Abstract
Genomics research has relied principally on the establishment and curation of a reference genome for the species. However, it is increasingly recognized that a single reference genome cannot fully describe the extent of genetic variation within many widely distributed species. Pangenome representations are based on high-quality genome assemblies of multiple individuals and intended to represent the broadest possible diversity within a species. A Bovine Pangenome Consortium (BPC) has recently been established to begin assembling genomes from more than 600 recognized breeds of cattle, together with other related species to provide information on ancestral alleles and haplotypes. Previously reported de novo genome assemblies for Angus, Brahman, Hereford, and Highland breeds of cattle are part of the initial BPC effort. The present report describes a complete single haplotype assembly at chromosome-scale for a fullblood Simmental cow from an F1 bison–cattle hybrid fetus by trio binning. Simmental cattle, also known as Fleckvieh due to their red and white spots, originated in central Europe in the 1830s as a triple-purpose breed selected for draught, meat, and dairy production. There are over 50 million Simmental cattle in the world, known today for their fast growth and beef yields. This assembly (ARS_Simm1.0) is similar in length to the other bovine assemblies at 2.86 Gb, with a scaffold N50 of 102 Mb (max scaffold 156.8 Mb) and meets or exceeds the continuity of the best Bos taurus reference assemblies to date.
Keywords: Fleckvieh, bison, F1 hybrid, pangenome, nanopore, trio binning
The current cattle reference genome, ARS-UCD1.2 was assembled from DNA sequence from a single inbred Hereford breed cow, L1 Dominette 01449 (Rosen et al. 2020). The intentional inbreeding of the “Line 1” Hereford cattle reduced heterozygosity between parental alleles, which was advantageous for genome assembly methods available at the time she was originally selected to be the reference (Bovine Genome Sequencing and Analysis Consortium et al. 2009). In addition, Hereford is one of the most common breeds in use for beef production globally. The original, Sanger sequence-based assembly of “Dominette” has been used to great effect for genome research including identification of millions of single nucleotide variants and development of highly parallel genotyping platforms (Bickhart et al. 2020).
The genomics research community has increasingly recognized that a single reference genome cannot fully describe the extent of genetic variation within a species, even in a relatively low-diversity species such as humans. This conclusion stems from the observation that as many as 10–15% of non-repetitive, high-quality sequence reads from an individual animal fail to map to the reference genome. This mapping inconsistency is true in humans as well as cattle and is the basis of the recently announced Human Pangenome Reference Sequence Project (HPRSP). The HPRSP aims to generate high-quality assemblies that reflect the diversity of human populations and has already identified hundreds of megabases of population-specific sequence that are not found in the human reference genome (Duan et al. 2019; Sherman et al. 2019). Concurrent pangenome efforts in plants have had great success, with more than 12 000 novel genes discovered in rice that were not observed in the reference genome and similar results in apple species (Sun et al. 2017, 2020). Even the recent long read-based update of the Hereford reference does not completely describe all genome segments present in the >600 cattle breeds in existence around the world, providing motivation for exploring methods to enhance the reference assembly to support genome research in identifying variation contributing to domestication, productivity, and health in cattle populations around the world. The reduced cost and improved ease of creating genome assemblies makes it now feasible and advantageous to transition toward a pangenome.
Trio binning genome assembly of offspring from interspecies crosses has recently produced some of the highest quality vertebrate genomes to date, including those from a domestic yak cow (Bos grunniens) and a Scottish Highland bull (Bos taurus; Rice et al. 2020). The power of this method is derived from heterozygosity of the F1 hybrid offspring, which has two haploid genomes separated by 5 million years since their most recent common ancestor (Kumar et al. 2017). The process uses parent-specific sequence alignments to sort the hybrid offspring’s long reads by parental haplotype (Koren et al. 2018). The resolution of sorting parent-of-origin reads is dependent on the heterozygosity of the parents, with higher heterozygous crosses requiring shorter read lengths to span the required variant sites. Phased assemblies can then be produced for each parent separately, resulting in two distinct species-specific reference genomes from a single hybrid individual and accelerating efforts of projects such as the Genome 10k and the cattle pangenome.
We present a fully phased reference genome from a North American Fleckvieh-type Simmental cow (B. taurus taurus), obtained through trio binning of long nanopore-derived sequence reads from an F1 cross with a Plains bison bull (Bison bison bison). Like yak and cattle, the most recent common ancestor between bison and cattle was about 5 million years ago, enhancing the sorting of sequence reads into bins of parental haplotype. Our report is presented in tandem with the accompanying bison reference genome article (Oppenheimer et al. submitted). The Fleckvieh Simmental genome sequence presented here (ARS_Simm1.0) is chromosome-scale, highly complete, and as contiguous as the best livestock and model organism reference genomes available.
