Abstract
Endometrial cancer (EC) is a major cause of death among gynecologic malignancies. To improve early detection of EC in patients, we carried out a large plasma-derived exosomal microRNA (miRNA) studies for diagnostic biomarker discovery in EC. Small RNA sequencing was performed to identify candidate exosomal miRNAs as diagnostic biomarkers in 56 plasma samples from healthy subjects and EC patients. These miRNA candidates were further validated in 202 independent plasma samples by droplet digital PCR (ddPCR), 32 pairs of endometrial tumors and adjacent normal tissues by quantitative real-time PCR (qRT-PCR), and matched plasma samples of 12 patients before and after surgery by ddPCR. miR-15a-5p, miR-106b-5p, and miR107 were significantly upregulated in exomes isolated from plasma samples of EC patients compared with healthy subjects. Particularly, miR-15a-5p alone yielded an AUC value of 0.813 to distinguish EC patients with stage I from healthy subjects. The integration of miR-15a-5p and serum tumor markers (CEA and CA125) achieved a higher AUC value of 0.899. There was also a close connection between miR-15a-5p and clinical manifestations in EC patients. Its exosomal expression was not only associated with the depth of muscular infiltration and aggressiveness of EC, but also correlated with levels of reproductive hormones such as TTE and DHEAS. Collectively, plasma-derived exosomal miR-15a-5p is a promising and effective diagnostic biomarker for the early detection of endometrial cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-021-01352-4.
Keywords: Endometrial cancer, Liquid biopsy, Plasma-derived exosomal miRNA, ddPCR, Cancer diagnosis, Early detection
Main text
Endometrial cancer (EC) is the second highest incidence of gynecologic cancer [1]. Patients have to undergo uterine apoxesis for accurate EC diagnosis, since there are no effective biomarkers [2]. Exosomes originate from the endosome, and then fuse with the plasma membrane under the traction of molecular motors, and are released to the extracellular environment [3, 4]. Exosomes are detected in body fluids such as plasma, urine, and amniotic fluid [5]. Exosomes encapsulate biomolecules such as proteins and miRNAs, maintain their integrity in the circulation, and transfer them to recipient cells [4]. MiRNA is the most abundant type in the RNA cargo of exosomes [6, 7], and exosomal miRNAs (exomiRs) are usually tumor-specific [8]. ExomiRs have received increasing attention in precision medicine, due to their non-invasiveness, and high accessibility and stability [2, 9]. Recent studies have shown that exomiRs have the potential to be efficient biomarkers for the screening, diagnosis, and monitoring of cancers [10–13]. However, exomiRs as biomarkers have not yet been reported in EC.
To improve early detection of EC patients, we carried out a large plasma-derived exosomal miRNA study for biomarker discovery in EC (Supplementary Methods). Candidates were identified by miRNA sequencing in plasma samples from healthy controls (HC) vs. EC patients, and were further validated in independent plasma samples and endometrial tumor tissues (Table S1 and Figure S1). Plasma-derived exosomal miR-15a-5p was identified as a promising diagnostic biomarker for early detection of endometrial cancer.
Identification of exomiRs for EC diagnosis
Exosomes were isolated from plasma of EC patients and age-matched HC subjects. The previously reported method to identify the shape and size of exosomes [5] was used with CD81, TSG101 and GM130 as positive or negative markers, respectively. Indeed, the fraction isolated from plasma was enriched in exosomes (Figure S2).
The miRNA sequencing was then performed in plasma-derived exosomes from 25 EC and 31 HC subjects. On average, approximately 50 million reads were generated in each library, and 384 exomiRs were detected in each sample (Table S2). Forty-nine miRNAs were differentially expressed between HC and EC groups (p < 0.01) (Fig. 1a and Table S3). Eighteen of them also differentially expressed between tumor and adjacent normal tissues in The Cancer Genome Atlas (TCGA) EC samples [14] (Table S4 and Figure S3). Next, a set of exomiRs from these 18 miRNAs were selected as a best panel to distinguish EC from HC subjects using random forest algorithm (Fig. 1b). Clustering analysis showed that these samples were largely divided into two distinct groups (EC vs HC) by these six exomiRs (Fig. 1c). The AUC for each exomiR ranged from 0.693 to 0.819 with a mean of 0.757. The AUC of the combined six exomiRs achieved 0.983 (Fig. 1d). miR-106b-5p, miR-107, miR-15a-5p, and miR-3615 were significantly up-regulated, while miR-139-3p and miR-574-3p were significantly down-regulated in plasma-derived exosomes of EC compared with HC (Fig. 1e). Consistent trends in expression of these exomiRs were observed in tumor tissues from TCGA EC patients (Fig. 1f).
