Abstract
The translocator protein (TSPO), previously known as the peripheral benzodiazepine receptor (PBR), is a membrane protein located on the outer mitochondrial membrane. Experimentally-derived structures of mouse TSPO (mTSPO) and its homologs from bacterial species have been determined by NMR spectroscopy and X-ray crystallography, respectively. These structures and ligand interactions within the TSPO binding pocket display distinct differences. Here, we leverage experimental and computational studies to derive a unified structural model of mTSPO in the presence and absence of the TSPO ligand, PK11195, and study the effects of DPC detergent micelles on the TSPO structure and ligand binding. From this work, we conclude that that the lipid-mimetic system used to solubilize mTSPO for NMR studies thermodynamically destabilizes the protein, introduces structural perturbations, and alters the characteristics of ligand binding. Furthermore, we used Rosetta to construct a unified mTSPO model that reconciles deviating features of the mammalian and bacterial TSPO. These deviating features are likely a consequence of the detergent system used for structure determination of mTSPO by NMR. The unified mTSPO model agrees with available experimental NMR data, appears to be physically realistic (i.e. thermodynamically not frustrated as judged by the Rosetta energy function), and simultaneously shares the structural features observed in sequence-conserved regions of the bacterial proteins. Finally, we identified the binding site for an imaging ligand VUIIS8310 that is currently positioned for clinical translation using NMR spectroscopy and propose a computational model of the VUIIS8310-mTSPO complex.
Keywords: Translocator protein (TSPO), NMR spectroscopy, Rosetta, Homology modeling, Protein folding, Ligand docking
Introduction
The 18-kDa TSPO, previously known as the peripheral benzodiazepine receptor (PBR), is an evolutionarily conserved five transmembrane (TM) span alpha-helical protein located in the mitochondrial outer membrane (Fan et al. 2012; Braestrup and Squires 1977). Research on TSPO spans decades and ranges from basic biochemical analysis to high-affinity ligand development. It has been found that TSPO is ubiquitously expressed in all human tissues, but is especially elevated in steroidogenic tissues and in certain pathological tissues such as cancer tumors and inflamed brain matter (Batarseh and Papadopoulos 2010; Ching et al. 2012). In vivo imaging suggests that TSPO is a promising target for cancer and is also a potential biomarker and therapeutic target for central nervous system (CNS) disorders (Galiegue et al. 2003; Fafalios et al. 2009). There is evidence that the majority of TSPO ligands interact with the loop between helices I and II, while cholesterol binding is mediated by the C-terminal cholesterol recognition/interaction amino acid consensus (CRAC) motif (Jamin et al. 2005; Gavish et al. 1999).
TSPO has been proposed to be a component of the mitochondrial permeability transition pore
TSPO, together with the voltage dependent anion channel (VDAC) and adenine nucleotide translocase (ANT), is a component of the mitochondrial permeability transition (MPT) pore during apoptosis (McEnery et al. 1992; Veenman and Gavish 2012). Moreover, TSPO-mediated cholesterol translocation has been proposed to be the rate-limiting step in progesterone synthesis in mitochondria, where cholesterol is further metabolized to steroid hormones (Lacapère and Papadopoulos 2003). Several studies have been conducted to probe the pharmacological or biochemical properties of TSPO involvement in hormone metabolism and apoptosis (Li and Papadopoulos 1998; Taketani et al. 1995; Verma et al. 1987). However, a number of recent in vitro and in vivo TSPO genetic knockout studies have challenged the paradigm that the function of TSPO plays a critical role in biological processes (Morohaku et al. 2014; Tu et al. 2014; Tu et al. 2015; Sileikyte et al. 2014). For example, it was reported that MPT pores still form in TSPO gene knockout mice (Sileikyte et al. 2014), where additional, possibly compensatory factors such as the F1F0-ATP synthase were found in the MPT pore complex (Giorgio et al. 2013). Moreover, some putative TSPO ligands were found to also inhibit F1F0-ATP synthase-mediated MPT (Cleary et al. 2007). Conditional or global knockouts of TSPO produced no apparent changes in viability or steroid hormone synthesis in mice (Morohaku et al. 2014; Tu et al. 2014), conflicting with the previous report that TSPO knockouts are embryonically lethal (although it must be noted that the methodology of generating TSPO null models were different). In vitro knockout of TSPO by siRNA also failed to display inhibition of the steroidogenic pathway (Tu et al. 2014). One report also suggested that the effect of early generation TSPO ligands on steroidogenesis might be the result of alteration of membrane properties by ligands or other off-target effects (Hatty et al. 2014). TSPO is highly expressed in activated microglia, and has been proposed as a biomarker of neuroinflammation (Liu et al. 2014). However, using the TSPO global knockout mice, it was subsequently demonstrated that activation of microglia after neuronal injury does not require TSPO, though microglia isolated from TSPO knockouts have altered oxygen consumption and ATP synthesis rates (Banati et al. 2014). At this point, it seems likely that compensating mechanisms exist in mitochondria for loss or malfunction of TSPO. Therefore, it seems possible that TSPO’s exact function in biological processes remains to be elucidated. Nevertheless, TSPO remains an ancient, well-conserved protein that is highly expressed under both physiological and disease states for which unambiguous structural insight is desirable.
TSPO is a target for imaging ligands
Despite the controversies surrounding TSPO function, TSPO has become a target for cancer imaging via positron emission tomography (PET). TSPO is overexpressed in several cancer types, including glioma (Black et al. 1989; Starostarubinstein et al. 1987), breast (Hardwick et al. 1999; Carmel et al. 1999), and colorectal (Deane et al. 2007; Maaser et al. 2001, 2002) cancers. TSPO overexpression in cancer cells has also been linked with disease progression and prognosis in patients with colorectal (Deane et al. 2007; Maaser et al. 2001, 2002), breast (Hardwick et al. 1999; Carmel et al. 1999), and brain (Black et al. 1989; Junck et al. 1989) cancers. Additionally, elevated TSPO levels appear to be associated with metastatic potential in breast and colorectal cancer cells (Hardwick et al. 1999, 2001; Han et al. 2003). Thus, TSPO is an important prognostic biomarker in oncology and is the target for tumor-selective TSPO PET ligands for cancer imaging. Second generation PET ligands derived from pyrazolopyrimidine such as [18F] DPA-714 (Tang et al. 2012) and [18F]VUIIS1008 (Tang et al. 2013, 2014) are used to visualize TSPO expression in tumors in preclinical settings. Recently, a new PET ligand 7-chloro-N,N,5-trimethyl-4-oxo-3(6-[F]fluoropyridin-2-yl)-3,5-dihydro-4H-pyridazino[4,5-b]indole-1-acetamide (VUIIS8310) was developed as a potential PET tracer for cancer imaging (Cheung et al. 2014). The structural determinants of the VUIIS8310 interaction with TSPO are unknown.
The fold of TSPO is conserved between mammalian and bacterial homologs
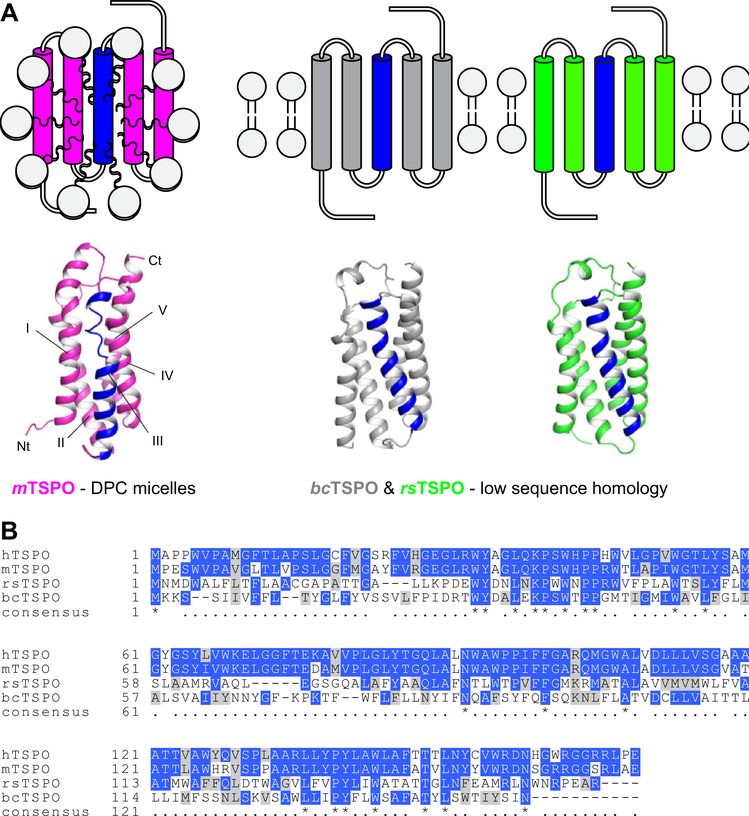
The current understanding of the TSPO structure at a molecular level stems from the solution NMR structure of mouse TSPO (mTSPONMR) (Jaremko et al. 2014, 2015) and two crystal structures of bacterial homologs from Rhodobacter sphaeroides (rsTSPOX-Ray) and Bacillus cereus (bcTSPOX-Ray) (Li et al. 2015; Guo et al. 2015) (Fig. 1A). mTSPONMR was determined in dodecylphosphocholine (DPC) detergent micelles under conditions where it was complexed with the imaging ligand PK11195 (Fig. 1A). The crystal structures of the two bacterial TSPO homologs were determined following lipidic cubic phase crystallization (Fig. 1A). All three structures share the same fold, despite bacterial TSPO having less than 30% sequence identity to human TSPO (hTSPO) (Fig. 1B). In these structures, the CRAC motif faces the lipid environment away from the dimerization interface, complicating the hypothesis that TSPO serves as a cholesterol transporter.
Fig. 1.
