Abstract
Background
In Uganda, artemether-lumefantrine is recommended for malaria treatment and sulfadoxine-pyrimethamine for chemoprevention during pregnancy, but drug resistance may limit efficacies.
Methods
Genetic polymorphisms associated with sensitivities to key drugs were characterized in samples collected from 16 sites across Uganda in 2018 and 2019 by ligase detection reaction fluorescent microsphere, molecular inversion probe, dideoxy sequencing, and quantitative polymerase chain reaction assays.
Results
Considering transporter polymorphisms associated with resistance to aminoquinolines, the prevalence of Plasmodium falciparum chloroquine resistance transporter (PfCRT) 76T decreased, but varied markedly between sites (0–46% in 2018; 0–23% in 2019); additional PfCRT polymorphisms and plasmepsin-2/3 amplifications associated elsewhere with resistance to piperaquine were not seen. For P. falciparum multidrug resistance protein 1, in 2019 the 86Y mutation was absent at all sites, the 1246Y mutation had prevalence ≤20% at 14 of 16 sites, and gene amplification was not seen. Considering mutations associated with high-level sulfadoxine-pyrimethamine resistance, prevalences of P. falciparum dihydrofolate reductase 164L (up to 80%) and dihydropteroate synthase 581G (up to 67%) were high at multiple sites. Considering P. falciparum kelch protein propeller domain mutations associated with artemisinin delayed clearance, prevalence of the 469Y and 675V mutations has increased at multiple sites in northern Uganda (up to 23% and 41%, respectively).
Conclusions
We demonstrate concerning spread of mutations that may limit efficacies of key antimalarial drugs.
Keywords: Plasmodium falciparum, drug resistance, PfCRT, PfMDR1, PfDHFR, PfDHPS, PfK13, Uganda
The prevalence of genetic polymorphisms associated with antimalarial drug resistance in Plasmodium falciparum collected from 16 sites across Uganda was studied. Most concerning was increasing prevalence of mutations associated with resistance to antifolates and artemisinins.
(See the Editorial Commentary by Sibley, on pages 927–9.)
Progress in malaria control has stalled in much of Africa and could further be challenged by the persistence, spread, or emergence of Plasmodium falciparum resistance to aminoquinolines, which are key partners in artemisinin-based combination therapies (ACTs); the antifolate sulfadoxine-pyrimethamine (SP), which remains a standard of care for chemoprevention; and artemisinins, which are the backbones of all ACTs [1, 2]. In Uganda and most other African countries, the ACT artemether-lumefantrine (AL) has been first-line therapy for uncomplicated malaria for about 15 years. Another ACT, artesunate-amodiaquine, is first-line therapy for uncomplicated malaria in a number of African countries. Considering chemoprevention, intermittent preventive treatment during pregnancy (IPTp) with SP is recommended in malaria-endemic regions across Africa, and seasonal malaria chemoprevention with amodiaquine-SP is recommended in areas of the Sahel subregion with seasonal transmission [3, 4]. Dihydroartemisinin-piperaquine is an alternative treatment and is under investigation as an alternative to SP for chemoprevention [5].
Resistance of P. falciparum to the aminoquinolines chloroquine and amodiaquine has been widespread and is mediated by polymorphisms in the P. falciparum chloroquine resistance transporter (PfCRT) and P. falciparum multidrug resistance protein 1 (PfMDR1) proteins [6]. PfCRT 76T is the principal resistance mediator, and the PfMDR1 86Y and 1246Y mutations modulate resistance in Africa [1]. Interestingly, wild-type PfCRT K76 and PfMDR1 N86 are associated with decreased sensitivity to lumefantrine [7] and are enriched in patients previously treated with AL [8, 9]. Although true failures after therapy with AL have been uncommon in Africa, in a pooled analysis, PfMDR1 N86 was independently associated with recrudescence following therapy [1, 10]. Consistent with decreased use of chloroquine and establishment of AL as a first-line therapy, PfCRT K76 and PfMDR1 N86 and D1246 wild-type prevalences have increased in many African countries [11–14].
Plasmodium falciparum resistance to SP is mediated by mutations in the target enzymes dihydrofolate reductase (PfDHFR) and dihydropteroate synthase (PfDHPS). A combination of 5 mutations (PfDHFR 51I, 59R, and 108N and PfDHPS 437G and 540E) mediates moderate SP resistance and is highly prevalent in much of Africa, except that PfDHPS 540E is uncommon in Central and West Africa [1, 15, 16]. Additional mutations in PfDHFR (164L) or PfDHPS (581G and 613S) mediate higher-level SP resistance and have been shown to be selected by IPTp with SP [16, 17]. Mutations that mediate higher-level SP resistance have been uncommon in Africa, but modest prevalence has been identified for PfDHFR 164L in southwestern Uganda and for PfDHPS 581G in northern Tanzania, eastern Democratic Republic of Congo (DRC), and southwestern Uganda [12, 14, 18, 19].