Methods
Ethics Statement
All cattle protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nebraska–Lincoln, an AAALAC International Accredited institution (IACUC Project ID 1697). Bison semen collections were approved by the IACUC at Colorado State University, IACUC protocol 17-7117A.
Animals, In Vitro Fertilization, Tissue Collection, and DNA Extraction
Semen from a Yellowstone bison bull (tag number 709, SAMN16823422) was collected 23 May 2013, dispensed into 0.5 mL straws and stored in liquid nitrogen for future use in artificial fertilization. A 4-year-old fullblood Simmental female (BHR Lady Sieg C235E, American Simmental Association registration number 3182916, SAMN16825967; Figure 1) was selected as the donor based on her being a fullblood representative of the breed and availability as a donor. She was evaluated for physical condition, reproductive health, and superovulation capacity with ultrasonography. Based on this evaluation a follicle-stimulating hormone (FSH) dosage was customized to allow for appropriate ovarian stimulation and superovulation. Five ova from the donor female were aspirated on 16 January 2019 and fertilized in vitro a day later with semen from Yellowstone bison 709. The same day, five Simmental heifers were selected as embryo recipients and underwent estrus synchronization using a combination of gonadotropin-releasing hormone, prostaglandin F2 alpha, and progesterone so that embryonic age was matched to estrus cycle of the recipients. Five embryos were implanted on 24 January 2019 and recipients were observed daily for repeat estrus cycling. Recipients were examined with ultrasonography at 28, 54, 75, and 105 days posttransplantation and controlled intravaginal drug release (CIDR) devices containing progesterone were replaced in the pregnant recipients at each event to help maintain pregnancies. Three pregnancies were confirmed at 22 and 54 days, two at 75 days, and one at 105 days post-transplantation. On 23 May 2019, the male F1 fetus was collected by cesarean at 119 days posttransplantation. Lung tissue was flash frozen in liquid nitrogen and stored at −80°C until DNA isolation and sequencing. Approximately 50 mg of frozen lung was crushed to powder in a cryopulverizer (Covaris Inc., Woburn, MA), followed by DNA extraction as described by Logsdon (2019). For chromatin conformation contact (Hi-C) analysis, approximately 50 mg of fetal lung tissue was cross-linked and processed using ProximoHi-Cv1.5 kit (Phase Genomics, Seattle, WA) as recommended in the protocol accompanying the kit.
Figure 1.
BHR LADY SIEG C235E (Reg 3182916) was selected as an available, fullblood, Fleckvieh-type donor female to represent Simmental.
Sequencing and Assembly
Template for long read sequencing on the PromethION instrument was prepared using the Ligation Sequencing Kit LSK-109 (Oxford Nanopore Technologies, Oxford, UK). Briefly, 10 μg of DNA was sheared by passing five times through a 26 gauge needle and concentrated by addition of 1 volume of AMPureXP bead solution. After collecting the beads by magnet, the beads were washed 2 times with 0.5 mL 80% ethanol and dried for 2 min. The beads were suspended in 51 μL EB and DNA eluted at 37°C for 5 min. The sheared, eluted DNA was then processed by ligation to AMX adapter as recommended by the manufacturer’s instructions in the LSK-109 product manual and sequenced in 16 flow cells with seven separate template preparations on a PromethION instrument using R9.4.1 flow cells. DNA template for ultra-long nanopore sequencing was prepared from the same DNA sample using a Ligation Sequencing Kit LSK109 (Oxford Nanopore, Oxford, UK) similar to the approach taken for PromethION sequencing. Notable deviations in this protocol include the use of a GridION ×5 sequencer and the use of a different protocol for DNA handling and cleanup (https://community.nanoporetech.com/posts/rocky-mountain-adventures). Briefly, sheared DNA was transferred to a tube with wide-bore pipette tips and mixed with a PEG/NaCl precipitation buffer (9% PEG8000, 1M NaCl and 10 mM Tris-Cl ph 8.0). The mixture was incubated at room temperature for 30 min and was centrifuged at 13.5 k RPM for 30 min. The supernatant was subjected to two, 200 μL, 70% ethanol precipitations with centrifugation, and the pellet was eluted in 41 μL of 10 mM Tris-Cl pH 8.0. All other methods in the LSK109 library preparation protocol were followed, apart from an increase in the room temperature incubation of the ligation reaction to 60 min at room temperature. Libraries were sequenced with 22 Min106 R9.4.1 flowcells on a GridION X5 sequencer and sequence read statistics were estimated using custom python scripts (https://github.com/njdbickhart/python_toolchain/blob/master/sequenceData/fastqStats.py). Fastq files were generated from fast5 raw data using the Guppy v3.1.5 Basecaller (available from Oxford Nanopore Technologies via their community site, https://community.nanoporetech.com). Median read length and N50 was determined by NanoStat 1.1.2.
