Abstract
Pathogenic missense mutations in the leucine-rich repeat kinase 2 gene, encoding LRRK2, results in the upregulation of Rab10 and Rab12 phosphorylation in different cells and tissues. Here, we evaluate levels of the LRRK2 kinase substrates pT73-Rab10 and pS106-Rab12 proteins in rat brain tissues from different genetic backgrounds. Whereas lines of Sprague Dawley rats have equivalent levels of pT73-Rab10 and pS106-Rab12 similar to Lrrk2 knockout rats, Long-Evans rats have levels of pT73-Rab10 and pS106-Rab12 comparable to G2019S-LRRK2 BAC transgenic rats. Strong LRRK2 kinase inhibitors are ineffective at reducing pT73-Rab10 and pS106-Rab12 levels in the Sprague Dawley rats, but potently reduce pT73-Rab10 and pS106-Rab12 levels in Long-Evans rats. Oral administration of the PFE-360 LRRK2 kinase inhibitor fails to provide neuroprotection from dopaminergic neurodegeneration caused by rAAV2/1-mediated overexpression of A53T-αsynuclein in Sprague Dawley rats. These results highlight substantial differences in LRRK2-mediated Rab10 and Rab12 phosphorylation in commonly utilized rat genetic backgrounds and suggest LRRK2 may not play a central role in Rab phosphorylation or mutant αsynuclein toxicity in Sprague Dawley rats.
Keywords: LRRK2, LRRK2 kinase inhibitor, Rab10, Rab12, neurodegeneration
1. Introduction
Pathogenic mutations in the leucine-rich repeat kinase 2 gene (LRRK2) are among the most common genetic causes of neurodegeneration and Parkinson’s disease (PD) (Trinh et al., 2014). LRRK2 mutations are thought to increase LRRK2 autophosphorylation and phosphorylation of Rab substrates (Alessi and Sammler, 2018; West, 2017). Novel therapeutics targeting LRRK2 kinase activity have emerged in clinical trials (Kelly and West, 2020). Owing to the genetic link between the overexpression of αsynuclein (αsyn) and PD (Singleton et al., 2003), a common pre-clinical model to explore novel therapeutics uses recombinant adeno-viral associated (rAAV) virus to acutely overexpress αsyn in rat dopaminergic neurons. Viral particles are typically administered via a single intracranial injection in the substantia nigra pars compacta (SNpc), with dopaminergic neurodegeneration observed weeks after injections (Kirik et al., 2002). In the Long-Evans strain of rat, neurodegeneration caused by rAAV2/1 WT-αsyn overexpression was attenuated with the knockout of Lrrk2 (rat) expression (Daher et al., 2014). Pharmacological Lrrk2 kinase inhibition with the PF-475 molecule, a small molecule with favorable pharmacokinetics in rats, also reduces rAAV2/1 WT-αsyn neurotoxicity in the Sprague Dawley genetic background (Daher et al., 2015).
Pharmacodynamic profiles used to study LRRK2 inhibitors in mice and rats have largely relied on measures of total LRRK2 protein and the pharmacodynamic LRRK2 phosphorylation site pS935-LRRK2 (Dzamko et al., 2010; Kelly et al., 2018; Kelly and West, 2020; Rideout et al., 2020). However, pS935-LRRK2 is not a direct measure of LRRK2 kinase activity and is regulated by different upstream kinases (Kelly and West, 2020). Recently, Rab10 and Rab12 have been identified as physiological LRRK2 substrates, and pathogenic LRRK2 mutations may increase Rab phosphorylation (Ito et al., 2016; Lis et al., 2018; Liu et al., 2018; Steger et al., 2017, 2016). Several reports have evaluated pT73-Rab10 levels in robust pharmacodynamic responses to LRRK2 inhibition in lung, kidney, and spleen tissue, with much lower apparent levels detected in brain tissues despite high levels of total Rab10 protein (Ito et al., 2016; Lis et al., 2018). Because of the apparent low levels of pT73-Rab10 in brain tissues, it is unclear whether pT73-Rab10 might serve as a useful pharmacodynamic marker for LRRK2 in inhibition in the brain.
In this study, we evaluated pT73-Rab10 as well as pS106-Rab12 levels in forebrain tissue extracts of several strains of rats used in past studies that include LRRK2 transgenic and knockout rats, in addition to Sprague Dawley and Long-Evans genetic backgrounds. The Long-Evans rat strain background includes the Lrrk2 knockout (LE) rat (Daher et al., 2014), whereas the Sprague Dawley strain includes the transgenic G2019S-LRRK2 BAC rat (Daher et al., 2015; Lee et al., 2015; Sloan et al., 2016; West et al., 2014). Our results highlight large genetic background differences driving LRRK2-dependent pT73-Rab10 and pS016-Rab12 levels. We further determined that the kinase inhibitor PFE-360, a more potent version of the PF-475 small molecule inhibitor used in the past, fails to block an aggressive lesion caused by mutant A53T-αsyn overexpression in Sprague Dawley rats. These results highlight a previously unknown complexity of apparent variable LRRK2 kinase regulation in different genetic backgrounds, and mutant A53T-αsyn neurotoxicity that is independent from LRRK2 kinase activity.
2. Results
2.1. pT73-Rab10 and pS106-Rab12 levels in the rat brain vary with genetic background
Previous immunohistochemical studies identified the highest levels of rat Lrrk2 expression in different neuronal subtypes within the dorsal striatum and frontal cortex in both Taconic non-transgenic (Tac(Sprague Dawley)) and transgenic G2019S-LRRK2 BAC (Tac(SD)) rat brains (West et al., 2014). To estimate how Lrrk2 kinase activity might vary depending on genetic background, we evaluated brain extracts from these and other strains of rats for levels of the novel LRRK2 kinase substrates pT73-Rab10 and pS106-Rab12 (Figure 1a). Homozygous Lrrk2 knockout (Long-Evans, LE) rats were bred from heterozygous Lrrk2 knockout (LE) rats on the Long-Evans genetic background. These rats completely lack Lrrk2 expression (Figure 1a–c). Total Lrrk2 expression levels in the brain did not vary between Long-Evans, Tac(Sprague Dawley), and CD(Sprague Dawley) rats (Figure 1b). Hemizygous G2019S-LRRK2 BAC (Tac(SD)) rats had 10-fold elevated total LRRK2 (human) compared to Tac(Sprague Dawley) littermates that lacked the transgene (Figure 1b). The ratio of endogenous rat pS935-Lrrk2 to total Lrrk2 protein was similar to pS935-LRRK2 to total LRRK2 protein in the G2019S-LRRK2 BAC (Tac(SD)) rats (Figure 1c). These results show that both total Lrrk2 protein levels do not vary in these genetic backgrounds. The ratio of pS935-LRRK2 to total protein also does not vary, even in the condition of over-expressed mutant LRRK2.
Figure 1. Upregulated pT73-Rab10 and pS106-Rab12 levels in Long-Evans and G2019S-LRRK2 BAC (Tac(SD)) rat brain tissue.
