Abstract
Wastewater is a pooled sampling instrument that may provide rapid and even early disease signals in the surveillance of COVID-19 disease at the community level, yet the fine-scale temporal dynamics of SARS-CoV-2 RNA in wastewater remains poorly understood. This study tracked the daily dynamics of SARS-CoV-2 RNA in the wastewater from two wastewater treatment plants (WWTPs) in Honolulu during a rapidly expanding COVID-19 outbreak and a responding four-week lockdown that resulted in a rapid decrease of daily clinical COVID-19 new cases. The wastewater SARS-CoV-2 RNA concentration from both WWTPs, as measured by three quantification assays (N1, N2, and E), exhibited both significant inter-day fluctuations (101.2-105.1 gene copies or GC/L in wastewater liquid fractions, or 101.4-106.2 GC/g in solid fractions) and an overall downward trend over the lockdown period. Strong and significant correlation was observed in measured SARS-CoV-2 RNA concentrations between the solid and liquid wastewater fractions, with the solid fraction containing majority (82.5%-92.5%) of the SARS-CoV-2 RNA mass and the solid-liquid SARS-CoV-2 RNA concentration ratios ranging from 103.6 to 104.3 mL/g. The measured wastewater SARS-CoV-2 RNA concentration was normalized by three endogenous fecal RNA viruses (F+ RNA coliphages Group II and III, and pepper mild mottle virus) to account for variations that may occur during the multi-step wastewater processing and molecular quantification, and the normalized abundance also exhibited similar daily fluctuations and overall downward trend over the sampling period.
Keywords: SARS-CoV-2, COVID-19, Wastewater, Temporal dynamics, Solid, Liquid
1. Introduction
The use of wastewater to monitor microbial infectious diseases in human communities dates back to the 1950s, and originally focused on enteric bacterial pathogen (in particular Salmonella) outbreaks (Shearer et al. 1959, Moore 1951). More recently, studies have explored the wastewater surveillance approach for many potential applications, including poliovirus eradication (Poyry et al. 1988), enteric disease outbreak detection (Hellmer et al. 2014, Diemert and Yan 2019), and to understand the diversity of microbial pathogens in human communities (Yang et al. 2014, Diemert and Yan 2020). The global pandemic of COVID-19 and the challenges in tracking its community transmission by the traditional clinical approaches have highlighted the unique potential advantages of the wastewater surveillance approach. Because wastewater consists of many types of human bodily wastes that can contain the SARS-CoV-2 virus, in particular feces (Cheung et al. 2020) and urine (Jones et al. 2020), SARS-CoV-2 viral RNA has been detected in raw wastewater and primary sludge samples at communities experiencing COVID-19 outbreaks (Medema et al. 2020, Randazzo et al. 2020, Ahmed et al. 2020a, Peccia et al. 2020).
Since municipal wastewater collects wastes from all COVID-19 infections (including asymptomatic, mildly symptomatic, pre-symptomatic, and symptomatic ones), it has the potential to provide comprehensive surveillance of disease transmission in a community. However, the wide distribution of SARS-CoV-2 viral RNA concentration in patients’ wastes (e.g. 103.4-7.6 genome copies/gram (GC/g) feces (Cheung et al. 2020),102.7-5.1 GC/g (Pan et al. 2020)) and zero to 109.3 GC/mL (Feng et al. 2021)) and changing viral load over the disease course (Wolfel et al. 2020) have presented challenges in using wastewater to estimate COVID-19 disease burden (Wu et al. 2020). Previous studies have reported rapid increasing of SARS-CoV-2 RNA concentration in wastewater at the rising limb of COVID-19 outbreaks in communities (Medema et al. 2020, Randazzo et al. 2020, Ahmed et al. 2020a, Peccia et al. 2020, Hata et al. 2021), while few studies have investigated the temporal dynamics of wastewater SARS-CoV-2 RNA during the recession limb of an outbreak (Graham et al. 2021, D'Aoust et al. 2021).
The fact that the sewer collection system carries a continuous stream of wastewater also means that it may be used as a near real-time pooled sampling instrument for the entire community, providing rapid disease signals and enabling quick responses. The symptomatic COVID-19 infections are now known to start shedding virus in upper respiratory during the incubation period, which typically lasts an average of 5.2 days (Li et al. 2020), and the highest viral load was generally reported at the time of symptom onset (Jones et al. 2020). Accordingly, pre-symptomatic transmission was estimated to account for about 44% of secondary cases (He et al. 2020), and could contribute to SARS-CoV-2 signals in wastewater. Asymptomatic infections, which were estimated to represent 40-45% of total SARS-CoV-2 infections and believed to play a significant role in viral transmission (Oran and Topol 2020), could generate SARS-CoV-2 signals in wastewater before emergence of clinical patients. However, our understanding of the temporal dynamics of SARS-CoV-2 RNA, especially at fine scales such as daily, in wastewater of communities that are experiencing ongoing COVID-19 outbreaks remains very limited.
In August 2020, Honolulu (Hawaii, USA) experienced a rapid surge of COVID-19 cases, which prompted a “stay-at-home” order that started on August 27, 2020 and lasted for four weeks. This provided a rare opportunity to investigate how SARS-CoV-2 viral RNA concentration in wastewater would respond to the expected rapid decreases of clinical cases in the community as a result of the emergency public health non-pharmacological intervention. In this study, daily flow-weighted composite raw wastewater samples were collected from the two largest wastewater treatment plants (WWTPs) in Honolulu, and each wastewater sample was separated into a solid fraction and a liquid fraction. The solid and liquid fractions of the wastewater samples were processed separately and the SARS-CoV-2 RNA concentration was measured by using three different RT-qPCR assays. Concentrations of three endogenous fecal RNA viruses which are usually used as surrogates for enteric viral pathogens in wastewater, including F+ RNA coliphages Group II (G2) and Group III (G3) (Friedman et al. 2011), and pepper mild mottle virus (PMMoV) (Rosario et al. 2009), in the solid and liquid fractions of the samples were also determined in parallel. The measured endogenous fecal RNA viruses were subsequently used as global normalization to account for variations in wastewater fecal strength and the multi-step quantification process.
