Abstract
Intracellular pH (pHi) is a crucial parameter in cell biology; thus, a series of pH probes have been developed to determine pHi changes in living cells. However, more sensitive and non-perturbing ratiometric pH probes are needed for accurate pHi measurements. While the fluorescence of circular permutated YFP (cpYFP) is hypersensitive to pH changes due to its intrinsic properties, the single excitation peak of this protein restricts its capacity of becoming a rational type of pH sensor. Herein, we collected several cpYFP-based probes with dual excitation peaks and constructed their corresponding loss-of-function mutants to screen for a potential competent pH probe. The most sensitive probe was named NocPer. NocPer consists of cpYFP inserted into inactive-mutated GAF and AAA+, which are two regulatory domains of E. coli NorR, a nitric oxide (NO)-specific transcription factor. Fluorescence emission of NocPer peaks at 517 nm while exhibiting dual excitation peaks at 420 and 495 nm, which can be used for ratiometric imaging. This new pH sensor has a large ratio response dynamic (pH range of 7.0–11.0), which covers the physiological pH range (pH 7.0–8.0), and exhibits an approximately 3-fold higher fluorescent signal in response to a pH increase from 7.0 to 8.0 than that of pHluorin. Using NocPer, we discovered a new biological phenomenon in which NO exposure decreases the E. coli pHi, which led to the hypothesis that pathogens decrease their own pHi during infection. Further, we elucidated that the NO-induced inhibition of cytochrome c oxidase in the respiratory chain is responsible for the decline in pHi, which might represent a protective strategy of E. coli under NO stress conditions. Our results demonstrated that NocPer is a ratiometric pH probe with high sensitivity for the physiological pH range.
Keywords: Fluorescent probe, cpYFP, pH indicator, Nitric oxide
Highlights
-
•
Circular permutated YFP was modified to be an a supersensitive and ratiometric pH probe NocPer.
-
•
Nitric oxide (NO) lowering intracellular pH (pHi) was discovered as a new biological phenomenon.
-
•
NO-induced inhibition of cytochrome c oxidase is responsible for the decline in pHi.
-
•
pHi decrease by NO might represent a bacterial protective mechanism.
1. Introduction
Intracellular pH (pHi) plays a crucial role in biochemical networks because changes in pHi affect cell functioning at different levels. It impacts protein folding, enzymatic activities, the protonation of biological macromolecules, lipids, and other metabolites [1]. Moreover, the proton electrochemical gradient across the cell membrane represents a key factor in the generation and conversion of cellular energy [2]. Recently, it has been reported that acidification of the cytoplasm drives its transition from a fluid-like to a solid-like state, promoting cellular survival under unfavourable conditions [3]. An increasing number of pH-sensing proteins have been identified in established signalling pathways, which supports the emerging view that protons can act as second messenger regulators of cell signalling, survival, and stress responses [4]. Therefore, pHi is a crucial parameter in cell biology.
To exactly understand the cellular function of pHi, a series of genetically encoded fluorescent proteins (FP)-based pH probes have been developed [[5], [6], [7]]. Among them, pHluorin is the most prevalent ratiometric type sensor, which is derived from a pH-sensitive GFP variant that contains clusters of mutations that direct pH sensitivity. It displays a proton dose-dependent fluorescence decrease in excitation at 395 nm with a concomitant increase in excitation at 475 nm. However, pHluorin does not exhibit high sensitivity for pH changes in the normal physiological pH range, an increase in the pH from 7.0 to 8.0 leading to less than 1.5-fold change in the fluorescence ratio (F410/F470) [8]. Thus, noise might have non-negligible effects on the measurements. While several GFP-based types and one RFP-based type of pH sensors have also been developed to meet different demands, the enhancement of their pH sensitivities were not significant [7,[9], [10], [11]]. Thus, using a responsive element far exceeding GFP's and its point mutant variants' pH sensitivity to develop a novel pH sensor for accurate observation of the pHi flux is needed. Circular permuted fluorescent proteins (cpFPs) including cpGFP, cpCFP, and cpYFP are the basis for many genetic sensors, such as Perceval [12], pericam [13], SoNar [14], and FHisJ [15]. cpFPs are derived from rearrangements of FPs, in which the amino and carboxyl portions are interchanged and re-joined with a short spacer connecting the original termini. cpFPs remain fluorescent while appearing more accessible to protons outside of the proteins due to the exposure of the pH-active phenoxy group of the chromophore lead by structural perturbation. In other words, cpFPs exhibit a higher pH change sensitivity than their native counterparts. Among cpFPs, cpYFP has the strongest response to acids [16]. It has been shown that cpYFP shows a >50-fold change in fluorescence intensity between pH 7 and 10, and minor pH perturbations elicit a pronounced signal response [17]. The extraordinary pH sensitivity of cpYFP makes it a potential pH sensor with better performance than GFP-based pH sensors such as pHluorin. However, the single fluorescence excitation peak of cpYFP (490 nm) restricts its capacity of becoming a rational pH probe. The first and unique cpYFP-based pH sensor, SypHer [18], was designed by Damon et al. by mutating HyPer [19], a well-known H2O2-sensing probe, at the H2O2-sensing cysteine residues. Compositionally, SypHer extended 127 and 106 amino acids at the N and C termini of cpYFP, respectively, which further enlarged the fluorescence changes upon pH change. However, although the 490/420 fluorescence ratio of this probe is sensitive to pH changes, this is mainly attributed to the marked fluorescence intensity decline at 490 nm rather than the weak fluorescence intensity increase at 420 nm, which remains the performance of the absolute quantitation-type probe. Notably, our recently reported H2O2 sensor, TScGP [20], composed of cpYFP sandwiched between a fungal peroxiredoxin and thioredoxin, shows two sharp excitation peaks at 415 and 500 nm, overcoming the inherent weaknesses of cpYFP as a pH probe. The responses of these modified cpYFPs to pH changes suggested that extending the N- and C-termini of cpYFP with appropriate peptides might optimize the performance of cpYFP-based pH probes.
Nitric oxide (NO) is an important antimicrobial molecule produced by human immune cells to eliminate invading microorganisms. However, pathogens have evolved various strategies to defend themselves against this immune effector. Considerable efforts have been made to identify and characterize NO detoxification enzymes and their corresponding regulatory mechanisms in response to NO [21,22]. However, there have been no studies that have focused on the changes in the cellular environment following bacterial NO stimulation or the consequential effects on the physiological functions of the cell. Nevertheless, sugars have been reported to act via the vacuolar ATPase (V-ATPase) and an ATP-driven proton pump (Pma1) to mediate pHi in yeast as a means of adapting to environmental stresses caused by hypoxia, ischemia, and diabetes [4,23]. In addition, exposure of E. coli to menadione (a known oxidizing agent) was reported to induce acidification of the cytoplasm of E. coli by an unknown pathway, and the resultant pHi change brought about an antibiotic-resistance phenotype [24]. Given that NO stimulation also causes pHi changes with biological functions, it will further illustrate that pHi change in response to the ex vivo stimulations is a conserved self-protection mechanism among living organisms. In this study, we screened several existing and newly constructed cpYFP-based pH probes that have two sharp excitation peaks and identified NocPer as the most sensitive pH probe among them. This new sensor has been validated to be more sensitively than pHluorin, and have a larger ratio response dynamic which well covers the physiological pH range; thus, demonstrating that NocPer is suitable for pHi sensing. Using NocPer, we noticed for the first time that NO exposure slightly decreases the pHi of E. coli. Furthermore, we demonstrated that NO-induced inhibition of the proton pump cytochrome c oxidase should be responsible for this pHi change, which might represent an efficient survival strategy employed by bacteria under NO stress conditions.