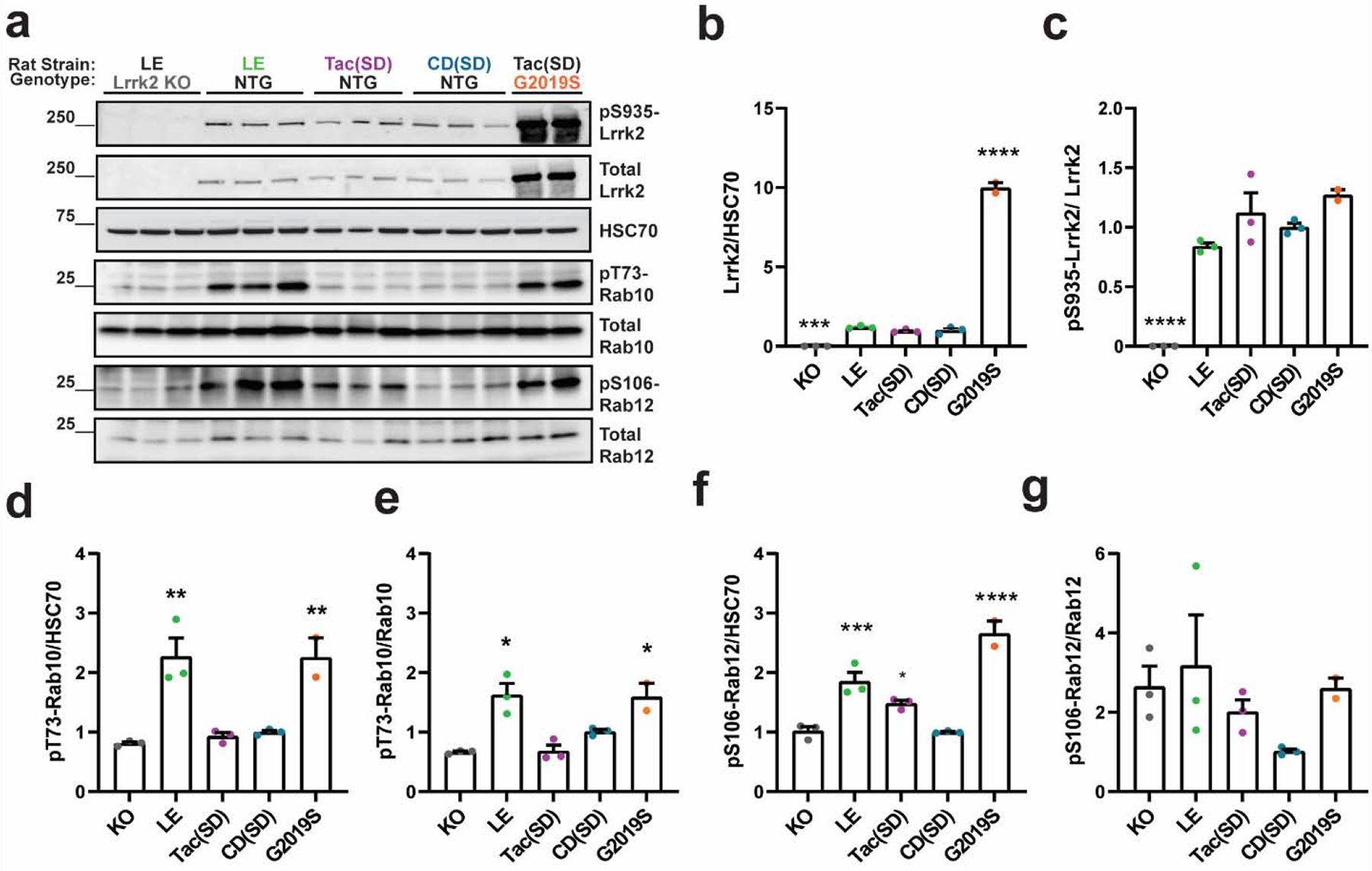
a. Representative immunoblots measuring Lrrk2 and Rab protein substrates in forebrain lysates from Lrrk2 knockout (KO, Long-Evans background), Long-Evans (LE), Tac(Sprague Dawley, (Tac(SD)), Charles Rivers(Sprague Dawley, (CD(SD)), and transgenic G2019S-LRRK2 BAC rats (on the Tac(SD) background). b. Quantification of total Lrrk2 protein levels normalized to HSC70 and c. pS935-Lrrk2 levels normalized to total Lrrk2 protein. Quantification of pT73-Rab10 levels normalized to d. HSC70 and e. total Rab10 protein. Quantification of pS106-Rab12 levels normalized to f. HSC70 and g. total Rab12. Data are graphed as average measures from triplicate western blots for biological replicates (N=3 rats per group, except for G2019S-LRRK2 BAC (N=2 rats)). Data are presented as fold changes of the mean value of the CD(SD) group. Bar graphs show group means ± SEM. Significance between groups was determined by one-way ANOVA with Dunnett’s multiple comparison test with respect to the indicated group and the CD(SD) group. *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.0001. All other comparisons not indicated by asterisks were not significant.
Unexpectedly, the Lrrk2 knockout (LE) rats had similar levels of pT73-Rab10 and pS106-Rab12 compared with Tac(Sprague Dawley) and CD(Sprague Dawley) rats, whether phospho-Rab levels were normalized to HSC70 (Figure 1d,f) or total Rab protein (Figure 1e,g). This finding suggests that the Sprague Dawley genetic background may have lowly active or inactive Lrrk2 protein in the healthy brain with respect to Rab10 and Rab12 phosphorylation, despite normal levels of pS935-Lrrk2. In contrast, Long-Evans rats with only endogenous Lrrk2 demonstrated pT73-Rab10 levels comparable to G2019S-LRRK2 BAC (Tac(SD)) rats (Figure 1d,e). The G2019S-LRRK2 BAC (Tac(SD)) rats and wild-type Long-Evans rats both had approximately two-fold increased pT73-Rab10 levels compared to the Sprague Dawley background. Similarly, pS106-Rab12 normalized to HSC70 was markedly increased in the brain extracts (Figure 1f). However, total Rab12 was more variable in expression as compared with total Rab10 between different rats (Supplemental Figure 1), and the ratio of pS106-Rab12 to total Rab12 was not significantly different between these strains or with overexpressed LRRK2 protein (Figure 1g). These results suggest that healthy Sprague Dawley strains have little or no measurable Lrrk2-dependent Rab10 and Rab12 phosphorylation in the brain, but LRRK2 activity can be recovered with G2019S-LRRK2 overexpression.
Efforts to evaluate pS1292-Lrrk2 in rat brain extracts, another marker of LRRK2 kinase activity and a marker of autophosphorylation (Kluss et al., 2018; Sheng et al., 2012), were attempted, but non-specific banding patterns prevented reliable discrimination of authentic signal outside of transgenic LRRK2 expression (Supplemental Figure 2). Similarly, no specific signals related to total Rab29 or pT71-Rab29 at expected molecular weights could be measured with the antibodies utilized here in any of the rat strains (Supplemental Figure 2).
2.2. Acute pharmacological LRRK2 kinase inhibition reduces elevated pT73-Rab10 and pS106-Rab12 levels in Long-Evans (but not Sprague Dawley) rats
To determine whether pT73-Rab10 and pS106-Rab12 levels in the different genetic background are dependent on Lrrk2 kinase activity, we first employed a potent and selective LRRK2 kinase inhibitor PF-475 that nearly ablates pS935-Lrrk2 levels in the brain without reducing total Lrrk2 protein levels (Daher et al., 2015). Two hours post oral gavage, a time point near plasma drug Cmax (Henderson et al., 2015), we observed very little remaining pS935-LrrK2 levels in the brain but normal total Lrrk2 protein levels (Figure 2a–c). Neither pT73-Rab10 levels nor pS106-Rab12 levels were reduced in CD(Sprague Dawley) rats when normalized to HSC70 levels after treatment with PF-475 (Figure 2d,f). Nominal reductions in the ratio of pT73-Rab10 and pS106-Rab12 levels to total Rab proteins were observed at the highest drug dose (Figure 2e,g). Unexpectedly, both total Rab10 and Rab12 protein levels were slightly higher in the drug treated groups compared to vehicle controls, potentially affecting interpretation of the apparent reduction in phospho-Rab levels when understood as a ratio of phosphorylated to non-phosphorylated protein (Supplemental Figure 1c,d). These results are potentially consistent with pT73-Rab10 and pS106-Rab12 levels comparable between Sprague Dawley rat strains and Lrrk2 knockout (LE) rats, where apparent pT73-Rab10 and pS106-Rab12 levels are largely unrelated to Lrrk2 kinase activity.
Figure 2. Variable pT73-Rab10 and pS106-Rab12 levels in CD(Sprague Dawley) rats treated with the LRRK2 kinase inhibitor PF-475.
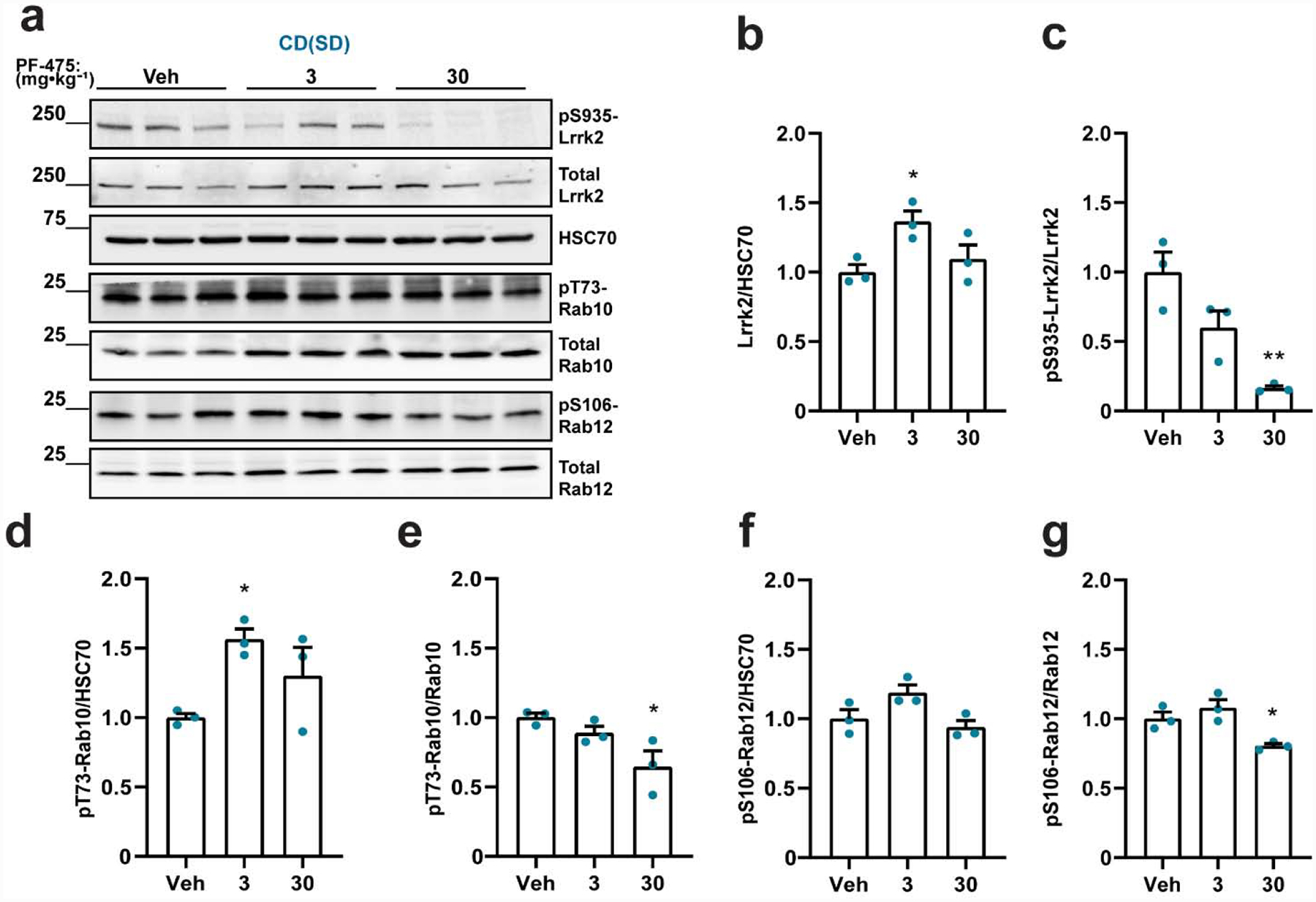
a. Representative immunoblots for detecting Lrrk2 and Rab proteins in forebrain lysates from Charles Rivers(Sprague Dawley, (CD(SD)) rats treated with PF-475 (3 or 30 mg·kg−1) and vehicle (Veh) controls. Tissue was collected two hours post-dosing. Quantification of b. total Lrrk2 protein levels normalized to HSC70, and c. pS935-Lrrk2 levels normalized to total Lrrk2 protein. Quantification of pT73-Rab10 levels normalized to d. HSC70 and e. total Rab10 protein. Quantification of pS106-Rab12 levels normalized to f. HSC70 and g. total Rab12. Data are graphed as average measures from triplicate western blots for biological replicates (N=3 rats per group). All values are presented as fold changes to the vehicle treated group. Bar graphs show group means ± SEM. Significance between groups was determined by one-way ANOVA with Dunnett’s multiple comparison test with respect to the indicated group and the vehicle treated group mean. *p< 0.05, **p< 0.01. All other comparisons not indicated by asterisks were not significant.