2. Materials and Methods
2.1. Wastewater sampling and pre-processing
Daily flow-weighted composite samples of influent raw wastewater from the Sand Island (SI) and the Honouliuli (HO) WWTPs in Honolulu were collected from 8/27/2020 to 10/4/2020 (i.e. Day 0 to Day 38, n = 39 for each WWTP). Honolulu is a consolidated City-County jurisdiction in the U.S. State of Hawaii, and includes the entire Island of Oʻahu and a residential population of ca. 969,000. There are ten WWTPs with separate sewer systems in Honolulu, with the SI and HO WWTPs being the first and the second largest, treating ca. 58% and 24% of total daily wastewater flow (serving ca. 39% and 33% of the total population) in Honolulu, respectively.
During the sampling period, daily influent flow rate of the two WWTPs remained at relatively stable levels (x̅ ± sx: 210.4 ± 3.8 and 102.8 ± 1.1 thousand cubic meter per day for SI and HO, respectively) (Figure S1), indicating limited impact of rainfall-derived infiltration and inflow (RDII) on the wastewater flow. The wastewater samples were collected in sterile plastic containers. Among samples from 39 sampling days, samples from seven sampling days were stored at 4°C for immediate processing within six hours and samples from 32 sampling days were frozen and stored at -20°C for weekly batch processing(Table S1). All frozen samples experienced only one cycle of freeze-thaw. This storage conditions showed no significant decay of SARS-CoV-2 RNA comparing with fresh samples (Hokajärvi et al. 2021).
Fresh wastewater or fully thawed frozen wastewater samples were first thoroughly mixed, and then separated into solid and liquid fractions by centrifugation, and the liquid fraction was further treated by polyethylene glycol (PEG) precipitation to collect suspended viral particles. 250 mL aliquots of wastewater samples were centrifuged at 13,000 × g for 30 min at 4°C to pellet suspended solids (referred to as solid fractions). After centrifugation, the wastewater supernatant of each wastewater sample was collected in a new sterile glass bottle for further viral precipitation by using the polyethylene glycol (PEG) method (Hjelmso et al. 2017). Briefly, 80 g/L of PEG 8000 (VWR; PA, USA) and 17.5 g/L of NaCl were added into the supernatant, mixed in an orbital shaker (New Brunswick Scientific; Edison, NJ, USA) at 100 rpm at 4°C for approximately 16 hours, and then centrifuged at 13,000 × g for 90 min at 4°C. After carefully decanting the supernatant, the viral pellet at the bottom of the centrifuge bottle was thoroughly resuspended in 500 µL of the supernatant and referred to as the liquid fraction of the wastewater sample.
2.2. Viral RNA extraction and reverse transcription
The solid fractions (300 mg, wet weight) and the liquid fractions (500 µL) were subjected to viral RNA extraction and eluted into 30 µL RNA products by using the QIAamp® Viral RNA Mini Kit, with appropriate scaling up factors (Qiagen; Valencia, CA, USA). Reverse transcription was performed to obtain complementary DNA (cDNA) by using random hexamers (Promega; Madison, WI, USA) and a highly inhibitor-resistant SuperScript® IV (SSIV) reverse transcriptase (Thermo Fisher Scientific; Waltham, MA, USA) according to manufacturers’ instructions. In short, 5 µL of RNA samples, 0.5 mM dNTP, 2.5 µM random hexamers, and nuclease free water were added to a volume of 13 µL. This RNA-primer mix were heated at 65°C for 5 min using GeneAmp® PCR System 9700 (Applied Biosystem; Beverly, MA, USA) and then incubated on ice for at least 1 min. A mixture of 1 × SSIV buffer, 5mM DTT, 2 U/µL of RNase inhibitor (Promega; Madison, WI, USA), and 200 U/µL of SSIV reverse transcriptase, which gave a total volume of 7 µL, were added to the prepared RNA-primer mix. The combined reaction mixtures were incubated at 23°C for 10 min, 55°C for 10 min, and 80°C for 10 min. The cDNA products from the reverse transcription reaction were then stored at -20°C to be used as DNA templates for subsequent real-time PCR (qPCR) quantification.
2.3. qPCR quantification
For each cDNA sample, qPCR assays for SARS-CoV-2 E gene (Corman et al. 2020), N gene (N1 and N2) (Lu et al. 2020), BCoV (Decaro et al. 2008), F+ RNA Coliphages Group II (G2) and Group III (G3) (Friedman et al. 2011), and pepper mild mottle virus (PMMoV) (Rosario et al. 2009) were performed in duplicate reactions in a ABI 7300 qPCR System (Applied Biosystem; Beverly, MA, USA). The N1, N2 and E gene assays were selected based on previous performance comparison on clinical specimen (Nalla et al. 2020a). The three types of fecal RNA viruses (G2, G3, and PMMoV) were selected as endogenous fecal viral RNA controls and used in global normalization to account for potential variations in wastewater fecal strength and in the multi-step quantification process. Each qPCR reaction mixture had a final volume of 20 µL and contained 1 × GoTaq® Probe qPCR Master Mix (Promega; Madison, MI, USA), a pair of forward and reverse primers, hydrolysis probe, and DNA template. Information of qPCR assays, thermal cycling conditions for each target gene were summarized in Table S2-S3.
The DNA standards for SARS-CoV-2 were generated with cDNA by reverse transcription with pure SARS-CoV-2 genomic RNA (Isolate USA_WA1/2020; BEI Resources; Manassas, VA, USA). The DNA standard for BCoV was generated with cDNA by reverse transcription with RNA extracts from bovine coronavirus vaccine (Zoetis; Kalamazoo, MI, USA). DNA standards for fecal RNA viruses (G2, G3 and PMMoV) were generated with cDNA by reverse transcription with RNA extracts from wastewater abundant with fecal RNA viruses. Target gene fragments in cDNA were firstly amplified by PCR with specific primer pairs and PCR amplicons were confirmed by 1.5% agarose gel electrophoresis with following illustration by an UVP GelStudio (Analytik Jena; Upland, CA, USA). Target DNA amplicons were excised and extracted from gel using a QIAquick® Gel Extraction Kit (Qiagen, Valencia, CA, USA), and quantified using Qubit™ 1 × dsDNA HS Assay Kit with a Qubit 4 Fluorometer (Invitrogen; Carlsbad, CA, USA). Target gene copy numbers in purified DNA amplicons were calculated based on the measured DNA quantity and the molecular weight of targeted PCR product (Staroscik 2004). The qPCR standard curves were generated using ten-fold serial dilutions of purified DNA amplicons (101 to 106 copies per reaction). The qPCR amplification efficiencies of calibration curves were in the range of 91% to 106.6% with R2 in the range of 0.991 to 0.999 for different target genes. Information of calibration curves was summarized in Table S4.
