Abstract
Background
Gastric cancer is the most common gastrointestinal tumor, and the rates of recurrence and metastasis are high. Research results on molecular biomarkers used for prognosis of gastric cancer remain inconclusive. This study aimed to explore the gene expression module of gastric cancer and to determine potential prognostic biomarkers.
Material/Methods
Three microarray datasets (GSE13911, GSE79973, and GSE29272) from Gene Expression Omnibus (GEO), including 206 pairs of gastric tumors and adjacent normal samples, were used for analysis of differentially expressed genes (DEGs). The 3 microarray datasets yielded 144 genes associated with the progression and prognosis of gastric cancer. After this, a risk score model was developed for result validation using an independent dataset from The Cancer Genome Atlas.
Results
The validation of the independent dataset showed significantly increased NID2, SPARC, and MFAP2 expression in gastric tumor tissues, which were associated with poor outcomes in gastric cancer patients. Moreover, the high risk score obtained was associated with poor overall survival (HR: 1.787; 1.069–2.986; P=0.027). Subgroup analyses revealed that these significant prognostic values were detected in patients aged <65.0 years, tumors in the antrum/distal colon, grade 3 tumors, or TNM-M0 stages of cancer.
Conclusions
The findings of this study show that NID2, SPARC, and MFAP2 are upregulated in gastric tumor tissues and are significantly associated with poor overall survival. Therefore, the predictive values of the risk score model employed for the prognosis of gastric cancer could be improved by using these 3 upregulated DEGs.
Keywords: Biological Markers; Models, Genetic; Stomach Neoplasms
Background
Gastric cancer is the third leading cause of cancer-related deaths in the world, accounting for 8.2% of all cancer-related deaths [1,2]. Effective preventive and treatment strategies are required to improve the treatment and prognosis of gastric cancer, especially in Asian countries. Currently, the overall survival of gastric cancer has already been improved due to the diagnosis of the disease at an early stage and the timely application of adjuvant chemotherapy [3–5]. Although the advances in the multidisciplinary approaches and the combination treatment regimen, the prognosis of advanced gastric cancer remains dismal. Moreover, the heterogeneity of somatic or germline changes in patients are associated with the prognosis of gastric cancer.
Earlier studies have already identified the potential value of genetic and epigenetic alterations for gastric cancer prognosis. These alterations affect cycle regulation, cell adhesion, angiogenesis, and tumor carcinogenesis, having a significant prognostic role in the survival outcome in gastric cancer patients [6–9]. Moreover, investigations have already evaluated the gene expression profile of gastric cancer based on DNA microarray data, and explored the potential role of differentially expressed genes (DEGs) in the prognosis of gastric cancer [10–12]. However, the results of the above studies are limited due to their small sample sizes and the lack of validation datasets established in clinical practice. Hence, the use of the identified DEGs for prognosis of gastric cancer has been limited. Therefore, potential novel DEGs should be identified whose role in the overall survival in gastric cancer patients should be assessed.
The potential role of genes in the progression and prognosis of gastric cancer could be revealed through microarray analysis [13,14]. Three microarray data (GSE13911, GSE79973, and GSE29272) were integrated and 144 DEGs were identified. After the validation of DEGs in The Cancer Genome Atlas (TCGA), we noted that NID2, SPARC, and MFAP2 were more significantly upregulated in gastric tissues than in their adjacent normal tissues. Therefore, the high expression of NID2, SPARC, and MFAP2 might affect the prognosis for GC, and the risk scores determined on the basis of NID2, SPARC, and MFAP2 expression on OS in patients with GC after adjustment for potential confounders should be explored.
Material and Methods
Gastric Cancer Datasets
The Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) was applied to obtain the discovery and validation datasets. Three independent gastric cancer microarray datasets – GSE13911, GSE79973, and GSE29272 – were used to identify the DEGs, with 206 pairs of gastric tumors and adjacent normal samples. These datasets were generated on the basis of GPL570 platforms (Affymetrix Human Genome U113 Plus 2.0 Array) and GPL6947 platforms (Illumina HumanHT-12 V3.0 expression beadchip). GSE13911 and GSE79973 datasets composed 62 and 10 pairs of matched primary gastric tumors and their adjacent normal tissues, respectively. The GSE29272 dataset contained 62 pairs of cardia and 72 pairs of non-cardia gastric cancer. Data related to gastric cancer gene expression and in clinical practice were obtained from The Cancer Genome Atlas-Stomach Adenocarcinoma (TCGA-STAD). A total of 761 samples were identified, and 333 cases were selected in the survival analysis after removing the normal sample data and the data from patients with insufficient follow-up.
Data Preprocessing
The raw probe-level data in this study were downloaded in CEL files, and the robust multi-array average algorithm RMA from the Affy package of R software was employed for processing the raw probe-level data [15]. The background correction, quantile normalization, and summarizing probe set values into 1 expression measure were processed for analysis of the data of gene expression. The annotations for the probe arrays were obtained from GEO, and the mean of the probe sets values was considered as a value of the expression when multiple probe sets were mapped to the same gene [16]. In our study, the log FC in datasets met the criteria for normal distribution.
Statistical Analysis
The identified DEGs were evaluated using the LIMMA package, with bayesian adjusted t-statistics from the linear models for microarray data [17]. Genes with |log2 fold change (FC)| >1 and P<0.05 were regarded as DEGs between tumors and normal tissues. We constructed volcano plots and Venn diagrams using ggplot2, and Venn diagram packages of R software were used to visualize the identified DEGs.
The GO and KEGG pathway analyses of functional enrichment analysis for 144 common DEGs was conducted by using the online software Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). All P values are 2-sided, and P<0.05 was considered to indicate statistically significant enrichment.