2. Materials and methods
2.1. Plasmid construction
The pHluorin coding gene sequence [8] was synthesized and inserted into the NdeI/BamHI sites of pET15b. The recombinant expression vectors pET28a-cpYFP, pET28a-HyPer, and pET28a-TScGP were constructed as previously described [16,19,20]. To construct the expression vector pET28a-GcpA, the cpYFP coding sequence was amplified from the pET28a-cpYFP plasmid using Pf1 and Pr1, and the gene encoding sequences of the GAF and AAA+ domains were amplified from E. coli genomic DNA using Pf2, Pr2 and Pf3, Pr3, respectively. Three PCR fragments were inserted into the EcoRI/HindIII sites of pET28a according to the protocol outlined in the ClonExpress MultiS One Step Cloning Kit (Vazyme, China). SypHer, TScGP (mut), and GcpA (mut) were generated by site-directed mutagenesis using the recombinant plasmids pET28a-HyPer, pET28a-TScGP, and pET28a-GcpA as the templates, respectively, and a QuikChange Site-Directed Mutagenesis Kit (Stratagene, USA). The mutated sites were in line with those presented in previous literature [18,20,25]. All amplification reactions were performed using the PrimeSTAR HS DNA Polymerase (Takara, Japan). The primers used in all amplification reactions are shown in Supplementary Table 1.
2.2. Protein expression and purification
All expression plasmids were transformed into E. coli BL21 (DE3). E. coli cells were grown in Luria-Bertani (LB) medium with 50 μg/mL of the corresponding antibiotics (kanamycin for pET28a and ampicillin for pET15b) and induced by 100 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 20 °C for 16 h. The cells were collected and then disrupted by sonication in buffer A (20 mM NaH2PO4 and 0.5 mM NaCl, pH 7.4). Proteins were purified using HiTrap Chelating HP (GE Healthcare, USA) and eluted with buffer B (buffer A supplemented with 0.5 M imidazole). The elution buffer in the purified protein solution was substituted with 50 mM sodium phosphate buffer (pH 7.4) using Amicon Ultra concentrators (Millipore, USA).
2.3. Fluorescence detection
Fluorescence detection was performed using an F-4600 fluorescence spectrophotometer (HITACHI, Japan) at room temperature (25 °C). For excitation and emission scans of the tested probes, the emission or excitation values were set according to previous literature [8,[17], [18], [19], [20]]. For NocPer, the excitation spectra were recorded from 380 to 510 nm with an emission wavelength of 525 nm, and the emission spectra were recorded from 500 to 600 nm with an excitation wavelength of 465 nm. Following initial fluorescence spectrum recording, various amounts of additives were added to the reaction mixture and the spectra were immediately remeasured. The excitation and emission slits were both set to 5 nm, the wavelength scan speed was 2400 nm/min, and the photomultiplier tube (PMT) voltage was 700 V.
2.4. In situ pH measurements and NocPer calibration
E. coli BL21 (DE3) expressing the probes were grown in LB media to an OD600 of approximately 1.0, collected by centrifugation, and rinsed twice prior to each assay. Cells were resuspended and treated with proton carriers (100 μM monensin, 10 μM nigericin) and 2% glucose in buffers from pH 6.0 to 12 containing 100 mM MES, 100 mM MOPS, 100 mM Tris-HCl, 100 mM CHES, 25 mM Na2CO3–NaOH, and 50 mM NaH2PO3–NaOH. After a 10 min incubation period, the ratio of emission intensity resulting from the excitation at 495 and 420 nm was calculated (ratio 495/420) and plotted against the corresponding buffer pH. A wildtype culture was simultaneously grown as a background fluorescence reference for ground subtraction at both separate excitation wavelengths. All pH determination experiments were repeated at least three times.
2.5. Fluorescence responses of pH probes to NO stimulation
To observe fluorescence responses of pH probes to NO derived from NO donor, a series concentrations of NO donors, including PROLI NONOate and MAHMA NONOate (Cayman Chemical, USA), were dropped into 100 mM MOPS (pH 7.4) and resuspended with appropriate concentrations of E. coli cells. Next, the fluorescence responses were recorded. As control for the NO treatment experiments, 100 μM carboxy-PTIO (NO scavenger) (Sigma, USA) was mixed with NO donors to block NO release.
The macrophage RAW 264.7 cell lines were also used to produce NO for the stimulation of the pH sensor fluorescence responses. RAW 264.7 cells were resuspended in fresh DMEM containing 1 μg/μL E. coli O111:B4 lipopolysaccharide (Sigma, USA) and 1 mM l-arginine (Sigma, USA) to induce NO production either with or without 300 μg/mL NG-monomethyl arginine citrate (L-NMMA; Sigma, USA), which is a known NO synthase inhibitor. The RAW 264.7 cells were seeded into culture dishes with a diameter of 34.8 mm at a concentration of 5 × 105 cells per well. After 20 h of incubation, 2.5 μmol/L NO specific probe DAF-FM DA (Beyotime, China) was added and incubated for another 30 min. RAW 264.7 cells were washed with PBS three times and the intracellular NO production was evaluated using confocal laser scanning microscope (Lecia TCS SP8, Germany). To observe the fluorescence responses of pH probe to NO released from RAW264.7 cells, E. coli strains expressing-pH probes were co-incubated with activated RAW 264.7 cells for 20 min. Lastly, cells were collected by centrifugation and then washed as well as resuspended with 20 mM MOPS buffer (pH 7.4) for fluorescence detection.
2.6. Detection of membrane potential
The cell membrane potential was detected using the fluorescent dye DiSC3(5) (AAT Bioquest, USA). E. coli cells were grown to the logarithmic growth stage and then suspended in MOPS buffer (100 mM MOPS, 2% glucose, pH 7.4) to adjust the cell density to 1 × 106/mL. The dye was added to the solution at a final concentration of 1 μM followed by incubation at 37 °C for 10 min. The excitation wavelength of DiSC3(5) was fixed at 590 nm, while the time-scan of the fluorescence intensity of the emission wavelength was fixed at 680 nm.
2.7. Determination of intracellular ATP levels
ATP concentration was measured using an ATP bioluminescent somatic cell assay kit (Sigma, USA) following the manufacturer's instructions. Bioluminescence was measured using a Multi-Mode microplate reader (BioTek, USA).
2.8. NO tolerance capability of E. coli
To explore the impact of the pHi decrease on the NO tolerance capacity of E. coli, the OD600 was monitored for their growth rate in different pH liquid medium in the presence of NO donor DETA NONOate (Sigma, USA). DH5α E. coli cells were pre-cultivated in 4 mL liquid LB medium for 10 h (37 °C, 220 rpm). Next, the cultures were equally distributed into four flasks with 100 mL fresh LB media. To modulate the pHi to pH 7.1 and pH 7.6, the media were prepared in 50 mM HEPES buffer with the indicated pH values containing proton carries (100 μM monensin, 10 μM nigericin). When the cells were grown to an OD600 = 0.5, 5 mM DETA NONOate was added to test the nitric oxide stress on growth rates evaluated using OD measurements.
