The enteric virome is composed of a staggering diversity of eukaryotic and prokaryotic viruses that can interact with the intestinal immune system. Li et al. discuss how resident enteric viruses directly or indirectly affect intestinal homeostasis and modulate innate and adaptive immune responses.
Abstract
The diverse enteric viral communities that infect microbes and the animal host collectively constitute the gut virome. Although recent advances in sequencing and analysis of metaviromes have revealed the complexity of the virome and facilitated discovery of new viruses, our understanding of the enteric virome is still incomplete. Recent studies have uncovered how virome–host interactions can contribute to beneficial or detrimental outcomes for the host. Understanding the complex interactions between enteric viruses and the intestinal immune system is a prerequisite for elucidating their role in intestinal diseases. In this review, we provide an overview of the enteric virome composition and summarize recent findings about how enteric viruses are sensed by and, in turn, modulate host immune responses during homeostasis and disease.
Introduction
The human gastrointestinal tract harbors diverse populations of bacteria, fungi, archaea, viruses, and eukaryotic microorganisms. This complex community of microbes, collectively termed the enteric microbiota, provides tremendous enzymatic capabilities and plays integral roles in host metabolism, immune system development, colonization resistance against pathogens, and local immune homeostasis (Belkaid and Hand, 2014; Buffie and Pamer, 2013; Gensollen et al., 2016; Rowland et al., 2018). Thus, the human enteric microbiota is considered to functionally serve as an essential “organ.” In addition to the well-appreciated contributions of the enteric bacterial microbiota to both health and disease, recent studies have begun to reveal that the enteric virome is also critical for homeostatic regulation and disease progression, acting through both virus–microbiome and virus–host interactions (Mirzaei and Maurice, 2017; Mukhopadhya et al., 2019; Pfeiffer and Virgin, 2016; Virgin, 2014).
The intestinal virome is composed of a staggering diversity of eukaryotic and prokaryotic viruses capable of infecting both host cells and a wide range of microorganisms. Human intestinal contents are estimated to contain at least 108–1010 virus-like particles per gram, the composition of which is partially defined by dietary habits and surrounding environment (Hoyles et al., 2014; Lepage et al., 2008). Gut viral diversity is age dependent across healthy, Western individuals, but in adults, viral intrapersonal variance remains low (Garmaeva et al., 2019; Gregory et al., 2020). Indeed, the adult human enteric virome is highly individual and temporally stable and correlates with the bacterial microbiome (Shkoporov et al., 2019). While gastrointestinal virus studies have historically focused on eukaryotic viruses due to their well-known pathogenic impact, such as acute gastroenteritis, on people of all ages around the world (Oude Munnink and van der Hoek, 2016), prokaryotic viruses, or bacteriophages, likely account for over 90% of the human enteric virome (Guerin et al., 2018). Our understanding of how enteric prokaryotic viruses directly or indirectly influence human health and disease is limited. However, owing to their sheer diversity and their intimate interaction with bacterial microbial ecology, enteric bacteriophages are likely to play numerous roles in promoting a healthy enteric ecosystem as well as participating in enteric microbiota-associated disease.
Compared with the bacterial component of the intestinal microbiota, study of the enteric virome is in its infancy, and much work needs to be done to fully elucidate its impact on human health and disease. Recent advances in high-throughput sequencing technologies and bioinformatic analysis of “metaviromes” have facilitated the identification of unknown viruses and added to our appreciation of the richness and complexity of the human enteric virome (Garmaeva et al., 2019; Handley et al., 2012; Norman et al., 2015; Santiago-Rodriguez and Hollister, 2019). Many virome studies have repeatedly shown that only a minor fraction of sequences can be classified and that up to 90% of virome sequences may go unannotated (Handley et al., 2016; Manrique et al., 2016; Minot et al., 2011). The rest of the virome, which cannot be assigned taxonomy, has been colloquially termed “viral dark matter” (Krishnamurthy and Wang, 2017). Elucidation of the full composition of the human virome will be paramount to our understanding of enteric viruses in health and disease.
Because the enteric virome is a complicated menagerie of both bacteriophages and eukaryotic viruses, as well as unresolved viral dark matter, it presents a substantial challenge to researchers. Accurate interpretation of virome data requires an understanding of bacteriophage and eukaryotic virology, bioinformatics, microbial ecology, and enteric immunology/physiology. While tools are improving to interrogate this complex network, much additional work remains to crystalize our understanding of the virome in health and disease. Many other reviews have surveyed the challenges presented by virome research (Garmaeva et al., 2019; Khan Mirzaei et al., 2021; Wang, 2020). However, less attention has been spent on how a virome, once characterized, can be integrated into classical models of viral pathogenesis. Here, we will review the current understanding of sensing and immune regulation of the human enteric virome and how this may lead to important consequences for health and disease.
Enteric virome composition
Bacteriophages
Metagenomic studies have shown that intestinal bacteriophages are the most abundant components of the enteric virome (Focà et al., 2015; Neil and Cadwell, 2018). Enteric phage populations in adult humans appear to not be largely affected by periodic fluctuations, remaining constant due to rapid adaptations to any changes by long-term members of the virome (Minot et al., 2013; Shkoporov et al., 2019). Early-life bacteriophage colonization appears to originate from a combination of maternal, dietary, and environmental sources and exists in a high predator–low prey dynamic (Lim et al., 2015; Minot et al., 2011). Some infant enteric microbiome studies have indicated early inverse correlations between infant bacteriophage communities and bacterial richness, with bacteriophage diversity rapidly decreasing in early life, while others have suggested rapid early expansion of phage communities from pioneer bacteria, followed by expansion of eukaryotic virus populations (Liang et al., 2020; Lim et al., 2015). Thus, the enteric virome, similar to the enteric bacterial microbiome, exhibits dynamism during early life but settles into a relatively stable configuration in adulthood.
Analysis of the enteric microbiome of healthy individuals has revealed the presence of a healthy enteric phageome composed of core bacteriophages common to healthy adults, which is likely globally distributed (Manrique et al., 2016). The healthy enteric phageome likely plays an important role in maintaining the enteric microbiome structure and function (Lin and Lin, 2019). Phages can be functionally grouped into two major categories based upon their replicative life cycle (Lawrence et al., 2019). Virulent phages are limited to a lytic lifestyle, whereby they hijack the host cell’s replication mechanism of viral packaging, ultimately lysing their bacterial host to release newly formed progeny. In contrast, temperate phages have lytic but also lysogenic life cycles, wherein they integrate their genetic material into their host cell’s chromosome as prophages or remain as circular or linear plasmids and passively replicate alongside the host bacteria (Girons et al., 2000). The human enteric virome, which exhibits a much lower ratio of phage:bacteria than other environments, is dominated by temperate phages, which play important roles in shaping the bacterial community and transferring genes among bacteria (Lin and Lin, 2019; Reyes et al., 2012). These concepts support a dynamic environment wherein bacteriophages participate in both taxonomic and functional construction of a host enteric microbiome with potential consequences of shaping either healthy or unhealthy enteric environments.
Defined members of intestinal phage communities are predominantly members of the Caudovirales order or the Microviridae family (Table 1; Callanan et al., 2018; Dion et al., 2020; Gregory et al., 2019 Preprint). The tailed double-stranded (ds) DNA viruses of Caudovirales include Siphoviridae (phages with long, noncontractile tails, largely temperate), Myoviridae (phages with long, contractile tails, strictly virulent), and Podoviridae (phages with short, contractile tails, largely virulent; Santiago-Rodriguez and Hollister, 2019). The single-stranded (ss) DNA members of the viral families Microviridae (phages with small icosahedral capsid, temperate or virulent; Krupovic and Forterre, 2011) and Inoviridae (phages with filamentous morphology, temperate; King et al., 2011) have also been identified as important constituents of fecal viromes. Recently, studies have identified DNA bacteriophages called crAssphages, which are ubiquitous in publicly available metagenomes and predominantly present in human feces (Dutilh et al., 2014). Human gut–associated crAssphages are not a single entity but rather a group of diverse crAss-like phages sharing genomic traits, which can be classified into 10 candidate genera that differ among human populations (Guerin et al., 2018). Based on read co-occurrence analysis, the bacteria of phylum Bacteroidetes, which dominate the enteric microbiome, have been pointed to as crAssphage hosts (Ahlgren et al., 2017; Dutilh et al., 2014), and indeed, crAssphage ΦCrAss001 has been shown to infect human gut symbiont Bacteroides intestinalis (Shkoporov et al., 2018). However, our understanding of the biological significance and role of crAssphage in the human intestine remains limited. Despite the significant advances in human enteric bacteriophage characterization made to date, only two RNA bacteriophage families, Leviviridae (ssRNA) and Cystoviridae (dsRNA), are currently known, and the role of RNA bacteriophages is poorly understood (Callanan et al., 2018). Whether this is secondary to a more limited variety of RNA bacteriophages being present in the intestine or to undersampling due to a frequent bias in virome library preparations toward exclusively analyzing the DNA component (Garmaeva et al., 2019) remains to be determined.
Table 1. Human gut virome.
| Viral type | Viral familial taxonomy | Host range | Definitive viral genera/species in human feces | Cellular tropism in humans | References | ||
|---|---|---|---|---|---|---|---|
| Eukaryotic virus | DNA virus | Single-stranded | Anelloviridae | Primates | Torque teno virus, Torque teno midi virus, Torque teno mini virus | Epithelial cells | Hino and Miyata, 2007; Spandole et al., 2015 |
| Circoviridae | Birds and mammals | Chicken anemia virus, porcine circoviruses | Monocytes, epithelial cells, and fibroblasts | Li et al., 2010 | |||
| Geminiviridae | Plants | Tomato yellow leaf curl virus | No evidence | Zhang et al., 2006 | |||
| Genomoviridae | Humans, mammals, birds, and fungi | Gemycircularvirus | Unclear | Gregory et al., 2020 | |||
| Parvoviridae | Vertebrates, insects | Human bocavirus 2-4 | Epithelial cells | Emlet et al., 2020; Guido et al., 2016 | |||
| Double-stranded | Adenoviridae | Vertebrates | Enteric adenovirus 40, 41 | Epithelial cells, fibroblasts, gut lymphocytes | Favier et al., 2004 | ||
| Herpesviridae | Vertebrates | Human CMV, EBV | CMV: Endothelial cells, leukocytes, and monocytes; EBV: endothelial cells, B cells | Emlet et al., 2020; Gerna et al., 2004; Stanfield and Luftig, 2017 | |||
| Iridoviridae | Amphibia, fish, invertebrates | Lymphocystis disease virus | No evidence | Gregory et al., 2020 | |||
| Papillomaviridae | Vertebrates | Human papillomavirus 6, 18, 66 | Epithelial cells | Egawa et al., 2015; Popgeorgiev et al., 2013 | |||
| Polyomaviridae | Mammals and birds | BK polyomavirus, JC polyomavirus, Human polyomavirus 9, 12 | Oligodendrocytes, epithelial cells | Cook, 2016; Emlet et al., 2020 | |||
| Rudiviridae | Thermophilic archaea from Crenarchaeota | Unclassified | No evidence | Gregory et al., 2020 | |||
| RNA virus | Single-stranded | Caliciviridae | Vertebrates | Human NoV, Sapovirus | Epithelial (enteroendocrine) cells, myeloid cells, and lymphoid cells in immunocompromised patients | Green et al., 2020; Karandikar et al., 2016 | |
| Astroviridae | Vertebrates | Human astrovirus | Enterocytes, goblet cells | Kolawole et al., 2019 | |||
| Virgaviridae | Plants | Pepper mild mottle virus, Tobacco mosaic virus | No evidence | Colson et al., 2010; Zhang et al., 2006 | |||
| Picornaviridae | Vertebrates | Human cosavirus, human klassevirus/ salivirus, Aichi virus, human enterovirus, human parechovirus, Saffold cardiovirus, human echovirus, human coxsackievirus, human poliovirus, hepatitis A | Epithelial cells, neuronal cells, hepatocytes | Krishnamurthy and Wang, 2018; Rivadulla and Romalde, 2020; Tapparel et al., 2013 | |||
| Retroviridae | Vertebrates | HIV-1 | T lymphocytes, macrophages | Gootenberg et al., 2017; Virgin, 2014 | |||
| Togaviridae | Humans, mammals, marsupials, birds, mosquitoes | Unclassified | Fibroblasts | Gregory et al., 2020 | |||
| Coronaviridae | Vertebrates | Severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, Middle East respiratory syndrome coronavirus (MERS-Cov) | Tracheo-bronchial epithelial cells, type 1 pneumocytes, intestinal epithelial cells | Corman et al., 2018 | |||
| Alphaflexiviridae | Plants, fungi | Unclassified | No evidence | Gregory et al., 2020 | |||
| Iflaviridae | Insects | Unclassified | No evidence | Gregory et al., 2020 | |||
| Hepeviridae | Humans, pigs, wild boars, sheep, cows, camels, monkeys, some rodents, bats, and chickens | Hepatitis E virus | Hepatocytes, intestinal epithelial cells | Oechslin et al., 2020 | |||
| Double-stranded | Picobirnaviridae | Unclear: mammals or bacteria | Human picobirnavirus | No evidence | Malik et al., 2014 | ||
| Reoviridae | Vertebrate, invertebrates, plants, fungi | Human rotavirus | Mature enterocytes | Komoto and Taniguchi, 2014 | |||
| Bacteriophages | DNA virus | Single-stranded | Microviridae | Bacteria | Chlamydia phage 1,3,4, Bdellovibrio phage phiMH2K, Chlamydia phage CPG1, Spiroplasma phage 4, Chlamydia phage CPAR39 | NA | Dion et al., 2020; Székely and Breitbart, 2016 |
| Inoviridae | Bacteria | Unclassified | NA | Dion et al., 2020; Székely and Breitbart, 2016 | |||
| Double-stranded | Myoviridae | Bacteria, archaea | phiBCD7, Bacillus phage G, phiP-SSM4 | NA | Barylski et al., 2020; Gregory et al., 2020; Guerin et al., 2018; Yutin et al., 2018 | ||
| Siphoviridae | Bacteria, archaea | Listeria phage A118, phiE125, Lactococcus phage bIL285, phiCP39-O, Clostridium phage phiCP39-O, Mycobacterium phage Athena, phage PA6, phage SM | NA | Barylski et al., 2020; Gregory et al., 2020; Guerin et al., 2018; Yutin et al., 2018 | |||
| Podoviridae | Bacteria | Enterobacteria phage P22, phage T3 | NA | Barylski et al., 2020; Gregory et al., 2020; Guerin et al., 2018; Yutin et al., 2018 | |||
| crAss-like phages | Bacteria | P-crAssphage | NA | Barylski et al., 2020; Gregory et al., 2020; Guerin et al., 2018; Yutin et al., 2018 | |||
| RNA virus | Single-stranded | Leviviridae | Bacteria | Enterobacteria phage MS-2 | NA | Callanan et al., 2018 | |
| Double-stranded | Cystoviridae | Bacteria | Pseudomonas phage | NA | Callanan et al., 2018 | ||
NA, not applicable.