To further investigate whether the elevated pT73-Rab10 and pS106-Rab12 levels found in Long-Evans rats are dependent on Lrrk2 kinase activity, we treated Long-Evans rats with PF-475 (Figure 3a). PF-475 did not alter total Lrrk2 protein levels but completely ablated pS935-Lrrk2 levels (Figure 3b,c), similar to effects observed in CD(Sprague Dawley) rats (Figure 2). pT73-Rab10 levels, normalized to HSC70 or to total Rab10 protein, were greatly reduced by PF-475 treatment in Long-Evans rats but not in littermate Lrrk2 knockout (LE) rats (Figure 3d,e). PF-475 treatment also caused a large reduction of pS106-Rab12 in Long-Evans rats but not in Lrrk2 knockout (LE) rats (Figure 3f,g). These results support the conclusion that elevated pT73-Rab10 and pS106-Rab12 levels in Long-Evans rats are due to elevated Lrrk2 kinase activity.
Figure 3. Reduced pT73-Rab10 and pS106-Rab12 levels in Long-Evans rats treated with the LRRK2 kinase inhibitor PF-475.
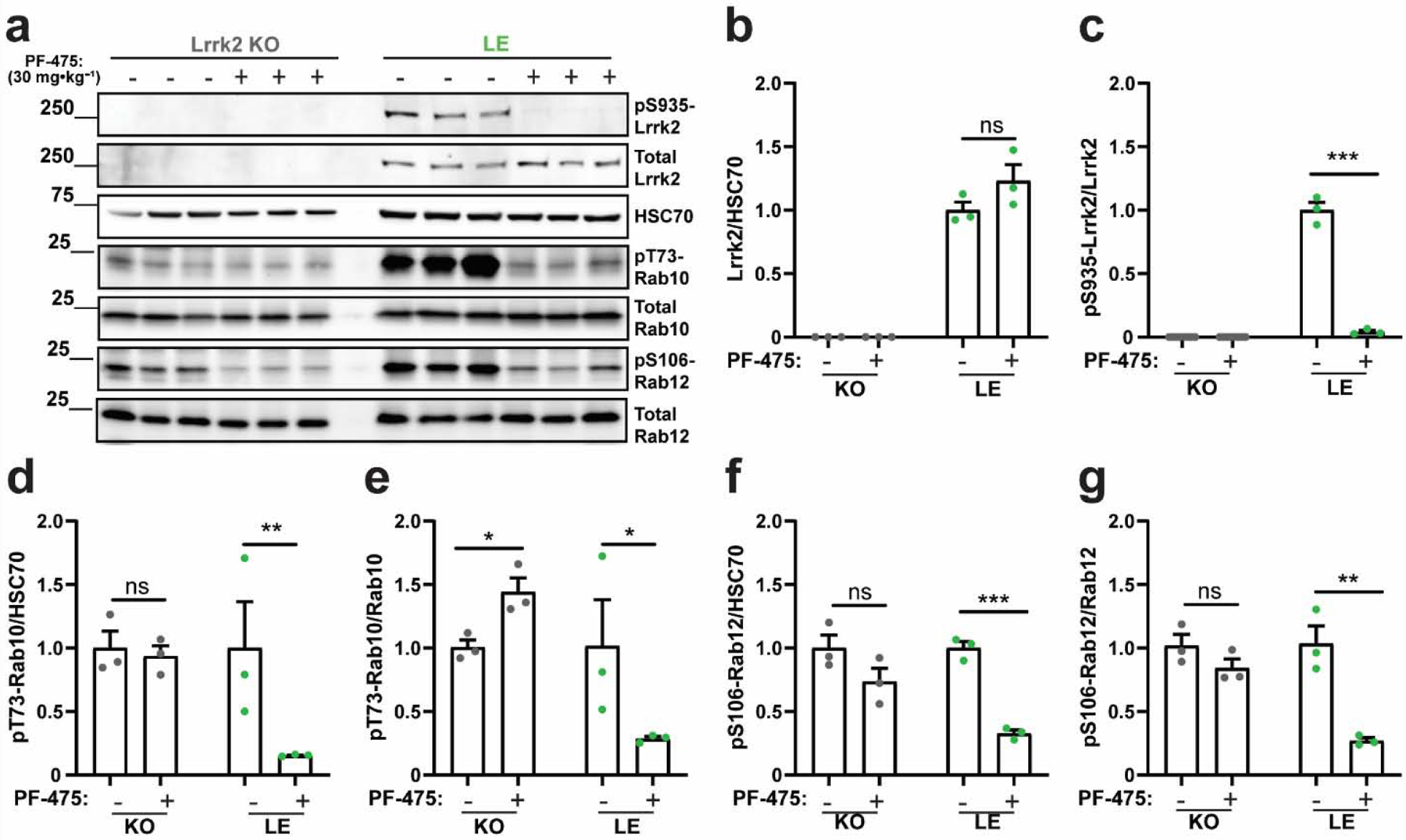
a. Representative immunoblots for detecting Lrrk2 and Rab proteins in forebrain lysates from Lrrk2 knockout (KO) and Long Evans (LE) rats treated with a high dose of PF-475 (30 mg·kg−1) or vehicle control. Tissue was collected two hours post-dosing. Quantification of b. total Lrrk2 protein levels normalized to HSC70 and c. pS935-Lrrk2 levels normalized to total Lrrk2 protein. Quantification of pT73-Rab10 levels normalized to d. HSC70 and e. total Rab10 protein. Quantification of pS106-Rab12 levels normalized to f. HSC70 and g. total Rab12. Data are graphed as average measures from triplicate western blots for biological replicates (N=3 rats per group). All values are presented as fold changes to the vehicle treated group. Bar graphs show group means ± SEM. Significance between treated and untreated (vehicle) controls was determined using two-tailed unpaired t-tests. Data in panels d and e were log transformed prior to t-tests to better approximate normal group distributions. *p< 0.05, **p< 0.01, ***p< 0.001, ns is not significant.