2.4. Quality assurance
Each batch of qPCR reactions for each gene assay contained at least one positive control and three no template controls (NTCs), and the results were accepted only when the positive control yield anticipated Ct values based on the calibration curves and all NTCs yield negative results. For each sample and target gene combination, duplicate qPCR reactions were performed, and arithmetic mean Ct values were used for analysis.
To test the method reproducibility, triplicate analyses of three wastewater samples were conducted for both the liquid and solid fractions, and the standard deviation (sx) of Ct values of triplicate analyses for individual samples based on the SARS-CoV-2 RNA E gene assay were compared. Amongst the three samples, the liquid fraction showed Ct values with a sx range of 0.5-1.0 and the solid fractions showed Ct values with a sx range of 0.4-1.3.
The exogenous process control bovine coronavirus (BCoV) (Zoetis; Kalamazoo, MI, USA) was spiked into the solid and liquid fractions of select wastewater samples to detect inhibition and assess recovery (three batches of samples, n = 40). BCoV was directly spiked into the solid fractions after collection and before viral RNA extraction; for the liquid fractions, BCoV was spiked into the supernatant after centrifugation and before PEG precipitation. The quantities seeded into different batches of samples were: 1.3 × 107 genome copy/spike (Batch 1; 9/8/2020 to 9/14/2020, n = 12), 3.7 × 107 genome copy/spike (Batch 2; 9/21/2020 to 9/28/2020, n = 14), and 2.5 × 107 genome copy/spike (Batch 3; 9/29/2020 to 10/4/2020, n = 14). Assessment of recovery based on the spiked BCoV showed that the average recovery ratios were 1.3% (sx =1.1%) for the liquid fractions (n=40) and 0.14% (sx =0.18%) for the solid fractions (n=40) (Figure S2). The recovery ratios in liquid fractions is significantly higher than that in solid fractions (paired t-test, P=0.05). No significant difference in recovery was observed between the different spiking concentrations (ANOVA test: P = 0.40 for liquid fractions and P = 0.08 for solid fractions).
2.5. Data analysis
Concentration of the three SARS-CoV-2 genes and the endogenous fecal RNA virus controls were calculated via mass balance to GC/L for the liquid fractions and GC/g (dry weigh) for the solid fractions. The dry weight of the solid fraction of a wastewater sample was calculated based on its water content, which was determined by measuring the weight difference before and after oven drying at 120°C overnight. Pearson's correlation analyses were conducted for SARS-CoV-2 RNA measurements between different assays (N1, N2, and E) or between the solid and liquid fractions of the same wastewater samples, Student's t-test was conducted to determine whether statistically significant differences existed. All statistical analyses were conducted in the R environment, and the default significance cut-off value was P=0.05. Dynamics of wastewater SARS-CoV-2 and daily COVID-19 were visualized by using the Seaborn functions in Jupyter notebook.
3. Results
3.1. Wastewater SARS-CoV-2 RNA during the lockdown
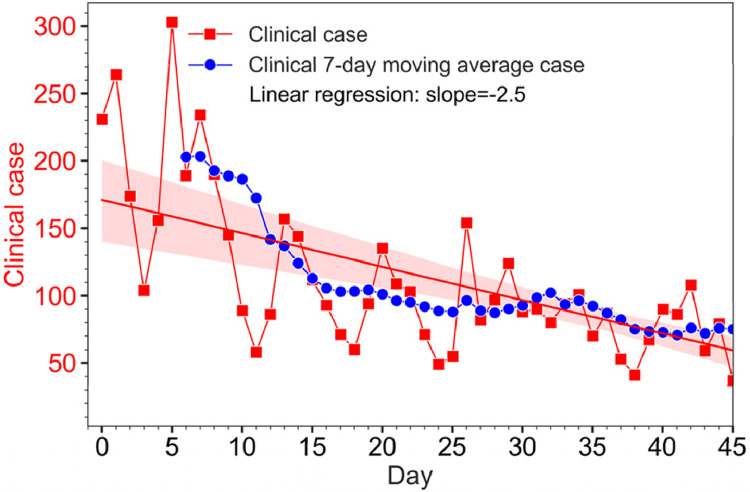
After the onset of the lockdown in Honolulu on August 27, 2020 (Day 0), the total daily new COVID-19 clinical case number in Honolulu reached its highest peak of 303 (7-day moving average of 217) on Day 5, and then started trending downward, reached a daily case of 82 (7-day moving average of 89) on Day 27 when the lockdown expired (Figure 1 ). On Day 38 when the wastewater sampling campaign stopped, the number of daily new cases was 41 (7-day moving average of 75).
Figure 1.
Daily new clinical COVID-19 case numbers and the 7-day moving average in Honolulu from the onset of lockdown on August 27, 2020 (Day 0) to Day 45. Lockdown ended on Day 27, and daily wastewater sampling ended on Day 38. The line and shading are linear regression and 95% confidence intervals, respectively.