Eukaryotic viruses
Although bacteriophages are the most abundant viral community in the human gastrointestinal tract, eukaryotic viruses are a critically important virome component. Human virome studies have revealed complex eukaryotic viral communities including ssRNA, ssDNA, dsRNA, and dsDNA viruses, many of which have a significant impact on human health (Table 1; Gregory et al., 2019 Preprint; Lozupone et al., 2012; Zhang et al., 2006). When mediating pathogenic effects, they can infect human intestinal cells and trigger immune responses, causing damage to the epithelial lining and the absorptive villi, leading to acute gastroenteritis, enteritis, colitis, or even oncogenesis (Emlet et al., 2020; Lopetuso et al., 2016). Viral pathogens of the families Parvoviridae (human bocavirus 2–4), Adenoviridae (adenovirus serotypes 40 and 41), Caliciviridae (norovirus [NoV] and sapovirus), Astroviridae (astrovirus), Picornaviridae (entero-, kobu-, and parechoviruses), Coronaviridae (severe acute respiratory syndrome [SARS] and SARS–CoV-2), Hepeviridae (hepatitis E virus), and Reoviridae (rotavirus) have all been implicated in acute gastrointestinal disease (Table 1). Most enteric eukaryotic viruses are RNA viruses, except for the parvoviruses and adenoviruses, and thus are often undetected in viral metagenomics studies due to the additional library preparation requirements needed to sequence viral cDNA. These viruses stimulate robust innate and adaptive immune responses from the host, which contribute to symptoms as well as to viral clearance and protection from reinfection.
In addition to eukaryotic virome members causing symptomatic infections, other enteric viruses are present in the intestine without clear roles as pathogens. Previous studies have documented persistent shedding of enteric eukaryotic viruses from Anelloviridae (Torque teno virus), Circoviridae (unclassified), Picobirnaviridae (picobirnavirus), and Picornaviridae (human parechovirus 1) families in healthy infants or children in longitudinal studies without causing symptomatic disease (Kapusinszky et al., 2012; Zhao et al., 2017). The absence of symptoms might reflect either a commensal viral infection with no apparent harm to the host or that the immune system predominantly suppresses detectable effects of infection (Focà et al., 2015; Okamoto, 2009; Virgin et al., 2009). In addition, pathogenic viruses are often detected in the absence of symptoms. For example, in the same cohorts, rotavirus and parvoviruses were also observed in healthy children. NoV, currently the leading cause of acute gastroenteritis outbreaks globally, can be shed asymptomatically over long periods of time (Newman et al., 2016). Even SARS–CoV-2, the viral pathogen responsible for the 2019 worldwide pandemic, can infect intestinal epithelium and is detectable in stool for at least 1–2 mo even with asymptomatic infection (Lamers et al., 2020; Lee et al., 2020; Xie et al., 2020; Zang et al., 2020). For many of these pathogens, it remains unclear whether asymptomatic infections are secondary to avirulent strains or host differences in susceptibility or responsiveness to infection. Other viruses from Herpesviridae (EBV), Papillomaviridae (human papillomavirus 6, 18, 66), Polyomaviridae (human polyomavirus), and Retroviridae (HIV) families can continuously replicate or reactivate from latency when host immunosurveillance fails, resulting in disease (Emlet et al., 2020; Gootenberg et al., 2017; Virgin, 2014). Though normally these asymptomatic viral infections don’t cause detectable phenotypes in their hosts, they are persistently interacting with the host immune system and may help maintain tonic levels of protective immunity or conversely increase host susceptibility to disease, depending upon context.
Interestingly, vertebrate, amoeba, insect, and plant viruses have also been found in human intestinal samples, likely derived from diet (Minot et al., 2011). An open discussion continues about whether these diet-derived viruses could contribute to gastrointestinal disease. For instance, pepper mild mottle virus, the most abundant pathogenic plant virus in human fecal samples, has been associated with specific immune responses and clinical symptoms in humans (Colson et al., 2010; Zhang et al., 2006). Further investigation is needed to define the role of these viruses in immune regulation and gastrointestinal disease.
Intestinal antiviral immune responses
The intestinal epithelium serves as a first line of defense, separating the underlying lamina propria from trillions of commensal microorganisms in the intestinal lumen. The intestinal epithelial cells (IECs) and mononuclear phagocytes as well as the gut-associated lymphoid tissues (GALTs) together maintain the intestinal barrier. These components play critical roles in spatial segregation, microbial sensing, and immunoregulatory responses to prevent inflammation (Peterson and Artis, 2014). Bacteriophages can affect the human host indirectly through interactions with bacteria, but new evidence suggests they also directly stimulate the immune system. Enteric eukaryotic viruses, due to their tropism for host cells, induce and modulate host immune responses directly. The general virus sensors, signaling pathways, and mononuclear phagocyte subsets of enteric virome sensing have been extensively reviewed elsewhere (Metzger et al., 2018), but here we provide a general overview to highlight key pathways.
Overview of enteric antiviral immune responses
As frontline sensors for microbial encounters, pattern recognition receptors (PRRs) recognize commensal or pathogenic microorganisms in the human gut and initiate innate immune and antigen-specific adaptive immune responses. PRRs are germline-encoded host sensors, including the membrane-bound TLRs, C-type lectin receptors (CLRs), and cytoplasmic cytosolic nucleotide-binding oligomerization domain–like receptors (NLRs), retinoic acid–inducible gene I (RIG-I)–like receptors, and DNA sensors (absent in melanoma 2 [AIM2]–like receptors) and cyclic guanosine monophosphate–adenosine monophosphate synthase (Kumar et al., 2011). Highly conserved microbial-associated molecular patterns, pathogen-associated molecular patterns (PAMPs), or damage-associated molecular patterns are identified by these sensors to trigger an array of canonical anti-microbial immune responses through the induction of various inflammatory cytokines, chemokines, and IFNs, which further induce IEC-secreted antimicrobial peptides and mucus. These responses also mediate recruitment and activation of intestinal mononuclear phagocytes and promote the development of pathogen-specific, long-lasting adaptive immunity through B and T lymphocytes. PRR sensing in the intestinal mucosa is strictly regulated to induce effective immune responses against invasive pathogenic viruses and to maintain immune tolerance to harmless commensals (Metzger et al., 2018). Thus, PRR sensing in the enteric tissue is essential to maintaining intestinal homeostasis, shaping the virome, and preventing intestinal pathology.
Depending on the cellular localization and recognition pattern of PRRs, viral sensing can be divided into two general categories. First, cell-extrinsic recognition, which does not require the cell-expressing PRRs to be infected, is mediated by transmembrane TLRs and CLRs when viral components attach to cell surfaces or are internalized into phagosomes or endosomes (Bermejo-Jambrina et al., 2018; Iwasaki and Medzhitov, 2015; Satoh and Akira, 2017). In general, recognition by TLRs and CLRs does not depend on the viability, replication, or invasiveness of foreign microbes and thus can detect a broad spectrum of microbial PAMPs regardless of their origin (Iwasaki and Medzhitov, 2015). In contrast, cell-intrinsic viral recognition relies on intracellular cytosolic sensors to detect ligands that emerge in infected cells. These include the RNA sensors of RIG-I–like receptors and cytosolic DNA sensors, which are ubiquitously expressed in both immune and nonimmune cells, as well as cytosolic NLRs (Iwasaki and Medzhitov, 2010; Schroder and Tschopp, 2010). Intracellular sensors are more restricted in their sensing spectrum, with PRR recognition potentially depending on microbial viability, invasiveness, and replication.
Despite differential expression and patterns recognized by individual PRRs, intestinal virus sensing triggers a cascade of signals leading to the activation of NF-κB–dependent production of proinflammatory cytokines including IL-1β, IL-6, IL-12, and TNF-α and the activation of IFN regulatory factor (IRF) 3– and/or IRF7-dependent transcription of type I and type III IFNs (Kawai and Akira, 2011; Lazear et al., 2019). In addition, the triggering of NLRs and AIM2 also leads to activation of the inflammasome, which induces caspase-1 to cleave immature forms of cytokines such as pro–IL-1β and pro–IL-18, resulting in the processing and secretion of mature cytokines IL-1β and IL-18 (Schroder and Tschopp, 2010). Type I IFNs, which include 13 members of the IFN-α family as well as IFN-β, IFN-ε, IFN-κ, and IFN-ω, bind to the IFN-α/-β receptor (IFNAR) to activate the Janus kinase-signal transduction and activation of transcription (JAK–STAT) signaling pathway. Signaling through IFNAR subsequently induces over 300 IFN-stimulated genes (ISGs), which play critical roles in viral control by interfering with multiple stages of viral life cycles. IFNAR signaling further induces the transcription of IRF7, leading to positive feedback of the IFN response (Honda et al., 2005). Type I IFNs also confer antiviral resistance to neighboring uninfected cells, activate the cytotoxic activity of natural killer cells and CTLs for clearance of virus-infected cells, and induce cytokines and chemokines for the initiation of adaptive immunity (Pang and Iwasaki, 2012). Type III IFNs can consist of up to four IFN-λ molecules, IFN-λ1 (IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B), and IFN-λ4. Similar to type I IFNs, IFN-λ cytokines signal through the IFN-λ receptor, which is composed of two subunits, IL28Rα and IL10Rβ, leading to activation of JAK–STAT signaling, expression of ISGs, and induction of an antiviral state. However, unlike type I IFNs, which mediate effects on virtually all cell types, IFN-λ primarily regulates antiviral immunity at barrier sites including the intestine (Sommereyns et al., 2008). Expression of the IL28Rα subunit is limited to specific cellular subsets, including epithelial cells of the gastrointestinal tract, respiratory tract, and reproductive tract and a specific subset of immune cells (Stanifer et al., 2019).
Interactions between enteric bacteriophages and the host immune system
Indirect influences of phages on immune responses
Intestinal bacteriophage communities are specifically related to and interdependent upon their bacterial host communities, and interactions between phages and bacterial hosts also influence the eukaryotic host intestinal immune system. The balance between lytic and lysogenic bacteriophage life cycles and differential spatial distribution of phages contributes to the eubiosis of the human microbiome, which is heavily implicated in the development of immune cells and the regulation of immune responses (De Paepe et al., 2014; Manrique et al., 2017). Bacteriophages accumulate on enteric mucosal surfaces and within mucus, near the frontline of defense against pathogens, at a high bacteriophage-to-bacteria ratio relative to the adjacent environment (Barr et al., 2013). Binding interactions between mucin and Ig-like protein domains exposed on phage capsids facilitate enrichment of phages at mucus glycoproteins, thereby providing ubiquitous non–host-derived immunity against bacterial invaders (Almeida et al., 2019; Barr et al., 2013).
