Abstract
Background
The current gold standard in coronavirus disease (COVID-19) diagnostics is the real-time reverse transcription–polymerase chain reaction (RT-PCR) assay for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in nasopharyngeal swab (NPS) samples. Alternatively, nasal swab (NS) or saliva swab (SS) specimens are used, although available data on test accuracy are limited. We examined the diagnostic accuracy of NPS/NS/SS samples for this purpose.
Methods
Ten patients were included after being tested positive for SARS-CoV-2 RT-PCR in NPS samples according to the National Institute of Infectious Disease guidelines. In comparison with this conventional diagnostic method, NPS/NS/SS samples were tested using the cobas 6800 systems RT-PCR device. To investigate the usefulness of the cobas method and the difference among sample types, the agreement and sensitivity were calculated. Five to six samples were collected over a total period of 5–6 d from each patient.
Results
Fifty-seven sets of NPS/NS/SS samples were collected, of which 40 tested positive for COVID-19 by the conventional method. Overall, the concordance rates using the conventional method were 86.0%/70.2%/54.4% for NPS/NS/SS samples (cobas); however, for samples collected up to and including on Day 9 after disease onset (22 negative and one positive specimens), the corresponding rates were 95.7%/87.0%/65.2%. The overall sensitivity estimates were 100.0%/67.5%/37.5% for NPS/NS/SS samples (cobas). For samples up to 9 d after onset, the corresponding values were 100.0%/86.4%/63.6%.
Conclusions
NS samples are more reliable than SS samples and can be an alternative to NPS samples. They can be a useful diagnostic method in the future.
Keywords: COVID-19 diagnostic test, nasopharyngeal swab, nasal swab, saliva
Introduction
Since December 2019, the coronavirus disease (COVID-19) has spread worldwide, and the number of associated deaths has been increasing gradually. The pathogen has been identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has a phylogenetic similarity to SARS-CoV [1]. The World Health Organisation (WHO) declared the outbreak of COVID-19 a public health emergency of international concern on 30 January 2020 [2]. As a result of these developments, there is a need for accurate and rapid diagnostic tests to aid infection control and prevention.
Kits for assay of SARS-CoV-2 RNA are commercially available. The current gold standard for COVID-19 confirmation is the real-time reverse transcription–polymerase chain reaction (RT-PCR) assay for the detection of SARS-CoV-2 from nasopharyngeal swab (NPS) samples. The WHO recommended NPS samples for detection of SARS-CoV-2 [3,4]. However, acquiring these samples is associated with significant patient discomfort and a high risk of infection transmission to healthcare staff; thus, testing methods using specimens that are easier to obtain, such as nostril/nasal wipes and saliva samples (including self-collection) are required [5,6].
The Centres for Disease Control and Prevention and the Infectious Diseases Society of America have recommended the testing of NS and SS samples for SARS-CoV-2 using RT-PCR [7,8]. RT-PCR assays using nasal swab (NS) and saliva swab (SS) have also been approved in Japan, following guidelines similar to those implemented overseas [9]; however, their diagnostic sensitivity is still debateable. Prior studies have found that NS and SS samples provide similar sensitivities to NPS [10,11]; however, it has not been demonstrated whether they are useful in clinical practice.
Providing evidence demonstrating that the rate of RNA virus detection using NS and SS samples is comparable to that using NPS samples may help resolve problems associated with the shortage of personal protective equipment available for sample collection, while helping increase the number of COVID-19 tests performed. To compare detection rates between samples, NPS, NS and SS samples were collected from patients diagnosed with COVID-19 and assayed using RT-PCR. Consecutive samples were also collected from the same patients to ascertain time-dependent changes in the SARS-CoV-2 viral load and to investigate their effect on the reliability of the test results obtained based on NPS, NS and SS samples.
Materials and methods
This was a single-centre, prospective study of patients diagnosed with COVID-19 and admitted to the National Centre for Global Health and Medicine (Tokyo, Japan) between July 18 and 3 August 2020. A ‘positive’ case of COVID-19 was defined as a positive RT-PCR test on NPS sample following the National Institute of Infectious disease guidelines (NIID). NPS, NS and SS samples were collected at the same time from patients over a total of 5–6 times at prescribed time intervals. The first three samples were collected every other day starting from the day after admission, and the fifth one was collected on the day of discharge; if the fifth sample was negative, a fifth one was collected the next day; otherwise, the samples were collected at intervals of more than 5 d. Similarly, if the fifth sample was negative, a sixth sample was taken the next day; otherwise, the samples were taken at intervals of 5 d or more. In case of non-working days, the schedule was shifted to the next working day.