The same DNA sample used for long read sequencing generated a short read library using the TruSeq PCR-Free Kit as recommended by the manufacturer (Illumina Inc., San Diego, CA), which was sequenced on a NextSeq500 platform using 2 × 150 base paired end reads. DNA extracted from a semen straw of the sire, and from a blood sample of the dam, were also processed into short read libraries and sequenced in the same way. Hi-C libraries were also sequenced using 2 × 150 base reads, to improve potential for assigning read pairs to haplotype prior to scaffolding. Quality of Hi-C library in terms of predicted distance between read pairs and other measures was generated using the hic_qc python script (Phase Genomics; https://github.com/phasegenomics/hic_qc downloaded June 20, 2019). First, read pairs were mapped to the ARS-UCD1.2 Hereford reference assembly using bwa v0.7.17 (Li and Durbin 2009) as recommended in the hic_qc documentation. The output bam file was used as input to estimate distance between read pairs, proportion of expected same-strand read pairs, and percentage of informative read pairs.
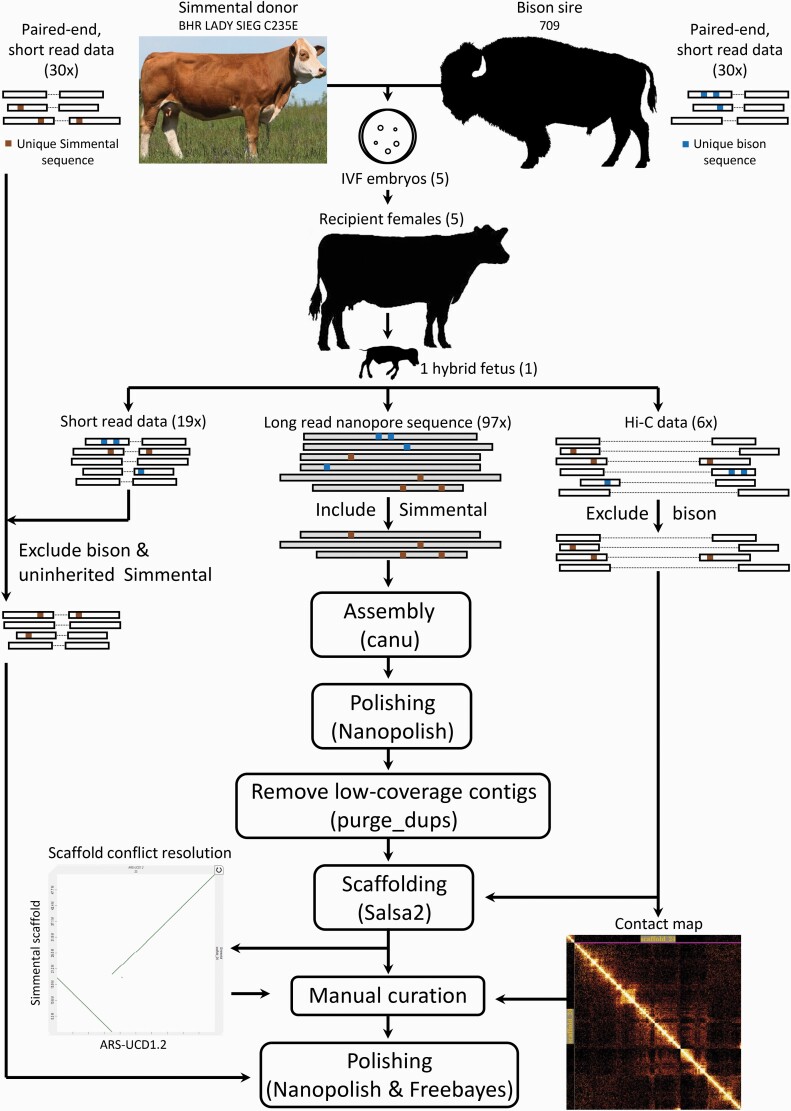
A comprehensive listing of software used in the assembly and analysis of this data is provided in Table 1 of the companion bison assembly manuscript Oppenheimer et al., in this issue and reproduced here as Supplemental Table S1. See Figure 2 for an overview of the assembly process. Prior to assembly, the parental Illumina data was cleaned using Trimmomatic v0.38 (Bolger et al. 2014) in paired end mode (LEADING:10 TRAILING:10 SLIDINGWINDOW:4:18 MINLEN:75) to trim low-quality sequence at the ends of the reads and discard reads below 75bp. Illumina sequence reads from the Sire and Dam were then decomposed into k-mers and filtered to identify 21-mers unique to each parent. Parental k-mers were used to separate long reads from the fetus into a pool of candidate “Simmental” origin reads (Koren et al. 2018). These reads were assembled into contigs. The trio binning, correction, and trimming stages of assembly were run with Canu v1.8. Unitigging was launched with Canu v1.9 to take advantage of a bug fix related to consensus generation. The command for launching the assembly was the same for both versions and used Canu’s recommended parameters for flip-flop basecalled reads sequenced on R9.4 flowcells and defined the parental haplotypes for read binning:
Table 1.
