Abstract
“Bryndza” cheese is an important Slovak traditional regional product. New knowledge on the role of microorganisms involved the “Bryndza” ripening process may provide valuable data on its quality and safety. In our study, the “Bryndza” made from pasteurized ewes milk was studied towards total count of bacteria, coliforms bacteria, enterococci, lactic acid bacteria, and microscopic filamentous fungi. All those groups of microbiota were detected using classical microbiological methods and identified using mass spectrometry. A total of 3,758 isolates were identified with score higher than 2.00. Altogether, 13 families, 24 genus, and 44 species of microbiota were identified in Slovak cheese “Bryndza.” The most often isolated species were yeasts Yarrowia lipolitica and Dipodascus geotrichum and the lactic acid bacteria Lactobacillus paracasei subsp. paracasei.
Keywords: gram-positive and gram-negative bacteria, microscopic filamentous fungi, sheep farms, ewes milk cheese “Bryndza”, mass spectrometry, microbiota identification
1. Introduction
Sheep herding and ewes milk production are important sectors of Slovak agriculture. The well-known product of ewes milk is a Slovak cheese named “Bryndza” or “Oštiepok” [1,2]. “Bryndza” was granted the Protected Geographic Indication (PGI) as it is produced in a defined mountainous regions of Slovakia [3,4]. “Bryndza” is a type of natural white, mature, and spreadable cheese made from matured 100% sheep lump cheese or mixed with up to 50% of cow lump cheese. First, ewes milk is sweetened. This raw material is then allowed to dry on the mountain hut to form lump sheep’s cheese. It is then transferred to a bryndziarna (cheese “Bryndza” production), where it is sorted and washed with water and then left to mature at 20°C in a cheese bath. The rind is then removed from the cheese, the excess liquid (whey) is expelled, and the cheese is crushed. The pulp is salted and spread on rollers to form a “Bryndza” [5]. The manufacturing has been done according to the traditional method. “Bryndza” is produced from raw ewes milk at 29–31°C for 30 min with chymosin or chymosin-identical enzymes [6]. In 2019, the production of ewes milk in Slovakia reached 13,524 tons, which is the highest level since joining the European Union. The annual production of “Bryndza” in Slovakia is almost 4,000 tons, whereas Slovaks consume about 0.6 kg of “Bryndza” per capita per year. The characteristics of the produced “Bryndza” cheese may vary under the climatic conditions and may be influenced by the feeding meant. The botanical composition of the plants in the sheep diet during pasture may affect the quality parameters of the milk used for production of the “Bryndza” cheese. Carpathian Mountains in Slovakia thus represent a specific habitat for sheep milk production by affecting the quality of raw milk [7] and the differences in ambient temperatures influence the ripening microbiota at the early cheese production steps [8]. In Slovak “Bryndza,” Hafnia alvei and Klebsiella oxytoca were the most abundant Gram-negative bacteria, whereas Lactococcus lactis and Lactobacillus paracasei the most abundant Gram-positive bacteria. Lactobacillus, Lactococcus, and Pediococcus were the main representatives of the lactic acid bacteria [8]. The aim of this study was to characterize microbiological variability of “Bryndza” cheese produced in summer of 2016–2019 in various geographical areas in Slovakia.
2. Materials and methods
2.1. “Bryndza” cheese samples
Samples of 80 unpasteurized ewes’ cheese “Bryndza” were provided by eight producers representing eight Slovak farms (F1 to F8). Slovak “Bryndza” is produced in the same way throughout the defined area. The same breed of sheep grazes in the defined area – Native Wallachian sheep, Improved Wallachian sheep, Domestic Tsigai, and East Friesian on pastures with the same flora and climatic conditions, which results in the same quality of the basic raw material – ewes milk [4]. The samples were collected between the end of April and August in the years 2016 to 2019. All samples were transported to the laboratory at 4°C and were analyzed immediately after delivery. Each sample was analyzed for total count of bacteria, coliform bacteria, enterococci, lactic acid bacteria, microscopic fungi, and yeasts MFF (microscopic filamentous fungi). The serial dilutions of the milk products in 0.89% sterile saline were made for plating out the sample material.
2.2. Microbiological analysis
The determination of total count of bacteria, coliforms, enterococci, lactic acid bacteria and fungi, and yeasts has been previously published [9,10]. The colonies from total count of bacteria, coliforms bacteria, enterococci, lactic acid bacteria, MFF, and yeasts were selected for further confirmation with MALDI-TOF MS Biotyper. Selected colonies were subcultured overnight on TSA agar aerobically or anaerobically and were used for identification.
2.3. Identification of bacteria and yeasts with MALDI-TOF MS Biotyper
Microbial isolates for MALDI-TOF MS Biotyper analysis were prepared in accordance with extraction procedure provided by the manufacturer (Bruker Daltonics, Bremen, Germany). The detailed procedure was previously published [11].
2.4. Identification of microscopic filamentous fungi with MALDI-TOF MS Biotyper
The detailed procedure of identification of fungal isolates was previously published [12]. Identification was done by MALDI-TOF MS Biotypes (Bruker Daltonics, Bremen, Germany) with Flex Control 3.4 software and Biotyper Realtime Classification 3.1 with BC specific software (Bruker Daltonics, Germany).
2.5. Krona charts
Krona charts showing automatically the taxonomic identification and relative abundance of the most abundant bacteria or microscopic filamentous fungi.
2.6. Statistical analysis
For microbial counts, lactic acid bacteria count, coliform bacteria, and microscopic filamentous fungi counts, the means and standard deviations were calculated. Krone diagrams were used for visualization of the relatedness of the identified microbial isolates.
3. Results
The number of coliform bacteria ranged from 3.67 in F1 to 3.84 log cfu/g in F2 and enterococci from 2.29 in F6 to 2.64 log cfu/g in F4. Total count of bacteria were from 4.38 in F1 to 4.73 log cfu/g in F4. The higher lactic acid bacteria counts were found in F7 (3.62 log cfu/g) and microscopic filamentous fungi in F6 (2.61 log cfu/g) (Table 1).
Table 1.
Microbial counts in sheep cheese “Bryndza” (average ± SD log cfu/g)
| Farm | CB | E | TCB | LAB | MFF |
|---|---|---|---|---|---|
| F1 | 3.67 ± 0.15 | 2.46 ± 0.13 | 4.38 ± 0.15 | 3.25 ± 0.12 | 2.42 ± 0.13 |
| F2 | 3.81 ± 0.19 | 2.42 ± 0.12 | 4.66 ± 0.11 | 3.37 ± 0.15 | 2.36 ± 0.12 |
| F3 | 3.79 ± 0.20 | 2.27 ± 0.17 | 4.64 ± 0.17 | 3.42 ± 0.17 | 2.15 ± 0.11 |
| F4 | 3.81 ± 0.19 | 2.64 ± 0.11 | 4.73 ± 0.12 | 3.43 ± 0.12 | 2.26 ± 0.12 |
| F5 | 3.76 ± 0.12 | 2.36 ± 0.10 | 4.48 ± 0.11 | 3.37 ± 0.15 | 2.41 ± 0.15 |
| F6 | 3.72 ± 0.12 | 2.29 ± 0.14 | 4.81 ± 0.12 | 3.26 ± 0.11 | 2.61 ± 0.12 |
| F7 | 3.74 ± 0.11 | 2.32 ± 0.11 | 4.64 ± 0.15 | 3.62 ± 0.12 | 2.41 ± 0.12 |
| F8 | 3.84 ± 0.14 | 2.46 ± 0.15 | 4.72 ± 0.13 | 3.38 ± 0.11 | 2.35 ± 0.15 |
CB – coliforms bacteria, E – enterococci, TCB – total count of bacteria, LAB – lactic acid bacteria, MFF – microscopic filamentous fungi, SD – standard deviation.
