Abstract
The complete chloroplast genomes of three species of Edgeworthia namely, Edgeworthia albiflora, Edgeworthia chrysantha, and Edgeworthia gardneri (Thymelaeaceae), are reported and characterized. The chloroplast genomes displayed a typical quadripartite structure with conserved genome arrangement and specific divergence. The genomes ranged in length from 172,708 to 173,621 bp and displayed similar GC content of 36.5–36.7%. A total of 138–139 genes were predicted, including 92–93 protein-coding, 38 tRNAs and eight rRNAs genes. Variation in the number of short simple repeats and inverted region boundaries of the three cp genomes were observed. A mutational hotspot was detected along the nucleotide sequence from the ndhF to the trnL-UAG genes. The chloroplast genome-based and internal transcribed spacer (ITS)-based phylogenetic analyses using maximum-likelihood (ML) and Bayesian inference (BI) revealed that E. albiflora diverged before E. chrysantha and E. gardneri and placed the Edgeworthia clade at the base of the Eurasian Daphne group with strong bootstrap support. With an effective taxonomic treatment of the species of Edgeworthia, further molecular analyses of their intra- and interspecific genetic variation are inclined to support the treatment of E. albiflora and E. gardneri as two natural groups. The genetic information obtained from this study will provide valuable genomic resources for the identification of additional species and for deducing the phylogenetic evolution of Edgeworthia.
Keywords: chloroplast genome, comparative analysis, Edgeworthia, internal transcribed spacers, phylogenetic relationship, Thymelaeaceae
Introduction
The family Thymelaeaceae is composed of about 900 species in 45 genera. The most recently proposed taxonomic classification based on palynological findings divided the family into two major subfamilies, Octolepidoideae and Thymelaeoideae. The latter consists of three tribes: Aquilarieae, Daphneae, and Synandrodaphneae (Herber, 2003). The Daphneae accounts for the most genera, which were further clustered into four different groups, Daphne, Gnidia, Linostoma, and Phaleria, with the Daphne group containing the most genera.
As a member of the Daphne group, Edgeworthia is reported to contain five species; E. albiflora Nakai, E. chrysantha Lindl., Edgeworthia eriosolenoides K. M. Feng & S. C. Huang, E. gardneri (Wall.) Meisn., and E. longipes Lace. They occur naturally in China, India, and nearby regions (The Plant List, 2013; Wang and Gilbert, 2017). Studies of the phytochemical and pharmacological properties of Edgeworthia have received much attention among researchers (Nan et al., 2018), as the inflorescences and stems of E. chrysantha and E. gardneri are regarded as effective folk medicines for muscle relaxation and to treat rheumatism (Xiao, 2002; Che et al., 2010). Rich in low-lignin fibers and their ease of propagation, plants of Edgeworthia are not only cultivated as ornamentals in urban areas (Clennett et al., 2002; Wang et al., 2017), but are also the preferred raw material for high quality paper products, such as banknotes, and artificial cotton production (Lan et al., 2013). For molecular information, genetic studies on Edgeworthia were confined to the molecular placement of E. chrysantha within the Thymelaeaceae using short gene sequences (Van der Bank et al., 2002; Beaumont et al., 2009; Motsi et al., 2010; Foster et al., 2016), while genetic information for other species of Edgeworthia is limited.
The chloroplast (cp) genome is responsible for photosynthesis (Leister, 2003; Wicke et al., 2011). In general, the cp is maternally inherited and consists of a quadripartite circular double-stranded DNA molecule that comprises two copies of inverted repeat (IRs) regions, a large single-copy (LSC) region, and a small single copy (SSC) region (Palmer, 1985). The length of a typical cp genome ranges between 120,000 and 160,000 bp but variations can occur. Due to its relatively small size, simple structure, conserved gene content, and order, cp genome sequences have been widely used in phylogenetic studies and provide valuable data for resolving complex evolutionary relationships (Jansen et al., 2007; Moore et al., 2010).
At present, there are only 25 complete cp genomes for taxa in the family Thymelaeaceae available publicly in the GenBank database (as of 1st December 2020), with Aquilaria sinensis as the first taxon reported (Wang et al., 2016). Due to the lack of molecular information on the genus Edgeworthia, we used next-generation sequencing technology to obtain the complete cp genomes of three species of Edgeworthia, including E. albiflora, E. chrysantha, and E. gardneri. We constructed and characterized the cp genome structure of these species and performed phylogenetic analyses at the genome-scale level. In addition, to expand the genomic resources from these valuable species, we also sequenced the nuclear ribosomal DNA internal transcribed spacer (ITS) region to reveal the phylogenetic relationships of these species of Edgeworthia to other closely related taxa in the Daphne group.
Materials and Methods
Plant Materials and DNA Extraction
Fresh leaf samples from three species of Edgeworthia namely, E. albiflora, E. chrysantha, and E. gardneri, were collected from plants in their natural habitat and were stored in Ziplock bags filled with silica gel beads prior to transportation to the laboratory. Voucher specimens of the three species were deposited in the Herbarium of Yunnan Normal University (YNUB) (Table 1). Total genomic DNA was extracted from the silica-dried leaves using the modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987) and was further purified using Wizard® DNA Clean-Up System (Promega, United States).
TABLE 1.
Basic characteristics of chloroplast genomes in three species of Edgeworthia.
| E. albiflora | E. chrysantha | E. gardneri | |
| Sample location | Miyi, Sichuan, China | Kunming Botanical Garden, Yunnan, China | Motuo, Tibet, China |
| Collector and collection number | Zhang Y.; ZYH11 | Zhang Y.; RXK26 | Zhang Y.; RXK53 |
| Chloroplast genome GenBank accession number | MW246180 | MN511715 | MW246181 |
| ITS GenBank accession number | MW255615 | MW255616 | MW255617 |
| LSC (bp) | 86,862 | 85,824 | 86,388 |
| SSC (bp) | 2,681 | 2,816 | 3,017 |
| IR (bp) | 42,039 | 42,034 | 41,952 |
| Total (bp) | 173,621 | 172,708 | 173,309 |
| Protein-coding genes | 93 | 93 | 92 |
| tRNA genes | 38 | 38 | 38 |
| rRNA genes | 8 | 8 | 8 |
| Total | 139 | 139 | 138 |
| GC content (%) | 36.5 | 36.7 | 36.5 |
Genome Sequencing, Assembly and Annotation
Next-generation sequencing was conducted on the Illumina HiSeq 2500 platform and a 350-bp paired-end library was prepared. The raw reads were filtered to obtain high-quality clean reads using NGS QC Toolkit v2.3.3 with default parameters (Patel and Jain, 2012). The cp genome was assembled using NOVOPlasty (Dierckxsens et al., 2017) with the rbcL gene of Daphne kiusiana Miq. extracted from the complete cp genome sequence (GenBank accession KY991380) as the seed sequence. The complete cp genome was annotated using Geneious v10.1.3 (Kearse et al., 2012) by referring to the cp genome sequence of D. kiusiana. Annotations on the protein-coding (CDS) sequences present in the genome were manually checked using the open reading frame (ORF) and the tRNA genes were verified using the online tRNAscan-SE web server with default settings (Lowe and Chan, 2016). The complete cp genome was visualized using OGDRAW v1.3.1 (Greiner et al., 2019); all cp genome sequences were deposited in the NCBI GenBank database under the accession numbers MW246180 (E. albiflora), MN511715 (E. chrysantha), MW246181 (E. gardneri) for future reference.