Fig. 1.
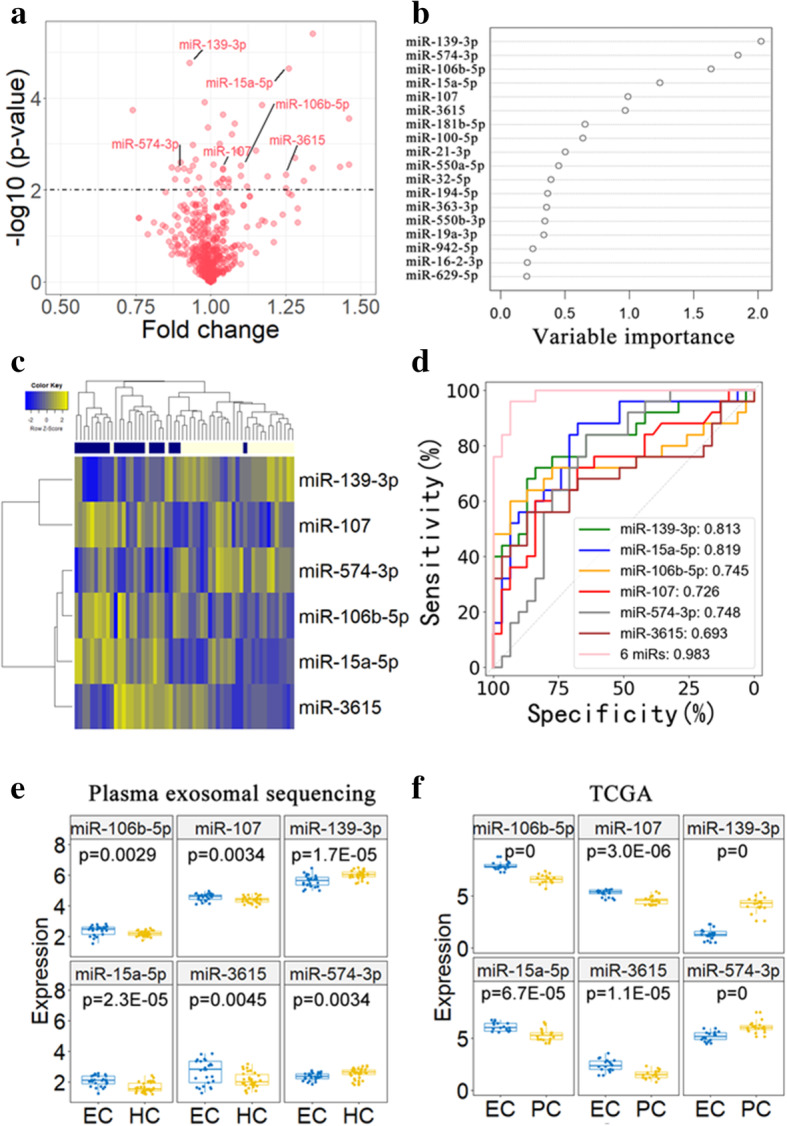
Dysregulated miRNAs in plasma exosomes of EC patients. a Volcano plot displaying differentially expressed miRNAs between plasma exosome of EC and HC. Small RNA sequencing was performed in plasma exosomes of 25 EC patients and 31 HC controls. There were 49 differentially expressed with p-value < 0.01 (i.e., above the dotted line). b Feature vectors forming the best panel to discriminate two different groups were determined by the random forest algorithm. Six miRNAs (miR-106b-5p, miR-107, miR-15a-5p, miR-139-3p, miR-3615, and miR-574-3p) that indicated higher variable importance in the random forest tree were identified as candidate biomarkers. c Hierarchical clustering analysis of 6 candidate miRNAs roughly divided plasma samples into two distinct groups (EC vs HC). d ROC curves to evaluate the sensitivity and specificity of 6 candidate miRNAs to discriminate EC and HC subjects. e The expression levels of 6 candidate miRNAs in plasma exosomes of EC (n = 25) and HC (n = 31) subjects. f The expression levels of 6 candidate miRNAs in EC tumor tissues and matched para-carcinoma tissues from TCGA (n = 18). HC: healthy controls; PC, para-carcinoma tissues
Validation of diagnostic exomiRs by ddPCR in independent plasma samples
Next, we applied droplet digital PCR (ddPCR) to verify these six exomiRs in an independent validation cohort including 115 EC and 87 HC plasma samples. Two stable high-abundance miRNAs (let-7b-5p and miR-26a-5p) were selected as endogenous references, due to their high consistency in expression across all samples (Figure S4). The miR-106b-5p, miR-107, and miR-15a-5p were consistently upregulated in plasma-derived exosomes from EC compared with HC (Fig. 2a). Their upregulation was further verified in 32 pairs of endometrial tumor tissues and adjacent normal tissues using qRT-PCR (Figure S5). While the other three exomiRs were not pursued because of no significant expression changes between EC and HC groups in the validation set or failure of primer design in ddPCR.