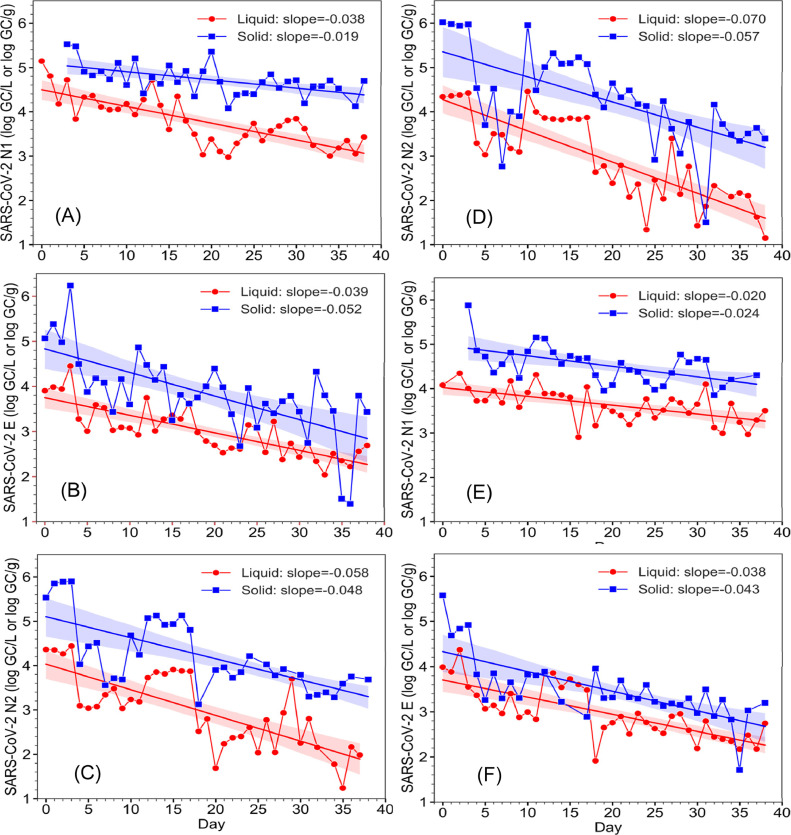
Corresponding to the decrease in new clinical cases resulting from the lockdown, the measured SARS-CoV-2 RNA concentration in the wastewater samples also exhibited an overall downward trend and significant concentration fluctuations in both WWTPs (Figure 2 ). For example, in the SI WWTP liquid fractions, the SARS-CoV-2 RNA concentration showed ranges of 103.0-105.1 GC/L, 101.2-104.5 GC/L, and 102.0-104.5 GC/L based on the N1, N2 and E gene assays, respectively. In the SI WWTP solid fractions, the SARS-CoV-2 RNA concentration also showed ranges of 104.1-105.5 GC/g, 101.5-106.0 GC/g, and 101.4-106.2 GC/g based on the N1, N2, and E gene assays respectively. Similar concentration ranges were also detected in the wastewater liquid and solid fractions from the HO WWTP. Linear regression of the measured SARS-CoV-2 RNA concentration over time showed negative slopes (range: -0.019 to –0.070; x̅ ± sx: -0.039 ± 0.019) for both liquid and solid fractions of wastewater samples from both WWTPs and by all three quantification assays. Slopes of linear regression and Pearson's r and P values are summarized in Table S5. Similar downward trends were also detected by calculating and plotting 7-day moving average (data not shown).
Figure 2.
Daily SARS-CoV-2 RNA concentration dynamics in the liquid (•) and solid (∎) wastewater fractions from the SI WWTP (A, B, C) and HO WWTP (D, E, F) as measured by the N1, N2 and E gene assays. The lines and shadings are linear regression and 95% confidence intervals, respectively.
3.2. Comparison of quantification assays
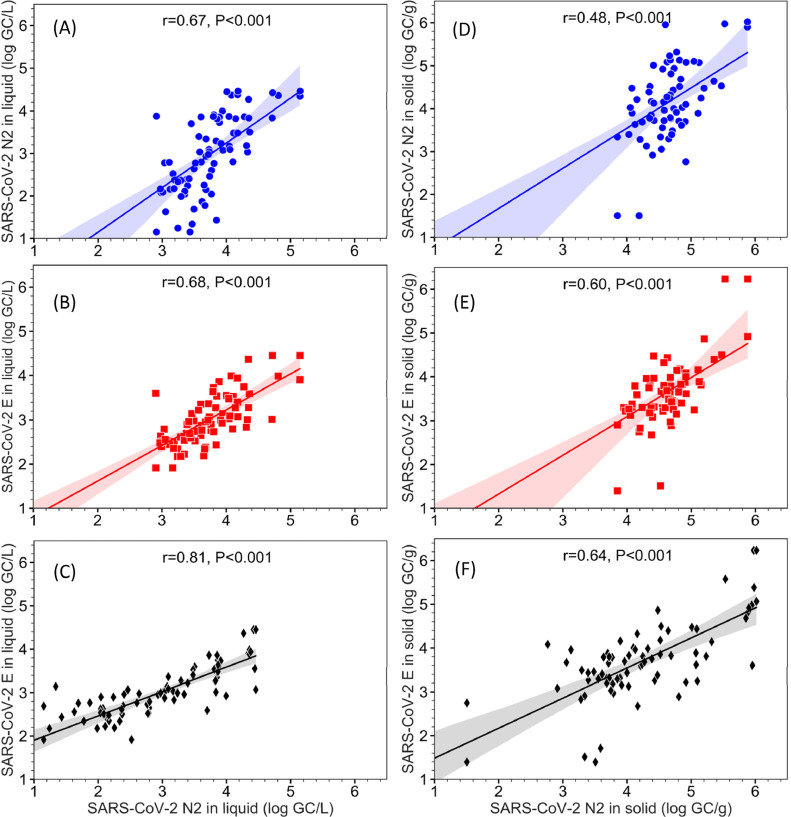
Measurements by the three quantification assays were plotted against each other and compared through correlation analysis (Figure 3 ). Although all comparisons showed significant correlation (Pearson's r: 0.65 - 0.81; P<0.001 for all comparisons), the best correlation was observed between N2 and E gene in the liquid fractions (Pearson's r=0.81, P<0.001). Between liquid and solid fractions, the liquid fraction showed better Pearson's r (x̅ ± sx: 0.72 ± 0.08) amongst the comparisons of three SARS-CoV-2 genes, and the Pearson's r values were significant higher (paired t-test P=0.049) than those of the solid fractions (x̅ ± sx: 0.57 ± 0.08)
Figure 3.
Comparison of log transformed SARS-CoV-2 RNA concentration based on N1, N2 and E gene in the liquid fraction (left pane: A, B, C) and solid fraction (right pane: D, E, F) of wastewater samples from both SI and HO WWTPs. The lines and shadings are linear regression and 95% confidence intervals, respectively.
