Abstract
Human respiratory syncytial virus (HRSV) constitutes one the main causes of respiratory infection in neonates and infants worldwide. Transcriptome analysis of clinical samples using high-throughput technologies remains an important tool to better understand virus-host complex interactions in the real-life setting but also to identify new diagnosis/prognosis markers or therapeutics targets. A major challenge when exploiting clinical samples such as nasal swabs, washes, or bronchoalveolar lavages is the poor quantity and integrity of nucleic acids. In this study, we applied a tailored transcriptomics workflow to exploit nasal wash samples from children who tested positive for HRSV. Our analysis revealed a characteristic immune signature as a direct reflection of HRSV pathogenesis and highlighted putative biomarkers of interest such as IP-10, TMEM190, MCEMP1, and TIMM23.
Keywords: respiratory syncytial virus, DNA microarray, transcriptome, nasal epithelium, NanoString assay
This study applied a tailored transcriptomics workflow to exploit nasal wash samples from HRSV-positive hospitalized children. The transcriptional immune signature appears as a direct reflection of HRSV pathogenesis and highlights putative biomarkers of interest.
Human respiratory syncytial virus (HRSV) constitutes one of the main causes of respiratory tract infection in newborns and young children worldwide [1] but also in the elderly, immunocompromised, and patients with chronic heart and lung conditions [2]. The global HRSV disease burden is estimated at approximately 200 000 deaths and more than 3 million hospitalizations per year [1, 3]. Despite numerous attempts and ongoing clinical trials, no efficacious HRSV vaccine is yet available, and the specific therapeutic arsenal currently available is very limited and remains relatively expensive [4]. In this context, we urgently need to increase our understanding of HRSV pathogenesis and the multiple facets of its virus-host interactions.
Much of the HRSV-induced disease is considered to be a reflection of the host innate immune response to infection [5, 6], with respiratory epithelial cells and monocytes/macrophages being the main actors in this response [7, 8]. Indeed, HRSV infection was previously shown to upregulate the expression of host genes involved in the antiviral and cell-mediated immune responses, such as genes coding for interferons (IFNs) and more largely several cytokines/chemokines such as CXCL10/IP-10, CXCL8/IL-8, MCP-1/CCL2, RANTES/CCL5, or interleukin-6 (IL-6) [8]. For example, we previously demonstrated that HRSV infection, alone or in the context of bacterial coinfection, strongly promotes CXCL10/IP-10 expression in human macrophages [9]. We also showed that HRSV infection of human respiratory epithelial cells induces a strong disequilibrium in the p53/nuclear factor-κB (NF-κB) balance, which appears to contribute to the upregulation of several proinflammatory cytokines and chemokines [10]. One limitation of these in vitro approaches is that they do not necessarily reflect the whole complexity of the in vivo environment. In this context, we advantageously investigated the HRSV-induced host response using an innovative and highly relevant primary human reconstituted airway epithelial model, cultivated at the air-liquid interface to assess previously undescribed facets of the HRSV biology, such as the impact of the infection on cilium mobility and morphogenesis [11].
The development of high-throughput “omics” approaches has contributed to deepen our understanding of the multiple levels of interplay between respiratory viruses and the host cell [12–15]. These approaches, in addition to being very informative about the dynamic interplay between the virus and the host and hence the pathogenesis mechanisms, could also constitute a powerful tool to identify new therapeutic targets and/or propose novel antiviral strategies. In the case of HRSV, few studies have investigated the transcriptomic host response using clinical specimens, and an even more limited number have exploited respiratory tract samples [16–18]. A major challenge associated with transcriptome analysis of clinical samples is the intrinsic low copy number and/or low integrity of the nucleic acids recovered. To tackle these hassles, several research groups, including ours, have proposed, developed, adapted and optimized sample processes [19–21].
In this study, we investigated the impact of the infection on the host cell using nasal washes from hospitalized children with laboratory-confirmed HRSV infection. The analysis of HRSV-induced gene expression signature validated the importance of several IFN and cytokine-related pathways, in line with previous studies, but also provided valuable insight on potential biomarkers of diagnostic interest or as surrogates for the evaluation of future innovative treatments.
METHODS
Clinical Samples and Ethical Considerations
Written consent was obtained from parents of the 3 hospitalized children with laboratory-confirmed HRSV infections. Control samples were from the collection of samples established by the Québec Centre Hospitalier Universitaire (CHU) in the context of the RespiVir surveillance study. The protocol was approved by the ethics committee of the CHU de Québec-Université Laval. RNAs were obtained from nasal washes (in 1.5 mL saline) stored at −80°C. After thawing, the cell pellets were transferred to RNAlater stabilization solution (Thermo Fisher Scientific).
RNA Extraction and Microarray Experiment
Isolation of total RNA from nasal washes was performed using the RNeasy Micro kit (QIAGEN) with Dnase I treatment following the manufacturer’s instructions. Samples were quantified using the Quantifluor RNA System (Promega) and qualified using an Agilent RNA 6000 Pico chip on Bioanalyzer 2100 (Agilent Technologies). Whole-RNA amplification using 3 rounds of in vitro transcription was then performed in 2 steps. First, the ExpressArt Trinucleotide mRNA amplification Pico kit (Amp Tec) was used for RNA amplification. Then, cDNA synthesis was performed with the GeneChip WT PLUS Reagent Kit (Affymetrix) with a minimal input requested of 5.5 μg. Antisense copy RNA (cRNA) made in vitro using T7 RNA polymerase was then hybridized to GeneChip Human Gene 2.0 ST Array (Affymetrix) for 16 hours at 45°C and scanned using an Affymetrix 3000 7G Scanner. CEL file generation and basic quality controls were performed with a GeneChip Command Console (Affymetrix).
