Abstract
Immortalized T cells such as T cell hybridomas, transfectomas, and transductants are useful tools to study trimolecular complexes consisting of peptide, MHC, and T cell receptor (TCR) molecules. These cells have been utilized for antigen discovery studies for decades due to simplicity and rapidness of growing cells. However, responsiveness to antigen stimulation is typically less sensitive compared to primary T cells, resulting in occasional false negative outcomes especially for TCRs having low affinity to a peptide-MHC complex (pMHC). To overcome this obstacle, we genetically engineered T cell hybridomas to express additional CD3 molecules as well as CD4 with two amino acid substitutions that increase affinity to MHC class II molecules. The manipulated T cell hybridomas that were further transduced with retroviral vectors encoding TCRs of interest responded to cognate antigens more robustly than non-manipulated cells without evoking non-antigen specific reactivity. Of importance, the manipulation with CD3 and mutated human CD4 expression was effective in increasing responsiveness of T cell hybridomas to a wide variety of TCR, peptide, and MHC combinations across class II genetic loci (i.e. HLA-DR, HLA-DQ, HLA-DP, and murine H2-IA) and species (i.e. both humans and mice), and thus will be useful to identify antigen specificity of T cells.
Keywords: Antigen discovery, T cell hybridomas, Genetic manipulation, CD4, MHC class II
1. Introduction
Identification of antigen specificity for self-reactive and tumor-specific T cells is important to develop antigen-specific biomarkers and immunotherapies for autoimmune diseases and cancers. One of most reliable ways to define antigens is to determine antigen specificity of tissue-targeting T cells such as tissue-derived T cell clones (Michels et al., 2017; Kent et al., 2005; Babon et al., 2016; Pathiraja et al., 2015). While T cell cloning is a powerful tool in this regard, it requires professional expertise, and T cell clones, in particular those derived from human peripheral blood cells tend to lose responsiveness to antigens after a long term cell culture and multiple freezing-and-thawing cycles. As an alternative to T cell cloning to overcome these technical difficulties, analysis of T cell hybridomas or T cell transductants that are genetically manipulated to express T cell receptors (TCR) on host T cells is now a common strategy to test antigen specificity (Scott-Browne et al., 2011; Bethune et al., 2016). Genetic manipulations include gene delivery using replication-incompetent retro/lentiviruses, which allows T cells to stably express TCRs of interest. While these T cell transductants are readily expanded without special skills and reagents, responsiveness to antigens by T cell transductants are generally not as robust as that by primary T cells and T cell clones. This is particularly a problem for autoreactive and tumor-specific T cells because their responsiveness to antigens is typically weak with in-vitro T cell stimulation assays (Tollefsen et al., 2006; Yang et al., 2014).
There are several possibilities that may cause insufficient responsiveness by T cell transductants. T cell transductants typically express low levels of TCRs on the cell surface, resulting in the low avidity between TCRs and peptide-MHC (pMHC) complexes. Alternatively, insufficient or lack of secondary molecules that support the TCR-pMHC interaction (e.g. CD28) and limited expression of other intra- and extra-cellular activation molecules may be a potential defect in T cell transductants. In this report, we focused on molecules that are required for the primary TCR-pMHC interaction to reinforce sensitivity and specificity to antigen stimulation rather than secondary or activation molecules which may increase T cell activity without antigen specificity. The primary TCR-pMHC interaction is supported by a CD4 or CD8 molecule on T cells, which directly binds to an MHC molecule and mediates antigen-specific stimulation signals in a T cell (Gay et al., 1987; Hampl et al., 1997). Recently, Mariuzza and his colleagues identified two amino acid residues in human CD4 that are critical for interaction with MHC class II molecules (Wang et al., 2011). They demonstrated that substitution of amino acids in CD4, glutamine to tyrosine at position 40 and threonine to tryptophan at position 45, greatly increases its affinity for HLA-DR1. Given this evidence, we hypothesized that expression of the mutant CD4 in T cell transductants greatly increases their antigen sensitivity.
Among numerous immortalized T cell lines to be used as host cells to express TCRs, we used a hybridoma T cell line derived from a mouse CD4 T cell, named 5KC cells (White et al., 1993), in this report. 5KC cells do not express their own endogenous TCR, therefore genetically introduced TCR genes are solely expressed without competition, resulting in the robust responsiveness to antigen stimulation, while pre-venting the occasional reactivity that may be induced by pairing with an endogenous TCR. Indeed, responses by 5KC T cell hybridomas are more potent and specific to antigen stimulation compared to other host cell lines expressing endogenous TCRs that we have studied simultaneously in our laboratories (i.e. Jurkat Clone E6-1 (ATCC TIB-152), J.RT3-T3.5 (ATCC TIB-153), SUP-T1 (ATCC CRL-1942), and primary T cells isolated from human peripheral blood). To further make 5KC cells more sensitive to antigen stimulation, in particular when testing TCRs derived from humans, we manipulated the cells with additional genes that are expected to augment the primary TCR-pMHC interaction. Here we show that introducing the high affinity mutant CD4 gene dramatically improved sensitivity to antigen stimulation without elevation of non-antigen specific responsiveness. Additional introduction of a retroviral vector expressing all the components of the CD3 complex further increased the intensity of response. Ultimately, this manipulated 5KC T cell line achieved several orders of magnitude higher sensitivity to antigen stimulation compared to non-manipulated cells.
2. Materials and methods
2.1. Retroviral vectors
2.1.1. Retroviral vectors encoding human CD4 genes with or without mutations and murine CD3 genes
The human CD4 gene fragment was synthesized based on the published amino acid sequences (NCBI NP_000607), and two mutations (Q40Y and T45W) were further introduced by elongation PCR using overlapping primers containing these mutations. The human CD4 gene constructs with or without the mutations were cloned into a murine stem cell virus (MSCV)-based retroviral vector encoding the hygro-mycin-resistant gene (Clontech). The retrovirus plasmid containing the murine CD3 genes (CD3γ, CD3δ, CD3ε, and the ζ chain (CD247) linked by 2A peptides) along with the ametrine fluorescent protein gene was kindly provided by Dr. Dario Vignali (Szymczak et al., 2004).
2.1.2. TCR gene-encoding retroviral vectors
Human TCR alpha and beta chain variable gene segments were amplified by RT-PCR using RNA that was isolated from each T cell clone (1B11 originally generated by Dr. Howard W. Davidson, T1D-3, T1D-4, and T1D-10 provided by Dr. William Kwok (Yang et al., 2014), clone 5 provided by Dr. Bart Roep (Eerligh et al., 2011), 489 and 233 provided by Dr. Ludvig Sollid (Tollefsen et al., 2006)) or synthesized based on the published sequence information (GSE.23G6 (Michels et al., 2017), 12-4.1 (Simone et al., 1997), Ob.3D1 (Wucherpfennig et al., 1994)). Each alpha or beta variable region gene segment was then connected to the mouse alpha or beta chain constant gene segment. Alpha and beta chain gene constructs from the TCR 1B11 were cloned into a MSCV-based vector encoding the neomycin-resistant or puromycin-resistant gene, respectively. For all others, alpha and beta chain gene fragments were connected with the porcine teschovirus-1 2A (P2A) peptide (Szymczak & Vignali, 2005) between them, followed by cloning into a MSCV-based retroviral vector encoding the neomycin-resistant gene, green fluorescent protein gene, or yellow fluorescent protein gene (Holst et al., 2006).