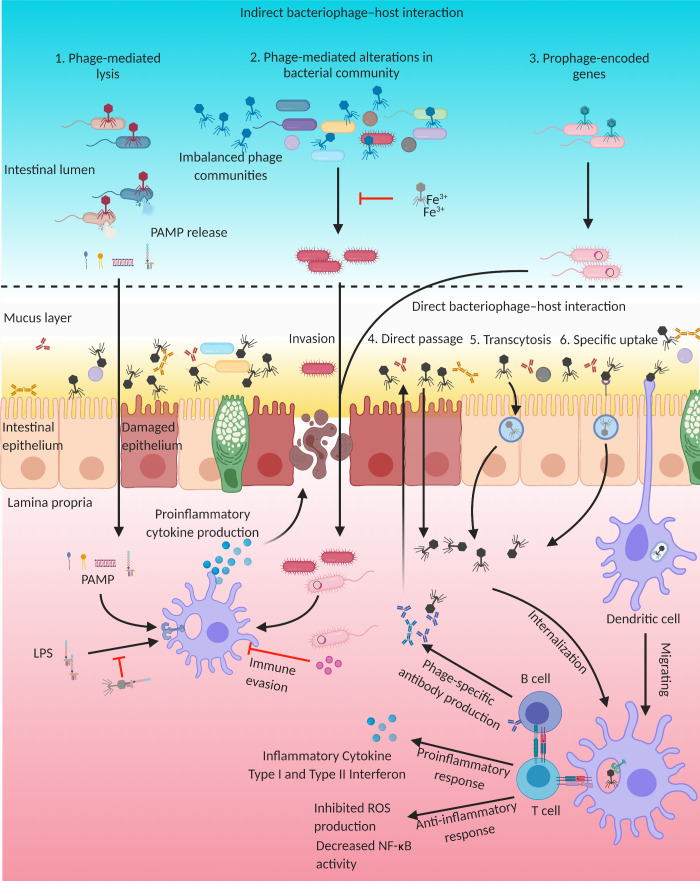
While this enrichment may provide a protective function, phage-mediated bacterial cell lysis can release PAMPs, such as bacterial DNA, LPS, peptidoglycan, and bacterial amyloid, which can induce proinflammatory responses in the gut (Fig. 1; Sinha and Maurice, 2019; Tetz and Tetz, 2018). In addition, a recent gnotobiotic mouse study reported that bacteriophage predation does not merely affect susceptible bacteria but also induces cascading disturbances to other bacterial species via interbacterial interactions (Hsu et al., 2019). Microbiome shifts caused by bacteriophage predation can have a direct impact on enteric metabolites, such as short-chain fatty acids, neurotransmitters, amino acids, and bile salts, which play key roles in neuro-immunoendocrine regulation (Hsu et al., 2019; Sokol et al., 2009).
Figure 1.
Indirect and direct influences of bacteriophages on immune responses. (1) Phage-mediated bacterial cell lysis can release PAMPs, which can transit through the intestinal epithelium and induce proinflammatory responses. Bacteriophage tail adhesins can bind LPS to dampen LPS-induced immune responses. (2) In the case of imbalanced phage communities, bacteriophage infection may dramatically influence the host bacterial community and lead to overgrowth of pathogens. The needle domain of the bacteriophage tail fiber region can bind and sequester iron ions and prevent pathogen overgrowth in the intestine. (3) Transfer of prophage-encoded genes can influence the pathogenicity of bacterial hosts and provide immune evasion capacity by directly inhibiting phagocytic cells. (4–6) Phages may also directly pass through damaged epithelial cells (4) or cross through the intestinal epithelium by nonspecific transcytosis (5) or via specific recognition of eukaryotic cells via structures that resemble bacterial receptors (6). These invading phages can interact with the intestinal immune system to induce pro- or anti-inflammatory responses and production of phage-specific neutralizing antibodies. Image created with BioRender.
In addition to directly affecting microbiome dynamics via lysis of bacterial hosts, bacteriophages dramatically influence host bacterial communities through horizontal transfer of novel genes, which can provide resistance to subsequent phage predation and alter host gene expression, as well as influence the physiology of the host (Fig. 1; Howard-Varona et al., 2017; Lekunberri et al., 2017; Touchon et al., 2017). As discussed above, prophages can integrate into bacterial chromosomes or be maintained as plasmids within host cells, and these acquired genes may contribute to bacterial adaptation (Modi et al., 2013). Bacteriophage-mediated genetic transfer can even occur between distinct species of bacteria; for example, phages from pathogenic bacteria can convert nonpathogenic bacteria to virulent strains by transferring phage-encoded virulence factors or other proteins that assist in immune evasion (Broudy and Fischetti, 2003; Chen and Novick, 2009; Faruque et al., 1999; Fillol-Salom et al., 2019). Carriage of Shiga toxin–encoding prophages not only disseminates toxin genes but also enhances antimicrobial tolerance of Escherichia coli by modifying the bacterium’s metabolism (Holt et al., 2017). Thus, phage-mediated horizontal transfer of genes can indirectly modulate immune responses to the intestinal microbial community.
Other indirect effects of bacteriophages on the intestinal environment have also been described, as phage tail fibers and exposed protein coats can serve as binding sites for other physiologically relevant products beyond mucus (Sinha and Maurice, 2019). The needle domain of the bacteriophage tail fiber region can bind and sequester iron ions, which can play important roles in virulence, tolerance, and replication of intestinal bacterial pathogens including Vibrio vulnificus, Samonella typhimurium, and Yersinia species (Kortman et al., 2014; Nazik et al., 2017; Penner et al., 2016). In addition, recent studies have also found that bacteriophage tail adhesins can bind LPS to dampen LPS-induced immune responses (Miernikiewicz et al., 2016). Thus, intestinal bacteriophage can serve as iron chelators or LPS adsorbers to modulate bacterial communities as well as inflammatory responses in this complex microbial ecosystem (Fig. 1). As we continue to uncover and characterize additional phages, it is likely that other mechanisms of modulation will also be discovered.
Direct influences of phages on immune responses
Until recently, intestinal bacteriophages were thought to only affect human immunity indirectly via effects on the microbiota. However, growing evidence suggests that phages can cross the intestinal epithelium to directly communicate with the enteric immune system. In addition to passing through the epithelial barrier when it is damaged due to inflammatory processes, phages can also be internalized by nonspecific transcytosis and/or by binding to moieties that resemble bacterial phage receptors (Sinha and Maurice, 2019). A Trojan horse hypothesis has also been presented, suggesting that infecting bacteria may act as delivery vehicles for phages to enter epithelial cells, although there is currently no evidence to support this hypothesis (Duerkop and Hooper, 2013; Sinha and Maurice, 2019). Traversing or internalized bacteriophages can directly interact with the epithelium, penetrate local dendritic cells (DCs), or drain into the GALT to stimulate innate immune responses in antigen-presenting cells to generate humoral immune responses (Fig. 1).
Previous studies have suggested that phages have either weak proinflammatory or immunomodulatory effects, making them potentially promising for medical application in phage therapies (Miernikiewicz et al., 2013; Principi et al., 2019). The production of ROS by immune cells, induced by LPS or E. coli, as well as NF-κB activity can be inhibited by T4 and Staphylococcus aureus phage (Górski et al., 2006; Miedzybrodzki et al., 2008; Zhang et al., 2018). Further, purified phage proteins have been shown to modulate anti-inflammatory responses similar to functional phage (Miernikiewicz et al., 2016). However, because there is such a vast diversity of phages, it is likely that a distinct phage can elicit either anti-inflammatory or proinflammatory immune responses (Górski et al., 2017; Van Belleghem et al., 2017). Studies in this area have been generally limited to cultivatable phage, and further studies on the immunobiological activities of bacteriophages and their proteins are greatly needed.
Gut-resident mononuclear phagocytes play central roles in the migration and activation of immune cells by sensing microbes to trigger cytokine and chemokine responses, as well as internalizing and digesting antigens for priming of intestinal T and B cell responses. A significant expansion of Caudovirales phages has been associated with inflammatory bowel disease (IBD), and increased phages that target the beneficial microbe Faecalibacterium prausnitzii have also been observed in IBD (Cornuault et al., 2018; Norman et al., 2015). Recently, bacteriophages isolated from IBD patients were found to elicit heightened T cell immune responses, dependent on interactions with DCs (Gogokhia et al., 2019). These phages are endocytosed by intestinal DCs and, via TLR9 within endosomes, stimulate both phage-specific as well as nonspecific IFN-γ–mediated immune responses, which can exacerbate colitis, supporting direct proinflammatory roles for phages in the context of IBD (Gogokhia et al., 2019). During bacterial infections, bacteriophages may also play proinflammatory roles. Phage produced by pathogenic bacteria can be internalized by human and murine immune cells, with phage RNA subsequently triggering TLR3- and Toll/IL-1 receptor–domain-containing adapter-inducing IFN-β (TRIF)–dependent type I IFN production while inhibiting phagocytosis and TNF production (Sweere et al., 2019). These antiviral responses suppress bacterial clearance, resulting in phage-facilitated bacterial infection (Sweere et al., 2019). Thus, direct interactions of phage with mammalian cells can have profound and direct effects on intestinal infection and inflammation.
Previous studies have demonstrated that bacteriophages are able to elicit specific humoral immune responses, which could limit further systemic phage dissemination (Fig. 1; Hodyra-Stefaniak et al., 2015; Pyun et al., 1989; Uhr and Finkelstein, 1963; Uhr et al., 1962). However, the factors that determine bacteriophage immunogenicity, as well as how specific humoral responses affect phages, are unclear. While antibodies that bind to the tail fiber region to inhibit the interaction of phage with bacterial hosts have long been understood to neutralize phage infectivity, antibody responses against the phage capsid protein are also capable of suppressing phage activity (Dąbrowska et al., 2014; Yanagida, 1977). Long-term exposure to high bacteriophage titers in drinking water induces both secretory anti-phage IgA antibodies in the gastrointestinal tract, which limit phage activity in the gut, as well as specific IgG, IgM, and IgA antibodies in blood (Majewska et al., 2015; Majewska et al., 2019). Additional exploration will be needed to fully define the effects of phage in modulating innate and adaptive immune responses, as well as in clarifying how the adaptive immune system may shape phage communities in the microbiota.
Eukaryotic viruses and the immune system
In the mammalian intestine, eukaryotic viruses interact directly with host cells to stimulate innate and adaptive responses. Much of our understanding of these intestinal immune responses relies upon murine models for human viral infections to effectively dissect pathways by which the host responds to viral challenge. Here, we describe recent findings related to host immune responses to murine enteric viral infections, which are likely to broadly reflect human enteric viral infections.
Cell intrinsic and innate responses to enteric eukaryotic viruses
Invading viral nucleic acids are sensed by cell intrinsic PRRs, which include endosomal TLRs that signal through adaptor protein Myeloid differentiation primary response 88 (MyD88; for TLR7/8/9) or TRIF (for TLR3/4), as well as the cytosolic sensors RIG-I and melanoma differentiation–associated protein 5 (MDA5) that signal through mitochondrial antiviral-signaling protein (MAVS). These PRR pathways ultimately stimulate the activation of IRF3/7 and subsequent production of types I and III IFNs and other antiviral or immunoregulatory genes (Fig. 2; Hartmann, 2017; Onoguchi et al., 2007). For example, the RIG-I/MDA5–MAVS pathway has been demonstrated to be essential for induction of IFNs in rotavirus-infected IECs and has a functionally important role in determining the magnitude of rotavirus replication in the intestinal epithelium (Broquet et al., 2011; Ding et al., 2018; Sen et al., 2011). Murine NoV (MNoV) infects both IECs and immune cells including macrophages, DCs, B cells, and T cells in GALT (Grau et al., 2017; Wilen et al., 2018), and sensing of these ssRNA viruses has been shown to rely on TLR3, TLR7, and MDA5, followed by phosphorylation of IRF3 and IRF7 through two independent pathways in infected cells (MacDuff et al., 2018; McCartney et al., 2008; Thackray et al., 2012). Cytosolic NLRs also contribute to control of enteric virus infections (Kanneganti, 2010). NLRP6 senses viral RNA via the RNA helicase Dhx15 and interacts with MAVS to induce both type I and type III IFNs, and Nlrp6-deficient mice exhibit increased intestinal viral loads of multiple enteric viruses (Wang et al., 2015). NLRP9b, which is specifically expressed in IECs, restricts the replication of rotavirus in IECs, indirectly binding short rotavirus-derived dsRNAs via the RNA helicase DHX9, leading to inflammasome formation, release of active IL-18, and pyroptosis of the infected cell (Zhu et al., 2017). It has been suggested that NLRP6 and NLRP9b, expressed in different regions of the intestine, may cooperate in defense against enteric viruses with distinct tropisms (Li and Zhu, 2020). Sensing of intestinal eukaryotic viruses is thus tightly controlled by a variety of PRRs.