Data Analysis
Data were analyzed using R software and its xps (eXpression Profiling System) package (version 1.32.0) downloaded from www.bioconductor.org. Quality controls were performed to assess technical bias, RNA degradation levels, and background noise. Preprocessing steps consisted of background correction, robust multichip average normalization, probe summarization, and log2 transformation. A linear model was used to assess differential expression with the limma (Linear Models for Microarray Data) R/Bioconductor software package [22]. Genes were considered for subsequent analysis if they exhibited at least a 2-fold change in expression levels compared to the control samples coupled with P values < .05. To further functionally characterize the patient transcriptomic signature, the web-based tool DAVID 6.8 was used to determine the enriched pathways [23]. Genes predicted by TargetScan 7.2 [24] to be targeted by the up- or downregulated microRNAs (miRNAs) with cumulative weighted score less than −0.5 were used for functional enrichment analysis using the same web-based tool. To further comprehend the connections between the modulated genes in our study, we chose to represent the interactome as a graph where nodes correspond with proteins and edges with pairwise interactions using the web-based tool STRING 11.0 [25] (https://string-db.org), paired with Markov clustering [26] to extract relevant modules from such graphs.
Pediatric mRNA Datasets
We chose 3 datasets from HRSV-infected host transcriptomic studies publicly available on the Gene Expression Omnibus database (GEO) and ArrayExpress, based on peripheral pediatric blood samples (GSE69606 and E-MTAB-5195) or peripheral blood mononuclear cell (PBMC) gene expression responses to infection (GSE34205, n = 51 HRSV-infected and n = 10 controls). We extracted raw data corresponding severe disease samples from the series GSE69606 (n = 8) and MTAB-5195 (n = 18) and the recovery corresponding samples or healthy control samples (each n = 8). Raw data were processed as previously described and differential analysis was performed according to the same thresholds (P value < .05 and absolute fold-change > 2).
Reconstituted Human Airway Epithelia and Viruses
The MucilAir human airway epithelia (HAE) model from Epithelix Sarl was used for validation purposes. Infections (multiplicity of infection [MOI] of 1) were performed as previously described [11, 21]. The HRSV (HRSV-A Long strain ATCC-VR26) was produced in LLCMK2 cells (ATCC CCL7) in Eagle’s minimum essential medium (EMEM) supplemented with 2 mM l-glutamine (Sigma Aldrich), penicillin (100 U/mL), and streptomycin (100 μg/mL; Lonza), at 37°C and 5% CO2.
NanoString nCounter Validation
The nCounter platform (NanoString Technologies) was used for mRNA detection of an 86-gene panel, according to manufacturer’s instructions [27]. This custom panel gathers immunity-related genes. Briefly, 300 ng of total RNA were hybridized to the probes at 67°C for 18 hours using a thermocycler (Biometra, Tprofesssional TRIO, Analytik Jena). After loading into the nCounter Prep Station (NanoString Technologies) for purification and immobilization, the sample cartridge was transferred and imaged on the nCounter Digital Analyzer (NanoString Technologies). Counts number were normalized by the geometric mean of HPRT1 (NM_000194.1), DECR1 (NM_001359.1), RPL19 (NM_000981.3), POLR2A (NM_000937.2), and TBP (NM_001172085.1) housekeeping genes count number, as well as the negative and positive control values using nSolver analysis software (version 4.0; NanoString Technologies). Gene expression results are expressed in fold change induction compared to the mock-infected condition.
RESULTS
Differential Gene Expression in HRSV-Infected Samples
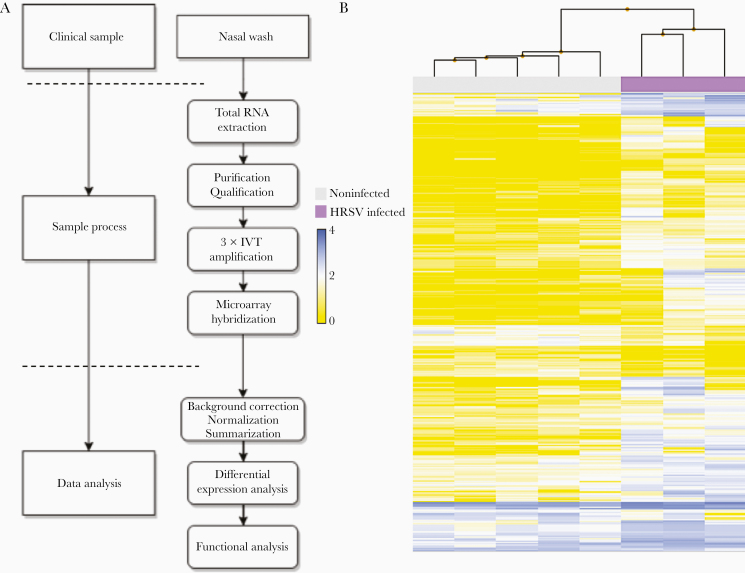
In this study, we assessed nasal airway gene expression on pediatric nasal wash samples (3 infected and 5 controls). Given the low quality and low integrity of these sensitive samples (quality status available in Supplementary Table 1), previously published adapted protocols were successfully used for their amplification and their subsequent hybridization on an Affymetrix GeneChip Human Gene 2.0ST [19]. An overview of the customized sample process and workflow is presented in Figure 1A and the subsequent hierarchical clustering of all analyzed samples is featured in Figure 1B. Despite the known heterogeneity of clinical samples, HRSV-infected and noninfected samples clustered appropriately to their corresponding experimental group.
