Abstract
The signature events caused by host-guest interactions in the nanopore system can be used as a novel and characteristic signal in quantitative detection and analysis of various molecules. However, the effect of several electrochemical factors on the host-guest interactions in nanopore still remains unknown. Here, we systematically studied host-guest interactions, especially oscillation of DNA-azide adamantane@cucurbit[6] in α-Hemolysin nanopore under varying pH, concentration of electrolytes and counterions (Li+, Na+, K+). Our results indicate correlations between the change of pH and the duration of the oscillation signal. In addition, the asymmetric electrolyte concentration and the charge of the counterions affects the frequency of signature events in oscillation signals, and even the integrity of the protein nanopore. This study provides insight into the design of a future biosensing platform based on signature oscillation signals of the host-guest interaction within a nanopore.
Keywords: Nanopore, Host-guest interaction, Biosensors, Electrochemistry
1. Introduction
Nanopore sensors are of great interest for researchers from a wide range of disciplines, including biomedical science, chemistry and engineering, because they provide unique chemical environments with nanoscale dimensions for analyzing single molecules that cannot be achieved in bulk systems.1,2 Over the past decade, significant progress has been made in the development of robust protocols for precise detection of various biomolecules (i.e. proteins, peptides, amino acids, etc.) using biological or solid-state nanopores.3–8
Recently, the study of host-guest interactions through nanopore attracted increasing attention because of their wide application in the fields of functional materials and nanomedicine.9–11 For example, the biomarkers in human bodily fluid can be quantified by establishing a correlation between the concentration of them and the frequency of unique signature signals generated by the host-guest interaction in the nanopore,12 which is particularly important for clinical diagnostic uses (e.g. early diagnosis and precise prediction of treatment outcomes) as a way to prevent false positives.13,14
In host-guest systems, host molecules, such as calixarenes or cyclodextrins cucurbiturils, bind with a guest molecule through noncovalent interactions.11,15–17 Bayley and co-workers pioneered the application of α-Hemolysin (α-HL) protein for monitoring host-guest interactions.18,19 In these studies, β-cyclodextrin (β-CD) was employed as host molecules as they can non-covalently lodge within nanopore due to their comparable diameters to that of the lumen of α-HL. The analytes as guest molecules bind with the β-CD through host-guest interactions, resulting in characteristic current signals (e.g., extent and level of current blockades) for analyte identification, including the different types of deoxyribonucleoside20, small drug molecules of ibuprofen and thalidomide21. Other host molecules, such as γ-cyclodextrin and cucurbituril have also been used for selectively detecting small organic molecules.15,16 These approaches enabled the detection of small molecules through nanopore.
More recently, Wu and co-workers developed a nanopore-based approach for quick and efficient detection of 5mC and 5hmC in ssDNA by generating characteristic current signatures of host-guest (ferrocene⊂cucurbit,7 Fc⊂CB7) during the translocation of modified DNA through α-HL.17 In this approach, the DNA strand containing 5mC or 5hmC is first modified with Fc⊂CB7 complex. Translocation of the DNA–Fc⊂CB7 hybrids through the nanopore produces highly characteristic signature events (i.e. oscillation signal) which correlate to the existence of 5mC and 5hmC. This feature allows detection at the single molecule level with high confidence, which further engendered the development of a versatile biosensing strategy by combining aptamers and host-guest interactions: an aptamer is first hybridized with a Fc⊂CB7 modified DNA probe, the presence of analytes causes the aptamer-probe duplex to unwind and release the DNA probe which can quantitatively produce signature current events during its translocation through α-HL nanopore.22 Furthermore, by incorporating a sandwich immunoassay involving CuO nanoparticles, a sensitive method to detect biomarkers with extremely low concentrations (sub-femtomolar level) was developed.12 In this approach, the released Cu2+ from immunoprecipitated CuO particles is taken to catalyze the “click” reaction which ligates a host-guest modified DNA probe, and the DNA probe is subjected to nanopore translocation measurement to afford quantification of biomarkers. All of these sensing strategies based on the host-guest interactions are not only beneficial to nanopore sensing, particularly in the detection of DNA modifications, but also suitable for the accurate detection of various biomarkers in clinical samples by eliminating false-positives from interferent biomolecules.23
Despite the abovementioned promising findings, many experimental conditions and mechanisms remain to be elucidated, as the interactions between the probe (i.e. DNA or other charged biomolecules), the host-guest complex and the nanopore can also be affected by the surrounding electrochemical environment including counterions, pH, concentration of electrolytes, etc. As the frequency of the signature oscillation events directly determines the limit of detection of the above strategy, it is essential to optimize the electrochemical conditions of the host-guest interaction and the subsequent translocation.24–28
Herein we report a study on the effects of electrochemical factors on the host-guest interaction in a α-HL nanopore. The alkynyl-modified DNA is covalently bonded to the cucurbit[6] (CB[6])-modified azide adamantane (AA) under the catalysis of copper ions to form a DNA-AA@CB[6] molecular structure. A characteristic signature event with oscillations on the resistive pulse recording is generated due to the hostguest interaction when the complex translocates the α-HL nanopore. Effects of different chemical environments, including counterions, pH, and concentration of electrolytes on the translocation behavior of this structure were systematically investigated.