2.1.3. MHC gene-encoding retroviral vectors
The HLA-DQ8 alpha (DQA1*0301) and beta (DQB1*0302), the HLA-DQ2 alpha (DQA1*0501) and beta (DQB1*0201), the HLA-DP4 alpha (DPA1*0103) and beta (DPB1*0401), and the HLA-DR15 alpha (DRA*0101) and beta (DRB1*1501) gene fragments were amplified from RNA isolated from peripheral blood of individuals having these alleles and were confirmed to be identical to sequences published in the IPD-IMGT/HLA Database (http://www.ebi.ac.uk/ipd/imgt/hla/). The H2-I-Ag7 alpha (I-Aαd) and beta (I-Aβg7) gene fragments were amplified from spleen cells of a non-obese diabetic mouse. The alpha and beta chain genes were connected by the P2A peptide, and cloned into a MSCV-based vector encoding the green fluorescent protein gene or the tag blue fluorescent protein 2 gene (Holst et al., 2006).
2.2. Generation of T and B cell transductants
Replication-incompetent retroviruses were generated by co-transfection of Phoenix-ECO cells (ATCC CRL-3214) with each MSCV-based retroviral plasmid and the pCL-Eco packaging plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). A T cell hybridoma line lacking TCR alpha and beta chain gene expression (5KC_73.8.20) (White et al., 1993) or a murine B cell line (M12C3) (Glimcher et al., 1985) were spin-infected with retroviruses carrying each gene of interest as described previously (Nakayama et al., 2012; Bettini et al., 2013). Transduced cells were selected by treatment with G418 sulfate (Thermo Fisher Scientific, 750 μg/ml for 5 days) and/or puromycin (Thermo Fisher Scientific, 3 μg/ml for 3 days) or flow cytometric cell sorting of fluorescence-positive cells according to the selection genes carried by the retroviral plasmids.
2.3. T cell stimulation assay
T cell responses were evaluated by IL-2 production measured by ELISA as described previously (Nakayama et al., 2012). Briefly, 1 × 105 5KC T cell transductants were cultured with various concentrations of peptides in the presence of 5 × 104 M12C3 B cell transductants expressing cognate MHC molecules in each well of 96-well plates for 16–20 h. Responsiveness to stimulation by anti-CD3ε antibody (Thermo Fisher Scientific, 145-2C11, 5 μg/ml) was assessed as well. IL-2 production into supernatant was measured by ELISA using the capture antibody (Thermo Fisher Scientific, JES6-1A12) and secondary biotinylated antibody (Thermo Fisher Scientific, JES6-5H4). Peptides (> 95% purity) were synthesized by Genemed Synthesis Inc. and were dissolved in phosphate-buffered saline at 2 mg/ml. Amino acid sequences of peptides used in this study are shown in Table 1.
Table 1.
Peptide amino acid sequences.
| Peptide name | Amino acid sequence |
|---|---|
| Insulin B:9–23 | SHLVEALYLVCGERG |
| Insulin B:9–23R22E | SHLVEALYLVCGEEG |
| Insulin B:13–23 | EALYLVCGERG |
| α-gliadin:228–240 | SGQGSFQPSQQNP |
| α-gliadin:62–73Q65E | PQPELPYPQPQL |
| MBP:85–99 | ENPVVHFFKNIVTPR |
2.4. Flow cytometry
5KC T cell transductants were stained with anti-murine TCRβ antibody (eBioscience, H57-597) and anti-CD3ε antibody (Thermo Fisher Scientific, 145-2C11) followed by analysis on FACS Calibur.
3. Results
3.1. Responsiveness to antigens by 5KC T cell hybridomas expressing endogenous murine CD4
We selected five TCRs expressed by T cell clones that were isolated from type 1 diabetes (T1D) patients, which is the immune-mediated form of diabetes, having the high genetic risk HLA-DQ8 gene (Yang et al., 2014; Eerligh et al., 2011) to be expressed on 5KC T cell hybridomas. All the T cell clones are reactive to an immunodominant insulin B chain peptide consisting of amino acid 9–23 (insulin B:9–23) presented by HLA-DQ8. The clone 5 TCR was isolated from a T cell clone that is derived from peripheral blood mononuclear cells pulsed with insulin B:6–22, whereas the other four clones (T1D-3, T1D-4, T1D-10, and 1B11) were established from peripheral blood mononuclear cells that were enriched based on specificity to a mimotope of insulin B:9–23 having a substitution of arginine with glutamic acid at position 22 (insulin B:9–23R22E, amino acid sequence shown in Table 1), therefore respond to insulin B:9–23R22E more strongly than the native insulin B:9–23 peptide (Yang et al., 2014). We expressed TCRs isolated from these five T cell clones on non-manipulated 5KC T cell hybridomas and tested them for the response to insulin B:9–23, insulin B:9–23R22E, or a truncated form of insulin B:9–23 (insulin B chain 13–23 amino acid peptide shown in Table 1, insulin B:13–23 hereafter) in the presence of M12C3 B cells expressing HLA-DQ8. 5KC cells expressing the TCRs from T1D-3, T1D-4, and T1D-10 minimally responded to insulin B:9–23 (Fig. 1A, C, and E, black diamonds) while responses to B:9–23R22E were detectable at moderate concentrations of peptide (Fig. 1B, D, and F, black diamonds). 5KC cells expressing the clone 5 TCR responded to insulin B:9–23 and B:13–23 only at the high concentration of peptides (Fig. 1I and J, black diamonds). On the other hand, responses by 5KC cells expressing the 1B11 TCR to either insulin B:9–23 or B:9–23R22E were undetectable (Fig. 1G and H, black diamonds) even at the highest peptide concentration (100 μg/ml).
Fig. 1.