Figure 1.
Sample processing workflow and transcriptomic hierarchical clustering. A, Adapted workflow for the processing and exploitation of clinical samples with low RNA quality/quantity. B, Hierarchical clustering of the signal intensities corresponding to all infected and noninfected clinical samples evaluated in the study. The resulting clusters are representative of the degree of similarity between samples and enable clustering into infected and noninfected experimental groups. The height of the y-axis at the branching points is a measure of similarity; y-axis units are arbitrary. For representation purposes, data was autoscaled and log2 transformed. Abbreviation: IVT, in vitro transcription.
For differential analysis, genes were considered significantly modulated if they exhibited at least a 2-fold change in expression levels compared to the control samples, with P values inferior or equal to .05. Using these criteria, we listed a total of 296 differentially expressed genes, 258 (87.16%) of them being upregulated (Supplementary Table 2). This unbalanced up- versus downregulated ratio was in line with previous observations [11]. As expected in the context of infected samples, many significantly upregulated genes with fold changes far above 5 were related to the immune and IFN responses, such as ISG15, OASL, CXCL10/IP-10, CCL2–3, IFITM1–3, and IRF1 (Supplementary Table 2). In contrast, among the 38 downregulated genes, we listed genes associated with protein heterodimerization activity (SRGAP2C, HIST3H2BB, and NTSR1), genes encoding zinc finger protein (ZNF439, ZNF28, ZNF286B, and ZNF500) or transmembrane proteins acting as receptors or T-cell coactivators (such as SLC7A5P1, MSLNL, and NTSR1). Of note, an important fraction (26%) of the downregulated genes was represented by miRNAs, such as hsa-miR-572, hsa-miR-486-2, hsa-miR-1229, and hsa-miR-663b (Supplementary Table 2). Aside from miRNAs, only 3 other downregulated genes (KIR3DL2, SLC7A5P1, and SRGAP2C) had fold changes lower than −3.
Gene Ontology-Based Functional Enrichment Analysis
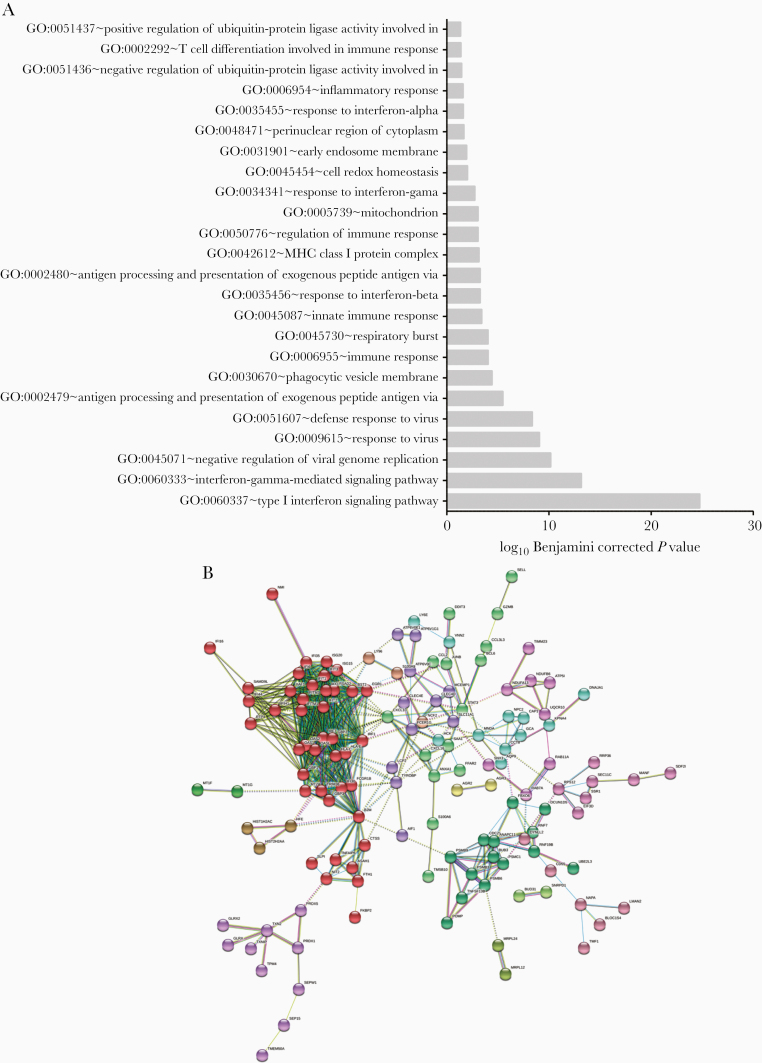
To provide further functional interpretation of these clinical transcriptomic signatures, we then performed a Gene Ontology (GO)-based functional enrichment analysis using the web-based DAVID version 6.8 toolkit (https://david.ncifcrf.gov/). GO terms, and particularly biological processes (BP), were considered enriched if their fold enrichment was higher than 2 and the Benjamini-Hochberg corrected enrichment P value was inferior to .05. This BP enrichment was based on the global list of deregulated genes (Figure 2). As anticipated, the most enriched BPs were primarily associated with interferon response (eg: GO:0060337; GO:0060333), response to virus (eg: GO:0009615; GO:0051607) or antigen processing (eg: GO:0002479; GO:0042612), which represent 16 of the 24 most enriched BPs listed (Figure 2A). Interestingly, the remaining GO terms were mainly related to mitochondria/respiratory burst (eg: GO:0005739; GO:0045730; GO:0045454) or ubiquitin ligase (eg: GO:0051437; GO:0051436).