3.3. SARS-CoV-2 RNA in solid and liquid fractions
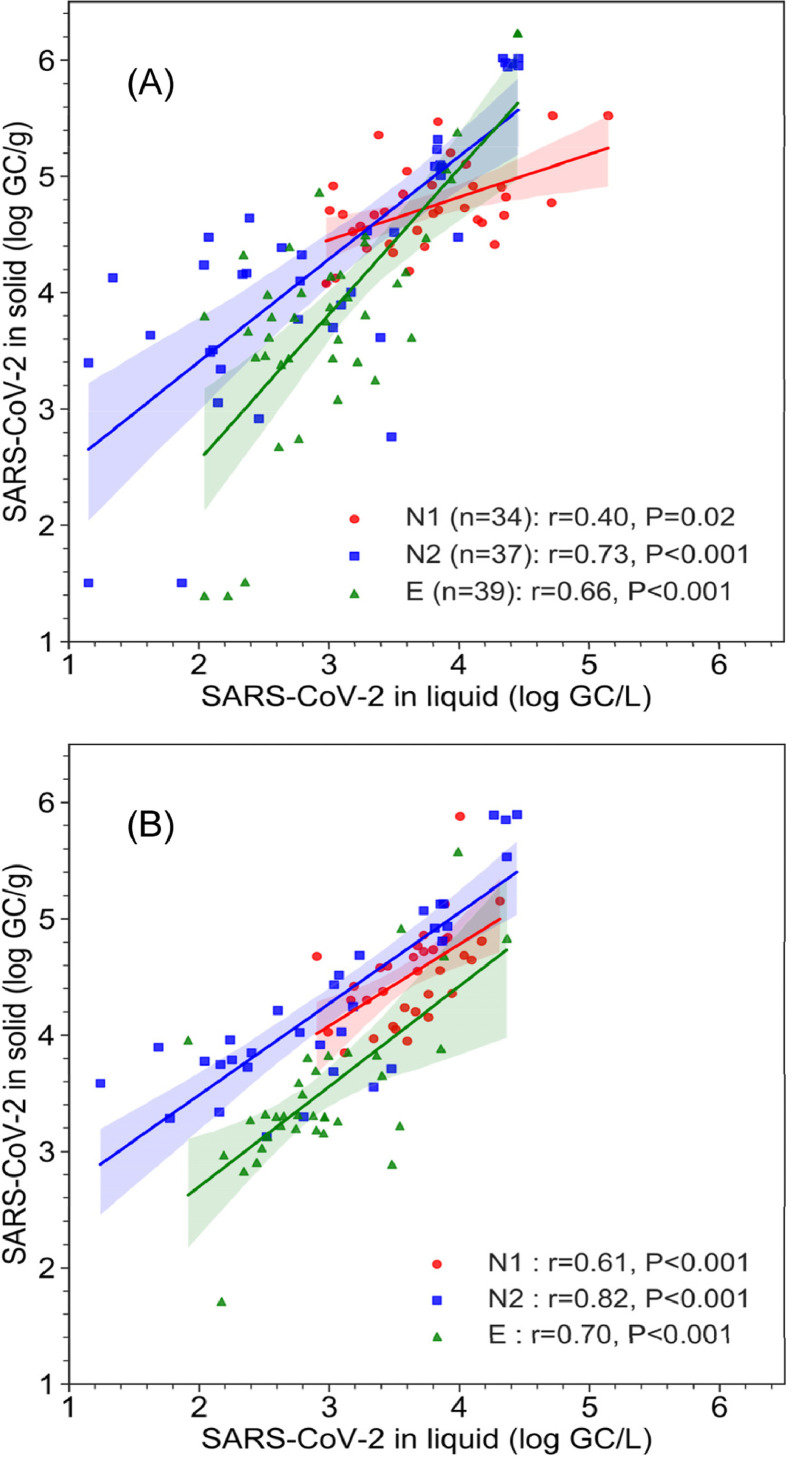
The time-series data and their linear regression showed that, in nearly all wastewater samples analyzed, the solid and liquid fractions appeared to exhibit similar temporal fluctuation patterns while the solid fraction contained significantly higher quantity of SARS-CoV-2 RNA than the corresponding liquid fractions (Figure 2). To further investigate these relationships, the measured SARS-CoV-2 RNA concentrations in the solid and liquid fractions of individual wastewater samples were plotted without the time variable and subjected to correlation analysis (Figure 4 ). Significant correlation was observed between the measurements of SARS-CoV-2 RNA in the solid and liquid fractions based on all three quantification assays and in both WWTPs. For samples from the SI WWTP (Figure 4A), Pearson's r values were 0.40 (P=0.02), 0.73 (P<0.001), and 0.66 (P<0.001) for N1, N2, and E gene assays, respectively. Similar correlation coefficients (r=0.61 (P<0.001), 0.82 (P<0.001), and 0.70 (P<0.001) for N1, N2, and E gene assays respectively) were observed for the samples from the HO WWTP (Figure 4B).
Figure 4.
Comparison of SARS-CoV-2 RNA concentration as measured by the N1, N2 and E gene assays between the solid fraction (GC/g) and the liquid fraction (GC/L) of the wastewater samples from SI (A) and HO (B) WWTPs. The lines and shadings are linear regression and 95% confidence intervals, respectively.
When compared on an equal mass basis, the measured concentrations of SARS-CoV-2 RNA in the solid fraction (GC/g) were significantly higher than that in the liquid fraction (GC/mL) (paired t-test, P<0.001). The N1 concentration ratios between the solid and liquid fractions (mL/g) were 104.00 (sx=100.47) and 103.89 (sx=100.35) for samples from the SI and HO WWTPs, respectively. The N2 concentration ratios between the solid and liquid fractions (mL/g) were 104.31 (sx=100.72) and 104.24 (sx=100.51) for samples from the SI and HO WWTPs, respectively. The E concentration ratios between the solid and liquid fractions (mL/g) were 103.83 (sx=100.67) and 103.58 (sx=100.51) for samples from the SI and HO WWTPs, respectively. Since there were no statistical difference in the solid-liquid concentration ratio between the SI and HO WWTP (t-Test, P=0.30, 0.65, and 0.08 for the N1, N2, and E assay results, respectively), data from the two WWTP plants were pooled to calculate the total mass distribution of SARS-CoV-2 RNA between the solid and liquid fractions of the wastewater samples after accounting for their respective mass percentages in individual samples. Average percentages of 90.5% (sx=8.1%) based on the N1 assay, 92.5% (sx=14.1%) based on the N2 assay, and 82.5% (sx=19.9%) based on the E assay of SARS-CoV-2 RNA resided in the solid fraction of the wastewater samples (Figure S3).
3.4. Normalization by endogenous fecal RNA viruses
To account for potential variations that may occur during the multi-step process, including fecal discharge, sewer collection, wastewater sample processing, and the reverse transcription step of molecular quantification, concentrations of three endogenous fecal RNA viruses (G2, G3, and PMMoV) in the same samples were determined and used to normalize the measured N1, N2, and E gene concentrations. No obvious trend in the concentrations of the endogenous fecal RNA viruses was observed in SI or HO WWTP (Figure S4). Overall the endogenous fecal RNA viruses showed relatively stable concentration levels, in spite of certain samples exhibiting significant variation. In the wastewater samples from the SI WWTP, the G2, G3 and PMMoV showed average concentrations in the liquid fractions of 104.1 (sx=100.6), 104.8 (sx=100.9), and 105.8 (sx=100.6) GC/L, respectively, and average concentrations in the solid fraction of 105.0 (sx=100.7), 105.6 (sx=100.9), and 106.2 (sx=100.5) GC/g respectively. In the wastewater samples from the HO WWTP, the G2, G3 and PMMoV showed average concentrations in the liquid fractions of 105.6 (sx=100.9), 106.6 (sx=100.8), and 106.5 (sx=100.9) GC/L, respectively, and average concentrations in the solid fraction of 104.3 (sx=100.5), 105.9 (sx=100.7), and 106.4 (sx=100.9) GC/g, respectively.