Genome assembly and annotations statistics for Simmental and Hereford genomes
| Statistic | Assembly | |
|---|---|---|
| ARS_Simm1.0 | ARS-UCD1.2 | |
| Breed | Simmental | Hereford |
| Assembly length (Gb) | 2.862 | 2.759 |
| No. of contigs | 1315 | 2212 |
| Contig N50 (Mb) | 70.8 | 25.9 |
| Contig L50 | 14 | 32 |
| Scaffold N50 (Mb) | 102.5 | 103.3 |
| Scaffold L50 | 12 | 12 |
| Number of gaps in chromosomes | 52 | 315 |
| BUSCO complete single copy genes | 90.8% | 90.6% |
| BUSCO complete duplicated genes | 1.1% | 1.1% |
| BUSCO fragmented genes | 2.3% | 2.3% |
| BUSCO missing genes | 5.8% | 6.0% |
| Simmental read-based kmer-estimated quality score | 36.5 | 30.0 |
| Hereford read-based kmer-estimated quality score | 28.4 | 32.0 |
| Simmental read-based kmer-estimated error rate | 0.0003 | 0.0013 |
| Hereford read-based kmer-estimated error rate | 0.0014 | 0.0006 |
| Simmental read-based kmer-estimated genome completeness | 95.6% | 93.8% |
| Hereford read-based kmer-estimated genome completeness | 93.0% | 95.4% |
Figure 2.
Schematic showing the Simmental trio binning and assembly process.
canu “corMhapOptions = --threshold 0.8 --ordered-sketch-size 1000 --ordered-kmer-size 14” saveOverlaps = true correctedErrorRate=0.105 “stageDirectory=&TMPDIR” -p BisonSimmental -d assembly genomesize = 2.7g -haplotypeBison_sire bison_sire.fastq -haplotypeSimmental_dam Simmental_dam.fq -nanopore-raw bison_simmental.ul.ont.fasta.gz
Assembly Polishing and Scaffolding
Assembled contigs were polished with Nanopolish v0.11.1 (Loman et al. 2015) which uses the raw signal data from nanopore reads to generate a more accurate consensus sequence. Purge_dups v1.0.1 (Guan et al. 2020), which uses long-read alignment read-depth and self-alignment to identify assembly artifacts, was then used to remove partially duplicated and low-coverage contigs, likely representing errors, to generate a final set of contigs. Prior to scaffolding, Hi-C reads from the male F1 fetus had to be classified into parental haplotypes. Due to the shorter read length of Illumina sequence, it is not possible to definitively assign Hi-C reads to either haplotype using unique parental k-mers. Instead, Hi-C reads containing unique Bison sire k-mers were excluded from the dataset with the following meryl command:
meryl-lookup -exclude -mers bison_sire.only.meryl -sequence hic_R1.fastq.gz -sequence2 hic_R2.fastq.gz -r2 Simmental_dam_hic-sire-excluded.R2.fastq.gz | pigz -c > Simmental_dam_hic-sire-excluded.R1.fastq.gz
Sire haplotype-excluded HiC reads were aligned to the polished contigs using bwa following the Arima mapping pipeline, which maps the ends of each paired read separately and trims chimeric reads (across ligation junctions) based on mapping orientation (https://github.com/ArimaGenomics/mapping_pipeline). Processed alignments were used as input to Salsa v2.2 (Ghurye et al. 2017) for scaffolding.
Scaffolds were assessed with the combination of Hi-C contact maps and alignments to existing cattle assemblies ARS-UCD1.2 and ARS_UNL_Btau-highland_paternal_1.0_alt. Hi-C data was realigned to the scaffolded assembly as above and processed with PretextMap v0.1 (PretextMap n.d.) and PretextView v0.01 (PretextView n.d.) to generate and visualize the Hi-C matrix and inspect the contigging and scaffolding results. Assembly to assembly alignments were produced with minimap2 using the parameters -cx asm5. PAF files were filtered to display only large alignment blocks (>~25 Mb) for easy viewing of large scale structural disagreements. Alignments were used to guide inspection of the Hi-C matrix and adjustments were made to the Salsa scaffolding when supported by the matrix. These corrections were made by breaking the assembly at existing scaffolding gaps and then properly joining and orienting contigs with the program CombineFasta (https://github.com/njdbickhart/CombineFasta).