Available TSPO structures have limitations as templates for human TSPO. A The structure of mouse TSPO (mTSPO) was determined by NMR in DPC micelles and conflicts with structural features seen in the crystal structures of TSPO from Rhodobacter sphaeroides (rsTSPO) and Bacillus cereus (bcTSPO), especially in helix III (colored blue). B Multiple sequence alignment of mammalian and bacterial TSPOs [generated with Clustal Omega (Larkin et al. 2007)]. Identical and similar residues are colored blue and gray, respectively. The sequence identity of rsTSPO and bcTSPO to hTSPO is as low as 30% and 25%, respectively
Ligand conformations observed in experimentally-determined structures differ
PK11195 co-crystalized with bcTSPO displays a different binding mode when compared to PK11195 positioned in the mTSPO structure determined by NMR spectroscopy (see Figure S1A + B). The ligand center-of-mass in bcTSPO is shifted 3Å away from that in mTSPO (after structural alignment of both proteins) and the chlorophenyl-N-methylisoquinoline group is rotated by 150° (compare with Figure S1C) while the position of the butanyl-carboxamide group between helix II and IV is almost unchanged. In bcTSPOX-Ray, the methylisoquinoline ring of PK11195 establishes π–π stacking interactions with F90 and W51 and the ligand’s carbonyl group makes polar contacts with the side-chains of W51 and W138 (Li et al. 2015; Guo et al. 2015) (Figure S1A). In contrast, in wild-type and in the A147T mutant of mTSPONMR, PK11195 is not engaged in polar contacts but the ligand binding pocket is lined with several hydrophobic residues: A23, V26, L49, I52, W95, W107, A110, L114, W143, F146, A147 and L150 (Jaremko et al. 2014, 2015) (Figure S1B). This leads to increased ligand burial in mTSPO (the solvent-accessible surface area of PK11195 is ~ 640 Å2 compared to ~ 710 Å2 in bcTSPO) and could explain the higher affinity of PK11195 to mTSPO which was reported to be a thousand-fold higher than the affinity to bcTSPO (Frison et al. 2017).
TSPO exists in an equilibrium of oligomeric states in biological membrane
While the mTSPONMR structure is monomeric in detergent systems, there are reports indicating that a fraction of mTSPO exists as oligomers in lipid bilayers (Teboul et al. 2012; Papadopoulos et al. 1994; Jaipuria et al. 2017). Bacterial TSPOs are homodimers under crystallization conditions, but different dimerization interfaces were observed. For bcTSPOX-Ray, helices I and II contribute to the main dimerization interface. The primary dimer interface for rsTSPOX-Ray is formed by helices I and III. mTSPO reconstituted in a lipid bilayer is shown to be in an equilibrium state of dimer and monomer mediated by the GxxxG motif on helix III, as demonstrated by solid state NMR studies (Jaipuria et al. 2017). Addition of cholesterol shifts the equilibrium towards the monomeric form and additionally induces structural changes of mTSPO distal to the cholesterol binding site.
An article from the original authors of the mTSPONMR study describes the conformational state of apo-mTSPO (Jaremko et al. 2015). In their findings, the apo-mTSPO NMR spectrum is not completely assigned and revealed pico- to nanosecond backbone motion of mTSPO in the absence of PK11195. Detergents of different acyl-chain length were used to demonstrate the conformational exchange is independent of the specific detergent system used. It is also shown that apo-mTSPO lacks tertiary interactions resulting in a loss in stability and local unfolding around the ligand-binding site (Jaremko et al. 2015). Even for the ligand-bound folded form of mTSPO, the high temperature at which NMR experiments were conducted and the lack of ligand titration data validating specific stoichiometric binding prompts questions regarding the structure’s physiological relevance. Understanding the conformational plasticity of mTSPO in a physiologically-relevant membrane mimetic is critical for reliably investigating the binding of PET imaging ligands to mTSPO with NMR spectroscopy.
Structural models for human TSPO (hTSPO) need additional experimental and computational validation before they can be employed to assist development of improved imaging agents and potential therapeutics
It is well established that only very high-accuracy structures can reliably be used for in silico ligand binding design efforts. Current models for hTSPO are based on the three experimental structures (mouse and two bacterial) (Fig. 1), which are unlikely to fulfill this criterion. The available structural snapshots for TSPO homologs contradict each other in important aspects, such that they cannot be expected to serve as reliable templates for ligand docking to hTSPO. However, these three experimental structures of TSPO do provide a critical starting point for understanding possible biological roles of the protein from a molecular level and serve as crucial stepping stones to a reliable integrative structural model for the human ortholog. In this study, we experimentally validate that mTSPO’s ligand binding properties under the originally-reported NMR conditions reflect native-like behavior. We also use integrated structural biology to derive a unified structural model for mTSPO based on all three experimentally determined structures in which the various non-physiological frustrations and contradictions between the original structures are resolved. The resulting integrative model for mTSPO can be expected to provide a reliable guide for future investigation and a template for the human TSPO ortholog. We also report the molecular interaction between the novel PET ligand VUIIS8310 and mTSPO based on computational docking and NMR experiments.
Materials and methods
Protein expression and purification
15N-labeled mTSPO was expressed in BL21 (DE3) E. coli cells in M9 minimal media, solubilized, and purified with DPC detergent, as adapted from previous publications (Jaremko et al. 2014; Murail et al. 2008). In short, the protein was induced at an OD600 of 0.8 with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12 h at 25 °C. The membrane-targeted mTSPO fraction was collected using ultracentrifugation after lysing cells by sonication. 1% (w/v) DPC was used to solubilize the membrane faction for Ni–NTA affinity chromatography. A concentration of 0.1% (w/v) of DPC was maintained throughout purification and mTSPO was eluted from the Ni resin using 300 mM imidazole. The purity of the protein was estimated on a SDS-PAGE gel stained with Coomassie blue to be > 95%. In cases when N-dodecyl β-D-maltoside (DDM) was used, 1% DDM (w/v) was used to solubilize the membrane fraction of mTSPO and 0.1% DDM (w/v) was used during the purification process. All final samples were buffer exchanged into 25 mM MES buffer, pH 6.2, and 100 mM NaCl for downstream NMR and fluorescence experiments.
Measurement of PK11195 binding to mTSPO by intrinsic tryptophan fluorescence
Quenching of the intrinsic fluorescence of tryptophan residues was used to study the ligand binding properties of PK11195 to mTSPO (Korkhov et al. 2010) under different experimental conditions. The quenching of protein fluorescence with the ligand PK11195 and 0.5 μM mTSPO reconstituted in either DPC or DDM micelles was monitored over a concentration range at both room temperature and 42 °C. For each titration point, TSPO tryptophan residues were excited at 290 nm and the fluorescence emission maximum was measured at 350 nm (spectral scan from 300 nm to 400 nm) on a Horiba Jobin–Yvon Fluoromax-3™ spectrofluorimeter. Control experiments were performed to correct for any absorption and emission contributions from the ligand, solvent and buffer. Since mTSPO contains multiple buried and solvent exposed tryptophan residues, the area under each emission spectrum was integrated to account for all tryptophan residues and a percentage fluorescence change for each concentration point was calculated. The binding curve was obtained by plotting the percent maximal change [i.e. %Change = (F − Fi)/(Ff − Fi)] against total ligand concentration [L] represented as the ligand-detergent molar ratio in Prism 6©, where F is the fluorescence at a specific titration point, Fi is initial fluorescence in the absence of ligand and Ff is the final fluorescence at the highest concentration of ligand. The solvent for membrane proteins is its lipid membrane mimetic (i.e. detergent micelles). Since fluorescence and NMR experiments operate at different protein and detergent concentrations, we used the ligand-detergent molar ratio (i.e. moles of ligand per moles of detergent that are in the micelle state) to allow for direct comparison of titration data across different conditions and spectroscopic techniques. It was seen that the quenching of tryptophan fluorescence was biphasic, with low and high concentration transition phases. The following equations were used to calculate the binding affinity for each transition (Tr1 and Tr2) assuming a 2:1 binding model without cooperativity between the two binding sites and taking into consideration ligand depletion ([Lfree] ≠ [Ltotal]).
| (1) |
| (2) |
| (3) |
Here [L] is the total ligand concentration, [P] is the total concentration of purified mTSPO protein (fixed at 0.5 μM in the experiment and during the fitting procedure), and are the apparent dissociation constant for the transition sites, %Change represents the calculated percentage quenching at each titration point and Offs (offset) allows a fit to go through zero. PK11195 was dissolved in DMSO as a stock solution and diluted with DMSO or buffer for fluorescence measurements. One representative binding curve is shown in each panel in Fig. 2, while the and values are reported as the average of three replicate measurements fit with Eq. 3 in Table S1.
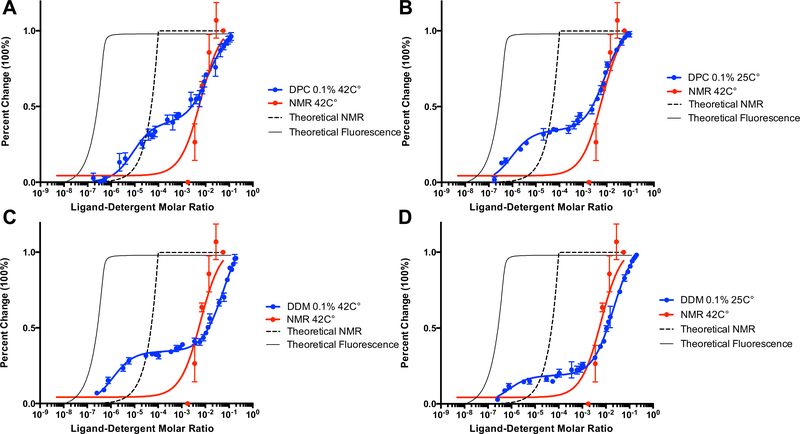
Fig. 2.
Characterization of PK11195 binding to mTSPO under different detergent and temperature conditions by intrinsic tryptophan fluorescence. The x-axis is plotted as log of the ligand-detergent molar ratio and the y-axis is plotted as percent maximal change to allow for direct comparison between datasets from different techniques. The NMR titration curve for PK11195 performed at 42 °C in the presence of 2% w/v DPC is plotted in all panels for comparison against fluorescence data. The theoretical titration curve of the reported binding affinity in the literature for binding of PK11195 to mTSPO is also plotted for both fluorescence (solid line) and NMR (dashed line). A Fluorescence quenching of mTSPO by PK11195 in 0.1% (w/v) DPC at 42 °C. B Fluorescence quenching of mTSPO by PK11195 in 0.1% (w/v) DPC at 25 °C. C Fluorescence quenching of mTSPO by PK11195 in 0.1% (w/v) DDM at 42 °C. D Fluorescence quenching of mTSPO by PK11195 in 0.1% (w/v) DDM at 25 °C. The average and standard deviations are represented as points and bars, respectively, and reflect at least three independent experiments
Measurement of PK11195 binding to mTSPO by NMR
2D 1H–15N transverse relaxation-optimized spectroscopy (TROSY) heteronuclear single quantum correlation (HSQC) spectra of mTSPO were recorded on an 800 MHz NMR spectrometer at 42 °C with 0.1 mM mTSPO in 25 mM MES and 100 mM NaCl buffer at pH 6.2 with 2% DPC (w/v). For the NMR titration experiments, 0.1 mM mTSPO was titrated with PK11195 over a range of concentrations (0.1 mM, 0.2 mM, 0.4 mM, 0.8 mM, 1.6 mM, 3.2 mM). The ligand concentration [L] was converted to mole fraction for data fitting. The spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed using Sparky (Lee et al. 2015). Assignment of the majority of peaks in the 1H–15N HSQC spectrum of mTSPO with PK11195 could be transferred from previous reports (Jaremko et al. 2014), as deposited in the Biological Magnetic Resonance Data Bank (BMRB ID: 19608), and only minimal peak position differences compared to the published assignment were noticed.