A total of 3,758 isolates from cheese “Bryndza” were identified using MALDI-TOF Biotyper (Table 2). The most abundant microbial species isolated from cheese were Yarrowia lipolitica (254 isolates), Lactobacillus paracasei subsp. paracasei (252 isolates), and Dipodascus geotrichum (Geotrichum candidum, 227 isolates). The most abundant family of cheese “Bryndza” was Lactobacillaceae (25.24%).
Table 2.
Number of isolated species from F1 to F8
| Species | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter baumannii | 10 | 14 | 15 | 18 | 12 | 11 | 9 | 13 | 102 |
| Acinetobacter tandoii | 15 | 18 | 10 | 16 | 15 | 14 | 11 | 10 | 109 |
| Aspergillus fumigatus | 12 | 15 | 14 | 18 | 10 | 12 | 16 | 13 | 110 |
| Bacillus pumilus | 8 | 9 | 7 | 8 | 6 | 8 | 7 | 9 | 62 |
| Candida catenulata | 4 | 6 | 5 | 8 | 9 | 11 | 12 | 8 | 63 |
| Candida krusei | 6 | 8 | 5 | 7 | 4 | 8 | 9 | 5 | 52 |
| Candida lusitaniae | 15 | 15 | 16 | 11 | 15 | 17 | 10 | 15 | 114 |
| Candida rugosa | 6 | 8 | 9 | 9 | 9 | 7 | 10 | 11 | 69 |
| Candida utilis | 8 | 5 | 8 | 5 | 9 | 6 | 8 | 10 | 59 |
| Citrobacter braakii | 8 | 6 | 6 | 8 | 7 | 8 | 5 | 5 | 53 |
| Citrobacter koseri | 5 | 6 | 8 | 9 | 5 | 4 | 5 | 6 | 48 |
| Dipodascus geotrichum | 25 | 30 | 28 | 32 | 26 | 28 | 30 | 28 | 227 |
| Dipodascus silvicola | 6 | 5 | 7 | 4 | 5 | 4 | 3 | 5 | 39 |
| Enterobacter cloacae | 2 | 4 | 8 | 9 | 5 | 6 | 4 | 10 | 48 |
| Enterobacter ludwigii | 8 | 9 | 7 | 5 | 6 | 8 | 9 | 5 | 57 |
| Enterococcus faecalis | 6 | 11 | 5 | 8 | 9 | 9 | 7 | 8 | 63 |
| Enterococcus faecium | 10 | 15 | 25 | 24 | 21 | 15 | 12 | 18 | 140 |
| Enterococcus hirae | 15 | 6 | 18 | 7 | 9 | 10 | 14 | 11 | 90 |
| Escherichia coli | 14 | 10 | 12 | 8 | 10 | 12 | 16 | 10 | 92 |
| Hafnia alvei | 5 | 6 | 8 | 7 | 8 | 9 | 10 | 8 | 61 |
| Klebsiella oxytoca | 10 | 5 | 6 | 8 | 9 | 7 | 6 | 8 | 59 |
| Klebsiella pneumoniae ssp. ozaenae | 6 | 4 | 8 | 7 | 8 | 8 | 5 | 6 | 52 |
| Klebsiella pneumoniae ssp. pneumoniae | 5 | 6 | 7 | 8 | 7 | 8 | 9 | 5 | 55 |
| Lactobacillus brevis | 12 | 15 | 18 | 21 | 16 | 18 | 21 | 18 | 139 |
| Lactobacillus harbinensis | 8 | 9 | 10 | 8 | 9 | 15 | 12 | 16 | 87 |
| Lactobacillus johnsonii | 5 | 8 | 8 | 6 | 12 | 4 | 11 | 8 | 62 |
| Lactobacillus plantarum | 23 | 21 | 25 | 18 | 26 | 22 | 20 | 32 | 187 |
| Lactobacillus paracasei ssp. paracasei | 28 | 31 | 39 | 45 | 36 | 15 | 28 | 30 | 252 |
| Lactobacillus paraplantarum | 8 | 15 | 15 | 16 | 15 | 17 | 14 | 10 | 110 |