Patients who had developed COVID-19 more than 8 d prior to enrolment and those aged less than 20 years were excluded. The date of onset was defined as the date of appearance of symptoms. Data on patient characteristics, symptoms at onset and during hospitalization, and the National Institutes of Health severity classification were collected [12]. Cardinal symptoms included fever (37.5 °C or higher), cough, sputum, haemoptysis, sore throat, nasal discharge, wheezing, dyspnoea, chest pain, muscle pain, joint pain, headache, fatigue, stomach-ache, nausea/vomiting and diarrhoea. In addition, patients were asked whether they had nasal discharge and the loss of the sense of taste or smell at each sample collection point. Finally, patients were asked whether they had eaten within 2 h or used any antibacterial toothpaste or mouthwash within 6 h prior to sample collection to investigate the impact of these factors on SS sample stability.
This study was approved by the National Centre for Global Health and Medicine Institutional Review Board (approval number: NCGM-G-003629-00). This study was conducted in compliance with the Declaration of Helsinki (2013 revised Forta Reza) and the Ethical Guidelines for Medical Research Involving Human Subjects (partially revised on 28 February 2017). All patients included in the study were required to provide informed consent themselves.
Sample collection
NPS samples were collected using Copan FLOQSwabs and a sterile tube containing Copan’s Universal Transport Medium (UTM, COPAN Diagnostics Inc., Murrieta, CA). NS samples were collected using dry swabs from the Cobas PCR media (CPM) kit (Roche Molecular Systems, South Branchburg, NJ) and a sterile tube containing UTM. The swab was inserted approximately 1–2 cm into each nostril and rotated for 5 s on each side to collect the sample. SS samples were collected by placing both a Copan FLOQSwab and a dry swab from the CPM kit under the tongue for 5 s, after which the Copan FLOQSwab was placed in a sterile tube containing UTM, and the dry swab was placed in a sterile tube containing CPM. To avoid contamination with respiratory specimens, saliva samples were collected with the swabs held beneath the tongue while the patient was instructed to refrain from coughing. The SS sample was collected first by the patient himself or herself (without physician supervision, unless the patient was intubated or otherwise incapable of collecting their own sample); a physician then collected the NS specimen, followed by the NPS specimen. Specimens were collected 5–6 times from each patient on different days, with the first three sets of specimens collected within 2 weeks of the disease onset.
Specimen processing
All the specimens were tested on their respective collection days, using cobas® SARS-CoV-2 on the cobas 6800 Systems RT-PCR device (Roche Molecular Systems, South Branchburg, NJ). The UTM containing NPS was stored at a temperature below −70 °C until being tested by SARS-CoV-2 RT-PCR according to the National Institute of Infectious Diseases guidelines (NIID) [13] (see Supplemental Table) as the conventional method. Using the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) or QuantStudio® 5 (Applied Biosystems), the test result is deemed to be positive when a rise in the amplification curve is observed by cycle 40 in either or both of the wells of the N2 set of specimens. Technicians who performed specimen processing and RT-PCR testing were unaware of participants’ names and hospital numbers. In the following sections, RT-PCR results of the NPS sample, NS sample and SS sample using cobas® are referred to as NPS sample (cobas), NS sample (cobas) and SS sample (cobas), respectively. The RT-PCR results of NPS sample using N and N2 primers and TaqMan assay are referred to as NPS sample (NIID).
Statistical analysis
Concordance rates were defined as the percentage of agreement of the RT-PCR results. By comparing the NPS sample (NIID) with the NPS/NS/SS sample (cobas) and by directly comparing the NPS sample (cobas) with the NS/SS sample (cobas), the cobas method was found to be non-inferior to the conventional method, and the diagnostic usefulness of each sample was evaluated. Sensitivity, specificity and concordance rates for the RT-PCR results, and their 95% confidence intervals (CIs), were calculated using Cohen’s kappa. Gwet’s AC1 statistic (AC1) was also calculated, as it is considered more robust than Cohen’s kappa, which may vary with prevalence estimates [14,15].