SPSS software (version 22.0, SPSS, Chicago, IL, USA) was used for statistical analysis. The risk score model consisted of gene expression, which could be validated in TCGA database. Next, the risk score model was constructed in TCGA-STAD, and the risk score of each individual patient was calculated. Moreover, the risk score was categorized into high and low, and the cutoff value was set to be the median of the risk score. The baseline characteristics between groups were compared using Kruskal-Wallis and chi-square tests based on the type of data. The propensity score analysis was used to adjust for imbalance in the baseline characteristics to avoid undue influences of confounding factors, which was analyzed using the MatchIt propensity score of R software, and the standardized mean difference for matching variables was defined as <20% between the groups. Kaplan-Meier and log-rank tests were employed for survival analysis. Subgroup analyses were also performed according to age, race, anatomic tumor site, grade, TNM-T, TNM-N, TNM-M, and stage. P<0.05 was considered to indicate statistical significance.
Results
Identification of DEGs Between Gastric Tumors and Adjacent Normal Samples
GSE13911, GSE79973, and GSE29272 were employed as the discovery datasets for the identified DEGs expressed in gastric tumors and their adjacent normal tissues. These 3 datasets included 206 pairs of gastric tumors and their adjacent normal samples. The DEGs were explored to evaluate the association between gene expression alteration and gastric cancer progression. The details regarding the expression data from primary gastric tumors and adjacent normal samples are shown in Figure 1. A total number of 144 DEGs were detected for the intersecting part of the 3 sets, which were generally related to gastric samples and potentially associated with the progression and prognosis of gastric cancer (Figures 2, 3). Detailed information of the 144 DEGs established is presented in Table 1.
Figure 1.
The details regarding the expression data from primary gastric tumors and adjacent normal samples in 4 subsets of 3 datasets.
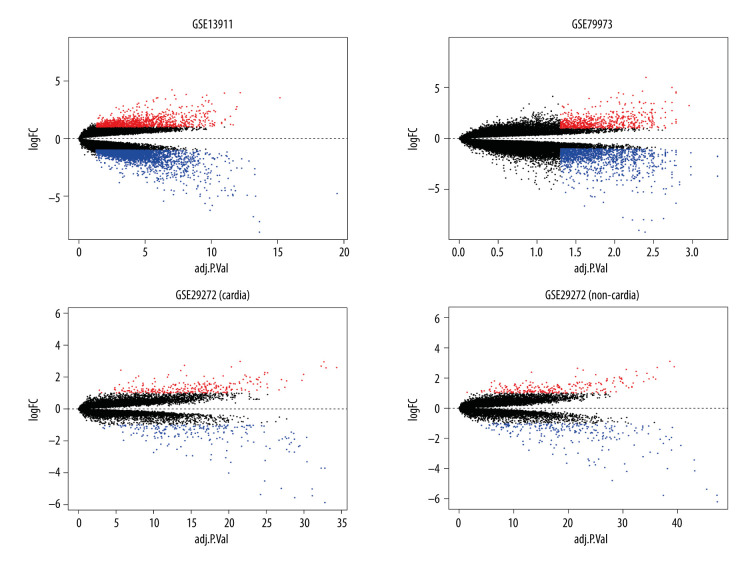
Figure 2.
Identification of differentially expressed genes. Visualization of the identified differentially expressed genes was performed by volcano plots. Dots represent genes with color coding: red indicates upregulated, blue indicates downregulated, and black indicates genes that are not differentially expressed.
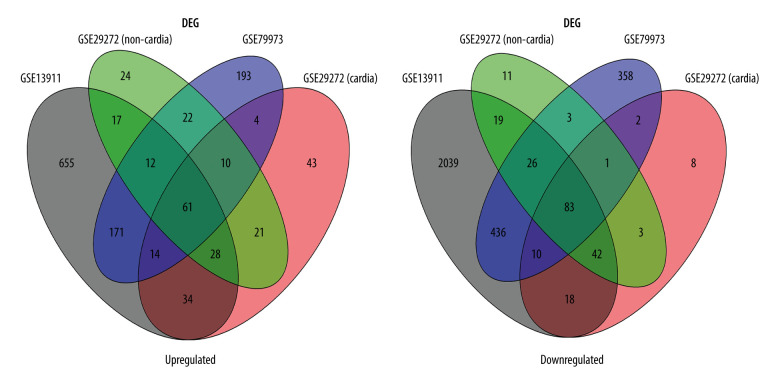
Figure 3.
Venn diagram of the overlapping parts of the 4 subsets of 3 datasets of differentially expressed genes. Sixty-one genes were upregulated and 83 were downregulated.
Table 1.
Common differentially expressed genes identified in gastric cancer.