Figure 2.
Gene Ontology-based functional enrichment and protein-protein interaction analyses. A, Enriched biological process terms corresponding to the upregulated gene list (enrichment score >2 and Benjamini-Hochberg corrected P value < .05). The downregulated gene list did not present sufficient enrichment to pass our thresholds. B, Evidence view of predicted protein associations associated with upregulated genes in the human respiratory syncytial virus-infected condition. Network nodes are host proteins and edges represent predicted functional associations. The color-coded lines correspond to the types of evidence supporting predicted associations (minimum required interaction score, 0.7). Node colors correspond to Markov clusters (inflation parameter, 1.5).
To better illustrate the impact of HRSV infection on the host immunity-related genes, we used the list of upregulated genes and explored the functional association networks of their protein products using the STRING [25] database (https://string-db.org). As presented in Figure 2B, this analysis highlighted a functional network based on 241 distinct proteins (nodes) and 637 protein-protein associations (edges). These associations, which highlight proteins sharing functions but not necessarily physical interaction, are categorized into 15 relevant clusters, among which 10 contained more than 3 proteins (each color = 1 cluster by Markov clustering [26]). Two major hubs concentrating a large number of edges were identified. The main hub consisted of proteins related to the immune response, with a central place for major actors like CXCL10, OASL, and ISG15 (Figure 2B, red dots). The second hub harbored proteins involved in the positive and negative regulation of ubiquitin-protein ligase activity during the mitotic cell cycle (GO:0051436 and GO:0051437, medium purple dots). The first hub would be expected in an infection context; however, the specific modulation of genes related to respiratory burst, cell redox homeostasis, or ubiquitin-protein ligase activity by HRSV infection has been less studied.
Because of the numerous miRNAs deregulated, we used a prediction algorithm (TargetScan 7.2) [24] for the identification of all genes targeted by the down- and upregulated miRNAs. The predicted genes targeted by the downregulated miRNAs were related to biological processes such as phagocytosis (GO:0006909), peptide cross-linking (GO:0018149), or positive regulation of release of cytochrome c from mitochondria (GO:0090200). Interestingly, the predicted targets of the upregulated miRNAs are linked to similar processes: negative regulation of nucleic acid-templated transcription (GO:1903507) and negative regulation of T cell proliferation (GO:0042130) for the most enriched BPs (Supplementary Figure 1).
Tissue-Specific Gene Expression
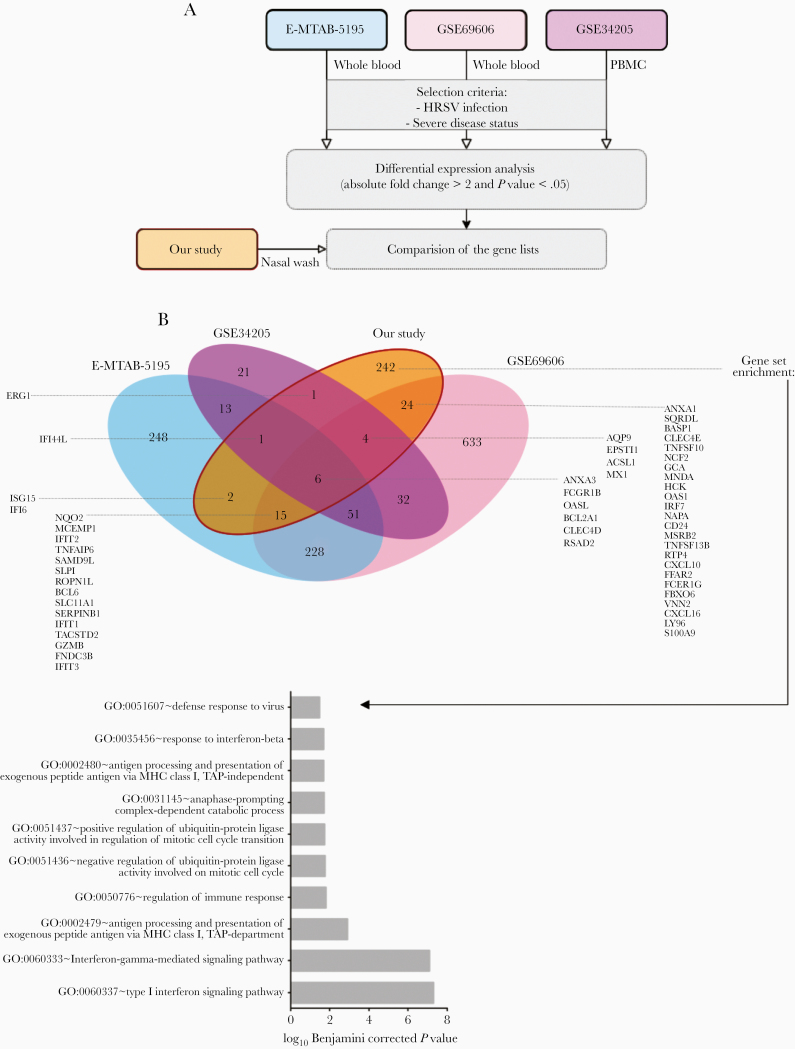
To describe tissue-specific HRSV infection signatures, differentially expressed gene lists were extracted from 3 pediatric mRNA array datasets [28–30] and compared to our data to highlight similarities and differences between blood and respiratory airway transcriptional profiles, respectively (Figure 3). As previously shown, overlap between the blood/PBMC and respiratory tract gene expression is scarce [29]. Only 6 genes (ANXA3, FCGR1B, OASL, BCL2A1, CLEC4D, and RSAD2) were upregulated in all analyzed datasets, mostly associated with the host immune response to the infection. As expected, both studies on peripheral blood shared a specific signature composed of 228 genes, whereas 51 additional genes were also modulated in PBMCs. These genes are mostly associated with the innate immune response of the host (GO:0045087). When comparing these signatures with the list of genes deregulated in our study, we highlighted 242 genes exclusively modulated in nasal washes, hence constituting specific drivers of the nasal epithelium signature. Among these tissue-specific modulated genes, were genes associated with the immune response regulation (GO:0050776) and, more precisely, with type I interferon signaling pathway (GO:0060337) or antigen processing and presentation (GO:0002479).