2. Methods
2.1. Materials
The DNA probe 5′-CCCCCCCCCCT*CCCCCCCCCC-3′ (T*—alkyne-modified thymine) was customized by Sangon Biotechnology Co. Ltd. (Shanghai). Micro Bio-Spin P6 gel columns (Tris buffer) were purchased from Bio-Rad (Hercules, CA). Cu(NO3)2 (99%), KCl (99.99%), NaCl (99.99%), LiCl (99.99%), TRIS hydrochloride (Tris·HCl), Azide Adamantane (97%), 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, ≥99.5%), (Ethylenedinitrilo) tetraacetic acid (EDTA, ≥99%), Ascorbic acid (≥99%), Acetonitrile (99.8%), Cucurbit[6]uril hydrate, α-HL from Staphylococcus aureus (lyophilized powder, Protein ~60% by Lowry, ≥10,000 units/mg protein) were purchased from Sigma-Alrich. All working solutions were prepared using deionized water from a Milli-Q water purification system (resistivity 18.2 MΩ/cm, 25 °C, Millipore Corporation) and were filtered through 0.02 μm filters before use.
2.2. Buffer preparation
KCl (22.37 g) and Tris·HCl (121 mg) were first dissolved in 80 mL deionized water. NaOH (2 M) and HCl (2 M) were used to adjust the pH to 8.0. The solution was then diluted with deionized water to 100 mL to afford the final buffer solution (3 M KCl and 10 mM Tris at pH 8.0). Other electrolyte solutions with various concentrations, pH and ions were prepared using similar methods.
2.3. Preparation of DNA probes
Alkyne-containing DNA (6 μL), deionized water (2 μL), Azide Adamantane (4 μL, AA, dissolved in acetonitrile, 200 mM), Ascorbic acid (3 μL, 20 mM), and copper nitrate (2.0 μL, 40 mM) were added to 4 μL HEPES (100 mM) buffer, with a final volume of 21 μL. The reaction was incubated on a shaker for 4 h at room temperature, followed by the addition of 4 μL EDTA solution (100 mM) to terminate the reaction. The DNA-AA product was purified with Micro Bio-spin P6 columns. Next, 10 μL CB[6] aqueous solution (5 mM) was added to the DNA solution and incubated for 8 h to afford the final DNA-AA@CB[6] probes.
2.4. Single-channel current recording
All resistive pulse recordings were performed on the Planar Lipid Bilayer workstation (Warner Instruments) at room temperature (~23 °C). Fabrication of α-HL nanopore devices follows traditional method previously reported. Briefly, an orifice (200 μm in diameter) punctured on a 25 μm thick Delrin wall that separates the cis (grounded) and the trans chambers of the flow cell was precoated with 1:10 hexadecane/pentane (Sigma-Aldrich). Then both chambers were filled with 1 mL of 3 M KCl solution buffered in 10 mM Tris-HCl (pH 8). To form a lipid bilayer membrane in the orifice, 20 μL (10 mg/mL) 1,2 diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) dissolved in pentane (Sigma-Aldrich) was added to the cis side of chambers to allow self-assembly. Following this, electrical potential was applied to the trans side using Ag/AgCl electrodes and slowly ramped up to examine the stability of the membrane at ±200 mV. To insert a single nanopore channel into the lipid bilayer, trans voltage was changed to 100 mV and a small amount (~2 μg) of α-HL protein (Sigma-Aldrich) were added from a monomeric stock solution made in 3 M KCl to the cis compartment. After a stable α-HL protein nanopore was inserted and confirmed by an open pore current, an analyte was added to the cis chamber at a bulk concentration of 0.5 μM from a 25 μM stock solution made in deionized water.
2.5. Data recording and analysis
Ionic current recordings were collected using a patch clamp amplifier (Warner Instruments) at a holding potential of +200 mV. After sample addition to the cis chamber, magnetic stirring was used to disperse the sample before characteristic signal was recorded. Each sample was measured in three replicates with a 30 min total duration. A fresh α-HL protein nanopore was used for each replicate. The raw data was analyzed using an in house Matlab based algorithm to find the current blockade and the dwell time of each eligible event, which are two commonly used properties for discriminating different molecules when they translocate nanopores. The current blockade (i.e. residual current) that represents the capture of single molecules and their translocation through the nanopore is defined as I/I0 (I: average current measured with the molecules inside the pore, I0: the average baseline value in absence of analytes). Dwell time (i.e. duration) represents the effective interaction time between the nanopore and a single molecule analyst, which was defined as the time difference between the start (when baseline drop to 1/2 blockade) and the end (when baseline return to 1/2 blockade) of the event. Results processed by the Matlab algorithm were confirmed by manual inspection. To profile each analyte, current blockade was plotted against dwell time using Python. The python modules used for scatter plots and contour plots were Matplotlib and Seaborn’s bivariate kernel density estimator.