| Regulation | DEGs (gene symbol) | ||
|---|---|---|---|
| Upregulated | APOC1 | CTSB | NEK2 |
| APOE | CXCL8 | NID2 | |
| ASPM | ECT2 | NT5DC2 | |
| ASPN | FAP | OLFML2B | |
| BGN | FBN1 | PMEPA1 | |
| CAMK2N1 | FCER1G | PRC1 | |
| CDH11 | FN1 | RAB31 | |
| CEMIP | GPNMB | RARRES1 | |
| CEP55 | HOXC6 | S100A10 | |
| COL10A1 | IGF2BP3 | SERPINH1 | |
| COL11A1 | IGFBP7 | SFRP4 | |
| COL1A1 | INHBA | SPARC | |
| COL1A2 | KPNA2 | SPP1 | |
| COL3A1 | LEF1 | SULF1 | |
| COL4A1 | LOC100129518///SOD2 | THBS1 | |
| COL4A2 | LOC101928916///NNMT | THBS2 | |
| COL5A1 | LY6E | THY1 | |
| COL5A2 | MEST | TIMP1 | |
| COL6A3 | MFAP2 | TOP2A | |
| CSE1L | CST1 | VCAN | |
| MIR1292///SNORD110///SNORD86///SNORD57///NOP56 | |||
| Downregulated | AADAC | COL2A1 | KCNE2 |
| ADAM28 | CPA2 | KCNJ15 | |
| ADGRG2 | CYP2C18 | KCNJ16 | |
| ADH1C | DGKD | KIAA1324 | |
| AKR1B10 | EPB41L4B | KLF4 | |
| AKR1C1 | ESRRG | KRT20 | |
| AKR7A3 | ETNPPL | LIPF | |
| ALDH1A1 | FA2H | LOC101930400///AKR1C2 | |
| ALDH3A1 | FBP2 | LTF | |
| ALDH6A1 | FCGBP | MAOA | |
| ATP4A | FMO5 | METTL7A | |
| ATP4B | FOLR1 | MT1E | |
| AZGP1 | GC | MT1F | |
| AZGP1P1///AZGP1 | GIF | MT1G | |
| CA2 | GKN1 | MT1H | |
| CA9 | GPRC5C | MT1HL1 | |
| CAPN9 | GSTA1 | MT1M | |
| CCKBR | HDC | MT1X | |
| CHGA | HPGD | MT2A | |
| CHIA | IGH | MUC5AC | |
| CKM | IGHA2///IGHA1///IGH | MYOC | |
| CKMT2 | JCHAIN | MYRF | |
| NEDD4L | PIK3C2G | SLC16A7 | |
| PBLD | PXMP2 | SLC28A2 | |
| PDGFD | RNASE1 | SOSTDC1 | |
| PGA4///PGA3///PGA5 | S100P | SULT1C2 | |
| PGC | UGT2B15 | TMPRSS2 | |
| XK | XYLT2 | ||
Functional Enrichment Analysis of DEGs
GO and KEGG pathway enrichment analyses were performed to investigate the biological roles of DEGs in gastric cancer progression, including cell cycle and cell adhesion. The enriched GO terms were mainly associated with the extracellular matrix of the cellular component, and the KEGG pathway analysis results showed that the most highly enriched pathway was ECM-receptor interaction. The results of the GO and KEGG pathway enrichment analyses are summarized and displayed in Tables 2 and 3.
Table 2.
GO analysis of the 144 differentially expressed genes.
| Category | Term | Count | P value | Benjamini | FDR | Genes |
|---|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO: 0030198 ~extracellular matrix organization | 21 | 5.81E-17 | 1.15E-13 | 1.78E-13 | COL4A2, COL4A1, OLFML2B, COL3A1, FBN1, COL2A1, SPARC, NID2, COL5A2, COL5A1, BGN, COL6A3, COL1A2, MFAP2, VCAN, COL1A1, THBS1, COL11A1, SPP1, FN1, COL10A1 |
| GOTERM_MF_DIRECT | GO: 0005201 ~extracellular matrix structural constituent | 13 | 9.89E-14 | 3.07E-11 | 1.33E-10 | COL4A2, BGN, COL4A1, COL3A1, FBN1, COL1A2, COL2A1, VCAN, COL1A1, MUC5AC, COL5A2, COL11A1, COL5A1 |
| GOTERM_CC_DIRECT | GO: 0005576 ~extracellular region | 42 | 4.25E-13 | 7.69E-11 | 5.24E-10 | GC, CHIA, IGFBP7, COL3A1, JCHAIN, APOC1, CXCL8, COL2A1, TIMP1, AZGP1, APOE, COL6A3, CPA2, LTF, PDGFD, THBS1, COL11A1, THBS2, SPP1, COL10A1, FN1, ADAM28, COL4A2, COL4A1, OLFML2B, FBN1, GIF, NID2, SPARC, COL5A2, COL5A1, INHBA, BGN, SFRP4, CEMIP, COL1A2, VCAN, MFAP2, COL1A1, CTSB, MUC5AC, LIPF |
| GOTERM_CC_DIRECT | GO: 0031012 ~extracellular matrix | 20 | 5.14E-13 | 4.65E-11 | 6.33E-10 | ASPN, COL4A2, COL4A1, IGFBP7, COL3A1, FBN1, COL2A1, NID2, COL5A2, COL5A1, BGN, APOE, COL6A3, COL1A2, VCAN, COL1A1, THBS1, THBS2, MYOC, FN1 |