To assess whether the NS and SS samples reflected the viral load obtained using NPS samples (NIID), cycle threshold (Ct) values and copy numbers were estimated and reported as the median value and an interquartile range and compared by the Wilcoxon rank-sum test, as these variables followed non-normal distributions. Univariate analyses were performed using Fisher’s exact test or Chi-square test. These statistical analyses were performed using JMP® 14.2 software (SAS Institute Inc., Cary, NC, USA). p Values of <.05 were considered statistically significant. In order to evaluate the time-dependent change in the Ct value for each sample, the Jonckheere–Terpstra test was performed with EZR [16], which is a modified version of R Commander designed to include statistical functions frequently used in biostatistics.
Results
This study included ten hospitalized patients diagnosed with COVID-19. All the patients were Japanese. Their median age was 47 years (range, 30–70 years). Eight were men, and two were women (one of whom was pregnant). All the COVID-19 patients were admitted to our hospital following a diagnosis using NPS specimens. Based on the National Institutes of Health severity classification, six and two patients had mild and moderate diseases, respectively, and one patient each had severe and critical diseases [17]. The patient with the severe disease was treated with high-flow nasal oxygen therapy, while the patient with the critical disease was placed on a ventilator. Patients’ characteristics are presented in Table 1.
Table 1.
Clinical characteristics of hospitalized patients with COVID-19 disease (n = 10).
| Age, years, median (range) | 47 (30–70) | |
|---|---|---|
| Sex (n) | ||
| Female | 8 | |
| Male | 2 | |
| Presenting symptoms | Day of symptom onset | During hospitalization |
| Fever (≥37.5 °C) | 6 | 5 |
| Cough | 6 | 8 |
| Sputum | 3 | 6 |
| Haemoptysis | 0 | 0 |
| Sore throat | 1 | 3 |
| Nasal mucus | 0 | 1 |
| Wheezing | 0 | 1 |
| Dyspnoea | 3 | 5 |
| Chest pain | 1 | 1 |
| Muscle pain | 3 | 0 |
| Joint pain | 4 | 2 |
| Headache | 2 | 4 |
| Fatigue | 6 | 7 |
| Stomach-ache | 0 | 0 |
| Vomiting/nausea | 1 | 1 |
| Diarrhoea | 2 | 2 |
| Past medical history | ||
| None | 3 | |
| Hypertension | 2 | |
| Hepatitis C | 2 | |
| Diabetes | 1 | |
| Asthma | 1 | |
| HIV infection | 1 | |
| Severity | ||
| Mild | 6 | |
| Moderate | 2 | |
| Severe | 1 | |
| Critical | 1 | |
| Antiviral agents | ||
| Used | 3 | |
| Not used | 7 | |
| Systemic steroids | ||
| Used | 4 | |
| Not used | 6 | |
Concordance rate, sensitivity and specificity
A total of 57 sets of NPS, NS, and SS samples were collected. A total of 40 specimens were positive in NPS samples (NIID), and a total of 48 specimens were positive for NPS samples (cobas). The concordance rate, sensitivity and specificity for each sample compared to NPS samples (NIID) are shown in Table 2 and those compared to NPS samples (cobas) are shown in Table 3. Concordance rates for all specimens (40 positive and 17 negative in NPS samples [NIID]) were 86.0% (κ 0.612, AC1 0.783), 70.2% (κ 0.382, AC1 0.437) and 54.4% (κ 0.285, AC1 0.158) for NPS, NS and SS samples (cobas), respectively. In the assessment of the specimens collected up to and including Day 9 after disease onset (22 positive and one negative in NPS samples [NIID]), the concordance rates for NPS, NS and SS samples (cobas) were 95.7% (κ 0.0, AC1 0.955), 87.0% (κ 0.355, AC1 0.838) and 65.2% (κ 0.132, AC1 0.473), respectively.
Table 2.