3. Results and discussion
3.1. Oscillating signature induced by the host-guest interaction
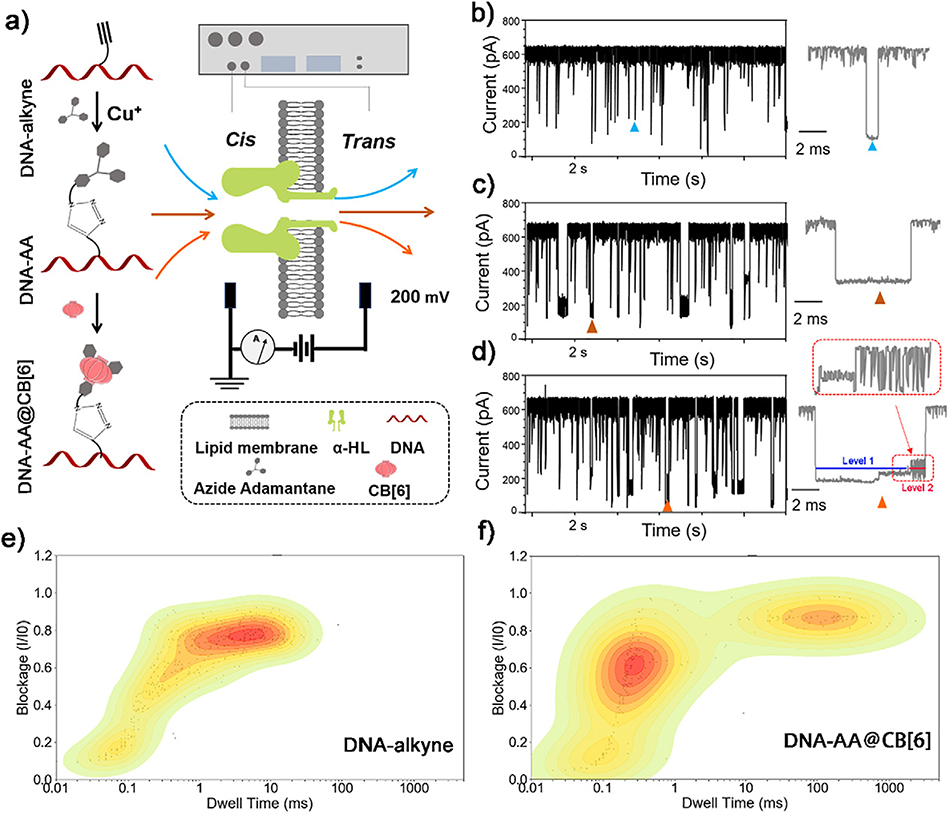
The DNA probe which will be used to generate characteristic oscillation events when translocating through α-HL was first constructed according to the strategy shown in Fig. 1a. We chose a commercially available alkyne containing thymine (T*) as the key modified base to construct DNA probes. The T* base was normally placed in the middle of the probe strand, and the rest of the sequence of the probe consists of cytosines (C). The terminal alkyne group on the side chain of T* enables “click” chemistry under the catalysis of Cu+ to attach a “guest” moiety (AA) which will then catch a “host” molecule (CB[6]) in solution to form the host-guest complex. The final DNA probe has a molecular structure of DNA-AA@CB[6].
Fig. 1.
(a) Schematic illustration of the construction of DNA probes through “click” chemistry and single-channel resistive pulse recordings of different samples through a α-HL nanopore. Legend is shown in the dashed box. Representative current traces of the translocation of DNA-alkyne (b), DNA-AA (c) and DNA-AA@CB[6] (d), respectively through α-HL nanopore. The corresponding contour plots depicting the blockade (I/I0) vs. dwell time distribution at the same conditions for DNA-alkyne (e) and DNA-AA@CB[6] (f), respectively. Data was acquired in 3 M KCl, 10 mM Tris buffer, pH 8.0, with the transmembrane potential held at +200 mV.
We first compared the changes of translocation signals before and after DNA modifications by testing DNA-alkyne, DNA-AA and DNA-AA@CB[6] using nanopore, respectively. As shown in Fig. 1b–d, although these three samples exhibited similar blockade (I/I0) (70.6%, 71.44% and 75.6%, respectively) and the event frequency, their dwell times and characteristics signals changed significantly. Comparing with the mean dwell time of 6.7 ms for signals of DNA-alkyne, the dwell time of characteristic signals for DNA-AA and DNA-AA@CB[6] are both above 10 ms, indicating reduced translocation speed of DNA by the conjugation with AA, which is beneficial for obtaining more information of these molecules. After the DNA-AA was incubated with CB[6], the noncovalent hybrid DNA-AA@CB[6] shows an obvious oscillating pattern at the end of the characteristic signal, dividing the event into two consecutive parts, with Level 1 featuring long and deep blockades and Level 2 featuring transient current oscillations caused by the host-guest interaction between CB[6] and AA10.