Figure 3.
Gene expression cross-analysis as a key to tissue-specific local reaction to infection. A, We selected 3 pediatric mRNA expression datasets for gene expression comparison across tissues. The GSE69606 dataset combines samples from 26 patients with acute HRSV infections, with symptoms spanning from mild to severe and the corresponding recovery paired samples. The E-MTAB-5195 dataset was originally used to investigate blood transcriptomics of 39 children during HRSV infection and for a longitudinal analysis to determine an 84-gene prognosis signature discriminating hospitalized infants with severe HRSV disease from infants with mild symptoms. The GSE34205 dataset is part of a wider study (GSE32140), aiming at establishing the signature induced by influenza and HRSV on PBMCs and primary airway epithelial cells. Because of the disease status of our study samples, we focused on severely ill children infected by HRSV. B, Comparative cross-analysis of the gene lists on the 4 datasets (3 external plus ours). Common and specific infection features and gene enrichment analysis applied to the 242 nasal-specific genes are shown. Abbreviations: HRSV, human respiratory syncytial virus; PBMC, peripheral blood mononuclear cell.
Validation of Differential Expression in Reconstituted Human Airway Epithelium
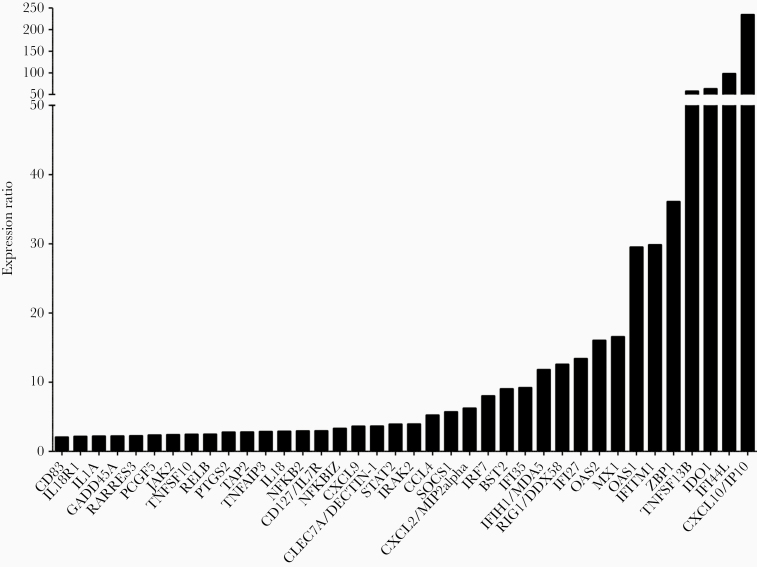
We then sought to validate these results in the context of experimental infections with the prototype HRSV A Long strain (MOI = 1) in a human reconstituted HAE model, as previously described [11]. This HAE model, originating from healthy donor biopsies, is composed of human primary ciliated columnar cells, mucus-secreting goblet cells, and basal cells cultivated at the air-liquid interface, and has been successfully used to study viral infections in previous studies [11, 21]. After 6 days of infection, HAE were lysed and total RNA was extracted and subsequently analyzed with the NanoString nCounter platform using a customized 94 immunity-related (cytokine production, T-cell proliferation, interferon-gamma-mediated signaling pathway, etc.) gene panel [27]. As shown in Figure 4, 39 of the 94 genes in the NanoString panel were differentially modulated in the HRSV-infected condition compared to the mock-infected control. Despite the differential nature of infectious samples, the comparison of global gene expression modulation results between Affymetrix microarray (clinical samples) and NanoString (experimental infections in HAE) assays showed a correlation coefficient of 0.63. This relatively high correlation could be explained by the fact that both types of samples contain cells of nasal origin. Unsurprisingly, the most upregulated gene in the infectious context was CXCL10/IP-10, followed by IFI44L, IDO1, and TNFSF13B, with expression ratios above 50 (Figure 4). The top 20 modulated genes are strongly linked to “type I interferon signaling pathway” (GO:0060337) or more widely to “response to virus” (GO:0009615 or GO:0051607). Altogether, the gene expression results observed in clinical samples were cross-validated using an alternative method and underline a global deregulation of the biological defenses of the host, notably in the case of interferon stimulated genes (ISGs) that constitute a hallmark of many infectious and/or autoimmune disease states.