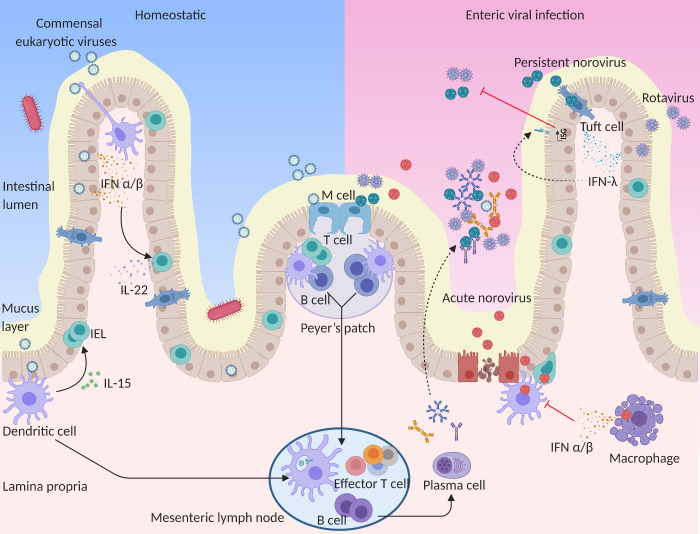
Figure 2.
Eukaryotic virus interactions with the intestinal immune system. In homeostatic states, commensal viruses stimulate basal type I IFN and IL-15 expression that maintains the antiviral state and sustains intestinal homeostasis. Type I IFNs stimulate IL-22 production from innate lymphoid cells to protect IECs, and IL-15 promotes IEL biogenesis. In the case of enteric viral infections, IFN-λ is induced, likely from epithelial cells, to protect IECs from epithelial tropic viruses including rotavirus, NoV, and astrovirus. Viral pathogens can also transit through the epithelium to infect or induce immune responses from lymphocytes or phagocytes in the lamina propria. The induction of type I IFNs mediates broad systemic control of infections but can also contribute to consequent immune pathology. During viral infection, intestinal phagocytes including lamina propria DCs, macrophages, and DCs in the epithelial dome of Peyer’s patches can also sample viral antigens at various sites. Antigen-laden DCs migrate to the mesenteric lymph nodes, where they present processed antigen and induce T and B cell responses. Induced B cells can differentiate into PCs, which home to the lamina propria and secrete antigen-specific IgG, IgM, and IgA. These Igs transcytose across the epithelial cell layer to provide protection against intestinal viral infection. Solid lines indicate the migration of immune cells, while dashed lines indicate secretion of antibodies and cytokines to the intestinal lumen. Image created with BioRender.
Ultimately, host sensing of eukaryotic virus sensing culminates in production of different types of IFN, which serve to control several aspects of enteric viral infection (Ingle et al., 2018). Although MNoV infections in immunocompetent mice are typically asymptomatic, mice lacking type I IFN signaling succumb to lethal infection by acute MNoV strains (e.g., CW3, MNV-1; Karst et al., 2003; McCartney et al., 2008; Nice et al., 2016; Thackray et al., 2012). However, for persistent MNoV infection (e.g., CR6), type I IFNs do not control viral loads in the intestine but prevent spread of virus from the intestine to systemic sites. Instead, IFN-λ has a profound antiviral effect on persistent and acute MNoV infection (Baldridge et al., 2017; Baldridge et al., 2015; Grau et al., 2020; Nice et al., 2015). Spatial as well as kinetic differences in type I and type III IFN responses highlight IFN-λ as important for front-line protection at the intestinal epithelial barrier, while type I IFNs affect broad systemic control of infections and consequent immune pathology (Ingle et al., 2018). For other enteric viruses such as rotavirus, type I and III IFNs both mediate development-specific roles in viral control (Lin et al., 2016; Pott et al., 2011).
Indeed, IFN responses are so central to regulation of enteric virus infection that many viruses have developed strategies to evade PRR detection, IFN production, and downstream activation of antiviral proteins (Beachboard and Horner, 2016; Ingle et al., 2018). For instance, secretion of nonstructural protein NS1 from a persistent MNV strain facilitates evasion of IFN-λ–mediated antiviral immunity and IEC tropism (Lee et al., 2019; Lee et al., 2017). The nonstructural protein 1 (NSP1) and structural protein VP3 of rotavirus also directly antagonize host innate immune responses. Putative E3 ubiquitin ligase NSP1 mediates degradation of a wide range of cellular factors including RIG-I and IRFs, while VP3 utilizes a 2H-phosphodiesterase domain to prevent activation of antiviral factors (Morelli et al., 2015).
Importantly, the innate response to intestinal viral infections does not only affect the inducing virus but may also confer interference against secondary enteric virus infections (Ingle et al., 2019; Wang et al., 2012). Although mechanisms of viral interference in the intestine have not yet been well studied, the presence of murine astrovirus in immunodeficient mice confers protection against other enteric viral challenges via the induction of IFN-λ (Fig. 2; Ingle et al., 2019). Conversely, it is possible that viral adaptations to evade immune responses may in some circumstances limit innate responses to multiple coinfecting viruses, though this remains to be explored.
Adaptive responses to enteric eukaryotic viruses
Adaptive cellular and humoral immune responses are essential for the control of commensal microbes and eradication of invading viruses (Da Silva et al., 2017; Spencer and Sollid, 2016). As the major antigen-presenting cells of the intestinal immune system, conventional DCs that have acquired antigens migrate to the intestinal-draining mesenteric lymph nodes in a CCR7-dependent manner, where they present processed antigen to cells of the adaptive immune system (Jang et al., 2006). Previous studies have found that both B cell and T cell responses are required for efficient clearance of NoV and rotavirus infection from the intestine and mesenteric lymph nodes (Fig. 2; Chachu et al., 2008a; Chachu et al., 2008b; Offit, 1994; Ray et al., 2007). MNoV-specific CD8+ T cells of the intestinal mucosa are a crucial component of NoV immunity for acute strains, though persistent strains induce fewer and less functional T cells and avoid adaptive immune responses in their immune-privileged niche (Tomov et al., 2013; Tomov et al., 2017). Functional human NoV-specific memory T cell responses have been demonstrated in healthy adults and children, and recent ex vivo expansion of these T cells has highlighted the potential of adoptively transferred T cell therapy for the treatment of chronic NoV infection (Hanajiri et al., 2020; Malm et al., 2019; Malm et al., 2016). Murine rotavirus–specific CD8+ T cells play an important role in the timely resolution of primary infection, and they can also provide short-term partial protection against reinfection (Franco et al., 2006). Cellular adaptive immune responses are thus important for pathogen clearance.
Intestinal humoral immune responses are driven chronically by enteric antigens, ultimately generating the largest population of antibody-producing plasma cells (PCs) in the body, which play crucial roles in shaping the persistent enteric humoral response and maintaining intestinal homeostasis (Fig. 2). The PCs of the gastrointestinal mucosa secrete IgA, IgM, and IgG to control and prevent infection; these antibodies may act in a variety of ways to neutralize viral infections. Secretory IgA is a key component in host–microbiota interactions, as defects in secretory IgA lead to overstimulation of innate immunity by IECs and dysbiosis of the intestinal microbiota (Kawamoto et al., 2012; Shulzhenko et al., 2011). Human NoV–specific IgA has been shown to confer protection from gastroenteritis and is also inversely correlated with both virus levels in stool and duration of virus shedding, indicating that IgA is likely important for viral clearance (Ramani et al., 2015). IgA-mediated protection against human NoV may rely specifically on blocking viral attachment to cellular receptors (Sapparapu et al., 2016). Previous studies have shown that rotavirus-specific B cell activation depends on plasmacytoid DCs and that rotavirus-primed memory B cells provide long-term protection against rotavirus infection (Deal et al., 2013; Franco et al., 2006). Anti-rotavirus non-neutralizing IgA antibodies, which can be developed by infants and young children with natural infections or following rotavirus vaccination, are protective in vivo (Afchangi et al., 2019; Burns et al., 1996) and, interestingly, can transmit into IECs via transcytosis to block rotavirus maturation inside cells (Aiyegbo et al., 2013). While Igs clearly play an important role in the host antiviral response, further investigation will be critical to fully understand the complex interactions between humoral immune responses and eukaryotic viral pathogens.
Adaptive immune responses have also been shown to be critical for maintenance of a homeostatic virome. In the context of dysfunctional T cell responses secondary to simian immunodeficiency virus (SIV) or HIV/AIDS, substantial virome alterations have been observed. Pathogenic SIV infection in rhesus monkeys is associated with a dramatically expanded enteric virome, including adenoviruses, caliciviruses, parvoviruses, and picornaviruses, and in gorillas with expansion of herpesviruses and reoviruses (D’Arc et al., 2018; Handley et al., 2012). Vaccination against SIV protects animals from emergence of these enteropathogens (Handley et al., 2016). In humans, low CD4+ T cell counts in HIV-infected individuals are similarly associated with expansion of enteric adenoviruses and anelloviruses (Monaco et al., 2016). Individuals with other sources of immunocompromise also exhibit increased prevalence, duration, and severity of enteric viral infections (Brown et al., 2016; Daniel-Wayman et al., 2018; Green, 2014). Enteric graft-versus-host disease patients, after allogeneic hematopoietic stem cell transplantation, exhibit a significant increase in the richness of persistent eukaryotic viruses (Legoff et al., 2017). Indeed, the presence of a dsRNA Picobirnaviridae species is predictive of later development of graft-versus-host disease, suggesting this could be an immune target associated with dysregulated inflammation (Legoff et al., 2017). Intriguingly, the human eukaryotic virome has also been associated with protection from disease. Specifically, Circoviridae were enriched in healthy controls compared with cases when the enteric virome was analyzed longitudinally in a cohort of children at risk for autoimmune disease type 1 diabetes (Zhao et al., 2017), suggesting the possibility that this viral family limits immune-mediated disease. Studies thus far emphasize the importance of adaptive immune control of enteric viral pathogens, but continued exploration of virome changes in the context of human immune disease will be critical to full understanding of virome effects.
Influences of the collective virome on immunity
Although PRRs can react to both intestinal bacteriophages and eukaryotic viruses, virome-wide sensing by multiple PRRs and the collective impact of these communities on intestinal homeostasis and inflammatory responses are just beginning to be explored. Several groups have used antiviral treatment of mice to deplete the virome to uncover two distinct pathways by which the virome mediates protective effects (Lee and Baldridge, 2019). In one study, broad antiviral treatment was found to reduce TLR3- or TLR7-driven production of tonic levels of type I IFN in the intestine (Yang et al., 2016), thereby sensitizing mice to colitis induced by chemical colitogen dextran sodium sulfate (DSS). Supporting this observation, TLR3, TLR7, and TLR9 agonists effectively improve DSS-induced colitis by stimulating type I IFN responses (Katakura et al., 2005; Sainathan et al., 2012; Vijay-Kumar et al., 2007). Similarly, MNoV infection alone can help to restore intestinal morphology and lymphocyte function in germ-free or antibiotic-treated mice by inducing type I IFN signaling (Kernbauer et al., 2014), indicating that a single virus may recapitulate effects of the collective virome. MNoV can also protect antibiotic-treated mice from severe infection by the bacterial pathogen Citrobacter rodentium and enhance colonization resistance to vancomycin-resistant Enterococcus (Abt et al., 2016; Kernbauer et al., 2014). Type I IFN signaling mediates these protective effects by stimulating IL-22 production from innate lymphoid cells, in turn promoting pSTAT3 signaling in IECs to protect from intestinal injury (Fig. 2; Neil et al., 2019). Similar signaling pathways may also contribute to protection of nonobese diabetic mice from development of type 1 diabetes with MNoV infection (Pearson et al., 2019). Importantly, human patients with combined TLR3 and TLR7 genetic variants display greater severity of ulcerative colitis (Yang et al., 2016), and in clinical trials, topically applied TLR9 agonists can induce remission in ulcerative colitis patients with moderate to severe activity (Atreya et al., 2016; Atreya et al., 2018; Schmitt et al., 2020). These results indicate sensing of the virome by TLRs and TLR-mediated type I IFN responses help protect against or ameliorate enteric inflammation.
The recent identification of a second pathway by which the virome regulates the intestinal immune system suggests multiple complex contributions of commensal viruses to intestinal homeostasis. Treatment of mice with an antiviral cocktail revealed that the commensal virome activates a type I and type III IFN–independent, unconventional RIG-I–MAVS-IRF1 signaling pathway to induce production of the cytokine IL-15 in DCs (Liu et al., 2019). IL-15 subsequently promotes the biogenesis of intraepithelial lymphocytes (IELs), which are the first line of immune cells in the intestinal epithelium and thus play an important role in safeguarding the integrity of the epithelial barrier (Fig. 2; Cheroutre et al., 2011; Liu et al., 2019). IELs are highly relevant to antiviral defense, as activation of IELs in vivo rapidly provokes type I and type III IFN responses to induce ISG expression in IECs (Swamy et al., 2015). Administration of IL-15 protects antiviral-treated mice, as well as Mavs-deficient mice, from sensitivity to DSS-induced colitis, providing another mechanism by which the virome promotes protective effects in the intestine (Liu et al., 2019). It is likely that other pathways by which the complex enteric virome controls and promotes intestinal homeostasis remain to be discovered.