Sensitivity, specificity and concordance rate (κ coefficient, Gwet’s AC1) for the NPS/NS/SS samples (cobas) compared with NPS samples (NIID) PCR.
| Day of specimen collection | Reference NPS (TaqMan) |
Sensitivity % (95% CI) | Specificity % (95% CI) | Cohen’s kappa (95% CI) | Gwet’s AC1 statistic | |||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||
| Overall | NPS | Positive | 40 | 8 | 100 (91.2–100.0) | 52.9 (31.0–73.8) | 0.612 (0.383-0.842) | 0.783 |
| Negative | 0 | 9 | ||||||
| NS | Positive | 27 | 4 | 67.5 (52.0–79.9) | 76.5 (52.7–90.4) | 0.382 (0.151–0.612) | 0.437 | |
| Negative | 13 | 13 | ||||||
| SS | Positive | 15 | 1 | 37.5 (24.2–53.0) | 94.1 (73.0–99.0) | 0.225 (0.067–0.383) | 0.088 | |
| Negative | 25 | 16 | ||||||
| Within 9 d after onset | NPS | Positive | 22 | 1 | 100.0 (85.1–100.0) | NA | 0.0 (0.0–0.0) | 0.955 |
| Negative | 0 | 0 | ||||||
| NS | Positive | 19 | 0 | 86.4 (66.7–95.3) | NA | 0.355 (−0.165 to 0.875) | 0.838 | |
| Negative | 3 | 1 | ||||||
| SS | Positive | 14 | 0 | 63.6 (43.0–80.3) | NA | 0.132 (−0.109 to 0.373) | 0.473 | |
| Negative | 8 | 1 | ||||||
| First collected specimens | NPS | Positive | 10 | 0 | 100.0 (72.2–100.0) | NA | NA | 1.0 |
| Negative | 0 | 0 | ||||||
| NS | Positive | 10 | 0 | 100.0 (72.2–100.0) | NA | NA | 1.0 | |
| Negative | 0 | 0 | ||||||
| SS | Positive | 6 | 0 | 60.0 (31.3–83.2) | NA | NA | 0.412 | |
| Negative | 4 | 0 | ||||||
NPS: nasopharyngeal swab; NS: nasal swab; SS: saliva specimen; PCR: polymerase chain reaction; NIID: National Institute of Infectious Diseases guidelines.
Table 3.
Sensitivity, specificity and concordance rate (κ coefficient, Gwet’s AC1) for the NS/SS samples (cobas) compared with NPS samples (cobas).
| Day of specimen collection | Reference NPS (cobas) |
Sensitivity % (95% CI) | Specificity % (95% CI) | Cohen’s kappa (95% CI) | Gwet’s AC1 statistic | |||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||
| Overall | NS | Positive | 30 | 1 | 62.5 (48.4–74.8) | 88.9 (56.5–98.0) | 0.291 (0.091–0.490) | 0.420 |
| Negative | 18 | 8 | ||||||
| SS | Positive | 16 | 0 | 33.3 (21.7–47.5) | 100.0 (70.1–100.0) | 0.136 (0.036–0.236) | −0.106 | |
| Negative | 32 | 9 | ||||||
| Within 9 d after onset | NS | Positive | 19 | 0 | 82.6 (62.9–93.0) | NA | NA | 0.793 |
| Negative | 4 | 0 | ||||||
| SS | Positive | 14 | 0 | 60.9 (40.8–77.8) | NA | NA | 0.429 | |
| Negative | 9 | 0 | ||||||
| First collected specimens | NS | Positive | 10 | 0 | 100.0 (72.2–100.0) | NA | NA | 1.0 |
| Negative | 0 | 0 | ||||||
| SS | Positive | 6 | 0 | 60.0 (31.3–83.2) | NA | NA | 0.412 | |
| Negative | 4 | 0 | ||||||
NPS: nasopharyngeal swab; NS: nasal swab; SS: saliva specimen.
As only one specimen tested negative, we were able to calculate sensitivity estimates only, which were 100.0, 86.4, and 63.6% for NPS, NS, and SS samples (cobas), respectively. The first ten specimens collected (median 6.5 d, range 3–9 d after onset) were all positive; the concordance rates and specificity estimates were not calculated, while the sensitivity estimates were 100.0, 100.0, and 60.0% for NPS, NS, and SS samples, respectively.
