Abstract
Endophyte mediated nanoparticles fabrication were emerging as a new frontier in nanomedicines that produce high biocompatible and functionalized silver nanoparticles. In this study, silver nanoparticles were successfully biosynthesized from the extracellular extract of endophytic bacterium Pantoea anthophila isolated from the stem of Waltheria indica for the first time. The synthesized nanoparticles showed a strong absorption band at 410 nm in the UV–Visible range. The dynamic light scattering and zeta potential analysis indicated that the average particle size was 16 nm at 5.30 mV. FTIR spectrum displayed the presence of various functional groups at 3423.65, 1633.71, 1022.27, 607.58 cm−1 that stabilised the nanoparticle. X-ray diffraction peaks were conferred to 100, 200, 220 and 311 planes of a face centred cubic structure. TEM and SEM micrograph revealed the spherical-shaped, polycrystalline nature with the presence of elemental silver analysed by EDAX. Selected area electron diffraction also confirms the orientation of silver nanoparticles with X-ray diffraction analysis. Antimicrobial activity against 10 different human pathogenic bacteria and fungi showed a broad spectrum inhibition against both Gram-positive and Gram-negative bacteria. Among the bacterial pathogens, B. Subtilis exhibited low activity compared to other pathogens. C. albicans was greatly controlled than other fungal species. A strong free radical scavenging activity of silver nanoparticles with IC50 values 31.29 ± 0.73, 19.83 ± 1.57, 35.64 ± 0.94, 42.07 ± 1.30, 29.70 ± 2.26, 29.10 ± 0.82, 36.80 ± 0.63 μg/ml was obtained in different antioxidant assays that were comparable to the reference. The study suggests that the silver nanoparticles can be biosynthesized from endophytic P. anthophila metabolites with significant therapeutic potential. With proper validation, the biosynthesized silver nanoparticles can be developed as a promising antiviral and anticancer drug candidate.
Keywords: Pantoea anthophila, Endophytic bacterium, Silver nanoparticles, Characterisation, Antimicrobial activity, Antioxidant activity
Introduction
The potential of human viral and bacterial pathogens causing diseases is rising alarmingly in recent years. The lethal condition caused by these etiological agents leads to a high death mortality rate. Scientific studies also proved that many of these infections trigger the production of reactive oxygen (ROS) and nitrogen species that can cause tissue damage resulting in inflammation. As a consequence, there is an increase in the risk of harmful diseases like acute lung injury, aging, acute renal failure, cardiovascular diseases, diabetes, neurodegenerative diseases, cancer etc. [1, 2]. Though wide-spectrum antibiotics are available they are least effective against infectious agents due to their increase in multi drug resistant nature. Products like carotenoids, vitamins and polyphenols from fruits and vegetables have the ability to modulate oxidative stress in human cells [3, 4]. Yet, low bioavailability and easy degradation during delivery limit their antioxidant activity [5]. The change in the way the prescribed drugs used for the treatment of various diseases or the development of novel candidates with high potency is the urgent need of the hour.
Nanotechnology, an emerging research area in science and technology has gained importance in biomedical, cosmetics, imaging, cancer therapy and targeted drug delivery [6, 7]. Nanostructures and nanodevices with controlled shape and size are designed and characterized for various applications by this innovative technique. Different types of metal nanoparticles like gold, silver, alginate, copper, titanium, zinc, magnesium, etc., are proved to have profound pharmacological activity [8]. Among the metallic nanoparticles, nanosilver is extensively researched for its wide-ranging applications in different fields. Sensors with silver-based nanocomposites and nanocatalysts have been developed for the removal of pollutants from environmental sample and health monitoring [9–11]. The good electrical conductivity and potential optical property of silver nanoparticles (AgNPs) attracted their role as catalysts in chemical reactions, as intercalating materials for electrical batteries, used in selective coatings for solar energy absorption, etc., [12, 13]. The broad-spectrum activity, low toxicity and unique physiological properties such as size, morphology and surface chemistry of AgNPs [14, 15] acquired an interest in the development of antimicrobial [16], anti-plasmodial [17], anti-platelet [18], anti-tumour [19], topical creams and wound healing [20] pharmaceutical products. Factors like cell type and reducing agents from which the NPs are produced also influences the biological activity of AgNPs that extends the applications in constructions, plant science, veterinary, food, ecology, and electronics. Its potency to rupture thick cell wall, inducing oxidative stress, inhibiting proteins, enzymes and DNA replication circumvented the infections and multi drug-resistant nature of microbial strains [21]. This antimicrobial property directed to the development of products like AgNPs integrated healing bandage, antiseptics and medical devices for sterilizing surfaces and air. AgNPs also proved to have significant cytotoxic activity in cancer cell lines and degenerative disease by reducing ROS [22, 23]. Its distinctive ability to cross various biological barriers, targeted delivery of drugs, increased synergistic effects of chemotherapeutic drugs with AgNPs, high biocompatibility with human cells lead to the design of Ag-based nanomaterials for medical applications [24].