Although the oscillating pattern in the signal events of hybrid DNA-AA@CB[6] is clearly different from the translocation signal of any known biomolecules tested with nanopores, blocking or binding to the α-HL by some large molecules (such as DNA-alkyne in this study) may also induce similar large current blockade that may interfere with the detection. In order to distinguish the true oscillation signals from these interferents, a contour plot was generated for each of DNA (Fig. 1e) and DNA-AA@CB[6] (Fig. 1f) from current blockade and dwell time values to profile their translocation behavior. By comparison, we can associate an event with a blocking time >10 ms to a modified DNA structure (DNA-AA or DNA-AA@CB[6]), which helps accurately calculate the event frequency of assigned samples.
3.2. Effect of α-HL nanopore orientation on the oscillating signature
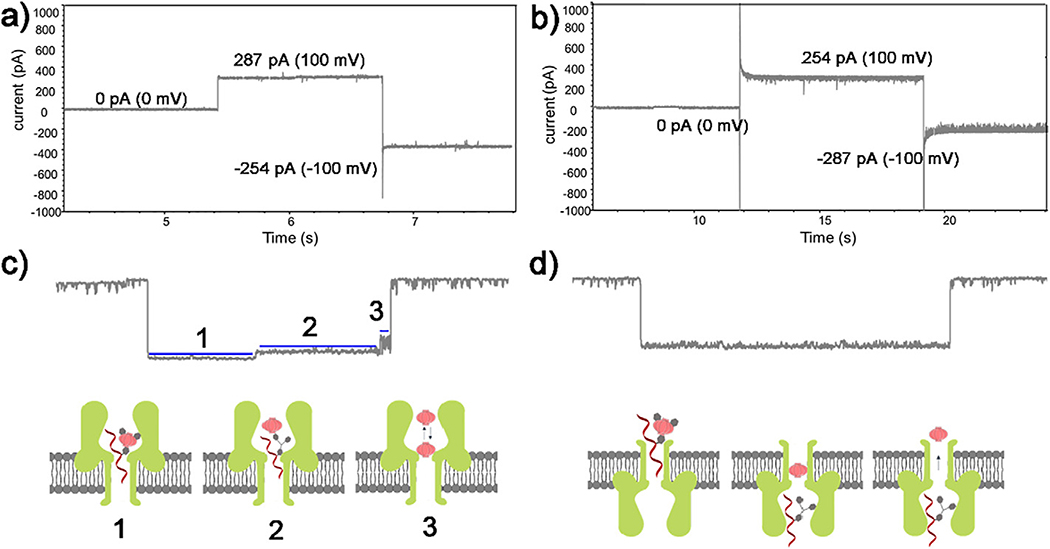
The mechanism for the oscillating current induced by hybrid DNA-AA@CB[6] translocation appears to be the trapping and oscillation of host CB[6] inside the vestibule of α-HL, which has an asymmetrical structure (Fig. 1a). In this method, negatively charged DNA-based probes should be added to the cis side and driven through the α-HL nanopore by a positive trans voltage, thus the direction of the α-HL in the lipid membrane should affect the signature events of DNA-AA@CB [6].
Fig. 2a and b show two different current traces with both positive and negative trans voltages after a nanopore was inserted into the lipid membrane with different direction. The nanopore in Fig. 2a exhibited larger ionic current under a positive trans voltage than it is under a negative voltage, while the adverse situation is shown in Fig. 2b. The test of the DNA-AA@CB[6] with these two setups further confirmed the direction of inserted α-HL protein: for the pore with larger current value under positive trans voltage, the oscillation signature events of CB[6] can be observed as shown in Fig. 2c with three characteristic stages: 1) trapping of DNA-AA@CB[6] in the pore, 2) dissociation of the DNA-AA@CB[6] and translocation of the DNA-AA through the pore, 3) oscillation of CB[6] in the cis vestibule of α-HL. However, no oscillating signal was found in the other setup with reversed pore direction (Fig. 2b and d), even with extended recording time and increased concentration of DNA-AA@CB[6]. Instead, a longer dwell time was observed, indicating that the CB[6] cannot enter the vestibule from the opposite direction. These results confirms the previously proposed10 molecular mechanism of the host-guest interaction in α-HL, and also provide a way to characterize the direction of the nanopore.
Fig. 2.
Change of current value with changing voltage bias (100 mV) direction after a α-HL nanopore inserted into the lipid membrane with the entrance towards the cis (a) and the trans (b) compartment, respectively. Corresponding signature events at each α-HL orientation when DNA-AA@CB[6] was added to the cis side (c, d). Inserts depict the step-by-step molecular process of DNA hybrid trapping, dissociation, and translocation, as well as CB[6] oscillation in the vestibule. Data was acquired in 3 M KCl, 10 mM Tris buffer, pH 8.0, with the transmembrane potential held at +200 mV.