Peak intensities in the 1H–15N TROSY-HSQC spectrum of mTSPO-PK11195 were monitored during ligand titration. The spectral changes upon addition of PK11195 were indicative of a typical slow-exchange process. Thus, the binding curve was obtained by plotting the percent change in peak intensity [i.e. %Change = (I − Ii)/(If − Ii)] against [L] as the ligand-detergent molar ratio, where I is the signal intensity at a specific titration point, Ii is the signal intensity in the absence of ligand and If is the final signal intensity at the highest concentration of ligand. The NMR titration experiment showed a one phase transition and was fit with Eq. 4 for a one-site binding event and taking into consideration ligand depletion ([Lfree] ≠ [Ltotal]).
| (4) |
The binding curve was then calculated by plotting the average of the %Change values of ten residues (W5, Y34, W42, W47, G72, F100, W107, S116, V118, W155) that are part of TSPO’s ligand binding pocket.
Measurement of VUIIS8310-induced NMR chemical shift changes in mTSPO
2D 1H–15N TROSY-HSQC spectra of mTSPO in the presence and absence of 5 mM VUIIS8310 were recorded on an 800 MHz NMR spectrometer at 42 °C with 0.1 mM mTSPO in 25 mM MES and 100 mM NaCl buffer at pH 6.2 with 2% DPC (w/v). The spectrum of mTSPO with VUIIS8310 was then compared to the previously recorded mTSPO-PK11195 spectrum in order to infer the VUIIS8310 binding mode with respect to the known location of PK11195, as detailed below. The weighted chemical shift change, ΔCS, was calculated relative to the 1H–15N TROSY-HSQC spectrum of mTSPO in the presence of 5 mM PK11195 using Eq. 5:
| (5) |
Amino acid residues whose NMR signal in the mTSPO-VUIIS8310 spectrum could be correlated with the corresponding signal in the mTSPO-PK11195 spectrum and for which ΔCS was > 0.5 ppm were considered as having a significant ΔCS. Residues whose signal disappeared in the mTSPO-VUIIS8310 spectrum were also marked as having a significant ΔCS. Residues with a ΔCS value between 0.1 ppm and 0.5 ppm were considered moderately perturbed. The resulting ΔCS mapping was used to identify the potential VUIIS8310 binding site on mTSPO.
RosettaCM modeling of mTSPO
Model 1 from the mTSPONMR structural ensemble (PDB 2GMY), the bcTSPOX-ray crystal structure (PDB 4RYI) and the rsTSPOX-Ray crystal structure (PDB 4UC2) were energy-minimized using the Rosetta FastRelax protocol (Conway et al. 2014) running alternating cycles of bond and angle optimization and side-chain repacking to produce mTSPONMR-opt, bcTSPOX-Ray-opt and rsTSPOX-Ray-opt models. The Rosetta comparative modeling (RosettaCM) protocol (Song et al. 2013) was then used to hybridize the five best energy-minimized models of either bcTSPOX-Ray-opt, rsTSPOX-Ray-opt, or a combination of bcTSPOX-Ray-opt, rsTSPOX-Ray-opt and mTSPONMR-opt models generating 2000 models of mTSPORosettaCM. The best 1% of models by Rosetta energy from each RosettaCM calculation were subjected to an additional round of energy minimization with Rosetta FastRelax. Final models were analyzed for convergence by calculating their Cα-atom root mean square distance (Cα-RMSD) relative to the lowest-energy mTSPORosettaCM model and inspecting the funnel-likeness of the Rosetta energy vs. Cα-RMSD plot. The lowest-energy mTSPORosettaCM model was selected as the representative model for further analysis. Its structural comparison and RMSD calculation to the experimentally determined structures of mTSPONMR, bcTSPOX-ray and rsTSPOX-Ray was done using Dali pairwise structural alignment (Holm and Rosenström 2010). Chemical shift predictions for mTSPORosettaCM were carried out using SPARTA + (Shen and Bax 2010). NOE distance deviations of mTSPORosettaCM were calculated by comparison with the reported long-range (|i–j| ≥ 4 residues) experimental NOEs (BMRB ID: 19608) and evaluated on a per residue basis. Validity of the model’s backbone conformation at the three Pro–Pro sites in mTSPO was evaluated by comparison with the φ/ψ dihedral angle distribution of Pro–Pro motifs observed in a database of membrane proteins downloaded from the PDB. The φ/ψ angles at positions P ro44–Pro45, Pro96–Pro97 and Pro131–Pro132 of the 20 top-energy models of mTSPONMR-opt and mTSPORosettaCM were used for the analysis.
Computational ligand docking
The ligand PK11195 was computationally docked into the bcTSPOX-Ray-opt crystal structure, and mTSPONMR-opt and mTSPORosettaCM models using RosettaLigand (Meiler and Baker 2006). Compound VUIIS8310 (Cheung et al. 2014) was computationally docked into mTSPONMR-opt and mTSPORosettaCM models. Ligand conformer libraries were generated prior to docking using MOE (Molecular Operating Environment 2013) (Molecular Operating Environment; Chemical Computing Group Inc., Montreal, QC, Canada). In the case of PK11195, the ligand was initially placed in the binding pocket at a position inferred from the experimentally determined PK11195 coordinates. In each docking calculation, 1000 models were constructed using maximal translational and rotational perturbations of 5Å and 360°, respectively, as described previously (Meiler and Baker 2006; Gregory et al. 2013). In the case of docking VUIIS8310, the ligand was placed into the protein center and in proximity to residues with pronounced ΔCS values. A total of 2000 docking models of VUIIS8310 in complex with mTSPORosettaCM and mTSPONMR-opt were constructed using maximal translational and rotational sampling magnitudes of 5Å and 360°, respectively. mTSPO-ligand models were ranked based on their Rosetta-predicted binding energy using terms for van der Waals attractive and repulsive forces, electrostatic interactions, hydrogen bonding, solvation, and likelihood of particular side-chain conformations. The top-scoring docked models were visually inspected for the nature of protein–ligand interactions using PyMol (Schrodinger 2015), and for the agreement with NMR ΔCS data.
Results
mTSPO reconstituted in DPC micelles has an NMR spectrum typical for a partially unfolded state
mTSPO was recombinantly expressed and purified using DPC detergent micelles as the membrane mimetic, the same system that was used to determine the mTSPONMR structure. The purity of mTSPO was > 95% according to SDS-PAGE analysis (Figure S2A). The NMR spectrum of apo-mTSPO in DPC micelles showed very low peak dispersion similarly to what has been reported before (Jaremko et al. 2014) (Fig. 3, top left panel). The spectral dispersion is rather low for a helical membrane protein. For example, there are no backbone amide peaks that exhibit 1H chemical shifts higher than 8.8 ppm. The suggests that apo-mTSPO adopts only a partially folded state in DPC detergent micelles at 42 °C.
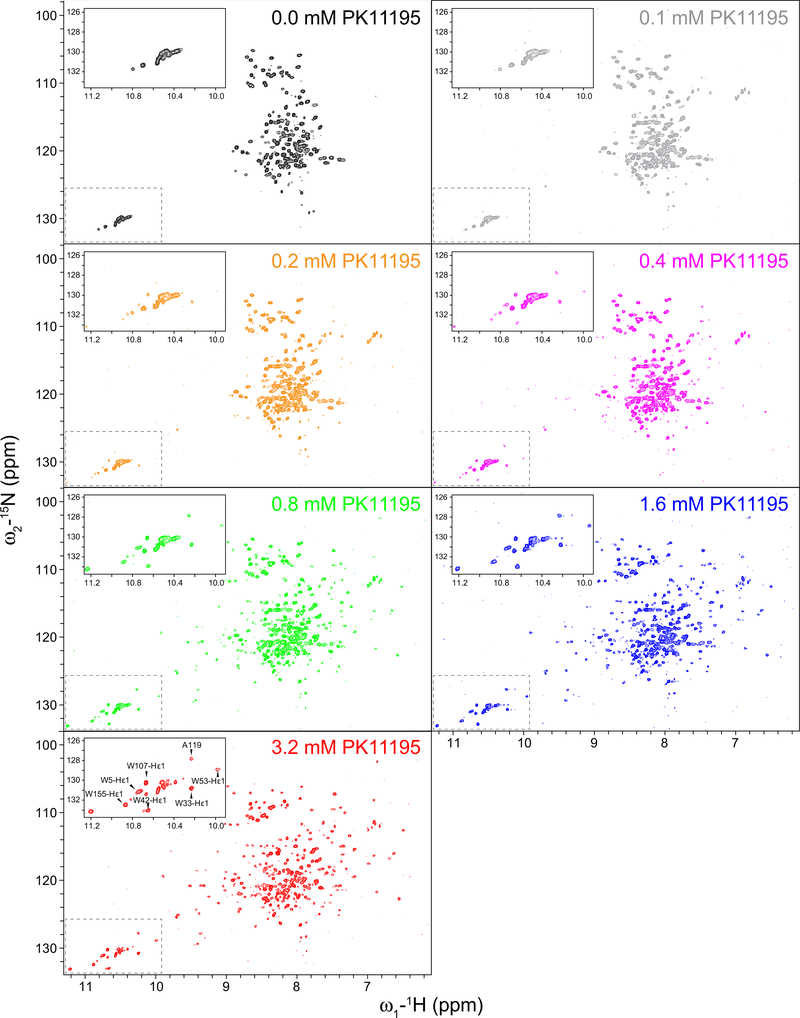
Fig. 3.