Repeats Analyses
Simple sequence repeats (SSRs) were identified using the MISA-web (Beier et al., 2017). The minimum number of repeats was set at 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. SSRs were manually checked for redundancy. Identification of the four different types (forward, palindromic, reverse, and complement) of large repeats were conducted with REPuter (Kurtz et al., 2001). The size and identity of the large repeats were limited to not less than 30 bp and 90%, respectively; while the Hamming distance was set at 3.0.
Genome Comparison
The junctions and borders of the inverted repeat (IR) regions were visualized using IRscope (Amiryousefi et al., 2018) and further edited using Adobe Photoshop CS6 (Adobe, United States). For comparative analysis, the three sequences of the Edgeworthia cp genomes were compared using mVISTA (Mayor et al., 2000) using Shuffle-LAGAN mode. The cp genome sequence of E. albiflora was selected as the reference genome. The output image was manually edited using Adobe Illustrator 2020 (Adobe, United States). All three genome sequences of the Edgeworthia cp were aligned using MAFFT v7.409 (Katoh and Standley, 2013). Highly divergent regions between the species were identified using DnaSP v5.10 (Librado and Rozas, 2009). The nucleotide divergence values of the cp genome sequence alignment were analyzed using the sliding window method, with a window length of 1000 bp and a 500-bp step size.
Codon Use Preference Analysis
All the CDS gene sequences were manually extracted from the chloroplast genome. The codon usage frequency in each of the three species of Edgeworthia was analyzed for all the PCGs using MEGA5 (Kumar et al., 2008). The relative synonymous codon usage (RSCU) was conducted to determine if the plastid genes were under selection.
Polymerase Chain Reaction and Sanger Sequencing
Polymerase chain reaction (PCR) amplification was conducted on a final reaction volume, with a 20 μL volume reaction consisting of 10 μL of 2× Taq PCR StarMix with loading dye (Genstar Biosolutions, China), 1 μL of each primer, 6 μL of distilled water, and 2 μL 5 ng genomic DNA as a template. The ITS universal primer set: ITS1, 5′-TCC GTA GGT GAA CCT GCG G-3′ (forward) and ITS4, 5′-TCC TCC GCT TAT TGA TAT GC-3′ (reverse), was used to obtain the ITS region (White et al., 1990). PCR amplification was programmed with thermal settings of an initial denaturation at 94°C for 5 min; denaturation at 94°C for 60 s, annealing at 55°C for 60 s, extension at 72°C for 60 s; and a final extension at 72°C for 7 min. Upon verification via electrophoresis on a 1.0% agarose gel and documented under the UV machine, the PCR products were sent for direct Sanger sequencing at both ends using an ABI 3730 DNA Analyzer (Applied Biosystems, United States). Results acquired from the Sanger sequencing were aligned and manually edited to obtain the clean sequences of the three species of Edgeworthia. The ITS sequences for E. albiflora (MW255615), E. chrysantha (MW255616), and E. gardneri (MW255617) were deposited in the NCBI GenBank database for future reference.
Phylogenetic Analyses
Complete cp genome sequences of 15 taxa from the family Thymelaeaceae were included for phylogenetic analyses using maximum likelihood (ML) methods and Bayesian inference (BI). Multiple sequence alignment was carried out using MAFFT v7.409 (Katoh and Standley, 2013). Based on the Akaike information criterion calculated from the Modeltest 3.7 (Posada and Crandall, 1998), the generalized-time-reversible (GTR) model with gamma (+G) and invariant sites included (+I) (=GTR + G + I) was the best-fitting substitution model for both the ML and BI analyses. The ML tree was constructed using RAxML 8.2.11, under 1,000 bootstrap replicates (Stamatakis, 2014). BI analysis was conducted using MrBayes 3.2.5 (Drummond and Rambaut, 2007), in which the Markov Chain Monte Carlo analysis was performed under 1,000,000 generations and four Markov chains. Samplings were conducted at every 1,000 generations. The first 25% of the trees was discarded as burn-in; the remaining trees were estimated using the 50% majority-rule consensus tree and Bayesian posterior probabilities. Two closely related species, Hibiscus hamabo (Malvaceae; KR259988) and Eugenia uniflora (Myrtaceae; KR867678) were included as outgroups.
A total of 23 ITS sequences from the members of Thymelaeaceae, representing 21 taxa from eight genera in the Daphne group of tribe Daphneae, two taxa from tribe Aquilarieae and one taxon from the subfamily Octolepidoideae, were retrieved from the NCBI GenBank database. The latter three taxa were then selected as outgroups. Along with the ITS sequences of the three Edgeworthia species, the sequences were MUSCLE-aligned using MEGA 5 (Kumar et al., 2008) and trimmed using trimAL v1.2 (Capella-Gutiérrez et al., 2009) with the gappyout method in order to reduce the systematic errors produced by poor alignment. Phylogenetic analyses were carried out using both the ML and BI method. For ML analysis, the optimal DNA substitution model for the ML analysis calculated using the “Find Best DNA/Protein Model (ML)” function embedded in MEGA 5 (Kumar et al., 2008) was the Kimura two-parameter (K2P) with discrete Gamma model (+G) and invariant included (+I) (=K2P + G + I). Calculation was conducted with 1,000 bootstrap replicates on each branch node and all gaps and missing data were included in the analysis. For BI analysis, calculation was performed using MrBayes v3.2.5 (Drummond and Rambaut, 2007) following the same parameters and settings as mentioned above.
Results
Chloroplast Genome Features
A total of 23,660,708 raw reads were obtained and the raw reads were directly fed into the assembly pipeline to obtained the maximum amount of useful data. Prior to genome assembly, a total of 374,342 aligned reads were acquired, and 222,318 assembled reads of an average coverage depth of 217 times per site were incorporated in the genome assembly. Three contigs representing three species of Edgeworthia were obtained at the end of the assembly process.