A recently described structurally related compound to PF-475 known as PFE-360 offers substantially improved selectivity and potency (see Supplemental Table 1, and (Henderson et al., 2015; Thirstrup et al., 2017)). Different from PF-475, PFE-360 binding to the Lrrk2 ATP pocket leads to destabilized total protein and accelerated turnover (Kelly et al., 2018). To determine whether an acute reduction of total Lrrk2 protein expression caused by PFE-360 treatment might affect pT73-Rab10 and pS106-Rab12 levels in the CD(Sprague Dawley) line, we utilized a high oral dose of PFE-360 and observed the expected acute reduction of total Lrrk2 protein in the brain (Figure 4a,b) accompanied by a total ablation of pS935-Lrrk2 levels (Figure 4c). However, similar to PF-475 treatment, PFE-360 treatment did not affect levels of pT73-Rab10 or pS106-Rab12 that were similar to levels found in Lrrk2 knockout (LE) rats (Figure 4d–g). These results support the conclusion that levels of pT73-Rab10 and pS106-Rab12 are not dependent on Lrrk2 kinase activity in Sprague Dawley rats. We next determined whether the higher potency associated with PFE-360 might further reduce pT73-Rab10 and pS106-Rab12 levels found in the Long-Evans rats compared to PF-475 treatment, given the larger effects on reducing total Lrrk2 protein (Figure 5a). PFE-360 treatment caused a loss of total Lrrk2 protein and pS935-Lrrk2 protein in Long-Evans rats as expected (Figure 5b,c). PFE-360 treatment also reduced both pT73-Rab10 and pS106-Rab12 levels in Long-Evans rats (Figure 5d–g), comparable to effects observed with PF-475 treatment in the same genetic background. Different from effects noted for PF-475, PFE-360 treatment decreased pS106-Rab12 levels in Lrrk2 knockout (LE) rats (Figure 5f,g). These results suggest that PFE-360 treatment did not substantially further reduce pT73- Rab10 and pS106-Rab12 levels compared to PF-475. PFE-360 activity in Lrrk2 knockout (LE) rats suggests unknown off-target interactions (Supplemental Table 1). Notably, the Lrrk2 knockout (LE) strain of rat utilized here has been extensively evaluated for traces of Lrrk2 protein in the brain with antibodies spanning the entirety of the protein, with no evidence for any Lrrk2 protein expression found (Davies et al., 2013).
Figure 4. The LRRK2 kinase inhibitor PFE-360 does not reduce pT73-Rab10 and pS106-Rab12 levels in CD(Sprague Dawley) rats.
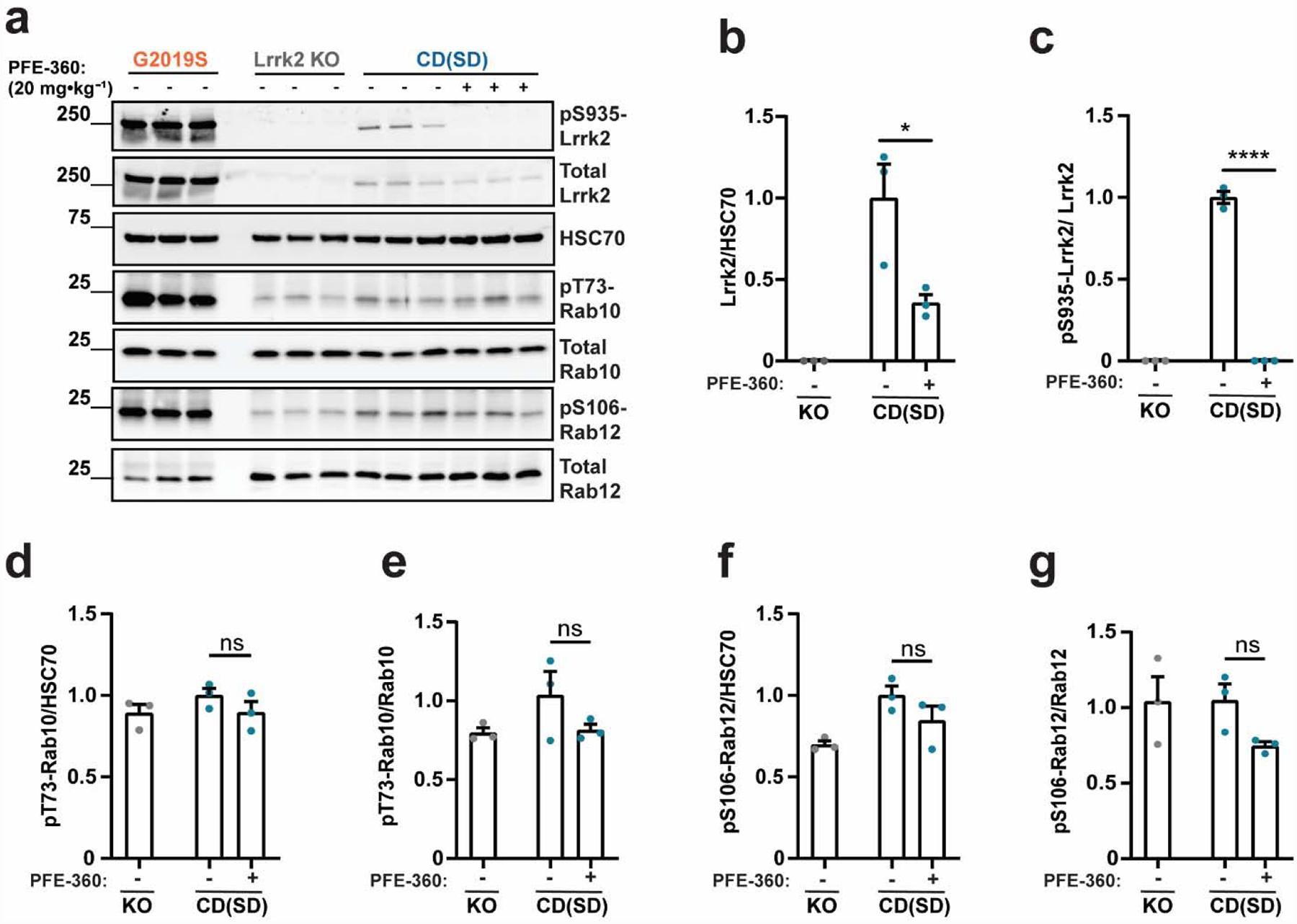
a. Representative immunoblots for detecting Lrrk2 and Rab proteins in forebrain lysates from Charles Rivers(Sprague Dawley, (CD(SD)) rats (shown in blue) treated with PFE-360 (20 mg·kg−1 or vehicle). Tissue was collected two hours post dosing. G2019S-LRRK2 BAC (Tac(SD)) and Lrrk2 knockout rats (Long-Evans background, KO, shown in grey) were included for comparison. Quantification of b. total Lrrk2 protein levels normalized to HSC70 and c. pS935-Lrrk2 levels normalized to total Lrrk2 protein. Quantification of pT73-Rab10 levels normalized to d. HSC70 and e. total Rab10 protein. Quantification of pS106-Rab12 levels normalized to f. HSC70 and g. total Rab12. Data are graphed as average measures from triplicate western blots for biological replicates (N=3 rats per group). All values are presented as fold changes to the vehicle treated group. Bar graphs show group means ± SEM. Significance between groups were determined with a two-tailed unpaired t-test and Welch’s correction. two-tailed unpaired t-test,. *p< 0.05, ****p< 0.0001, ns is not significant.
Figure 5. Reduced pT73-Rab10 and pS106-Rab12 levels in Long-Evans rats treated with the LRRK2 kinase inhibitor PFE-360.
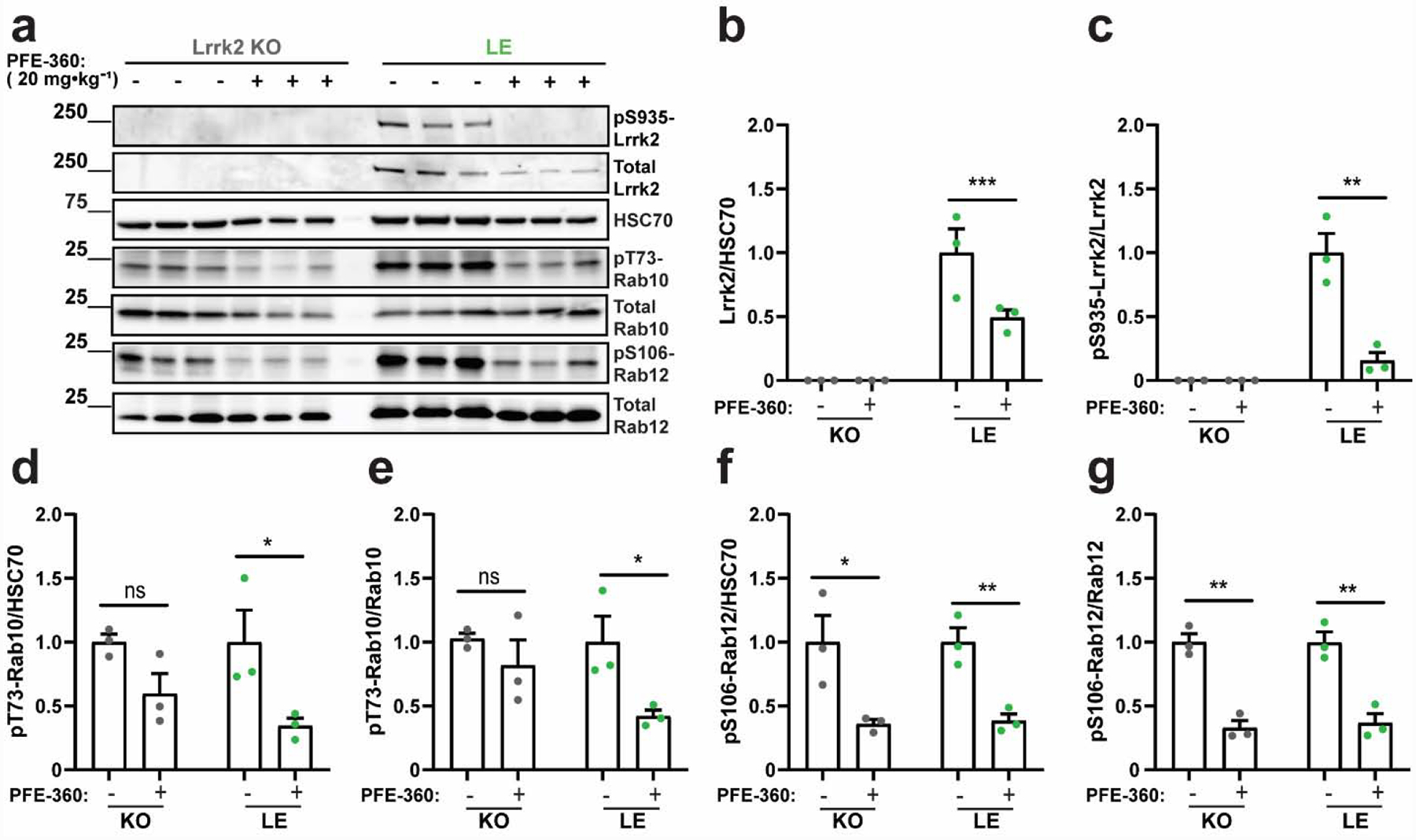
a. Representative immunoblots for detecting Lrrk2 and Rab proteins in forebrain lysates from Lrrk2 knockout (KO) and Long-Evans (LE) rats treated with PFE-360 (20 mg·kg−1) or vehicle control. Tissue was collected two hours post dosing. Quantification of b. total Lrrk2 protein levels normalized to HSC70 and c. pS935-Lrrk2 levels normalized to total Lrrk2 protein. Quantification of pT73-Rab10 levels normalized to d. HSC70 and e. total Rab10 protein. Quantification of pS106-Rab12 levels normalized to f. HSC70 and g. total Rab12. Data are graphed as average measures from triplicate western blots for biological replicates (N=3 rats per group). All values are presented as fold changes to the vehicle treated group. Bar graphs show group means ± SEM. Significance between groups were determined by two-tailed unpaired t-test. Data in panels b and d were log transformed prior to t-tests to better approximate normal group distributions. *p< 0.05, **p< 0.01., ***p< 0.001, ns is not significant.