Nanoparticles are synthesized by physicochemical methods that are costly, highly toxic and time-consuming. An eco-friendly, sustainable technique has to be developed for the production of AgNPs for various medical challenges. Biosynthesis of metal nanoparticles gained considerable attention in the past decade. Moreover, it has been proposed as a less toxic, cost-effective, environmentally friendly alternative to chemical and physical methods. Biological sources such as plants, microbes (bacteria and fungi) and biomolecules (amino acids and vitamins) are recognized to reduce metal ions to nano metals [25, 26]. The biomoieties in the green extracts such as proteins, enzymes, vitamins, alcoholic compounds, alkaloids, flavonoids, phenols, quinones, terpenoids, tannins, saponins, sugars, sulfhydryls, pigments, amino acids, etc., influence the fabrication of nanoparticles that increases its microbial inhibitory and cellular toxicity nature, yet it is least toxic to human and environment [27].
Waltheria indica, a tropical medicinal plant is used traditionally in the treatment of cough, skin diseases, microbial infections and inflammatory diseases [28–30]. Endophytes have opened a new direction of exploration in nanoparticle synthesis. They colonize the internal tissues of plants, diversify between species, organs and play a key role in plant growth and defence. They are well known for the inimitable intracellular and extracellular bioactive metabolites that act as the major source of drugs against human diseases. Endophytic fungi from different plants are extensively studied for nanoparticle synthesis yet the potency of endophytic bacteria in nano research is at the primitive stage [31, 32]. We explored the diversity of endophytic bacteria in the stem parts of W. indica and identified a novel bacterium Pantoea anthophila for the first time. The multidimensional property of endophytes may produce stable nanoparticles with high medicinal potency. This bottom-up approach may generate targeted nanomaterials at very high efficiency.
Nanoparticles biosynthesized extracellularly requires simple downstream processing and is cost-effective while intracellular synthesis involves additional purification steps to release the synthesized nanoparticles. The negligible use of toxic chemicals and sustainable large scale production of AgNPs is an added advantage to explore its potential applications. Hence current research widely focuses on the extracellular synthesis of AgNPs stabilised by bacterial metabolites that control their size, shape and dispersity [33, 34]. Based on these considerations, the present study has been designed to undertake the extracellular biosynthesis of silver nanoparticles (Pa-AgNPs) from P. anthophila and evaluate its therapeutic potential. To the best of our knowledge, this study is the first report on biofabrication, characterization, antimicrobial and antioxidant potentials of AgNPs biosynthesized from the bacterium P. anthophila isolated from W.indica plant. The study would also recommend the application of endophytic bacteria for nanoparticles synthesis in pharmaceutical applications as an alternative greener approach.
Materials and Methods
Chemicals, Endophytic Bacteria, and Microbial Strains
Analytical grade silver nitrate (AgNO3, 99% pure), tetracycline, kanamycin, nutrient agar, potato dextrose agar, Mueller–Hinton agar used in the present study are purchased from Himedia Lab, Ltd., Mumbai, India. The endophytic bacteria Pantoea anthophila (GenBank accession no. MN077163) identified earlier by 16S ribosomal RNA gene sequencing from W. indica were pure cultured and stored in the Department of Biotechnology, Vinayaka Mission's Kirupananda Variyar Engineering College, Salem. Bacterial pathogens such as Gram-positive Staphylococcus epidermidis (MTCC737), Bacillus subtilis (MTCC1133), Staphylococcus aureus (MTCC2940), Gram-negative Escherichia coli (MTCC40), Proteus mirabilis (MTCC425), Salmonella typhi (MTCC733), Klebsiella pneumoniae (MTCC2405) and fungal strains of Aspergillus niger (MTCC404), Candida albicans (MTCC183) and Penicillium chrysogenum (MTCC947) were obtained from Microbial Type Culture Collection (MTCC), Chandigarh.