| Lactobacillus suebicus | 5 | 4 | 6 | 5 | 8 | 7 | 9 | 6 | 50 |
| Lactococcus lactis ssp. lactis | 18 | 22 | 25 | 19 | 16 | 8 | 15 | 21 | 144 |
| Lactococcus lactis | 15 | 10 | 10 | 12 | 14 | 10 | 18 | 14 | 103 |
| Microbacterium liquefaciens | 8 | 9 | 8 | 9 | 11 | 6 | 4 | 5 | 60 |
| Mucor circinelloides | 8 | 4 | 6 | 10 | 9 | 8 | 7 | 9 | 61 |
| Pediococcus acidilactici | 6 | 4 | 8 | 11 | 10 | 5 | 8 | 9 | 61 |
| Penicillium sp. | 7 | 5 | 8 | 8 | 6 | 4 | 5 | 8 | 51 |
| Pichia cactophila | 4 | 5 | 6 | 6 | 7 | 8 | 5 | 6 | 47 |
| Raoultella ornithinolytica | 5 | 7 | 8 | 9 | 10 | 12 | 8 | 5 | 64 |
| Rhizopus sp. | 8 | 7 | 5 | 4 | 2 | 4 | 6 | 5 | 41 |
| Serratia liquefaciens | 6 | 7 | 5 | 8 | 5 | 4 | 6 | 5 | 46 |
| Staphylococcus aureus ssp. aureus | 6 | 2 | 5 | 4 | 5 | 3 | 4 | 6 | 35 |
| Staphylococcus pasteuri | 2 | 4 | 6 | 7 | 5 | 4 | 6 | 3 | 37 |
| Stenotrophomonas maltophilia | 5 | 4 | 6 | 5 | 8 | 6 | 4 | 5 | 43 |
| Yarrowia lipolytica | 25 | 28 | 30 | 28 | 39 | 45 | 28 | 31 | 254 |
| Total | 421 | 442 | 499 | 503 | 493 | 455 | 466 | 479 | 3,758 |
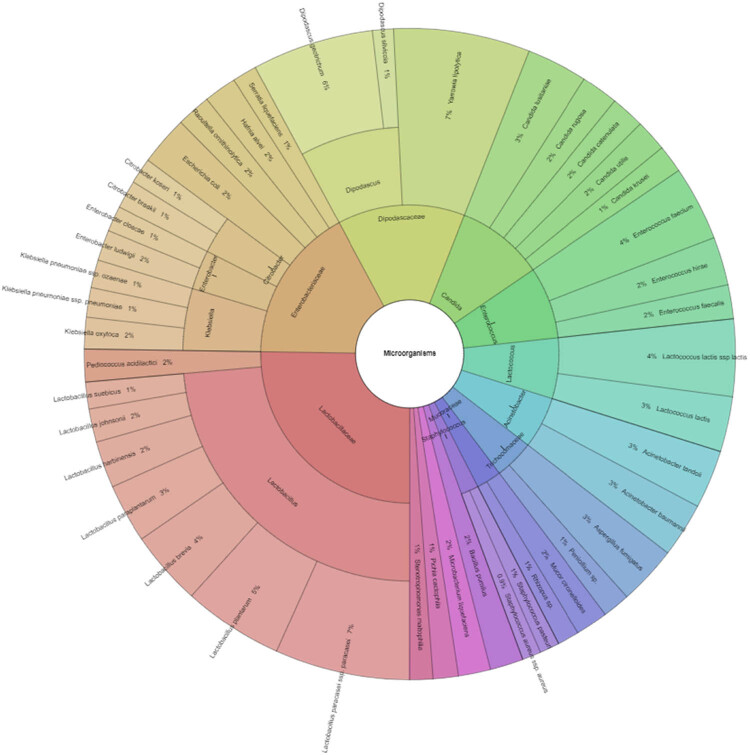
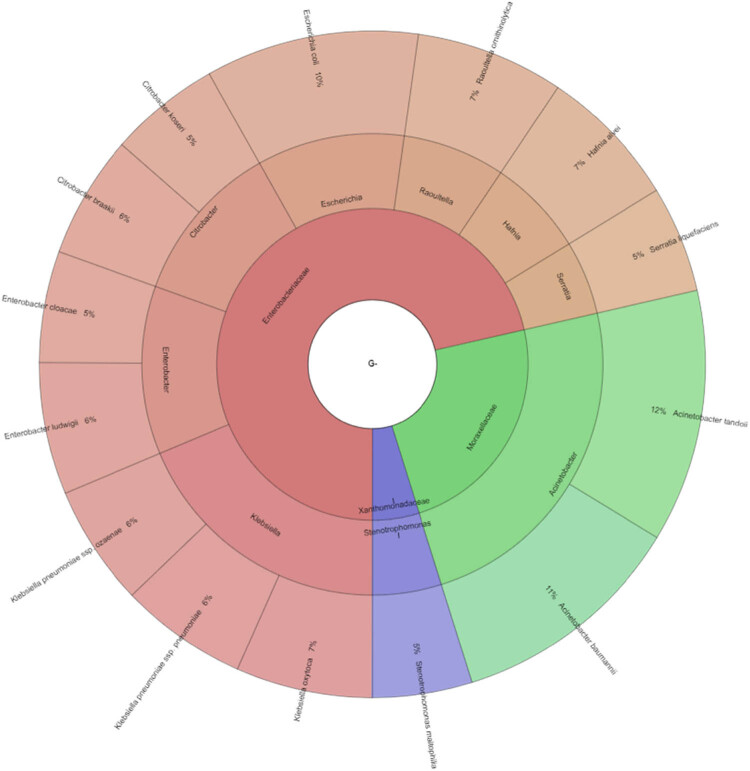
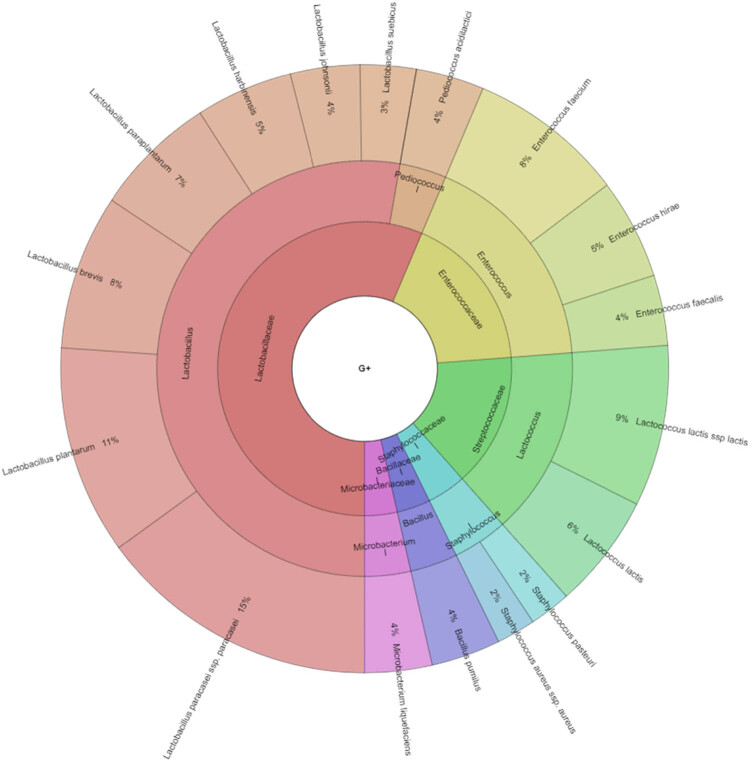
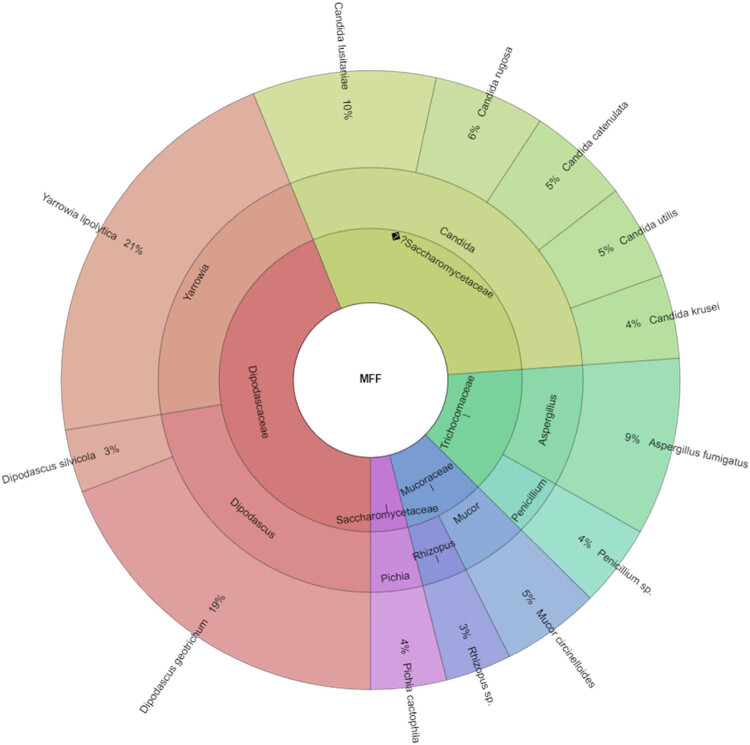
In our study, Gram-negative, Gram-positive bacteria, and microscopic filamentous fungi accounted for majority of the identified microbiota in “Bryndza.” The Enterobacteriaceae family was the most abundant family of Gram-negative bacteria isolated. Percentage of all isolated family, genera, and species indicates Krona (Figure 1). Altogether 889 isolates belonged to Gram-negative bacteria group. The Lactobacillaceae family was the most abundant among Gram-positive bacteria (Figure 2). Altogether 1,682 isolates of Gram-positive bacteria were identified. Yarrowia lipolitica was the most abundant microscopic fungi (21%) (Figures 3 and 4).
Figure 1.
Diversity microorganisms isolated from ewes’ cheese “Bryndza” (outermost ring: species, middle ring: genus, innermost ring: family).
Figure 2.
Diversity of Gram-negative bacteria isolated from ewes’ cheese “Bryndza” (outermost ring: species, middle ring: genus, innermost ring: family).
Figure 3.
Diversity of Gram-positive bacteria isolated from ewes’ cheese “Bryndza” (outermost ring: species, middle ring: genus, innermost ring: family).
Figure 4.
Krona chart for microscopic filamentous fungi isolated from ewes’ cheese “Bryndza” (outermost ring: species, middle ring: genus, innermost ring: family).