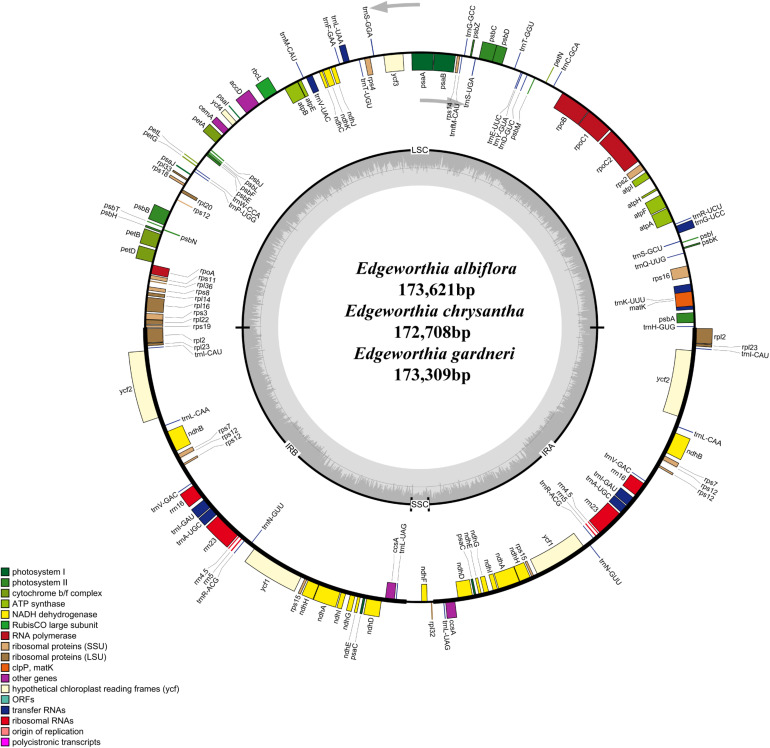
The cp genomes of the species of Edgeworthia were typical quadripartite structures that ranged in size from 172,708 bp (E. chrysantha) to 173,621 bp (E. albiflora) (Figure 1). All genomes contained a pair of IRs (41,952–42,039 bp), separated by a large single-copy (LSC) region (85,824–86,862 bp) and a small single-copy (SSC) region (2,681–3,017 bp) (Table 1). A total of 139 genes were predicted for the cp genomes of E. albiflora and E. chrysantha, including 93 CDS genes, 38 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. However, only 138 genes were recorded in the cp genome of E. gardneri, in which only 92 CDS genes, 38 tRNA genes, and eight rRNA genes were predicted. In the LSC region, E. albiflora and E. chrysantha each had 61 CDS genes, but only 60 CDS genes were reported in E. gardneri. The CDS gene, cemA, was presumably a pseudogene in the cp genome of E. gardneri; while only two CDS genes were recorded in all three species of Edgeworthia. All three cp genomes of Edgeworthia had 27 genes replicated in both IR regions, including 15 CDS genes (ccsA, ndhA, ndhB, ndhD, ndhE, ndhG, ndhH, ndhI, rpl2, rpl23, rps12, rps15, psaC, ycf1, and ycf2), eight tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnL-UAG, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) (Table 2). Of the 19 intron-containing genes, 17 of them (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, whereas two of them (rps12 and ycf3) contained two introns (Table 3). The GC content of E. chrysantha and E. albiflora was 36.5%, while the GC content of E. chrysantha was 36.7%.
FIGURE 1.
Gene map for chloroplast genomes of three species of Edgeworthia. Annotated genes are colored according to functional categories. Genes placed outside the outer circle were transcribed clockwise; genes placed inside the circle were transcribed counterclockwise. Dark gray in inner circle represents GC content whereas light gray corresponds to AT content.
TABLE 2.
Gene contents in three Edgeworthia species chloroplast genome.
| Classification | Genes |
| Self-replication | |
| Large ribosomal subunits | rpl2*(×2), rpl14, rpl16*, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| Small ribosomal subunits | rps2, rps3, rps4, rps7, rps8, rps11, rps12**a(×2), rps14, rps15(×2), rps16*, rps18, rps19 |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 |
| Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn23(×2) |
| Transfer RNAs | trnA-UGC*(×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU(×2), trnI-GAU*(×2), trnK-UUU*, trnL-CAA(×2), trnL-UAA*, trnL-UAG(×2), trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC*, trnW-CCA, trnY-GUA |
| Photosynthesis-related genes | |
| Photosystem I | psaA, psaB, psaC(×2), psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| NAD(P)H dehydrogenase complex | ndhA*(×2), ndhB*(×2), ndhC, ndhD(×2), ndhE(×2), ndhF, ndhG(×2), ndhH(×2), ndhI(×2), ndhJ, ndhK |
| F-type ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| Cytochrome b6/f complex | petA, petB*, petD*, petG, petL, petN |
| Rubisco | rbcL |
| Other genes | |
| Envelope membrane protein | cemA |
| Maturase | matK |
| Cytochrome c biogenesis protein | ccsA(×2) |
| Subunit of acetyl-CoA-carboxylase | accD |
| Unknown | |
| Conserved open reading frames | ycf1(×2), ycf2(×2), ycf3**, ycf4 |
*Gene containing one intron, **gene containing two introns, atrans-splinting gene, (×2) shows gene shave two copies.
TABLE 3.
Locations and sizes of the intron-containing genes recorded in the chloroplast genome of the three Edgeworthia species.
| Species | Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
| Edgeworthia albiflora | atpF | LSC | 144 | 864 | 411 | na | na |
| ndhA | IR | 558 | 1165 | 540 | na | na | |
| ndhB | IR | 777 | 686 | 756 | na | na | |
| petB | LSC | 6 | 770 | 698 | na | na | |
| petD | LSC | 8 | 786 | 475 | na | na | |
| rpl2 | IR | 393 | 724 | 391 | na | na | |
| rpl16 | LSC | 9 | 1080 | 399 | na | na | |
| trnA-UGC | IR | 38 | 812 | 35 | na | na | |
| trnG-UCC | LSC | 23 | 720 | 49 | na | na | |
| trnI-GAU | IR | 37 | 955 | 35 | na | na | |
| trnK-UUU | LSC | 37 | 2559 | 35 | na | na | |
| trnL-UAA | LSC | 34 | 546 | 50 | na | na | |
| trnV-UAC | LSC | 38 | 615 | 35 | na | na | |
| ycf3 | LSC | 126 | 732 | 228 | 761 | 153 | |
| Edgeworthia chrysantha | atpF | LSC | 144 | 867 | 411 | na | na |
| ndhA | IR | 558 | 1163 | 540 | na | na | |
| ndhB | IR | 777 | 690 | 756 | na | na | |
| petB | LSC | 6 | 759 | 698 | na | na | |
| petD | LSC | 8 | 790 | 475 | na | na | |
| rpl2 | IR | 391 | 683 | 434 | na | na | |
| rpl16 | LSC | 9 | 1066 | 399 | na | na | |
| rpoC1 | LSC | 432 | 767 | 1617 | na | na | |
| rps16 | LSC | 40 | 934 | 206 | na | na | |
| trnA-UGC | IR | 38 | 811 | 35 | na | na | |
| trnG-UCC | LSC | 23 | 718 | 49 | na | na | |
| trnI-GAU | IR | 37 | 955 | 35 | na | na | |
| trnK-UUU | LSC | 37 | 2555 | 35 | na | na | |
| trnL-UAA | LSC | 34 | 551 | 50 | na | na | |
| trnV-UAC | LSC | 38 | 621 | 35 | na | na | |
| ycf3 | LSC | 126 | 732 | 228 | 761 | 153 | |
| Edgeworthia gardneri | atpF | LSC | 144 | 856 | 411 | na | na |
| ndhA | IR | 558 | 1162 | 540 | na | na | |
| ndhB | IR | 777 | 690 | 756 | na | na | |
| petB | LSC | 6 | 766 | 698 | na | na | |
| petD | LSC | 8 | 781 | 475 | na | na | |
| rpl2 | IR | 393 | 681 | 434 | na | na | |
| rpl16 | LSC | 9 | 1076 | 399 | na | na | |
| rpoC1 | LSC | 432 | 757 | 1617 | na | na | |
| rps16 | LSC | 40 | 946 | 200 | na | na | |
| trnA-UGC | IR | 38 | 812 | 35 | na | na | |
| trnG-UCC | LSC | 23 | 725 | 49 | na | na | |
| trnI-GAU | IR | 37 | 955 | 35 | na | na | |
| trnK-UUU | LSC | 37 | 2531 | 35 | na | na | |
| trnL-UAA | LSC | 34 | 547 | 50 | na | na | |
| trnV-UAC | LSC | 38 | 620 | 35 | na | na | |
| ycf3 | LSC | 126 | 727 | 228 | 762 | 153 |
na, not available.