Plasmodium falciparum resistance to artemisinin derivatives, manifested as delayed clearance after therapy, is associated with any of a number of propeller domain mutations in the P. falciparum kelch protein (PfK13), and key propeller domain mutations are now widespread in the Greater Mekong subregion of southeast Asia [20–22]. In Africa, PfK13 propeller domain mutations have been uncommon, mutations shown to be associated with artemisinin resistance in Asia have been rare, and clinical artemisinin resistance is not confirmed [1, 20, 23]. However, recent reports suggest emergence of parasites in East Africa with mutations that may mediate artemisinin resistance. First, the 561H mutation, which was previously associated with artemisinin resistance in southeast Asia [20, 24], was identified in 7% of isolates collected in Rwanda [25], and in another study, in 17%–20% of isolates from 2 sites in Rwanda, including 50% of parasites persisting 3 days after initiation of therapy [26]. The 561H mutation has also been detected in parasites from DRC and Tanzania [1, 27]. Second, the 675V mutation, also associated with delayed clearance after artemisinin therapy [20, 24], was identified in an isolate collected in Rwanda in 2015, an isolate with delayed in vitro parasite clearance collected in northern Uganda in 2016, and 6% of isolates collected from 3 sites in northern Uganda in 2017 [12, 28, 29]. Third, the 469Y mutation, also associated with artemisinin delayed clearance [24], was seen in parasites collected from 4 different sites in Uganda in 2017 [12]. Another mutation at the same locus, 469F, was seen in parasites from Equatorial Guinea [1]. Of note, sequencing of 1183 samples collected from 7 sites across Uganda from 1999 to 2015 did not identify any isolates with PfK13 469F/Y, 561H, or 675V mutations [30].
Additional polymorphisms mediate P. falciparum resistance to ACT partner drugs. Pfmdr1 gene amplification mediates P. falciparum resistance to mefloquine and was seen in Southeast Asia after use of that drug [31]. In pooled analyses, pfmdr1 amplification was found to be associated with uncommon AL treatment failures in Africa and Asia [10, 32]. Amplification of plasmepsin-2/3 and novel mutations in PfCRT have been associated with piperaquine resistance and dihydroartemisinin-piperaquine treatment failure in southeast Asia [33–35]. To date, these polymorphisms have been rare in African parasites [1, 7, 12, 36, 37].
Clearly, it is important to maintain surveillance for changes in antimalarial drug sensitivity in Africa. Characterization of the ex vivo sensitivity of parasites is valuable, but in Uganda has been limited to a few locations [7, 29, 38]. Surveillance for P. falciparum genetic polymorphisms associated with resistance is simpler, and presents an opportunity for broad characterization of parasite genotypes at greater spatiotemporal scale [12, 14]. Here we report on surveillance in 2018 and 2019 of key polymorphisms associated with drug sensitivity at 16 sites across Uganda with varied malaria epidemiology.
MATERIALS AND METHODS
Collection of Samples
Building on a prior 10-site study [11], we expanded to 16 sites to represent varied malaria epidemiology across Uganda (Figure 1), where we set out to collect 50 samples per site per year in both 2018 and 2019. At each site, as part of routine care, healthcare personnel evaluated individuals >6 months of age (up to age 10 years for 2018; all ages for 2019) with clinical syndromes suggestive of malaria using either Giemsa-stained blood smears or histidine-rich protein 2 (HRP2)–based rapid diagnostic tests, following national guidelines and depending on local test availability. Subjects or their parents or guardians were approached for enrollment, and if consent was obtained, blood was collected as blood spots dried on Whatman 3MM filter paper. Filter paper samples were stored in zip-lock storage bags with desiccant at room temperature and transported to laboratories in Kampala and the United States for processing and evaluation. Samples were collected in April–June of each year, except in Kabale, where sample collection was extended to August 2019. This study was approved by the Makerere University Research and Ethics Committee, the Uganda National Council of Science and Technology, and the University of California, San Francisco Committee on Human Research.
Figure 1.
Map of Uganda. The districts where samples were collected are shown.
Sample Preparation
Genomic DNA was extracted from blood spots using Chelex 100 as previously described [38], except that later isolations for molecular inversion probe (MIP) capture and dideoxy sequencing replaced saponin lysis buffer with 0.01% Tween 20.
Ligase Detection Reaction Fluorescent Microsphere Assay
Samples collected in 2018 were characterized for pfmdr1, pfcrt, pfdhfr, and pfdhps polymorphisms of interest by a ligase detection reaction fluorescent microsphere (LDR-FM) assay, as previously described [39], with minor modifications to incorporate nested polymerase chain reaction (PCR).
MIP Capture and Next-Generation Sequencing
All samples were characterized by MIP capture and next-generation sequencing. We designed a MIP panel consisting of probes targeting P. falciparum genes associated with antimalarial drug resistance (Supplementary Table 1) using MIPTools software (version 0.19.12.13; https://github.com/bailey-lab/MIPTools). The panel covered 43 known polymorphisms in the pfcrt, pfmdr1, pfdhfr, and pfdhps genes; the full sequence of PfK13; and consideration of copy number of the plasmepsin-2/3 and pfmdr1 genes (Supplementary Table 2). MIP capture, library preparation, and sequencing were carried out as published previously [19] and are detailed in the Supplementary Methods. Sequencing reads are available in the National Center for Biotechnology Information (NCBI) under accession number PRJNA655702. Individual sample genpotypes are provided in Supplementary Table 3.