While thus far we have discussed the beneficial effects of the enteric virome on the host, it must be emphasized that these protective effects are context dependent. It has also been reported that, in the setting of host genetic risk factors or immunodeficiency, commensal viruses can contribute to pathological effects in the intestine (Cadwell et al., 2010; Chamaillard et al., 2014). For instance, a common variant of ATG16L1, a gene that mediates cellular autophagy, is associated with increased susceptibility to Crohn’s disease and with morphological defects in Paneth cells (Cadwell et al., 2010). MNoV infection of Atg16L1-mutant mice enhances intestinal pathology, sensitizing Paneth cells to TNF-α–mediated necroptosis in response to viral infection (Matsuzawa-Ishimoto et al., 2017). Similarly, mice deficient in Il10 are also susceptible to MNoV-triggered inflammation (Basic et al., 2014; Bolsega et al., 2019). Further, though MNoV may protect susceptible nonobese diabetic mice from type 1 diabetes, rotavirus infection has the opposite effect, instead accelerating disease, acting through TLR7 to trigger type I IFN signaling to activate lymphocytes (Pane et al., 2016; Pane et al., 2014). Thus, both host-specific and virome-specific factors can ultimately contribute to observed disease phenotypes.
Conclusions and future directions
Over the last two decades, our understanding of the interplay between the virome and the immune system has increased dramatically, but there is still much dark matter to explore. Improved understanding of the fundamental components of the virome, in both health and disease and in human cohorts as well as animal models, is crucial. Further refinement of viral databases and improvements in long- and short-read sequencing approaches, as well as improved capture of RNA genomes, will greatly facilitate resolution of metaviromes. Ongoing efforts to cultivate diverse phage collections will hopefully yield model phage communities for future experimental application, and recognition and cultivation of individual novel eukaryotic viruses will also be critically important to our understanding of how the virome mediates effects.
Many of our experimental models to test the role of the virome can also be expanded, with more specific antiviral treatments or use of germ-free mice for colonization with defined viral species. Much intestinal virome research has relied upon single virus infection models, and while these have proven incredibly informative, expansion into coinfection models or use of a greater diversity of virus types will help to separate common from unique consequences of infections. Further, it will continue to be important to consider the intestinal—and extraintestinal—effects that the virome has on innate and adaptive immune responses, with an appreciation that host context is key.
Finally, while reviewed extensively elsewhere (Robinson, 2019; Walker and Baldridge, 2019), better understanding of the interactions between the prokaryotic and eukaryotic components of the virome and the bacteria, fungi, and other organisms comprising the microbiota is a critical future direction. Identifying how the virome is regulated by and exploits the rest of the intestinal ecosystem will facilitate our understanding of how these communities contribute to intestinal immune regulation and pathophysiology. Virome–immune interactions is a new and rapidly growing field likely to facilitate development of antiviral therapies and vaccines. Further studies are greatly needed to fully define the contribution of viruses to intestinal immunity, as well as to the development of intestinal diseases and other complex inflammatory disorders.
Acknowledgments
The authors thank Forrest Walker, Sanghyun Lee, and Heyde Makimaa for helpful comments on the manuscript.
M.T. Baldridge was supported by National Institutes of Health grants R01 AI127552, R01 AI139314, and R01 AI141478, a Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital Interdisciplinary Research Initiative grant (MI-II-2019–790), the Pew Biomedical Scholars Program, and the Mathers Foundation. S.A. Handley was supported by National Institutes of Health grants RC2 DK116713-01A1 and U01 AI151810.
Author contributions: Y. Li, S.A. Handley, and M.T. Baldridge wrote, read, and edited the manuscript. All authors read and have agreed to the final version of the manuscript.
References
- Abt, M.C., Buffie C.G., Sušac B., Becattini S., Carter R.A., Leiner I., Keith J.W., Artis D., Osborne L.C., and Pamer E.G.. 2016. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci. Transl. Med. 8:327ra25. 10.1126/scitranslmed.aad6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afchangi, A., Jalilvand S., Mohajel N., Marashi S.M., and Shoja Z.. 2019. Rotavirus VP6 as a potential vaccine candidate. Rev. Med. Virol. 29:e2027. 10.1002/rmv.2027 [DOI] [PubMed] [Google Scholar]
- Ahlgren, N.A., Ren J., Lu Y.Y., Fuhrman J.A., and Sun F.. 2017. Alignment-free $d_2^*$ oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 45:39–53. 10.1093/nar/gkw1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyegbo, M.S., Sapparapu G., Spiller B.W., Eli I.M., Williams D.R., Kim R., Lee D.E., Liu T., Li S., Woods V.L. Jr., et al. 2013. Human rotavirus VP6-specific antibodies mediate intracellular neutralization by binding to a quaternary structure in the transcriptional pore. PLoS One. 8:e61101. 10.1371/journal.pone.0061101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, G.M.F., Laanto E., Ashrafi R., and Sundberg L.R.. 2019. Bacteriophage Adherence to Mucus Mediates Preventive Protection against Pathogenic Bacteria. MBio. 10:e01984-19. 10.1128/mBio.01984-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, R., Bloom S., Scaldaferri F., Gerardi V., Admyre C., Karlsson Å., Knittel T., Kowalski J., Lukas M., Löfberg R., et al. 2016. Clinical Effects of a Topically Applied Toll-like Receptor 9 Agonist in Active Moderate-to-Severe Ulcerative Colitis. J. Crohn’s Colitis. 10:1294–1302. 10.1093/ecco-jcc/jjw103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, R., Reinisch W., Peyrin-Biroulet L., Scaldaferri F., Admyre C., Knittel T., Kowalski J., Neurath M.F., and Hawkey C.. 2018. Clinical efficacy of the Toll-like receptor 9 agonist cobitolimod using patient-reported-outcomes defined clinical endpoints in patients with ulcerative colitis. Dig. Liver Dis. 50:1019–1029. 10.1016/j.dld.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Baldridge, M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., Diamond M.S., Ivanova Y., Artyomov M., and Virgin H.W.. 2015. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 347:266–269. 10.1126/science.1258025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge, M.T., Lee S., Brown J.J., McAllister N., Urbanek K., Dermody T.S., Nice T.J., and Virgin H.W.. 2017. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J. Virol. 91:e02079-16. 10.1128/JVI.02079-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, J.J., Auro R., Furlan M., Whiteson K.L., Erb M.L., Pogliano J., Stotland A., Wolkowicz R., Cutting A.S., Doran K.S., et al. 2013. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA. 110:10771–10776. 10.1073/pnas.1305923110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylski, J., Enault F., Dutilh B.E., Schuller M.B., Edwards R.A., Gillis A., Klumpp J., Knezevic P., Krupovic M., Kuhn J.H., et al. 2020. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Phages. Syst. Biol. 69:110–123. 10.1093/sysbio/syz036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic, M., Keubler L.M., Buettner M., Achard M., Breves G., Schröder B., Smoczek A., Jörns A., Wedekind D., Zschemisch N.H., et al. 2014. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm. Bowel Dis. 20:431–443. 10.1097/01.MIB.0000441346.86827.ed [DOI] [PubMed] [Google Scholar]
- Beachboard, D.C., and Horner S.M.. 2016. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 32:113–119. 10.1016/j.mib.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid, Y., and Hand T.W.. 2014. Role of the microbiota in immunity and inflammation. Cell. 157:121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Jambrina, M., Eder J., Helgers L.C., Hertoghs N., Nijmeijer B.M., Stunnenberg M., and Geijtenbeek T.B.H.. 2018. C-Type Lectin Receptors in Antiviral Immunity and Viral Escape. Front. Immunol. 9:590. 10.3389/fimmu.2018.00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolsega, S., Basic M., Smoczek A., Buettner M., Eberl C., Ahrens D., Odum K.A., Stecher B., and Bleich A.. 2019. Composition of the Intestinal Microbiota Determines the Outcome of Virus-Triggered Colitis in Mice. Front. Immunol. 10:1708. 10.3389/fimmu.2019.01708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet, A.H., Hirata Y., McAllister C.S., and Kagnoff M.F.. 2011. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 186:1618–1626. 10.4049/jimmunol.1002862 [DOI] [PubMed] [Google Scholar]
- Broudy, T.B., and Fischetti V.A.. 2003. In vivo lysogenic conversion of Tox(-) Streptococcus pyogenes to Tox(+) with Lysogenic Streptococci or free phage. Infect. Immun. 71:3782–3786. 10.1128/IAI.71.7.3782-3786.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.R., Shah D., and Breuer J.. 2016. Viral gastrointestinal infections and norovirus genotypes in a paediatric UK hospital, 2014-2015. J. Clin. Virol. 84:1–6. 10.1016/j.jcv.2016.08.298 [DOI] [PubMed] [Google Scholar]
- Buffie, C.G., and Pamer E.G.. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13:790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, J.W., Siadat-Pajouh M., Krishnaney A.A., and Greenberg H.B.. 1996. Protective effect of rotavirus VP6-specific IgA monoclonal antibodies that lack neutralizing activity. Science. 272:104–107. 10.1126/science.272.5258.104 [DOI] [PubMed] [Google Scholar]
- Cadwell, K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., and Virgin H.W.. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141:1135–1145. 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callanan, J., Stockdale S.R., Shkoporov A., Draper L.A., Ross R.P., and Hill C.. 2018. RNA Phage Biology in a Metagenomic Era. Viruses. 10:386. 10.3390/v10070386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachu, K.A., LoBue A.D., Strong D.W., Baric R.S., and Virgin H.W.. 2008a. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 4:e1000236. 10.1371/journal.ppat.1000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachu, K.A., Strong D.W., LoBue A.D., Wobus C.E., Baric R.S., and Virgin H.W. IV. 2008b. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 82:6610–6617. 10.1128/JVI.00141-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard, M., Cesaro A., Lober P.E., and Hober D.. 2014. Decoding norovirus infection in Crohn’s disease. Inflamm. Bowel Dis. 20:767–770. 10.1097/01.MIB.0000440613.83703.4a [DOI] [PubMed] [Google Scholar]
- Chen, J., and Novick R.P.. 2009. Phage-mediated intergeneric transfer of toxin genes. Science. 323:139–141. 10.1126/science.1164783 [DOI] [PubMed] [Google Scholar]
- Cheroutre, H., Lambolez F., and Mucida D.. 2011. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11:445–456. 10.1038/nri3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P., Richet H., Desnues C., Balique F., Moal V., Grob J.J., Berbis P., Lecoq H., Harlé J.R., Berland Y., and Raoult D.. 2010. Pepper mild mottle virus, a plant virus associated with specific immune responses, Fever, abdominal pains, and pruritus in humans. PLoS One. 5:e10041. 10.1371/journal.pone.0010041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, L.2016. Polyomaviruses. Microbiol. Spectr. 4. 10.1128/microbiolspec.DMIH2-0010-2015 [DOI] [PubMed] [Google Scholar]
- Corman, V.M., Muth D., Niemeyer D., and Drosten C.. 2018. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 100:163–188. 10.1016/bs.aivir.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuault, J.K., Petit M.A., Mariadassou M., Benevides L., Moncaut E., Langella P., Sokol H., and De Paepe M.. 2018. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 6:65. 10.1186/s40168-018-0452-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’arc, M., Furtado C., Siqueira J.D., Seuánez H.N., Ayouba A., Peeters M., and Soares M.A.. 2018. Assessment of the gorilla gut virome in association with natural simian immunodeficiency virus infection. Retrovirology. 15:19. 10.1186/s12977-018-0402-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, C., Wagner C., Bonnardel J., Gorvel J.P., and Lelouard H.. 2017. The Peyer’s Patch Mononuclear Phagocyte System at Steady State and during Infection. Front. Immunol. 8:1254. 10.3389/fimmu.2017.01254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dąbrowska, K., Miernikiewicz P., Piotrowicz A., Hodyra K., Owczarek B., Lecion D., Kaźmierczak Z., Letarov A., and Górski A.. 2014. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 88:12551–12557. 10.1128/JVI.02043-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Wayman, S., Fahle G., Palmore T., Green K.Y., and Prevots D.R.. 2018. Norovirus, astrovirus, and sapovirus among immunocompromised patients at a tertiary care research hospital. Diagn. Microbiol. Infect. Dis. 92:143–146. 10.1016/j.diagmicrobio.2018.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe, M., Leclerc M., Tinsley C.R., and Petit M.A.. 2014. Bacteriophages: an underestimated role in human and animal health? Front. Cell. Infect. Microbiol. 4:39. 10.3389/fcimb.2014.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, E.M., Lahl K., Narváez C.F., Butcher E.C., and Greenberg H.B.. 2013. Plasmacytoid dendritic cells promote rotavirus-induced human and murine B cell responses. J. Clin. Invest. 123:2464–2474. 10.1172/JCI60945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S., Zhu S., Ren L., Feng N., Song Y., Ge X., Li B., Flavell R.A., and Greenberg H.B.. 2018. Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. eLife. 7:e39494. 10.7554/eLife.39494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M.B., Oechslin F., and Moineau S.. 2020. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18:125–138. 10.1038/s41579-019-0311-5 [DOI] [PubMed] [Google Scholar]