Sequence Repeats
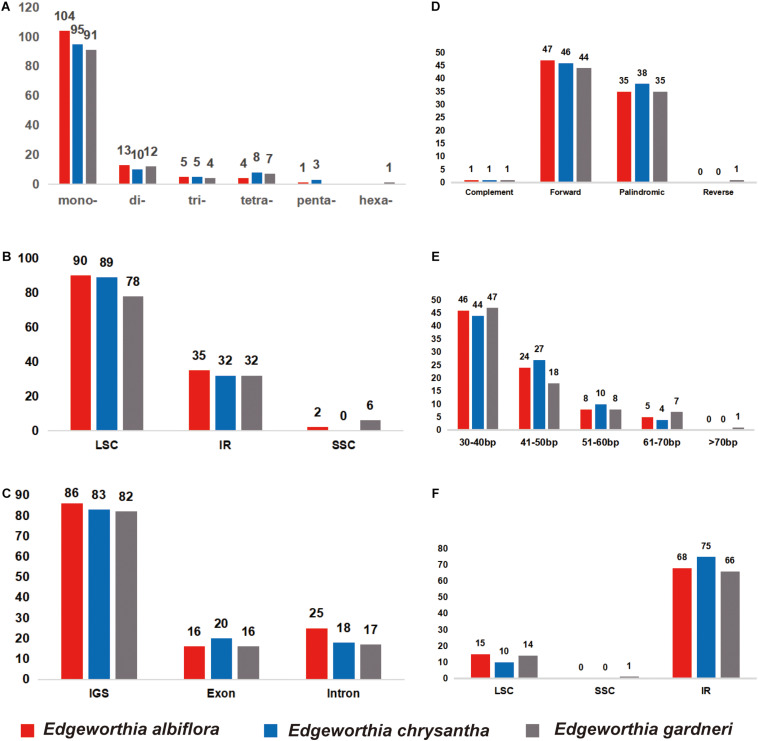
Simple sequence repeat analysis detected 127, 121, and 115 SSRs in E. albiflora, E. chrysantha, and E. gardneri, respectively (Figure 2A). Most of the SSRs were in the LSC regions when compared to the SSC and IR regions (Figure 2B). The SSRs were more abundant in the intergenic spacer region when compared to both the intronic and exon regions; more than 80 SSRs were detected in the intergenic spacer regions of the three species of Edgeworthia, and the number of SSRs in the intronic and exon regions were only recorded between 16 and 25 (Figure 2C). Cp genomes of all three Edgeworthia species contained mono-, di-, tri-, and tetranucleotide SSRs; while E. albiflora and E. chrysantha had one and three pentanucleotide SSRs, respectively, none were recorded in E. gardneri. Yet, E. gardneri was recorded with one hexanucleotide SSR, which was not present in the other two species. Considering sequence complementary, eight classified repeat types were found present in all three species of Edgeworthia (data not shown). The repeat type C/G were only detected in E. albiflora and E. chrysantha; while the repeat type AATT/AATT were only detected in E. chrysantha and E. gardneri. The repeat types AAG/GTT and ACAT/ATGT were exclusive to E. chrysantha, and E. gardneri was the only species recorded with the repeat types AATTC/AATTG and ACCCC/GGGGT.
FIGURE 2.
Distribution of small sequence repeats (SSRs) in the chloroplast genomes of three species of Edgeworthia. (A) Number of different SSR types detected in chloroplast genomes of three species of Edgeworthia; (B) Frequencies of identified SSRs in large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRs) regions; (C) Frequencies of identified SSRs in the gene coding and intergenic region. (D) Frequencies of four different large repeat types (complement, forward, palindromic, and reverse) in the chloroplast genomes of three species of Edgeworthia; (E) Frequencies of large repeats based on different sequence length (bp) groups; (F) Frequencies of large repeats in the LSC, SSC, and IRs regions.
For large repeats, forward repeats were recorded most abundant in the cp genome of the three Edgeworthia species, ranging from 44 to 47, followed by the palindromic repeats that ranged from 35 to 38 (Figure 2D). However, there was no records for reverse repeats in E. albiflora and E. chrysantha, but one in E. gardneri. The large repeats were recorded mostly in sequence length of 30–40 bp (Figure 2E). Edgeworthia gardneri was recorded with one large repeat with sequence length over 70 bp, but only large repeats with length from 60 to 70 bp were recorded longest for E. albiflora and E. chrysantha. The large repeats were recorded mostly distributed in the IR region, followed by the LSC region; at least one large repeat was recorded in the SSC region in E. gardneri (Figure 2F).
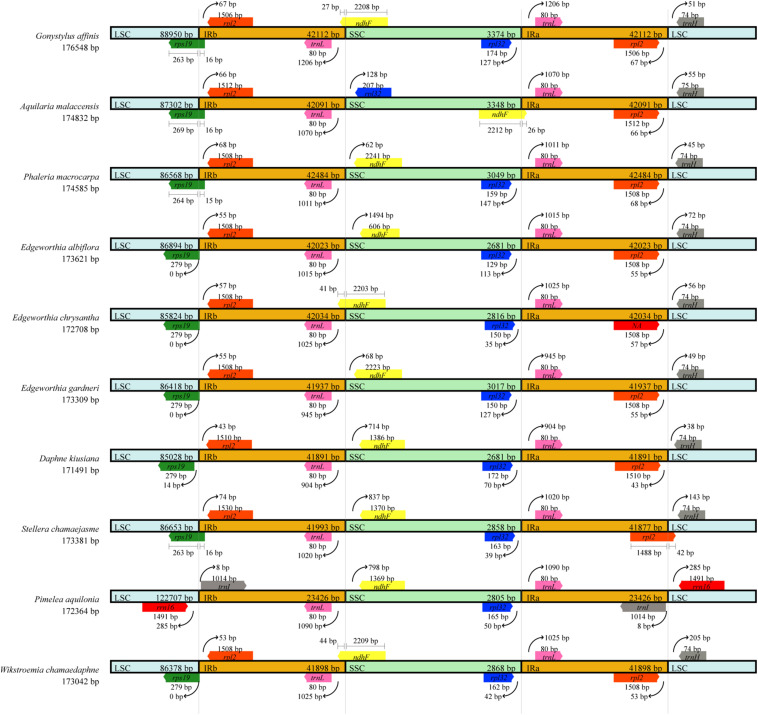
Contraction and Expansion of the IR Regions
Chloroplast genome structure and the junction positions between IR regions were well-conserved among the three species of Edgeworthia, but structural variation was present in the IRs/SC borders (Figure 3). The ndhF gene extended to the IRB region in the cp genome of E. chrysantha, but not for E. albiflora and E. gardneri. The ndhF gene in the latter two species was located in the SSC region. The rps19 gene that was located in the LSC region in E. chrysantha extended into the IRB region in E. albiflora and E. gardneri. When compared to other seven closely related genera in the family Thymelaeaceae, the placement of genes adjacent to the IR junctions were identical to those in the cp genome of Gonystylus affinis, D. kiusiana, Phaleria macrocarpa, Stellera chamaejasme, and Wikstroemia chamaedaphne.
FIGURE 3.