PK11195 binding to mTSPO reconstituted in DPC detergent micelles followed by NMR. Series of 1H–15N TROSY-HSQC NMR spectra of apo-mTSPO in the absence of PK11195 (black) and with increasing concentrations of PK11195. An enlarged view of the tryptophan sidechain region is shown in the upper left corner to signify peak changes upon the addition of ligand. The assignment of selected NMR peaks that appear upon addition of PK11195 is shown in the lower spectrum corresponding to a PK11195 concentration of 3.2 mM
PK11195 binding affinity to mTSPO is weaker in micelles compared to membrane preparations monitored by NMR spectroscopy
mTSPO was titrated with PK11195 under the conditions reported in the original NMR study to probe the effect of detergent on the binding affinity. Once saturation with PK11195 was reached, we were able to transfer the assignment for the majority of peaks in the 1H–15N TROSY-HSQC spectrum using the chemical shift assignment of PK11195-bound mTSPO (BMRB ID: 19608) (Fig. 3, Figure S5). It is striking that ligand binding dramatically enhances the dispersion of the NMR spectrum of TSPO (Fig. 3), strongly suggesting that the ligand-free protein is partially unfolded, but that ligand binding is coupled to the stabilization of the fully folded protein conformation.
The spectral changes observed during ligand titration were typical for a slow-exchange process on the NMR timescale. Peaks characteristic for the apo-mTSPO spectrum diminished with increasing ligand concentration while peaks associated with the PK11195-bound state surfaced in the HSQC spectrum (Fig. 3). The changes in peak intensity with increasing concentration of PK11195 showed a single transition as a response of ligand binding (red curves in Fig. 2). The lower limit of the apparent binding constant ( Kapp) determined from the NMR ligand titration binding curve was 5.7 × 10−3 (ligand-detergent molar ratio, Fig. 2, red line), while PK11195 was previously reported to bind mTSPO with nanomolar affinity (Lacapère and Papadopoulos 2003; Papadopoulos et al. 1994; Owen et al. 2012). A nanomolar binding affinity would be equivalent to an approximate mole fraction concentration of ~ 10−7 (calculated assuming a ligand to lipid/detergent ratio of 1 to 100). A theoretical NMR titration binding curve assuming such high nanomolar affinity and a 1:1 binding stoichiometry is plotted in Fig. 2. The resulting titration curve accounts for excess protein concentration (i.e. free ligand does not equal total ligand concentration) and compared to the experimental NMR titration curve, is right shifted. The relatively high apparent K d for PK11195, reflective of weak binding to mTSPO under NMR conditions relative to bilayer conditions (TSPO in proteoliposomes and human platelets at lower temperatures) is likely an attribute to the experimental temperature and the destabilizing effects of the DPC detergent micelles.
Intrinsic tryptophan fluorescence reveals the effects of detergent and temperature on the PK11195 binding affinity to mTSPO
To determine the effect of detergent concentration, detergent type and temperature on the binding affinity of PK11195 to mTSPO, an intrinsic tryptophan fluorescence quenching assay was used to complement the NMR binding studies. The ligand-induced quenching of intrinsic tryptophan fluorescence was used to monitor ligand binding (Fig. 2). Different concentrations of DPC were tested for the mTSPO-DPC detergent micelle system at room temperature to compare against the NMR mTSPO solution structure determined at 42 °C. mTSPO reconstituted in DDM detergent micelles (mTSPO-DDM) was also included to determine the effect of detergent type on the PK11195 binding affinity. For both detergent conditions, the fluorescence quenching was biphasic with a transition at low and high ligand concentration (Fig. 2 and Figure S3). Without prior assumption of possible non-specific binding, the data was fit to a 2:1 model for binding. For all samples of mTSPO-DPC at 42 °C, the high-affinity apparent binding constant for Tr1 was around ~ 10−6 mol fraction units and was independent of the DPC concentration (Figure S3A). The low-affinity apparent binding constant for Tr2 was determined to be 5.4·10−3 mol fraction at 0.4% DPC, 6.8·10−3 mol fraction at 0.2% DPC and 12.0·10−3 mol fraction at 0.1% DPC. This concentration phase in the fluorescence titration experiments correlated with the binding transition observed under NMR conditions (Fig. 2). Additionally, we plotted theoretical titration curves for the fluorescence quenching experiments. For comparison, mTSPO in a DPC detergent system showed a shifted binding equilibrium in contrast to a theoretical binding curve assuming nanomolar affinity of PK11195 to mTSPO. Thus, fluorescence-monitored titrations confirm the binding event observed by NMR, but also suggest some additional, high affinity, binding process to the mTSPO-micelle complex.
To assess temperature effects on the Tr1 and Tr2 transitions, we performed the same set of fluorescence experiments at room temperature for mTSPO in DPC micelles and in DDM micelles (Fig. 2 and S3). At room temperature, the value (i.e. low-affinity apparent binding constant) of mTSPO in DPC micelles appeared to increase (i.e. weaker affinity) slightly by half an order of magnitude relative to the binding affinity at 42 °C (Fig. 2, Table S1). The value (i.e. high-affinity apparent binding constant) displayed a decrease (i.e. higher affinity) at room temperature under the conditions of 0.1% DPC and 0.2% DPC, but was not significantly changed at 0.4% DPC. For mTSPO in DDM micelles at 42 °C, the value showed a two order of magnitude increase (i.e. weaker affinity) relative to all DPC conditions (Table S1, Figure S3), whereas the change in the high-affinity binding transition phase was negligible. Taken together, our data show that PK11195 binding to mTSPO is dependent both on temperature (specifically transition Tr1) and on the detergent system (most notably for transition Tr2).
Structural templates of mammalian TSPO in biologically relevant conformations are lacking
The available structural models of TSPOs obtained by NMR and X-ray crystallography have several limitations (Fig. 1): The mTSPONMR structure was determined under detergent conditions at high temperature where the energy landscape is substantially perturbed. Additionally, some features of the NMR structure conflict with conformations observed in sequence-conserved regions of bcTSPOX-Ray and rsTSPOX-Ray, specifically in helix III. At the same time, these bacterial homologs, although determined in a more native-like lipidic cubic phase environment, have low homology to mTSPO; the sequence identity is only 25% (bcTSPOX-ray) and 30% (rsTSPOX-ray). This low homology prevents bcTSPOX-Ray and rsTSPOX-Ray from being directly used to model structural interactions of small molecule probes, imaging agents or therapeutics to mammalian TSPO. Although, bcTSPOX-ray was co-crystallized with the same PK11195 ligand as mTSPONMR, less than 40% of the sequence-aligned residues in the binding pocket are identical, leading to changes in the ligand binding mode (compare with Figure S1).
An integrated comparative model of mTSPO is physically realistic and reconciles conflicting features from bcTSPOX-Ray, rsTSPOX-Ray, and mTSPONMR
Homology modeling and geometry optimization for all TSPO structural models was carried out in RosettaMembrane. This relaxed the detergent-determined mTSPO structure towards a conformation adjusted to a membrane bilayer and generated a unified model of hTSPO by comparative modeling from the mouse and bacterial homologs. In order for the modeling to capture the physical and chemical characteristics of the membrane environment, yet being computationally efficient for many of Rosetta’s Monte Carlo protocols, RosettaMembrane (Lazaridis 2003; Yarov-Yarovoy et al. 2006) introduces an implicit model of the membrane bilayer model with a layered structure. In the high-resolution mode, the membrane bilayer is modeled as fixed size slab with a hydrophobic core of 18 Å and transition region of 6 Å on either side (Barth et al. 2007). The RosettaMembrane score function combines physical and knowledge-based score terms that evaluate a given protein conformation (e.g. Lennard-Jones potential, electrical potential, Ramachandran potential) with a membrane-specific solvation term, a membrane-depth dependent environment term and a correction term for the strength of hydrogen bonds in the membrane. The solvation term is calculated with the IMM1 model(Lazaridis 2003) which is an extension of the EEF1 model (Lazaridis and Karplus 1999) for soluble proteins and very efficient to compute, even more than other implicit solvent models (e.g. generalized Born) usually applied in MD simulations. In the IMM1 model, the solvation free energy is modeled via Gaussian exclusion due to surrounding atoms.
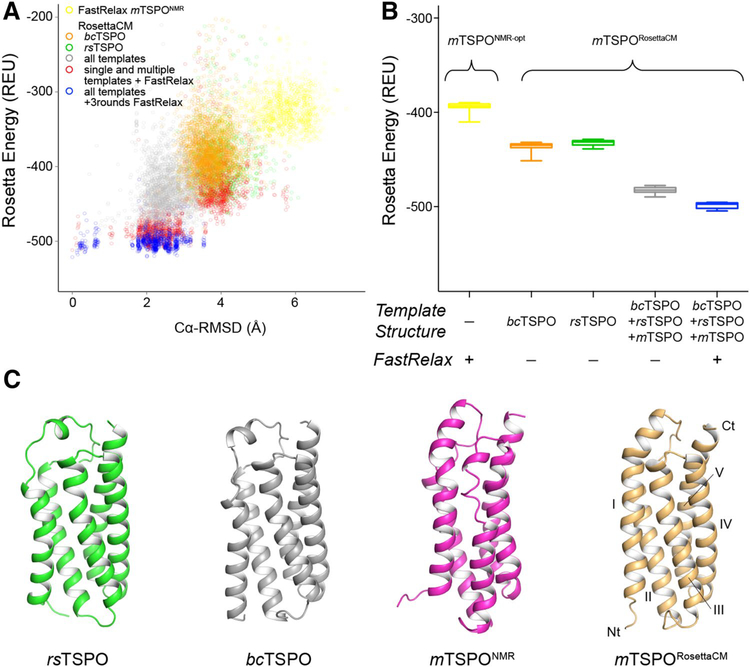
To assess how well-suited the available structures as templates of mammalian TSPO are and to combine their structural features, we decided to construct homology models of mTSPO, termed mTSPORosettaCM, using either single templates (bcTSPOX-Ray or rsTSPOX-Ray) or multiple templates (bcTSPOX-Ray, rsTSPOX-Ray, and mTSPONMR), which were hybridized with RosettaCM. Homology modeling used the multiple sequence alignment shown in Fig. 1B to infer the mTSPO structure from template coordinates. We also performed energy optimization on the mTSPONMR structure with the RosettaMembrane score function to computationally evaluate its energy landscape. The mTSPORosettaCM model produced by multiple template homology modeling had the lowest energy amongst other strategies after one round of RosettaCM and three rounds of energy optimization with Rosetta FastRelax (Fig. 4). The mTSPONMR-opt structural model had a higher energy compared to any of the single and multiple template homology models. The average Rosetta energy of models obtained from the different sampling strategies decreased in the following order: mTSPONMR-opt models > single template mTSPORosettaCM models > multiple template mTSPORosettaCM models (Fig. 4B). This result suggests that the NMR structure when inserted into a bilayer model of the biological membrane is in a high energy state, i.e. a physically unrealistic state with many local unfavorable interactions. Similarly, the bacterial templates failed to guide sampling towards low-energy conformations of mTSPO indicating their limited utility for homology modeling. However, when structural features from all three templates were combined using RosettaCM, the energy of mTSPO was seen to be optimized. The mTSPORosettaCM model obtained from multiple template modeling was considered the final mTSPORosettaCM model and used for further analysis. It shares the same global fold with the experimentally determined TSPO structures (Fig. 4C) and has a similar backbone conformation: mTSPONMR (Cα-RMSD = 4.1Å), bcTSPOX-Ray (Cα-RMSD = 3.3Å), rsTSPOX-Ray (Cα-RMSD = 3.5Å).