2.9. Quantification of NO release from NO donors
NO released from PROLI NONOate and MAHMA NONOate was measured amperometrically using an ISO–NOP NO electrode (World Precision Instruments, USA) as previously described. The NO electrode was calibrated prior to each experiment by detection of known amounts of NO produced from the reaction between sodium nitrite and potassium iodide under acidified conditions (Supplementary Fig. 2).
2.10. Statistical analysis
All results in this study are presented either as a representative example of a single experiment that was repeated at least in triplicate or as at least three independent experiments. All experimental data are presented as the mean ± SEM. One-way ANOVA followed by Bonferroni's post-test were performed. P-values <0.05 were considered statistically significant (*p < 0.05, **p < 0.01, and ***p < 0.001).
3. Results
3.1. Comparison of cpYFP and pHluorin responses to pH
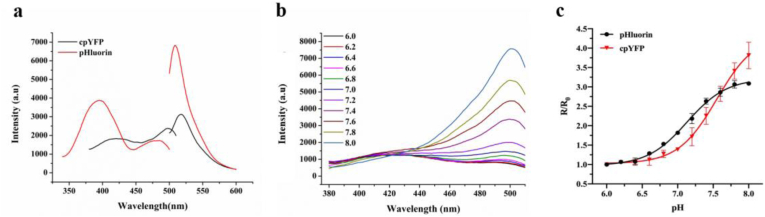
To compare the performance of pHluorin and cpYFP during pH measurement, we first prepared both recombinant proteins using E. coli expression systems and then observed their responses to changes in pH. Purified cpYFP possesses a bimodal excitation profile with a major peak at 495 nm and a minor peak at 420 nm measured with an emission maximum at 517 nm (Fig. 1a). Alternatively, pHluorin shows two excitation peaks at 395 and 475 nm and one emission peak at 508 nm (Fig. 1a). The fluorescence spectra of both proteins tested here are consistent with observations from previous reports [8,17]. Changes in the ratio of excitation intensities at 495 nm and 420 nm (F495/F420) upon pH fluctuation were used to reflect the pH dependency of cpYFP fluorescence rate changes. As shown in a previous report and our present work, the excitation peak at 420 nm is too broad and weak to be read under most pH conditions (Fig. 1b), which is the major defect of cpYFP as a reliable rational pH indicator [17]. In comparison, upon acidification, the excitation peak of pHluorin at 395 nm decreases with a corresponding increase in the excitation peak at 475 nm, showing a ratiometric signal change [8].
Fig. 1.
Fluorescence spectroscopic analysis of the cpYFP and pHluorin response to pH changes. (a) Fluorescence excitation and emission spectra of purified cpYFP and pHluorin in 50 mM phosphate buffer (pH 7.4). (b) Fluorescence excitation spectra (emission at 515 nm) of purified cpYFP in response to different pH values. (c) Fluorescence ratio changes of intracellular pHluorin and cpYFP in response to different pH values. Prior to fluorescence analysis, cells were suspended in buffers with pH ranging between 6.0 and 8.0 containing 2% glucose and treated with monensin and nigericin for 10 min. R represents the F495 nm/F420 nm ratio for cpYFP or the F475 nm/F395 nm ratio for pHluorin at various pH values and was normalized to their corresponding values at pH 6.0 (R0).
Next, we compared the pH sensitivities of cpYFP and pHluorin expressed in E. coli cells. To manipulate the pHi, living cells were incubated in buffers with different pH values ranging from 6.0 to 8.0-containing H+ ionophores (100 μM monensin, 10 μM nigericin) to allow for equilibration of the extracellular and intracellular pH. For sensitivity comparison convenience, the excitation fluorescence rate changes of both probes at different pH values (R) were normalized to their respective values at pH 6.0 (R0). Between pH 6.0 and 8.0, both the F495/F420 of cpYFP and F395/F475 of pHluorin increased in a pH-dependent manner (Fig. 1c). Notably, the fluorescence signal change level of pHluorin was higher than that of cpYFP for pH ranging between 6.5 and 7.0, while for pH ranging between 7.0 and 8.0, the fluorescence signal change of cpYFP was more pronounced (Fig. 1c). This indicates that cpYFP exhibits a better match with the cytosol pH range (fluctuating around approximately 7.5), which makes cpYFP a potential highly sensitive pH probe capable to measure the changes in pHi.
3.2. Identification of the new pH probe NocPer
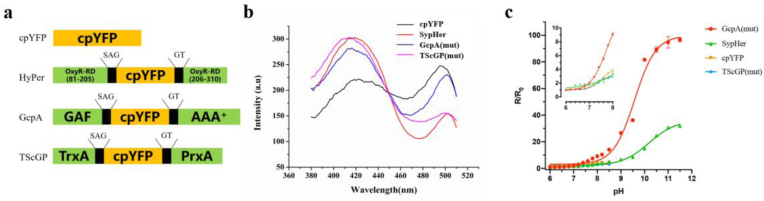
While the fluorescence peak of cpYFP excited at 495 nm sensitively responds to pH changes, the absence of another exact excitation peak makes cpYFP an absolute quantitation-type probe rather than a ratiometric-type probe. Previous studies showed that several cpYFP-based probes, such as HyPer and TScGP, have bimodal excitation peaks [19,20], which led us to propose that extending each terminus of cpYFP might be an effective method to generate a cpYFP-based ratiometric probe. HyPer, consisting of cpYFP inserted into the regulatory domain of a prokaryotic H2O2-sensing protein (Fig. 2a), is one of the most prevalent H2O2-sensing probes. TScGP is another highly sensitive H2O2 probe developed by our group. Structurally, TScGP comprises of a cpYFP sandwiched between a peroxiredoxin and a thioredoxin (Fig. 2a). GcpA is a new unpublished probe exhibiting bimodal excitation peaks (Fig. S1), which was designed in our lab to detect NO. In GcpA, cpYFP is sandwiched between the GAF and AAA+, which are the two domains of the E. coli NO-specific transcriptional factor NorR [26,27].
Fig. 2.
Comparison of the responses of cpYFP, SypHer, GcpA (mut), and TScGP (mut) to pH changes. (a) Schematic models for cpYFP chimaeras. (b) Fluorescent excitation spectra of purified cpYFP, SypHer, GcpA (mut), and TScGP (mut) in 50 mM phosphate buffer (pH 7.4). (c) Fluorescence ratio changes of cpYFP, TScGP (mut) in response to intracellular pH values ranging between 6.0 and 8.0 as well as fluorescence ratio changes of SypHer, GcpA (mut) in response to intracellular pH values ranging between 6.0 and 11.5. Inset, dynamic ratio ranges of four proteins at physiological pH values (6.0–8.0). The F495 nm/F420 nm ratio of these four fluorescent proteins at different pH values (R) were normalized to their corresponding values at pH 6.0 (R0).
Ratiometric pH sensors. To generate a pH-specific version of these cpYFP-based sensors, we mutated H2O2-sensing residues of HyPer and TScGP as previously described by Belousov et al. and Yang et al. respectively, and mutated the NO-binding residue of GcpA (R75 deletion, R81G) as described by Eroglu et al. [18,20,25]. The resulting probes were SypHer [18], TScGP (mut), and GcpA (mut), respectively. As expected, the first two probes expressed in E. coli cells showed bimodal excitation peaks (Fig. 2b), which were in line with those observed in previous studies [18,20]. GcpA (mut) showed similar excitation and emission spectra to those of its parent variant GcpA, which exhibited two exact excitation peaks at 420 and 495 nm and one emission peak at 517 nm (Fig. S1).