3.3. Effect of electrolyte pH on the oscillation signature
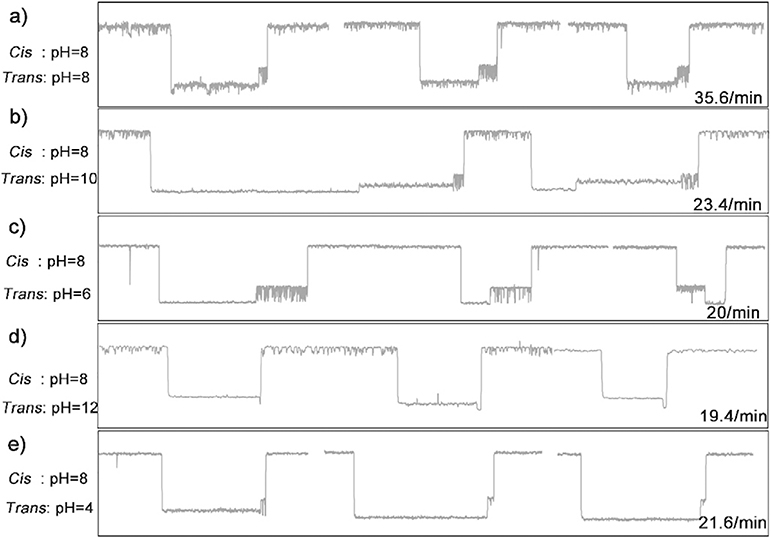
The electrophoretic force acting on DNA backbone of the hybrid DNA-AA@CB[6] molecule is the main driving force to pull the host-guest complex into the vestibule of nanopore, where it can be captured by the α-HL. Thus, the translocation behavior of the DNA under different electrochemical environments may influence the interaction between host and guest. We further studied the effect of different pH of the KCl electrolytes on the oscillating signal. As shown in Fig. 3, as DNA-AA@ CB[6] was added to the cis compartment with pH of 8, different oscillating signals can be observed by changing the pH of trans compartment. First, highly characteristic current events at a frequency of 35.6 events/min can be observed with the pH of trans compartment at 8 (Fig. 3a). The oscillating events frequency was significantly reduced to around 20 events/min when the pH of trans compartment was increased or decreased (Fig. 3b–e). Additionally, in comparison to the setup with equal cis and trans pH, increasing the trans pH to 10 appears to decelerate the dissociation of the DNA-AA@CB[6] and translocation of the DNA-AA through the pore (Fig. 3b), while decreasing the trans pH to 6 can extend the oscillation time of CB[6] (Fig. 3c). However, further increasing (Fig. 3d) or decreasing (Fig. 3e) the trans pH leads to reduced duration of the oscillation or even disappearing of the oscillating signal. Although more investigation on the mechanism of this observation is needed, the above results provide a viable pH range (6–10) for studying oscillating signals based on host-guest interactions.
Fig. 3.
Signature events generated during the translocation of DNA-AA@CB[6] through α-HL with various pH in cis/trans chambers. Data was acquired in 3 M KCl and 10 mM Tris buffer with the transmembrane potential held at +200 mV. The concentration of hybrid DNA is 0.5 μM.
3.4. Effect of electrolyte concentration on the oscillation signature
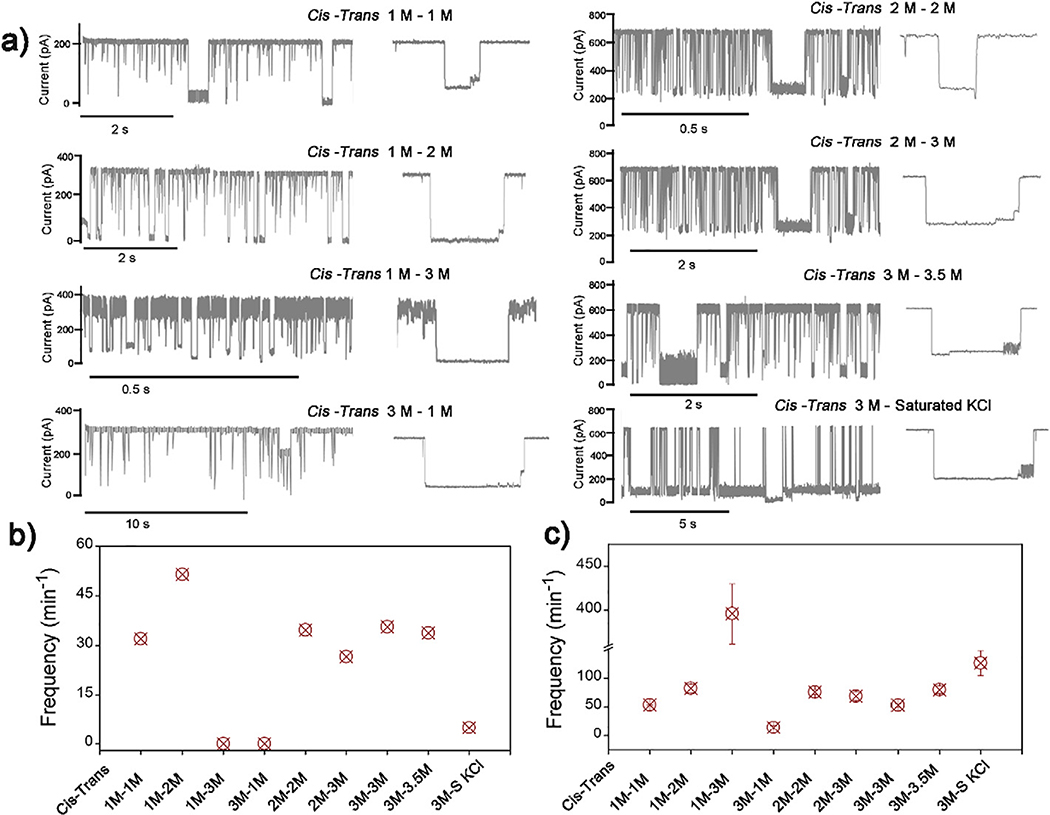
Next we studied the effects of electrolyte (KCl) concentrations on the oscillating signal of DNA-AA@CB[6] with identical cis and trans pH (8/8). Fig. 4 a shows the continuous ionic current traces with DNA-AA@CB[6] translocation across the nanopore with different combinations of KCl concentrations. With the cis electrolyte concentration fixed at 1 M, translocation event frequency of modified DNA including DNA-AA@CB[6] and DNA-AA (dwell time ≥ 10 ms) increased while the trans electrolyte concentration was raised from 1 M to 3 M, (Fig. 4c). Similar trends can also be found in the other two concentration gradients: fixed cis concentration at 2 M and 3 M, with varying trans concentration from 2 M to 3 M, and from 3 M to saturated solution, respectively. Results indicate that increasing the electrolyte concentration in the trans side can accelerate DNA translocation through the nanopore, which can be attributed to the enhancement of ionic current with asymmetric electrolyte concentration25. Whereas decreasing the trans electrolyte concentration prohibits the translocation, as shown by the lowest events frequency of DNA-AA@CB[6] and DNA-AA in the test with cis/trans concertation of 3 M /1 M with (Fig. 4c).