Fig. 2.
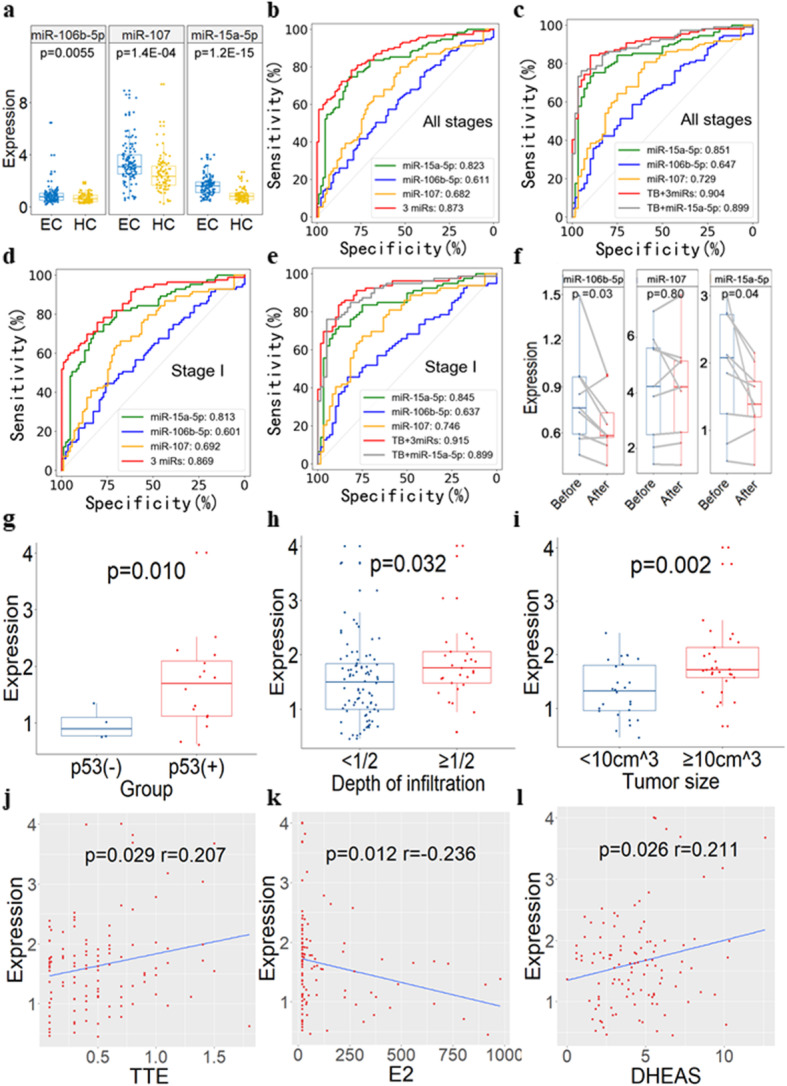
ddPCR validation of plasma exosmal miR-15a-5p, miR-106b-5p and miR-107 as diagnostic markers in 202 independent plasma samples. a The expression level of exosomal miR-15a-5p, miR-106b-5p, and miR-107 measured by ddPCR in independent validation samples. b ROC curves to validate the discrimination efficiency (for all EC with different stages vs HC) of exosomal miR-15a-5p, miR-106b-5p, miR-107, and their combinations (AUC = 0.832). n = 202. c ROC curves to validate the discrimination efficiency (for all EC with different stages vs HC) of exosomal miR-15a-5p, miR-106b-5p, miR-107, tumor biomarkers (TB), and their combinations (AUC = 0.885). CEA and CA125 were analyzed in TB analysis. n = 169. d ROC curves to validate the discrimination efficiency (for stage I EC vs HC) of exosomal miR-15a-5p, miR-106b-5p, miR-107, and their combinations (AUC = 0.815). Only EC patients with stage I in validation samples were analyzed for ROC curves. n = 170. e ROC curves to validate the discrimination efficiency (for stage I EC vs HC) of exosomal miR-15a-5p, miR-106b-5p, miR-107, TB (i.e., CEA and CA125), and their combinations (AUC = 0.875). n = 139. f The expression level of plasma-derived exosomal miRNAs from the same EC patients (n = 12) before and after surgery quantified by ddPCR. g-l The relationship between exosomal miR-15a-5p and clinical manifestations. Plasma-derived exosomal miR-15a-5p is differentially expressed between EC patients with p53 positive and negative staining (g), EC patients with depth of muscular infiltration < 1/2 and ≥ 1/2 (h), and EC patients with tumor size < 10 cm3 and ≥ 10 cm3 (i). Exosomal miR-15a-5p is positively correlated with TTE (j) and DHEAS (k), while is negatively correlated with E2 (l). TTE: testosterone; DHEAS: dehydroepiandrosterone sulfate; E2: estradiol