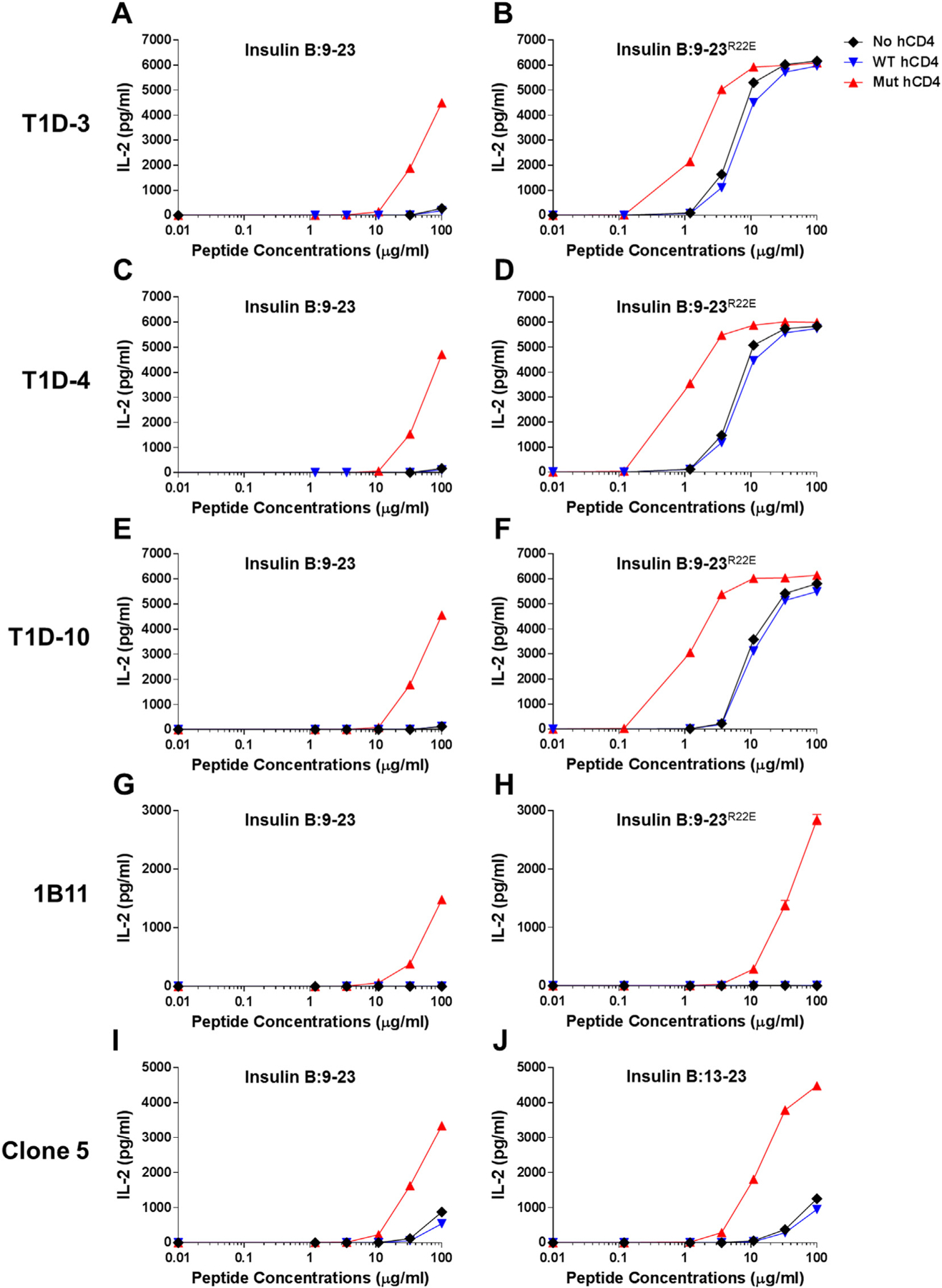
Responses to the insulin B chain peptides by 5KC T cell transductants expressing the native or mutated human CD4 molecule. 5KC cells expressing the T1D-3 (A and B), T1D-4, (C and D), T1D-10 (E and F), 1B11 (G and H), and clone 5 (I and J) were tested for the response to the insulin B:9–23 peptide (A, C, E, G, and I), the insulin B:9–23R22E mimotope (B, D, F, and H), or the truncated insulin B:13–23 peptide (J). Non-manipulated 5KC cells (black diamonds), those expressing native human CD4 (blue inverse-triangles) or mutated human CD4 [Q40Y, T45W] (red triangles) were studied in parallel. 5KC T cell transductants expressing the mutated human CD4 robustly responded to the cognate peptides at lower concentrations than non-manipulated or those expressing the native human CD4. All data shown are representative of at least three independent experiments.
3.2. No significant effects on sensitivity to antigen stimulation by expression of the native human CD4 molecule
5KC T cell hybridomas are derived from spleen cells of a B10.BR mouse (White et al., 1993), and thus these cells express the mouse CD4 molecule. Functional compatibility between mouse CD4 and human HLA molecules is still inconclusive (Yamamoto et al., 1994; Altmann et al., 1995; Pan et al., 1998; Taneja & David, 1998), partly because affinity of CD4 for pMHCII is extremely low (Xiong et al., 2001; Davis et al., 2003; Jonsson et al., 2016). Therefore, we tested whether expression of human CD4 exaggerates reactivity to antigens by the five TCRs described above. As shown in Fig. 1, response levels by all TCR transductants were similar whether or not cells express human CD4 (Fig. 1, blue inverse-triangles [expressing human CD4] vs. black diamonds [no manipulation]). Expression of the native human CD4 does not improve responsiveness by 5KC cells to cognate antigens.
3.3. Expression of the mutated human CD4 increases responsiveness to antigens
Given the study by Mariuzza et al. that two amino acid substitutions, Q40Y and T45W, in the human CD4 molecule dramatically increase the affinity between CD4 and HLA-DR1 (Wang et al., 2011), we examined whether expression of the human CD4 molecule with these mutations exaggerates responsiveness to antigens by 5KC cells. Of note, all amino acid residues in HLA molecules, DR1 and DQ8, that interact with CD4 are identical (Table 2), including those residues that contact the two mutated amino acids in CD4. Strikingly, 5KC cells expressing the mutated human CD4 responded to the insulin peptides more robustly compared to non-manipulated 5KC cells and those expressing the native human CD4. IL-2 secretion upon stimulation by insulin B:9–23 and B:13–23 by the clone 5 5KC cells were significantly stronger when expressing the mutated human CD4 than non-manipulated cells (Fig. 1I and J, red triangles). Likewise, 5KC cells with T1D-3, T1D-4, and T1D-10 responded to insulin B:9–23R22E several orders of magnitude more robustly (Fig. 1B, D, and F, red triangles), and responses to the native insulin B:9–23 were detectable only when expressing the mutated human CD4 (Fig. 1A, C, and E, red triangles). Importantly, not only were the intensities of the responses elevated, but also responsiveness was seen to lower concentrations of peptides. Indeed, while responses of the 1B11 TCR to the insulin B:9–23 and B:9–23R22E peptides were undetectable in non-manipulated and the native human CD4-expressing 5KC cells, responsiveness to those peptides was notably detected by expressing the mutated human CD4 (Fig. 1G and H, red triangles). In addition, it is noteworthy that IL-2 secretion from cells cultured without antigens did not elevate when expressing the mutated human CD4, implying that reactivity is specific for the peptide component of the antigen. In summary, expression of the mutated human CD4, which binds to HLA-DQ8 with greater affinity and thereby enhances affinity of the primary TCR-pMHC interaction, increased sensitivity to antigen stimulation as measured by IL-2 secretion by 5KC T cell transductants.
Table 2.