Fig. 4.
(a) Signature events generated during the translocation of DNA-AA@CB[6] through α-HL with various concentration of KCl in cis/trans compartments. (b) Oscillation events frequency and (c) translocation events frequency with dwell time >10 ms in each sample with different electrolyte combination. Data was acquired in 10 mM Tris buffer, pH 8.0, with the transmembrane potential held at +200 mV. The concentration of hybrid DNA is 0.5 μM.
However, the frequency of the oscillating signal is not directly proportional to the translocation event frequency of the modified DNA. As shown in Fig. 4b, frequencies of oscillating signal events observed in 3 M/S (cis/trans, S represents a saturated solution of KCl) and 1 M/3 M (cis/trans) concentration combinations are much lower than the total translocation event frequencies in these experiments. The highest oscillating event frequency (51.4 events/min) was obtained with the 1 M/2 M (cis/trans) setup. This optimized condition can be used for the future development of biosensing assay to achieve lower limit of detection.
3.5. Effect of electrolyte counterions on the oscillation signature
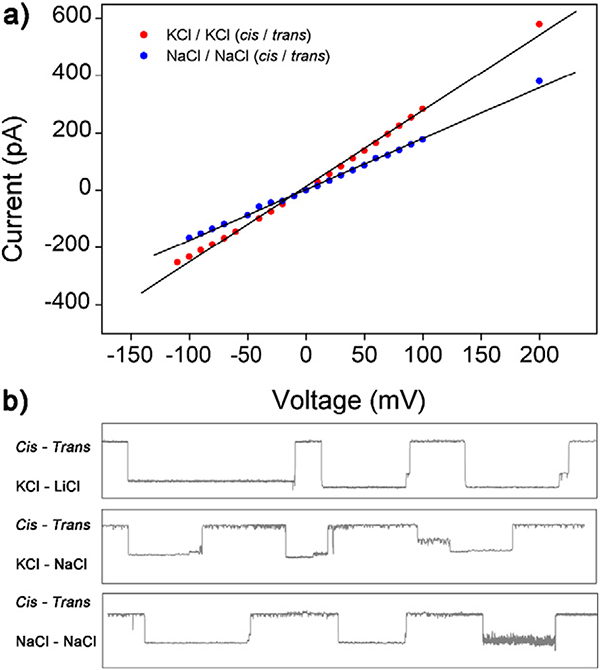
The effects of different counterions (K+, Na+, and Li+) on the signature events of DNA-AA@CB[6] through the nanopore were investigated as counterions can profoundly affect physical properties of DNA.24 First, the stability of nanopore in different electrolyte solution with the same ions at the cis and trans sides was demonstrated. As presented in Fig. 5a, in KCl and NaCl sensing system, the α-HL nanopore showed high stability in a wide voltage range from −200 mV to +200 mV, meanwhile, the absolute current value in KCl system is higher than the value in NaCl system at the same voltage. Unfortunately, we were not able to obtain a stable nanopore when the cis/trans sides are both filled with LiCl solution, which may be attributed to the weak metality and nuclear charge of Li ions. Due to the decreasing number of nuclear charges of Li+ and Na+, characteristics of the oscillating signal were also weakened comparing to the KCl / KCl(cis/trans)system. As shown in Fig. 5b, with the cis electrolyte solution fixed to KCl and the trans electrolyte varying from LiCl to NaCl, more obvious oscillating signal events of DNA-AA@ CB[6] were observed. No oscillating events were detected in the case of NaCl/NaCl (cis/trans) system. These results indicate that the nuclear charge number of counterions has a decisive influence on the detection of oscillating signals originated from the host-guest interaction in α-HL nanopore.