The average AUC of these three verified exomiRs was 0.705, ranging from 0.611 to 0.823, in the independent plasma samples. The combination of these exomiRs yielded a much higher AUC than tumor biomarkers (TB, including CEA and CA125) alone (0.873 vs. 0.736) (Figs. 2b and S6). Integration of these exomiRs into routine TB test models significantly improved the diagnostic performance, yielding an AUC of 0.904 (Fig. 2c). To evaluate the efficiency of these exomiRs in the early detection of EC, we specifically analyzed stage I EC patients in the independent plasma samples. Interestingly, the three exomiRs performed reasonably well with an average AUC of 0.702, ranging from 0.601 to 0.813. The AUC of the combined three exomiRs achieved 0.869 (Fig. 2d), while the AUC of TB alone is 0.760 (Figure S6). Integrating these exomiRs into routine TB test models yielded an AUC of 0.915 (Fig. 2e). Particularly, miR-15a-5p yielded an AUC values of 0.823 and 0.813 to distinguish between EC and HC subjects, for all EC patients and stage I EC patients only, respectively. The integration of miR-15a-5p and TB could achieve a higher AUC of 0.899 (Fig. 2b-e).
We also assessed the expression changes of these diagnostic exomiRs after surgery by comparing plasma samples from same patients before and after surgery (n = 12). As expected, the expression of miR-15a-5p and miR-106b-5p tended to decrease in EC patients after tumor resection surgery (Fig. 2f), which may be due to the massive reduction of exosomes secreted by tumor cells. Furthermore, there was no significant difference in the exosomal expression of miR-15a-5p, miR-106b-5p, and miR-107 between types I and II of EC patients (p > 0.05) (Figure S7).