Comparison of borders between large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR) regions across ten chloroplast genomes of Thymelaeaceae.
Comparative Genomic Analysis
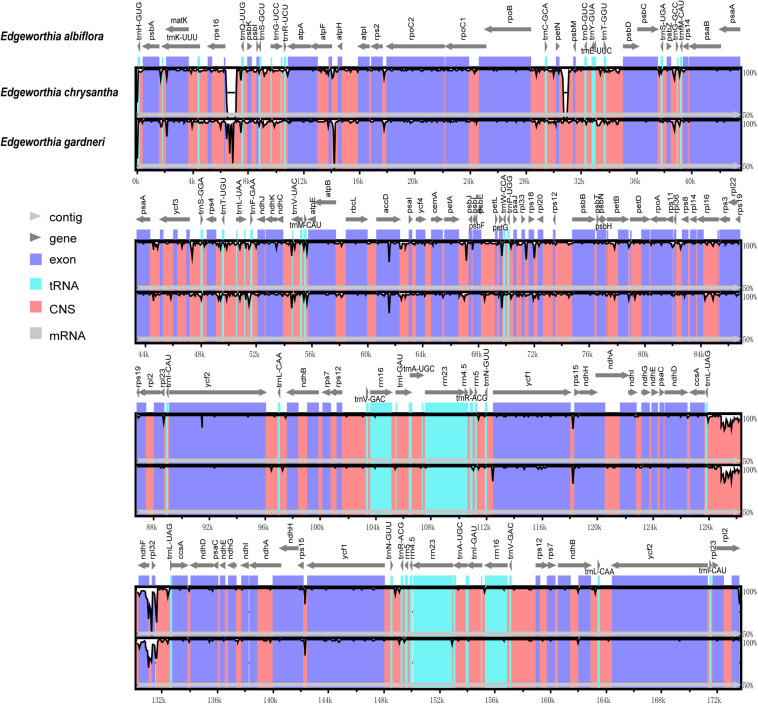
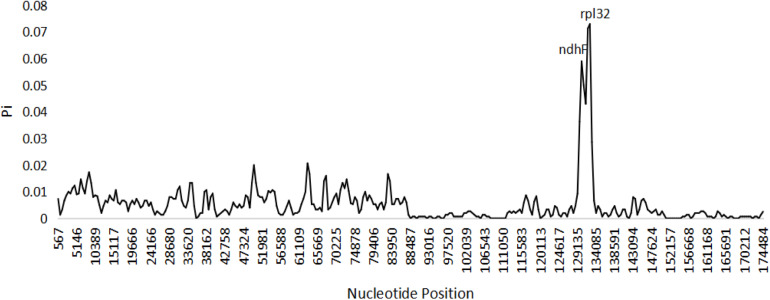
Based on the genome sequence alignment of the three species of Edgeworthia, distinct sequence variation was detected in three gene regions; petN-psbM, trnL-UAG-rpl32, and rps16-trnQ-UUG (Figure 4). With a nucleotide diversity (Pi) cut-off point set at Pi ≥ 0.04, the sliding window analysis detected three highly variable regions in the genome sequence alignment of the three species of Edgeworthia (Figure 5). The highly variable regions were all located within the protein-coding genes, ndhF and mainly manifested in the SSC region, between ndhF and rpl32 of the cp genome.
FIGURE 4.
Complete chloroplast genome comparison of three species of Edgeworthia using mVISTA alignment program. Genome regions are color-coded and described in the figure legend.
FIGURE 5.
Sliding window analysis of the complete chloroplast genome sequences among three species of Edgeworthia (Window length: 1000 bp; step size: 500 bp).
Codon Use Preference Analysis
A total of 29,529–30,093 codons of the CDS genes were recorded in the three cp genomes of Edgeworthia. The RSCU value for each species exhibited similar codon preference in the 64 codons in the CDS genes (Supplementary Table 1). As a result, 30 of them exhibited greater preference (RSCU > 1); 32 of them were least preferred (RSCU < 1); two of them displayed no preferences (RSCU = 1). The isoleucine (Ile)-encoded codon AUU exhibited the greatest occurrence (n = 1,269); while the Ile-encoded codon UGA exhibited the least occurrence (n = 20). Among the preferred codons, 27 of them were A/U-ended. Among the three stop codons, UAA was recorded to be more abundant than UAA and UGA, thus displaying higher preferences. There were no rare codons (RSCU < 0.1) found in the CDS genes of the three cp genomes of Edgeworthia.
Phylogenetic Analysis
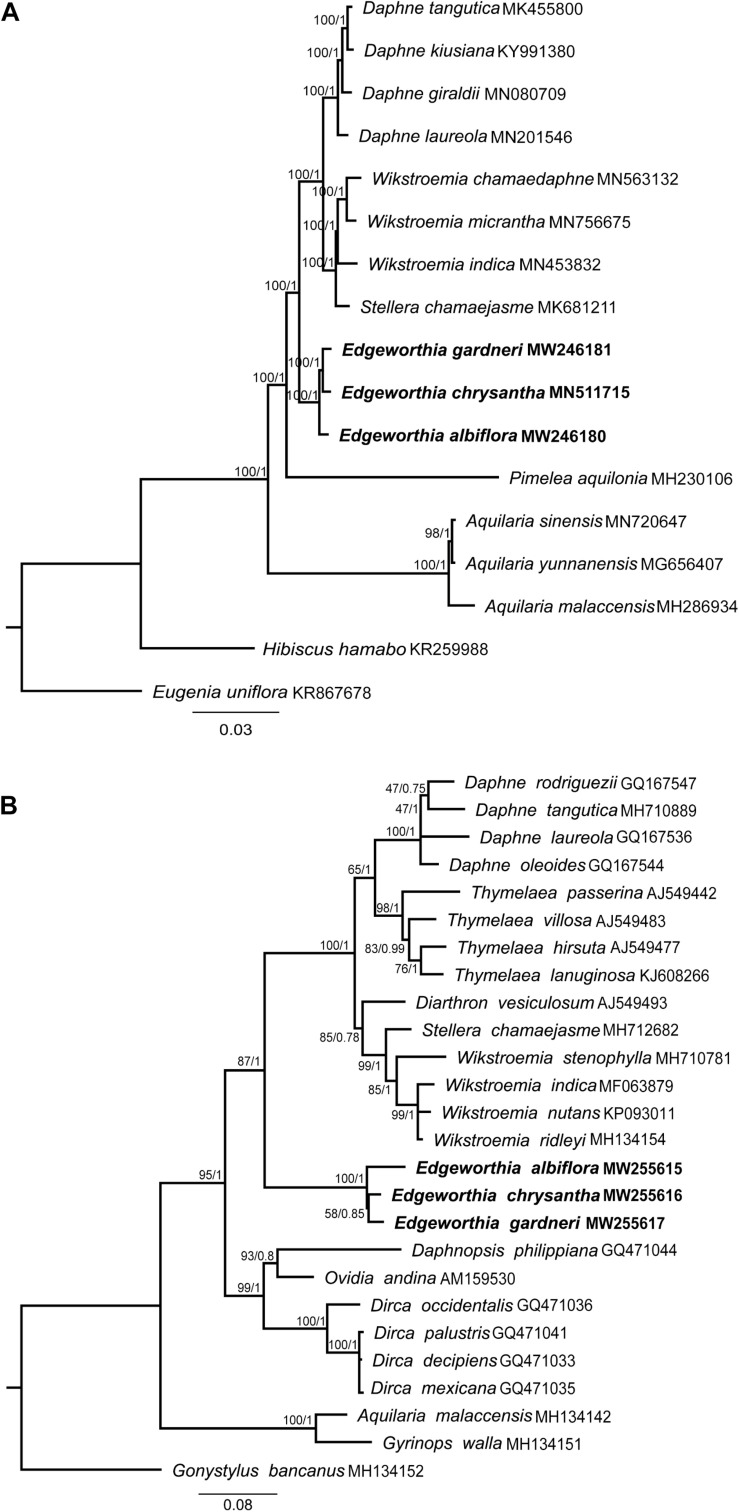
Phylogenetic analyses using the complete cp genome sequence for both ML and BI methods revealed similar topological structure between the two phylogenetic trees (Figure 6A). Strong bootstrap support and high posterior probabilities were recorded at all branch nodes. All taxa included in this study displayed monophyletic relationships. In the Daphneae, Edgeworthia diverged before Daphne, Stellera, and Wikstroemia. The three species of Edgeworthia formed a monophyletic clade, with E. albiflora diverging before E. chrysantha and E. gardneri.