Further comparison of the performance of these proteins as pH probes indicated that SypHer, TScGP (mut), and GcpA (mut) were all responsive to pH changes, while GcpA (mut) showed the highest sensitivity among the tested probes especially for samples with a pH over 7.0 (Fig. 2c). Moreover, for pH changes from 7.0 to 8.0, the R/R0 of GcpA (mut) increased 4.4-fold and was 2.6-fold higher than that of pHluorin expressed in E. coli (compared with Fig. 1c); thus, exhibiting a great advantage in pH sensing. In situ calibration of the GcpA (mut) ratio against pH showed a pKa value of 9.6, far exceeding the cytosolic physiological pH range (pH 7–8). Nevertheless, a nearly linear calibration curve can be obtained during the physiological pH range (Fig. 2c, inset), providing convenience for pHi measurements.
We evaluated the selectivity of GcpA (mut) as a pH sensor (Fig. S2b). The addition of the indicated concentrations of impurities, including several common metal ions and some oxidizing as well as reducing agents, did not induce drastic changes in the purified GcpA (mut) fluorescence, which is indicative of the high pH specificity of GcpA (mut). In general, the output signal stability is regarded as an essential characteristic of probes for accurate measurements. Purified GcpA (mut) showed no fluctuation in F495/F420 within 10 min at pH 7.0, 7.5, and 7.8, respectively (Fig. S2a), ensuring good output signal stability during pHi measurement. Thus, we named the new GcpA (mut) pH probe, NocPer, and used it for following pHi measurements.
3.3. Effects of NO stimulation on pHi
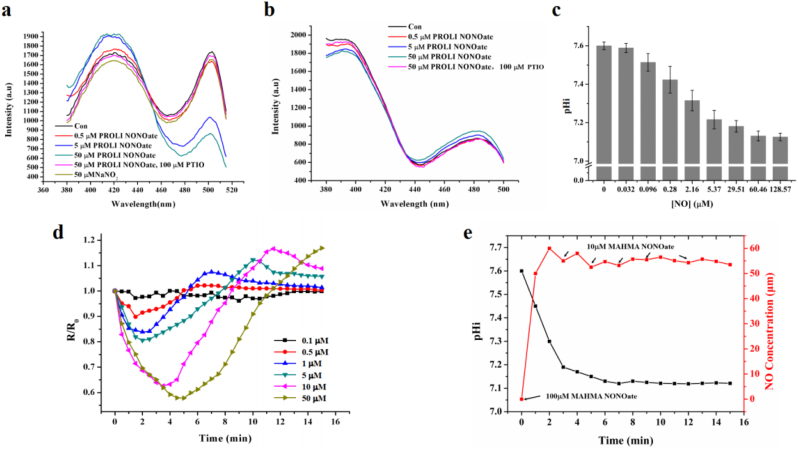
While purified NocPer did not react with NO (Fig. S2b), the treatment of E. coli expressing NocPer with an NO donor led to changes in the fluorescent signal, suggesting that the intracellular NocPer may respond to NO exposure (Fig. 3a). The NO donor, PROLI NONOate (half-time 2 s), at concentrations as low as 0.5 μM, was capable to initiate a NocPer response and the changes in the F495/F420 ratio became significant following NO donor concentration increase from 0.5 to 5 and 50 μM (Fig. 3a). Moreover, NO donor mixed with PTIO (NO scavenger) abolished the changes in NocPer signal, thus, excluding the possibility of a NO donor skeleton-dependent response of the pH probe (Fig. 3a). NO is known to autoxidize into nitrite under aerobic conditions, which might indicate that the pHi decline could be caused by the NO oxidation product rather than NO. However, exposing E. coli to 50 μM NO-equivalent nitrite did not yield any changes in the F495/F420 ratio (Fig. 3a), indicating that the intracellular NocPer does not respond to nitrite. Therefore, we concluded that although NocPer is not a NO probe, it can sense the intracellular changes triggered by NO stimulation in a NO-dose dependent manner.
Fig. 3.
NO stimulation decreases the pHi of E. coli. (a and b) Fluorescence responses (excitation spectra) of NocPer (a) and pHluorin (b) in E. coli to the indicated concentrations of PROLI NONOate. As control, 100 μM carboxy-PTIO (NO scavenger) and 50 μM NaNO2 was added, respectively. (c) pHi of E. coli following exposure to the indicated concentrations of NO. The concentration of NO released by PROLI NONOate was quantified using an ISO–NOP NO electrode. (d) Time course for the fluorescence ratio of intracellular NocPer following stimulation with the indicated concentrations of PROLI NONOate. F495 nm/F420 nm of intracellular NocPer following NO stimulation (R) was normalized to the corresponding value without NO stimulation (R0). (e) Constant pH decline was maintained by a continuous supply of NO donor. To maintain NO concentrations in reaction buffer, an appropriate amount of MAHMA NONOate was added at the points indicated by arrows.
Considering the molecular constitution of NocPer (Fig. 2a, GcpA), we hypothesized that only the cpYFP segment within NocPer might respond to the intracellular changes associated with NO stimulation, and the most likely intracellular change may be the pHi changes. To test this hypothesis, we investigated whether the fluorescence signal of intracellular pHluorin also changes upon NO exposure. While pHluorin did not respond to low doses of NO released by 0.5 μM NO donor, elevating the concentrations of the NO donor to 5 μM and especially to 50 μM caused a very slight increase in the excitation peak value at 475 nm and a decrease in the excitation peak value at 395 nm (Fig. 3b). Given the responsiveness of the two probes to NO treatment and the pH sensitivity of cpYFP, we demonstrated that NO treatment decreases the pHi of E. coli.
Based on the pH sensitivity distinction between NocPer and pHluorin (Fig. 1c and 2c), combined with the weak fluorescence changes of pHluorin following NO treatment, we hypothesized that the NO-induced pHi fluctuation was within a narrow range even for high NO concentration stimuli. To confirm this hypothesis, we used NocPer to quantitatively analyse the correlation between the pHi change and the NO exposure concentration. We estimated the amount of NO released from PROLI NONOate using the NO detection electrode ISO–NOP based on the standard calibration curve (Fig. S3). As previously mentioned, NO is a chemically unstable gaseous molecule which is released from NO donor while simultaneously being oxidized to nitrite; thus, we used the highest concentrations of NO accumulated in the reaction buffer measured by the NO electrode to indicate the NO concentrations used for cell treatments (Table S2). As shown in Fig. 3c, after treating the cells with NO at concentrations ranging between 0.1 μM and 60.5 μM, the pHi declined in a dose-dependent manner. The pHi changes induced by NO concentrations lower than 0.03 μM could not be detected by NocPer, while those induced by concentrations greater than 60 μM saturated the effects on pHi change. These results indicated that the NO-dependent pHi change is so subtle that they can only be captured by highly sensitive probes, such as NocPer, but not pHluorin.