In the comparison between the same RT-PCR methods, concordance rates for all specimens (48 positive and nine negative in NPS samples [cobas]) were 66.7% (κ 0.291, AC1 0.420) and 43.9% (κ 0.136, AC1 −0.106) for NS and SS samples (cobas), respectively. The concordance rate was 82.6% (AC1 0.793) and 60.9% (AC1 0.429) for NS and SS samples (cobas), respectively, up to Day 9 of onset. Since the comparisons were made using the same PCR tests, the difference was purely by specimen type, indicating that the NS specimen had higher reliability than the SS specimen.
Ct values for specimens with consistent results
For the 40 NPS specimens that tested positive by the TaqMan assay, the results of the corresponding NS and SS samples tended to be more consistent when the number of copies was higher and the Ct value was lower (Figure 1).
Figure 1.
Correlation between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cycle threshold (Ct) value and reverse transcription–polymerase chain reaction (RT-PCR) results of each sample. We examined the differences between the Ct value and copy number of nasopharyngeal swab (NPS) and whether the NPS and the corresponding nasal swab (NS)/saliva specimen (SS) results were consistent. Comparisons were performed with the Wilcoxon rank-sum test.
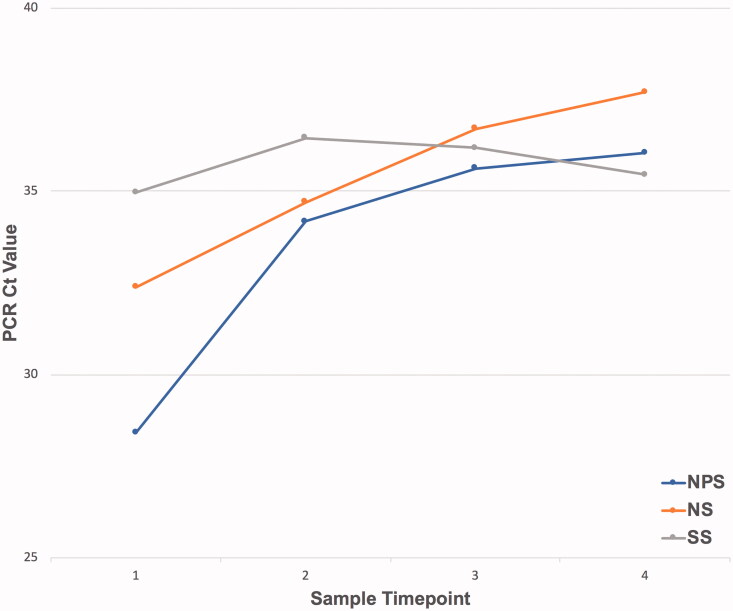
The median Ct values of NPS samples (NIID) paired with positive NPS/NS/SS samples (cobas), respectively, were calculated to be 33.65 (23.16–38.54) for NPS, 34.12 (22.79–38.78) for NS and 35.34 (29.89–38.01) for SS, with the Ct value increasing in order from NPS through NS to SS samples (cobas). In addition, among positive-testing specimens obtained from NPS and NS samples (cobas), the corresponding Ct values increased over time (Figure 2). A significant time-dependent increase in the Ct values of NPS and NS samples was observed, but no such significant increase was evident for SS samples (NPS, p < .001; NS, p = .003; SS, p = .585, Jonckheere–Terpstra test).
Figure 2.
Changes to cycle threshold (Ct) values of nasopharyngeal swab/nasal swab/saliva specimen (NPS/NS/SS) samples since onset, shown in the graph of the median Ct value per days classification. The sample timepoint was classified as 1 within 7 d, 2 within 8–1 d, 3 within 12–16 d and 4 after 17 d. NPS, p<.001; NS, p=.003; SS, p=.585.