Figure 4.
Experimental validation of gene expression results by NanoString assay in human airway epithelia (HAE). The expression of immunity-related genes in infected nasal HAE was validated using Nanostring nCounter technology. Data processing and normalization were performed with nSolver 4.0 analysis software and significant results (absolute fold change > 2) are expressed in fold change induction compared to the mock-infected condition.
Discussion
In the particular context of HRSV infections, most of the respiratory samples collected for clinical studies are originating from pediatric patients, from whom only limited amounts of material are obtained. Moreover, given the fact that the main purpose of patient sampling is usually clinical analysis rather than research, the quantity and quality of exploitable material is far from optimal. In this study, we showed that these hurdles for the exploitation of highly degraded clinical samples could be mitigated by using adapted protocols and microarray, such as the Affymetrix GeneChip Human Gene 2.0 ST. Thus, despite starting from a limited number of nasal washes from children presenting an acute HRSV infection, our adapted sample-processing pipeline enabled the determination and characterization of robust pediatric HRSV-induced nasal transcriptome signatures. In this study, patient-derived samples were processed with adapted protocols and transcriptomic signatures obtained by hybridization on the HuGene 2.0 st Affymetrix microarray and subsequent processing of the data. We compared our results to published pediatrics blood microarray datasets for the establishment of a nasal-specific signature. We also included biological results obtained using our previously described relevant reconstituted HAE model of HRSV infection [11] for a deeper comprehension of the virus impact on the host epithelium.
Initial signature analysis enabled the identification of well-known markers of HRSV infection, namely the upheaval of the immune cascade [15–17], notably highlighting the strong up-regulation of CXCL10/IP-10 gene expression. We then advantageously used a biologically relevant reconstituted HAE model to reproduce and validate these results by NanoString nCounter assay. IP-10 has already been described as the most abundant cytokine in bronchoalveolar lavages collected from HRSV-infected patients [31]. We also identified upregulated genes that seem to be commonly modulated in different infection scenarios. For example, HRASLS2 is highly induced in rhinovirus infection [32], 4FFAR2 promotes internalization during influenza A virus entry [33], and IFI6 has pleiotropic functions in HRSV, Dengue, and hepatitis C virus infections [34–36]. Conversely, our analysis revealed TMEM190 and MCEMP1 as potential specific biomarkers of HRSV infection. Although, TMEM190 expression was largely decreased in small airway epithelium by smoking [37] and MCEMP1 was proposed as a biomarker for stroke prognosis [38], a particular expression profile of these genes has not yet been described in other viral infections, for which they underscore further study as potential biomarkers. A similar rationale supports the study of TIMM23, which was strongly upregulated in our study. A previous study conducted by Zaas et al [15] identified a panel of 15 genes specifically modulated in HRSV-infected adults, among which FCGR1B, GBP1, RTP4, RSAD2, ISG15, and IFIT2 were also significantly deregulated in our study. Despite the different nature of the biological samples used (children nasal washes versus adult blood), the high degree of concordance observed between our results and theirs supports a distinctive HRSV infection signature.
In addition to genes related to the immune response, our transcriptomics results also highlighted the modulation of genes and pathways related to global mitochondrion cellular process disruption. This suggests that HRSV infection could alter less-studied biological processes related to the cAMP cascade, the redox complexes of mitochondrial respiratory chain (respiratory burst). Although this impact on the respiratory chain is a less-explored aspect of HRSV pathogenesis in the respiratory tract, a study by Bataki et al [39] investigated whether HRSV can directly signal activation or deactivation of neutrophil cytotoxic function or not in the context of infant bronchiolitis. Besides HRSV infections, the respiratory burst is known to be defective in influenza-infected neutrophils and during coinfections [40, 41]. The disruption of such metabolic processes could be a first clue regarding prognosis in children infected by HRSV.