4. Discussion
The total count of bacteria in ewes’ cheese “Bryndza” samples was from 4.38 ± 0.15 log cfu/g for F1 to 4.81 ± 0.12 log cfu/g for F6 total count of bacteria. The microbial counts similar to detected here were described previously for ewes’ cheese “Bryndza,” where number of total count of bacteria ranged from 3.87 to 4.32 log cfu/g [10]. Our findings on coliform bacteria counts (3.84 log cfu/g) were in line with Pangallo et al. [13] who reported their counts in the spring “Bryndza” at 3.87 log cfu/g. The counts of coliform and Staphylococcus spp. in Šaková et al.’s [3] study reached 105 to 106 cfu/g in “Bryndza” cheese; moreover, the majority of Staphylococcus spp. isolates were coagulase-positive (104 cfu/g). Within this study, the coliforms counts were less than 105 cfu/g. Coliforms were frequently identified in dairy products and contamination of dairy products may occur during milk storage, transportation, and processing, if the hygienic requirements are not met [14,15,16,17]. Intrinsic and extrinsic factors such as low pH, salt concentration, and water activity all affect the survival of coliforms and a decline in coliforms counts was detected during the ripening of other cheese types [18,19,20]. Escherichia coli, Klebsiella sp., and Enterobacter sp. were isolated from local Nigerian soft and semisoft cheese [21], whereas Citrobacter braakii, Enterobacter sakazakii, and E. coli were identified in the local Italian cheese [22,23]. High microbial counts of 9.0 log cfu/g were detected in raw ewes milk after 30 days of maturation [24]. The most important bacteria for ripening of ewes’ cheese are lactic acid bacteria and several these bacteria in “Bryndza” were considered as potentially probiotic in character [25]. Lactic acid bacteria up to 10.95 log cfu/g in “Bryndza” was reported previously [13] in comparison to 3.62 log cfu/g identified in our study. The microbiological quality of dairy products can be improved by adding a starter culture such as lactic acid bacteria, which will prevent the growth of pathogenic microorganisms [26].
Yeasts counts (2.61 log cfu/g) and Enterococcus counts (2.64 log cfu/g) in our study were lower than 5.97 log cfu/g and 7–8 log cfu/g, respectively, detected in other studies [13,14]. Yeasts belong to the natural microbiota of “Bryndza” cheese and contribute to the ripening of the cheese. The differences in yeasts counts in “Bryndza” cheese from raw and pasteurized milk were not significant. Dipodascus geotrichum was the predominant yeast identified in the present study in “Bryndza cheese.” The functional significance of this yeast is related to a breakdown of sugars, milk fat, and proteins [27,28]. Some strains of Dipodascus geotrichum were reported to form esters and sulfur compounds, important for the development of typical aroma and other characteristics of cheese [29].
Candida catenulata, C. krusei, C. lusitaniae, C. rugosa, C. utilis, Dipodascus geotrichum, D. silvicola, Pichia cactophila, and Yarrowia lipolitica were isolated in the present study. Galactomyces/Geotrichum (Dipodascus geotrichum) was reported in “Bryndza” of Slovak origin [30]. Yeasts as compounds of the milk microbiota are important in the agri-food industry as they are involved in biodegradation and depollution, may act as contaminants. Dipodascus geotrichum was found as a commensal of gut of humans and animals [31] and is important for milk production since it is naturally present in raw milk [32,33,34,35].
A total of 3758 isolates were identified to species level in cheese “Bryndza” and the most frequently isolated species were Yarrowia lipolitica, Lactobacillus paracasei subsp. paracasei, and Dipodascus geotrichum (Geotrichum candidum). Lactobacillaceae (25.24%) was the most frequently isolated family of the cheese “Bryndza.” Among the isolates, 889 were Gram-negative, 1,682 were Gram-positive, and 1,187 were microscopic filamentous fungi. In other study [10], a total of 1,175 isolates were identified by mass spectrometry and included Gram-negative bacteria, Gram-positive bacteria, and microscopic filamentous fungi. In the same study [10], 199 isolates were isolated and identified from Gram-negative, 599 isolates from Gram-positive, and 377 isolates of yeast and molds.
Beside the abundance of Lactobacillaceae (25.24%), other isolated families were Bacillaceae (1.65%), Dipodascaceae (13.84%), Enterobacteriaceae (16.89%), Enterococaceae (7.8%), Microbacteriaceae (1.6%), Moraxellaceae (5.61%), Mucoraceae (2.71%), Saccharomycetaceae (10.75%), Staphylococcaceae (1.91%), Streptococaceae (6.57%), Trichocomaceae (4.29%), and Xanthomonadaceae (1.14%). Similar results of identified genera were found [10].
In our study, 44 microbial species representing 24 genera were identified using MALDI-TOF MS Biotyper with score higher than 2.00. Lactobacillus paracasei subsp. paracasei was identified as most often identified species in the present study. Lactococcus, Lactobacillus, Streptococcus, Leuconostoc, and Enterococcus were members of lactic acid bacteria group commonly found in cheeses [36]. Needs for characterization of complex population and discovery of new lactic acid bacteria strains facilitate the research on the microbiota present in raw milk cheese and other traditional dairy products [37]. The genus Enterococcus was frequently isolated from cheese and identified as the prevalent among the lactic acid bacteria isolated from Coalho cheese in Brazil [38]. Several species of Enterococcus genus such as E. faecium, E. faecalis, E. italicus, E. durans, E. casseliflavus, and E. gallinarum were isolated from raw milk and cheese [38,39]. The dominant group of bacteria in “Bryndza” was lactic acid bacteria, mainly Lactobacillus species [10]. Lactococcus, Pediococcus, Enterococcus, and Streptococcus were abundant in “Bryndza” from different Slovak regions [3,40,41]. Previous study [42] identified Klebsiella (65%), Escherichia (20%), Serratia (10%), and Enterobacter (5%) in cheese samples. Klebsiella oxytoca, K. pneumoniae and K. ornithinolytica, and E. coli were identified among Enterobacteriaceae.
Aspergilus fumigatus, Mucor circeneloides, Rhizopus sp., and Penicillium sp. from micromycetes were isolated in “Bryndza” in our study. “Bryndza” could serve as a natural habitat for microscopic fungi, but their role in cheese ripening needs to be studied. Mucor spp. has been occasionally isolated from sheep cheese, accompanied by recognized human and plant pathogens [43,44,45,46]. Various applications of M. circinelloides were reported previously: starter cultures in China and Vietnam, and a contributor to organoleptic proprieties in study in Poland [47,48]. Mucor strains are accumulated in lipid bodies rich in polyunsaturated fatty acids [49].
In contrast to industrial food products, traditional cheese is of particular interest to consumers who care about the nature, origin, and nutritional value of foods [50]. Much of their reputation is attributed to the unique organoleptic properties and to the indigenous microorganisms living in raw milk or natural starters [51]. However, raw milk can also contain pathogenic microorganisms that have been raising public health concerns since the beginnings of the dairy industry [27].
5. Conclusion
The microbiota of traditional ewes’ cheese produced in Slovakia include diverse families of Bacillaceae, Dipodascaceae, Enterobacteriaceae, Enterococaceae, Lactobacillaceae, Microbacteriaceae, Moraxellaceae, Mucoraceae, Saccharomycetaceae, Staphylococcaceae, Streptococaceae, Trichocomaceae, and Xanthomonadaceae. The most abundant microbial species were Yarrowia lipolitica, Lactobacillus paracasei subsp. paracasei, and Dipodascus geotrichum. The unique sensory properties of the Slovak “Bryndza” are believed to be attributed to microbial activity of the microbiota primary present in the lump ewes’ cheese – Lactobacillus spp., Lactococcus spp., Streptococcus spp., Enterococcus spp., Kluyveromyces marxianus, and Dipodascus geotrichum. The results of this study contribute to better understanding of the microbiota of the local cheese produced in Slovakia. The study of “Bryndza” microbiota is important for development of processing technology and improvements in safety of products.
Acknowledgments
Dr. Marcin Nowicki (University of Tennessee, Knoxville, TN) is gratefully acknowledged for critical reading and linguistic copy editing of an early version of this study’s report.