Concordance rates under different conditions
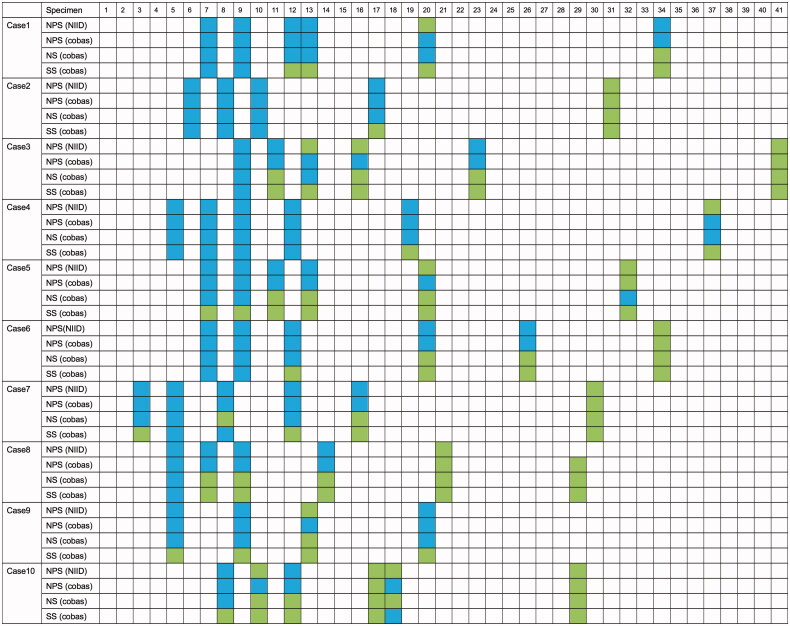
In univariate analysis, there was no difference in concordance rates between NPS, NS, or SS samples (cobas), regardless of the accompanying symptoms. The timing of SS collection relative to eating or using a mouthwash (within 2 and 6 h, respectively) did not affect the associated concordance rates. Figure 3 presents results per patient and per type of specimen, stratified by time since disease onset.
Figure 3.
RT-PCR results from 10 patients, according to the number of days since symptom onset. NPS (NIID): nasopharyngeal swab tested by SARS-CoV-2 RT-PCR according to the National Institute of Infectious Diseases guidelines as reference. NPS: nasopharyngeal swab; NS: nasal swab; SS: saliva specimen; RT-PCR: reverse transcription–polymerase chain reaction.
Discussion
In our study of paired NPS/NS/SS samples from COVID-19 patients, high positive concordance was found between NPS samples (cobas) and NPS samples (NIID) and between NS samples (cobas) and NPS samples (cobas) within 9 d. For most specimens with inconsistent results, an initial rise in the growth curve was seen, but the Ct value was ≥40, resulting in a negative assessment, indicating that the virus was detected below the cut-off value. This finding suggests that RT-PCR using a Cobas 6800 System is a more sensitive test than the conventional NIID guideline-based method. As specimens with high Ct values may be obtained from pre-symptomatic or asymptomatic patients [18], high sensitivity of RT-PCR may help identify such patients at an early stage. Nevertheless, it should be noted that even if the Cobas 6800 Systems RT-PCR device detects a positive result, NPS samples collected at late disease stages may only capture the non-infectious state with viable viral shedding.
Positive RT-PCR findings obtained from patients tested more than 10 d after disease onset, who are less likely to infect others, have been reported [19,20]. In cases with a positive test in this context, isolation and treatment may not be required. These findings suggest the importance of interpreting RT-PCR results of NPS samples in the broader context of patients’ clinical characteristics, including disease symptoms and the number of days since disease onset. In addition, the overall specificity estimates for NS and SS samples (cobas) were high, but their concordance rates and sensitivity estimates were low.
However, as diagnostic specimen collection typically takes place within 9 d after disease onset, we compared specimens collected within this time period, showing that the concordance rate and sensitivity estimates were higher for NS samples (cobas) than for SS samples (cobas). Only one NPS sample (NIID) collected within 9 d after symptom onset tested negative; when the apparent prevalence is high, the reliability of the κ coefficient is low.
The result consistency was, therefore, compared using the AC1 statistic, which was 0.838 for NS samples collected within 9 d after disease onset, indicating high consistency. The concordance rate and sensitivity between NPS samples (cobas) and NS/SS samples (cobas) were also similar, with NS samples (cobas) being higher than SS samples (cobas) for specimens collected within 9 d of onset. Moreover, for the first set of specimens collected (ten specimens), the sensitivity of NS samples was 100%, highlighting its value as an initial test for symptomatic patients.