2.3. Chronic PFE-360 treatment does not prevent dopaminergic neurodegeneration caused by AAV2/1-A53T-αsyn in CD(Sprague Dawley) rats
AAV-mediated overexpression of human αsyn in the midbrain has been shown to induce degeneration of dopaminergic neurons in the substantia nigra pars compacta (SNpc) in Tac(Sprague Dawley) rats (Daher et al., 2015; Ip et al., 2017; Kirik et al., 2002). We utilized a viral vector recently developed by the Michael J. Fox Foundation (Baptista et al., 2013; Padmanabhan et al., 2020) for broad industry and academic usage to test PFE-360 potency in blocking neurotoxicity caused by mutant A53T-αsyn expression. Four weeks post-AAV injection, we utilized the monoclonal antibody SYN514, a marker of misfolded and nitrosylated/oxidized αsyn, to determine that PFE-360 treatment did not appear to grossly affect viral expression at endpoints (Figure 6a). The AAV-null group (virus-only control, no αsyn expression, but equivalent viral particles) did not cause a loss of tyrosine-hydroxylase (TH+) cells in the SNpc, but the AAV-A53T-αsyn vehicle treated group demonstrated an approximate 50% lesion of TH+ neurons four weeks post viral injection (Figure 6b). Continuous PFE-360 treatment had no effect on the development of this lesion and loss of TH+ neurons. TH+ fibers in the dorsal striatum were unaffected by AAV-null exposure, whereas AAV-mediated A53T-αsyn expression also caused an approximate 50% loss of fibers. PFE-360 treatment likewise did not change the magnitude of the loss of TH+ fibers in the dorsal striatum (Figure 6c). These results suggest that the potent PFE-360 inhibitor does not protect dopaminergic neurons from mutant αsyn expression in CD(Sprague Dawley) rats.
Figure 6. PFE-360 treatment does not protect CD(Sprague Dawley) rats from AAV-A53T-αsyn induced dopaminergic neurodegeneration.
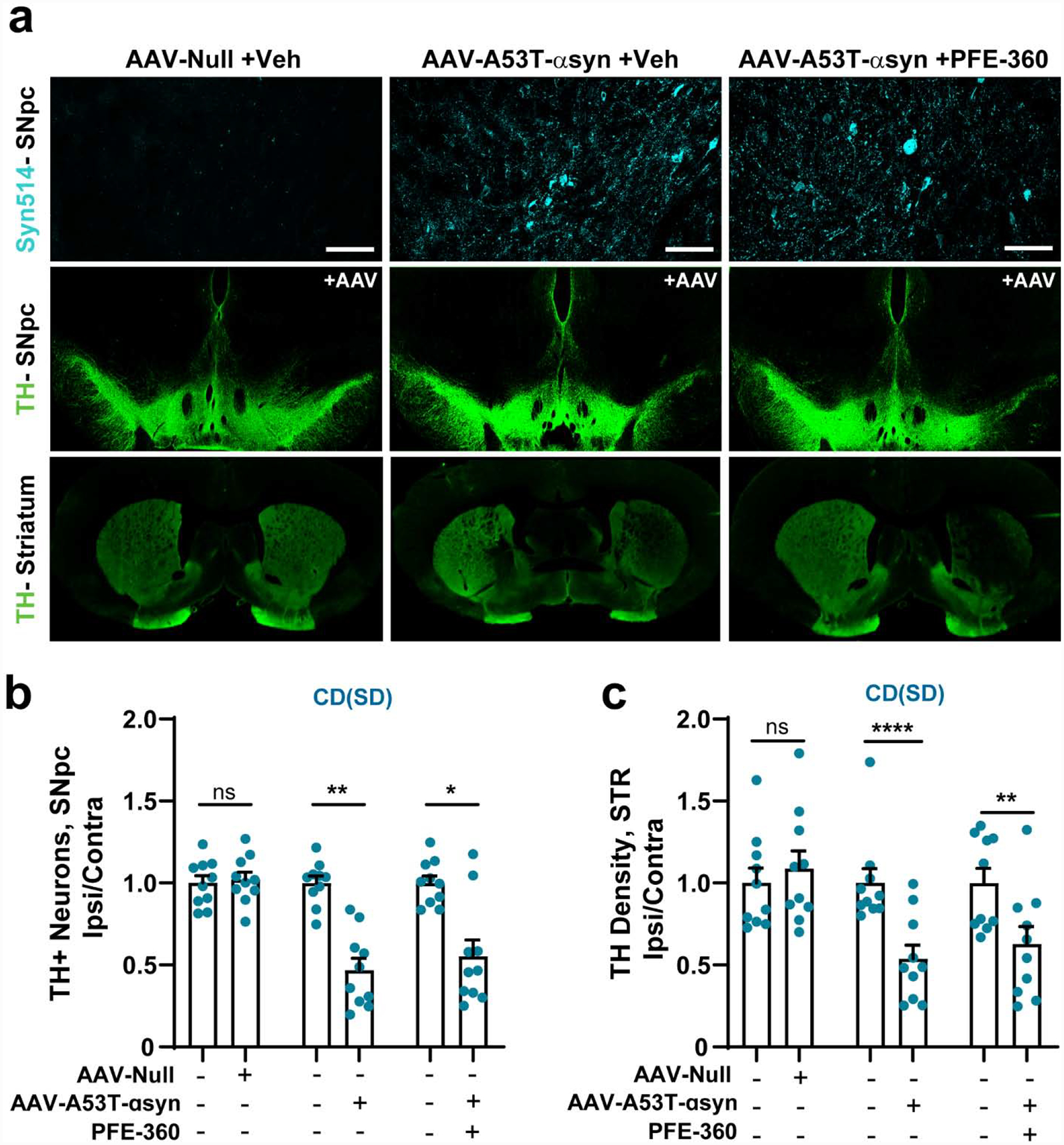
Brain tissues were harvested 28-days post injection of A53T-αsyn AAV or control in the SNpc. Male CD(Sprague Dawley) rats were treated with PFE-360 (10 mg·kg−1 BID), or vehicle, for the full 28 days post-virus injection. a. Representative images of immunofluorescence from coronal sections that highlight αsynuclein deposition in the SNpc, tyrosine hydroxylase (TH+) cells in the SNpc, and TH+ fibers in the dorsal striatum (STR). b. Unbiased stereological counts of TH+ cells in the contralateral (−) and ipsilateral (+) SNpc with vehicle (−) or PFE-360 (+) treatment. Ten rats were included per group, with individual data points representing the analysis of the full SNpc from each rat. c. Immunofluorescent density analysis of TH+ fibers in the contralateral (−) and ipsilateral (+) dorsal striatum with vehicle (−) or PFE-360 (+) treatment. Data are graphed with each data point indicating the mean of ~6 sections through the dorsal striatum analyzed from each rat, where N=10 rats per group. Data are presented normalized to the contralateral (uninjected) side (left hemisphere). Bar graphs show group means ± SEM. Significance was determined by a two-way ANOVA (i.e., virus and drug) with Sidak’s multiple comparison test used to detect ipsilateral versus contralateral differences in group mean values. *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.0001, ns is not significant.