| GOTERM_CC_DIRECT | GO: 0005615 ~extracellular space | 38 | 8.88E-13 | 5.36E-11 | 1.10E-09 | GC, CHIA, PGC, IGFBP7, COL3A1, JCHAIN, CXCL8, COL2A1, SERPINH1, ALDH3A1, TIMP1, AZGP1, APOE, SOSTDC1, FAP, COL6A3, CPA2, LTF, PDGFD, THBS1, MYOC, SPP1, FN1, ATP4A, FBN1, GIF, CST1, SPARC, CHGA, SFRP4, SULF1, COL1A2, VCAN, COL1A1, CTSB, MUC5AC, CA2, GKN1 |
| GOTERM_BP_DIRECT | GO: 0030574 ~collagen catabolic process | 12 | 2.42E-12 | 1.26E-09 | 3.85E-09 | COL4A2, COL4A1, COL3A1, COL6A3, COL1A2, COL2A1, COL1A1, CTSB, COL11A1, COL5A2, COL5A1, COL10A1 |
| GOTERM_CC_DIRECT | GO: 0070062 ~extracellular exosome | 51 | 3.44E-10 | 1.56E-08 | 4.24E-07 | RARRES1, PGC, IGFBP7, KIAA1324, AZGP1, APOE, LTF, AKR7A3, PDGFD, AKR1C1, ALDH6A1, FBP2, METTL7A, THY1, BGN, AKR1B10, COL1A2, CTSB, CA2, GC, GPRC5C, JCHAIN, APOC1, SERPINH1, PBLD, TIMP1, ALDH1A1, CSE1L, FOLR1, COL6A3, SULT1C2, NEDD4L, THBS1, MYOC, SPP1, FN1, MEST, GSTA1, TMPRSS2, RNASE1, COL4A2, S100P, FBN1, S100A10, NID2, ADGRG2, COL5A1, MUC5AC, FCGBP, HPGD, CDH11 |
| GOTERM_CC_DIRECT | GO: 0005581 ~collagen trimer | 11 | 1.32E-09 | 4.79E-08 | 1.63E-06 | COL3A1, COL6A3, COL1A2, COL2A1, COL1A1, COL11A1, SERPINH1, COL5A2, COL5A1, COL10A1, TIMP1 |
| GOTERM_CC_DIRECT | GO: 0005788 ~endoplasmic reticulum lumen | 14 | 1.55E-09 | 4.68E-08 | 1.91E-06 | COL4A2, COL4A1, COL3A1, COL6A3, COL1A2, COL2A1, COL1A1, PDGFD, THBS1, SERPINH1, COL5A2, COL11A1, COL5A1, COL10A1 |
| GOTERM_BP_DIRECT | GO: 0071294~cellular response to zinc ion | 7 | 5.25E-09 | 1.82E-06 | 8.34E-06 | MT1M, MT2A, MT1E, MT1H, MT1X, MT1G, MT1F |
| GOTERM_BP_DIRECT | GO: 0045926~negative regulation of growth | 7 | 5.25E-09 | 1.82E-06 | 8.34E-06 | MT1M, MT2A, MT1E, MT1H, MT1X, MT1G, MT1F |
| GOTERM_CC_DIRECT | GO: 0005578 ~proteinaceous extracellular matrix | 15 | 1.01E-08 | 2.62E-07 | 1.25E-05 | ASPN, OLFML2B, FBN1, SPARC, COL5A2, COL5A1, TIMP1, BGN, COL6A3, COL1A2, VCAN, COL11A1, MYOC, FN1, COL10A1 |
| GOTERM_MF_DIRECT | GO: 0048407 ~platelet-derived growth factor binding | 6 | 1.07E-08 | 1.66E-06 | 1.44E-05 | COL4A1, COL3A1, COL1A2, COL2A1, COL1A1, COL5A1 |
| GOTERM_BP_DIRECT | GO: 0030199~collagen fibril organization | 8 | 1.97E-08 | 5.12E-06 | 3.13E-05 | COL3A1, COL1A2, COL2A1, COL1A1, COL11A1, SERPINH1, COL5A2, COL5A1 |
| GOTERM_MF_DIRECT | GO: 0050840 ~extracellular matrix binding | 6 | 1.39E-06 | 1.44E-04 | 0.001872262 | BGN, OLFML2B, SPARC, THBS1, COL11A1, SPP1 |
| GOTERM_CC_DIRECT | GO: 0005604 ~basement membrane | 8 | 1.76E-06 | 3.99E-05 | 0.002172174 | COL4A1, FBN1, COL2A1, NID2, SPARC, THBS2, COL5A1, TIMP1 |
| GOTERM_BP_DIRECT | GO: 0007155 ~cell adhesion | 16 | 3.26E-06 | 6.77E-04 | 0.005182285 | ATP4B, IGFBP7, NID2, COL5A1, THY1, AZGP1, FAP, COL6A3, VCAN, COL1A1, THBS1, GPNMB, THBS2, SPP1, FN1, CDH11 |
| GOTERM_BP_DIRECT | GO: 0071276~cellular response to cadmium ion | 5 | 8.02E-06 | 0.001386589 | 0.012731623 | MT1E, MT1H, MT1X, MT1G, MT1F |
| GOTERM_BP_DIRECT | GO: 0007586~digestion | 7 | 9.91E-06 | 0.001467951 | 0.015725541 | CHIA, CCKBR, PGC, AKR1B10, CAPN9, GKN1, AKR1C1 |
| GOTERM_BP_DIRECT | GO: 0001501~skeletal system development | 9 | 1.26E-05 | 0.001632159 | 0.019983654 | COL3A1, FBN1, COL1A2, COL2A1, VCAN, COL1A1, COL5A2, COL10A1, CDH11 |
| GOTERM_MF_DIRECT | GO: 0005178 ~integrin binding | 8 | 1.58E-05 | 0.001223983 | 0.021220906 | FAP, COL3A1, FBN1, THBS1, GPNMB, COL5A1, FN1, THY1 |
Table 3.
KEGG pathway enrichment analysis of the 144 differentially expressed genes.