3.4. Dynamic pHi changes in E. coli cells after NO exposure
Next, we tracked the dynamic pHi changes in E. coli cells following NO exposure. The addition of a series of concentrations of PROLI NONOate into the reaction buffer led to a rapid decline in R/R0 values in a NO donor dose-dependent manner (Fig. 3d). After decreasing into a valley, the pHi values began to recover, indicating that the decline in pHi was reversible. Moreover, we used another type of NO donor, MAHMA NONOate, which exhibits a slower NO release due to its longer half-life (3 min) compared to those of PROLI NONOate, to reproduce the effects of NO on the dynamic changes in pHi. The R/R0 changes in NocPer upon treatment with the two NO donors were similar, except for the time lag to reach the valley value of pHi following treatment with MAHMA NONOate (Fig. S4), which should be attributed to the longer half-life of the NO donor. Therefore, we concluded that NO induces a rapid and reversible decline of the pHi in E. coli.
Is the pHi recovery the result of NO exhaustion or self-adaptation of the cells to NO stress? To answer this question, we maintained the NO concentration at approximately 50 μM in reaction buffer by continuously re-adding an NO donor and simultaneously monitoring the pHi changes (Fig. 3e). We found that the pHi dropped from pH 7.6 to 7.1 within 5 min and then remained at this low level under these conditions. Together with the results presented in Fig. 3c, we concluded that this decline in pHi following NO stimulation is not a transient reaction but a constant effect.
3.5. pHi of E. coli responds to macrophage-derived NO
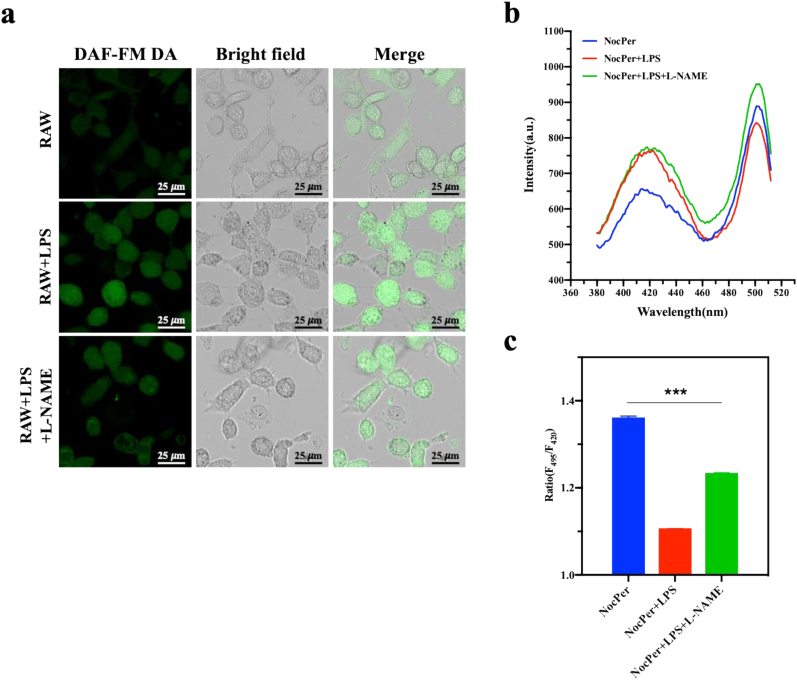
Following infection occurrence, microphages generate NO to defend against pathogens [28], which lead us to wonder whether bacteria decrease their pHi during the infection procedure. To address this question, we co-cultivated macrophage cells with E. coli harbouring NocPer to record the changes in pH. First, we confirmed that NO generation by macrophage RAW 264.7 cells using the NO specific probe DAF-FM DA. Only basal levels of NO were detected in 24-h cultured RAW cells without any treatments (Fig. 4a). As expected, E. coli O111:B4 lipopolysaccharide (LPS)-activated RAW cells produced more intracellular NO, while the well-known NO synthase inhibitor, NG-monomethyl arginine citrate (L-NMMA), decreases the NO levels in RAW cells (Fig. 4a). The following exposure of E. coli to RAW cells in distinct states yielded distinct NocPer fluorescent signals (Fig. 4b). The excitation fluorescence ratio F495/F420 followed the order of RAW > LPS and LNAME-treated RAW > LPS-treated RAW (Fig. 4c), which just reflected the order of NO levels in RAW cells and the opposite order of pHi in E. coli. Taken together, our work might have revealed a novel biological phenomenon including the capacity of pathogens to decrease their own pHi during infection.
Fig. 4.
The pHi of E. coli responding to macrophage-derived NO. (a) Fluorescence microscopy imaging of NO production in LPS-treated RAW 264.7 macrophage cell line and the effect of NO inhibitor L-NAME. LPS and L-NAME were used to activate and inhibit the NO production in RAW 264.7 cells. Intracellular NO production was evaluated using DAF-FM diacetate by confocal laser scanning microscopy. (b) Fluorescent spectra changes of NocPer in E. coli as a response to RAW 264.7 cells in different states. Blue line, RAW 264.7 without any treatment; red line, LPS-activated RAW 264.7; green line, LPS and L-NAME treated RAW 264.7. (c) The quantified relative fluorescence intensity ratio in E. coli corresponding to (b). Data are expressed as the mean ± standard error. (one-way ANOVA, Bonferroni's post-test; ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. NO decreases E. coli pHi by inhibiting cytochrome c oxidase
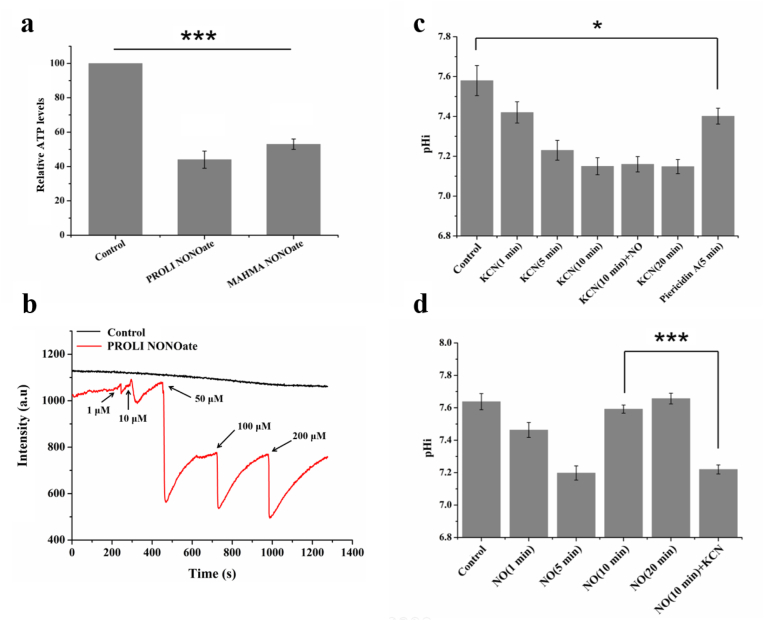
Next, we provided insight into the underlying mechanism through which NO stimulation leads to a decline in E. coli pHi. The water-dissolved NO quickly and spontaneously produces nitrite under aerobic conditions, whose high acidity might be responsible for the pHi decline. However, this possibility can be excluded since the pHi decline reached a saturation point at approximately pH 7.1 in the presence of an excessive amount of NO (Fig. 3c), which is not characteristic of the catalytic chemical reaction. Next, we shifted our focus to ATPase, since it synthesizes cellular ATP coupling proton uptake into the cytoplasm leading to pHi decline. Moreover, it has been reported that ATPase can receive a variety of post-translational modiflcations, including S-nitrosation (NO binding to cysteines), to alter its function [29]. Thus, we wondered whether NO could activate the ATPase in E. coli, which ultimately induces a decline in pHi. Therefore, we investigated the influence of NO exposure on cellular ATP production. As shown in Fig. 5a, 50 μM NO released by both NO donors, which has been shown to cause significant pHi decline, greatly decreased the ATP levels compared to those observed in the control condition. Thus, the possibility of NO decreasing the pHi by activating ATP synthase to accelerate proton influx was excluded.