Discussion
Novel therapeutics targeting LRRK2 kinase activity have recently advanced to clinical trials with the hopes of slowing or halting ongoing neurodegenerative processes (Kelly and West, 2020). LRRK2 kinase activity appears central to LRRK2-linked disease (Alessi and Sammler, 2018; West, 2017). LRRK2 phosphorylates Rab-GTPase substrates in different cells and tissues, with notable evidence for LRRK2 phosphorylation of Rab8a, Rab10, Rab12, Rab29, and Rab35 (Liu et al., 2018; Steger et al., 2016). With specific and sensitive monoclonal antibodies recently available, we sought to evaluate the status of Rab LRRK2 kinase substrates in rat strains commonly used in pre-clinical research. Few studies have yet validated tissue phospho-Rab levels as authentic pharmacodynamic markers in the brain. For example, a recent large study in non-human primates evaluating three different LRRK2 kinase inhibitors (PFE-360, MLi2, and GNE-7951) did not report whether inhibitor treatments altered phospho-Rab substrates in the brain (Baptista et al., 2020).
Our conclusions in this study center on three main findings. First, we found that the commonly used Sprague Dawley strain of rat shows low pT73-Rab10 and pS106-Rab12 levels in the adult brain, indistinguishable from those found in Lrrk2 knockout (LE) rats. In contrast, the G2019S-LRRK2 BAC (Tac(SD)) brain demonstrates elevated levels of pT73-Rab10 and pS106-Rab12 that are comparable to those found in healthy Long-Evans rats. Second, we found that the elevated pT73-Rab10 and pS106-Rab12 levels in the Long-Evans rats can be reduced to levels found in Lrrk2 knockout (LE) rats with strong LRRK2 kinase inhibitors. These results highlight the disconnect between total Lrrk2 expression and pS935-Lrrk2 levels in predicting levels of phospho-Rab proteins, and the lack of utility of pT73-Rab10 and pS106-Rab12 levels as Lrrk2 pharmacodynamic markers in certain genetic backgrounds. Notably, the ratio of pS935-LRRK2 to total LRRK2 was similar in all strains even in the condition of over-expressed mutant G2019S-LRRK2 protein. Third, we found that the novel and potent PFE-360 inhibitor failed to provide neuroprotection in a standardized rat model of A53T-αsyn-mediated neurodegeneration originally developed in Sprague Dawley rats. These observations suggest further work is needed to isolate a suitable standardized in vivo pre-clinical model that might facilitate the identification of the most efficacious LRRK2-targeting therapeutics. Together, our results suggest that genetic background may critically determine LRRK2 kinase activity with respect to Rab substrate phosphorylation.
Owing in part to the successful development of highly sensitive and specific monoclonal antibodies, pT73-Rab10 protein has been proposed as a robust pharmacodynamic marker of LRRK2 kinase activity (Atashrazm et al., 2019; Ito et al., 2016; Thirstrup et al., 2017). Reductions in pT73-Rab10 levels in response to LRRK2 kinase inhibition have been assessed in peripheral cells (Baptista et al., 2020). Similar to reports in mice, treatment with LRRK2 kinase inhibitor GNE-7915 (30 mg per kg BID) reduces pT73-Rab10 levels in macaque lung tissue without altering total Rab10 levels (Baptista et al., 2020). Our recent results in urinary exosomes collected from non-human primates acutely dosed with PFE-360 or MLi2 also demonstrate reduced pT73-Rab10 levels (Wang et al., 2020). However, in stimulated human peripheral blood mononuclear cells (PBMCs), LRRK2 dependent phosphorylation of Rab10 and Rab12 are differentially affected by PFE-360 treatment, where pT73-Rab10 levels are acutely reduced and pS106-Rab12 levels may be unaltered (Thirstrup et al., 2017). We were unable to reliably measure other LRRK2-Rab substrates via western blot including Rab8a, Rab29, and Rab35 due to ambiguous staining patterns on membranes that seemed insensitive to either LRRK2 kinase inhibition or mutant G2019S-LRRK2 over-expression. As newer monoclonal antibodies become available to additional Rab targets, panels of brain extracts from different rat strains, treated with potent kinase inhibitors or vehicle control, may provide valuable additional insights.
Collectively, our data show that Lrrk2 kinase activity in the brain, assessed through measures of pT73-Rab10 and pS106-Rab12 levels, is different between Long-Evans and Sprague Dawley rats despite comparable total Lrrk2 protein levels. Sprague Dawley rats align with Lrrk2 knockout (LE) rats with respect to phospho-Rab levels, whereas Long-Evans rats align with G2019S-LRRK2 BAC (Tac(SD)) rats that overexpress mutant G2019S-LRRK2 protein ten-fold. All of our analyses were performed using tissue from the dorsal striatum and the frontal cortex because these areas have been previously shown to harbor the highest levels of Lrrk2 protein in the rat brain (West et al., 2014). Clearly, total Lrrk2 protein expression or pS935-Lrrk2 levels do not predict the extent of Rab phosphorylation, and Lrrk2-mediated Rab phosphorylation may be higher in other parts of the brain not evaluated here that have lower total Lrrk2 levels. The activity of phosphatases acting on Rab10 and Rab12 may also play a role in determining phosphorylated Rab levels, as well as the possibility of differential LRRK2-activating proteins at the endosome where Rab phosphorylation occurs. The data herein favor the latter possibility, since low phospho-Rab expression in the Sprague Dawley background can be recovered with LRRK2 overexpression. Further, basal levels of phospho-Rabs are similar between Lrrk2 knockout (LE) strains and Long-Evans strains treated with LRRK2 inhibitors. If variable phosphatase activity were central in driving apparent differences in phosphorylated Rabs between the genetic backgrounds, we would have expected phospho-Rab levels to be inefficiently recovered with LRRK2 overexpression in the Sprague Dawley line, and we would have predicted that basal levels of phospho-Rabs would be different between the strains irrespective of Lrrk2 activity. Since neither of these suppositions appear true, our results favor the presence of differential LRRK2 activating co-factors that can vary greatly between different genetic backgrounds. Further studies are required to identify such factors.
The PFE-360 molecule utilized here was designed to have improved selectivity, potency, and bioavailability compared to PF-475 and outperforms MLi2 in potency in vivo in both rats and non-human primates at equivalent doses (Andersen et al., 2018; Kelly et al., 2018; Wang et al., 2020). PF-475 and PFE-360 have different off-target binding profiles with no known overlaps in off-targets (Henderson et al., 2015; Thirstrup et al., 2017). The main off-target of PF-475 likely to be inhibited with oral dosing is the MST class of kinases that are also suspected to phosphorylate Rab proteins, at least in vitro (Vieweg et al., 2020). Instead, we had the opposite result, where PFE-360, but not PF-475, demonstrated measurable activity in Lrrk2 knockout (LE) rats. In interpretation of phosphorylated Rab proteins as pharmacodynamic markers of LRRK2 kinase activity in the brain, it has been suggested that there may be preferential recognition of non-phosphorylated Rab10 with total Rab10 antibodies (Karayel et al., 2020). In comparing the robustness of our two measures used to evaluate phosphorylated Rab proteins in pharmacodynamic responses, either measuring the ratio of phosphorylated Rab to total Rab protein or measuring phosphorylated Rab normalized to a housekeeping protein, our data favor the latter approach as a more reliable method less susceptible to bias. The normalization of the phospho-Rab protein fraction to a house-keeping protein may provide a more accurate measure of inhibition, especially when total Rab proteins may decrease in abundance with a loss of Lrrk2 activity.
We previously found that Lrrk2 knockout (LE) rats are protected from a mild lesion (~20–30%) caused by AAV2/1-WT-αsyn vectors in Long-Evans rats (Daher et al., 2014), and that PF-475 oral administration provides robust protection from neurodegeneration caused by AAV2/1-WT-αsyn vectors in Tac(Sprague Dawley) rats (Daher et al., 2015). In utilization of a different viral vector, AAV2/1 A53T-αsyn, the resultant lesion size was nearly doubled in a shorter period of time of 4-weeks. We hypothesized LRRK2-activation in the SNpc may occur in the context of this aggressive disease model even in the context of low or no activity associated with healthy Sprague Dawley rats. However, we were unable to measure Lrrk2 activation in the Sprague Dawley rats in the context of the lesion, either through immunohistochemical analysis with phospho-Rab antibodies or immunoblot analysis of brain extracts. Further, we were unable to measure a neuroprotective effect associated with PFE-360 treatment in the context of mutant αsyn dopaminergic neurotoxicity. Given these results, we would predict that future studies that prioritize Long-Evans rats with a milder αsyn lesion caused by WT-αsyn expression may be better suited to study the effects of novel LRRK2 therapeutics as well as disease-associated phosphorylated Rab changes.