MHC Amino acid residues interacting with CD4.
| Alpha gene | Position | |||
|---|---|---|---|---|
| 88 | 90 | 176 | ||
| DR1/15 | DRA*0101 | E | T | K |
| DQ8 | DQA1*0301 | E | T | K |
| DQ2 | DQA1*0501 | E | T | K |
| DP4 | DPA1*0103 | E | T | K |
| I-Ag7 | I-Ag7 alpha | Q | T | K |
| Beta gene | Position | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 104 | 114 | 116 | 142 | 143 | 144 | 145 | 148 | 158 | 160 | 162 | ||
| DR1 | DRB1*0101 | S | L | V | V | V | S | T | I | L | M | E |
| DR15 | DRB1*1501 | S | L | V | M | V | S | T | I | L | M | E |
| DQ8 | DQB1*0302 | S | L | V | V | V | S | T | I | L | M | E |
| DQ2 | DQB1*0201 | S | L | V | V | V | S | T | I | L | M | E |
| DP4 | DPB1*0401 | S | L | V | V | V | S | T | I | L | M | E |
| I-Ag7 | I-Ag7 beta | S | T | V | V | S | S | T | I | L | M | E |
The previous study by Mariuzza and his colleagues determined amino acid residues in HLA-DR1 that interact with the mutated human CD4 having Q40Y and T45W (Wang et al., 2011). The table shows amino acids of other MHC class II alleles in these positions.
3.4. Additional CD3 expression partly increases responsiveness to antigens
TCR alpha and beta chain molecules are expressed on the surface of T cells as a complex with CD3γ, CD3δ, CD3ε, and ζ chain. Therefore, insufficient supply of CD3 molecules may result in the few number of TCR/CD3 complex on the cell surface, leading to the low avidity and weak responsiveness. To test this idea, we introduced additional CD3 genes into 5KC cells that have been transduced with the mutated human CD4 gene and evaluated the expression of TCR/CD3 complex by staining with either anti-CD3 antibody or anti-TCRβ antibody. As expected, TCR/CD3 expression significantly increased when transduced with the CD3 genes (Fig. 2A, B, C, and D). Consistent with the higher TCR/CD3 expression, 5KC cells transduced with the CD3 genes responded to stimulation by anti-CD3 antibody more robustly compared to those without additional CD3 (Fig. 2E). However, influence to antigen stimulation by expressing additional CD3 was limited. Responses by 5KC cells expressing the clone 5 TCR (Fig. 3G and H) but not the other three (Fig. 3A–3F) elevated when transduced with the CD3 genes. In addition, the minimum peptide concentration by which IL-2 production is detectable was unaltered by introducing the additional CD3 genes. Therefore, while the introduction of additional CD3 components into these transductants successfully increased the level of surface TCR expression, improvement in antigen sensitivity was limited.
Fig. 2.
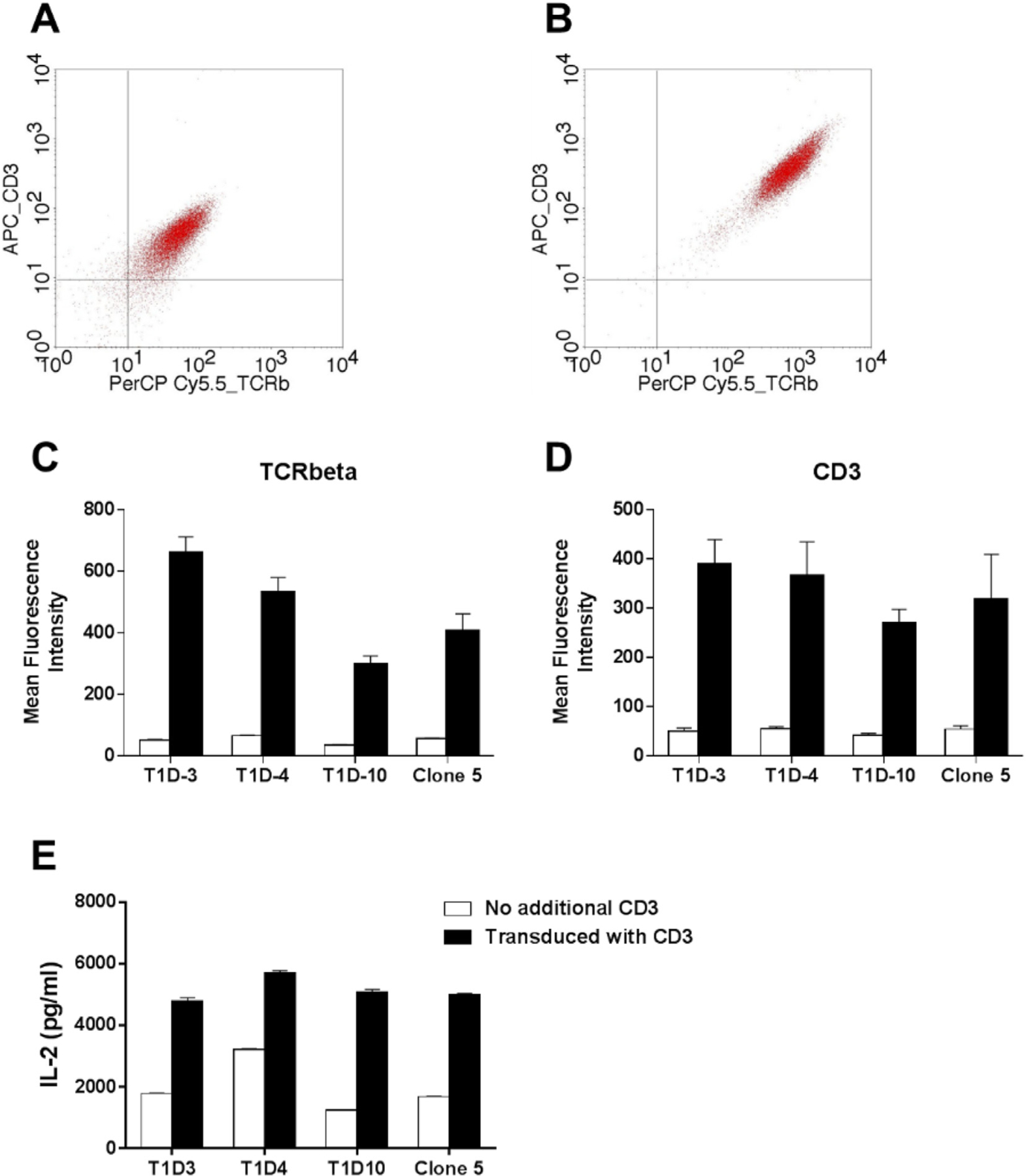
Increased responsiveness by 5KC T cell transductants with additional CD3 expression. 5KC cells were transduced with the retroviral vector encoding the CD3γ, CD3δ, CD3ε, and CD3ζ genes. Cells before (A) and after (B) transduction were stained with anti-TCRbeta (X-axis) and anti-CD3 (Y-axis) antibodies. Panels (C) and (D) show mean fluorescence intensity of staining with anti-TCRbeta (C) or anti-CD3 (D) antibody (white bars: pre transduction, black bars: post transduction). Data are shown as mean ± S.E.M of three independent experiments. Panel (E) shows IL-2 secretion upon stimulation by anti-CD3 antibody (white bars: pre transduction, black bars: post transduction). Data shown in panel E are representative of at least three independent experiments.