Fig. 4.
Rosetta homology modeling of mTSPO improves model energy over available structural templates. A Rosetta energy vs. Cα-RMSD plot for energy-minimized mTSPONMR-opt and mTSPORosettaCM models created with different sampling strategies. The RMSD was calculated relative to the final lowest-energy mTSPORosettaCM model after one round of RosettaCM and three rounds of energy minimization with FastRelax (Conway et al. 2014). The colors refer to the following sampling strategies: optimization of mTSPONMR with Rosetta FastRelax (yellow), RosettaCM based on bcTSPOX-Ray (orange), RosettaCM based on rsTSPOX-Ray (green), RosettaCM based on bcTSPOX-ray, rsTSPOX-ray and mTSPONMR (gray); additional energy minimization of single and multiple template mTSPORosettaCM models with FastRelax (red), three rounds of FastRelax on multiple template mTSPORosettaCM models (blue). B Box plot of the Rosetta energy of the top 20 models produced by each sampling strategy. The color scheme follows that of panel A. The relaxed mTSPORosettaCM model has improved energy over other Rosetta models. c Cartoon representation of experimental TSPO structures and the mTSPORosettaCM model. The flexible tail region (mTSPO residues 160–169, rsTSPO residues 151–157) is omitted for clarity. The N- and C-terminus and TM helix regions are labeled. The Cα-RMSD of mTSPORosettaCM relative to the experimental templates was calculated after sequence-independent structural alignment with DALI (Holm and Rosenström 2010): 4.1Å (mTSPONMR), 3.3Å (bcTSPOX-Ray) and 3.5Å (rsTSPOX-Ray)
The mTSPORosettaCM model combines features of experimentally determined structures and agrees with experimental NMR data
The mTSPORosettaCM model is energetically favored over the mTSPONMR structure in an implicit bilayer membrane. Globally, the model resembled the conformations observed in the bacterial proteins more closely. Hence, we wondered if this model is still generally consistent with the restraints obtained by NMR spectroscopy, keeping in mind that the NMR restraints were derived on the thermodynamically destabilized micelle conformation of mTSPO whereas the homology model was constructed in an implicit model membrane. Our expectations were that the overall protein fold as well as secondary structure are conserved between mTSPO in a micelle and in a membrane. On the other hand, we expected slightly larger deviations in the tertiary structure due to changes in the rotation and packing of TM helix segments.
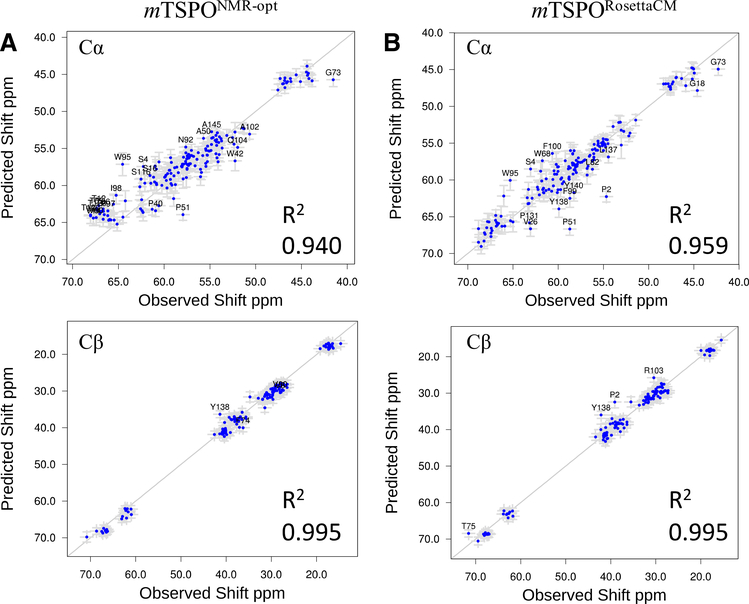
As sensitive probes of protein secondary structure we calculated 13Cα and 13Cβ chemical shifts from our mTSPORosettaCM model and optimized mTSPONMR-opt model using SPARTA + (Shen and Bax 2010), and compared those to the experimental chemical shifts of mTSPO retrieved from the BMRB deposition 19608 (see Fig. 5). Both models agree well with the experimental chemical shift data indicating that many local structural features are consistent between both models and with the mTSPONMR structure. Note that chemical shifts were not used as restraints in the RosettaCM calculation, but only employed in the form of backbone segments with defined φ/ψ angles taken from the mTSPONMR template.
Fig. 5.
Back-calculated chemical shift of the mTSPORosettaCM model correlates with observed chemical shift from NMR data. A Back-calculated chemical shifts for 13Cα (top) and 13Cβ (bottom) from experimental mTSPONMR-opt structures correlates with the observed NMR data. B Back-calculated chemical shift for 13Cα (top) and 13Cβ (bottom) from mTSPORosettaCM model correlates with the observed NMR data. R2 values were calculated and noted in each case. Error bars represent the predicted shift error
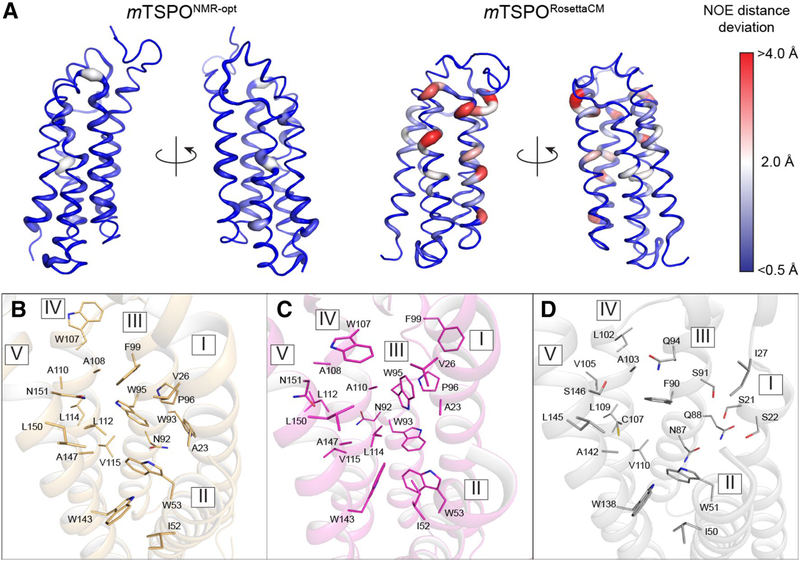
As a measure of global protein structure similarity, we calculated NOE distance deviations of our mTSPORosettaCM and mTSPONMR-opt models to the experimentally determined long-range (|i–j| ≥ 4 residues) NOE restraints (see Figs. 6 and S4A) (Jaremko et al. 2014). Figure 5A displays the per-residue distance deviations mapped onto the mTSPORosettaCM and mTSPONMR-opt models. The energy-minimized mTSPONMR-opt model showed very small deviations, below 1Å for all residues except for W53, Y62, Q104. The mTSPORosettaCM model differed more strongly from the experimental NOE data: 16 residues had an average distance deviation of more than 2Å. Those residues are located in helices III, IV, and V which adopt alternative backbone and side chain conformations in our mTSPORosettaCM model. Helix III is a continuous α-helix and three turns longer than in the mTSPONMR structure where it is partially unstructured. Helix IV undergoes a one-residue phase shift compared to mTSPONMR which accounts for the increased distance deviations, and leads to a change in the ligand binding pocket configuration (see below). The majority of residues with NOE deviations are located in the upper part of the protein close to the PK11195 binding site, while other parts of mTSPORosettaCM agree well with the majority of long-range NOE restraints. We interpret this finding as consistent with our experimental observations: even with ligand saturation the structure of mTSPO is somewhat distorted in micelles. Moreover, the PK11195 binding pocket only fully forms at ligand concentrations sufficiently high to induce protein folding. The induced tertiary structure in this region is the most stable conformation in micelles but deviates from the low-energy conformation in a membrane bilayer.
Fig. 6.
The mTSPONMR-opt model and mTSPORosettaCM model generally agree with experimentally measured long-range NOEs (Jaremko et al. 2014), but differ in ligand binding pocket configuration. A Per residue distance deviation between experimental NOE restraints and model-measured distances. Left: mTSPONMR-opt, Right: mTSPORosettaCM. The magnitude of the deviation is represented by the width of the ribbon diagram and color coded (blue: < 0.5Å, white: 2.0Å, red: > 4.0Å). Panels B-D display the ligand binding pockets of B mTSPORosettaCM, C mTSPONMR and D bcTSPOX-ray. Binding site residues are depicted as sticks and labeled by their one-letter amino acid code and residue number
The ligand binding pocket of mTSPORosettaCM shares similarity to that of the bacterial homologs
The ligand binding pocket of the mTSPORosettaCM model is displayed in Fig. 6B and is compared to that one of mTSPONMR in Fig. 6C and to bcTSPOX-ray in Fig. 6D. The mTSPORosettaCM ligand binding pocket has lower structural similarity to mTSPONMR than it has to bcTSPOX-ray where sequence-conserved residues are also spatially aligned. For example, in addition to residues W53 and W143 (W51 and W138 in bcTSPO) on helix II and V, two other conserved residues, N92 and A108 (N87 and A103 in bcTSPO), occupy identical positions in mTSPORosettaCM and bcTSPOX-ray and are facing the same side on helix III and IV. In contrast, those residues are rotated out of the binding pocket in mTSPONMR due to differences in helix conformation and phase. Another bulky residue, W93, is solvent-exposed in mTSPORosettaCM but pointing towards the protein core in mTSPONMR which alters the chemical nature of its ligand binding pocket.