Preparation of Cell-Free Endophytic Culture
Pantoea anthophila (Pa) isolated from W. indica are subcultured and maintained in nutrient agar plates. A loopful of Pa culture is inoculated in 100 ml of Luria–Bertani broth at 37 °C and placed in an orbital shaker at 200 rpm overnight. When the optical density (OD) of the culture was 0.242 at 600 nm, it was taken in 50 ml falcon vials and centrifuged at 10,000 rpm for 10 min in a high-speed centrifuge (RM-03 Plus). The extracellular broth devoid of cells were collected in a sterile beaker and used for AgNP synthesis.
Biosynthesis of Pa-AgNPs
The 10 ml of bacterial cell-free suspension was mixed with 90 ml of 1 mM (2%, v/v) AgNO3 solution in deionized water for the biosynthesis of AgNPs at neutral pH. The reaction mixture was incubated at 30 °C for 24 h in dark to avoid any photochemical reactions [35, 36]. Simultaneously, the cell-free supernatant without AgNO3 was maintained as control. The formation of Pa-AgNPs was monitored by the colour change in the bacterial extract and separated by centrifuging at 10,000 rpm for 15 min. The water-soluble biological molecules and other impurities are removed by washing the samples repeatedly (× 3) with deionized water. The final mass of the AgNPs was collected and freeze-dried.
Biophysical Characterization of Pa-AgNPs
The Pa-AgNPs are further characterized by UV–Visible spectroscopy, Dynamic Light scattering analysis (DLS), Zeta potential, Fourier Transform Infrared Spectroscopy (FTIR), X-ray Diffraction (XRD), Transmission Electron Microscopy (TEM), Scanning Electron Microscopy (SEM), Energy Dispersive of X-Ray spectrum (EDAX).
The formation of AgNPs was confirmed by measuring the absorption spectra in UV–Visible spectroscopy (SHIMADZU 1800). The reaction mixture was scanned at the speed of 300 nm min−1 in 200–600 nm range. DLS analysis determines the particle size and distribution pattern of Pa-AgNP. Zeta sizer (Malvern Instruments Ltd., U.K ZS90) was used to determine the surface charge potential, the magnitude of charge attraction or repulsion and electrophoretic stability of Pa-AgNP.
The FTIR analysis was carried out to identify the functional biomolecules in the endophytic extract that reduces Ag+ to AgNPs and the capping agents that stabilize the nanoparticles. The Pa-AgNPs is scanned at the transmission mode ranging from 500 to 4000 cm−1 in a solid phase, at a resolution of 1 cm−1 using FTIR spectrophotometer (SHIMADZU, IR PRESTIGE 21). XRD pattern provides the details of the structure and composition of Pa-AgNPs. The sample was analyzed by Shimadzu XRD 6000 diffractometer operated at 40 kV voltage, 30 mA current in a scanning mode range of θ–2θ between 10° and 80° with sampling pitch of 0.1000° equipped with a Cu Kα radiation.
The crystallite structure of the Pa-AgNP was measured using TEM (JEOL JEM 2100). The sample was placed on copper grids coated carbon films, dried at room temperature and analysed at 200 kV. SEM shows the size, shape and surface morphological properties of Pa-AgNP synthesized from the endophytic extract. The nanoparticle was observed in SEM (JEOL-JSM 6390) at a voltage of 15–20 kV at different magnifications. The atomic composition of Pa-AgNP was confirmed by EDAX analysis (Oxford instrument, INCA PentaFET × 3) coupled with SEM.
Antimicrobial Screening of Pa-AgNPs
The antimicrobial activity of the synthesized Pa-AgNPs was analysed by Agar well diffusion method [37] in triplicates. The bacterial strains S. epidermidis, B. subtilis, S. aureus, E. coli, P. mirabilis, S. typhi, K. pneumonia were inoculated in nutrient media and incubated at 37 °C for 2–4 h. At the exponential growth phase, the organisms are cultured in Muller Hinton agar plates in which six wells of about 5 mm diameter were bored. Different concentrations of Pa-AgNPs (25 µg/ml, 50 µg/ml, 75 µg/ml, 100 µg/ml), 10 µg/ml of dimethyl sulfoxide (DMSO) (negative control) and Tetracycline (positive control) were added to the wells.