- Duerkop, B.A., and Hooper L.V.. 2013. Resident viruses and their interactions with the immune system. Nat. Immunol. 14:654–659. 10.1038/ni.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh, B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K., et al. 2014. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5:4498. 10.1038/ncomms5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa, N., Egawa K., Griffin H., and Doorbar J.. 2015. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses. 7:3863–3890. 10.3390/v7072802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlet, C., Ruffin M., and Lamendella R.. 2020. Enteric Virome and Carcinogenesis in the Gut. Dig. Dis. Sci. 65:852–864. 10.1007/s10620-020-06126-4 [DOI] [PubMed] [Google Scholar]
- Faruque, S.M., Rahman M.M., Asadulghani K.M., Nasirul Islam K.M., and Mekalanos J.J.. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXPhi. Infect. Immun. 67:5723–5729. 10.1128/IAI.67.11.5723-5729.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier, A.L., Burmeister W.P., and Chroboczek J.. 2004. Unique physicochemical properties of human enteric Ad41 responsible for its survival and replication in the gastrointestinal tract. Virology. 322:93–104. 10.1016/j.virol.2004.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillol-Salom, A., Alsaadi A., Sousa J.A.M., Zhong L., Foster K.R., Rocha E.P.C., Penadés J.R., Ingmer H., and Haaber J.. 2019. Bacteriophages benefit from generalized transduction. PLoS Pathog. 15:e1007888. 10.1371/journal.ppat.1007888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focà, A., Liberto M.C., Quirino A., Marascio N., Zicca E., and Pavia G.. 2015. Gut inflammation and immunity: what is the role of the human gut virome? Mediators Inflamm. 2015:326032. 10.1155/2015/326032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, M.A., Angel J., and Greenberg H.B.. 2006. Immunity and correlates of protection for rotavirus vaccines. Vaccine. 24:2718–2731. 10.1016/j.vaccine.2005.12.048 [DOI] [PubMed] [Google Scholar]
- Garmaeva, S., Sinha T., Kurilshikov A., Fu J., Wijmenga C., and Zhernakova A.. 2019. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 17:84. 10.1186/s12915-019-0704-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensollen, T., Iyer S.S., Kasper D.L., and Blumberg R.S.. 2016. How colonization by microbiota in early life shapes the immune system. Science. 352:539–544. 10.1126/science.aad9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna, G., Baldanti F., and Revello M.G.. 2004. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65:381–386. 10.1016/j.humimm.2004.02.009 [DOI] [PubMed] [Google Scholar]
- Girons, I.S., Bourhy P., Ottone C., Picardeau M., Yelton D., Hendrix R.W., Glaser P., and Charon N.. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700–5705. 10.1128/JB.182.20.5700-5705.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogokhia, L., Buhrke K., Bell R., Hoffman B., Brown D.G., Hanke-Gogokhia C., Ajami N.J., Wong M.C., Ghazaryan A., Valentine J.F., et al. 2019. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe. 25:285–299.e8. 10.1016/j.chom.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg, D.B., Paer J.M., Luevano J.M., and Kwon D.S.. 2017. HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation. Curr. Opin. Infect. Dis. 30:31–43. 10.1097/QCO.0000000000000341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski, A., Kniotek M., Perkowska-Ptasińska A., Mróz A., Przerwa A., Gorczyca W., Dabrowska K., Weber-Dabrowska B., and Nowaczyk M.. 2006. Bacteriophages and transplantation tolerance. Transplant. Proc. 38:331–333. 10.1016/j.transproceed.2005.12.073 [DOI] [PubMed] [Google Scholar]
- Górski, A., Dąbrowska K., Międzybrodzki R., Weber-Dąbrowska B., Łusiak-Szelachowska M., Jończyk-Matysiak E., and Borysowski J.. 2017. Phages and immunomodulation. Future Microbiol. 12:905–914. 10.2217/fmb-2017-0049 [DOI] [PubMed] [Google Scholar]
- Grau, K.R., Roth A.N., Zhu S., Hernandez A., Colliou N., DiVita B.B., Philip D.T., Riffe C., Giasson B., Wallet S.M., et al. 2017. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat. Microbiol. 2:1586–1591. 10.1038/s41564-017-0057-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau, K.R., Zhu S., Peterson S.T., Helm E.W., Philip D., Phillips M., Hernandez A., Turula H., Frasse P., Graziano V.R., et al. 2020. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 5:84–92. 10.1038/s41564-019-0602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, K.Y.2014. Norovirus infection in immunocompromised hosts. Clin. Microbiol. Infect. 20:717–723. 10.1111/1469-0691.12761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, K.Y., Kaufman S.S., Nagata B.M., Chaimongkol N., Kim D.Y., Levenson E.A., Tin C.M., Yardley A.B., Johnson J.A., Barletta A.B.F., et al. 2020. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat. Commun. 11:2759. 10.1038/s41467-020-16491-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, A.C., Zablocki O., Howell A., Bolduc B., and Sullivan M.B.. 2019. The human gut virome database. bioRxiv. 655910 (Preprint posted May 31, 2019).
- Gregory, A.C., Zablocki O., Zayed A.A., Howell A., Bolduc B., and Sullivan M.B.. 2020. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe. 28:724–740.e8. 10.1016/j.chom.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin, E., Shkoporov A., Stockdale S.R., Clooney A.G., Ryan F.J., Sutton T.D.S., Draper L.A., Gonzalez-Tortuero E., Ross R.P., and Hill C.. 2018. Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut. Cell Host Microbe. 24:653–664.e6. 10.1016/j.chom.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Guido, M., Tumolo M.R., Verri T., Romano A., Serio F., De Giorgi M., De Donno A., Bagordo F., and Zizza A.. 2016. Human bocavirus: Current knowledge and future challenges. World J. Gastroenterol. 22:8684–8697. 10.3748/wjg.v22.i39.8684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanajiri, R., Sani G.M., Saunders D., Hanley P.J., Chopra A., Mallal S.A., Sosnovtsev S.V., Cohen J.I., Green K.Y., Bollard C.M., and Keller M.D.. 2020. Generation of Norovirus-Specific T Cells From Human Donors With Extensive Cross-Reactivity to Variant Sequences: Implications for Immunotherapy. J. Infect. Dis. 221:578–588. 10.1093/infdis/jiz491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley, S.A., Thackray L.B., Zhao G., Presti R., Miller A.D., Droit L., Abbink P., Maxfield L.F., Kambal A., Duan E., et al. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 151:253–266. 10.1016/j.cell.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley, S.A., Desai C., Zhao G., Droit L., Monaco C.L., Schroeder A.C., Nkolola J.P., Norman M.E., Miller A.D., Wang D., et al. 2016. SIV Infection-Mediated Changes in Gastrointestinal Bacterial Microbiome and Virome Are Associated with Immunodeficiency and Prevented by Vaccination. Cell Host Microbe. 19:323–335. 10.1016/j.chom.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, G.2017. Nucleic Acid Immunity. Adv. Immunol. 133:121–169. 10.1016/bs.ai.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino, S., and Miyata H.. 2007. Torque teno virus (TTV): current status. Rev. Med. Virol. 17:45–57. 10.1002/rmv.524 [DOI] [PubMed] [Google Scholar]
- Hodyra-Stefaniak, K., Miernikiewicz P., Drapała J., Drab M., Jończyk-Matysiak E., Lecion D., Kaźmierczak Z., Beta W., Majewska J., Harhala M., et al. 2015. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep. 5:14802. 10.1038/srep14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, G.S., Lodge J.K., McCarthy A.J., Graham A.K., Young G., Bridge S.H., Brown A.K., Veses-Garcia M., Lanyon C.V., Sails A., et al. 2017. Shigatoxin encoding Bacteriophage ϕ24B modulates bacterial metabolism to raise antimicrobial tolerance. Sci. Rep. 7:40424. 10.1038/srep40424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., Yanai H., Takaoka A., and Taniguchi T.. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367–1378. 10.1093/intimm/dxh318 [DOI] [PubMed] [Google Scholar]
- Howard-Varona, C., Hargreaves K.R., Abedon S.T., and Sullivan M.B.. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11:1511–1520. 10.1038/ismej.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyles, L., McCartney A.L., Neve H., Gibson G.R., Sanderson J.D., Heller K.J., and van Sinderen D.. 2014. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 165:803–812. 10.1016/j.resmic.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Hsu, B.B., Gibson T.E., Yeliseyev V., Liu Q., Lyon L., Bry L., Silver P.A., and Gerber G.K.. 2019. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe. 25:803–814.e5. 10.1016/j.chom.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle, H., Peterson S.T., and Baldridge M.T.. 2018. Distinct Effects of Type I and III Interferons on Enteric Viruses. Viruses. 10:46. 10.3390/v10010046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle, H., Lee S., Ai T., Orvedahl A., Rodgers R., Zhao G., Sullender M., Peterson S.T., Locke M., Liu T.C., et al. 2019. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 4:1120–1128. 10.1038/s41564-019-0416-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., and Medzhitov R.. 2010. Regulation of adaptive immunity by the innate immune system. Science. 327:291–295. 10.1126/science.1183021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., and Medzhitov R.. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16:343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, M.H., Sougawa N., Tanaka T., Hirata T., Hiroi T., Tohya K., Guo Z., Umemoto E., Ebisuno Y., Yang B.G., et al. 2006. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J. Immunol. 176:803–810. 10.4049/jimmunol.176.2.803 [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.D.2010. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10:688–698. 10.1038/nri2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusinszky, B., Minor P., and Delwart E.. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J. Clin. Microbiol. 50:3427–3434. 10.1128/JCM.01589-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandikar, U.C., Crawford S.E., Ajami N.J., Murakami K., Kou B., Ettayebi K., Papanicolaou G.A., Jongwutiwes U., Perales M.A., Shia J., et al. 2016. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol. 97:2291–2300. 10.1099/jgv.0.000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst, S.M., Wobus C.E., Lay M., Davidson J., and Virgin H.W. IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 299:1575–1578. 10.1126/science.1077905 [DOI] [PubMed] [Google Scholar]
- Katakura, K., Lee J., Rachmilewitz D., Li G., Eckmann L., and Raz E.. 2005. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J. Clin. Invest. 115:695–702. 10.1172/JCI22996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T., and Akira S.. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Kawamoto, S., Tran T.H., Maruya M., Suzuki K., Doi Y., Tsutsui Y., Kato L.M., and Fagarasan S.. 2012. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 336:485–489. 10.1126/science.1217718 [DOI] [PubMed] [Google Scholar]
- Kernbauer, E., Ding Y., and Cadwell K.. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 516:94–98. 10.1038/nature13960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Mirzaei, M., Xue J., Costa R., Ru J., Schulz S., Taranu Z.E., and Deng L.. 2021. Challenges of Studying the Human Virome - Relevant Emerging Technologies. Trends Microbiol. 29:171–181. 10.1016/j.tim.2020.05.021 [DOI] [PubMed] [Google Scholar]
- King, A.M., Lefkowitz E., Adams M.J., and Carstens E.B.. 2011. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, London. [Google Scholar]
- Kolawole, A.O., Mirabelli C., Hill D.R., Svoboda S.A., Janowski A.B., Passalacqua K.D., Rodriguez B.N., Dame M.K., Freiden P., Berger R.P., et al. 2019. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 15:e1008057. 10.1371/journal.ppat.1008057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoto, S., and Taniguchi K.. 2014. [Rotaviruses]. Uirusu. 64:179–190. 10.2222/jsv.64.179 [DOI] [PubMed] [Google Scholar]
- Kortman, G.A., Raffatellu M., Swinkels D.W., and Tjalsma H.. 2014. Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol. Rev. 38:1202–1234. 10.1111/1574-6976.12086 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, S.R., and Wang D.. 2017. Origins and challenges of viral dark matter. Virus Res. 239:136–142. 10.1016/j.virusres.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, S.R., and Wang D.. 2018. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology. 516:108–114. 10.1016/j.virol.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Krupovic, M., and Forterre P.. 2011. Microviridae goes temperate: microvirus-related proviruses reside in the genomes of Bacteroidetes. PLoS One. 6:e19893. 10.1371/journal.pone.0019893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, H., Kawai T., and Akira S.. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16–34. 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- Lamers, M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science. 369:50–54. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, D., Baldridge M.T., and Handley S.A.. 2019. Phages and Human Health: More Than Idle Hitchhikers. Viruses. 11:587. 10.3390/v11070587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear, H.M., Schoggins J.W., and Diamond M.S.. 2019. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 50:907–923. 10.1016/j.immuni.2019.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., and Baldridge M.T.. 2019. Viruses RIG up intestinal immunity. Nat. Immunol. 20:1563–1564. 10.1038/s41590-019-0530-y [DOI] [PubMed] [Google Scholar]