Dideoxy Sequencing
For 2019 samples, the gene encoding the PfK13 propeller domain was PCR amplified and sequenced as previously described [12]. Sequences were evaluated using CodonCode Aligner version 9.0.1 (CodonCode Corporation) and are available in NCBI under accession numbers MT857288–MT857721.
Gene Copy Number Determination
Copy numbers for pfmdr1 and plasmepsin-2/3 were determined from MIP data based on the depth of coverage, as detailed in the Supplementary Methods. Plasmepsin-2/3 gene copy number was also measured using quantitative PCR (qPCR) for a subset of samples collected in 2019, as previously described [7].
Data Analysis
The raw MIP sequencing data were analyzed using MIPTools (https://github.com/bailey-lab/MIPTools) as previously described [39], and detailed in the Supplementary Methods. Downstream analysis was performed using R version 3.6.2. Prevalence data for different polymorphisms were summarized using ggplot2 and tmap packages in R version 3.6.2 software. Kappa analyses were done in rstudio using the kappa command in the vcd package (version 1.4–7 Meyer, 2020).
RESULTS
Study Population and Samples
Our goal was to collect 50 samples per site each year, but fewer samples were available at some sites due to technical challenges, and at the Kabale and Kapchorwa sites due to low malaria incidence (Supplementary Table 4). Samples were characterized for genetic polymorphisms associated with drug resistance using LDR-FM, MIP, dideoxy sequencing, and qPCR assays. The results presented are based on MIP capture next-generation sequencing, with mixed and mutant results combined for each locus, and available data from the other assays are provided for validation.
Prevalence of Polymorphisms That Mediate Aminoquinoline Resistance
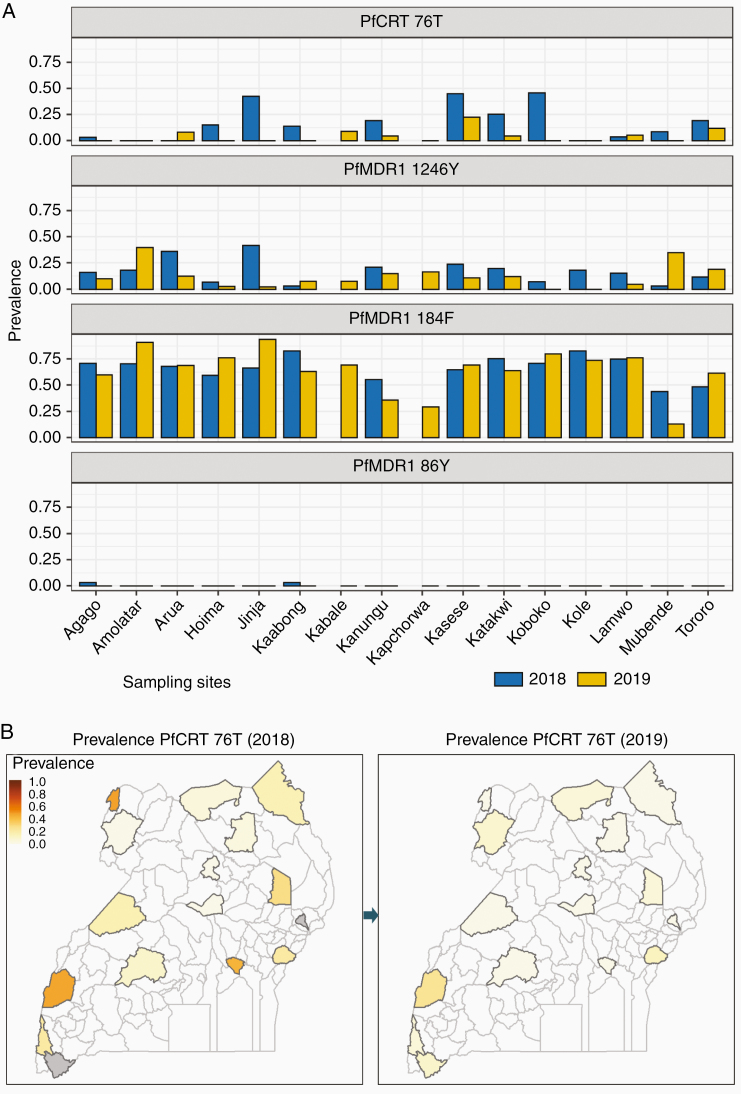
The prevalence of polymorphisms in PfCRT and PfMDR1 associated with aminoquinoline resistance varied between sites and over time (Figure 2). PfCRT 76T, the principal mediator of resistance to chloroquine and amodiaquine, had prevalences <10% at most sites (prevalence 0% at 9 of 16 sites in 2019), but considerably higher (up to 45.8% in 2018 and 22.7% in 2019) at a few sites. Consistent with prior results [11, 14], prevalence of previously common mutations in PfMDR1 was low, with the 86Y mutation completely absent in 2019, and prevalence of the 1246Y mutation ≤20% at all except 2 sites in 2019 (Supplementary Table 5). As seen previously, prevalence of the 184F mutation, which does not clearly impact on drug sensitivity, was >50% at most sites. Increased gene copy number was not seen by MIP analysis in any of 520 isolates evaluated for pfmdr1 or 462 isolates evaluated for plasmepsin-2/3. PfCRT mutations linked to piperaquine resistance in southeast Asia (93S, 97Y, 145I, 218F, 343L, 353V) [35, 40] were absent among the 695 isolates assessed. Analysis of samples from 2018 by LDR-FM and 2019 for plasmepsin-2/3 gene copy number by qPCR yielded similar results (Supplementary Table 6).