The Pro–Pro motif conformation in the TM region is reasonable based on database values
There are three Pro–Pro motifs in mTSPO, at positions Pro44–Pro45, Pro96–Pro97 and Pro131–Pro132, which border TM regions II, III and IV. The Pro45–Pro45 site is sequence-conserved between mammalian and bacterial TSPOs while the other two sites are not (compare with Fig. 1B). In the mTSPONMR structure, the Pro96–Pro97 site is part of a region in helix III that lacks regular secondary structure whereas in the mTSPORosettaCM model these two proline residues adopt a slightly perturbed but still helical conformation. To test whether this distorted helix is indeed a likely conformation for a Pro–Pro motif in a membrane protein, we compared its φ/ψ angles to a Ramachandran diagram obtained for all Pro–Pro motifs in the TM regions of membrane proteins with known structure (Figure S4B). The φ/ψ angles of residues Pro96 and P ro97 for both mTSPORosettaCM model and mTSPONMR-opt fell into allowed regions of the Ramachandran diagram, supportive of the accuracy of the structural models.
Computational ligand docking into TSPO recaptures the ligand interactions observed in bcTSPO crystal structure
We employed computational ligand docking with RosettaLigand (Meiler and Baker 2006) to interrogate the ligand-binding conformation of our structural models. Docking of PK11195 to the energy-minimized bcTSPOX-ray-opt structure yielded a low energy model similar to the ligand binding pose observed in the crystal structure (Figure S6A and S7A + B). Docking of PK11195 into the binding pocket of mTSPONMR-opt failed to converge to the published NMR observed ligand pose (Figure S6B and S7C + D). It is possible that geometry optimization of mTSPONMR-opt in the Rosetta implicit bilayer model reshaped the configuration of the ligand binding pocket to favor a different binding mode than the one observed by NMR in micelles. Docking of PK11195 into the mTSPORosettaCM model (Figures S6C and S7E) converged to a low-energy ligand conformation that was however, different from both of the bcTSPOX-ray and mTSPONMR structures. It is likely that structure hybridization during RosettaCM caused rearrangements in the protein backbone and side-chains around the ligand binding pocket that can explain the deviation in the PK11195 binding mode compared to the two experimental structures. However, the same bulky residues W53 and W95 (W51 and F90 in bcTSPO) were still engaged in binding of the methylisoquinoline and chlorophenyl rings of PK11195 as in mTSPONMR and bcTSPOX-ray. In summary, ligand docking recaptured the PK11195 binding mode for bcTSPO, but yielded a binding mode for mTSPO that was different from the experimentally observed bound conformation. We attribute this finding to changes in the ligand binding pocket configuration upon relaxation of the mTSPONMR structure in the Rosetta membrane model. Additionally, variation in the ligand bound conformation may reflect the lack of polar residues in the mTSPO binding pocket such that few specific directional contacts become possible, but most of the ligand binding energy stems from non-directional van-der-Waals interactions (e.g. with W53 and W95).
VUIIS8310 interaction with mTSPO interrogated by NMR and computational ligand docking
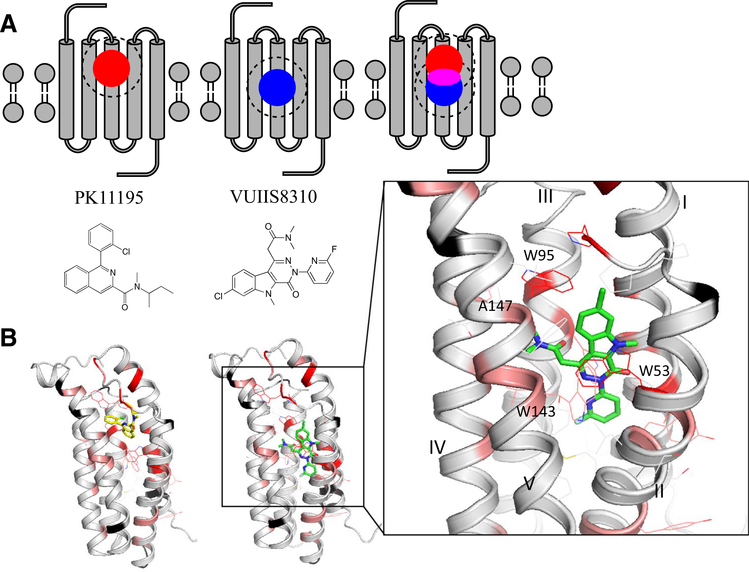
We studied the interaction of mTSPO with the small PET ligand VUIIS8310 by measuring ligand-induced chemical shift changes (ΔCS) in the 2D 1H–15N TROSY-HSQC NMR spectrum. Because the apo-mTSPO state in DPC micelles is partially unfolded, this complicates distinguishing peak changes due to a direct protein–ligand contact from those that are caused by induced protein folding. We therefore decided to modify the previously described titration experiment. The 1H–15N TROSY-HSQC spectrum of mTSPO in the presence of saturating concentrations of VUIIS8310 was compared to the spectrum of mTSPO in the presence of excess PK11195. Based on this comparison, the approximate location of the VUIIS8310 binding site can still be deduced (see schema in Fig. 7A). For residues belonging to either the VUIIS8310 or PK11195 binding sites, we expected to find the most pronounced ΔCS values. For residues that are part of both the VUIIS8310 and PK11195 binding site ΔCS values can be reduced if the two ligands interact with mTSPO in a similar manner giving rise to a similar chemical environment. Residues with unaltered chemical shifts will most likely not be part of either binding pocket. Using the measured ΔCS values between the VUIIS8310- and PK11195-bound spectrum of mTSPO and the information about where PK11195 binds, we were able to narrow down the location of the VUIIS8310 binding site.
Fig. 7.
Mapping of VUIIS8310 and PK11195 binding sites onto mTSPO identified by chemical shift perturbations. A Schematic representations of the PK11195- (left, red circle) and VUIIS8310-induced (middle, blue circle) chemical shift perturbations on mTSPO. When the NMR spectra of TSPO complexed with these two ligands are compared, the largest ΔCS are observed in regions that are exclusive to either PK11195 or VUIIS8310, whereas residues that fall in both binding sites (magenta) typically exhibit only reduced perturbations. B VUIIS8310-induced ΔCS values mapped onto the mTSPORosettaCM model (Black: no ΔCS; Gray: ΔCS < 0.1 ppm or unassigned peak; Pink: 0.5 ppm > ΔCS > 0.1 ppm; Red: ΔCS > 0.5 ppm). Left: PK11195 docked to mTSPORosettaCM model. Middle: VUIIS8310 docked to mTSPORosettaCM model. VUIIS8310 binds deeper in the mTSPO binding pocket compared to PK11195. Right: enlarged representation of the VUIIS8310 binding site, with mTSPO side chains and the VUIIS8310 molecule depicted as sticks
The 1H–15N TROSY-HSQC spectra of mTSPO in the presence of VUIIS8310 and PK11195 are shown in Figure S6, and the ΔCS values are mapped onto the mTSPORosettaCM model in Fig. 7B. ΔCSs clustered in two areas: one cluster is located in the upper half of mTSPO and coincides with the PK11195 binding site whereas the second cluster is located in the center of the protein and was interpreted as the VUIIS8310 binding pocket. Computational docking models of VUIIS8310 with mTSPORosettaCM and mTSPONMR-opt agreed qualitatively with the ΔCS data and are shown in Fig. 7B and S6E. VUIIS8310 penetrates deep into mTSPO; the approximate distance between the VUIIS8310 and PK11195 center-of-mass is 3Å. VUIIS8310 engages in π-π stacking interactions between its elongated conjugated ring system and residues W53 and W96 on mTSPO (see right panel in Fig. 7B and S7F), and makes several hydrophobic contacts with A22, L89, L112, Y140, L144 and A147.
Discussion
Current micelle preparations of mTSPO display reduced ligand affinity, likely through a thermodynamic destabilization of the protein
mTSPO solubilized in detergent micelles displays a starkly reduced binding affinity to PK11195 that contrasts the previously reported high nanomolar affinity of reconstituted TSPO in lipid bilayers (Lacapere et al. 2001). Differences in ligand binding affinity have been reported by other authors, too (Lacapère and Papadopoulos 2003; Papadopoulos et al. 1994; Owen et al. 2012). Based on our fluorescence and NMR results, we hypothesize an initial interaction of PK11195 to a partially folded state of the mTSPO-micelle complex and a subsequent slow folding of the protein. The first transition phase (Tr1) observed in the fluorescence ligand titration experiment is sensitive to temperature and detergent conditions, and could represent nonspecific interactions of PK11195 with the detergent micelle. It is possible that this first transition phase represents PK11195 binding to the micelle triggering a structural change in mTSPO that alters protein fluorescence. The second transition phase (Tr2) in the fluorescence binding curve coincides with the transition observed in the NMR titration experiment (i.e. happens at the same concentration of PK11195) and likely represents folding of mTSPO upon ligand binding as revealed by NMR. In the absence of PK11195, the NMR spectrum of apo-mTSPO in DPC micelles at 42 °C clearly indicates lack of a compact three-dimensional structure. Increasing concentrations of PK11195 induce folding of mTSPO into a more compact conformation that can be followed by the appearance of new peaks in the NMR spectrum revealing a slow exchange process on the NMR timescale. Ligand-induced changes in the secondary structure of mTSPO were also detected by CD (Murail et al. 2008) and IF (Lacapere et al. 2014) spectroscopy. The ligand-stabilized mTSPO structure may still bear structural frustrations from its biologically relevant conformation due to the altered and thermodynamically destabilizing energy landscape in the micelle environment.
In a recent solid-state NMR study of mTSPO reconstituted in DMPC bicelles (Jaipuria et al. 2017) it became also necessary to stabilize the protein by adding a high-affinity ligand (DAA1106) because apo-mTSPO is flexible and lacks a compact tertiary structure, even in bicelles. This suggests that even in membrane bilayers mTSPO exists in a dynamic, partially folded but ligand-binding state. It is likely that specific lipids or environmental conditions contribute to the stability and function of TSPO in the mitochondrial membrane, which has an asymmetric lipid composition and where TSPO is found in complex with VDAC, ANT, and possibly other proteins in the mitochondrial permeability transition pore (McEnery et al. 1992; Veenman and Gavish 2012). Such interactions seem likely to contribute to stability of mammalian TSPO.