| Category | Term | Count | P value | FDR | Genes |
|---|---|---|---|---|---|
| KEGG_PATHWAY | hsa04512: ECM-receptor interaction | 14 | 3.93E-12 | 4.53E-09 | COL4A2, COL4A1, COL3A1, COL2A1, COL5A2, COL5A1, COL6A3, COL1A2, COL1A1, THBS1, COL11A1, THBS2, SPP1, FN1 |
| KEGG_PATHWAY | hsa04974: Protein digestion and absorption | 12 | 1.48E-09 | 1.70E-06 | COL4A2, COL4A1, COL3A1, COL6A3, COL1A2, CPA2, COL2A1, COL1A1, COL11A1, COL5A2, COL5A1, COL10A1 |
| KEGG_PATHWAY | hsa04510: Focal adhesion | 15 | 2.34E-08 | 2.69E-05 | COL4A2, COL4A1, COL3A1, COL2A1, COL5A2, COL5A1, COL6A3, COL1A2, PDGFD, COL1A1, THBS1, COL11A1, THBS2, SPP1, FN1 |
| KEGG_PATHWAY | hsa05146: Amoebiasis | 11 | 1.36E-07 | 1.57E-04 | COL4A2, COL4A1, COL3A1, COL1A2, CXCL8, COL2A1, COL1A1, COL11A1, COL5A2, COL5A1, FN1 |
| KEGG_PATHWAY | hsa04978: Mineral absorption | 7 | 5.95E-06 | 0.006851 | MT1M, MT2A, MT1E, MT1H, MT1X, MT1G, MT1F |
| KEGG_PATHWAY | hsa04151: PI3K-Akt signaling pathway | 15 | 1.22E-05 | 0.014013 | COL4A2, COL4A1, COL3A1, COL2A1, COL5A2, COL5A1, COL6A3, COL1A2, PDGFD, COL1A1, THBS1, COL11A1, THBS2, SPP1, FN1 |
| KEGG_PATHWAY | hsa04971: Gastric acid secretion | 7 | 1.13E-04 | 0.129558 | KCNJ16, KCNJ15, CCKBR, ATP4A, ATP4B, KCNE2, CA2 |
| KEGG_PATHWAY | hsa04611: Platelet activation | 8 | 4.23E-04 | 0.486191 | COL3A1, COL1A2, FCER1G, COL2A1, COL1A1, COL11A1, COL5A2, COL5A1 |
| KEGG_PATHWAY | hsa00982: Drug metabolism – cytochrome P450 | 6 | 7.27E-04 | 0.833392 | GSTA1, FMO5, MAOA, ADH1C, UGT2B15, ALDH3A1 |
| KEGG_PATHWAY | hsa00980: Metabolism of xenobiotics by cytochrome P450 | 6 | 0.001069 | 1.223788 | GSTA1, ADH1C, AKR7A3, UGT2B15, AKR1C1, ALDH3A1 |
| KEGG_PATHWAY | hsa05204: Chemical carcinogenesis | 5 | 0.010061 | 10.99085 | GSTA1, CYP2C18, ADH1C, UGT2B15, ALDH3A1 |
| KEGG_PATHWAY | hsa00340: Histidine metabolism | 3 | 0.022373 | 22.93516 | HDC, MAOA, ALDH3A1 |
| KEGG_PATHWAY | hsa00830: Retinol metabolism | 4 | 0.030117 | 29.67766 | ALDH1A1, CYP2C18, ADH1C, UGT2B15 |
| KEGG_PATHWAY | hsa04966: Collecting duct acid secretion | 3 | 0.032861 | 31.93507 | ATP4A, ATP4B, CA2 |
Validation of DEGs in an Independent Database
TCGA-STAD included 333 GC patients, who were regarded as a validation cohort, which was assessed to verify the expression of DEGs. The results indicated that NID2, SPARC, and MFAP2 were the 3 top-ranked upregulated genes for the risk of GC. Further, we developed a risk score model described by the following formula: risk score=0.005974532×ExpNID2+ 0.004623909×ExpSPARC+ 0.054586198×ExpMFAP2. The categories of high and low risk scores were based on the median values of the of risk scores.
Risk Score and Overall Survival for Patients with Gastric Cancer
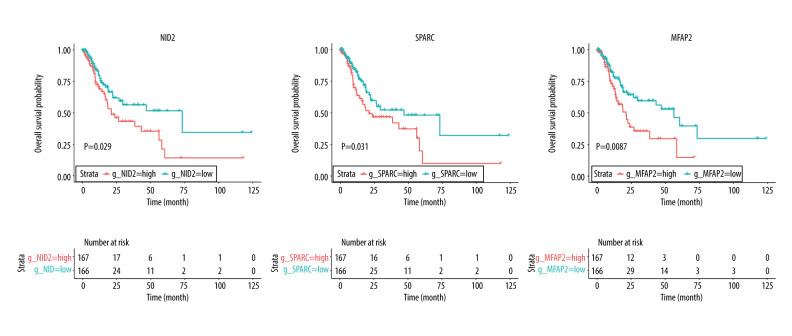
The baseline characteristics of the high (n=166) and low (n=167) risk score groups are presented in Table 4. Significant differences were observed between groups in terms of race and tumor stages, whereas no significant differences were established for age, sex, anatomic tumor site, tumor grade, TNM-T, TNM-N, and TNM-M. Overall, we noted that a high risk score was obviously associated with poor overall survival (HR: 2.041; 95% CI: 1.272–3.274; P=0.003; Figure 4). Significant associations were observed mainly in the following patients: <65.0 years, with a tumor in the antrum/distal colon, with a grade 3 tumor, irrespective of the TNM-T stage, TNM-N2-3, TNM-M0, and stage III and IV (Table 2). After propensity score analysis, the higher risk scores were associated with poorer overall survival (HR: 1.787; 1.069–2.986; P=0.027; Figure 5). Subgroup analysis showed that high risk scores were associated with poor overall survival in patients age <65.0 years and if they had a tumor in the antrum/distal colon, grade 3 tumor, or TNM-M0 stages gastric cancer (Table 5).
Table 4.
Baseline characteristics of patients in high and low risk score groups.