Final polishing of the manually curated assembly proceeded with another round of Nanopolish and two rounds of polishing with short read Illumina data using Freebayes (version v1.3.1-1-g5eb71a3-dirty; Garrison and Marth 2012). Variants called for polishing with both methods were screened with Merfin (https://github.com/arangrhie/merfin, version downloaded October 20, 2020) which predicts the k-mer consequences of variant calls and validates supported variants. Only K-mers inherited from the Simmental dam haplotype were included for consideration. For Freebayes polishing, exclusion of k-mers not inherited from the dam with Merfin enabled us to combine the short read data from the dam and F1 hybrid, increasing the coverage of homozygous sites considered by Freebayes without risking haplotype conversion. The polished consensus was generated with bcftools 1.9 (Li et al. 2009) by selecting homozygous ALT and heterozygous non-REF variants that passed quality filtering [“QUAL>1 && (GT=‘AA’ || GT=‘Aa’)”] and choosing the longest variant at heterozygous non-REF sites.
Annotation
Liftoff 1.5.1 (Shumate and Salzberg 2020) was used to lift over the ARS-UCD1.2 genome annotation (GCF_002263795.1) to obtain a preliminary annotation for the ARS_Simm1.0 assembly prior to final annotation by the NCBI Eukaryotic Genome Annotation Pipeline. The -chrom option was used to apply the annotation chromosome-by-chromosome and -sc 0.95 was used to identify extra gene copies present in ARS_Simm1.0 with a minimum identity of 0.95. RepeatMasker v4.1.0 was used with the -e nbci and -species cow options to identify repetitive content present in the assembly.
Assembly Evaluation
Assembly completeness and accuracy were measured through k-mer content estimates and read mapping statistics of the Simmental parent’s short reads against the assembly sequence. Read mapping statistics were generated using Lumpy-sv 0.3.0 (Layer et al. 2014), FRC_align 1.0.0 (Vezzi 2012), and Freebayes v1.3.1-1-g5eb71a3-dirty (Garrison and Marth 2012) as previously described (Bickhart et al. 2017). Feature response curves including all of the errors discovered by the aforementioned tools were plotted in python (Supplementary Figure S1). K-mer completeness estimates and spectral plots were generated using the Merqury (version 1.0) pipeline (Rhie et al. 2020). Single copy gene completeness was estimated by BUSCO v4 analysis against the Mammalia_od10 database (9226 genes) using the standard lineage workflow (Seppey et al. 2019). Comparative assembly plots (Supplementary Figure S2) and variant histograms (Supplementary Figure S3) were generated from assembly-to-assembly minimap2 alignments. General assembly statistics were calculated using custom python and R scripts available in open-source github repositories (https://github.com/njdbickhart/Themis-ASM). A comprehensive list of assembly quality metrics can be found in Supplemental Table S2. Individual missing BUSCO genes were examined by searching the GenBank Gene database for bovine versions of transcripts reported as missing and examining the annotation provided for those genes in the ARS-UCD1.2 reference at NCBI.
Results
The 16 PromethION flow cells produced 487.5 Gb of sequence in 14.2 million reads according to the PromethION software (mean 23.7 kb), with 424.8 Gb present in the fastq_pass files produced by the basecaller. The estimated read length median varied between cells from 4.63 to 37.0 kb, with read length N50 ranging from 25.5 to 53.0 kb. Ultra-long read libraries were also created and sequenced on MinION flow cells. In total, 22 cells were sequenced and 3.7 Gb of reads larger than 100 kb in length were produced from 31.1 total Gb of sequence data (an 11% efficiency). Total genome coverage of pass filter reads going into the assembly, assuming the 2.7 Gb size of the Hereford reference, was 193.8× (approximately 97× coverage of each haplotype). The distribution of read lengths is shown in Supplemental Figure S4. The Simmental dam and the bison sire short read data included 980M reads (147 Gb; 54× coverage) and 675M reads (102 Gb; 38× coverage), respectively. Additionally, 346M Illumina reads (52 Gb; 19× coverage) were generated from the same F1 hybrid DNA.