Fig. 3.
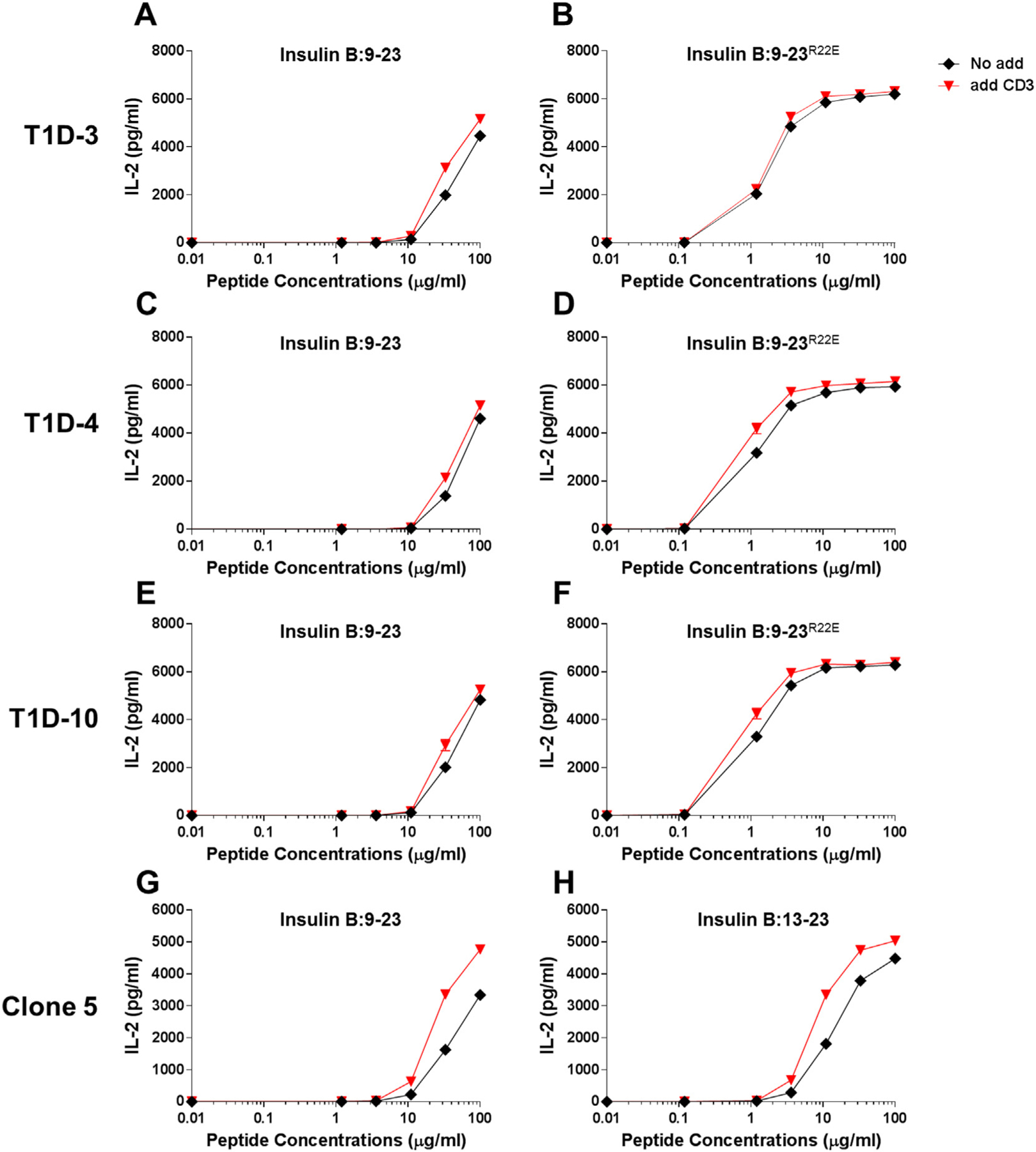
Minimal modulation of responses to the insulin B chain peptides by 5KC T cell transductants with or without additional CD3 expression. 5KC cells expressing the T1D-3 (A and B), T1D-4, (C and D), T1D-10 (E and F), and clone 5 (G and H) were tested for the response to the insulin B:9–23 peptide (A, C, E, and G), the insulin B:9–23R22E mimotope (B, D, and F), or the truncated insulin B:13–23 peptide (H). 5KC cells transduced with the retroviral vector encoding the CD3γ, CD3δ, CD3ε, and CD3ζ genes (red inverse-triangles) and non-transduced cells (black diamonds) were studied in parallel. All data shown are representative of at least three independent experiments.
3.5. Addition of the mutated human CD4 molecule increases responsiveness to diverse pMHC complexes
To test whether the mutated human CD4 expression is globally effective in improving sensitivity of responsiveness to antigens presented by variety of MHC molecules, we analyzed responses by 5KC T cells to several different pMHC complexes. We prepared 5KC cells expressing or not expressing either the native or mutated human CD4 along with five different TCRs that are reactive to a peptide presented by HLA-DQ8 (TCR 489) (Tollefsen et al., 2006), HLA-DQ2 (TCR 233) (Tollefsen et al., 2006), HLA-DP4 (TCR GSE.23G6) (Michels et al., 2017), HLA-DR15 (TCR Ob.3D1) (Wucherpfennig et al., 1994), or mouse MHC class II I-Ag7 (TCR 12-4.1) (Simone et al., 1997). These 5KC cells were transduced with the CD3 complex genes as well. Fig. 4 shows IL-2 secretion by each 5KC cell line when culturing with peptides in the presence of antigen presenting cells expressing a cognate MHC molecule. For all five TCR-pMHC combinations studied, 5KC cells expressing the mutated human CD4 responded more remarkably to lower concentrations of peptides than those without the mutated human CD4 (Fig. 4, red triangles vs. black diamonds and blue inverse triangles). In particular, responses by the Ob.3D1 TCR transductants were detectable only when expressing the mutated human CD4 (Fig. 4D). Moreover, it is noteworthy that expression of the mutated human CD4 exaggerated responses to a peptide presented by a mouse MHC molecule (Fig. 4E) even though multiple amino acids in I-Ag7 that directly interact with CD4 are different from those of HLA molecules (Table 2). These results demonstrate that expression of human CD4 with amino acid substitutions that increase affinity to diverse MHC molecules enhances sensitivity to antigen stimulation detected by T cell transductants.
Fig. 4.