These types of membrane protein distortions, especially in the TM helix regions, by non-native mimetics like detergent micelles is well documented (Cross et al. 2011, 2013; Zhou and Cross 2013). There are three main sources of possible distortions for a multispan helical membrane protein. First, the hydrophobic span of protein-empty micelle is typically smaller than the hydrophobic diameter of a real membrane, leading to possible hydrophobic mismatch between the TM span of a membrane protein and the micelle. This sometimes will destabilize the protein (particularly water-exposed ends of what would otherwise be buried TM helices) and/or promote its formation of oligomers or aggregates, which both seem to be a serious problem for TSPO. The micelle interior is also much more hydrated than the interior of bilayers, which can destabilize otherwise very stable TM helices, as well as any intramembrane tertiary interactions that involve electrostatic or H-bonding interactions, which could be the case for TSPO. Finally, the geometry of detergents in micelles is different than for lipids in a bilayer, the latter of which are uniformly aligned along the bilayer normal, where they exert lateral pressure on transmembrane helices, which also align with the bilayer normal. This potential is not present in micelles, which means that the orientation of TM helices may sometimes be distorted, as seems likely to be the case for the published mTSPO NMR structure. In micelles, there is also a tendency for TM helices to bow towards the always-near quasi-spherical micelle surface. Some distortion of these classes may pertain to TSPO in this study. However, the fact that in our study TSPO can bind cognate ligands with high affinity suggests that micelle-induced distortions are modest. It should be kept in mind that while micelles can distort membrane protein structures, the native conformational folding stability of membrane proteins can force distortion of micelles in such a way that, the micelle structure adapts to the membrane protein’s native structure.
An integrated mTSPORosettaCM model provides a physically realistic working mTSPO model that is consistent with all available experimental data
While the available TSPO structures are considered low-energy states under their specific experimental conditions, care must be taken when interpreting their biological function since structural characteristics may be the consequence of the micelle environment. Herein we employed RosettaCM homology modeling to develop a model of mTSPO in the bilayer membrane. RosettaCM leverages known structural features of the mTSPONMR structure and homologous bacterial proteins (bcTSPO and rsTSPO) and allows efficient exploration of the conformational space around those templates guided by the Rosetta energy function. Unlikely conformations with high energy are rejected. The resulting mTSPORosettaCM model reconciles the structural features of mTSPONMR and bacterial TSPOs, while being consistent with the experimental NMR data collected on mTSPO in micelles.
We also employed computational docking to assess the models’ potential usage for future structure-based therapeutic design. The mTSPORosettaCM model was able to produce meaningful binding poses for the PET tracer molecule VUIIS8310, which agrees with our ΔCS data. Benzodiazepines and imaging agents are known to bind to the inner core of mTSPO (Jaremko et al. 2014; Tang et al. 2016) and often interact with all five TM helices. We found that VUIIS8310 occupies a site that partially overlaps with the position of PK11195 but penetrates deeper into TSPO to maximize the overlap between its conjugated ring system and the indole side-chain of W53. Additional contacts are made with hydrophobic residues A22, L89, L112, Y140, L144 and A147. The A147 position is a well-known site of TSPO and the A147T variant is implicated in rs6971 polymorphism (Kreisl et al. 2013).
In addition, we found that re-docking of PK11195 into structures of bcTSPOX-ray and mTSPONMR after energy minimization in the Rosetta implicit membrane model recapitulates ligand binding mode observe by crystallography for bcTSPOX-ray but fails to converge to the NMR-determined ligand binding mode for mTSPONMR. We attribute this to remaining micelle-induced energetic frustrations in the mTSPONMR structure that cannot be remodeled by simple energy minimization. This is a significant limitation associated with mTSPONMR for structure-based drug discovery.
The ligand binding mechanism and its effect on TSPO function requires rigorous validation
The functional role of TSPO in biological processes is still not fully understood, particularly in light of the recent paradigm shift of TSPO’s involvement in cholesterol biosynthesis pathways. While this ancient protein has been proposed to be involved in alternative biological processes such as heme and porphyrin transport (Taketani et al. 1995; Verma et al. 1987) and oxidative stress sensing (Ginter et al. 2013), TSPO’s function remains poorly understood. Despite the controversies regarding its biological function, mammalian TSPO is an attractive target for imaging molecules and possibly cancer therapeutics. We think that mTSPORosettaCM provides a useful working model for future development of potent mammalian TSPO ligands.
The information that residue A147 is part of the VUIIS8310 binding site of TSPO is an important piece of information for determining the molecular mechanism of rs6971 polymorphism and assessing ligand potency (Owen et al. 2012; Kreisl et al. 2013). In individuals who are homozygote carriers of the A147T variant, TSPO has low binding affinity for radioligands, meaning that carriers of this mutant may be unresponsive to drugs that target TSPO for diagnostic or therapeutic purposes. Using the mTSPO homology model and computational ligand docking, however, allows rapid and systematic testing of ligand binding affinity and their molecular mechanism(s) of binding.
It remains unclear, which role ligand binding and accompanying protein stabilization play for TSPO’s biological activity. It has been demonstrated that ligand-bound mTSPO retains its cholesterol binding ability and exists in a dynamic monomer–dimer equilibrium in lipid membranes (Jaipuria et al. 2017). Future experimental studies are required to illuminate the mechanistic interplay of small molecule ligand binding and TSPO’s interaction with cholesterol and proteins of the mitochondrial permeability transition pore.
Conclusions
In conclusion, we show by our NMR and fluorescence experiments that the structure of mTSPO in DPC micelles is destabilized, but can be induced to fold via titration with ligands such as PK11195. However, even then the folded structure is different from the native (bilayer conditions) structure. Our homology model of mTSPO in an implicit membrane bilayer model reconciles conflicting features between the structures of mTSPO and bacterial TSPOs. It provides the starting point for structural studies of human TSPO and serves as a tool for the structure-based design of new TSPO ligands as cancer imaging probes. Using computational docking and chemical shift perturbations, we have also proposed a model for the interaction of mTSPO with the novel PET imaging ligand VUIIS8310.
Supplementary Material
Acknowledgements
This work was conducted using the computing cluster of the Advanced Center for Research and Education (ACCRE) at Vanderbilt University, TN. Work in the Meiler laboratory is supported through NIH (R01 GM080403, R01 GM099842) and NSF (CHE 1305874). CRS was supported by NIH RF1 AG056147. VUIIS8310 was developed with support from NIH R01 CA163806 (HCM) and the Kleberg Foundation (HCM). The authors would also like to thank Dr. Markus Voehler and the Biomolecular NMR Facility at Vanderbilt for support and advice regarding the NMR data acquisition.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10858-019-00257-1) contains supplementary material, which is available to authorized users.
References
- Banati RB, Middleton RJ, Chan R, Hatty CR, Kam WW, Quin C, Graeber MB, Parmar A, Zahra D, Callaghan P, Fok S, Howell NR, Gregoire M, Szabo A, Pham T, Davis E, Liu GJ (2014) Positron emission tomography and functional characterization of a complete PBR/TSPO knockout. Nat Commun 5:5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P, Schonbrun J, Baker D (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA 104:15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batarseh A, Papadopoulos V (2010) Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol Cell Endocrinol 327:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KL, Ikezaki K, Toga AW (1989) Imaging of brain tumors using peripheral benzodiazepine receptor ligands. J Neurosurg 71:113–118 [DOI] [PubMed] [Google Scholar]
- Braestrup C, Squires RF (1977) Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc Natl Acad Sci USA 74:3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel I, Fares FA, Leschiner S, Scherubl H, Weisinger G, Gavish M (1999) Peripheral-type benzodiazepine receptors in the regulation of proliferation of MCF-7 human breast carcinoma cell line. Biochem Pharmacol 58:273–278 [DOI] [PubMed] [Google Scholar]
- Cheung Y-Y, Buck J, Nickels M, Tang D, Manning HC (2014) Preclinical evaluation of 7-chloro-N, N,5-trimethyl-4-oxo-3(6-[18F] fluoropyridin-2-yl)-3,5-dihydro-4H-pyridazino[4,5-b]indole-1-acetamide: a novel pyridazinoindole ligand for PET imaging of TSPO in cancer. J Nucl Med 55:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching AS, Kuhnast B, Damont A, Roeda D, Tavitian B, Dolle F (2012) Current paradigm of the 18-kDa translocator protein (TSPO) as a molecular target for PET imaging in neuroinflammation and neurodegenerative diseases. Insights Imaging 3:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary J, Johnson KM, Opipari AW Jr, Glick GD (2007) Inhibition of the mitochondrial F1F0-ATPase by ligands of the peripheral benzodiazepine receptor. Bioorg Med Chem Lett 17:1667–1670 [DOI] [PubMed] [Google Scholar]
- Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D (2014) Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci 23:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Sharma M, Yi M, Zhou H-X (2011) Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci 36:117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Murray DT, Watts A (2013) Helical membrane protein conformations and their environment. Eur Biophys J 42:731–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane NG, Manning HC, Foutch AC, Washington MK, Aronow BJ, Bornhop DJ, Coffey RJ (2007) Targeted imaging of colonic tumors in smad3−/− mice discriminates cancer and inflammation. Mol Cancer Res 5:341–349 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- Fafalios A, Akhavan A, Parwani AV, Bies RR, McHugh KJ, Pflug BR (2009) Translocator protein blockade reduces prostate tumor growth. Clin Cancer Res 15:6177–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Lindemann P, Feuilloley MG, Papadopoulos V (2012) Structural and functional evolution of the translocator protein (18 kDa). Curr Mol Med 12:369–386 [DOI] [PubMed] [Google Scholar]
- Frison M, Mallach AK, Kennedy E, Campanella M (2017) The 18 kDa Translocator protein (TSPO): cholesterol trafficking and the biology of a prognostic and therapeutic mitochondrial target. In: Rostovtseva TK (ed) Molecular basis for mitochondrial signaling. Springer, Cham, pp 285–315 [Google Scholar]
- Galiegue S, Tinel N, Casellas P (2003) The peripheral benzodiazepine receptor: a promising therapeutic drug target. Curr Med Chem 10:1563–1572 [DOI] [PubMed] [Google Scholar]
- Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A (1999) Enigma of the peripheral benzodiazepine receptor. Pharmacol Rev 51:629–650 [PubMed] [Google Scholar]
- Ginter C, Kiburu I, Boudker O (2013) Chemical catalysis by the translocator protein (18 kDa). Biochemistry 52:3609–3611 [DOI] [PubMed] [Google Scholar]
- Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabo I, Lippe G, Bernardi P (2013) Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci USA 110:5887–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Nguyen ED, Reiff SD, Squire EF, Stauffer SR, Lindsley CW, Meiler J, Conn PJ (2013) Probing the metabotropic glutamate receptor 5 (mGlu(5)) positive allosteric modulator (PAM) binding pocket: discovery of point mutations that engender a “molecular switch” in PAM pharmacology. Mol Pharmacol 83:991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kalathur RC, Liu Q, Kloss B, Bruni R, Ginter C, Kloppmann E, Rost B, Hendrickson WA (2015) Protein structure: Structure and activity of tryptophan-rich TSPO proteins. Science 347:551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Slack RS, Li W, Papadopoulos V (2003) Expression of peripheral benzodiazepine receptor (PBR) in human tumors: relationship to breast, colorectal, and prostate tumor progression. J Recept Signal Transduct Res 23:225–238 [DOI] [PubMed] [Google Scholar]
- Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V (1999) Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res 59:831–842 [PubMed] [Google Scholar]
- Hardwick M, Rone J, Han ZQ, Haddad B, Papadopoulos V (2001) Peripheral-type benzodiazepine receptor levels correlate with the ability of human breast cancer MDA-MB-231 cell line to grow in SCID mice. Int J Cancer 94:322–327 [DOI] [PubMed] [Google Scholar]
- Hatty CR, Le Brun AP, Lake V, Clifton LA, Liu GJ, James M, Banati RB (2014) Investigating the interactions of the 18 kDa translocator protein and its ligand PK11195 in planar lipid bilayers. Biochim Biophys Acta 1838:1019–1030 [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaipuria G, Leonov A, Giller K, Vasa SK, Jaremko L, Jaremko M, Linser R, Becker S, Zweckstetter M (2017) Cholesterol-mediated allosteric regulation of the mitochondrial translocator protein structure. Nat Commun 8:14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin N, Neumann JM, Ostuni MA, Vu TK, Yao ZX, Murail S, Robert JC, Giatzakis C, Papadopoulos V, Lacapere JJ (2005) Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol Endocrinol 19:588–594 [DOI] [PubMed] [Google Scholar]
- Jaremko Ł, Jaremko M, Giller K, Becker S, Zweckstetter M (2014) Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science 343:1363–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremko M, Jaremko L, Giller K, Becker S, Zweckstetter M (2015a) Structural integrity of the A147T polymorph of mammalian TSPO. ChemBioChem 16:1483–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremko L, Jaremko M, Giller K, Becker S, Zweckstetter M (2015b) Conformational flexibility in the transmembrane protein TSPO. Chemistry 21:16555–16563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junck L, Olson JMM, Ciliax BJ, Koeppe RA, Watkins GL, Jewett DM, Mckeever PE, Wieland DM, Kilbourn MR, Starostarubinstein S, Mancini WR, Kuhl DE, Greenberg HS, Young AB (1989) Pet imaging of human gliomas with ligands for the peripheral benzodiazepine binding-site. Ann Neurol 26:752–758 [DOI] [PubMed] [Google Scholar]
- Korkhov VM, Sachse C, Short JM, Tate CG (2010) Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure 18:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl WC, Jenko KJ, Hines CS, Lyoo CH, Corona W, Morse CL, Zoghbi SS, Hyde T, Kleinman JE, Pike VW, McMahon FJ, Innis RB (2013) A genetic polymorphism for translocator protein 18 kDa affects both in vitro and in vivo radioligand binding in human brain to this putative biomarker of neuroinflammation. J Cereb Blood Flow Metab 33:53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacapère J-J, Papadopoulos V (2003) Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 68:569–585 [DOI] [PubMed] [Google Scholar]
- Lacapere JJ, Delavoie F, Li H, Peranzi G, Maccario J, Papadopoulos V, Vidic B (2001) Structural and functional study of reconstituted peripheral benzodiazepine receptor. Biochem Biophys Res Commun 284:536–541 [DOI] [PubMed] [Google Scholar]
- Lacapere J-J, Iatmanen-Harbi S, Senicourt L, Lequin O, Tekely P, Purusottam RN, Hellwig P, Kriegel S, Ravaud S, Juillan-Binard C (2014) Structural studies of TSPO, a mitochondrial membrane protein. In: Mus-Veteau I (ed) Membrane proteins production for structural analysis. Springer, New York, pp 393–421 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- Lazaridis T (2003) Effective energy function for proteins in lipid membranes. Proteins 52:176–192 [DOI] [PubMed] [Google Scholar]
- Lazaridis T, Karplus M (1999) Effective energy function for proteins in solution. Proteins 35:133–152 [DOI] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics (Oxford, England) 31:1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Papadopoulos V (1998) Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus patterns. Endocrinology 139:4991–4997 [DOI] [PubMed] [Google Scholar]
- Li F, Liu J, Zheng Y, Garavito RM, Ferguson-Miller S (2015) Protein structure: Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism. Science 347:555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GJ, Middleton RJ, Hatty CR, Kam WW, Chan R, Pham T, Harrison-Brown M, Dodson E, Veale K, Banati RB (2014) The 18 kDa translocator protein, microglia and neuroinflammation. Brain Pathol 24:631–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaser K, Hèopfner M, Jansen A, Weisinger G, Gavish M, Kozikowski AP, Weizman A, Carayon P, Riecken EO, Zeitz M, Scherèubl H, Medical Clinic I (2001) Specific ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in human colorectal cancer cells. Br. J Cancer 85(11):1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaser K, Grabowski P, Sutter AP, Hopfner M, Foss HD, Stein H, Berger G, Gavish M, Zeitz M, Scherubl H (2002) Overexpression of the peripheral benzodiazepine receptor is a relevant prognostic factor in stage III colorectal cancer. Clin Cancer Res 8:3205–3209 [PubMed] [Google Scholar]
- McEnery MW, Snowman AM, Trifiletti RR, Snyder SH (1992) Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA 89:3170–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J, Baker D (2006) ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins 65:538–548 [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment, 2013.08; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2013 [Google Scholar]
- Morohaku K, Pelton SH, Daugherty DJ, Butler WR, Deng W, Selvaraj V (2014) Translocator protein/peripheral benzodiazepine receptor is not required for steroid hormone biosynthesis. Endocrinology 155:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murail S, Robert JC, Coic YM, Neumann JM, Ostuni MA, Yao ZX, Papadopoulos V, Jamin N, Lacapere JJ (2008) Secondary and tertiary structures of the transmembrane domains of the translocator protein TSPO determined by NMR. Stabilization of the TSPO tertiary fold upon ligand binding. Biochim Biophys Acta 1778:1375–1381 [DOI] [PubMed] [Google Scholar]
- Owen DR, Yeo AJ, Gunn RN, Song K, Wadsworth G, Lewis A, Rhodes C, Pulford DJ, Bennacef I, Parker CA, StJean PL, Cardon LR, Mooser VE, Matthews PM, Rabiner EA, Rubio JP (2012) An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab 32:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos V, Boujrad N, Ikonomovic MD, Ferrara P, Vidic B (1994) Topography of the Leydig cell mitochondrial peripheral-type benzodiazepine receptor. Mol Cell Endocrinol 104:R5–R9 [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC (2015) The PyMOL Molecular Graphics System, Version 1.8
- Shen Y, Bax A (2010) SPARTA + : a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR 48:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sileikyte J, Blachly-Dyson E, Sewell R, Carpi A, Menabo R, Di Lisa F, Ricchelli F, Bernardi Forte M (2014) Regulation of the mitochondrial permeability transition pore by the outer membrane does not involve the peripheral benzodiazepine receptor (TSPO). J Biol Chem 289:13769–13781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D (2013) High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostarubinstein S, Ciliax BJ, Penney JB, Mckeever P, Young AB (1987) Imaging of a glioma using peripheral benzodiazepine receptor ligands. Proc Natl Acad Sci USA 84:891–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketani S, Kohno H, Furukawa T, Tokunaga R (1995) Involvement of peripheral-type benzodiazepine receptors in the intracellular transport of heme and porphyrins. J Biochem 117:875–880 [DOI] [PubMed] [Google Scholar]
- Tang D, Hight MR, McKinley ET, Fu A, Buck JR, Smith RA, Tantawy MN, Peterson TE, Colvin DC, Ansari MS, Nickels M, Manning HC (2012) Quantitative preclinical imaging of TSPO expression in glioma using N, N-diethyl-2-(2-(4-(2–18F-fluoroethoxy) phenyl)-5,7-dimethylpyrazolo[1,5-a]pyrimi din-3-yl)acetamide. J Nucl Med 53:287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, McKinley ET, Hight MR, Uddin MI, Harp JM, Fu A, Nickels ML, Buck JR, Manning HC (2013) Synthesis and structure-activity relationships of 5,6,7-substituted pyrazolopyrimidines: discovery of a novel TSPO PET ligand for cancer imaging. J Med Chem 56:3429–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Nickels ML, Tantawy MN, Buck JR, Manning HC (2014) Preclinical imaging evaluation of novel TSPO-PET ligand 2-(5,7-Diethyl-2-(4-(2-[F]fluoroethoxy)phenyl)pyrazolo[1,5-a]pyrimidin-3-yl)-N, N- diethylacetamide ([F]VUIIS1008) in Glioma. Mol Imaging Biol 16:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Li J, Buck JR, Tantawy MN, Xia Y, Harp JM, Nickels ML, Meiler J, Manning HC (2016) Evaluation of TSPO PET ligands [18F]VUIIS1009A and [18F]VUIIS1009B: tracers for cancer imaging. Mol Imaging Biol 19:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul D, Beaufils S, Taveau J-C, Iatmanen-Harbi S, Renault A, Venien-Bryan C, Vie VR, Lacapere J-J (2012) Mouse TSPO in a lipid environment interacting with a functionalized monolayer. Biochim Biophys Acta 1818:2791–2800 [DOI] [PubMed] [Google Scholar]
- Tu LN, Morohaku K, Manna PR, Pelton SH, Butler WR, Stocco DM, Selvaraj V (2014) Peripheral benzodiazepine receptor/translocator protein global knock-out mice are viable with no effects on steroid hormone biosynthesis. J Biol Chem 289:27444–27454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu LN, Zhao AH, Stocco DM, Selvaraj V (2015) PK11195 effect on steroidogenesis is not mediated through the translocator protein (TSPO). Endocrinology 156:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenman L, Gavish M (2012) The role of 18 kDa mitochondrial translocator protein (TSPO) in programmed cell death, and effects of steroids on TSPO expression. Curr Mol Med 12:398–412 [DOI] [PubMed] [Google Scholar]
- Verma A, Nye JS, Snyder SH (1987) Porphyrins are endogenous ligands for the mitochondrial (peripheral-type) benzodiazepine receptor. Proc Natl Acad Sci USA 84:2256–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D (2006) Multipass membrane protein structure prediction using Rosetta. Proteins 62:1010–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Cross TA (2013) Influences of membrane mimetic environments on membrane protein structures. Ann Rev Biophys 42:361–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.