Fig. 5.
NO decreases the pHi by inhibiting cytochrome c oxidase. (a) Effects of NO stimulation on intracellular ATP levels. The ATP levels in E. coli in the logarithmic growth phase without NO treatment was set to 100. (b) Time course of the electric potential (ΔΨ) in E. coli treated with the indicated concentrations of PROLI NONOate. ΔΨ was detected using the membrane potential fluorescent probe DiSC3(5). (c) Effects of respiratory chain inhibitors on the pHi of E. coli. KCN (10 μM) and piericidin A (20 μM) were used for the indicated times to inhibit respiratory chain activity. NO (100 μM) was added after 10 min of KCN treatment. (d) pHi of E. coli after NO treatment for the indicated time. KCN (10 μM) was added after 10 min of NO treatment. Asterisks over the columns indicate significant differences compared to the control values (a, c) or another group value (d) (one-way ANOVA, Bonferroni's post-test; *P < 0.05, ***P < 0.001).
Bacteria commonly maintain pHi homeostasis by exploring proton transporters in the respiratory chain to direct the active uptake or efflux of protons [30]. Thus, we hypothesized that invading NO may inhibit the E. coli proton extrusion pump in the respiratory chain to redistribute the transmembrane proton gradient. In bacteria, the proton motive force (PMF) is subsequently necessary for ATP synthesis by F1F0-ATPase and is thus critical to bacterial survival. PMF represents the sum of two parameters: the electric potential (ΔΨ) and the transmembrane proton gradient (ΔpH). A decrease in either component is compensated for by a counteracting increase in the other [31,32]. ΔΨ can be detected by the membrane potential fluorescent probe DiSC3(5), whose fluorescence intensity is inversely related to ΔΨ [32]. Thus, based on the fluorescence intensity change profile of DiSC3(5), the ΔpHi trend can be discerned. As shown in Fig. 5b and Fig. S5, both NO donor treatments resulted in rapid and partially reversible increases in ΔΨ in a dose-dependent manner, based on which an accompanying decline in the pHi caused by NO treatment could be deduced. This result confirmed the conclusion that the pHi decline caused by NO is a result of the alteration to the membrane proton gradient induced by NO, which easily led us to hypothesise that proton extrusion pumps were blocked by NO.
Among the complexes in the electron transfer chain, complex IV is the primary proton pump that is susceptible to NO inhibition, which is attributed to the binding and inhibition of NO to cytochrome c oxidase [[33], [34], [35], [36], [37]]. To verify whether the pHi decline was a result of NO-induced inhibition of complex IV, we treated E. coli with KCN, a specific complex IV inhibitor, to examine the effects of the complex IV inhibitor on pHi. The addition of KCN quickly decreased the pHi from pH 7.7 to pHi 7.4 within 1 min and then slowly dropped to pHi 7.1 after another 10 min (Fig. 5c). This KCN-induced pHi decline seemed irreversible since no signs of recession of the influence of KCN on pHi were observed within 20 min. Similarly, NO induced a quick and significant decrease in pHi; however, the NO-induced inhibition was reversible (Fig. 5d), which was consistent with the results shown in Fig. 3d. Further KCN exposure in NO-treated but pHi-recovering cells decreased their pHi to 7.2 (Fig. 5d). Moreover, neither the exposure of KCN-treated cells to NO, nor exposure of consecutively NO-treated cells to KCN yielded any pHi changes (Fig. 5c and d), suggesting that KCN and NO should act on the same cellular target (cytochrome c oxidase) in complex IV. To confirm the role of respiratory chain proton pumps on pHi control, we treated E. coli cells with piericidin A (a specific inhibitor of complex I) and found that the pHi decreased rapidly to pH 7.4 (Fig. 5c), underscoring that the decline in pHi is a non-negligible effect of a disturbance within the respiratory chain.
3.7. Potential survival advantage by pHi decline in defence against NO
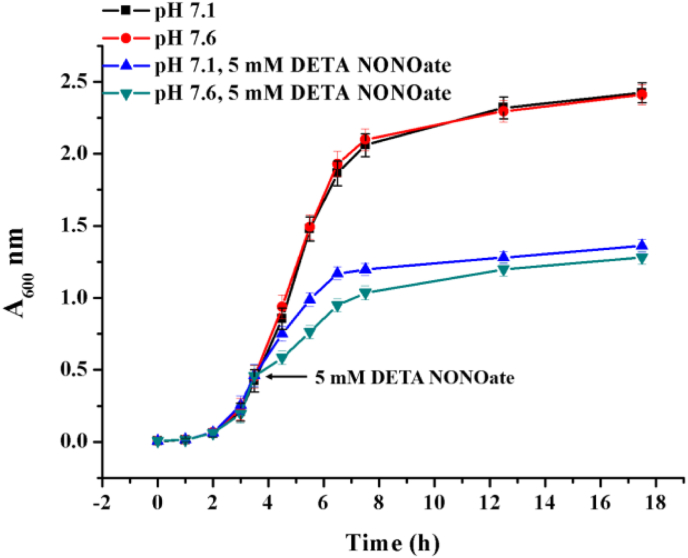
It has been widely accepted that NO kills cells or inhibits their growth by its direct cytotoxic effects, including respiratory inhibition, thiol nitrosation, iron–sulphur cluster ([Fe–S]) destruction, DNA deamination, and tyrosine nitration/nitrosylation [[38], [39], [40], [41]]. We wondered whether pHi decline is accompanied by cytotoxic effects. To confirm that, we adjusted the pHi of E. coli to pH 7.1 and pH 7.6 by treating the cells with nigericin as well as monensin and compared their growth speeds while cultivating them in media with corresponding pH values. As shown in Fig. 6, altering pHi value from pH 7.6 to pH 7.1 did not result in any growth differences of E. coli. Next, we investigated whether the pHi alterations affected the cellular NO resistance. We used 5 mM DETA NONOate, an NO donor with a long half-time (20 h) which is capable to maintain NO concentration (approximate 5 μmol/L) in neutral media and exert constant NO stress to cells [42]. Unexpectedly, the inhibition of cellular growth rate induced by NO stress was attenuated when the pHi decreased from 7.6 to 7.1 (Fig. 6), leading us to consider the possibility that the pHi decline induced by NO exposure might represent an unknown bacterial protection mechanism against NO toxicity.
Fig. 6.
Acidification of pHi provides advantage in NO resistance of E. coli strains. To modulate pHi to pH 7.1 and pH 7.6, the LB media were prepared in 50 mM HEPES buffer with the indicated pH values containing 100 μM monensin and 10 μM nigericin. When necessary, 5 mM DETA NONOate (a long-acting NO donor) was added at the points indicated by arrows. Cell viability assay was performed by monitoring the optical density at 600 nm (A600 nm). Standard deviations were calculated from three independent biological replicates.