| Pre-propensity score matching | Post-propensity score matching | |||||
|---|---|---|---|---|---|---|
| Low | High | P | Low | High | P | |
| Age, median (Q1, Q3) | 67.00 (58.00,72.00) | 67.00 (58.00,72.00) | 0.960 | 67.00 (58.00,72.00) | 68.00 (59.00,72.00) | 0.617 |
| Gender | ||||||
| Female | 55 (33.13) | 62 (37.13) | 0.445 | 49 (35.51) | 50 (36.23) | 0.900 |
| Male | 111 (66.87) | 105 (62.87) | 89 (64.49) | 88 (63.77) | ||
| Race | ||||||
| White | 96 (57.83) | 114 (68.26) | 0.031 | 89 (64.49) | 92 (66.67) | 0.932 |
| Asian | 38 (22.89) | 33 (19.76) | 34 (24.64) | 27 (19.57) | ||
| Others | 32 (19.28) | 20 (11.98) | 15 (10.87) | 19 (13.77) | ||
| Anatomic tumor site | ||||||
| Antrum/distal | 61 (36.75) | 63 (37.72) | 0.394 | 54 (39.13) | 51 (36.96) | 0.955 |
| Fundus/body | 56 (33.73) | 62 (37.13) | 47 (34.06) | 51 (36.96) | ||
| Cardia/proximal | 24 (14.46) | 20 (11.98) | 19 (13.77) | 17 (12.32) | ||
| Gastroesophageal junction | 18 (10.84) | 20 (11.98) | 13 (9.42) | 18 (13.04) | ||
| Others | 7 (4.22) | 2 (1.20) | 5 (3.62) | 1 (0.72) | ||
| Grade | ||||||
| G1 | 4 (2.41) | 4 (2.40) | 0.930 | 4 (2.90) | 4 (2.90) | 0.917 |
| G2 | 60 (36.14) | 55 (32.93) | 48 (34.78) | 43 (31.16) | ||
| G3 | 98 (59.04) | 103 (61.68) | 83 (60.14) | 87 (63.04) | ||
| Gx | 4 (2.41) | 5 (2.99) | 3 (2.17) | 4 (2.90) | ||
| TNM-T | ||||||
| T1–2 | 44 (26.51) | 38 (22.75) | 0.427 | 35 (25.36) | 36 (26.09) | 0.891 |
| T3–4 | 122 (73.49) | 129 (77.25) | 103 (74.64) | 102 (73.91) | ||
| TNM-N | ||||||
| N0 | 53 (31.93) | 55 (32.93) | 0.533 | 44 (31.88) | 47 (34.06) | 0.419 |
| N1 | 38 (22.89) | 46 (27.54) | 33 (23.91) | 40 (28.99) | ||
| N2 | 41 (24.70) | 31 (18.56) | 35 (25.36) | 24 (17.39) | ||
| N3 | 34 (20.48) | 35 (20.96) | 26 (18.84) | 27 (19.57) | ||
| TNM-M | ||||||
| M0 | 157 (94.58) | 153 (91.62) | 0.287 | 131 (94.93) | 128 (92.75) | 0.453 |
| M1 | 9 (5.42) | 14 (8.38) | 7 (5.07) | 10 (7.25) | ||
| Stage | ||||||
| I | 29 (17.47) | 15 (8.98) | 0.029 | 22 (15.94) | 15 (10.87) | 0.073 |
| II | 42 (25.30) | 62 (37.13) | 38 (27.54) | 55 (39.86) | ||
| III | 75 (45.18) | 67 (40.12) | 63 (45.65) | 50 (36.23) | ||
| IV | 14 (8.43) | 20 (11.98) | 10 (7.25) | 16 (11.59) | ||
Figure 4.
Overall survival according to the expression of NID2, SPARC, and MFAP2. Red line indicates high expression and blue line indicates low expression.
Figure 5.
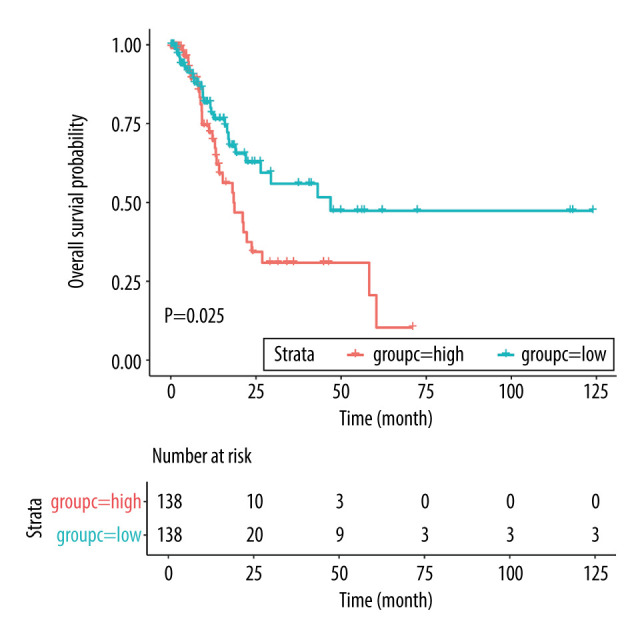
Overall survival according to the risk scores after propensity score analysis. Red line indicates high risk score and blue line indicates low risk score.
Table 5.
Subgroup analyses for overall survival before and after propensity score analysis.
| Factors | Group | Before propensity score | Propensity score analysis | ||
|---|---|---|---|---|---|
| HR and 95% CI | P-value | HR and 95%CI | P-value | ||
| Age (years) | <65.0 | 2.949 (1.283–6.774) | 0.011 | 2.840 (1.161–6.945) | 0.022 |
| >65.0 | 1.581 (0.890–2.808) | 0.118 | 1.363 (0.725–2.562) | 0.337 | |
| Race | White | 1.700 (0.907–3.186) | 0.098 | 1.530 (0.781–2.997) | 0.215 |
| Asian | 8.072 (0.823–79.156) | 0.073 | 6.308 (0.571–69.650) | 0.133 | |
| Anatomic tumor site | Antrum/distal | 3.278 (1.553–6.922) | 0.002 | 3.018 (1.353–6.732) | 0.007 |
| Fundus/body | 1.904 (0.805–4.504) | 0.143 | 1.392 (0.563–3.438) | 0.474 | |
| Position-others | 1.082 (0.423–2.770) | 0.869 | 1.133 (0.408–3.150) | 0.810 | |
| Grade | 1–2 | 1.933 (0.904–4.131) | 0.089 | 1.320 (0.592–2.942) | 0.497 |
| 3 | 2.376 (1.275–4.428) | 0.006 | 2.576 (1.267–5.238) | 0.009 | |
| TNM-T | T1–2 | 3.838 (1.130–13.038) | 0.031 | 3.590 (0.937–13.760) | 0.062 |
| T3–4 | 1.856 (1.103–3.124) | 0.020 | 1.639 (0.926–2.901) | 0.090 | |
| TNM-N | N0–1 | 1.873 (0.923–3.802) | 0.082 | 1.746 (0.840–3.627) | 0.135 |
| N2–3 | 2.127 (1.114–4.064) | 0.022 | 1.695 (0.812–3.539) | 0.160 | |
| TNM-M | M0 | 2.132 (1.274–3.567) | 0.004 | 1.843 (1.055–3.217) | 0.032 |
| M1 | 1.103 (0.333–3.651) | 0.872 | 1.149 (0.305–4.329) | 0.837 | |
| Stage | I and II | 2.305 (0.855–6.215) | 0.099 | 2.291 (0.782–6.712) | 0.131 |
| III and IV | 2.095 (1.191–3.685) | 0.010 | 1.652 (0.893–3.055) | 0.110 | |
CI – confidence interval; HR – hazard ratio; M – metastasis; N – node; T – tumor.