4. Experimental Procedures
4.1. Animals:
All study protocols were approved by local Institutional Review Boards and Institutional Animal Care and Use Committees. Male non-transgenic Sprague Dawley (SD) rats utilized in this study were either purchased from Charles River (CD(SD) #001) or were littermates (Tac(Sprague Dawley)) from G2019S-LRRK2 human-BAC (Tac(SD)) rats (Taconic #10681), as indicated in the figure legends and results text. Non-transgenic and Lrrk2 knockout Long-Evans (LE) rats were purchased from Envigo (formerly Sage Biosciences), with the Long-Evans rat line used by Sage originating from Charles Rivers. All rats were ~10 weeks of age at initial treatment.
4.2. Small molecules and administration:
Small molecule LRRK2 kinase inhibitors PF-475 and PFE-360 were synthesized in-house as previously described (Daher et al., 2015; Kelly et al., 2018). All compounds were >98.5% pure as assessed by LC/MS and NMR. Oral administration (p.o.) of compounds were delivered in a 0.5% methylcellulose vehicle. Tissue samples for generating protein lysates were collected 2 hours post dosing for PF-475 (3 or 30 mg per kg) and PFE-360 (20 mg per kg). CD(Sprague Dawley) rats undergoing small molecule administration for evaluating neuroprotection were evaluated four weeks post injection of AAV-A53T-αsyn.
4.3. Immunoblotting:
Forebrain rat tissue samples were dissected following transcardial perfusion with cold 1x PBS. Tissue was homogenized by probe-tip sonication in a RIPA lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton, and 0.1% SDS supplemented with 1x protease and phosphatase complete inhibitor cocktails (Roche). Homogenates were centrifuged at 10,000g for ten minutes at 4°C to precipitate any contaminating cellular debris. The supernatant was analyzed using a BCA assay (Pierce) to determine total protein concentration and was further diluted 1:1 with 2x Laemmeli sample buffer supplemented with 40 mM NaF and 10% dithiothreitol (DTT). Brain samples containing 50 μg of total protein were electrophoresed on SDS-PAGE 4–20% gradient mini-PROTEAN TGX stain-free gels (BioRad) and transferred to Immobilon-FL PVDF membrane (Millipore), followed by immunoblotting with indicated primary antibodies and secondary antibodies. The following antibodies were used in this study: N241/34 anti-LRRK2 (Antibodies Inc), phospho-S935-LRRK2 (Abcam), phospho-T73-Rab10 (MJF-R21, Abcam), Rab10 antibody (Cell Signaling), phospho-S106-Rab12 (MJF-R25–9, Abcam), Rab12 (Thermofisher), loading control HSC70 (HSPA8, Cell Signaling), Donkey anti-mouse 680-LT (LI-COR Biosciences), donkey anti-mouse 800CW (LI-COR Biosciences), and Goat anti-rabbit HRP (Jackson Immunoresearch). Western blot images were captured digitally using a Chemidoc MP Imaging System Touch (BioRad) where chemiluminescent signals were developed with Crescendo ECL reagent (Millipore) and fluorescent signals were detected using the IRdye680 and IRdye800 channels. Quantifications were performed using Image Lab 6.0.1 software (BioRad). Uncropped images for all immunoblots in the main figures are presented in Supplemental Figure 3.
4.4. Unilateral rAAV2/1 infusions into SNpc:
All surgical procedures were performed using aseptic conditions and sterile materials, solutions, and equipment. Rats were anesthetized with 5% isoflurane (in 70% N2O and 30% O2; flow 300 mL per min) and placed in a stereotactic frame. During the operation, the concentration of anesthetic was reduced to 1–1.5%. The rectal temperature was maintained at 37.0 ± 1.0°C with a homeothermic blanket system. The skin was opened by a medial incision and retracted laterally so that the right brain hemisphere could be exposed through a small craniectomy to the skull. The dura mater was carefully removed with fine forceps and a stereotaxic injection was performed with either empty AAV2/1 or AAV2/1 expressing human A53T αsyn (1.7 * 1012 viral-genomes per mL). A total of 4 μl of AAV-vector was infused at a speed of 0.4 μl per min at the following coordinates: AP −5.2, ML 2.1, DV −7.5 mm from the skull surface. The infusion cannula remained in place for an additional 5 min before being withdrawn. The skin was sutured and disinfected following the procedure. During recovery from anesthesia, the rats were carefully monitored for possible post-surgical complications and were then returned to their home cages with ad libitum access to food and water.
At time of sacrifice, rats were deeply anesthetized with isoflurane and transcardially perfused with cold PBS (pH 7.4). Brains were extracted and dissected for immunoblot or continued to perfusion with 4% PFA buffered in PBS for immunohistochemistry. Brains were post-fixed for 24 hours in 4% PFA and were then transferred to 30% sucrose PBS solution for up to three days. Brain tissue was cryopreserved in 2-methylbutane solution (−50°C) and stored at −80°C until they were sectioned at 40 mm thickness on a freezing sliding microtome (Leica).
4.5. Immunohistochemistry and Immunofluorescence:
For the immunohistochemical detection of tyrosine hydroxylase (TH, Millipore) in the SNpc, free-floating tissue sections were first quenched in 0.6% H2O2 in methanol. After copious washing, sections were then incubated in an antigen retrieval buffer (10mM Na Citrate + 0.05% Tween-20, pH 6.0) at 37°C. Brain tissue sections were permeabilized (5% goat serum, 0.3% TX-100) at room temperature for one hour, then probed with primary antibody for 48 hours followed by secondary antibody (biotinylated goat anti-rabbit IgG, Vector Laboratories) for 48 hours. An avidin-biotin complex (ABC) kit was utilized to amplify the sensitivity of detection (Vector Laboratories) before exposing the tissue sections to 3,3’-diaminobenzidine (DAB) substrate to develop the signal. Sections were then mounted onto glass coverslips, underwent standard histological processing, and was then coverslipped using Permount mounting medium (Electron Microscopy Sciences).
Immunofluorescent detection of TH in the dorsal striatum and nitrated αsyn (Syn514, BioLegend) in the SNpc was performed similarly. Free-floating tissue sections were first incubated in an antigen retrieval buffer as described above. Brain tissue sections were permeabilized in 5% donkey serum, 0.1%TX-100, then incubated in respective primary antibody for 48–72 hours followed by secondary antibody for 48–72 hours. IRDye 800CW donkey anti-rabbit IgG (LiCOR) and donkey anti-mouse IgG Alexa Fluor 647 (Thermofisher) were used for TH and Syn514 detection, respectively. Sections were mounted onto glass slides with ProLong Gold reagent (Thermofisher). TH+ fibers in the dorsal striatum were imaged on an Odyssey CLx (LI-COR Biosciences) and quantified using Image Studio Lite Ver 5.2 software. Syn514+ cells were imaged on a Leica SP5 confocal microscope.
4.6. Stereological Assessments:
Unbiased stereological counts for the total number of TH+ cells in the SNpc were estimated using an optical fractionator probe (Stereologer software, Stereology Resource Center) by an investigator blinded to group identities. Brain sections used for counting were equally spaced and cut 120 μm apart. The border of the SNpc was outlined as the counting reference space for all midbrain levels at 4x magnification. Counting frames were then generated on a sampling grid size of 200 μm covering the ROI. A minimum of three counting frames were used for analysis. Cell counts were performed at 60x magnification, beginning with the first randomly assigned frame and systematically progressed through remaining counting frames until the entire space had been assessed.
4.7. Statistics:
Statistical analyses were conducted using GraphPad Prism 8 software. Specific statistical tests for datasets are outlined in figure descriptions. All tests were performed using significance level of α=0.05 with 95% confidence to determine significant group mean differences (*p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.0001). Data are presented as mean values with standard error of the mean (SEM).
Supplementary Material
Highlights.
Phospho-Rab10 and Rab12 levels in Sprague Dawley resemble Lrrk2 knockout
Phospho-Rab10 and Rab12 levels are elevated in Long-Evans, like G2019S-BAC rats
LRRK2 kinase inhibitors reliably reduce phospho-Rab10 and Rab12 in Long-Evans rats
Potent LRRK2 kinase inhibition does not protect dopamine neurons from A53T-αsyn overexpression
Acknowledgements
The authors thank Charles River Discovery Research Services, Finland, for assistance in injecting rAAV viruses and dosing rats with LRRK2 kinase inhibitors.
Funding
This work was supported by NINDS grants R01NS064934 and P50NS108675 to A.B.W.