4. Discussion
In this study, we developed a novel cpYFP-based pH sensor, NocPer, which originated from an invalid NO sensor. The dynamic response range of NocPer covered the physiological pH range of the cytosol, and its sensitivity is higher than that of the most commonly used pH sensor, pHluorin. This makes NocPer an appealing alternative for pHi reporting. While this new sensor could not directly detect NO, it could sense the pHi changes resulting from NO exposure in E. coli cells, which led to the discovery and elucidation of a new biological phenomenon in which NO decreases the E. coli pHi. The slight pHi decrease induced by NO stimulation might represent a self-protection strategy for pathogen infection; however, such an important phenomenon is difficult to detect without using a highly sensitive pH sensor such as NocPer.
Although there is considerable controversy regarding the capacity of cpYFP to detect superoxide, due to its intrinsic properties, there is no doubt that the fluorescent signal of cpYFP is hypersensitive to pH change [16]. The unique constraint for cpYFP to become a good pH sensor is due to its single excitation peak, which makes it an absolute quantitative-type sensor rather than a rational-type sensor. We noticed that some cpYFP-based sensors, including ratiometric pericam [13], SoNar [14], FHisJ [15], and HyPer [19] exhibit two clear-cut excitation peaks, while other sensors, such as FY5a+5a [43], and Frex [44], do not (or have a tiny hump besides the major peak), indicating that extending the C- and N-termini of cpYFP with various amino acid fragments makes it possible to obtain derivates of cpYFP with two peaks. Directly comparing the performance of HyPer, TScGP, and our novel sensor NocPer as pH probes indicated that NocPer exhibits the highest sensitivity to pH changes (Fig. 2). These results demonstrated that the C- and N-terminal fragments determine not only the excitation peak profiles but also the pH sensitivities of these cpYFP-based sensors. However, due to limited examples, it is not well known how these fragments affect the characteristics of the sensor. Nevertheless, it can be speculated that the surrounding microenvironment of the chromophore might interfere with the protonation or deprotonation of the chromophore and determine the performance of the sensors. These interfering factors include the length, conformation of the fused fragments and the amino acid residues around the chromophore. However, determining the exact molecular basis for the rational design of cpYFP-based pH sensors deserves further screening and analysis.
Protons play a crucial role in biochemical networks; therefore, it is not surprising that the cytoplasmic pH of all living organisms is tightly controlled. Failure to achieve pHi homeostasis is accompanied by the cessation of cell division, and the recovery of pHi homeostasis always precedes the resumption of cell growth [30,45]. Thus, there might be a tight connection between the two processes. When E. coli cultures were subjected to a rapid decrease in the external pH, from 7.6 to 5.5, the cytoplasmic pH decreased rapidly, followed by a noticeable recovery within a few minutes [46]. During the rapid pH change, the genes that participated in acid stress resistance were upregulated. Thus, transient pHi homeostasis failure may be an on switch for acid stress resistance gene induction. Within a few minutes, E. coli induced gene expression as well as pHi recovery to maintain normal cell growth [45]. In contrast to the transient pHi decrease caused by the external pH change, NO exposure resulted in a sustained pHi decrease until NO dissipation (Fig. 3d). Is there any physiological significance to maintain a lower pHi under NO stress? Our present results indicated that decreasing the pHi below 7.2 promoted cellular growth rate under NO stress conditions, which led us to hypothesise that E. coli takes advantage of the NO-induced pHi decrease to reduce NO damage to cells (Fig. 6). Recently, acidification of pHi induced via the activation of the marRAB response was reported to be an efficient strategy employed by E. coli to facilitate antibiotic resistance [24]. However, the exact mechanism through which pHi acidification provides advantage in NO resistance of E. coli has not been known. Whether protons act as an intracellular messenger to induced NO detoxification enzymes or other cellular impaired repair systems to support E. coli growth require further investigation. Taken together, we hypothesise that this ability to adjust cytoplasmic pH is an essential feature of cellular physiology that enables a response to adverse growth conditions.
NO is widespread in mammalian and plant cells. In mammalian cells, NO is an essential biological signalling molecule playing an important role in the regulation of nervous, vascular, and immune systems [47]. In plant cells, NO is also involved in many physiological processes such as plant-microbe interactions, seed germination, root and pollen tube growth, stomatal closure, flowering, iron homeostasis, programmed cell death, and adaptive responses to biotic as well as abiotic stressors [48]. Mammalian NO is generated by NO synthases (NOSs) located in different subcellular compartments including the cytosol, nuclear and plasma membrane, as well as the peroxisome [49]. Furthermore, the differential subcellular location of NO synthesis might contribute to its diverse functions [1,2,50,51]. While the subcellular localization of endogenous NO has also been intensively investigated, the origin of NO in plants remains unclear. Previous studies reported that NO could be identified in plant mitochondria, plasma membranes, peroxisomes [49,52], cytosol, chloroplasts [53], plastids, and microsome fractions [6]. It has been widely accepted that NO participates in biological events by modification of biomacromolecule such as proteins, lipids, and DNA strands. However, whether NO generated in these organelles alters their pHi and whether NO partially exerts its signal function by increasing intracellular protons, remains elusive. As a simple cpYFP-based biosensor, easily expressing and efficiently targeted NocPer to different subcellular compartments by concatenating cellular organelle localization sequences with NocPer can be expected. Therefore, NocPer has wide prospective applications in the field of pHi detection and is suitable for all cell types. Furthermore, one of NocPer's greatest advantages could be observed in real-time pHi monitoring resulting from NO emergence in various subcellular compartments, and the further exploration of the physiological significances raised by pHi changes.