Discussion
The gene expression modules at the genome-wide scale in gastric cancer were investigated in our study through integrating multiple gastric cancer transcriptome microarray datasets. Our findings provide information on alterations at the molecular level; we achieved higher robustness than that of data from a single dataset. We screened 144 DEGs in gastric tumors and adjacent normal samples and discovered that the expression levels of NID2, SPARC, and MFAP2 were the 3 top-ranked upregulated genes. Next, a risk score model based on 3 DEGs was constructed (risk score=0.005974532×ExpNID2+ 0.004623909×ExpSPARC+ 0.054586198×ExpMFAP2), which was significantly associated with poor overall survival in patients with GC, based on data from TCGA database. Furthermore, using propensity score analysis, we observed these associations mainly in patients younger than 65.0 years, with a tumor in the antrum/distal colon, with a grade 3 tumor, or with TNM-M0 stages of GC.
The results of this study indicated that GC is involved in cell cycle, cell adhesion, and the extracellular matrix; these processes were found in patients with upregulated NID2, SPARC, and MFAP2. The cell adhesion dysfunction was significantly associated with gastric cancer metastasis, which could be considered to represent multiple activated signaling pathways in the malignancy [18]. Moreover, the common characteristics of gastric cancer were the dense stroma with enormous quantities of extracellular matrix in the surrounding area [19,20]. The gene annotation analysis results support our findings on the enriched cellular components of extracellular matrix and ECM-receptor interaction pathway.
We noted the expression of NID2, SPARC, and MFAP2 in gastric tumors was upregulated compared with adjacent normal tissue samples. The role of abnormal NID2 methylation in cancer prognosis at various sites has already been highlighted in previous research [21–25]. NID2, which is a member of the nidogen protein family, has been reported to maintain the stability and integrity of the basement membrane. Moreover, the involvement of SPARC in the prognosis of gastric cancer has also been confirmed in many studies [26–28]. The study conducted by Liao et al identified 4 microarray datasets and found SPARC is significantly upregulated in gastric tissues, which was associated with poor prognosis [26]. Evidence has shown that cell adhesion, proliferation, migration, and tissue remodeling are regulated by SPARC during cell development and the extracellular matrix turnover processes [29,30]. Recently, MFAP2 was found to modulate tropoelastin deposition into micro-fibrils, which participates in the formation of elastic fibers [31]. Moreover, it was considered as the co-expressed gene of the NF-κB/Snail/YY1/RKIP circuitry, which was upregulated in tumor tissues; the extent of this upregulation was specific evidence of lymph node metastasis [32,33]. In the present study, we constructed a risk score model for predicting overall survival of gastric cancer patients, which showed that a high risk score was associated with poor overall survival. Moreover, stratified analyses of patients’ characteristics also confirmed our findings.
Several limitations to this study should be acknowledged: (1) The interpretation of the results should be cautions due to the collection of data from different platforms; (2) Bioinformatics analysis was applied, whose findings should be verified in further research to clarify the mechanisms of the association between these genes and poor GC survival; (3) The range of the analyses was limited due to variations in the characteristics of the patients; and (4) The role of the expression of the studied 3 genes associated with other survival outcomes in patients with GC should be further explored, including the determination of progression-free survival.
Conclusions
In conclusion, the findings of this study suggest the upregulation of NID2, SPARC, and MFAP2 is strongly associated with overall survival in patients with gastric cancer. Moreover, the risk score of the overall survival of gastric cancer patients is affected by age, the anatomic tumor site, tumor grade, and TNM-M. Further research should be conducted in laboratory settings to explore the underlying molecular mechanisms and to translate these research findings into the development of novel targeted-treatment strategies.
Abbreviations
- DEGs
differentially expressed genes
- GEO
Gene Expression Omnibus
Footnotes
Conflict of Interest
None.