Taken together, we have identified and validated three plasma-derived exomiRs (miR-106b-5p, miR-107, and miR-15a-5p), which can be served as potential diagnostic biomarkers for both early- and late-stage EC patients.
Pathway enrichment analysis of diagnostic exomiRs
KEGG pathway enrichment analysis for target genes of the three diagnostic miRNAs were performed to investigate their potential functions involvement in EC (Figure S8). Among the 20 significant pathways (FDR < 0.05), most of them are cancer-related, such as TGF-beta, Hippo, MAPK, p53, FoxO, Wnt, mTOR, and ErbB signaling pathways. The other pathways are related to fatty acid metabolism and biosynthesis, whereas prolactin signaling, oocyte meiosis, and endocytosis pathways are known to be associated with exosome biogenesis and release [14]. These results suggested that these miRNAs can not only serve as diagnostic biomarkers, but also are potentially involved in various steps in EC carcinogenesis and progression.
Exosomal miR-15a-5p associated with clinicopathologic characteristics
We assessed the relationship between three diagnostic exomiRs and clinicopathologic characteristics in EC patients (Fig. 2g-i). The plasma-derived exosomal miR-15a-5p expression was significantly higher in patients with p53 positive than p53 negative staining (p = 0.010). The exosomal miR-15a-5p level increased with the increase of muscular infiltration depth in EC patients (p = 0.032). Patients with large tumor had higher exosomal miR-15a-5p expression compared with small tumor (p = 0.002). These results implies that exosomal miR-15a-5p expression is strongly predictive of the aggressiveness and p53 mutation status of EC tumors.
Exosomal miR-15a-5p was also associated with testosterone (TTE), dehydroepiandrosterone sulfate (DHEAS), and estradiol (E2) (p < 0.05). However, its expression was not associated with normal menstrual cycles (Figure S9). Given a positive association of elevated circulating levels of TTE and DHEAS with EC risk [15], exosomal miR-15a-5p may also be a valuable clinical indicator for EC patients.
Tissue specificity of miR-15a-5p expression
Finally, we analyzed miR-15a-5p expression in other cancer types, including cervical, breast, ovarian and lung cancer. The miR-15a-5p expression is much more abundant (7–19 times) in EC tumor tissues than that in the other cancer types (Figure S10A). Compared with adjacent tissues, the miR-15a-5p expression was increased to more than 7 times in EC tumor tissues, while the miR-15a-5p was either downregulated in tumor tissues or showed small difference between tumor and adjacent tissues or normal controls of the other cancer types (Figure S10B).
Conclusions
Our study identified plasma-derived exosomal miR-15a-5p as a valuable diagnostic biomarker for the early detection of EC. Compared with uterine apoxesis, blood extraction is more convenient and carries less risk of vaginal/uterine cervix infection. It has the potential to be incorporated into routine blood examinations for screening endometrial cancer in the general population. Further validation of miR-15a-5p in large sample sizes is warranted before the clinical use as a diagnostic biomarker. Functional investigation of these diagnostic exomiRs will help reveal the mechanisms that underlie the occurrence and development of EC.
Supplementary Information
Acknowledgments
We thank the Core Facility at Zhejiang University School of Medicine for providing technical support, Dr. Anna Williams and anonymous reviewers for reading and commenting on the manuscript.
Abbreviations
- AUC
Area under receiver operating characteristics curve
- CA125
Cancer antigen 125
- CEA
Carcinoembryonic antigen
- ddPCR
Droplet digital PCR
- DHEAS
Dehydroepiandrosterone sulfate
- E2
Estradiol
- EC
Endometrial cancer
- exomiRs
Exosomal miRNAs
- FDR
False discover rate
- FSH
Follicle stimulating hormone
- HC
Healthy controls
- LH
Luteinizing hormone
- miRNA
microRNA
- NTA
Nanoparticle tracking analysis
- P
Progesterone
- PRL
Prolactin
- qRT-PCR
Quantitative real-time PCR
- RPM
Reads Per Million mapped reads
- TB
Tumor biomarker
- TCGA
The Cancer Genome Atlas
- TTE
Testosterone
Authors’ contributions
YL, WL and PL considered and designed the study. Lanyun Zhou, WW and FW performed experiments. Lanyun Zhou and JH did data analyses. SY, BL, ZP, YM, SL, PS and WL collected plasma samples, tissue specimens and clinical data. BL reviewed HE stained slides and confirmed the diagnosis. MZ and Liyuan Zhou provided assistance in the data analyses. Lanyun Zhou, PL and YL wrote the manuscript. All of the authors discussed and commented the study. The author(s) read and approved the final manuscript.