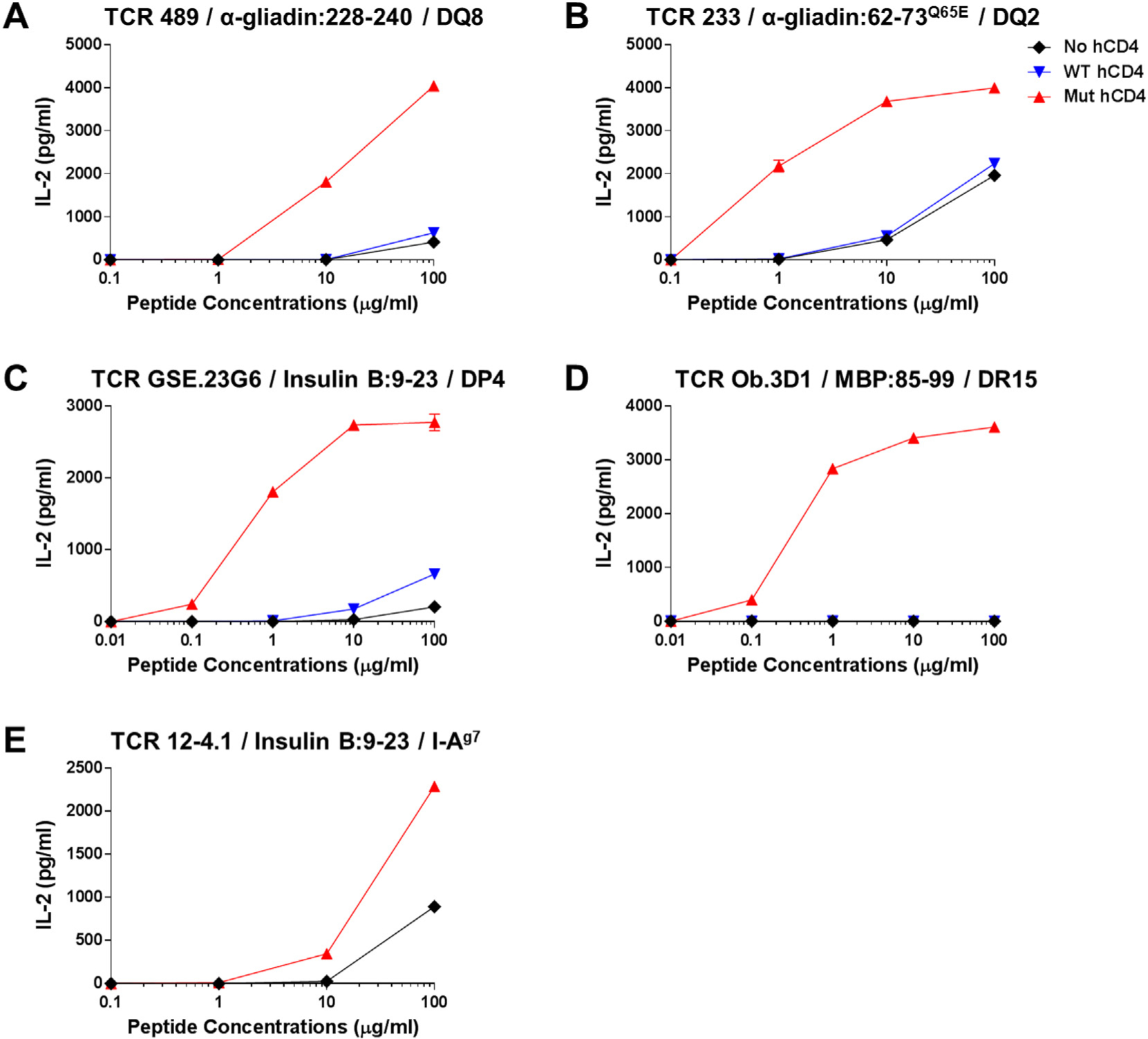
Responses to peptides presented by various MHC molecules by 5KC T cell transductants. 5KC cells expressing the 489 (A), 233, (B), GSE.23G6 (C), Ob.3D1 (D), and 12–4.1 (E) were tested for the response to a cognate peptide designated in each panel in the presence of antigen presenting cells expressing HLA-DQ8 (A), DQ2 (B), DP4 (C), DR15 (D), and mouse MHC class II I-Ag7 (E). Non-manipulated 5KC cells (black diamonds), those expressing native human CD4 (blue inverse-triangles) or mutated human CD4 [Q40Y, T45W] (red triangles) were studied in parallel. All 5KC cell lines were transduced with the CD3 genes as well. All data shown are representative of at least three independent experiments.
4. Discussion
We established a host cell line for T cell transductants that is highly sensitive to antigen stimulation by expressing the human CD4 having two amino acid substitutions [Q40Y, T45W] that magnify the binding to an MHC class II molecule (Wang et al., 2011). Importantly, the expression of the mutated human CD4 is effective in potentiating antigen stimulation for TCRs that are restricted to a variety of MHC class II molecules.
Identification of antigens for pathogenic T cells such as autoreactive, tumor-specific, and virus-specific T cells is crucial for developing biomarkers and therapeutic reagents. Quantitative and qualitative evaluation of antigen-specific T cells in peripheral blood may serve as biomarkers to assess prognosis and to identify individuals responding to therapies. Immunotherapies targeting antigen-specific T cells minimize impact on the whole immune system and may reduce undesirable side effects. While large numbers of antigens have been identified for individual diseases for these purposes, there remain critical questions that need to be addressed to develop robust immunodiagnostic and therapeutic tools. For instance: What is the range of antigens targeted by disease-specific T cells in multiple patients? How frequently are tissue-specific T cells present in peripheral blood? Do the antigens that initiate the T cell immune response differ from those recognized by T cells that participate in disease development in a later phase? Are the antigens for regulatory T cells distinct from those for conventional T cells? T cell transductants expressing disease-relevant TCRs allow us to investigate these questions. In particular, because of the recent advancements in sequencing technologies that facilitate identifying disease-relevant TCRs, the T cell transductant system becomes an effective strategy to study antigen specificity of T cells expressing these TCRs.