Similarly, the fungal strains A. niger, C. albicans and P. chrysogenum were inoculated in potato dextrose agar in which the antifungal activity of Pa-AgNPs at different concentrations (25 µg/ml, 50 µg/ml, 75 µg/ml, 100 µg/ml) are evaluated with Kanamycin (10 µg/ml) as a positive control. The plates were incubated for 24 h at 37 °C to observe the clear zones of inhibition (ZOI). The diameter of clear zones was measured and recorded.
Antioxidant Efficacy of Pa-AgNPs
1, 1-Diphenyl-2-picrylhydrazyl (DPPH) Radical Scavenging Assay
DPPH radical scavenging assay potential of Pa-AgNPs was assayed [38] at different concentrations (10, 20, 30, 40 & 50 μg/ml) of Pa-AgNPs and standard ascorbic acid. In methanol solution, dissolved 50 μl of 0.659 mM DPPH added to the samples and the volume is made up to one with double distilled water. The tubes were incubated in dark at 25 °C for 20 min and the absorbance was recorded at 510 nm using Shimadzu UV 1800 spectrophotometer. The % inhibition (I%) was calculated as (I%) = 100 × (A0–A1)/A0, Where A0 is the absorbance of the control, A1 is the absorbance of the Pa-AgNPs and standard.
Nitric Oxide Radical Scavenging Assay
At physiological pH, in an aqueous solution, sodium nitroprusside generates nitric oxide that interacts with oxygen to produce nitrite ions, which can be measured in the presence of Griess reagent [38]. To various concentrations (10–50 μg/ml) of Pa-AgNPs and standard ascorbic acid, added 50 μl of 10 mM sodium nitroprusside dissolved in 0.5 M phosphate buffer (pH 7.4) and incubated under fluorescent light at room temperature for 15 min. Added 125 μl of Griess reagent, the tubes were incubated again at room temperature for 10 min and the absorbance was recorded at 546 nm.
Hydrogen Peroxide (H2O2) Scavenging Assay
The ability of the Pa-AgNPs to scavenge H2O2 was determined according to the method of Nabavi et al. [39]. 0.6 ml of 40 mM of H2O2 was prepared using 50 mM phosphate buffer (pH 7.4) and added to varied concentrations (10–50 μg/ml) of Pa-AgNPs and standard ascorbic acid. The tubes were incubated for 10 min and the absorbance was noted at 230 nm.
Total Antioxidant Capacity Assay
The total antioxidant capacity assay was determined [40] by adding 1 ml of reagent solution containing sulphuric acid (0.6 M), sodium phosphate (28 mM) and ammonium molybdate (4 mM) to Pa-AgNPs and standard ascorbic acid (10–50 μg/ml). The tubes were capped, incubated at 95 °C for 90 min in a thermal block and then cooled to room temperature, the absorbance was measured at 695 nm.
2,2′-Azino-bis-3-Ethyl Benzothiozoline-6-Sulfonic Acid (ABTS) Radical Scavenging Activity
The assay is based on the scavenging of light by ABTS radicals. An antioxidant that donates a hydrogen atom will quench the stable free radical which can be quantified spectrometrically at 734 nm [38]. 200 μl of 70 mM potassium persulphate and 50 ml of 2 mM ABTS were mixed before 2hours. To the 0.5 ml of various concentrations (10-50 μg/ml) of Pa-AgNPs and standard ascorbic acid, 0.3 ml of ABTS radical cation and 1.7 ml of phosphate buffer (pH 7.4) was added and the absorbance was measured.
Reducing Power Assay
Different concentrations (10–50 μg/ml) of Pa-AgNPs solution were mixed with 2.5 ml of 200 mM phosphate buffer (pH 6.6) and 1% potassium ferricyanide each. Incubated at 50 °C for 20 min, the mixture was cooled rapidly. Subsequently, added 2.5 ml of 10% Trichloroacetic acid and centrifuged at 3000 rpm for 10 min. The supernatant (5 ml) with an equal amount of distilled water were mixed and added to 1 ml of 0.1% ferric chloride. Using ascorbic acid as a standard, the absorbance was measured at 700 nm [41].
Superoxide (O2−) Radical Scavenging Assay
The superoxide scavenging activity of the Pa-AgNPs was assayed by the reduction of nitro blue tetrazolium (NBT) [42]. To Pa-AgNPs solution (10-50 μg/ml), 3 ml Tris–HCl buffer (16 mM, pH 8), 1 ml NBT (50 μM), 1 ml Nicotinamide Adenine Dinucleotide (78 μM) and 1 ml phenazine methosulfate (PMS) solution (10 μM) were mixed and kept for 5 min at 25 °C. The absorbance was recorded at 560 nm. Ascorbic acid was used as standard.