Footnotes
Author contributions: Conceptualization, M.K. and S.K.; methodology, M.K. and S.K.; validation, M.K., M.T., S.K., P.H., and J.Š.; formal analysis, M.K., M.T., S.K., and J.Š.; investigation, M.K., and S.K; resources, M.K., S.K., and J.Š; data curation, M.K., S.K., and J.Š.; writing – original draft preparation, M.K., M.T., S.K., J.Š., P.H., and P.Ł.K.; writing – review and editing, M.T., and P.Ł.K.; visualization, P.Ł.K.; supervision, M.K. All authors have read and agreed to the published version of the manuscript.
Funding: This publication was supported by the Operational Program Integrated Infrastructure within the projects: Sustainable smart farming systems taking into account the future challenges 313011W112, co-financed by the European Regional Development Fund and Demand-driven research for the sustainable and innovative food, Drive4SIFood 313011V336, co-financed by the European Regional Development Fund.
Conflict of interest: Przemysław Łukasz Kowalczewski who is the co-author of this article is a current Editorial Board member of Open Life Sciences. This fact did not affect the peer-review process.
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Contributor Information
Miroslava Kačániová, Email: miroslava.kacaniova@gmail.com.
Margarita Terentjeva, Email: margarita.terentjeva@llu.lv.
Simona Kunová, Email: simona.kunova@uniag.sk.
Peter Haščík, Email: peter.hascik@uniag.sk.
Przemysław Łukasz Kowalczewski, Email: przemyslaw.kowalczewski@up.poznan.pl.
Jana Štefániková, Email: jana.stefanikova@uniag.sk.
References
- [1].Zajác P, Martišová P, Čapla J, Čurlej J, Golian J. Characteristics of textural and sensory properties of oštiepok cheese. Potravin Slovak J Food Sci. 2019;13:116–30. 10.5219/855. [DOI]; Zajác P, Martišová P, Čapla J, Čurlej J, Golian J. Characteristics of textural and sensory properties of oštiepok cheese. Potravin Slovak J Food Sci. 2019;13:116–30. doi: 10.5219/855. [DOI] [Google Scholar]
- [2].Šnirc M, Árvay J, Král M, Jančo I, Zajác P, Harangozo Ľ, et al. Content of mineral elements in the traditional oštiepok cheese. Biol Trace Elem Res. 2020;196:639–45. 10.1007/s12011-019-01934-w. [DOI] [PubMed]; Šnirc M, Árvay J, Král M, Jančo I, Zajác P, Harangozo Ľ. et al. Content of mineral elements in the traditional oštiepok cheese. Biol Trace Elem Res. 2020;196:639–45. doi: 10.1007/s12011-019-01934-w. [DOI] [PubMed] [Google Scholar]
- [3].Šaková N, Sádecká J, Lejková J, Puškárová A, Koreňová J, Kolek E, et al. Characterization of May bryndza cheese from various regions in Slovakia based on microbiological, molecular and principal volatile odorants examination. J Food Nutr Res. 2015;54:239–51.; Šaková N, Sádecká J, Lejková J, Puškárová A, Koreňová J, Kolek E. et al. Characterization of May bryndza cheese from various regions in Slovakia based on microbiological, molecular and principal volatile odorants examination. J Food Nutr Res. 2015;54:239–51. [Google Scholar]
- [4].Commission Regulation (Ec) No 676/2008 of 16 July 2008 Registering certain names in the register of protected designations of origin and protected geographical indications (Ail De La Drôme (Pgi), Všestarská Cibule (Pdo), Slovenská Bryndza (Pgi), Ajo Morad. J Eur Union. 2008;L189:19–20.; Commission Regulation (Ec) No 676/2008 of 16 July 2008 Registering certain names in the register of protected designations of origin and protected geographical indications (Ail De La Drôme (Pgi), Všestarská Cibule (Pdo), Slovenská Bryndza (Pgi), Ajo Morad. J Eur Union. 2008;L189:19–20. [Google Scholar]
- [5].Semjon B, Reitznerová A, Poláková Z, Výrostková J, Maľová J, Koréneková B, et al. The effect of traditional production methods on microbial, physico-chemical and sensory properties of “Slovenská bryndza” protected geographical indication cheese. Int J Dairy Tech. 2018;71(3):709–16.; Semjon B, Reitznerová A, Poláková Z, Výrostková J, Maľová J, Koréneková B. et al. The effect of traditional production methods on microbial, physico-chemical and sensory properties of “Slovenská bryndza” protected geographical indication cheese. Int J Dairy Tech. 2018;71(3):709–16. [Google Scholar]
- [6].Sádecká J, Šaková N, Pangallo D, Koreňová J, Kolek E, Puškárová A, et al. Microbial diversity and volatile odour-active compounds of barrelled ewes’ cheese as an intermediate product that determines the quality of winter bryndza cheese. LWT. 2016;70:237–44. 10.1016/j.lwt.2016.02.048. [DOI]; Sádecká J, Šaková N, Pangallo D, Koreňová J, Kolek E, Puškárová A. et al. Microbial diversity and volatile odour-active compounds of barrelled ewes’ cheese as an intermediate product that determines the quality of winter bryndza cheese. LWT. 2016;70:237–44. doi: 10.1016/j.lwt.2016.02.048. [DOI] [Google Scholar]
- [7].Ostrovský I, Pavlíková E, Blaško J, Górová R, Kubinec R, Margetín M, et al. Variation in fatty acid composition of ewes’ milk during continuous transition from dry winter to natural pasture diet. Int Dairy J. 2009;19:545–9. 10.1016/j.idairyj.2009.03.006. [DOI]; Ostrovský I, Pavlíková E, Blaško J, Górová R, Kubinec R, Margetín M. et al. Variation in fatty acid composition of ewes’ milk during continuous transition from dry winter to natural pasture diet. Int Dairy J. 2009;19:545–9. doi: 10.1016/j.idairyj.2009.03.006. [DOI] [Google Scholar]
- [8].Sádecká J, Kolek E, Pangallo D, Valík L, Kuchta T. Principal volatile odorants and dynamics of their formation during the production of May Bryndza cheese. Food Chem. 2014;150:301–6. 10.1016/j.foodchem.2013.10.163. [DOI] [PubMed]; Sádecká J, Kolek E, Pangallo D, Valík L, Kuchta T. Principal volatile odorants and dynamics of their formation during the production of May Bryndza cheese. Food Chem. 2014;150:301–6. doi: 10.1016/j.foodchem.2013.10.163. [DOI] [PubMed] [Google Scholar]
- [9].Kačániová M, Kunová S, Štefániková J, Felšӧciová S, Godočíková L, Horská E, et al. Microbiota of the traditional Slovak sheep cheese “Bryndza”. J Microbiol Biotechnol Food Sci. 2019;9:482–6. 10.15414/jmbfs.2019.9.special.482-486. [DOI]; Kačániová M, Kunová S, Štefániková J, Felšӧciová S, Godočíková L, Horská E. et al. Microbiota of the traditional Slovak sheep cheese “Bryndza”. J Microbiol Biotechnol Food Sci. 2019;9:482–6. doi: 10.15414/jmbfs.2019.9.special.482-486. [DOI] [Google Scholar]