Source of support: Departmental sources
References
- 1.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v10, Cancer Incidence and Mortality Worldwide:IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; [accessed on 06/018/2019]. http://globocan.iarc.fr. [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Information Committee of Korean Gastric Cancer Association. Korean Gastric Cancer Association Nationwide Survey on Gastric Cancer in 2014. J Gastric Cancer. 2016;16(3):131–40. doi: 10.5230/jgc.2016.16.3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang YJ, Kim YW, Yang HK, et al. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC) : A phase 3 open-label, randomised controlled trial. Lancet. 2012;379(9813):315–21. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 5.Sasako M, Sakuramoto S, Katai H, et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29(33):4387–93. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 6.Akama Y, Yasui W, Yokozaki H, et al. Frequent amplification of the cyclin E gene in human gastric carcinomas. Jpn J Cancer Res. 1995;86(7):617–21. doi: 10.1111/j.1349-7006.1995.tb02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graziano F, Mandolesi A, Ruzzo A, et al. Predictive and prognostic role of E-cadherin protein expression in patients with advanced gastric carcinomas treated with palliative chemotherapy. Tumour Biol. 2004;25(3):106–10. doi: 10.1159/000079141. [DOI] [PubMed] [Google Scholar]
- 8.Tanigawa N, Amaya H, Matsumura M, et al. Correlation between expression of vascular endothelial growth factor and tumor vascularity, and patient outcome in human gastric carcinoma. J Clin Oncol. 1997;15(2):826–32. doi: 10.1200/JCO.1997.15.2.826. [DOI] [PubMed] [Google Scholar]
- 9.Sanz-Ortega J, Steinberg SM, Moro E, et al. Comparative study of tumor angiogenesis and immunohistochemistry for p53, c-ErbB2, c-myc and EGFr as prognostic factors in gastric cancer. Histol Histopathol. 2000;15(2):455–62. doi: 10.14670/HH-15.455. [DOI] [PubMed] [Google Scholar]
- 10.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–57. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CN, Lin JJ, Chen JJ, et al. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23(29):7286–95. doi: 10.1200/JCO.2004.00.2253. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Liu Y, Niu Z, et al. Prognostic value of a 25-gene assay in patients with gastric cancer after curative resection. Sci Rep. 2017;7(1):7515. doi: 10.1038/s41598-017-07604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang W, Ma L, Lin L, et al. Identification of novel hub genes associated with liver metastasis of gastric cancer. Int J Cancer. 2009;125(12):2844–53. doi: 10.1002/ijc.24699. [DOI] [PubMed] [Google Scholar]
- 14.Zhu T, Gao YF, Chen YX, et al. Genome-scale analysis identifies GJB2 and ERO1LB as prognosis markers in patients with pancreatic cancer. Oncotarget. 2017;8(13):21281–89. doi: 10.18632/oncotarget.15068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Li K, Zhao L, et al. Bioinformatics analysis reveals disturbance mechanism of MAPK signaling pathway and cell cycle in Glioblastoma multiforme. Gene. 2014;547(2):346–50. doi: 10.1016/j.gene.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 17.Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JP, Liu D, Gu JF, et al. Shikonin inhibits the cell viability, adhesion, invasion and migration of the human gastric cancer cell line MGC-803 via the Toll-like receptor 2/nuclear factor-kappa B pathway. J Pharm Pharmacol. 2015;67(8):1143–55. doi: 10.1111/jphp.12402. [DOI] [PubMed] [Google Scholar]
- 19.Gan L, Meng J, Xu M, et al. Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin beta4/FAK/SOX2/HIF-1alpha signaling pathway in gastric cancer. Oncogene. 2018;37(6):744–55. doi: 10.1038/onc.2017.363. [DOI] [PubMed] [Google Scholar]
- 20.Wu Q, Li X, Yang H, et al. Extracellular matrix protein 1 is correlated to carcinogenesis and lymphatic metastasis of human gastric cancer. World J Surg Oncol. 2014;12:132. doi: 10.1186/1477-7819-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Zhao Y, Xu H, et al. Silencing NID2 by DNA hypermethylation promotes lung cancer. Pathol Oncol Res. 2020;26(2):801–11. doi: 10.1007/s12253-019-00609-0. [DOI] [PubMed] [Google Scholar]
- 22.Wu Q, Zhang B, Wang Z, et al. Integrated bioinformatics analysis reveals novel key biomarkers and potential candidate small molecule drugs in gastric cancer. Pathol Res Pract. 2019;215(5):1038–48. doi: 10.1016/j.prp.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 23.van der Heijden AG, Mengual L, Ingelmo-Torres M, et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics. 2018;10:71. doi: 10.1186/s13148-018-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai AWY, Cheung AKL, Dai W, et al. Elevated levels of serum nidogen-2 in esophageal squamous cell carcinoma. Cancer Biomark. 2018;21(3):583–90. doi: 10.3233/CBM-170484. [DOI] [PubMed] [Google Scholar]
- 25.Torky HA, Sherif A, Abo-Louz A, et al. Evaluation of serum Nidogen-2 as a screening and diagnostic tool for ovarian cancer. Gynecol Obstet Invest. 2018;83(5):461–65. doi: 10.1159/000481798. [DOI] [PubMed] [Google Scholar]
- 26.Liao P, Li W, Liu R, et al. Genome-scale analysis identifies SERPINE1 and SPARC as diagnostic and prognostic biomarkers in gastric cancer. Onco Targets Ther. 2018;11:6969–80. doi: 10.2147/OTT.S173934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Li AD, Xu L, et al. SPARC expression in gastric cancer predicts poor prognosis: Results from a clinical cohort, pooled analysis and GSEA assay. Oncotarget. 2016;7(43):70211–22. doi: 10.18632/oncotarget.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Hao B, Yang Y, et al. Prognostic role of SPARC expression in gastric cancer: A meta-analysis. Arch Med Sci. 2014;10(5):863–69. doi: 10.5114/aoms.2014.46207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funk SE, Sage EH. The Ca2(+)-binding glycoprotein SPARC modulates cell cycle progression in bovine aortic endothelial cells. Proc Natl Acad Sci USA. 1991;88(7):2648–52. doi: 10.1073/pnas.88.7.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8(2):163–73. [PubMed] [Google Scholar]
- 31.Mecham RP, Gibson MA. The microfibril-associated glycoproteins (MAGPs) and the microfibrillar niche. Matrix Biol. 2015;47:13–33. doi: 10.1016/j.matbio.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaravinos A, Kanellou P, Lambrou GI, et al. Gene set enrichment analysis of the NF-kappaB/Snail/YY1/RKIP circuitry in multiple myeloma. Tumour Biol. 2014;35(5):4987–5005. doi: 10.1007/s13277-014-1659-9. [DOI] [PubMed] [Google Scholar]
- 33.Silveira NJ, Varuzza L, Machado-Lima A, et al. Searching for molecular markers in head and neck squamous cell carcinomas (HNSCC) by statistical and bioinformatic analysis of larynx-derived SAGE libraries. BMC Med Genomics. 2008;1:56. doi: 10.1186/1755-8794-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]