Previous studies that have directly compared the diagnostic utility of NPS and NS samples have estimated the sensitivity of RT-PCR findings using NS specimens at 82.5–95.8% in the initial test performed for patients with suspected SARS-CoV-2 infection [10,21,22]; the corresponding values in this study were higher than those previously reported. This discrepancy may have been due to between-study differences in the patient populations, the type of PCR assays, or the collection media used.
Additionally, in this study, the higher the copy number of NPS samples and the lower the Ct value, the more positive the paired NS and SS samples tended to be. The decrease in the viral load of both NPS and NS samples was correlated with the time elapsed since the onset of illness. These results suggest that the viral load of NS samples is likely to reflect the viral load of NPS samples. Pinninti et al. investigated the viral load in NPS and NS sample pairs and observed that particularly when the Ct value was ≤30 for an NPS sample, the corresponding NS-based finding was almost always positive; meanwhile, the concordance rate for NS samples decreased with an increase in the number of days since onset [23], a result similar to that of the present study. These facts support the idea that NS specimens are a suitable substitute for NPS specimens, especially in the early stages of the disease.
NS-based tests are not currently recommended for use with asymptomatic patients [9]. However, in this study, there was no difference in the concordance rates of NPS- and NS-based test results, regardless of symptom presence. The similarity of concordance rates between NPS- and NS-based tests regardless of the presence of nasal discharge may indicate that these tests detect viral load in the mucosa rather than in nasal mucus. Previous studies reported that NSs in asymptomatic and mildly-symptomatic patients showed similar sensitivity to NPSs [24], and that nasopharyngeal and NSs were equally sensitive regardless of symptom presence [25]. Although no specimens were collected from any asymptomatic patients in this study, the result of this study and previous reports suggest that NS samples may be useful in the assessment of both asymptomatic and symptomatic patients. Overall, their use for screening asymptomatic patients should be considered, in part because they cause less discomfort to patients than NPS samples [26].
Although many studies have reported the sensitivity of SS samples in the range of 85–100%, which makes their validity equivalent to that of NPS samples [6,27,28], in this study, their concordance rate and sensitivity estimates were both low. This discrepancy may be due to between-study differences in testing methods, including sample collection and dilution. The SS samples used in this study constituted sublingual swabs; tests based on larger amounts of saliva might have yielded different results.
In addition, previous studies have shown that saliva collected after coughing is a reliable specimen [29], and since the patients in this study were instructed to refrain from coughing to minimize the risk of contamination with respiratory specimens, this approach may have affected the results. There was also a report of low sensitivity (66%) with self-collected saliva specimens in the past [25], suggesting that the method of instruction when collecting saliva specimens is important. Currently, evidence for testing in saliva specimens is accumulating [11], but nasal specimens were more reliable in this study. Further studies are required to ascertain the optimum testing method for RT-PCR using SS samples, such as assays and sample collection.
This study had some limitations. The sample size of this study was small and only symptomatic patients were enrolled. It is unclear whether the testing of asymptomatic or pre-symptomatic patients (such as those in quarantine and undergoing preoperative tests) would have yielded similar results. In addition, in this study, saliva was collected by patients themselves, whereas NS samples were collected by physicians; the impact of self-collection on sensitivity and specificity estimates is unknown, but the results would have been more real-world based. Moreover, NS sample collection is a simple procedure easily taught to patients and tests based on self-collected samples are considered highly reliable [5,30]. Self-collection may also reduce the risk of spreading infection to healthcare workers and should be investigated.
In conclusion, RT-PCR results based on NS samples collected from patients within 9 d since the onset of symptoms showed a high concordance rate with those based on NPS samples, indicating that NS samples may provide an alternative to NPS samples. Furthermore, these results also showed greater reliability than did those associated with SS samples. Particular attention should be paid to the low sensitivity of self-collected SS specimens. The use of NS sampling as an alternative to NPS sampling may reduce the physical burden for patients and the transmission risk among medical professionals, and their positive use is desirable.
Supplementary Material
Acknowledgements
We thank all patients who participated in this study. We also thank Mrs. Shimada for her excellent technical assistance. Finally, we thank Mr. Kubota for managing data.