- [10].Kačániová M, Nagyová Ľ, Štefániková J, Felsöciová S, Godočíková L, Haščík P, et al. The characteristic of sheep cheese “Bryndza” from different regions of Slovakia based on microbiological quality. Potravin Slovak J Food Sci. 2020;14:69–75. 10.5219/1239. [DOI]; Kačániová M, Nagyová Ľ, Štefániková J, Felsöciová S, Godočíková L, Haščík P. et al. The characteristic of sheep cheese “Bryndza” from different regions of Slovakia based on microbiological quality. Potravin Slovak. J Food Sci. 2020;14:69–75. doi: 10.5219/1239. [DOI] [Google Scholar]
- [11].Kačániová M, Kunova S, Horská E, Nagyová Ľ, Puchalski C, Haščík P, et al. Diversity of microorganisms in the traditional Slovak cheese. Potravin Slovak J Food Sci. 2019;13(1):532–7. 10.5219/1061. [DOI]; Kačániová M, Kunova S, Horská E, Nagyová Ľ, Puchalski C, Haščík P. et al. Diversity of microorganisms in the traditional Slovak cheese. Potravin Slovak J Food Sci. 2019;13(1):532–7. doi: 10.5219/1061. [DOI] [Google Scholar]
- [12].Singh A, Singh PK, Kumar A, Chander J, Khanna G, Roy P, et al. Molecular and matrix-assisted laser desorption ionization – time of flight mass spectrometry-based characterization of clinically significant melanized fungi in India. J Clin Microbiol. 2017;55(4):1090–103. 10.1128/jcm.02413-16. [DOI] [PMC free article] [PubMed]; Singh A, Singh PK, Kumar A, Chander J, Khanna G, Roy P. et al. Molecular and matrix-assisted laser desorption ionization – time of flight mass spectrometry-based characterization of clinically significant melanized fungi in India. J Clin Microbiol. 2017;55(4):1090–103. doi: 10.1128/jcm.02413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pangallo D, Šaková N, Koreňová J, Puškárová A, Kraková L, Valík Ľ, et al. Microbial diversity and dynamics during the production of May bryndza cheese. Int J Food Microbiol. 2014;170:38–43. 10.1016/j.ijfoodmicro.2013.10.015. [DOI] [PubMed]; Pangallo D, Šaková N, Koreňová J, Puškárová A, Kraková L, Valík Ľ. et al. Microbial diversity and dynamics during the production of May bryndza cheese. Int J Food Microbiol. 2014;170:38–43. doi: 10.1016/j.ijfoodmicro.2013.10.015. [DOI] [PubMed] [Google Scholar]
- [14].Vrabec M, Lovayová V, Dudriková K, Gallo J, Dudríková E. Antibiotic resistance and prevalence of enterococcus spp. and escherichia coli isolated from Bryndza Cheese. Ital J Anim Sci. 2016;14(4):3968. 10.4081/ijas.2015.3968. [DOI]; Vrabec M, Lovayová V, Dudriková K, Gallo J, Dudríková E. Antibiotic resistance and prevalence of enterococcus spp. and escherichia coli isolated from Bryndza Cheese. Ital J Anim Sci. 2016;14(4):3968. doi: 10.4081/ijas.2015.3968. [DOI] [Google Scholar]
- [15].Hatzikamari M, Litopoulou‐Tzanetaki E, Tzanetakis N. Microbiological characteristics of Anevato: a traditional Greek cheese. J Appl Microbiol. 1999;87:595–601. 10.1046/j.1365-2672.1999.00857.x. [DOI] [PubMed]; Hatzikamari M, Litopoulou‐Tzanetaki E, Tzanetakis N. Microbiological characteristics of Anevato: a traditional Greek cheese. J Appl Microbiol. 1999;87:595–601. doi: 10.1046/j.1365-2672.1999.00857.x. [DOI] [PubMed] [Google Scholar]
- [16].Macedo AC, Malcata FX, Hogg TA. Microbiological profile in Serra ewe’s cheese during ripening. J Appl Bacteriol. 1995;79:1–11. 10.1111/j.1365-2672.1995.tb03117.x. [DOI]; Macedo AC, Malcata FX, Hogg TA. Microbiological profile in Serra ewe’s cheese during ripening. J Appl Bacteriol. 1995;79:1–11. doi: 10.1111/j.1365-2672.1995.tb03117.x. [DOI] [Google Scholar]
- [17].Nikolaou E, Tzanetakis N, Litopoulou‐Tzanetaki E, Robinson RK. Changes in the microbiological and chemical characteristics of an artisanal, low‐fat cheese made from raw ovine milk during ripening. Int J Dairy Technol. 2002;55:12–7. 10.1111/j.1365-2672.1995.tb03117.x. [DOI]; Nikolaou E, Tzanetakis N, Litopoulou‐Tzanetaki E, Robinson RK. Changes in the microbiological and chemical characteristics of an artisanal, low‐fat cheese made from raw ovine milk during ripening. Int J Dairy Technol. 2002;55:12–7. doi: 10.1111/j.1365-2672.1995.tb03117.x. [DOI] [Google Scholar]
- [18].Zarate V, Belda F, Pérez C, Cardell E. Changes in the microbial flora of Tenerife goat’s milk cheese during ripening. Int Dairy J. 1997;7:635–41. 10.1016/s0958-6946(97)00065-4. [DOI]; Zarate V, Belda F, Pérez C, Cardell E. Changes in the microbial flora of Tenerife goat’s milk cheese during ripening. Int Dairy J. 1997;7:635–41. doi: 10.1016/s0958-6946(97)00065-4. [DOI] [Google Scholar]
- [19].Dahl S, Tavaria FK, Malcata FX. Relationship between flavour and microbiological profiles in Serra da Estrela cheese throughout ripening. Int Dairy J. 2000;10:255–62. 10.1016/s0958-6946(00)00042-x. [DOI]; Dahl S, Tavaria FK, Malcata FX. Relationship between flavour and microbiological profiles in Serra da Estrela cheese throughout ripening. Int Dairy J. 2000;10:255–62. doi: 10.1016/s0958-6946(00)00042-x. [DOI] [Google Scholar]
- [20].Medina M, Fernandez Del Pozo BM, Rodríguez‐Marin A, Gaya P, Nuñez M. Effect of lactic starter inoculation on chemical, microbiological, rheological and sensory characteristics of La Serena cheese. J Dairy Res. 1991;58:355–61. 10.1017/s0022029900029939. [DOI]; Medina M, Fernandez Del Pozo BM, Rodríguez‐Marin A, Gaya P, Nuñez M. Effect of lactic starter inoculation on chemical, microbiological, rheological and sensory characteristics of La Serena cheese. J Dairy Res. 1991;58:355–61. doi: 10.1017/s0022029900029939. [DOI] [Google Scholar]
- [21].Sangoyomi A, Owoseni A, Okerokun O. Prevalence of enteropathogenic and lactic acid bacteria species in wara: a local cheese from Nigeria. Afr J Microbiol Res. 2010;4:1624–30.; Sangoyomi A, Owoseni A, Okerokun O. Prevalence of enteropathogenic and lactic acid bacteria species in wara: a local cheese from Nigeria. Afr J Microbiol Res. 2010;4:1624–30. [Google Scholar]
- [22].Aureli P, Costntini A, Felicia L, Gianfraceschi M, Rainakli L. Occurrence of pathogenic Escherichia coli in available Italian soft cheeses. Arch Food Hyg. 1992;43:1–2.; Aureli P, Costntini A, Felicia L, Gianfraceschi M, Rainakli L. Occurrence of pathogenic Escherichia coli in available Italian soft cheeses. Arch Food Hyg. 1992;43:1–2. [Google Scholar]
- [23].Chaves‐López C, De Angelis M, Martuscelli M, Serio A, Paparella A, Suzzi G. Characterization of the Enterobacteriaceae isolated from an artisanal Italian ewe’s cheese (Pecorino Abruzzese). J Appl Microbiol. 2005;101:353–60. 10.1111/j.1365-2672.2006.02941.x. [DOI] [PubMed]; Chaves‐López C, De Angelis M, Martuscelli M, Serio A, Paparella A, Suzzi G. Characterization of the Enterobacteriaceae isolated from an artisanal Italian ewe’s cheese (Pecorino Abruzzese) J Appl Microbiol. 2005;101:353–60. doi: 10.1111/j.1365-2672.2006.02941.x. [DOI] [PubMed] [Google Scholar]