T cell hybridomas, TCR transfectomas and transductants are useful tools to analyze antigen specificity for T cells. However, in general, these T cell “avatars” are less sensitive to antigen stimulation than primary T cells. Among molecules that are involved in the antigen-specific T cell response, we chose those composing the primary TCR-pMHC complex, i.e. CD3 and CD4, to improve the response (Wang et al., 2009). When pMHC is recognized by a TCR, CD4 binds to MHCII, resulting in recruitment of lymphocyte-specific protein tyrosine kinase to the cytoplasmic tail domain of CD4, which then phosphorylates CD3 molecules (Liu et al., 1997; Palacios & Weiss, 2004; Smith-Garvin et al., 2009). Thus, CD4 and CD3 play an essential role in TCR signal transduction mediated by the engagement of TCR to the pMHC complex, and therefore we expected that addition or modification of these molecules improves responsiveness by T cell transductants in an antigen-specific manner. Expression of native human CD4 in 5KC cells, which endogenously express mouse CD4, was ineffective in improving responsiveness. This may be because the majority of amino acid residues in CD4 that interact with MHCII are homologous between humans and mice (Table 4) (Wang et al., 2011), and the affinity of native human CD4 for MHCII is unmeasurable (> 400 μM) (Wang et al., 2011). Indeed, the results are consistent with our previous study showing that addition of native CD4 has no influence on the binding of MHCII tetramers to T cells (Crawford et al., 1998). This contrasts with the binding of CD8 that has a measurable affinity for MHCI, and the presence of CD8 greatly improves MHCI tetramer binding especially to T cells that have weak TCR/pMHCI affinity (Wooldridge et al., 2005). In contrast to native human CD4, expression of mutated human CD4 with two amino acid substitutions that engender higher affinity between CD4 and MHCII dramatically increased reactivity. This suggests that increased affinity between CD4 and MHC class II molecules resulted in stabilization of the TCR-pMHC interaction and vigorous TCR signal-transduction, leading to potent reactivity to antigen stimulation. Consequently, enhancement of T cell reactivity is limited to when TCRs bind to pMHC complexes, thereby eliciting only minimal non antigen-specific responses while preserving robust responsiveness to a cognate antigen. It is notable that expression of mutated human CD4 was effective in various TCRs that are restricted by a wide range of MHCIIs. This may be because amino acid residues in MHC molecules that interact with the two mutated amino acids in CD4 (Wang et al., 2011) are extremely conserved across different HLA alleles, genes, and even species (Table 2). When analyzing amino acid sequences of all HLA class II alleles that have been reported to date (ebi.ac.uk/ipd/imgt/hla/) (Robinson et al., 2015), nearly 100% of alleles share conserved amino acid residues at these positions (Table 3). Thus, the mutated CD4 expression is expected to be practical to enhance the reactivity of the majority of TCRs derived from CD4 T cells.
Table 4.
CD4 Amino acid residues interacting with MHC.
| Position | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | |
| Mouse CD4 | K | I | L | G | Q | H | G | G | V | L | I | R | G | P |
| Human CD4 | K | I | L | G | N | Q | G | S | F | L | T | K | G | P |
| Human CD4 with mutations | K | I | L | G | N | Y | G | S | F | L | W | K | G | P |
| Position | |||||
|---|---|---|---|---|---|
| 59 | 60 | 61 | 62 | 63 | |
| Mouse CD4 | K | G | A | W | E |
| Human CD4 | R | S | L | W | D |
| Human CD4 with mutations | R | S | L | W | D |
The previous study by Mariuzza and his colleagues identified two regions (residues 35–48 and 59–63) in the mutated human CD4 with Q40Y and T45W that interact with HLA-DR1 (Wang et al., 2011). The table shows amino acid residues of mouse and native human CD4 in these regions.
Table 3.
Conservation of HLA Amino acid residues interacting with CD4.
| Alpha gene | Position | ||
|---|---|---|---|
| 88 | 90 | 176 | |
| DRA*0101 | E | T | K |
| DRA | 2/2 | 2/2 | 2/2 |
| (100%) | (100%) | (100%) | |
| DQA1 | 32/32 | 32/32 | 32/32 |
| (100%) | (100%) | (100%) | |
| DPA1 | 14/14 | 14/14 | 14/14 |
| (100%) | (100%) | (100%) |
| Beta gene | Position | |||||
|---|---|---|---|---|---|---|
| 104 | 114 | 116 | 142 | 143 | 144 | |
| DRB1*0101 | S | L | V | V | V | S |
| DRB1 | 249/319 | 316/317 | 316/317 | 281/314 | 314/314 | 314/314 |
| (78%)* | (99%) | (99%) | (89%)** | (100%) | (100%) | |
| DQB1 | 355/357 | 357/357 | 356/357 | 346/351 | 351/351 | 351/351 |
| (99%) | (100%) | (99%) | (99%) | (100%) | (100%) | |
| DPB1 | 178/178 | 178/178 | 178/178 | 175/177 | 174/177 | 177/177 |
| (100%) | (100%) | (100%) | (99%) | (98%) | (100%) |
| Beta Gene | Position | ||||
|---|---|---|---|---|---|
| 145 | 148 | 158 | 160 | 162 | |
| DRB*10101 | T | I | L | M | E |
| DRB1 | 314/314 | 313/313 | 312/313 | 313/313 | 313/313 |
| (100%) | (100%) | (99%) | (100%) | (100%) | |
| DQB1 | 348/351 | 350/351 | 351/351 | 349/351 | 351/351 |
| (99%) | (99%) | (100%) | (99%) | (100%) | |
| DPB1 | 177/177 | 177/177 | 177/177 | 176/177 | 177/177 |
| (100%) | (100%) | (100%) | (99%) | (100%) |
The previous study by Mariuzza and his colleagues determined amino acid residues in HLA-DR1 that interact with the mutated human CD4 having Q40Y and T45W (Wang et al., 2011). The table shows the number of alleles that have an identical amino acid residue to HLA-DR1/the number of alleles of which amino acid residue is determined at the designated position.
69 alleles (22%) have alanine rather than serine.
33 alleles (11%) have methionine rather than valine.
Similar efforts have been given to restoring reactivity by CD8 T cell hybridomas and transductants that recognize peptides presented by MHC class I molecules (Wooldridge et al., 2010; Wooldridge et al., 2007; Devine et al., 2006). Devine and colleagues demonstrated that responses of the mouse 2C T cell hybridoma to H2-Ld presenting the cognate, but not to an unrelated peptide, were enhanced when co-expressing mouse CD8-beta having certain mutations (Devine et al., 2006). If these mutations in CD8-beta prove to enhance the response of the mouse and human CD8 T cells to peptide presented by other mouse or even human MHC class I alleles, T cell transductants with the mutated CD8-beta may be broadly usable for antigen discovery studies for CD8 T cells. Affinity between CD8 and MHCI is naturally higher compared to that of CD4/MHCII (Xiong et al., 2001; Wyer et al., 1999), thereby leaving only small room to improve this interaction before losing antigen specificity that is determined by TCR/pMHC interaction (Wooldridge et al., 2010). Thus, it will be a key whether the mutations provoke non-specific reactivity by CD8 T cells expressing a wide range of pMHCI-reactive TCRs.
Extra CD3 expression had the minimal effect to improve responsiveness to antigens, while it improved the anti-CD3 response. Compared to the anti-CD3 response that anti-CD3 antibodies capture all available TCR/CD3, reactivity to antigens is controlled by the availability of pMHC on antigen presenting cells, therefore the extra TCR/CD3 may not lead to additional TCR engagement. Antigen presenting cells that solely express a single MHC without endogenous MHC expression and those that express the high number of MHC molecules on cell surface by augmenting MHC turnover or using a robust gene promoter may be able to present antigens effectively. Using such antigen presenting cells may further improve reactivity by T cell transductants that are additionally transduced with the CD3 component genes.