FIGURE 6.
Maximum-likelihood (ML) and MrBayes (BI) tree analyses were based on 1,000 bootstrap replicates. Bootstrap support values and Bayesian posterior probabilities are indicated at each branch nodes. Three species of Edgeworthia used in this study are highlighted in bold. (A) Combined phylogenetic tree of Edgeworthia and allied genera based on the chloroplast genome sequences of 15 taxa from Thymelaeaceae. Two related species, Hibiscus hamabo (Malvaceae; KR259988) and Eugenia uniflora (Myrtaceae; KR867678) were included as outgroups. (B) Phylogenetic analyses of Thymelaeaceae based on nuclear ribosomal DNA internal transcribed spacer (ITS) gene sequences. A total of 23 ITS sequences from the members of Thymelaeaceae, representing 21 taxa from eight genera in the Daphne group of tribe Daphneae, two taxa from tribe Aquilarieae and one taxon from subfamily Octolepidoideae.
For the ITS-based ML and BI analyses, both phylogenetic trees exhibited identical tree structure and species placement (Figure 6B). The Daphne group displayed a paraphyletic relationship, in which the genera Daphnopsis, Dirca, and Ovidia formed a cluster; while Edgeworthia was placed at the base of the latter cluster, with strong bootstrap support and Bayesian posterior probability (ML ≥ 75, BI ≥ 0.95) that also consisted of Daphne, Diarthron, Stellera, Thymelaea, and Wikstroemia. In the Edgeworthia clade, the three Edgeworthia species formed a monophyletic clade, with E. albiflora diverging before E. chrysantha and E. gardneri under strong bootstrap support and Bayesian posterior probability.
Discussion
Similar to other members of the Thymelaeaceae, the cp genome of Edgeworthia was rather well-conserved across the three species analyzed in this study. By comparing published cp genome reports on members of the Thymelaeaceae, it was determined that the complete cp genome sequence of Edgeworthia is shorter than those of G. affinis of subfamily Octolepidoideae (176,548 bp) and nine species of Aquilaria (Aquilaria beccariana, Aquilaria crassna, Aquilaria hirta, Aquilaria malaccensis, Aquilaria microcarpa, Aquilaria rostrata, A. sinensis, Aquilaria subintegra, and Aquilaria yunnanensis; 174,693–174,907 bp) from Aquilarieae of subfamily Thymelaeoideae (Hishamuddin et al., 2020), but was longer than the four species of Daphne (Daphne giraldii, D. kiusiana, Daphne laureola, and Daphne tangutica; 169,944–171,643 bp) (Cho et al., 2018; Könyves et al., 2019; Yan et al., 2019a,b), two species of Wikstroemia (W. chamaedaphne and Wikstroemia indica; 151,731–173,042 bp) from the Daphne group (Qian and Zhang, 2020; Qian et al., 2020), and Pimelea aquilonia (172,364 bp) from the Gnidia group (Foster et al., 2018), all belonging to the Daphneae of subfamily Thymelaeoideae. Stellera chamaejasme, from the monospecific Stellera, was the only species from the Daphne group with a total cp genome size within the cp genome size range of Edgeworthia, which was 173,381 bp (Yun et al., 2019). Eventually, rather small size of the SSC region was observed in the cp genome of most species from Thymelaeaceae. In general, the expansion, shrinkage and loss of the IR regions are some of the known reasons to variations in cp genome sizes of seed plants (Jansen and Ruhlman, 2012). Since information on complete cp genomes in members of the Thymelaeaceae is limited, we could not infer any significance between the cp genome size and the systematic position in the Thymelaeaceae.
In this study, the three species of Edgeworthia shared almost the same number of genes in their complete cp genomes; E. gardneri was recorded as having one gene fewer when compared to the other two species – the protein-coding gene, cemA, was not annotated. The cemA gene of E. gardneri was thought to be a pseudogene as the loci of the premature stop codons differ from the others and could be causing the gene to be non-functional; while the location of the stop codons in cemA could be useful for distinguishing E. gardneri from E. albiflora and E. chrysantha. The cemA gene encodes the chloroplast membrane protein, which may play an important role in plastid maintenance and intracellular communication (Sasaki et al., 1993; Sonoda et al., 1997). The cemA homolog is also recorded to be essential for carbon dioxide (CO2) transfer in cyanobacteria (Katoh et al., 1996). The stomatal density of E. gardneri is lower than that in E. chrysantha (Zhang et al., 2015) and the deletion of cemA gene may be related to its adaptation in its habitat, which has an average altitude of above 1,000 m a.s.l.
Based on literature reports, members of the Daphne group can be divided into two natural groups due to the effects on geographical separation and by minor morphological differences (Herber, 2003). The seven American genera (Daphnopsis, Dirca, Funifera, Goodallia, Lagetta, Ovidia, and Schoenobiblus) have petals and/or long filaments, whereas the seven Eurasian genera (Daphne, Diarthron, Edgeworthia, Rhamnoneuron, Stellera, Thymelaea, and Wikstroemia) lack petals, but are reported to have sessile or subsessile anthers (Herber, 2003). Eventually, the most comprehensive phylogenetic study conducted on Thymelaeaceae using the combined rbcL, trnL-trnF datasets and ITS sequences revealed that members of the Daphne group were actually divisible into two different clades in which Daphne, Diarthron, Edgeworthia, Stellera, and Wikstroemia formed one clade; while Ovidia and Dirca were in another clade with Peddiea (Phaleria group) and Stephanodaphne (Linostoma group) (Beaumont et al., 2009). Similarly, a non-monophyletic relationship was observed in the Daphne group in this study based on the ITS sequences. In our study, the three American genera, Dirca, Ovidia, and Daphnopsis formed a clade independent from the Eurasian genera (Figure 6B). Both the phylogenetic analyses based on cp genome sequences and the nuclear ribosomal DNA ITS sequences, or a combination of both the cp and nuclear gene sequences, placed Edgeworthia at the base of the Daphne group among the Eurasian genera.