Fig. 5.
(a) I-V curves of a single open α-HL nanopore in NaCl and KCl. Data was obtained in 3 M M+Cl− (M+ = Na+, K+), 10 mM Tris buffer, pH 8.0 without any analytes in both sides of the nanopore. (b) Signature events generated during the translocation of DNA-AA@CB [6] through α-HL with various counterions (Li+, Na+, K+) in cis/trans chambers. Data was acquired in 10 mM Tris buffer, pH 8.0, with the transmembrane potential held at +200 mV. The concentration of hybrid DNA is 0.5 μM.
4. Conclusions
In summary, the effects of electrochemical environments on host-guest interactions, especially characteristic signals in α-HL nanopore by varying counterions (Li+, Na+, K+), pH, and concentration of electrolytes were systematically studied. The results demonstrated that the change of pH shortens or prolongs the duration of the oscillating signal events. The asymmetric concentration affects the frequency of the signature event, and the nuclear charge number of the counterions affect the generation of the oscillating signal and even the insertion of the nanopore. We thus believed that the findings herein will benefit the further study of the host-guest interaction induced current oscillations in the nanopore technology.
Acknowledgment
C. Liu acknowledges supports from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number K22AI136686, and the South Carolina IDeA Networks of Biomedical Research Excellence Developmental Research Project funded by the National Institute of General Medical Sciences of the National Institutes of Health.
Biography
 Xiaojun Wei received his Ph.D. (2018) in Physical Chemistry at the Institute of Chemistry, Chinese Academy of Sciences. He is currently working as a Postdoctoral Fellow at the College of Engineering and Computing, University of South Carolina. His research focus is the application of nanopore for biomarker detection.
Xiaojun Wei received his Ph.D. (2018) in Physical Chemistry at the Institute of Chemistry, Chinese Academy of Sciences. He is currently working as a Postdoctoral Fellow at the College of Engineering and Computing, University of South Carolina. His research focus is the application of nanopore for biomarker detection.
 Chang Liu is an Assistant Professor of Biomedical Engineering and Chemical Engineering at the University of South Carolina. Prior to this appointment, he served as an Assistant Research Scientist at the Biodesign Institute of Arizona State University. Dr. Liu completed his Postdoctoral Fellowship in the Department of Nanomedicine at the Research Institute of Houston Methodist Hospital. He received his Ph.D. (2013) in Biomedical Engineering from Florida International University. Dr. Liu’s research focuses on applying nanotechnology on the development of biosensors for disease diagnosis.
Chang Liu is an Assistant Professor of Biomedical Engineering and Chemical Engineering at the University of South Carolina. Prior to this appointment, he served as an Assistant Research Scientist at the Biodesign Institute of Arizona State University. Dr. Liu completed his Postdoctoral Fellowship in the Department of Nanomedicine at the Research Institute of Houston Methodist Hospital. He received his Ph.D. (2013) in Biomedical Engineering from Florida International University. Dr. Liu’s research focuses on applying nanotechnology on the development of biosensors for disease diagnosis.
Footnotes
The authors declare no competing financial interest.