Funding Statement
Y.T. has received research grants from Roche Diagnostic Inc. for the submitted work. Funding for this study was provided by Roche Diagnostics K.K. The study number CTRP no. 20-06-22.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Guan W, Ni Z, Hu Y, et al. . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Statement on the third meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of coronavirus disease (COVID-19) [Internet]; 2021. [cited 2021 Jan 14]. Available from: https://www.who.int/news-room/detail/01-05-2020-statement-on-the-third-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-coronavirus-disease-(covid-19).
- 3.World Health Organization . Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Geneva, Switzerland: World Health Organization; 2020. p. 1–7. [Google Scholar]
- 4.Zou L, Ruan F, Huang M, et al. . SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wehrhahn MC, Robson J, Brown S, et al. . Self-collection: an appropriate alternative during the SARS-CoV-2 pandemic. J Clin Virol. 2020;128:104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KKW, Tsang OTY, Leung WS, et al. . Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19). Atlanta, Georgia: Centers for Disease Control and Prevention; 2019. [Google Scholar]
- 8.Hanson KE, Caliendo AM, Arias CA, et al. . Infectious Diseases Society of America guidelines on the diagnosis of coronavirus disease 2019. Clin Infect Dis. 2020;ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Japanese Ministry of Health L and W . Guidelines for the testing of novel coronavirus pathogens (covid-19) (Version 1). Available from: https://www.mhlw.go.jp/content/000678571.pdf.
- 10.Tu YP, Jennings R, Hart B, et al. . Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020;383(5):494–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler-Laporte G, Lawandi A, Schiller I, et al. . Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. 2021;181(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization COVID-19 Treatment Guidelines Panel. Coronavirus diseases 2019 (COVID-19) treatment guidelines. National Institutes of Health [Internet]; 2021. [cited 2021 Jan 14]. Available from: https://www.covid19treatmentguidelines.nih.gov/whats-new/.
- 13.National Institute of Infectious Disease . Manual for the detection of pathogen 2019–nCoV Ver.2.6; 2020. [cited 2021 Jan 14]. Available from: https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf.
- 14.Gwet KL. Intrarater reliability. Wiley encyclopedia of clinical trials. Hoboken (NJ): Wiley; 2008. [Google Scholar]
- 15.Nishiura H. A robust statistic AC for assessing inter-observer agreement in reliability studies. Nihon Hoshasen Gijutsu Gakkai Zasshi. 2010;66(11):1485–1491. [DOI] [PubMed] [Google Scholar]
- 16.Kanda Y. Investigation of the freely available easy-to-use software “'EZR' for medical statistics”. Bone Marrow Transplant. 2013;48(3):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.COVID-19 Treatment Guidelines Panel. Coronavirus diseases 2019 (COVID-19) treatment guidelines. National Institutes of Health [Internet]; 2021. [cited 2021 Jan 14]. Available from: https://www.covid19treatmentguidelines.nih.gov/whats-new/ [PubMed]
- 18.Long QX, Tang XJ, Shi QL, et al. . Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti L, Wymant C, Kendall M, et al. . Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491):eabb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Chen L, Deng Q, et al. . The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92(7):833–840. [DOI] [PubMed] [Google Scholar]
- 21.Péré H, Podglajen I, Wack M, et al. . Nasal swab sampling for SARS-CoV-2: a convenient alternative in times of nasopharyngeal swab shortage. J Clin Microbiol. 2020;58(6):2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berenger BM, Conly JM, Fonseca K, et al. . Saliva collected in universal transport media is an effective, simple and high-volume amenable method to detect SARS-CoV-2. Clin Microbiol Infect. 2020;S1198-743X(20)30689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinninti S, Trieu C, Pati SK, et al. . Comparing nasopharyngeal and mid-turbinate nasal swab testing for the identification of severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2020;ciaa882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBlanc JJ, Heinstein C, MacDonald J, et al. . A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Virol. 2020;128:104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima N, Turner F, Slepnev V, et al. . Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis. 2020;ciaa1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazee BW, Rodríguez-Hoces de la Guardia A, Alter H, et al. . Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med. 2018;71(4):509–517.e1. [DOI] [PubMed] [Google Scholar]
- 27.Williams E, Bond K, Zhang B, et al. . Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azzi L, Carcano G, Gianfagna F, et al. . Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To KKW, Tsang OTY, Yip CCY, et al. . Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCulloch DJ, Kim AE, Wilcox NC, et al. . Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open. 2020;3(7):e2016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.