Taxonomic controversy is also present in Edgeworthia. Based on the Flora of China, there are at least five species, whereas four species (E. albiflora, E. chrysantha, E. eriosolenoides, and E. gardneri) are in the Asia region (Wang and Gilbert, 2017). The recent revision by The Plant List committee recognizes only four species; E. eriosolenoides, E. gardneri, E. longipes, and Edgeworthia tomentosa (The Plant List, 2013). The former synonymized E. chrysantha with E. tomentosa, as the name E. tomentosa was considered invalid in a revision conducted on the collections of Thunberg on Magnolia tomentosa (Hamaya, 1955), and also E. albiflora with E. gardneri (Wang and Gilbert, 2017). Edgeworthia albiflora, a species treated as a distinct since it was first discovered in 1924 (Nakai, 1924; Duncan and Mellinger, 1972), was later regarded as a synonym of E. gardneri (Clennett et al., 2002). The synonymy was not accepted in the Flora of China, where E. albiflora was treated as distinct (Wang and Gilbert, 2017). It is noteworthy that we also failed to recover information to synonymize E. albiflora under E. gardneri. Based on our field observations, E. albiflora and E. gardneri can be differentiated through their morphological features and do not pose a challenge in species recognition. The interior of the calyx of E. gardneri is yellow, the ovary uniformly hairy, the stigma rounded; the interior of the calyx of E. albiflora is white, the base of the ovary is glabrous and the apex hairy and the stigma is clavate (Wang and Gilbert, 2017). Meanwhile, the leaf epidermis is entirely different in E. albiflora and E. gardneri, with paracytic stomatal types and cyclocytic stomatal types, respectively (Zhang et al., 2015). From a molecular perspective, it is generally accepted that species are delimited when the interspecific variation is greater than intraspecific variation (Lim et al., 2012). Thus, to further strengthen the case for recognizing E. albiflora, we compared the genetic information and found that the alignment between E. albiflora, E. chrysantha, and E. gardneri is consisted of greater interspecific variation (cp genome: pairwise distance = 0.0045–0.0061, containing 1038 singletons; ITS: pairwise distance = 0.0226–0.0365, containing 29 singletons) than its intraspecific variation that was based on the alignment between our collection of E. chrysantha and another published genome of E. chrysantha (cp genome: MT135125; ITS: AJ744932) (cp genome: pairwise distance = 0.0004, containing 70 singletons; ITS: pairwise distance = 0.0030, containing two singletons) (data not shown). Furthermore, the molecular placement with strong bootstrap support based on the ML and BI trees using both the complete cp genome sequences and ITS sequence, analyzed separately, revealed that E. albiflora and E. gardneri should be treated separately (Figure 6B). Unless there is a stronger case to synonymize the two species via morphological features, judging from the molecular evidence and personal field observation, they should be regarded as two natural groups. On the other hand, two species, E. eriosolenoides and E. longipes were not included in this study; no specimens had been collected since they were first described.
Conclusion
The entire cp genomes of E. albiflora, E. chrysantha, and E. gardneri were sequenced and analyzed in this study. We obtained such comprehensive molecular information as SSRs, IR contraction and expansion, codon usage and phylogenomic placement through explicit bioinformatic analyses of the cp genome. Furthermore, the addition of the ITS sequences for the understudied species of Edgeworthia provided insight for the first time on the phylogenetic relationships of the three species of Edgeworthia at the nuclear gene level. The data obtained from this study will likely provide a powerful genetic resource for future studies on population genetics, biological functions, molecular phylogeny, as well as evolution of Edgeworthia.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi. nlm.nih.gov/genbank/, MW246180; https://www.ncbi.nlm.nih.gov/genbank/, MN511715; https://www.ncbi.nlm.nih.gov/genbank/, MW246181; https://www.ncbi.nlm.nih.gov/genbank/, MW255615; https://www.ncbi.nlm.nih.gov/genbank/, MW255616; and https://www.ncbi.nlm.nih.gov/genbank/, MW255617.
Author Contributions
SQ performed the experiments, analyzed the data, and wrote the manuscript. YZ and SL conceived the research and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31760048) and the Fundamental Research Funds for the Central Universities (33000-31611215).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.643552/full#supplementary-material
References
- Amiryousefi A., Hyvönen J., Poczai P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34 3030–3031. 10.1093/bioinformatics/bty220 [DOI] [PubMed] [Google Scholar]
- Beaumont A. J., Edwards T. J., Manning J., Maurin O., Rautenbach M., Motsi M. C., et al. (2009). Gnidia (Thymelaeaceae) is not monophyletic: taxonomic implications for Thymelaeoideae and a partial new generic taxonomy for Gnidia. Bot. J. Linn. Soc. 160 402–417. 10.1111/j.1095-8339.2009.00988.x [DOI] [Google Scholar]
- Beier S., Thiel T., Münch T., Scholz U., Mascher M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics. 33 2583–2585. 10.1093/bioinformatics/btx198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che C. T., Cheung H. Y., Law R., Luo G. A., Man R. Y., Tsim K. W., et al. (2010). Buddlejae Flos.,” in Hong Kong Chinese materia medica standards. Hong Kong, China: The Department of Health, The Government of the Hong Kong Special Administrative Region, 179–187. [Google Scholar]
- Cho W. B., Han E. K., Choi G., Lee J. H. (2018). The complete chloroplast genome of Daphne kiusiana, an evergreen broad-leaved shrub on Jeju Island. Conserv. Genet. Resour. 10 103–106. 10.1007/s12686-017-0774-5 [DOI] [Google Scholar]
- Clennett C., Harvey Y., Pearman G. (2002). Plate 433. Edgeworthia chrysantha. Curtis’S Bot. Magazine 19 19–27. 10.1111/1467-8748.00325 [DOI] [Google Scholar]
- Dierckxsens N., Mardulyn P., Smits G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. 10.1093/nar/gkw955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19 11–15. 10.1016/0031-9422(80)85004-7 [DOI] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan W. H., Mellinger M. (1972). Edgeworthia (Thymelaeaceae) new to the western hemisphere. Rhodora. 74 436–479. 10.2307/23310953 [DOI] [Google Scholar]
- Foster C. S. P., Cantrill D. J., James E. A., Syme A. E., Jordan R., Douglas R., et al. (2016). Molecular phylogenetics provides new insights into the systematics of Pimelea and Thecanthes (Thymelaeaceae). Aust. Syst. Bot. 29 185–196. 10.1071/SB16013 [DOI] [Google Scholar]
- Foster C. S. P., Henwood M. J., Ho S. Y. W. (2018). Plastome sequences and exploration of tree-space help to resolve the phylogeny of riceflowers (Thymelaeaceae: Pimelea). Mol. Phylogenet. Evol. 127 156–167. 10.1016/j.ympev.2018.05.018 [DOI] [PubMed] [Google Scholar]
- Greiner S., Lehwark P., Bock R. (2019). Organellar genomedraw (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47 W59–W64. 10.1093/nar/gkz238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaya T. (1955). A dendrological monograph on the thymelaeaceae plants of japan. Univ. Tokyo Faculty Agric. Exp. Forest Rep. 50 45–96. [Google Scholar]