The Canu assembler assigned 7 888 850 nanopore reads to the Simmental (dam) haplotype containing 238.9 Gb (46%) of the total sequence (only 1822 reads were not assigned to a parental haplotype, with an average length of 1459 bases). Average read length was 30.3 kb and N50 read length was 49.5 kb for the dam haplotype bin. Subsequent assembly steps used 1 954 154 corrected reads to assemble the Simmental haplotype, representing 108.5 Gb (approximately 40× coverage; the default of Canu). Initial contig formation generated 1803 contigs of total length 2.91 Gb with NG50 of 71.2 Mb and LG50 of 14. Purge_dups then removed 438 low-coverage contigs and trimmed 26 partially duplicated contigs reducing the contigs to 1365.
The Hi-C library used for scaffolding the contigs produced slightly more than 99 million read pairs of which 70% were high quality, with 11% of pairs mapping >10 kb apart and 7% of high quality read pairs mapping to separate contigs according to the hic_qc estimate. There were 8% duplicate reads and 22% of pairs had zero map distance. Salsa broke 9 contigs prior to scaffolding and joined the resulting 1374 contigs into 1345 scaffolds. Salsa appears to be highly conservative in the way it joins contigs as manual inspection of the Hi-C matrix clearly supported 30 additional joins and 7 inversions and rearrangements (data for generating the PretextView in Supplemental File S1, manual edits in Supplemental File S2). The final 1315 scaffolds represent the 29 autosomes, chromosome X, the mitochondria, and 1284 unassigned scaffolds/contigs. Unassigned scaffolds contained a total of 217 Mb of sequence (7.4% of total assembly) and ranged from 1 kb to 1.68 Mb, with 13 that were >1 Mb in length.
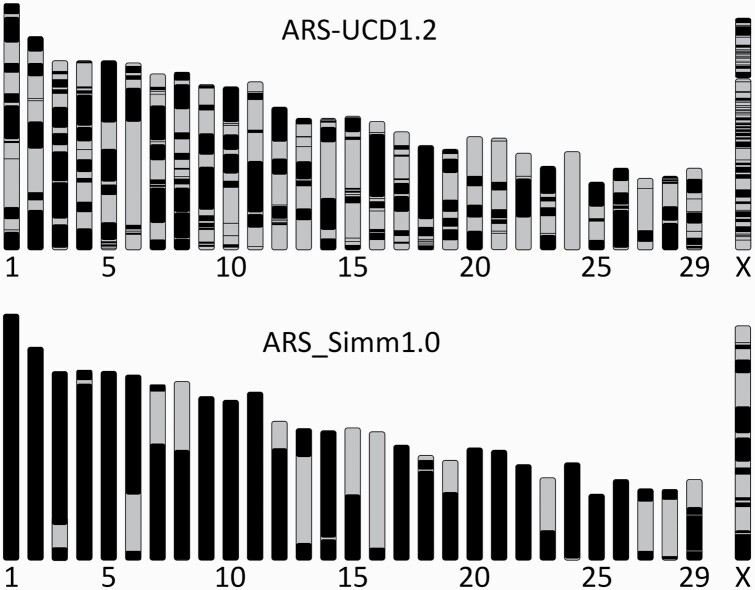
Final assembly statistics after scaffolding, chromosome assignment by alignment to the Hereford reference, and polishing are shown in Table 1 with the statistics of the reference genome for comparison. There were 52 gaps present in chromosome scaffolds with 13 chromosomes contained entirely within a single contig, and 6 chromosomes with only one gap (Figure 3), compared with a single ungapped chromosome in the Hereford reference. Chromosome X accounted for 38% (20 gaps) of all gaps in the chromosome scaffolds. Analysis of “lift over” annotation from ARS-UCD1.2 to the Simmental assembly showed that 20 800 of 21 039 protein coding genes (98.86%) were successfully lifted over. In addition, there were 331 protein coding genes in ARS_Simm1.0 with extra copies not found in ARS-UCD1.2.
Figure 3.
Ideograms of Hereford (ARS-UCD1.2) and Simmental (ARS_Simm1.0) genome assembly karyotypes. Chromosomes 1 through 29 and X are shown to approximate scale by scaffold/contig length. Each chromosome-assigned contig is shown as a solid color block along the chromosome, with gaps indicated by the change to a different block color. Simmental chromosomes that are spanned by a single contig with no gaps are solid black.
The quality of the final assembly was estimated in two ways. First, representation of genes contained in the Mammalia_od10 BUSCO database indicated approximately equal number of complete, fragmented, and missing genes in the Simmental and Hereford assemblies (Table 1). There were 540 genes of the 9226 total in the database that were categorized as missing by BUSCO in the ARS_Simm1.0, of which 508 were also reported as missing in the ARS-UCD1.2 assembly. An arbitrary selection of 20 genes that BUSCO listed as missing from both assemblies was examined in the Hereford reference assembly at NCBI. All 20 genes were properly annotated in the reference with corresponding protein translations, indicating that the BUSCO result was in error and underscoring that the process is only a useful metric as a relative indicator, sensitive to the version of the software and to the database of genes examined by the program.