Using the pHluorin probe, a previous study showed that other extracellular treatments, such as centrifugation, temperature shifts, prolonged suspension at high density, or changes in medium osmolarity, might result in slight changes in the pHi [45]. Finally, NocPer should be capable of accurately analysing the relationship between pHi changes and stimulation strength and further elucidating the physiological significance of pHi changes under these conditions.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by the National Major Science and Technology Projects of China (2019ZX09739001), the National Natural Science Foundation of China (22077032, 21672065 and 21636003), the Project Funded by the International S&T Innovation Cooperation Key Project (2017YFE0129600), the Fundamental Research Funds for the Central Universities (22221818014), and the 111 Project (B18022).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101943.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lagadic-Gossmann D., Huc L., Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls D.G. The influence of respiration and ATP hydrolysis on the proton-electrochemical gradient across the inner membrane of rat-liver mitochondria as determined by ion distribution. Eur. J. Biochem. 1974;50:305–315. doi: 10.1111/j.1432-1033.1974.tb03899.x. [DOI] [PubMed] [Google Scholar]
- 3.Munder M.C. A pH-driven transition of the cytoplasm from a fluid-to a solid-like state promotes entry into dormancy. elife. 2016;5 doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dechant R. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V‐ATPase. EMBO J. 2010;29:2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J., Ling P., Gao F. A mitochondria-targeted ratiometric biosensor for pH monitoring and imaging in living cells with Congo-red-Functionalized dual-emission semiconducting polymer dots. Anal. Chem. 2017;89:11703–11710. doi: 10.1021/acs.analchem.7b03154. [DOI] [PubMed] [Google Scholar]
- 6.Marín M.J., Galindo F., Thomas P., Russell D.A. Localized intracellular pH measurement using a ratiometric photoinduced electron-transfer-based nanosensor. Angew. Chem. Int. Ed. Engl. 2012;124:9795–9799. doi: 10.1002/anie.201203866. [DOI] [PubMed] [Google Scholar]
- 7.Tantama M., Hung Y.P., Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem. Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miesenböck G., De Angelis D.A., Rothman J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 9.Burgstaller S. pH-Lemon, a fluorescent protein-based pH reporter for acidic compartments. ACS Sens. 2019;4:883–891. doi: 10.1021/acssensors.8b01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinoda H., Shannon M., Nagai T. Fluorescent proteins for investigating biological events in acidic environments. Int. J. Mol. Sci. 2018;19:1548. doi: 10.3390/ijms19061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benčina M. Illumination of the spatial order of intracellular pH by genetically encoded pH-sensitive sensors. Sensors. 2013;13:16736–16758. doi: 10.3390/s131216736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg J., Hung Y.P., Yellen G. A genetically encoded fluorescent reporter of ATP: ADP ratio. Nat. Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai T., Sawano A., Park E.S., Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. U. S. A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y. SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metabol. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H. A genetically encoded toolkit for tracking live-cell histidine dynamics in space and time. Sci. Rep. 2017;7:1–9. doi: 10.1038/srep43479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baird G.S., Zacharias D.A., Tsien R.Y. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzländer M. The ‘mitoflash’probe cpYFP does not respond to superoxide. Nature. 2014;514:E12−E14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belousov V.V. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y. Novel peroxiredoxin-based sensor for sensitive detection of hydrogen peroxide. Biochem. Biophys. Res. Commun. 2019;517:260–265. doi: 10.1016/j.bbrc.2019.07.062. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y., Lim J., Xu J., Yu J.H., Zheng W. Nitric oxide as a developmental and metabolic signal in filamentous fungi. Mol. Microbiol. 2020;113:872–882. doi: 10.1111/mmi.14465. [DOI] [PubMed] [Google Scholar]
- 22.Poole R. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 23.Isom D.G. Coordinated regulation of intracellular pH by two glucose-sensing pathways in yeast. J. Biol. Chem. 2018;293:2318–2329. doi: 10.1074/jbc.RA117.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes-Fernández E.Z., Schuldiner S. Acidification of cytoplasm in Escherichia coli provides a strategy to cope with stress and facilitates development of antibiotic resistance. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-66890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eroglu E. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bush M., Ghosh T., Tucker N., Zhang X., Dixon R. Transcriptional regulation by the dedicated nitric oxide sensor, NorR: a route towards NO detoxification. Biochem. Soc. Trans. 2011;39:289–293. doi: 10.1042/BST0390289. [DOI] [PubMed] [Google Scholar]
- 27.D'Autréaux B., Tucker N.P., Dixon R., Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 28.Fang F.C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 29.Nesci S., Trombetti F., Ventrella V., Pagliarani A. Post-translational modifications of the mitochondrial F1FO-ATPase. Biochim. Biophys. Acta Gen. Subj. 2017;1861:2902–2912. doi: 10.1016/j.bbagen.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Krulwich T.A., Sachs G., Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker E.P., Mangerich W.E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 1981;147:820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farha M.A., Verschoor C.P., Bowdish D., Brown E.D. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem. Biol. 2013;20:1168–1178. doi: 10.1016/j.chembiol.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Antunes F., Boveris A., Cadenas E. On the mechanism and biology of cytochrome oxidase inhibition by nitric oxide. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16774–16779. doi: 10.1073/pnas.0405368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason M.G., Nicholls P., Wilson M.T., Cooper C.E. Nitric oxide inhibition of respiration involves both competitive (heme) and noncompetitive (copper) binding to cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:708–713. doi: 10.1073/pnas.0506562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper C.E. Nitric oxide and cytochrome oxidase: substrate, inhibitor or effector? Trends Biochem. Sci. 2002;27:33–39. doi: 10.1016/s0968-0004(01)02035-7. [DOI] [PubMed] [Google Scholar]
- 36.Cassina A., Radi R. Differential inhibitory action of nitric oxide and peroxynitrite on mitochondrial electron transport. Arch. Biochem. Biophys. 1996;328:309–316. doi: 10.1006/abbi.1996.0178. [DOI] [PubMed] [Google Scholar]
- 37.Poderoso J.J., Helfenberger K., Poderoso C. The effect of nitric oxide on mitochondrial respiration. Nitric Oxide. 2019;88:61–72. doi: 10.1016/j.niox.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Zhou S. NO-inducible nitrosothionein mediates NO removal in tandem with thioredoxin. Nat. Chem. Biol. 2013;9:657–663. doi: 10.1038/nchembio.1316. [DOI] [PubMed] [Google Scholar]
- 39.Zhou S. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 2012;78:103–109. doi: 10.1128/AEM.06195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman L.A., McLean S., Poole R.K., Fukuto J.M. The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins. Adv. Microb. Physiol. 2011;59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 41.Robinson J.L., Brynildsen M.P. Discovery and dissection of metabolic oscillations in the microaerobic nitric oxide response network of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1757–E1766. doi: 10.1073/pnas.1521354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones-Carson J., Laughlin J.R., Stewart A.L., Voskuil M.I., Vázquez-Torres A. Nitric oxide-dependent killing of aerobic, anaerobic and persistent Burkholderia pseudomallei. Nitric Oxide. 2012;27:25–31. doi: 10.1016/j.niox.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianchi-Smiraglia A. Internally ratiometric fluorescent sensors for evaluation of intracellular GTP levels and distribution. Nat. Methods. 2017;14:1003–1009. doi: 10.1038/nmeth.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metabol. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karagiannis J., Young P.G. Intracellular pH homeostasis during cell-cycle progression and growth state transition in Schizosaccharomyces pombe. J. Cell Sci. 2001;114:2929–2941. doi: 10.1242/jcs.114.16.2929. [DOI] [PubMed] [Google Scholar]
- 46.Kannan G. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 2008;8:1–13. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gantner B.N., LaFond K.M., Bonini M.G. Nitric oxide in cellular adaptation and disease. Redox Biol. 2020:101550. doi: 10.1016/j.redox.2020.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santolini J., André F., Jeandroz S., Wendehenne D. Nitric oxide synthase in plants: where do we stand? Nitric Oxide. 2017;63:30–38. doi: 10.1016/j.niox.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Stolz D.B. Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology. 2002;36:81–93. doi: 10.1053/jhep.2002.33716. [DOI] [PubMed] [Google Scholar]
- 50.Talapka P., Bódi N., Battonyai I., Fekete É., Bagyánszki M. Subcellular distribution of nitric oxide synthase isoforms in the rat duodenum. World J. Gastroenterol. 2011;17:1026. doi: 10.3748/wjg.v17.i8.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez F.A. Functional significance of cytosolic endothelial nitric-oxide synthase (eNOS): regulation of hyperpermeability. J. Biol. Chem. 2011;286:30409–30414. doi: 10.1074/jbc.M111.234294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corpas F.J. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004;136:2722–2733. doi: 10.1104/pp.104.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jasid S., Simontacchi M., Bartoli C.G., Puntarulo S. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 2006;142:1246–1255. doi: 10.1104/pp.106.086918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.