- Lee, S., Wilen C.B., Orvedahl A., McCune B.T., Kim K.W., Orchard R.C., Peterson S.T., Nice T.J., Baldridge M.T., and Virgin H.W.. 2017. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell Host Microbe. 22:449–459.e4. 10.1016/j.chom.2017.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Liu H., Wilen C.B., Sychev Z.E., Desai C., Hykes B.L. Jr., Orchard R.C., McCune B.T., Kim K.W., Nice T.J., et al. 2019. A Secreted Viral Nonstructural Protein Determines Intestinal Norovirus Pathogenesis. Cell Host Microbe. 25:845–857.e5. 10.1016/j.chom.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.J., Kopetz S., Vilar E., Shen J.P., Chen K., and Maitra A.. 2020. Relative Abundance of SARS-CoV-2 Entry Genes in the Enterocytes of the Lower Gastrointestinal Tract. Genes (Basel). 11:645. 10.3390/genes11060645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoff, J., Resche-Rigon M., Bouquet J., Robin M., Naccache S.N., Mercier-Delarue S., Federman S., Samayoa E., Rousseau C., Piron P., et al. 2017. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat. Med. 23:1080–1085. 10.1038/nm.4380 [DOI] [PubMed] [Google Scholar]
- Lekunberri, I., Subirats J., Borrego C.M., and Balcázar J.L.. 2017. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 220(Pt B):981–984. 10.1016/j.envpol.2016.11.059 [DOI] [PubMed] [Google Scholar]
- Lepage, P., Colombet J., Marteau P., Sime-Ngando T., Doré J., and Leclerc M.. 2008. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 57:424–425. 10.1136/gut.2007.134668 [DOI] [PubMed] [Google Scholar]
- Li, R., and Zhu S.. 2020. NLRP6 inflammasome. Mol. Aspects Med. 76:100859. 10.1016/j.mam.2020.100859 [DOI] [PubMed] [Google Scholar]
- Li, L., Kapoor A., Slikas B., Bamidele O.S., Wang C., Shaukat S., Masroor M.A., Wilson M.L., Ndjango J.B., Peeters M., et al. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84:1674–1682. 10.1128/JVI.02109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G., Zhao C., Zhang H., Mattei L., Sherrill-Mix S., Bittinger K., Kessler L.R., Wu G.D., Baldassano R.N., DeRusso P., et al. 2020. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature. 581:470–474. 10.1038/s41586-020-2192-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M., Warner B.B., Tarr P.I., Wang D., and Holtz L.R.. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21:1228–1234. 10.1038/nm.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.M., and Lin H.C.. 2019. A theoretical model of temperate phages as mediators of gut microbiome dysbiosis. F1000 Res. 8:997. 10.12688/f1000research.18480.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.D., Feng N., Sen A., Balan M., Tseng H.C., McElrath C., Smirnov S.V., Peng J., Yasukawa L.L., Durbin R.K., et al. 2016. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 12:e1005600. 10.1371/journal.ppat.1005600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Gong T., Tao W., Lin B., Li C., Zheng X., Zhu S., Jiang W., and Zhou R.. 2019. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat. Immunol. 20:1681–1691. 10.1038/s41590-019-0513-z [DOI] [PubMed] [Google Scholar]
- Lopetuso, L.R., Ianiro G., Scaldaferri F., Cammarota G., and Gasbarrini A.. 2016. Gut Virome and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 22:1708–1712. 10.1097/MIB.0000000000000807 [DOI] [PubMed] [Google Scholar]
- Lozupone, C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., and Knight R.. 2012. Diversity, stability and resilience of the human gut microbiota. Nature. 489:220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff, D.A., Baldridge M.T., Qaqish A.M., Nice T.J., Darbandi A.D., Hartley V.L., Peterson S.T., Miner J.J., Iwai K., and Virgin H.W.. 2018. HOIL1 Is Essential for the Induction of Type I and III Interferons by MDA5 and Regulates Persistent Murine Norovirus Infection. J. Virol. 92:e01368-18. 10.1128/JVI.01368-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska, J., Beta W., Lecion D., Hodyra-Stefaniak K., Kłopot A., Kaźmierczak Z., Miernikiewicz P., Piotrowicz A., Ciekot J., Owczarek B., et al. 2015. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses. 7:4783–4799. 10.3390/v7082845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska, J., Kaźmierczak Z., Lahutta K., Lecion D., Szymczak A., Miernikiewicz P., Drapała J., Harhala M., Marek-Bukowiec K., Jędruchniewicz N., et al. 2019. Induction of Phage-Specific Antibodies by Two Therapeutic Staphylococcal Bacteriophages Administered per os. Front. Immunol. 10:2607. 10.3389/fimmu.2019.02607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, Y.S., Kumar N., Sharma K., Dhama K., Shabbir M.Z., Ganesh B., Kobayashi N., and Banyai K.. 2014. Epidemiology, phylogeny, and evolution of emerging enteric Picobirnaviruses of animal origin and their relationship to human strains. BioMed Res. Int. 2014:780752. 10.1155/2014/780752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm, M., Tamminen K., Vesikari T., and Blazevic V.. 2016. Norovirus-Specific Memory T Cell Responses in Adult Human Donors. Front. Microbiol. 7:1570. 10.3389/fmicb.2016.01570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm, M., Hyöty H., Knip M., Vesikari T., and Blazevic V.. 2019. Development of T cell immunity to norovirus and rotavirus in children under five years of age. Sci. Rep. 9:3199. 10.1038/s41598-019-39840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique, P., Bolduc B., Walk S.T., van der Oost J., de Vos W.M., and Young M.J.. 2016. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA. 113:10400–10405. 10.1073/pnas.1601060113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique, P., Dills M., and Young M.J.. 2017. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses. 9:141. 10.3390/v9060141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa-Ishimoto, Y., Shono Y., Gomez L.E., Hubbard-Lucey V.M., Cammer M., Neil J., Dewan M.Z., Lieberman S.R., Lazrak A., Marinis J.M., et al. 2017. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214:3687–3705. 10.1084/jem.20170558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, S.A., Thackray L.B., Gitlin L., Gilfillan S., Virgin H.W., and Colonna M.. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108. 10.1371/journal.ppat.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, R.N., Krug A.B., and Eisenächer K.. 2018. Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses. 10:146. 10.3390/v10040146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedzybrodzki, R., Switala-Jelen K., Fortuna W., Weber-Dabrowska B., Przerwa A., Lusiak-Szelachowska M., Dabrowska K., Kurzepa A., Boratynski J., Syper D., et al. 2008. Bacteriophage preparation inhibition of reactive oxygen species generation by endotoxin-stimulated polymorphonuclear leukocytes. Virus Res. 131:233–242. 10.1016/j.virusres.2007.09.013 [DOI] [PubMed] [Google Scholar]
- Miernikiewicz, P., Dąbrowska K., Piotrowicz A., Owczarek B., Wojas-Turek J., Kicielińska J., Rossowska J., Pajtasz-Piasecka E., Hodyra K., Macegoniuk K., et al. 2013. T4 phage and its head surface proteins do not stimulate inflammatory mediator production. PLoS One. 8:e71036. 10.1371/journal.pone.0071036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernikiewicz, P., Kłopot A., Soluch R., Szkuta P., Kęska W., Hodyra-Stefaniak K., Konopka A., Nowak M., Lecion D., Kaźmierczak Z., et al. 2016. T4 Phage Tail Adhesin Gp12 Counteracts LPS-Induced Inflammation In Vivo. Front. Microbiol. 7:1112. 10.3389/fmicb.2016.01112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot, S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., and Bushman F.D.. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–1625. 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot, S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., and Bushman F.D.. 2013. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA. 110:12450–12455. 10.1073/pnas.1300833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei, M.K., and Maurice C.F.. 2017. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15:397–408. 10.1038/nrmicro.2017.30 [DOI] [PubMed] [Google Scholar]
- Modi, S.R., Lee H.H., Spina C.S., and Collins J.J.. 2013. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 499:219–222. 10.1038/nature12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco, C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S., Lankowski A., Baldridge M.T., Wilen C.B., Flagg M., et al. 2016. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 19:311–322. 10.1016/j.chom.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli, M., Ogden K.M., and Patton J.T.. 2015. Silencing the alarms: Innate immune antagonism by rotavirus NSP1 and VP3. Virology. 479-480:75–84. 10.1016/j.virol.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhya, I., Segal J.P., Carding S.R., Hart A.L., and Hold G.L.. 2019. The gut virome: the ‘missing link’ between gut bacteria and host immunity? Therap. Adv. Gastroenterol. 12. 10.1177/1756284819836620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazik, H., Joubert L.M., Secor P.R., Sweere J.M., Bollyky P.L., Sass G., Cegelski L., and Stevens D.A.. 2017. Pseudomonas phage inhibition of Candida albicans. Microbiology (Reading). 163:1568–1577. 10.1099/mic.0.000539 [DOI] [PubMed] [Google Scholar]
- Neil, J.A., and Cadwell K.. 2018. The Intestinal Virome and Immunity. J. Immunol. 201:1615–1624. 10.4049/jimmunol.1800631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil, J.A., Matsuzawa-Ishimoto Y., Kernbauer-Hölzl E., Schuster S.L., Sota S., Venzon M., Dallari S., Galvao Neto A., Hine A., Hudesman D., et al. 2019. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat. Microbiol. 4:1737–1749. 10.1038/s41564-019-0470-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, K.L., Moe C.L., Kirby A.E., Flanders W.D., Parkos C.A., and Leon J.S.. 2016. Norovirus in symptomatic and asymptomatic individuals: cytokines and viral shedding. Clin. Exp. Immunol. 184:347–357. 10.1111/cei.12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice, T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., and Virgin H.W.. 2015. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 347:269–273. 10.1126/science.1258100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice, T.J., Osborne L.C., Tomov V.T., Artis D., Wherry E.J., and Virgin H.W.. 2016. Type I Interferon Receptor Deficiency in Dendritic Cells Facilitates Systemic Murine Norovirus Persistence Despite Enhanced Adaptive Immunity. PLoS Pathog. 12:e1005684. 10.1371/journal.ppat.1005684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, J.M., Handley S.A., Baldridge M.T., Droit L., Liu C.Y., Keller B.C., Kambal A., Monaco C.L., Zhao G., Fleshner P., et al. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 160:447–460. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oechslin, N., Moradpour D., and Gouttenoire J.. 2020. Hepatitis E virus finds its path through the gut. Gut. 69:796–798. 10.1136/gutjnl-2019-320206 [DOI] [PubMed] [Google Scholar]
- Offit, P.A.1994. Rotaviruses: immunological determinants of protection against infection and disease. Adv. Virus Res. 44:161–202. 10.1016/S0065-3527(08)60329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, H.2009. History of discoveries and pathogenicity of TT viruses. Curr. Top. Microbiol. Immunol. 331:1–20. 10.1007/978-3-540-70972-5_1 [DOI] [PubMed] [Google Scholar]
- Onoguchi, K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., and Fujita T.. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 282:7576–7581. 10.1074/jbc.M608618200 [DOI] [PubMed] [Google Scholar]
- Oude Munnink, B.B., and van der Hoek L.. 2016. Viruses Causing Gastroenteritis: The Known, The New and Those Beyond. Viruses. 8:42. 10.3390/v8020042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, J.A., Webster N.L., and Coulson B.S.. 2014. Rotavirus activates lymphocytes from non-obese diabetic mice by triggering toll-like receptor 7 signaling and interferon production in plasmacytoid dendritic cells. PLoS Pathog. 10:e1003998. 10.1371/journal.ppat.1003998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, J.A., Fleming F.E., Graham K.L., Thomas H.E., Kay T.W., and Coulson B.S.. 2016. Rotavirus acceleration of type 1 diabetes in non-obese diabetic mice depends on type I interferon signalling. Sci. Rep. 6:29697. 10.1038/srep29697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, I.K., and Iwasaki A.. 2012. Control of antiviral immunity by pattern recognition and the microbiome. Immunol. Rev. 245:209–226. 10.1111/j.1600-065X.2011.01073.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, J.A., Tai N., Ekanayake-Alper D.K., Peng J., Hu Y., Hager K., Compton S., Wong F.S., Smith P.C., and Wen L.. 2019. Norovirus Changes Susceptibility to Type 1 Diabetes by Altering Intestinal Microbiota and Immune Cell Functions. Front. Immunol. 10:2654. 10.3389/fimmu.2019.02654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner, J.C., Ferreira J.A.G., Secor P.R., Sweere J.M., Birukova M.K., Joubert L.M., Haagensen J.A.J., Garcia O., Malkovskiy A.V., Kaber G., et al. 2016. Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology (Reading). 162:1583–1594. 10.1099/mic.0.000344 [DOI] [PubMed] [Google Scholar]