Figure 2.
Prevalence of mutations associated with aminoquinoline resistance. Gray, no samples obtained. Abbreviations: PfCRT, Plasmodium falciparum chloroquine resistance transporter; PfMDR1, Plasmodium falciparum multidrug resistance protein 1.
Prevalence of Polymorphisms in Folate Pathway Enzymes That Mediate Antifolate Resistance
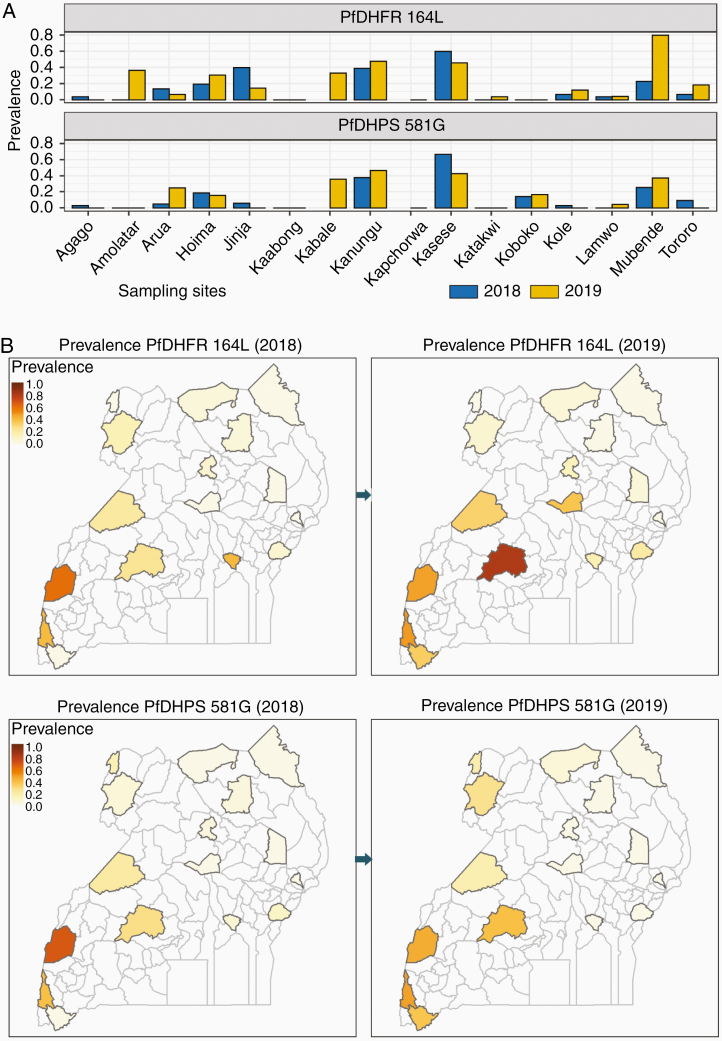
Similar to what we previously reported for earlier surveys [12, 14], 5 antifolate mutations (PfDHFR 51I, 59R, 108N; PfDHPS 437G, 540E) were very common across Uganda, with prevalences of each mutation >80% at all sites (Supplementary Figure 1). Two additional mutations associated with high-level SP resistance (PfDHFR 164L, PfDHPS 581G) showed increasing prevalence compared with earlier reports (Figure 3 and Supplementary Table 5) [12, 14]. Prevalence of these 2 mutations varied greatly across the country. As seen previously in samples from 2016–2017 [12], the highest prevalence for both mutations was in central and western Uganda, but the mutations were more broadly distributed than in the prior surveys. The PfDHPS 613S mutation was not identified in any samples. Analysis of samples from 2018 by LDR-FM yielded similar results (Supplementary Table 6).
Figure 3.
Prevalence of Plasmodium falciparum dihydrofolate reductase (PfDHFR) 164L and Plasmodium falciparum dihydropteroate synthase (PfDHPS) 581G mutations. Gray, no samples obtained.