The second estimate of assembly quality was performed by a strategy that identifies kmers in the short read data and assesses the accuracy and completeness based on the content of those kmers in the final, polished assembly (Rhie et al. 2020). This evaluation is sensitive to the source of the input short read data, so the evaluation was run twice on both the Simmental assembly and the Hereford assembly using either the Simmental dam or the Hereford short read data. The overall assembly quality score and the per-base average quality score were both a half order of magnitude improved in the Simmental assembly compared with the Hereford reference (36.5 versus 30.0 on log scale; Table 1), and completeness was higher and error rate lower, when Simmental short read data was used to generate the kmer list. Conversely when short read data generated from the Hereford were used, the Hereford assembly had higher completeness and read and kmer-based quality values, and reduced error rate relative to the Simmental assembly.
Discussion
The Simmental breed of cattle has become one of the most influential seedstock and commercial breeds in the United States. The North American population of Simmental is descended from those imported in the late 1960s from Central Europe. The breed’s history dates back to the Middle Ages and crosses between large German cattle and those of a smaller Swiss breed indigenous to the Simme Valley, in the Bern Switzerland. As late as the 1950s, top priorities of the breed were milk, meat, and work (fitness). The Simmental genome assembly presented here (ARS_Simm1.0) is a continuous chromosome-scale haplotype with quality rivalling the best livestock and model organism reference genomes available anywhere. The final Simmental assembly had only 52 chromosome gaps compared to: 315 for Hereford, 277 for Angus, 216 for Brahman, and 40 for Highland (Low et al. 2020; Rice et al. 2020; Rosen et al. 2020). Thus, a trio binning approach with F1 species hybrids continues to produce outstanding novel genome assemblies. Moreover, the use of in vitro fertilization and recipient females, combined with early fetal collection, increases the ability to safely and efficiently produce novel F1 hybrids from domestic and wild species that do not naturally interbreed. This combined approach allows the best reference assemblies to be produced for all breeds of cattle and related species and would pave the way for understanding functional genetic differences between them.
The pangenome effort for cattle will provide a resource for identification of breeds with unique genetic complements and support efforts to maintain biodiversity within the species. As cattle were domesticated at least twice from aurochs 8000–12 000 years ago in geographically disperse regions, the global diversity reflected in modern breeds may be extensive (Pitt et al. 2019). Some breeds are maintained as very small populations in resource-stressed locations, and it is imperative to assess the extent to which they harbor unique genome segments and haplotypes for prioritization of conservation efforts. Moreover, the wide variety of environments in which cattle are raised may well have selected for unique alleles and haplotypes best suited to those environments. Comparative whole genome studies with structural and small nucleotide variants will aid in identifying genetic elements that influence environmental adaptation. This may enhance our ability to select for production traits in target populations without disturbing the selective advantage that may have been accumulated over millennia.
Supplementary Material
Acknowledgments
The results reported here were made possible with resources provided by the USDA shared compute cluster (Ceres) as part of the ARS SciNet initiative. We thank the USMARC Core Facility staff for outstanding technical assistance. Also thank Dr. A. Bassett, B. Lee, J. Carlson, K. McClure, H. Clark, M. Pelster, H. Sadd, M. Sadd, and B. Shuck for outstanding technical support. We thank the American Simmental Association for their enthusiastic support and assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer. The funding bodies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding
U.S. Department of Agriculture, Agricultural Research Service appropriated projects (3040-31000-100-00D to T.P.L.S. and L.A.K., and 5438-32000-034-00D to M.P.H.); The University of Nebraska Great Plains Veterinary Educational Center project (2162390003 to B.L.V.L.); the Nebraska Beef Industry Endowment (2662390323001 to B.L.V.L.); USDA, ARS appropriated project (5090-31000-026-00D to D.M.B. and J.C.M.); National Institutes of Health (T32 HG008345-01 to J.O.); U.S. National Science Foundation (DEB-1754451 to B.S. and J.O.). The reproductive and animal husbandry portions of the project were supported by the American Simmental Association.