Funding
This work has been supported in part by the Medical Health Science and Technology Key Project of Zhejiang Provincial Health Commission (WKJ-ZJ-2007), National Natural Science Foundation of China (82072857, 81772766, 81871864, and 81872112), National Key R&D Program of China (2019YFC1315700), Key Program of Zhejiang Provincial Natural Science Foundation of China (LZ20H160001), and the Key R&D Program of Zhejiang Province (2021C03126C and 2019C03010).
Availability of data and materials
All data generated during this study are included in this published article and its supplementary files. The sequencing data were deposited in the Genome Sequence Archive (GSA) under accession HRA000737 (http://bigd.big.ac.cn/gsa-human).
Declarations
Ethics approval and consent to participate
This study was reviewed and approved by the Ethnics Committees of Women’s Hospital of Zhejiang University School of Medicine (Hangzhou, China; ID: 20170142). The study was conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects. All samples have been collected and utilized following strict human subjects protection guidelines, written informed consent and IRB review of protocols.
Consent for publication
Not applicable.
Competing interests
No potential conflicts of interest were disclosed.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lanyun Zhou, Wei Wang and Fenfen Wang contributed equally to this work.
Contributor Information
Pengyuan Liu, Email: pyliu@zju.edu.cn.
Weiguo Lu, Email: lbwg@zju.edu.cn.
Yan Lu, Email: yanlu76@zju.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Muinelo-Romay L, Casas-Arozamena C, Abal M. Liquid biopsy in endometrial Cancer: new opportunities for personalized oncology. Int J Mol Sci. 2018;19(8). 10.3390/ijms19082311. [DOI] [PMC free article] [PubMed]
- 3.Elsherbini A, Bieberich E. Ceramide and Exosomes: a novel target in Cancer biology and therapy. Adv Cancer Res. 2018;140:121–154. doi: 10.1016/bs.acr.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Wang Y, Wang Q, Liu Y, Bao W, Wu S. Exosomes in cancer: small transporters with big functions. Cancer Lett. 2018;435:55–65. doi: 10.1016/j.canlet.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 5.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136(2):169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falcone G, Felsani A, D'Agnano I. Signaling by exosomal microRNAs in cancer. J Exp Clin Cancer Res. 2015;34(1):32. doi: 10.1186/s13046-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in Cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks JJ, Thaxton CS, Mirkin CA. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci U S A. 2016;113(38):10655–10660. doi: 10.1073/pnas.1611596113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjornetro T, Redalen KR, Meltzer S, Thusyanthan NS, Samiappan R, Jegerschold C, Handeland KR, Ree AH. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles. 2019;8(1):1567219. doi: 10.1080/20013078.2019.1567219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, Su M, Pan H, Shen L, Xie D, Xie C. Evaluation of tumor-derived Exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung Cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 13.Zheng M, Hou L, Ma Y, Zhou L, Wang F, Cheng B, Wang W, Lu B, Liu P, Lu W, Lu Y. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol Cancer. 2019;18(1):76. doi: 10.1186/s12943-019-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito K, Miki Y, Suzuki T, McNamara KM, Sasano H. In situ androgen and estrogen biosynthesis in endometrial cancer: focus on androgen actions and intratumoral production. Endocr Relat Cancer. 2016;23(7):R323–R335. doi: 10.1530/ERC-15-0470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article and its supplementary files. The sequencing data were deposited in the Genome Sequence Archive (GSA) under accession HRA000737 (http://bigd.big.ac.cn/gsa-human).