Abbreviations:
- αsyn
αsynuclein
- CD(Sprague Dawley)
Charles River Sprague Dawley
- G2019S-LRRK2 BAC (Tac(SD))
Bacterial artificial overexpression of human G2019S-LRRK2 on the Taconic Sprague Dawley strain background
- KO
Lrrk2 knockout on the Long-Evans strain background
- Lrrk2 (rodent)/LRRK2 (human)
Leucine-rich repeat kinase 2 protein
- LE
Long-Evans
- PD
Parkinson’s disease
- SD
Sprague Dawley
- Tac(Sprague Dawley)
Taconic Sprague Dawley
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alessi DR, Sammler E, 2018. LRRK2 kinase in Parkinson’s disease. Science 360, 36–37. [DOI] [PubMed] [Google Scholar]
- Andersen MA, Wegener KM, Larsen S, Badolo L, Smith GP, Jeggo R, Jensen PH, Sotty F, Christensen KV, Thougaard A, 2018. PFE-360-induced LRRK2 inhibition induces reversible, non-adverse renal changes in rats. Toxicology 395, 15–22. [DOI] [PubMed] [Google Scholar]
- Atashrazm F, Hammond D, Perera G, Bolliger MF, Matar E, Halliday GM, Schüle B, Lewis SJG, Nichols RJ, Dzamko N, 2019. LRRK2-mediated Rab10 phosphorylation in immune cells from Parkinson’s disease patients. Mov. Disord 34, 406–415. [DOI] [PubMed] [Google Scholar]
- Baptista MAS, Dave KD, Sheth NP, De Silva SN, Carlson KM, Aziz YN, Fiske BK, Sherer TB, Frasier MA, 2013. A strategy for the generation, characterization and distribution of animal models by The Michael J. Fox Foundation for Parkinson’s Research. Dis. Model. Mech 6, 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista MAS, Merchant K, Barrett T, Bhargava S, Bryce DK, Ellis JM, Estrada AA, Fell MJ, Fiske BK, Fuji RN, Galatsis P, Henry AG, Hill S, Hirst W, Houle C, Kennedy ME, Liu X, Maddess ML, Markgraf C, Mei H, Meier WA, Needle E, Ploch S, Royer C, Rudolph K, Sharma AK, Stepan A, Steyn S, Trost C, Yin Z, Yu H, Wang X, Sherer TB, 2020. LRRK2 inhibitors induce reversible changes in nonhuman primate lungs without measurable pulmonary deficits. Sci. Transl. Med 12. 10.1126/scitranslmed.aav0820 [DOI] [PubMed] [Google Scholar]
- Daher JPL, Abdelmotilib HA, Hu X, Volpicelli-Daley LA, Moehle MS, Fraser KB, Needle E, Chen Y, Steyn SJ, Galatsis P, Hirst WD, West AB, 2015. Leucine-rich Repeat Kinase 2 (LRRK2) Pharmacological Inhibition Abates α-Synuclein Gene-induced Neurodegeneration. J. Biol. Chem 290, 19433–19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher JPL, Volpicelli-Daley LA, Blackburn JP, Moehle MS, West AB, 2014. Abrogation of α-synuclein–mediated dopaminergic neurodegeneration in LRRK2-deficient rats. Proc. Natl. Acad. Sci. U. S. A 111, 9289–9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P, Hinkle KM, Sukar NN, Sepulveda B, Mesias R, Serrano G, Alessi DR, Beach TG, Benson DL, White CL, Cowell RM, Das SS, West AB, Melrose HL, 2013. Comprehensive characterization and optimization of anti-LRRK2 (leucine-rich repeat kinase 2) monoclonal antibodies. Biochem. J 453, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ, 2010. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J 430, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JL, Kormos BL, Hayward MM, Coffman KJ, Jasti J, Kurumbail RG, Wager TT, Verhoest PR, Noell GS, Chen Y, Others, 2015. Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7 H-pyrrolo [2, 3-d] pyrimidin-5-yl] benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor. J. Med. Chem 58, 419–432. [DOI] [PubMed] [Google Scholar]
- Ip CW, Klaus L-C, Karikari AA, Visanji NP, Brotchie JM, Lang AE, Volkmann J, Koprich JB, 2017. AAV1/2-induced overexpression of A53T-α-synuclein in the substantia nigra results in degeneration of the nigrostriatal system with Lewy-like pathology and motor impairment: a new mouse model for Parkinson’s disease. Acta neuropathologica communications 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito G, Katsemonova K, Tonelli F, Lis P, Baptista MAS, Shpiro N, Duddy G, Wilson S, Ho PW-L, Ho S-L, Reith AD, Alessi DR, 2016. Phos-tag analysis of Rab10 phosphorylation by LRRK2: a powerful assay for assessing kinase function and inhibitors. Biochem. J 473, 2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayel Ö, Tonelli F, Virreira Winter S, Geyer PE, Fan Y, Sammler EM, Alessi DR, Steger M, Mann M, 2020. Accurate MS-based Rab10 Phosphorylation Stoichiometry Determination as Readout for LRRK2 Activity in Parkinson’s Disease. Mol. Cell. Proteomics 19, 1546–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K, Wang S, Boddu R, Liu Z, Moukha-Chafiq O, Augelli-Szafran C, West AB, 2018. The G2019S mutation in LRRK2 imparts resiliency to kinase inhibition. Exp. Neurol 309, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K, West AB, 2020. Pharmacodynamic Biomarkers for Emerging LRRK2 Therapeutics. Front. Neurosci 14, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Burger C, Lundberg C, Johansen TE, Muzyczka N, Mandel RJ, Björklund A, 2002. Parkinson-Like Neurodegeneration Induced by Targeted Overexpression of α-Synuclein in the Nigrostriatal System. The Journal of Neuroscience. 10.1523/jneurosci.22-07-02780.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluss JH, Conti MM, Kaganovich A, Beilina A, Melrose HL, Cookson MR, Mamais A, 2018. Detection of endogenous S1292 LRRK2 autophosphorylation in mouse tissue as a readout for kinase activity. NPJ Parkinsons Dis 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-W, Tapias V, Di Maio R, Timothy Greenamyre J, Cannon JR, 2015. Behavioral, neurochemical, and pathologic alterations in bacterial artificial chromosome transgenic G2019S leucine-rich repeated kinase 2 rats. Neurobiology of Aging. 10.1016/j.neurobiolaging.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis P, Burel S, Steger M, Mann M, Brown F, Diez F, Tonelli F, Holton JL, Ho PW, Ho S-L, Chou M-Y, Polinski NK, Martinez TN, Davies P, Alessi DR, 2018. Development of phospho-specific Rab protein antibodies to monitor in vivo activity of the LRRK2 Parkinson’s disease kinase. Biochem. J 475, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bryant N, Kumaran R, Beilina A, Abeliovich A, Cookson MR, West AB, 2018. LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet 27, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Fiske BK, Baptista MAS, 2020. The Michael J. Fox Foundation’s Strategies for Accelerating Translation of LRRK2 into Therapies for Parkinson Disease. Cells 9. 10.3390/cells9081878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout HJ, Chartier-Harlin M-C, Fell MJ, Hirst WD, Huntwork-Rodriguez S, Leyns CEG, Mabrouk OS, Taymans J-M, 2020. The Current State-of-the Art of LRRK2-Based Biomarker Assay Development in Parkinson’s Disease. Frontiers in Neuroscience. 10.3389/fnins.2020.00865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Zhang S, Bustos D, Kleinheinz T, Le Pichon CE, Dominguez SL, Solanoy HO, Drummond J, Zhang X, Ding X, Cai F, Song Q, Li X, Yue Z, van der Brug MP, Burdick DJ, Gunzner-Toste J, Chen H, Liu X, Estrada AA, Sweeney ZK, Scearce-Levie K, Moffat JG, Kirkpatrick DS, Zhu H, 2012. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med 4, 164ra161. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K, 2003. alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841. [DOI] [PubMed] [Google Scholar]
- Sloan M, Alegre-Abarrategui J, Potgieter D, Kaufmann A-K, Exley R, Deltheil T, Threlfell S, Connor-Robson N, Brimblecombe K, Wallings R, Cioroch M, Bannerman DM, Bolam JP, Magill PJ, Cragg SJ, Dodson PD, Wade-Martins R, 2016. LRRK2 BAC transgenic rats develop progressive, L-DOPA-responsive motor impairment, and deficits in dopamine circuit function. Hum. Mol. Genet 25, 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, Karayel O, Tonelli F, Martinez TN, Lorentzen E, Pfeffer SR, Alessi DR, Mann M, 2017. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. Elife 6. 10.7554/eLife.31012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M, Tonelli F, Ito G, Davies P, Trost M, Vetter M, Wachter S, Lorentzen E, Duddy G, Wilson S, Baptista MAS, Fiske BK, Fell MJ, Morrow JA, Reith AD, Alessi DR, Mann M, 2016. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife. 10.7554/elife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirstrup K, Dächsel JC, Oppermann FS, Williamson DS, Smith GP, Fog K, Christensen KV, 2017. Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci. Rep 7, 10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh J, Guella I, Farrer MJ, 2014. Disease penetrance of late-onset parkinsonism: a meta-analysis. JAMA Neurol. 71, 1535–1539. [DOI] [PubMed] [Google Scholar]
- Vieweg S, Mulholland K, Bräuning B, Kachariya N, Lai Y-C, Toth R, Singh PK, Volpi I, Sattler M, Groll M, Others, 2020. PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72. Biochem. J 477, 1651–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kelly K, Brotchie JM, Koprich JB, West AB, 2020. Exosome markers of LRRK2 kinase inhibition. npj Parkinson’s Disease 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, 2017. Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson disease. Experimental Neurology. 10.1016/j.expneurol.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Cowell RM, Daher JPL, Moehle MS, Hinkle KM, Melrose HL, Standaert DG, Volpicelli-Daley LA, 2014. Differential LRRK2 expression in the cortex, striatum, and substantia nigra in transgenic and nontransgenic rodents. J. Comp. Neurol 522, 2465–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