In conclusion, we demonstrated that increased avidity between the TCR/CD3 complex and pMHC led to enhanced antigen stimulation in the T cell transductant system. When co-expressing TCRs of interest, 5KC T hybridoma cells that are engineered to express additional CD3 as well as the mutated human CD4 with two amino acid substitutions that elicit affinity to MHC molecules are highly responsive to antigens presented by a variety of MHC class II molecules without increasing non-antigen specific reactivity. We propose this cell line, 5KC T cells transduced with the mutated human CD4 and the additional mouse CD3 genes, as a potent tool for antigen discovery studies using a T cell transductant system.
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers R01DK099317, R01DK032083, R01DK108868, DP3DK110845, UC4DK104223, 5U01DK104162-02, R21AI126189-01, R21AI124076-02, AI18785, P01AI118688), JDRF (grant numbers 1-INO-2014-173-A-V, 2-SRA-2018-480-S-B), American Diabetes Association (grant number 1-17-ICTS-074), and the Culshaw Family Junior Investigator Award.
Abbreviations:
- TCR
T cell receptor
- pMHC
peptide-MHC complex
- B:9–23
Insulin B chain 9–23 amino acids peptide
- B:9–23R22E
Insulin B chain 9–23 amino acids peptide having a substitution of arginine with glutamic acid at position 22
- B:13–23
Insulin B chain 13–23 amino acids peptide
References
- Altmann DM, et al. , 1995. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J. Exp. Med 181 (3), 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon JA, et al. , 2016. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med 22 (12), 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune MT, et al. , 2016. Domain-swapped T cell receptors improve the safety of TCR gene therapy. elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini ML, et al. , 2013. Generation of T cell receptor-retrogenic mice: improved retroviral-mediated stem cell gene transfer. Nat. Protoc 8 (10), 1837–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford F, et al. , 1998. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8 (6), 675–682. [DOI] [PubMed] [Google Scholar]
- Davis SJ, et al. , 2003. The nature of molecular recognition by T cells. Nat. Immunol 4 (3), 217–224. [DOI] [PubMed] [Google Scholar]
- Devine L, et al. , 2006. Mapping the binding site on CD8 beta for MHC class I reveals mutants with enhanced binding. J. Immunol 177 (6), 3930–3938. [DOI] [PubMed] [Google Scholar]
- Eerligh P, et al. , 2011. Functional consequences of HLA-DQ8 homozygosity versus heterozygosity for islet autoimmunity in type 1 diabetes. Genes Immun. 12 (6), 415–427. [DOI] [PubMed] [Google Scholar]
- Gay D, et al. , 1987. Functional interaction between human T-cell protein CD4 and the major histocompatibility complex HLA-DR antigen. Nature 328 (6131), 626–629. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, et al. , 1985. Complex regulation of class II gene expression: analysis with class II mutant cell lines. J. Immunol 135 (5), 3542–3550. [PubMed] [Google Scholar]
- Hampl J, Chien YH, Davis MM, 1997. CD4 augments the response of a T cell to agonist but not to antagonist ligands. Immunity 7 (3), 379–385. [DOI] [PubMed] [Google Scholar]
- Holst J, et al. , 2006. Generation of T-cell receptor retrogenic mice. Nat. Protoc 1 (1), 406–417. [DOI] [PubMed] [Google Scholar]
- Jonsson P, et al. , 2016. Remarkably low affinity of CD4/peptide-major histocompatibility complex class II protein interactions. Proc. Natl. Acad. Sci. U. S. A 113 (20), 5682–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent SC, et al. , 2005. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435 (7039), 224–228. [DOI] [PubMed] [Google Scholar]
- Liu CP, et al. , 1997. Development and function of T cells in T cell antigen receptor/CD3 zeta knockout mice reconstituted with Fc epsilon RI gamma. Proc. Natl. Acad. Sci. U. S. A 94 (2), 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AW, et al. , 2017. Islet-Derived CD4 T Cells Targeting Proinsulin in Human Autoimmune Diabetes. Diabetes 66 (3), 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, et al. , 2012. Germline TRAV5D-4 T-cell receptor sequence targets a primary insulin peptide of NOD mice. Diabetes 61 (4), 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios EH, Weiss A, 2004. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene 23 (48), 7990–8000. [DOI] [PubMed] [Google Scholar]
- Pan S, et al. , 1998. HLA-DR4 (DRB1*0401) transgenic mice expressing an altered CD4-binding site: specificity and magnitude of DR4-restricted T cell response. J. Immunol 161 (6), 2925–2929. [PubMed] [Google Scholar]
- Pathiraja V, et al. , 2015. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+T cells infiltrate islets in type 1 diabetes. Diabetes 64 (1), 172–182. [DOI] [PubMed] [Google Scholar]
- Robinson J, et al. , 2015. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–D431 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, et al. , 2011. Evolutionarily conserved features contribute to alphabeta T cell receptor specificity. Immunity 35 (4), 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone E, et al. , 1997. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proc. Natl. Acad. Sci. U. S. A 94 (6), 2518–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS, 2009. T cell activation. Annu. Rev. Immunol 27, 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak AL, Vignali DA, 2005. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert. Opin. Biol. Ther 5 (5), 627–638. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, et al. , 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol 22 (5), 589–594. [DOI] [PubMed] [Google Scholar]
- Taneja V, David CS, 1998. HLA transgenic mice as humanized mouse models of disease and immunity. J. Clin. Invest 101 (5), 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen S, et al. , 2006. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Invest 116 (8), 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. , 2009. A conserved CXXC motif in CD3epsilon is critical for T cell development and TCR signaling. PLoS Biol. 7 (12), e1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XX, et al. , 2011. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc. Natl. Acad. Sci. U. S. A 108 (38), 15960–15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, et al. , 1993. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J. Exp. Med 177 (1), 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, et al. , 2005. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J. Biol. Chem 280 (30), 27491–27501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, et al. , 2007. Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur. J. Immunol 37 (5), 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, et al. , 2010. MHC class I molecules with Superenhanced CD8 binding properties bypass the requirement for cognate TCR recognition and nonspecifically activate CTLs. J. Immunol 184 (7), 3357–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, et al. , 1994. Clonal expansion and persistence of human T cells specific for an immunodominant myelin basic protein peptide. J. Immunol 152 (11), 5581–5592. [PubMed] [Google Scholar]
- Wyer JR, et al. , 1999. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity 10 (2), 219–225. [DOI] [PubMed] [Google Scholar]
- Xiong Y, et al. , 2001. T Cell Receptor Binding to a pMHCII Ligand is Kinetically Distinct from and Independent of CD4. J. Biol. Chem 276 (8), 5659–5667. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, et al. , 1994. Functional interaction between human histocompatibility leukocyte antigen (HLA) class II and mouse CD4 molecule in antigen recognition by T cells in HLA-DR and DQ transgenic mice. J. Exp. Med 180 (1), 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. , 2014. Autoreactive T cells specific for insulin B:11–23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc. Natl. Acad. Sci. U. S. A 111 (41), 14840–14845. [DOI] [PMC free article] [PubMed] [Google Scholar]


