. First-in-human phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br. J. Cancer. 2013;109:2579–2586. doi: 10.1038/bjc.2013.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Regulus Therapeutics . PRNewswire; La Jolla, CA: 2017. Regulus Announces Pipeline Updates and Advancements. [Google Scholar]
- 224.Tanowitz M., Hettrick L., Revenko A., Kinberger G.A., Prakash T.P., Seth P.P. Asialoglycoprotein receptor 1 mediates productive uptake of N- acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res. 2017;45:12388–12400. doi: 10.1093/nar/gkx960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kandimalla E.R., Bhagat L., Wang D., Yu D., Sullivan T., La Monica N., Agrawal S. Design, synthesis and biological evaluation of novel antagonist compounds of Toll-like receptors 7, 8 and 9. Nucleic Acids Res. 2013;41:3947–3961. doi: 10.1093/nar/gkt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Miller C.M., Donner A.J., Blank E.E., Egger A.W., Kellar B.M., Ostergaard M.E., Seth P.P., Harris E.N. Stabilin-1 and stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res. 2016;44:2782–2794. doi: 10.1093/nar/gkw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kotula J.W., Pratico E.D., Ming X., Nakagawa O., Juliano R.L., Sullenger B.A. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012;22:187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Castanotto D., Lin M., Kowolik C., Wang L., Ren X.Q., Soifer H.S., Koch T., Hansen B.R., Oerum H., Armstrong B., Wang Z., Bauer P., Rossi J., Stein C.A. A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 2015;43:9350–9361. doi: 10.1093/nar/gkv964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Abdul-Manan N., Williams K.R. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Weidner D.A., Valdez B.C., Henning D., Greenberg S., Busch H. Phosphorothioate oligonucleotides bind in a non sequence-specific manner to the nucleolar protein C23/nucleolin. FEBS Lett. 1995;366:146–150. doi: 10.1016/0014-5793(95)00517-d. [DOI] [PubMed] [Google Scholar]
- 231.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016;44:731. doi: 10.1093/nar/gkw595. [DOI] [PMC free article] [PubMed] [Google Scholar]