Data Availability
The datasets produced during this study are available in the NCBI BioProject repository under accessions PRJNA677947, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA677947 (Simmental) and PRJNA677946, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA677946 (bison). Fastq files for Simmental cow BHR Lady Sieg C235E (SAMN16825967, https://www.ncbi.nlm.nih.gov/biosample/16825967) and Yellowstone bison bull 709 (SAMN16823422) are deposited in the NCBI Short Read Archive under SRX9528670, https://www.ncbi.nlm.nih.gov/sra/SRX9528670/ and SRX9528561, https://www.ncbi.nlm.nih.gov/sra/SRX9528561 accessions, respectively.
References
- Bickhart DM, McClure JC, Schnabel RD, Rosen BD, Medrano JF, Smith TPL. 2020. Symposium review: advances in sequencing technology herald a new frontier in cattle genomics and genome-enabled selection. J Dairy Sci. 103:5278–5290. [DOI] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 49:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine Genome Sequencing and Analysis Consortium, Elsik C G, Tellam R L, Worley K C, Gibbs R A, Muzny D M, Weinstock G M, Adelson D L, Eichler E E, Elnitski L, Guigó R, et al. 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 324:522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Qiao Y, Lu J, Lu H, Zhang W, Yan F, Sun C, Hu Z, Zhang Z, Li G, et al. 2019. HUPAN: a pan-genome analysis pipeline for human genomes. Genome Biol. 20:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. Available from: http://arxiv.org/abs/1207.3907
- Ghurye J, Pop M, Koren S, Bickhart D, Chin CS. 2017. Scaffolding of long read assemblies using long range contact information. BMC Genomics. 18:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36: 2896– 2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Rhie A, Walenz BP, Dilthey AT, Bickhart DM, Kingan SB, Hiendleder S, Williams JL, Smith TPL, Phillippy AM. 2018. De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol. 36, 1174– 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Blair Hedges S. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon G. 2019. HMW gDNA Purification and ONT Ultra-Long-Read Data Generation v1 (protocols.io.bchhit36). Protocols.io. doi: 10.17504/protocols.io.bchhit36. [DOI] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 12:733–735. [DOI] [PubMed] [Google Scholar]
- Low WY, Tearle R, Liu R, Koren S, Rhie A, Bickhart DM, Rosen BD, Kronenberg ZN, Kingan SB, Tseng E, et al. 2020. Haplotype-resolved genomes provide insights into structural variation and gene content in angus and brahman cattle. Nat Communications. 11:2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, Martinez R, Bruford MW, Orozco-terWengel P. 2019. Domestication of cattle: two or three events? Evol Appl. 12:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PretextMap . n.d. https://github.com/wtsi-hpag/PretextMap.
- PretextView . n.d. https://github.com/wtsi-hpag/PretextView.
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice ES, Koren S, Rhie A, Heaton MP, Kalbfleisch TS, Hardy T, Hackett PH, Bickhart DM, Rosen BD, Ley BV, et al. 2020. Continuous chromosome-scale haplotypes assembled from a single interspecies F1 hybrid of yak and cattle. GigaScience. 9:giaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, Rowan TN, Low WY, Zimin A, Couldrey C, et al. 2020. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience. 9:giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Sherman RM, Forman J, Antonescu V, Puiu D, Daya M, Rafaels N, Boorgula MP, Chavan S, Vergara C, Ortega VE, et al. 2019. Author correction: assembly of a pan-genome from deep sequencing of 910 humans of african descent. Nat Genet. 51:364. [DOI] [PubMed] [Google Scholar]
- Shumate A, Salzberg S. 2020. Liftoff: an accurate gene annotation mapping tool. BioRxiv. https://www.biorxiv.org/content/10.1101/2020.06.24.169680v1.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hu Z, Zheng T, Lu K, Zhao Y, Wang W, Shi J, Wang C, Lu J, Zhang D, et al. 2017. RPAN: rice pan-genome browser for ∼3000 rice genomes. Nucleic Acids Res. 45:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jiao C, Schwaninger H, Chao CT, Ma Y, Duan N, Khan A, Ban S, Xu K, Cheng L, et al. 2020. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat Genet. 52:1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzi F, Narzisi G, Mishra B. 2012. Reevaluating assembly evaluations with feature response curves: GAGE and assemblathons. PLoS One. 7:e52210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets produced during this study are available in the NCBI BioProject repository under accessions PRJNA677947, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA677947 (Simmental) and PRJNA677946, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA677946 (bison). Fastq files for Simmental cow BHR Lady Sieg C235E (SAMN16825967, https://www.ncbi.nlm.nih.gov/biosample/16825967) and Yellowstone bison bull 709 (SAMN16823422) are deposited in the NCBI Short Read Archive under SRX9528670, https://www.ncbi.nlm.nih.gov/sra/SRX9528670/ and SRX9528561, https://www.ncbi.nlm.nih.gov/sra/SRX9528561 accessions, respectively.