Prevalence of PfK13 Polymorphisms Associated With Artemisinin Resistance
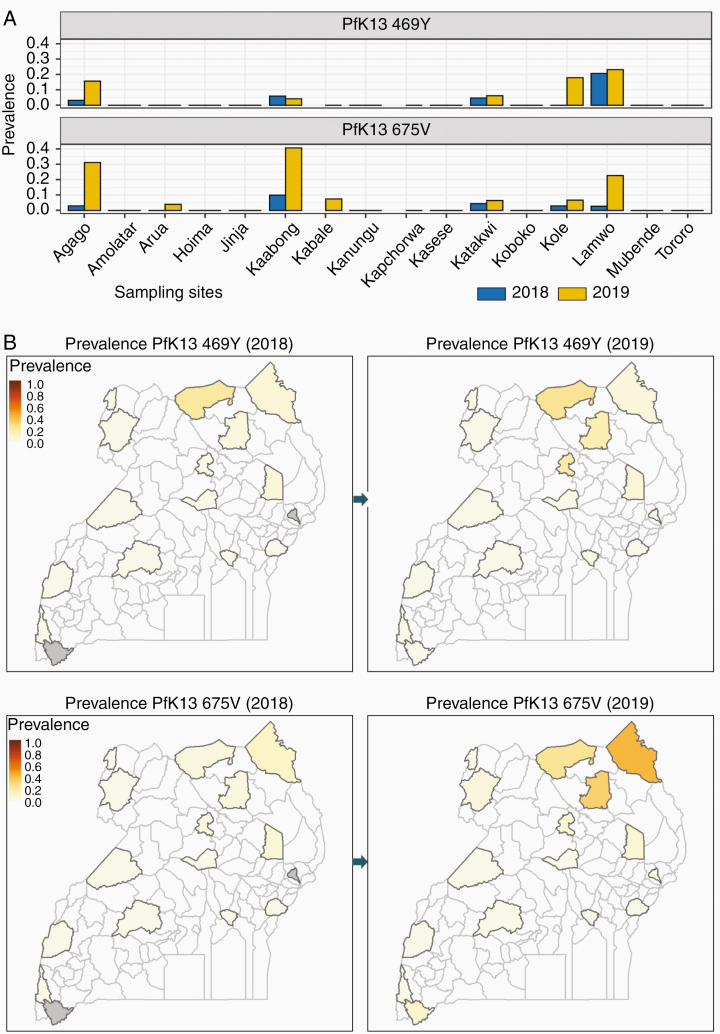
Among the 796 samples for which PfK13 sequences were obtained by MIP assays, mutations were detected at 16 loci, including 8 in the propeller domain, detected in 93 samples (Supplementary Table 7). Polymorphisms at 5 of these propeller domain loci (442L, 469Y/F, 539K/I/T, 578S/T/D, and 675V) were previously reported in Africa [1]. Two polymorphisms, each associated with delayed clearance after artemisinin therapy in Asia [20, 24], and identified previously in Uganda in samples from 2016–2017 [12], had overall prevalence ≥3.9% in 2019 (469Y, 3.9%; 675V, 6.0%), each with prevalence >15% at 3 sites in northern Uganda (Figure 4 and Supplementary Table 8). The 561H mutation, which was recently identified in Rwanda [25, 26], was detected in a single isolate from Jinja. Only a small number of samples was available from Kabale, the site near the Rwandan border, due to low malaria incidence. Dideoxy sequencing of PfK13 yielded similar results, but generally with lower prevalences compared to MIP sequencing, presumably due to lower sensitivity for minority alleles (Supplementary Table 6).
Figure 4.
Prevalence of Plasmodium falciparum kelch protein (PfK13) 469Y and 675V mutations. Gray, no samples obtained.
DISCUSSION
We offer a comprehensive evaluation of the prevalence of P. falciparum genetic polymorphisms associated with altered antimalarial drug sensitivity in 2018–2019 at 16 sites of varied malaria transmission intensity across Uganda. We found a continuation of trends described earlier, with decreased prevalence of polymorphisms associated with aminoquinoline resistance (PfCRT 76T; PfMDR1 86Y and 1246Y), increased prevalence of polymorphisms that mediate high-level SP resistance (PfDHFR 164L and PfDHPS 581G), and emergence of PfK13 propeller domain mutations (469Y and 675V) associated with delayed clearance after artemisinin-based therapy in southeast Asia. These polymorphisms demonstrated marked geographic variation across Uganda, with varied prevalence of the PfCRT 76T mutation, highest prevalence of PfDHFR 164L and PfDHPS 581G in western Uganda, and highest prevalence of the PfK13 469Y and 675V mutations in northern Uganda. These results raise concerns regarding the activity of artemisinins and treatment efficacy of ACTs, and regarding the preventive efficacy of SP, the only recommended regimen for IPT.