- [24].Pintado AIE, Pinho O, Ferreira IM, Pintado MME, Gomes AMP, Malcata FX. Microbiological, biochemical and biogenic amine profiles of Terrincho cheese manufactured in several dairy farms. Int Dairy J. 2008;8(6):631–40.; Pintado AIE, Pinho O, Ferreira IM, Pintado MME, Gomes AMP, Malcata FX. Microbiological, biochemical and biogenic amine profiles of Terrincho cheese manufactured in several dairy farms. Int Dairy J. 2008;8(6):631–40. [Google Scholar]
- [25].Kačániová M, Borotová P, Terenjeva M, Kunová S, Felsöciová S, Haščík P, et al. Bryndza cheese of Slovak origin as potential resources of probiotic bacteria. Potravin Slovak J Food Sci. 2020;14:641–6. 10.5219/1413. [DOI]; Kačániová M, Borotová P, Terenjeva M, Kunová S, Felsöciová S, Haščík P. et al. Bryndza cheese of Slovak origin as potential resources of probiotic bacteria. Potravin Slovak J Food Sci. 2020;14:641–6. doi: 10.5219/1413. [DOI] [Google Scholar]
- [26].Frece J, Vrdoljak M, Filipčić M, Jelić M, Čanak I, Jakopović Ž, et al. Microbiological quality and variability of natural microbiota in croatian cheese maturing in lambskin sacks. Food Technol Biotechnol. 2016;54(2):129–34. 10.17113/ftb.54.02.16.4418. [DOI] [PMC free article] [PubMed]; Frece J, Vrdoljak M, Filipčić M, Jelić M, Čanak I, Jakopović Ž. et al. Microbiological quality and variability of natural microbiota in croatian cheese maturing in lambskin sacks. Food Technol Biotechnol. 2016;54(2):129–34. doi: 10.17113/ftb.54.02.16.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Koňuchová M, Liptáková D, Šípková A, Valík Ľ. Role of geotrichum candidum in dairy industry. Chem Listy. 2016;110:491–7.; Koňuchová M, Liptáková D, Šípková A, Valík Ľ. Role of geotrichum candidum in dairy industry. Chem Listy. 2016;110:491–7. [Google Scholar]
- [28].Jaster H, Judacewski P, Ribeiro JCB, Zielinski AAF, Demiate IM, Los PR, et al. Quality assessment of the manufacture of new ripened soft cheese by geotrichum candidum: physico-chemical and technological properties. Food Sci Technol. 2018;39:50–8.; Jaster H, Judacewski P, Ribeiro JCB, Zielinski AAF, Demiate IM, Los PR. et al. Quality assessment of the manufacture of new ripened soft cheese by geotrichum candidum: physico-chemical and technological properties. Food Sci Technol. 2018;39:50–8. [Google Scholar]
- [29].Štefániková J, Ducková V, Miškeje M, Kačániová M, Čanigová M. The impact of different factors on the quality and volatile organic compounds profile in “Bryndza” cheese. Foods. 2020;9:1195. 10.3390/foods9091195. [DOI] [PMC free article] [PubMed]; Štefániková J, Ducková V, Miškeje M, Kačániová M, Čanigová M. The impact of different factors on the quality and volatile organic compounds profile in “Bryndza” cheese. Foods. 2020;9:1195. doi: 10.3390/foods9091195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Laurencik M, Sulo P, Slavikova E, Pieckova E, Seman M, Ebringer L. The diversity of eukaryotic microbiota in the traditional Slovak sheep cheese – Bryndza. Int J Food Microbiol. 2008;127(1–2):176–9. 10.1016/j.ijfoodmicro.2008.06.016. [DOI] [PubMed]; Laurencik M, Sulo P, Slavikova E, Pieckova E, Seman M, Ebringer L. The diversity of eukaryotic microbiota in the traditional Slovak sheep cheese – Bryndza. Int J Food Microbiol. 2008;127(1–2):176–9. doi: 10.1016/j.ijfoodmicro.2008.06.016. [DOI] [PubMed] [Google Scholar]
- [31].Gente S, Sohier D, Coton E, Duhamel C, Gueguen M. Identification of geotrichum candidum at the species and strain level: proposal for a standardized protocol. J Ind Microbiol Biotechnol. 2006;33(12):1019–31. 10.1007/s10295-006-0130-3. [DOI] [PubMed]; Gente S, Sohier D, Coton E, Duhamel C, Gueguen M. Identification of geotrichum candidum at the species and strain level: proposal for a standardized protocol. J Ind Microbiol Biotechnol. 2006;33(12):1019–31. doi: 10.1007/s10295-006-0130-3. [DOI] [PubMed] [Google Scholar]
- [32].Desmasures N, Bazin F, Gueguen M. Microbiological composition of raw milk from selected farms in the Camembert region of Normandy. J Appl Microbiol. 1997;83(1):53–8. 10.1046/j.1365-2672.1997.00166.x. [DOI] [PubMed]; Desmasures N, Bazin F, Gueguen M. Microbiological composition of raw milk from selected farms in the Camembert region of Normandy. J Appl Microbiol. 1997;83(1):53–8. doi: 10.1046/j.1365-2672.1997.00166.x. [DOI] [PubMed] [Google Scholar]
- [33].Boutrou R, Guéguen M. Interests in geotrichum candidum for cheese technology. Int J Food Microbiol. 2005;102(1):1–20. 10.1016/j.ijfoodmicro.2004.12.028. [DOI] [PubMed]; Boutrou R, Guéguen M. Interests in geotrichum candidum for cheese technology. Int J Food Microbiol. 2005;102(1):1–20. doi: 10.1016/j.ijfoodmicro.2004.12.028. [DOI] [PubMed] [Google Scholar]
- [34].Marcellino N, Beuvier E, Grappin R, Guéguen M, Benson DR. Diversity of geotrichum candidum strains isolated from traditional cheese making fabrications in France. Appl Env Microbiol. 2001;67(10):4752–9. 10.1128/aem.67.10.4752-4759.2001. [DOI] [PMC free article] [PubMed]; Marcellino N, Beuvier E, Grappin R, Guéguen M, Benson DR. Diversity of geotrichum candidum strains isolated from traditional cheese making fabrications in France. Appl Env Microbiol. 2001;67(10):4752–9. doi: 10.1128/aem.67.10.4752-4759.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gaborit P, Menard A, Morgan F. Impact of ripening strains on the typical flavour of goat cheeses. Int Dairy J. 2001;11(4–7):315–25. 10.1016/s0958-6946(01)00061-9. [DOI]; Gaborit P, Menard A, Morgan F. Impact of ripening strains on the typical flavour of goat cheeses. Int Dairy J. 2001;11(4–7):315–25. doi: 10.1016/s0958-6946(01)00061-9. [DOI] [Google Scholar]
- [36].Fox PF, Guinee TP, Cogan TM, Mcsweeney PLH. Fundamentals of cheese science. Gaithersburg: Aspen Publishers; 2000. p. 54–97 (Cap. 5).; Fox PF, Guinee TP, Cogan TM, Mcsweeney PLH. Fundamentals of cheese science. Gaithersburg: Aspen Publishers; 2000. pp. p. 54–97. (Cap. 5). [Google Scholar]
- [37].Wouters JTM, Ayad EHE, Hugenholtz J, Smit G. Microbes from raw milk for fermented dairy products. Int Dairy J. 2002;12(2–3):91–109. 10.1016/S0958-6946(01)00151-0. [DOI]; Wouters JTM, Ayad EHE, Hugenholtz J, Smit G. Microbes from raw milk for fermented dairy products. Int Dairy J. 2002;12(2–3):91–109. doi: 10.1016/S0958-6946(01)00151-0. [DOI] [Google Scholar]