- Peterson, L.W., and Artis D.. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14:141–153. 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- Pfeiffer, J.K., and Virgin H.W.. 2016. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 351:aad5872. 10.1126/science.aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popgeorgiev, N., Temmam S., Raoult D., and Desnues C.. 2013. Describing the silent human virome with an emphasis on giant viruses. Intervirology. 56:395–412. 10.1159/000354561 [DOI] [PubMed] [Google Scholar]
- Pott, J., Mahlakõiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., and Hornef M.W.. 2011. IFN-λ determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA. 108:7944–7949. 10.1073/pnas.1100552108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principi, N., Silvestri E., and Esposito S.. 2019. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 10:513. 10.3389/fphar.2019.00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyun, K.H., Ochs H.D., Wedgwood R.J., Yang X.Q., Heller S.R., and Reimer C.B.. 1989. Human antibody responses to bacteriophage phi X 174: sequential induction of IgM and IgG subclass antibody. Clin. Immunol. Immunopathol. 51:252–263. 10.1016/0090-1229(89)90024-X [DOI] [PubMed] [Google Scholar]
- Ramani, S., Neill F.H., Opekun A.R., Gilger M.A., Graham D.Y., Estes M.K., and Atmar R.L.. 2015. Mucosal and Cellular Immune Responses to Norwalk Virus. J. Infect. Dis. 212:397–405. 10.1093/infdis/jiv053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, P.G., Kelkar S.D., Walimbe A.M., Biniwale V., and Mehendale S.. 2007. Rotavirus immunoglobulin levels among Indian mothers of two socio-economic groups and occurrence of rotavirus infections among their infants up to six months. J. Med. Virol. 79:341–349. 10.1002/jmv.20804 [DOI] [PubMed] [Google Scholar]
- Reyes, A., Semenkovich N.P., Whiteson K., Rohwer F., and Gordon J.I.. 2012. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10:607–617. 10.1038/nrmicro2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadulla, E., and Romalde J.L.. 2020. A Comprehensive Review on Human Aichi Virus. Virol. Sin. 35:501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.M.2019. Enteric viruses exploit the microbiota to promote infection. Curr. Opin. Virol. 37:58–62. 10.1016/j.coviro.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, I., Gibson G., Heinken A., Scott K., Swann J., Thiele I., and Tuohy K.. 2018. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57:1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainathan, S.K., Bishnupuri K.S., Aden K., Luo Q., Houchen C.W., Anant S., and Dieckgraefe B.K.. 2012. Toll-like receptor-7 ligand Imiquimod induces type I interferon and antimicrobial peptides to ameliorate dextran sodium sulfate-induced acute colitis. Inflamm. Bowel Dis. 18:955–967. 10.1002/ibd.21867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rodriguez, T.M., and Hollister E.B.. 2019. Human Virome and Disease: High-Throughput Sequencing for Virus Discovery, Identification of Phage-Bacteria Dysbiosis and Development of Therapeutic Approaches with Emphasis on the Human Gut. Viruses. 11:656. 10.3390/v11070656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapparapu, G., Czakó R., Alvarado G., Shanker S., Prasad B.V., Atmar R.L., Estes M.K., and Crowe J.E. Jr. 2016. Frequent Use of the IgA Isotype in Human B Cells Encoding Potent Norovirus-Specific Monoclonal Antibodies That Block HBGA Binding. PLoS Pathog. 12:e1005719. 10.1371/journal.ppat.1005719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, T., and Akira S.. 2017. Toll-Like Receptor Signaling and Its Inducible Proteins. InMyeloid Cells in Health and Disease. Gordon S., editor. ASM Press, Washington, DC. pp. 447–453. 10.1128/9781555819194.ch24 [DOI] [Google Scholar]
- Schmitt, H., Ulmschneider J., Billmeier U., Vieth M., Scarozza P., Sonnewald S., Reid S., Atreya I., Rath T., Zundler S., et al. 2020. The TLR9 Agonist Cobitolimod Induces IL10-Producing Wound Healing Macrophages and Regulatory T Cells in Ulcerative Colitis. J. Crohn’s Colitis. 14:508–524. 10.1093/ecco-jcc/jjz170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, K., and Tschopp J.. 2010. The inflammasomes. Cell. 140:821–832. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Sen, A., Pruijssers A.J., Dermody T.S., García-Sastre A., and Greenberg H.B.. 2011. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J. Virol. 85:3717–3732. 10.1128/JVI.02634-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkoporov, A.N., Khokhlova E.V., Fitzgerald C.B., Stockdale S.R., Draper L.A., Ross R.P., and Hill C.. 2018. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9:4781. 10.1038/s41467-018-07225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkoporov, A.N., Clooney A.G., Sutton T.D.S., Ryan F.J., Daly K.M., Nolan J.A., McDonnell S.A., Khokhlova E.V., Draper L.A., Forde A., et al. 2019. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe. 26:527–541.e5. 10.1016/j.chom.2019.09.009 [DOI] [PubMed] [Google Scholar]
- Shulzhenko, N., Morgun A., Hsiao W., Battle M., Yao M., Gavrilova O., Orandle M., Mayer L., Macpherson A.J., McCoy K.D., et al. 2011. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 17:1585–1593. 10.1038/nm.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, A., and Maurice C.F.. 2019. Bacteriophages: Uncharacterized and Dynamic Regulators of the Immune System. Mediators Inflamm. 2019:3730519. 10.1155/2019/3730519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol, H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., and Doré J.. 2009. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15:1183–1189. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- Sommereyns, C., Paul S., Staeheli P., and Michiels T.. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. 10.1371/journal.ppat.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandole, S., Cimponeriu D., Berca L.M., and Mihăescu G.. 2015. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch. Virol. 160:893–908. 10.1007/s00705-015-2363-9 [DOI] [PubMed] [Google Scholar]
- Spencer, J., and Sollid L.M.. 2016. The human intestinal B-cell response. Mucosal Immunol. 9:1113–1124. 10.1038/mi.2016.59 [DOI] [PubMed] [Google Scholar]
- Stanfield, B.A., and Luftig M.A.. 2017. Recent advances in understanding Epstein-Barr virus. F1000 Res. 6:386. 10.12688/f1000research.10591.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer, M.L., Pervolaraki K., and Boulant S.. 2019. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 20:1445. 10.3390/ijms20061445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy, M., Abeler-Dörner L., Chettle J., Mahlakõiv T., Goubau D., Chakravarty P., Ramsay G., Reis E Sousa C., Staeheli P., Blacklaws B.A., et al. 2015. Intestinal intraepithelial lymphocyte activation promotes innate antiviral resistance. Nat. Commun. 6:7090. 10.1038/ncomms8090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere, J.M., Van Belleghem J.D., Ishak H., Bach M.S., Popescu M., Sunkari V., Kaber G., Manasherob R., Suh G.A., Cao X., et al. 2019. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science. 363:eaat9691. 10.1126/science.aat9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely, A.J., and Breitbart M.. 2016. Single-stranded DNA phages: from early molecular biology tools to recent revolutions in environmental microbiology. FEMS Microbiol. Lett. 363:fnw027. 10.1093/femsle/fnw027 [DOI] [PubMed] [Google Scholar]
- Tapparel, C., Siegrist F., Petty T.J., and Kaiser L.. 2013. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 14:282–293. 10.1016/j.meegid.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Tetz, G., and Tetz V.. 2018. Bacteriophages as New Human Viral Pathogens. Microorganisms. 6:54. 10.3390/microorganisms6020054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray, L.B., Duan E., Lazear H.M., Kambal A., Schreiber R.D., Diamond M.S., and Virgin H.W.. 2012. Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J. Virol. 86:13515–13523. 10.1128/JVI.01824-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov, V.T., Osborne L.C., Dolfi D.V., Sonnenberg G.F., Monticelli L.A., Mansfield K., Virgin H.W., Artis D., and Wherry E.J.. 2013. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J. Virol. 87:7015–7031. 10.1128/JVI.03389-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov, V.T., Palko O., Lau C.W., Pattekar A., Sun Y., Tacheva R., Bengsch B., Manne S., Cosma G.L., Eisenlohr L.C., et al. 2017. Differentiation and Protective Capacity of Virus-Specific CD8+ T Cells Suggest Murine Norovirus Persistence in an Immune-Privileged Enteric Niche. Immunity. 47:723–738.e5. 10.1016/j.immuni.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon, M., Moura de Sousa J.A., and Rocha E.P.. 2017. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 38:66–73. 10.1016/j.mib.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Uhr, J.W., and Finkelstein M.S.. 1963. Antibody formation. IV. Formation of rapidly and slowly sedimenting antibodies and immunological memory to bacteriophage phi-X 174. J. Exp. Med. 117:457–477. 10.1084/jem.117.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhr, J.W., Finkelstein M.S., and Baumann J.B.. 1962. Antibody formation. III. The primary and secondary antibody response to bacteriophage phi X 174 in guinea pigs. J. Exp. Med. 115:655–670. 10.1084/jem.115.3.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem, J.D., Clement F., Merabishvili M., Lavigne R., and Vaneechoutte M.. 2017. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 7:8004. 10.1038/s41598-017-08336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar, M., Wu H., Aitken J., Kolachala V.L., Neish A.S., Sitaraman S.V., and Gewirtz A.T.. 2007. Activation of toll-like receptor 3 protects against DSS-induced acute colitis. Inflamm. Bowel Dis. 13:856–864. 10.1002/ibd.20142 [DOI] [PubMed] [Google Scholar]
- Virgin, H.W.2014. The virome in mammalian physiology and disease. Cell. 157:142–150. 10.1016/j.cell.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin, H.W., Wherry E.J., and Ahmed R.. 2009. Redefining chronic viral infection. Cell. 138:30–50. 10.1016/j.cell.2009.06.036 [DOI] [PubMed] [Google Scholar]
- Walker, F.C., and Baldridge M.T.. 2019. Interactions between noroviruses, the host, and the microbiota. Curr. Opin. Virol. 37:1–9. 10.1016/j.coviro.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.2020. 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog. 16:e1008318. 10.1371/journal.ppat.1008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Moon S., Wang Y., and Jiang B.. 2012. Multiple virus infection alters rotavirus replication and expression of cytokines and Toll-like receptors in intestinal epithelial cells. Virus Res. 167:48–55. 10.1016/j.virusres.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Wang, P., Zhu S., Yang L., Cui S., Pan W., Jackson R., Zheng Y., Rongvaux A., Sun Q., Yang G., et al. 2015. Nlrp6 regulates intestinal antiviral innate immunity. Science. 350:826–830. 10.1126/science.aab3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen, C.B., Lee S., Hsieh L.L., Orchard R.C., Desai C., Hykes B.L. Jr., McAllaster M.R., Balce D.R., Feehley T., Brestoff J.R., et al. 2018. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 360:204–208. 10.1126/science.aar3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Long X., Ren C., He R., Yan X., Li W., Luo Z., Li Q., Xu H., and Liu E.. 2020. Follow-up Study of Long-time Positive RT-PCR in Stool Specimens From Asymptomatic Children Infected With SARS-CoV-2. Pediatr. Infect. Dis. J. 39:e315–e317. 10.1097/INF.0000000000002837 [DOI] [PubMed] [Google Scholar]
- Yanagida, M.1977. Molecular organization of the shell of T-even bacteriophage head. II. Arrangement of subunits in the head shell of giant phages. J. Mol. Biol. 109:515–537. 10.1016/S0022-2836(77)80089-2 [DOI] [PubMed] [Google Scholar]
- Yang, J.Y., Kim M.S., Kim E., Cheon J.H., Lee Y.S., Kim Y., Lee S.H., Seo S.U., Shin S.H., Choi S.S., et al. 2016. Enteric Viruses Ameliorate Gut Inflammation via Toll-like Receptor 3 and Toll-like Receptor 7-Mediated Interferon-β Production. Immunity. 44:889–900. 10.1016/j.immuni.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Yutin, N., Makarova K.S., Gussow A.B., Krupovic M., Segall A., Edwards R.A., and Koonin E.V.. 2018. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 3:38–46. 10.1038/s41564-017-0053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. 2020. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5:eabc3582. 10.1126/sciimmunol.abc3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W., Hibberd M.L., Liu E.T., Rohwer F., and Ruan Y.. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Hou X., Sun L., He T., Wei R., Pang M., and Wang R.. 2018. Corrigendum: Staphylococcus aureus Bacteriophage Suppresses LPS-Induced Inflammation in MAC-T Bovine Mammary Epithelial Cells. Front. Microbiol. 9:2511. 10.3389/fmicb.2018.02511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, G., Vatanen T., Droit L., Park A., Kostic A.D., Poon T.W., Vlamakis H., Siljander H., Härkönen T., Hämäläinen A.M., et al. 2017. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. USA. 114:E6166–E6175. 10.1073/pnas.1706359114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S., Ding S., Wang P., Wei Z., Pan W., Palm N.W., Yang Y., Yu H., Li H.B., Wang G., et al. 2017. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 546:667–670. 10.1038/nature22967 [DOI] [PMC free article] [PubMed] [Google Scholar]