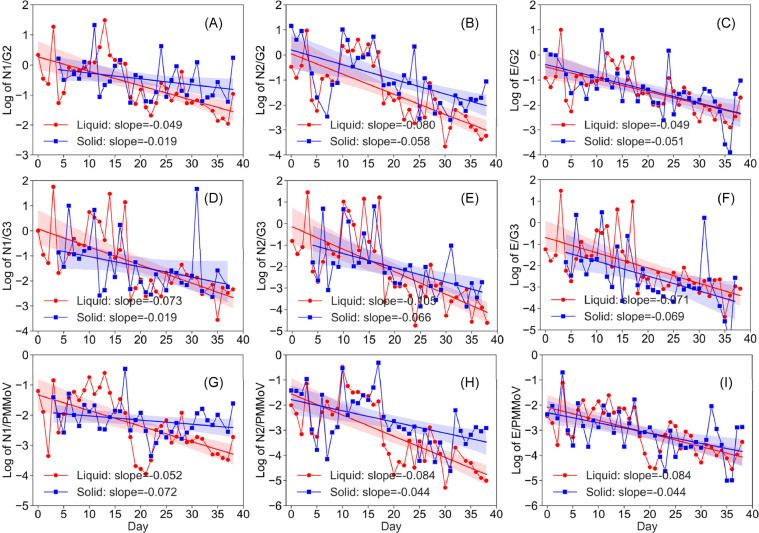
The normalized abundance of SARS-CoV-2 RNA still exhibited significant daily fluctuation and an overall downward trend over the sampling period in the wastewater samples from the SI WWTP (Figure 5 ). Using the E gene results as example, in the liquid fractions from the SI WWTP, E/G2 showed a range of 10−2.9-101.0 (x̅±sx:10−1.4±0.9), E/G3 showed a range of 10−4.4-101.5(x̅±sx:10−2.1±1.2), and E/PMMoV showed a range of 10−4.5-10−1.1(x̅±sx:10−3.1±0.9). In the corresponding solid subsamples from the SI WWTP, E/G2 showed a range of 10−2.9-101.0 (x̅±sx:10−1.4±1.0), E/G3 showed a range of 10−5.6-100.5 (x̅±sx:10−2.5±1.4), and E/PMMoV showed a range of 10−5.0-10−0.7(x̅±sx:10−3.1±0.9). A downward trend was observed in all normalized abundance in both liquid and solid fractions based on all three SARS-CoV-2 RNA quantification assays and three endogenous fecal RNA viruses, as indicated by the negative slopes of linear regression. The slopes of the linear regression for the liquid fractions exhibited a range from -0.049 to -0.105, and the slopes of the linear regression for the solid fractions exhibited a range from -0.019 to -0.072. Similar observations were made in the normalized abundance of SARS-CoV-2 RNA in the wastewater samples from the HO WWTP (Figure S5). Slopes of linear regression and Pearson's r and P values of normalized abundances are summarized in Table S6.
Figure 5.
Daily dynamics of normalized abundance of SARS-CoV-2 RNA based on N1, N2, E gene and G2, G3, and PMMoV in the liquid and solid fractions of wastewater samples from the SI WWTP. The lines and shadings are linear regression and 95% confidence intervals, respectively.
4. Discussion
The effectiveness of the non-pharmacological intervention through a public health lockdown in controlling the COVID-19 outbreak was indicated by the decreasing new clinical case numbers (Figure 1) and the decreasing of measured SARS-CoV-2 RNA concentration in the wastewater samples (Figure 2). Similar trends were observed in both the solid and liquid fractions, at both WWTPs, and by all three quantification assays, which corroborated and supported the potential of using wastewater SARS-CoV-2 concentration to monitor community disease burden. This is congruent with previous observations where increasing wastewater SARS-CoV-2 RNA concentration corresponded to rapidly expanding COVID-19 outbreaks (Medema et al. 2020, Randazzo et al. 2020, Ahmed et al. 2020a, Peccia et al. 2020) and observations where reduction of clinical cases corresponded with decreasing wastewater SARS-CoV-2 RNA concentration (Graham et al. 2021).
The fine-scale temporal dynamics enabled by the daily sampling in this study detected significant inter-day fluctuation of the wastewater SARS-CoV-2 RNA abundance in both the liquid and solid fractions, even within the same weeks. For example, the SARS-CoV-2 RNA concentration measured by the E gene assay in the liquid and solid fractions from the SI WWTP decreased from 104.5 GC/L and 106.2 GC/g on Day 3 to 103.6 GC/L and 104.2 GC/g on Day 6, respectively. Recent studies also observed significant daily fluctuation of SARS-CoV-2 RNA concentration in sludge samples (Peccia et al. 2020). Many factors could have contributed to the observed inter-day fluctuation, including decay, random measurement errors, and fundamental underlying factors. The relatively short hydraulic retention times of the wastewater collection systems (i.e. a few hours) in relation to the decay kinetics of SARS-CoV-2 RNA in untreated wastewater (e.g. average T90 was shown to range from 8.04 to 27.8 days (Ahmed et al. 2020b) suggests decay may not be an important factor to the observed inter-day variation. Since similar inter-day fluctuations were observed by the three different assays (Figure 3) and between both solid and liquid fractions of wastewater samples (Figure 4), and the sample processing and molecular quantification methods used showed reasonable reproducibility (i.e. sx of Ct values of triplicate analysis less than 1.0 and 1.3 for liquid and solid fractions, respectively), random measurement errors in wastewater sampling and molecular analysis were unlikely to be the primary contributor to the significant inter-day fluctuations observed.