The biological interpretation of the 38 downregulated genes is not as straightforward. Indeed, no biological process or function was significantly enriched in our study, only modulations of individual gene were highlighted. Some markers, already described in the context of other viral infections, were found in the top downregulated genes [42, 43]. This protection is known to be mediated partially by the inhibitory natural killer (NK) cell receptor KIR3DL2, whose gene expression was strongly inhibited in our HRSV infection context. The SLC7A5P1 gene is also predicted to be linked to NK cells (GO:0032825~positive regulation of natural killer cell differentiation) whereas SRGAP2C has already been linked to HRSV bronchiolitis. Interestingly, we also observed the significant downregulation of several miRNAs. Among them, some have already been described in the literature, such as miRNA-572 a prognosis biomarker for renal cell carcinoma and sclerosis [44, 45], and miRNA-769, which was included in an miRNA panel for discrimination between Mycobacterium tuberculosis infected and healthy individuals [46].
Regardless of the tissue studied, HRSV is reported to be a major disruptor of the host immune response [47–49]. Here, comparing our signatures with 3 others from pediatric whole-blood transcriptomic analyses [28–30], we highlighted the common deregulation of 7 genes, independent of the tissue, and, interestingly, 242 genes that seemed to be specific to nasal epithelium HRSV-induced gene expression, in line with data from the literature [50].
Collectively, the transcriptomic analysis of nasal wash samples highlights the qualitative importance of such clinical samples, particularly in the context of their limited availability. The results obtained with a complementary approach such as the reconstituted HAE greatly contribute to bridge the knowledge gap in the understanding of the specific effects of HRSV on the host respiratory tissue and pave the way for several, so far undescribed, avenues of investigation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Sophie Assant for her help with the NanoString nCounter assay and Epithelix (Switzerland) for help with MucilAir human airway epithelia.
Author contributions. C. N. D. L., A. P., B. L., G. B., C. L. L., O. T., and M. R. C. participated in conception and coordination of the study. C. N. D. L., A. P., B. P., J. C., E. O., T. J., A. T., B. L., M. E. H., M. R., J. T., G. B., C. L. L., O. T., and M. R. C. carried out the experiments and analysis of the results. C. N. D. L., J. D., O. T., A. P., and M. R. C. designed the study and wrote the manuscript.
Disclaimer. Funding institutions had no participation in the design of the study, collection, analysis and interpretation of data, or in the writing of the manuscript.
Financial support. This work was supported by Région Auvergne Rhône-Alpes (CMIRA grant number 14007029 and AccueilPro COOPERA grant number 15458); and Canadian Institutes of Health Research (grant numbers 229733 and 230187). C. N. D. L. was funded by the National Association for Research in Technology.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 2010; 375:1545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 2005; 352:1749–59. [DOI] [PubMed] [Google Scholar]
- 3. Hall CB, Simőes EAF, Anderson LJ Clinical and epidemiologic features of respiratory syncytial virus. Berlin, Heidelberg: Springer, 2013:39–57. [DOI] [PubMed] [Google Scholar]
- 4. Griffiths C, Drews SJ, Marchant DJ. Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin Microbiol Rev 2017; 30:277–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Durbin JE, Johnson TR, Durbin RK, et al. The role of IFN in respiratory syncytial virus pathogenesis. J Immunol 2002; 168:2944–52. [DOI] [PubMed] [Google Scholar]
- 6. Ramaswamy M, Shi L, Monick MM, Hunninghake GW, Look DC. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am J Respir Cell Mol Biol 200430:893–900. [DOI] [PubMed] [Google Scholar]
- 7. Tripp RA, Oshansky C, Alvarez R. Cytokines and respiratory syncytial virus infection. Proc Am Thorac Soc 2005; 2:147–9. [DOI] [PubMed] [Google Scholar]
- 8. Russell CD, Unger SA, Walton M, Schwarze J. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev 2017; 30:481–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Machado D, Hoffmann J, Moroso M, et al. RSV infection in human macrophages promotes CXCL10/IP-10 expression during bacterial co-infection. Int J Mol Sci 201718:2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Machado D, Pizzorno A, Hoffmann J, et al. Role of p53/NF-κB functional balance in respiratory syncytial virus-induced inflammation response. J Gen Virol 2018; 99:489–500. [DOI] [PubMed] [Google Scholar]
- 11. Nicolas de Lamballerie C, Pizzorno A, Dubois J, et al. Characterization of cellular transcriptomic signatures induced by different respiratory viruses in human reconstituted airway epithelia. Sci Rep 2019; 9:11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parnell GP, McLean AS, Booth DR, et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 2012; 16:R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hancock AS, Stairiker CJ, Boesteanu AC, et al. Transcriptome analysis of infected and bystander type 2 alveolar epithelial cells during influenza a virus infection reveals in vivo Wnt pathway downregulation. J Virol 2018; 92:e01325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Troy NM, Bosco A. Respiratory viral infections and host responses; insights from genomics. Respir Res 2016; 17:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009; 6:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levitz R, Gao Y, Dozmorov I, Song R, Wakeland EK, Kahn JS. Distinct patterns of innate immune activation by clinical isolates of respiratory syncytial virus. PLoS One 2017; 12:e0184318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Do LAH, Pellet J, van Doorn HR, et al. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J Infect Dis 2017; 217:134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ampuero S, Andaur R, Milano M, et al. Time-course of transcriptome response to respiratory syncytial virus infection in lung epithelium cells. Acta Virol 2018; 62:310–25. [DOI] [PubMed] [Google Scholar]