Inhibition % versus concentration curve was plotted for each assay and the concentration of sample required for 50% inhibition was determined and expressed as IC50 value. The lower IC50 value indicates a high antioxidant capacity.
Results and Discussion
Biosynthesis and Characterization Studies of Pa-AgNPs
UV–Visible Spectroscopy
The yellow colour of bacterial extract, when mixed with AgNO3 turned dark brown after 24 h (Fig. 1b), at the same time, no colour change was detected in the control (Fig. 1a). This colour change is due to the reduction of Ag+ to Ag0 in the aqueous solution that occurs due to excitation of surface plasmon resonance (SPR) in AgNPs [43]. The free electrons of Ag metal oscillate in resonance with reflected light and induce this phenomenon. Also, the shape, size and the interaction of bacterial metabolites with AgNPs influences this property that can be measured by spectroscopic methods. Usually, the SPR peak of AgNPs lies in the range of 400–500 nm. Pa-AgNPs exhibited a maximum absorption at the wavelength 410 nm that confirms the characteristics of AgNPs (Fig. 2). Khatoon et al. [44] reported similar UV absorption spectra for chemically synthesized AgNPs that had promising antifungal activity. Similar peaks were also reported in many studies [35, 45] relevant to the biosynthesis of AgNPs.
Fig. 1.
a Endophytic bacterial extract (Control). b Endophytic bacterial extract mixed with AgNO3 after 24 h
Fig. 2.
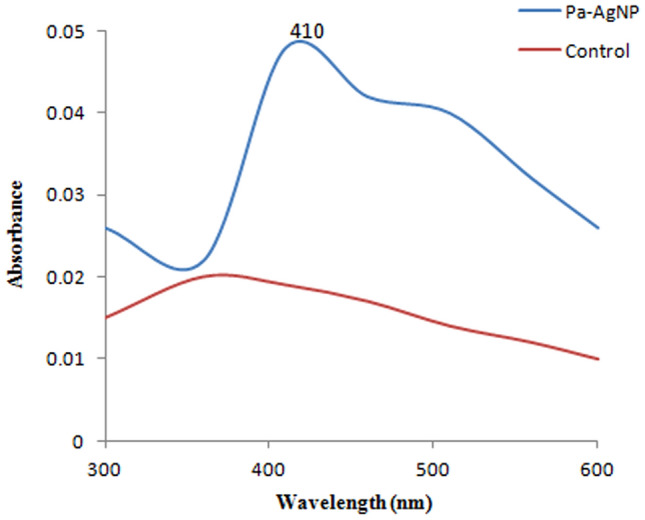
UV–visible absorption spectrum of Pa-AgNPs
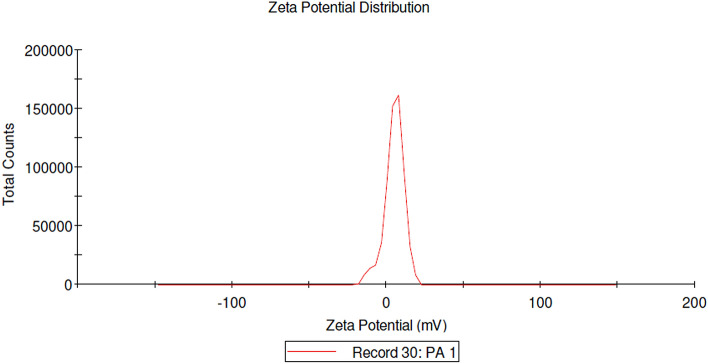
DLS and Zeta Potential Analysis
The bioefficiency of the AgNPs depends on the size and surface charge that prevents agglomeration. The DLS analysis (Fig. 3) showed that the average size of Pa-AgNPs was 16 nm and the size varies from 10 to 24 nm. The zeta potential of the Pa-AgNPs was found as a sharp peak at 5.30 mV (Fig. 4) which indicates that the surface of synthesized nanoparticles was positively charged. This positive charge may be due to the capping of bioactive metabolites on the surface. Ali et al. [46] showed zeta potential value of AgNPs synthesized from apple extract was 5.68 ± 3.28 mV that had strong agglomeration and precipitation. Prakasham et al. [47] also synthesized AgNPs from marine Streptomyces albidoflavus that had − 8.5 mV zeta potential with strong antimicrobial activity. In agreement with the above reports, Pa-AgNPs had the same zeta potential for more than 100 days that proved their stability with strong antimicrobial and antioxidant activities.