The decreasing prevalence of polymorphisms associated with aminoquinoline resistance is not a new observation. In the 1990s, the replacement of chloroquine to treat uncomplicated malaria in Malawi led to reversion to P. falciparum with the wild-type PfCRT K76 sequence [41]. With more recent replacement of chloroquine with ACTs to treat malaria across Africa, loss of the PfCRT 76T and PfMDR1 86Y and 1246Y mutations has been documented in many countries, including Uganda, where prevalence of parasites with the PfCRT 76T allele decreased markedly over the last decade [11, 12, 14, 42, 43]. However, unlike the situation documented in Malawi, mutant parasites have persisted in Uganda over a decade after replacement of chloroquine. Our survey of 16 sites identified PfCRT 76T mutant parasites at 11 sites in 2018 and 7 sites in 2019, albeit with decreased prevalence of this mutation at 12 of 14 evaluable sites between 2018 and 2019. Changes have been more marked for PfMDR1 resistance markers, with the PfMDR1 86Y mutation rare in 2018 and absent in 2019, and prevalence of the 1246Y mutation also decreased in recent years. In contrast, the PfMDR1 184F mutation, which has an uncertain impact on drug sensitivity, has remained common in Ugandan parasites, with prevalence >50% at most sites. Amplification of pfmdr1, which is associated with decreased sensitivity to lumefantrine and mefloquine [10, 31, 32], was not seen, consistent with older studies [12, 14, 36, 37]. Amplification of plasmepsin-2/3 genes and novel PfCRT mutations associated with resistance to piperaquine and dihydroartemisinin-piperaquine failures in southeast Asia [33–35] were not seen in Ugandan samples. Taken together, our results suggest P. falciparum sensitivity to aminoquinolines, without the worrisome polymorphisms suggestive of resistance to ACT partner drugs in Uganda. These results and recent evidence of continued excellent ex vivo activity of amodiaquine, piperaquine, and lumefantrine against cultured fresh isolates of P. falciparum [7] suggest that partner drugs for ACTs commonly used in Uganda remain active against malaria parasites currently circulating in the country.
Resistance of P. falciparum to SP has been widespread in Uganda for many years, with near-fixation of 5 PfDHFR/PfDHPS mutations that mediate an intermediate level of resistance [12, 14, 16]. SP is no longer recommended to treat malaria, but remains the World Health Organization–recommended regimen for IPTp. Of concern has been selection of additional PfDHFR/PfDHPS mutations that mediate high-level resistance [16]. Our results show high prevalence of the PfDHFR 164L and PfDHPS 581G mutations in western and central Uganda, possibly due to the selective pressures of SP used for IPTp and the antifolate trimethoprim-sulfamethoxazole used for bacterial infections and prophylaxis in human immunodeficiency virus–infected individuals. Maintenance of the PfDHFR 164L mutation might be associated with amplification of the gene encoding another folate enzyme, GTP cyclohydrolase-1 [44]; studies of GTP cyclohydrolase amplification in these samples are planned. Prevalence of these mutations, especially PfDHPS 581G, was previously noted to be high near the DRC border [12], adjacent to areas of DRC with high prevalence of 581G [19], but by 2019 both the PfDHFR 164L and PfDHPS 581G mutations were at quite high prevalence in southwestern and central Uganda. Thus, it can be anticipated that the antimalarial protective efficacy of SP is now poor in much of Uganda. However, despite its limitations as an antimalarial, IPTp with SP offered similar protective efficacy against low birth weight to that of dihydroartemisinin-piperaquine, which offers far superior antimalarial preventive efficacy, presumably due to nonmalarial benefits of SP [45]. Thus, though use of SP to prevent malaria in Uganda should be discouraged, it remains the standard of care for IPTp. Studies of the efficacy of SP for IPTp in the setting of the newly emerged PfDHFR/PfDHPS mutations are a high priority.
ACTs rely on artemisinin activity for rapid parasite clearance. Delayed clearance after therapy with ACTs has been associated with treatment failure when there is resistance to partner drugs, notably mefloquine and piperaquine [46, 47]. More than 100 PfK13 propeller domain mutations have been identified in P. falciparum, and approximately 20 of them have been associated with clinical delayed clearance [20, 24]. Many PfK13 mutations have been identified in Africa [1], but generally at low prevalence, with mutations confirmed to be associated with delayed clearance very uncommon. However, recent findings challenge the longstanding impression that artemisinin resistance is absent in Africa. First, in Rwanda, a PfK13 mutation previously associated with artemisinin resistance in Southeast Asia, 561H, was recently identified [25, 26]; we identified this mutation in a single isolate. Second, we and others have seen emergence in northern Uganda of another PfK13 mutation, 675V, that was also associated with delayed clearance after artemisinin therapy in southeast Asia [12, 20, 29]. This mutation was first identified in northern Uganda, when 1 of 194 isolates collected in 2016 had the mutation and delayed in vitro clearance in a dihydroartemisinin ring survival assay [29]. Subsequently, we identified 675V in 5.5% of isolates causing uncomplicated malaria at 3 sites in northern Uganda in 2017 [12]. Third, another PfK13 mutation, 469Y, which was also associated with delayed artemisinin clearance, was seen at 4 sites in northern Uganda in 2017 [12]. Our current results show apparent spread of the 469Y and 675V mutations in northern Uganda over 2018–2019, with prevalence of both mutations at ≥15% at 3 northern sites in 2019. Despite limited data to date on the biological significance of PfK13 mutations in Africa, the recent increases in prevalence and geographical spread suggest selection of PfK13 polymorphisms that may endanger the antimalarial activity of artemisinins. With some decrease in antimalarial activity of lumefantrine due to disappearance of the PfCRT 76T and PfMDR1 86Y mutations [7], the emergence of artemisinin resistance in Uganda may threaten the antimalarial efficacy of AL, the first-line treatment regimen.