How the fundamental underlying factors, in particular varying viral shedding by infected individuals and daily fluctuations in disease burden in the community, might have contributed to the observed inter-day fluctuation of wastewater SARS-CoV-2 RNA concentration requires further investigation. The wide range of SARS-CoV-2 viral RNA concentration detected in different patients’ wastes (e.g. 102.7-7.6 GC/g (Cheung et al. 2020, Pan et al. 2020)) and the rapid changing of fecal viral load in individual patients over the disease course (Jones et al. 2020, Wolfel et al. 2020) can generate different fluxes of total SARS-CoV-2 RNA mass in the sewer systems. Significant daily fluctuation of new clinical cases in Honolulu was also observed (Figure 1), which was similar to weekly inter-day oscillations widely observed in other communities experiencing COVID-19 disease outbreaks (Bukhari et al. 2020). Although this phenomenon has been considered by many to be primarily a reflection of diagnostic and reporting biases (Bergman et al. 2020), contribution from fluctuations in disease burden remains as a possibility (Ricon-Becker et al. 2020), as individual communities can experience different disease transmission dynamics. Nevertheless, the observed significant inter-day fluctuation could have significant implications in designing and conducting wastewater surveillance of COVID-19 diseases in communities. For example, since daily flow-weighted composite samples of influent raw wastewater could contain levels of SARS-CoV-2 RNA that differ by up to 101.7 folds (as exampled above by the E gene results in the SI WWTP), one single daily flow-weighted composite sample for a time interval (e.g. a week) may introduce significant variations in surveillance outcome. Although there has been no reported study on the concentration dynamics of SARS-CoV-2 RNA at even finer temporal scale (e.g. intra-day or hourly), by inference, wastewater grab samples are expected to experience even larger variations than daily flow-weighted composite samples. Therefore, composite samples over longer periods of time (e.g. weekly composites) may be needed to alleviate inter-day fluctuation and provide smoothed depiction of disease burden in the community.
Many RT-qPCR assays with high specificity to the SARS-CoV-2 RNA have been developed to target different regions of the viral genome, including E gene (Corman et al. 2020), N gene (Lu et al. 2020), ORF1ab (ChinaCDC 2020), and previous studies on wastewater have adopted these assays for detection in wastewater samples (Medema et al. 2020, Peccia et al. 2020, Graham et al. 2021, Sherchan et al. 2020). Since wastewater samples are much more complex than clinical samples, this complexity needs to be taken into consideration. In this study, three different assays (N1, N2, and E) were used to provide independent analysis for redundancy and corroboration. The significant correlation observed between results by the three assays (Figure 3) illustrated their general utility for wastewater samples. Interestingly, the results also showed that the N2 and E gene assay results presented the best correlation coefficient than their respective correlation coefficients with the N1 assay results. This is in line with previous observation that the N2 and E gene assays exhibited higher sensitivity than the N1 assay in clinical COVID-19 samples (Nalla et al. 2020b). On the other hand, different concentration results were obtained for the same samples by the different RT-qPCR assays (Figure 2), which was also illustrated by the linear regression lines not passing through the origin (Figure 3). This can be attributed to the inherence variation in the RT-qPCR processes, which were also observed previously in clinical specimen (Nalla et al. 2020a) and in wastewater samples (Medema et al. 2020).
The separation of wastewater samples into solid and liquid fractions in this study not only provided a quasi-replicate analysis of each wastewater sample, but also allowed for side-by-side comparison of SARS-CoV-2 RNA concentration and calculation of its mass distribution between the solid and liquid fractions. The significant correlation observed between the solid and liquid fractions by all three assays and in both WWTPs (Figure 4) further supported the time-series data observations of both the overall trend and the inter-day fluctuation of the SARS-CoV-2 RNA concentration in wastewater (Figure 2). In spite of their different sensitivity and performance, the three assays all reported significantly higher concentration of SARS-CoV-2 in the solid fractions than in the liquid fractions, with the solid-liquid SARS-CoV-2 RNA concentration ratios ranging from 103.6 to 104.3 mL/g. Several other studies have reported high levels of SARS-CoV-2 RNA in primary sludge samples (Peccia et al. 2020, D'Aoust et al. 2021), and also high ratios of SARS-CoV-2 RNA concentrations between primary sludge and raw influent samples (e.g. 102.5-103.5 mL/g (Graham et al. 2021)). Previous laboratory experiments showed that enveloped mouse hepatitis virus (MHV) and bacteriophage Phi6 exhibited partition coefficients of 103.2 and 103.1 mL/g, respectively (Ye et al. 2016).
The highly complex nature of the wastewater matrix also means that the resulting nucleic acid samples could potentially contain chemical inhibitors and a dominant background of non-target nucleic acids, which could introduce additional variations in both the reverse transcription and subsequent qPCR steps. Since chemical inhibition is most likely to occur in the reverse transcription step, a highly inhibitor-resistant reverse transcriptase SSIV, which was shown to tolerate high level of chemical inhibitors (e.g. 10% isopropanol and 0.05% of bile salts), was used in this study to alleviate potential chemical inhibition. The effectiveness of this strategy was supported by the detection of endogenous fecal indicator viruses in the wastewater samples in ranges similar to those reported in literature. For example, the measured PMMoV concentration in the liquid and solid fractions of the wastewater samples in this study averaged at 106.5 GC/L (sx=100.9 GC/L) and 106.4 GC/g (sx=100.9 GC/g); similar concentrations were reported in the literature (e.g. 106.6 GC/L in raw wastewater (Kitajima et al. 2014)) and 106.3 GC/g in wastewater sludge (Graham et al. 2021)). Since the solid fraction of wastewater samples are more likely to suffer from chemical inhibition, the detection of high solid-liquid ratios of SARS-CoV-2 in the wastewater samples (up to 104.3 mL/g) further indicated that chemical inhibition, although cannot be completely ruled out, had limited impact on the sample analysis and particularly on the overall trend observed assuming any potential impact is consistent amongst the samples.
Another source of potential variation was the multi-step process that starts from varying fecal discharge by the community, dilution and decay during sewer collection, to different recovery during wastewater sample processing. To globally account for the variations in the process, this study also tested the feasibility of using three groups of endogenous fecal RNA viruses (G2, G3, and PMMoV) to normalize the measured SARS-CoV-2 RNA concentration. G2 and G3 are inherently linked with fecal coliforms (Friedman et al. 2011), while PMMoV is highly abundant in wastewater due to human dietary consumption of pepper, the latter of which has been used for SARS-CoV-2 RNA normalization in wastewater (Graham et al. 2021). A previous study have shown that normalization of eae gene (genetic marker for enteropathogenic E. coli) by uidA gene (genetic marker for general E. coli) in the same wastewater samples produced normalized abundance with much reduced variation over time (Yang et al. 2014). Although G2, G3, and PMMoV are all non-enveloped viruses, which may exhibit different behaviors than enveloped viruses such as SARS-CoV-2 during wastewater pre-processing (Ye et al. 2016) and even RNA extraction, their RNA undergoes the same molecular quantification process and the normalized abundance is expected to remove impacts from potential chemical inhibition and/or process efficiency variation within individual samples. Similar to the observations made with measured SARS-CoV-2 RNA concentration, the normalized abundance still exhibited downward trend and inter-day fluctuation over the sampling period (Figure 5), providing further support to the observations made by the direct concentration measurements.