- 19. Dupinay T, Nguyen A, Croze S, et al. Next-generation sequencing of ultra-low copy samples: from clinical FFPE samples to single-cell sequencing. Curr Top Virol 2012; 10:63–83. [Google Scholar]
- 20. Degletagne C, Keime C, Rey B, et al. Transcriptome analysis in non-model species: a new method for the analysis of heterologous hybridization on microarrays. BMC Genomics 2010; 11:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pizzorno A, Terrier O, Nicolas de Lamballerie C, et al. Repurposing of drugs as novel influenza inhibitors from clinical gene expression infection signatures. Front Immunol 2019; 10:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4:P3. [PubMed] [Google Scholar]
- 24. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115:787–98. [DOI] [PubMed] [Google Scholar]
- 25. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Dongen SD Graph clustering by flow simulation [PhD dissertation]. Utrecht, the Netherlands: University of Utrecht, 2000. [Google Scholar]
- 27. Tsang H-F, Xue VW, Koh S-P, Chiu Y-M, Ng LP-W, Wong S-CC. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn 2017; 17:95–103. [DOI] [PubMed] [Google Scholar]
- 28. Jong VL, Ahout IM, van den Ham HJ, et al. Transcriptome assists prognosis of disease severity in respiratory syncytial virus infected infants. Sci Rep 2016; 6:36603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ioannidis I, McNally B, Willette M, et al. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol 2012; 86:5422–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brand HK, Ahout IM, de Ridder D, et al. Olfactomedin 4 serves as a marker for disease severity in pediatric respiratory syncytial virus (RSV) infection. PLoS One 2015; 10:e0131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McNamara PS, Flanagan BF, Hart CA, Smyth RL. Production of chemokines in the lungs of infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis 2005; 191:1225–32. [DOI] [PubMed] [Google Scholar]
- 32. Chen Y, Hamati E, Lee PK, et al. Rhinovirus induces airway epithelial gene expression through double-stranded RNA and IFN-dependent pathways. Am J Respir Cell Mol Biol 2006; 34:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang G, Jiang L, Wang J, et al. The G protein-coupled receptor FFAR2 promotes internalization during influenza A virus entry. J Virol 2019; 94:e01707-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mejias A, Dimo B, Suarez NM, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi Y, Li Y, Zhang Y, et al. IFI6 inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2 infected vascular endothelial cells. PLoS One 2015; 10:e0132743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer K, Kwon YC, Liu S, Hagedorn CH, Ray RB, Ray R. Interferon-α inducible protein 6 impairs EGFR activation by CD81 and inhibits hepatitis C virus infection. Sci Rep 2015; 5:9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hackett NR, Butler MW, Shaykhiev R, et al. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genomics 2012; 13:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raman K, O’Donnell MJ, Czlonkowska A, et al. Peripheral blood MCEMP1 gene expression as a biomarker for stroke prognosis. Stroke 2016; 47:652–8. [DOI] [PubMed] [Google Scholar]
- 39. Bataki EL, Evans GS, Everard ML. Respiratory syncytial virus and neutrophil activation. Clin Exp Immunol 2005; 140:470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heo JY, Song JY, Noh JY, et al. Effects of influenza immunization on pneumonia in the elderly. Hum Vaccin Immunother 2018; 14:744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 2013; 9:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hansasuta P, Dong T, Thananchai H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol 2004; 34:1673–9. [DOI] [PubMed] [Google Scholar]
- 43. Cosman D, Fanger N, Borges L, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 1997; 7:273–82. [DOI] [PubMed] [Google Scholar]
- 44. Pan X, Li Z, Zhao L, et al. microRNA‑572 functions as an oncogene and a potential biomarker for renal cell carcinoma prognosis. Oncol Rep 2018; 40:3092–101. [DOI] [PubMed] [Google Scholar]
- 45. Mancuso R, Hernis A, Agostini S, Rovaris M, Caputo D, Clerici M. MicroRNA-572 expression in multiple sclerosis patients with different patterns of clinical progression. J Transl Med 2015; 13:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cui JY, Liang HW, Pan XL, et al. Characterization of a novel panel of plasma microRNAs that discriminates between Mycobacterium tuberculosis infection and healthy individuals. PLoS One 2017; 12:e0184113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP, Cormier SA. Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front Immunol 2019; 10:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heinonen S, Rodriguez-Fernandez R, Diaz A, Oliva Rodriguez-Pastor S, Ramilo O, Mejias A. Infant immune response to respiratory viral infections. Immunol Allergy Clin North Am 2019; 39:361–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carvajal JJ, Avellaneda AM, Salazar-Ardiles C, Maya JE, Kalergis AM, Lay MK. Host components contributing to respiratory syncytial virus pathogenesis. Front Immunol 2019; 10:2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pennings JLA, Schuurhof A, Hodemaekers HM, et al. Systemic signature of the lung response to respiratory syncytial virus infection. PLoS One 2011; 6:e21461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.