- [38].Medeiros RS, Araújo LM, Queiroga Neto V, Andrade PP, Melo MA, Gonçalves MMBP. Identification of lactic acid bacteria isolated from artisanal Coalho cheese produced in the Brazilian Northeast. CyTA – J Food. 2016;14(4):613–20. 10.1080/19476337.2016.1185468. [DOI]; Medeiros RS, Araújo LM, Queiroga Neto V, Andrade PP, Melo MA, Gonçalves MMBP. Identification of lactic acid bacteria isolated from artisanal Coalho cheese produced in the Brazilian Northeast. CyTA – J Food. 2016;14(4):613–20. doi: 10.1080/19476337.2016.1185468. [DOI] [Google Scholar]
- [39].Gaaloul N, Braiek OB, Berjeaud JM, Arthur T, Cavera VL, Chikindas ML, et al. Evaluation of antimicrobial activity and safety aspect of enterococcus italicus ggn10 strain isolated from tunisian bovine raw milk. J Food Saf. 2014;34:300–11. 10.1111/jfs.12126. [DOI]; Gaaloul N, Braiek OB, Berjeaud JM, Arthur T, Cavera VL, Chikindas ML. et al. Evaluation of antimicrobial activity and safety aspect of enterococcus italicus ggn10 strain isolated from tunisian bovine raw milk. J Food Saf. 2014;34:300–11. doi: 10.1111/jfs.12126. [DOI] [Google Scholar]
- [40].Sádecká J, Čaplová Z, Tomáška M, Šoltys K, Kopuncová M, Budiš J, et al. Microorganisms and volatile aroma-active compounds in bryndza cheese produced and marketed in Slovakia. J Food Nutr Res. 2019;58(4):382–92.; Sádecká J, Čaplová Z, Tomáška M, Šoltys K, Kopuncová M, Budiš J. et al. Microorganisms and volatile aroma-active compounds in bryndza cheese produced and marketed in Slovakia. J Food Nutr Res. 2019;58(4):382–92. [Google Scholar]
- [41].Berta G, Chebeňová V, Brežná B, Pangallo D, Valík Ľ, Kuchta T. Identification of lactic acid bacteria in Slovakian bryndza cheese. J Food Nutr Res. 2009;48(2):65–71.; Berta G, Chebeňová V, Brežná B, Pangallo D, Valík Ľ, Kuchta T. Identification of lactic acid bacteria in Slovakian bryndza cheese. J Food Nutr Res. 2009;48(2):65–71. [Google Scholar]
- [42].Mladenović KG, Muruzović MŽ, Žugić Petrović T, Stefanović OD, Čomić LR. Isolation and identification of Enterobacteriaceae from traditional Serbian cheese and their physiological characteristics. J Food Saf. 2017;38(1), 10.1111/jfs.12387. [DOI]; Mladenović KG, Muruzović MŽ, Žugić Petrović T, Stefanović OD, Čomić LR. Isolation and identification of Enterobacteriaceae from traditional Serbian cheese and their physiological characteristics. J Food Saf. 2017;38(1) doi: 10.1111/jfs.12387. [DOI] [Google Scholar]
- [43].Hayaloglu AA, Kirbag S. Microbial quality and presence of moulds in Kuflu cheese. Int J Food Microbiol. 2007;115(3):376–80. 10.1016/j.ijfoodmicro.2006.12.002. [DOI] [PubMed]; Hayaloglu AA, Kirbag S. Microbial quality and presence of moulds in Kuflu cheese. Int J Food Microbiol. 2007;115(3):376–80. doi: 10.1016/j.ijfoodmicro.2006.12.002. [DOI] [PubMed] [Google Scholar]
- [44].Schwarz P, Bretagne S, Gantier J-C, Garcia-Hermoso D, Lortholary O, Dromer F, et al. Molecular identification of zygomycetes from culture and experimentally infected tissues. J Clin Microbiol. 2006;44(2):340–9. 10.1128/jcm.44.2.340-349.2006. [DOI] [PMC free article] [PubMed]; Schwarz P, Bretagne S, Gantier J-C, Garcia-Hermoso D, Lortholary O, Dromer F. et al. Molecular identification of zygomycetes from culture and experimentally infected tissues. J Clin Microbiol. 2006;44(2):340–9. doi: 10.1128/jcm.44.2.340-349.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rakeman JL, Bui U, LaFe K, Chen Y-C, Honeycutt RJ, Cookson BT. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol. 2005;43(7):3324–33. 10.1128/jcm.43.7.3324-3333.2005. [DOI] [PMC free article] [PubMed]; Rakeman JL, Bui U, LaFe K, Chen Y-C, Honeycutt RJ, Cookson BT. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol. 2005;43(7):3324–33. doi: 10.1128/jcm.43.7.3324-3333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chayakulkeeree M, Ghannoum MA, Perfect JR. Zygomycosis: the re-emerging fungal infection. Eur J Clin Microbiol Inf Dis. 2006;25(4):215–29. 10.1007/s10096-006-0107-1. [DOI] [PubMed]; Chayakulkeeree M, Ghannoum MA, Perfect JR. Zygomycosis: the re-emerging fungal infection. Eur J Clin Microbiol Inf Dis. 2006;25(4):215–29. doi: 10.1007/s10096-006-0107-1. [DOI] [PubMed] [Google Scholar]
- [47].Han B-Z, Rombouts FM, Nout MJR. A Chinese fermented soybean food. Int J Food Microbiol. 2001;65(1–2):1–10. 10.1016/s0168-1605(00)00523-7. [DOI] [PubMed]; Han B-Z, Rombouts FM, Nout MJR. A Chinese fermented soybean food. Int J Food Microbiol. 2001;65(1–2):1–10. doi: 10.1016/s0168-1605(00)00523-7. [DOI] [PubMed] [Google Scholar]
- [48].Szczęsna-Antczak M, Antczak T, Piotrowicz-Wasiak M, Rzyska M, Binkowska N, Bielecki S. Relationships between lipases and lipids in mycelia of two Mucor strains. Enz Microb Technol. 2006;39(6):1214–22. 10.1016/j.enzmictec.2006.03.008. [DOI]; Szczęsna-Antczak M, Antczak T, Piotrowicz-Wasiak M, Rzyska M, Binkowska N, Bielecki S. Relationships between lipases and lipids in mycelia of two Mucor strains. Enz Microb Technol. 2006;39(6):1214–22. doi: 10.1016/j.enzmictec.2006.03.008. [DOI] [Google Scholar]
- [49].Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86(11):807–15. 10.1016/j.biochi.2004.09.017. [DOI] [PubMed]; Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86(11):807–15. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- [50].Shahab Lavasani AR, Ehsani MR, Mirdamadi S, Mousavi MAEZ. Changes in physicochemical and organoleptic properties of traditional Iranian cheese Lighvan during ripening. Int J Dairy Technol. 2012;65:64–70. 10.1111/j.1471-0307.2011.00724.x. [DOI]; Shahab Lavasani AR, Ehsani MR, Mirdamadi S, Mousavi MAEZ. Changes in physicochemical and organoleptic properties of traditional Iranian cheese Lighvan during ripening. Int J Dairy Technol. 2012;65:64–70. doi: 10.1111/j.1471-0307.2011.00724.x. [DOI] [Google Scholar]
- [51].Serhan M, Mattar J. Characterization of four Lebanese artisanal goat milk cheeses: Darfi yeh, Aricheh, Shankleesh and Serdale by physico-chemical, microbiological and sensory analyses. J Food Agric Environ. 2013;11:97–101.; Serhan M, Mattar J. Characterization of four Lebanese artisanal goat milk cheeses: Darfi yeh, Aricheh, Shankleesh and Serdale by physico-chemical, microbiological and sensory analyses. J Food Agric Environ. 2013;11:97–101. [Google Scholar]