5. Conclusion
Overall, the study observed a downward trend of SARS-CoV-2 RNA abundance (both measured concentrations and normalized abundance) in the wastewater samples, which corresponded to the decrease of clinical COVID-19 new case numbers as a result of the public health lockdown in response to a community outbreak. The fine-scale temporal dynamics enabled by the daily sampling in this study detected significant daily fluctuation of the wastewater SARS-CoV-2 RNA abundance, even within the same weeks, indicating the fine-scale temporal dynamics of SARS-CoV-2 RNA in wastewater needs to be taken into consideration in designing and implementing wastewater-based surveillance. The parallel quantification in solid and liquid fractions of the same wastewater samples indicated that the SARS-CoV-2 viral RNA is much more abundant in the solid fraction than in the liquid fraction. Because the wastewater solid fraction contains majority of the SARS-CoV-2 mass in a wastewater sample and can be processed in a more time-efficient fashion, wastewater solids may be a more convenient sample matrix. Further research is needed to address challenges associated with potential higher likelihood of chemical inhibition and the presence of a more complex nucleic acid background in the wastewater solid matrix.
Declaration of Competing Interest
The authors declare no competing interest.
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant No. (CBET-2027059). Partial financial support was provided by the City and County of Honolulu for analyzing its wastewater samples for SARS-CoV-2 RNA. The authors would like to thank Lyle Shizumura, Gayatri Vithanage, and Sherilyn Soriano for assistance with wastewater sample collection, Joshua Stanbro for assistance with obtaining epidemiological data, and Dr. Mark Borchardt and Susan Spencer (Agricultural Research Service, USDA) for providing the bovine coronavirus vaccine.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117093.
Appendix. Supplementary materials
References
- Shearer L.A., Browne A.S., Gordon R.B., Hollister A.C., Jr. Discovery of typhoid carrier by sewage sampling. J Am Med Assoc. 1959;169(10):1051–1055. doi: 10.1001/jama.1959.03000270033008. [DOI] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.H., Fung A.Y., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.H., Yuen K.Y., Leung W.K. Gastroenterology; 2020. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environmental Science & Technology Letters. 2020;7:7. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Poyry T., Stenvik M., Hovi T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl Environ Microbiol. 1988;54(2):371–374. doi: 10.1128/aem.54.2.371-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemert S., Yan T. Clinically Unreported Salmonellosis Outbreak Detected via Comparative Genomic Analysis of Municipal Wastewater Salmonella Isolates. Appl Environ Microbiol. 2019;85(10) doi: 10.1128/AEM.00139-19. 0013900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemert S., Yan T. Municipal Wastewater Surveillance Revealed a High Community Disease Burden of a Rarely Reported and Possibly Subclinical Salmonella enterica Serovar Derby Strain. Appl Environ Microbiol. 2020;86(17) doi: 10.1128/AEM.00814-20. e00814-e00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Xu K., Gu S., Zheng S., Zou Q., Xu Y., Yu L., Lou F., Yu F., Jin T. Multi-route transmission potential of SARS-CoV-2 in healthcare facilities. Journal of hazardous materials. 2021;402 doi: 10.1016/j.jhazmat.2020.123771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Science of The Total Environment. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ Sci Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.-J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann Intern Med. 2020 doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.D., Cooper E.M., Calci K.R., Genthner F.J. Design and assessment of a real time reverse transcription-PCR method to genotype single-stranded RNA male-specific coliphages (Family Leviviridae) J Virol Methods. 2011;173(2):196–202. doi: 10.1016/j.jviromet.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol. 2009;75(22):7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Science of The Total Environment. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmer M., Paxeus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergstrom T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl Environ Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmso M.H., Hellmer M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsasser D., Aarestrup F.M., Lofstrom C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of Methods for the Concentration and Extraction of Viruses from Sewage in the Context of Metagenomic Sequencing. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Mari V., Radogna A., Colaianni M.L., Cirone F., Tempesta M., Buonavoglia C. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J Virol Methods. 2008;151(2):167–171. doi: 10.1016/j.jviromet.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. The detection of enteric carriers in towns by means of sewage examination. J R Sanit Inst. 1951;71(1):57–60. doi: 10.1177/146642405107100109. [DOI] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. Journal of clinical microbiology. 2020;58(6) doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroscik A. Calculator for determining the number of copies of a template. URI Genomics & Sequencing Center. 2004;19:2012. [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari Q., Jameel Y., Massaro J.M., D'Agostino R.B., Sr., Khan S. Periodic Oscillations in Daily Reported Infections and Deaths for Coronavirus Disease 2019. JAMA Netw Open. 2020;3(8) doi: 10.1001/jamanetworkopen.2020.17521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A., Sella Y., Agre P., Casadevall A. Oscillations in U.S. COVID-19 Incidence and Mortality Data Reflect Diagnostic and Reporting Factors. mSystems. 2020;5(4) doi: 10.1128/mSystems.00544-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricon-Becker I., Tarrasch R., Blinder P., Ben-Eliyahu S. medRxiv; 2020. A seven-day cycle in COVID-19 infection and mortality rates: Are inter-generational social interactions on the weekends killing susceptible people? [Google Scholar]
- ChinaCDC . Specific primers and probes for detection 2019 novel coronavirus; 2020. National Institute for Viral Disease Control and Prevention.http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana. USA. Sci Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative Performance of SARS-CoV-2 Detection Assays using Seven Different Primer/Probe Sets and One Assay Kit. J Clin Microbiol. 2020 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Pagaling E., Yan T. Estimating the Prevalence of Potential Enteropathogenic Escherichia coli and Intimin Gene Diversity in a Human Community by Monitoring Sanitary Sewage. Appl Environ Microbiol. 2014;80(1):119–127. doi: 10.1128/AEM.02747-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ Sci Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes–identification of potential viral indicators. Sci Total Environ. 2014:290–296. doi: 10.1016/j.scitotenv.2014.04.087. 488-489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.