References
- 1.Lee K, Park KB, Kim HJ, et al. Recent progress in solid-state nanopores. Adv Mater 2018;30(42). 10.1002/adma.201704680. [DOI] [PubMed] [Google Scholar]
- 2.Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton JM, Pelta J. Sensing proteins through nanopores: fundamental to applications. ACS Chem Biol 2012;7(12):1935–49. 10.1021/cb300449t. [DOI] [PubMed] [Google Scholar]
- 3.Robertson JWF, Reiner JE. The utility of nanopore technology for protein and peptide sensing. PROTEOMICS 2018;18(18):1800026. 10.1002/pmic.201800026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K, Park K-B, Kim H- J, et al. Recent progress in solid-state nanopores. Adv Mater 2018;30(42):1704680. 10.1002/adma.201704680. [DOI] [PubMed] [Google Scholar]
- 5.Wilson J, Sloman L, He ZR, Aksimentiev A. Graphene nanopores for protein sequencing. Adv Funct Mater 2016;26(27):4830–8. 10.1002/adfm.201601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM. Nanoscopic porous sensors. Annu Rev Anal Chem 2008;1:737–66. 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 7.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev 2009;38(8):2360–84. 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 8.Wanunu M Nanopores: a journey towards DNA sequencing. Phys Life Rev 2012;9 (2):125–58. 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JH, Liu XL, Hu ZL, Ying YL, Long YT. Intelligent identification of multi-level nanopore signatures for accurate detection of cancer biomarkers. Chem Commun 2017;53(73):10176–9. 10.1039/c7cc04745b. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Wu HC. DNA-based nanopore sensing. Angewandte Chemie-International Edition 2016;55(49):15216–22. 10.1002/anie.201604405. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y, Ying YL, Gao R, Long YT. Single-molecule sensing with nanopore confinement: from chemical reactions to biological interactions. Chem Eur J 2018;24(50): 13064–71. 10.1002/chem.201800669. [DOI] [PubMed] [Google Scholar]
- 12.Zhang ZH, Li T, Sheng YY, Liu L, Wu HC. Enhanced sensitivity in nanopore sensing of cancer biomarkers in human blood via click chemistry. Small 2019; 15(2), 10.1002/smll.201804078. [DOI] [PubMed] [Google Scholar]
- 13.Liotta LA, Ferrari M, Petricoin E. Written in blood. Nature 2003;425(6961):905. 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 14.Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst 2010;135(10): 2496–511. 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asandei A, Apetrei A, Luchian T. Uni-molecular detection and quantification of selected beta-lactam antibiotics with a hybrid alpha-hemolysin protein pore. J Mol Recognit 2011;24(2):199–207. 10.1002/jmr.1038. [DOI] [PubMed] [Google Scholar]
- 16.Braha O, Webb J, Gu LQ, Kim K, Bayley H. Carriers versus adapters in stochastic sensing. Chemphyschem 2005;6(5):889–92. 10.1002/cphc.200400595. [DOI] [PubMed] [Google Scholar]
- 17.Zeng T, Liu L, Li T, Li YR, Gao J, Zhao YL. Wu HC. Detection of 5-methylcytosine and 5-hydroxymethylcytosine in DNA via host-guest interactions inside alpha-hemolysin nanopores. Chem Sci 2015;6(10):5628–34. 10.1039/c5sc01436k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 1999;398(6729):686–90. [DOI] [PubMed] [Google Scholar]
- 19.Gu LQ, Bayley H. Interaction of the noncovalent molecular adapter beta-cyclodextrin with the staphylococcal alpha-hemolysin pore. Biophys J 2000;79(4):1967–75. 10.1016/s0006-3495(00)76445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5 ‘-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc 2006;128(5):1705–10. 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 21.Kang XF, Cheley S, Guan XY, Bayley H. Stochastic detection of enantiomers. J Am Chem Soc 2006;128(33):10684–5. 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Liu L, Li YR, Xie JN, Wu HCA. Universal strategy for Aptamer-based Nanopore sensing through host-guest interactions inside -Hemolysin. Angewandte Chemie-International Edition 2015;54(26):7568–71. 10.1002/anie.201502047. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Li T, Zhang SW, et al. Simultaneous quantification of multiple Cancer biomarkers in blood samples through DNA-assisted Nanopore sensing. Angewandte Chemie-International Edition 2018;57(37):11882–7. 10.1002/anie.201803324. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA translocation through a Nanopore in Lithium chloride. Nano Lett 2012;12(2):1038–44. 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Wu GS, Si W, et al. Ionic current modulation from DNA translocation through nanopores under high ionic strength and concentration gradients. Nanoscale 2017;9(2):930–9. 10.1039/c6nr08123a. [DOI] [PubMed] [Google Scholar]
- 26.Bell NAW, Chen K, Ghosal S, Ricci M, Keyser UF. Asymmetric dynamics of DNA entering and exiting a strongly confining nanopore. Nat Commun 2017;8. 10.1038/s41467-017-00423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Tian K, Du X, Shi RC. Gu LQ. Remote activation of a Nanopore for high-performance genetic detection using a pH taxis-mimicking mechanism. Anal Chem 2017;89(24):13039–43. 10.1021/acs.analchem.7b03979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu ZL, Li MY, Liu SC, Ying YL, Long YT. A lithium-ion-active aerolysin nanopore for effectively trapping long single-stranded DNA. Chem Sci 2019;10(2):354–8. 10.1039/c8sc03927e. [DOI] [PMC free article] [PubMed] [Google Scholar]