Our studies allowed us to compare different modalities for testing genotypes of P. falciparum field isolates. Dideoxy sequencing of gene fragments of interest, a well-established LDR-FM assay, and a multiplex MIP capture next-generation sequencing assay yielded similar, but not identical data. Differences between assays is not surprising, as many study samples were expected to be polyclonal, as is typical for high-transmission African sites, and assays with different sensitivities for identification of low abundance strains would be expected to differ in prevalence results. Most commonly, MIP analyses yielded higher prevalences for mutations, suggesting greater sensitivity for minority clones in parasite mixtures. However, MIP assays were less often successful, presumably due to the need for greater quantities of DNA compared to the other assays. Overall, our results offer support for any of these 3 methods for surveillance for P. falciparum genetic polymorphisms of interest. In addition, the MIP analysis afforded high-throughput data on gene copy number, with results consistent with prior and concurrent data using qPCR.
Our study had some limitations. First, limited capacity allowed us to study only 50 isolates per site each year. Second, assay performance limited capture of data for different polymorphisms. Together these concerns led to limited ability to identify uncommon polymorphisms. Third, clinical and ex vivo drug sensitivity data were not available from this limited surveillance study, although recent studies in Uganda have shown continued excellent clinical efficacy of the major ACTs [48, 49] and good ex vivo efficacy of artemisinins and leading ACT partner drugs [7, 38]. Fourth, our sequencing strategy did not consider sequences of a number of additional P. falciparum genes whose products have been suggested as potential mediators of artemisinin resistance [50]. Despite these limitations, the overall sample size and temporal and geographic framework of this study allowed a comprehensive assessment of P. falciparum genetic polymorphisms associated with drug resistance recently circulating in Uganda.
In summary, we found decreasing prevalence of transporter mutations associated with aminoquinoline resistance, increasing prevalence of PfDHFR/PfDHPS mutations associated with high-level resistance to SP, and increasing prevalence of PfK13 propeller domain mutations that may mediate delayed parasite clearance after artemisinin therapy. These results have important implications for choosing appropriate drugs to treat and prevent malaria in Uganda and suggest that continued monitoring for evolution of antimalarial drug resistance in Africa should be a high priority.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the participants and staff members of the facilities where samples were collected.
Financial support. This work was supported by the National Institutes of Health (grant numbers R01AI075045, U19AI089674, R01AI139520).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Conrad MD, Rosenthal PJ. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis 2019; 19:e338–51. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization. World malaria report 2019. Geneva, Switzerland: WHO, 2019. [Google Scholar]
- 3. Bardají A, Bassat Q, Alonso PL, Menéndez C. Intermittent preventive treatment of malaria in pregnant women and infants: making best use of the available evidence. Expert Opin Pharmacother 2012; 13:1719–36. [DOI] [PubMed] [Google Scholar]
- 4. Dicko A, Diallo AI, Tembine I, et al. Intermittent preventive treatment of malaria provides substantial protection against malaria in children already protected by an insecticide-treated bednet in Mali: a randomised, double-blind, placebo-controlled trial. PLoS Med 2011; 8:e10000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gutman J, Kovacs S, Dorsey G, Stergachis A, Ter Kuile FO. Safety, tolerability, and efficacy of repeated doses of dihydroartemisinin-piperaquine for prevention and treatment of malaria: a systematic review and meta-analysis. Lancet Infect Dis 2017; 17:184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci 2006; 27:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasmussen SA, Ceja FG, Conrad MD, et al. Changing antimalarial drug sensitivities in Uganda. Antimicrob Agents Chemother 2017; 61:e01516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sisowath C, Petersen I, Veiga MI, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis 2009; 199:750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Somé AF, Séré YY, Dokomajilar C, et al. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob Agents Chemother 2010; 54:1949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Venkatesan M, Gadalla NB, Stepniewska K, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 2014; 91:833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Achieng AO, Muiruri P, Ingasia LA, et al. Temporal trends in prevalence of Plasmodium falciparum molecular markers selected for by artemether-lumefantrine treatment in pre-ACT and post-ACT parasites in western Kenya. Int J Parasitol Drugs Drug Resist 2015; 5:92–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Asua V, Vinden J, Conrad MD, et al. Changing molecular markers of antimalarial drug sensitivity across Uganda. Antimicrob Agents Chemother 2019; 63:e01818-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Somé AF, Sorgho H, Zongo I, et al. Polymorphisms in K13, pfcrt, pfmdr1, pfdhfr, and pfdhps in parasites isolated from symptomatic malaria patients in Burkina Faso. Parasite 2016; 23:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tumwebaze P, Tukwasibwe S, Taylor A, et al. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 2017; 215:631–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 2005; 57:117–45. [DOI] [PubMed] [Google Scholar]
- 16. Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol 2013; 29:505–15. [DOI] [PubMed] [Google Scholar]