- Herber B. E. (2003). “Thymelaeaceae,” in The Families and Genera of Vascular Plants, eds Kubitzki K., Bayer C. (Berlin, Heidelberg: Springer; ), 373–396. [Google Scholar]
- Hishamuddin M. S., Lee S. Y., Ng W. L., Ramlee S. I., Lamasudin D. U., Mohamed R. (2020). Comparison of eight complete chloroplast genomes of the endangered Aquilaria tree species (Thymelaeaceae) and their phylogenetic relationships. Sci. Rep. 10:13034. 10.1038/s41598-020-70030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Cai Z., Raubeson L. A., Daniell H., Depamphilis C. W., James L. M., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A. 104 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Ruhlman T. A. (2012). “Plastid genomes of seed plants,” in Genomics of Chloroplasts and Mitochondria, eds Bock R., Knoop V. (New York: Springer; ), 103–126. [Google Scholar]
- Katoh A., Lee K. S., Fukuzawa H., Ohyama K., Ogawa T. (1996). CemA homologue essential to CO2 transport in the cyanobacterium synechocystis PCC6803. Proc. Natl. Acad. Sci. U. S. A. 93 4006–4010. 10.1073/pnas.93.9.4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Steven S. H., Cheung M., Sturrock S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könyves K., Yooprasert S., Culham A., David J. (2019). The complete plastome of Daphne laureola L. (Thymelaeaceae). Mitochondrial DNA B Resour. 4 3364–3365. 10.1080/23802359.2019.1674201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinf. 9 299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29 4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C. N., Fan W. G., Bao X. M. (2013). Cultivation techniques for Edgeworthia chrysantha Lindl.——a wood plant with high economic value. J. Forestry Engin. 4 119–121. 10.3969/j.issn.1000-8101.2013.04.033 [DOI] [Google Scholar]
- Leister D. (2003). Chloroplast research in the genomic age. Trends Genet. 19 47–56. 10.1016/s0168-9525(02)00003-3 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lim G. S., Balke M., Meier R. (2012). Determining species boundaries in a world full of rarity: singletons, species delimitation methods. Syst. Biol. 61 165–169. 10.1093/sysbio/syr030 [DOI] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., et al. (2000). VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 16 1046–1047. 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- Moore M. J., Soltis P. S., Bell C. D., Burleigh J. G., Soltis D. E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U. S. A. 107 4623–4628. 10.1073/pnas.0907801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motsi M. C., Moteetee A., Beaumont A. J., Rye B. L., Van der Bank M. (2010). A phylogenetic study of Pimlea and Thecanthes (Thymelaeaceae): Evidence from plastid and nuclear ribosomal DNA sequence data. Aust. Syst. Bot. 23 270–284. 10.1071/SB09002 [DOI] [Google Scholar]
- Nakai T. (1924). Some new and noteworthy ligneous plants of eastern Asia. J. Arnold Arbor., Harv. Univ. 5 72–83. [Google Scholar]
- Nan C. Y., Lu Y. X., Zhu J. X., Jiang W., Zhong G. Y., Li M. (2018). Advances in research on chemical constituents and pharmacological effects of Edgeworthia plants. Chinese Trad. Patent Med. 1 166–171. 10.3969/j.issn.1001-1528.2018.01.034 [DOI] [Google Scholar]
- Palmer J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19 325–354. 10.1146/annurev.ge.19.120185.001545 [DOI] [PubMed] [Google Scholar]
- Patel R. K., Jain M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PloS One. 7:e30619. 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics. 14 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Qian S. J., Zhang Y. H. (2020). Characterization of the complete chloroplast genome of a medicinal plant, Wikstroemia indica (Thymelaeaceae). Mitochondrial DNA B Resour. 5 83–84. 10.1080/23802359.2019.1696249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S. J., Zhang Y. H., Li G. D. (2020). The complete chloroplast genome of a medicinal plant, Wikstroemia chamaedaphne (Thymelaeaceae). Mitochon. DNA B Resour. 5 648–649. 10.1080/23802359.2019.1711228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Sekiguchi K., Nagano Y., Matsuno R. (1993). Chloroplast envelope protein encoded by chloroplast genome. Febs. Lett. 316 93–98. 10.1016/0014-5793(93)81743-J [DOI] [PubMed] [Google Scholar]
- Sonoda M., Katoh H., Ohkawa H., Ogawa T. (1997). Cloning of the cotA gene of Synechococcus PCC7942 and complementation of a cotA-less mutant of synechocystis PCC6803 with chimeric genes of the two strains. Photosynth. Res. 54 99–105. 10.1023/A:1005830303453 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Plant List. (2013). The plant list. Available Online at: http://www.theplantlist.org/ (accessed December 1st, 2020). [Google Scholar]
- Van der Bank M., Fay M. F., Chase M. W. (2002). Molecular phylogenetics of thymelaeaceae with particular reference to african and australian genera. Taxon 51 329–339. 10.2307/1554930 [DOI] [Google Scholar]
- Wang X. L., Luo W. C., Dong Y. C., Yan G. F. (2017). Preliminary study on the ornamental characteristics and landscape application of Edgeworthia chrysantha. Anhui Agric. Sci. Bull. 9 124–126. 10.16377/j.cnki.issn1007-7731.2017.09.052 [DOI] [Google Scholar]
- Wang Y., Gilbert M. (2017). “Edgeworthia,” in Flora of China, eds Raven P. H., Wu Z. Y., Hong D. Y. (Beijing: Science Press; ), 247–248. [Google Scholar]
- Wang Y., Zhan D. F., Jia X., Mei W. L., Dai H. F., Chen X. T., et al. (2016). Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) gilg and evolution analysis within the malvales order. Front. Plant Sci. 7:280. 10.3389/fpls.2016.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T. D., Lee S. B., Taylor J. W., Innis M. A., Gelfand D. H., et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 31 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wicke S., Schneeweiss G. M., Depamphilis C. W., Kai F. M., Quandt D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76 273–297. 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P. (2002). Modern Chinese Materia Medica. Beijing: Chemical Industry Press. [Google Scholar]
- Yan F., Tao X., Wang Q. L., Zhang Y. J., Zhang C. M., Yu H. L. (2019a). The complete chloroplast genome sequence of the medicinal shrub Daphne giraldii Nitsche. (Thymelaeaceae). Mitochondrial DNA B Res. 4 2685–2686. 10.1080/23802359.2019.1644233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Wang Q. L., Zhang Y. J., Zhang C. M., Chen Y. (2019b). The complete chloroplast genome sequence of medicinal plant, Daphne tangutica Maxim. (Thymelaeaceae). Mitochon. DNA B Res. 4 1776–1777. 10.1080/23802359.2019.1611397 [DOI] [Google Scholar]
- Yun N., Park J., Oh S. H. (2019). The complete chloroplast genome of the traditional medicinal plant Stellera chamaejasme L. (Thymelaeaceae). Mitochondrial DNA B Resour. 4 1796–1797. 10.1080/23802359.2019.1612296 [DOI] [Google Scholar]
- Zhang Y. Z., Sun W. G., Gao C. Z., Cao W. J., Zhang Y. H., Li Z. M. (2015). Character of leaf epidermis and their systematic significance of Daphne and Wikstroemia. Plant Diversity Res. 37 493–512. 10.7677/ynzwyj201515019 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi. nlm.nih.gov/genbank/, MW246180; https://www.ncbi.nlm.nih.gov/genbank/, MN511715; https://www.ncbi.nlm.nih.gov/genbank/, MW246181; https://www.ncbi.nlm.nih.gov/genbank/, MW255615; https://www.ncbi.nlm.nih.gov/genbank/, MW255616; and https://www.ncbi.nlm.nih.gov/genbank/, MW255617.