- 17. Harrington WE, Mutabingwa TK, Muehlenbachs A, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci U S A 2009; 106:9027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alifrangis M, Lusingu JP, Mmbando B, et al. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg 2009; 80:523–7. [PubMed] [Google Scholar]
- 19. Aydemir O, Janko M, Hathaway NJ, et al. Drug-resistance and population structure of Plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J Infect Dis 2018; 218:946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amaratunga C, Hanitriniaina VA, Ashley E, et al. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments—a WWARN individual patient data meta-analysis. BMC Med 2019; 17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashley EA, Dhorda M, Fairhurst RM, et al. ; Tracking Resistance to Artemisinin Collaboration (TRAC) . Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2014; 371:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamilton WL, Amato R, van der Pluijm RW, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis 2019; 19:943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor SM, Parobek CM, DeConti DK, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 2015; 211:680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization. Artemisinin and artemisinin-based combination therapy resistance. Geneva, Switzerland: WHO, 2017. [Google Scholar]
- 25. Uwimana A, Legrand E, Stokes BH, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020; 20:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uwimana A, Umulisa N, Venkatesan M, et al. Presence of k13 561H artemisinin resistance mutations in Plasmodium falciparum infection in Rwanda, 2018 [abstract LB 5295]. In: Program and Abstracts of the 68th Annual Meeting of the American Society for Tropical Medicine and Hygiene, National Harbor, MD, 2019. [Google Scholar]
- 27. Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in southeast of Tanzania. Sci Rep 2020; 10:3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tacoli C, Gai PP, Bayingana C, et al. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in southern Rwanda, 2010–2015. Am J Trop Med Hyg 2016; 95:1090–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikeda M, Kaneko M, Tachibana SI, et al. Artemisinin-resistant Plasmodium falciparum with high survival rates, Uganda, 2014–2016. Emerg Infect Dis 2018; 24:718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conrad MD, Nsobya SL, Rosenthal PJ. The diversity of the Plasmodium falciparum K13 propeller domain did not increase after implementation of artemisinin-based combination therapy in Uganda. Antimicrob Agents Chemother 2019;63:12345-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 2004; 364:438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Price RN, Uhlemann AC, van Vugt M, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis 2006; 42:1570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amato R, Lim P, Miotto O, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 2017; 17:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Witkowski B, Duru V, Khim N, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 2017; 17:174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ross LS, Dhingra SK, Mok S, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 2018; 9:3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holmgren G, Björkman A, Gil JP. Amodiaquine resistance is not related to rare findings of pfmdr1 gene amplifications in Kenya. Trop Med Int Health 2006; 11:1808–12. [DOI] [PubMed] [Google Scholar]
- 37. Witkowski B, Nicolau ML, Soh PN, et al. Plasmodium falciparum isolates with increased pfmdr1 copy number circulate in West Africa. Antimicrob Agents Chemother 2010; 54:3049–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tumwebaze P, Conrad MD, Walakira A, et al. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 2015; 59:3018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Verity R, Aydemir O, Brazeau NF, et al. The impact of antimalarial resistance on the genetic structure of Plasmodium falciparum in the DRC. Nat Commun 2020; 11:2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hamilton WL, Amato R, van der Pluijm RW, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis 2019; 19:943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003; 187:1870–5. [DOI] [PubMed] [Google Scholar]
- 42. Mbogo GW, Nankoberanyi S, Tukwasibwe S, et al. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg 2014; 91:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malmberg M, Ngasala B, Ferreira PE, et al. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J 2013; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kümpornsin K, Modchang C, Heinberg A, et al. Origin of robustness in generating drug-resistant malaria parasites. Mol Biol Evol 2014; 31:1649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kajubi R, Ochieng T, Kakuru A, et al. Monthly sulfadoxine-pyrimethamine versus dihydroartemisinin-piperaquine for intermittent preventive treatment of malaria in pregnancy: a double-blind, randomised, controlled, superiority trial. Lancet 2019; 393:1428–39. [DOI] [PubMed] [Google Scholar]
- 46. Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis 2008; 14:716–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Amaratunga C, Lim P, Suon S, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 2016; 16:357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeka A, Kigozi R, Conrad MD, et al. Artesunate/amodiaquine versus artemether/lumefantrine for the treatment of uncomplicated malaria in Uganda: a randomized trial. J Infect Dis 2016; 213:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yeka A, Wallender E, Mulebeke R, et al. Comparative efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for the treatment of uncomplicated malaria in Ugandan children. J Infect Dis 2019; 219:1112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ross LS, Fidock DA. Elucidating mechanisms of drug-resistant Plasmodium falciparum. Cell Host Microbe 2019; 26:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.