Fig. 3.
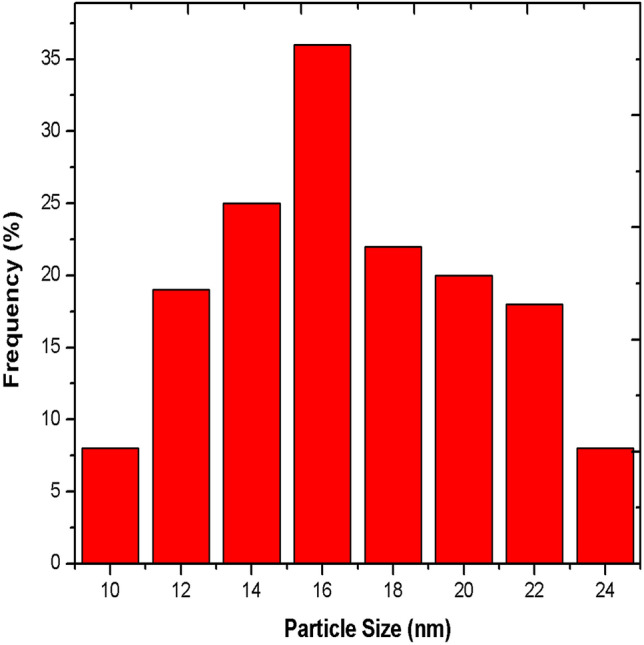
Size distribution of Pa-AgNPs by Dynamic Light scattering analysis technique
Fig. 4.
Graphical presentation of Zeta potential of Pa-AgNPs
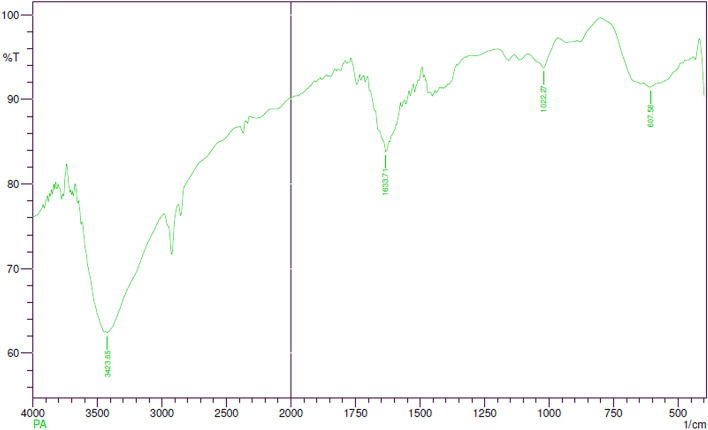
FTIR Spectroscopy Study
FTIR spectrum displayed the presence of predominant bands at 3423.65, 1633.71, 1022.27, 607.58 cm−1 (Fig. 5). The peak at 3423.65 cm−1 corresponds to the O–H stretching vibrations of water molecules. The bands at 1633.71 cm−1 show C=C stretching vibrations of the non-conjugated, disubstituted alkene group. The band at 1022.27 cm−1 can be assigned to medium C–N stretching vibrations of amines signifying the presence of amino acid. The peak obtained at 607.58 cm−1 regions could be attributed to alkyl halides stretching. The results obtained coincide with the earlier findings of similar AgNPs FTIR prediction [36, 48] that revealed the presence of hydroxyl, alkene and amine groups in the bacterial extract acting as reducing, capping and stabilizing agents of silver ions.
Fig. 5.
Fourier Transform Infrared spectrum of Pa-AgNPs
XRD Analysis
XRD analysis showed the strongest Braggs peaks at 2θ values of 38.30°, 44.62°, 65.01° and 77.27° (Fig. 6). These peaks are conferred to 100, 200, 220 and 311 planes of a face centred cubic structure (FCC) of Pa-AgNPs that agrees with the JCPDS file no. 87-0720 for silver [49]. The crystallite domain sizes of Pa-AgNPs were calculated using Debye–Scherrer formula [50] D = 0.94 λ/βcosθ, where D is the average crystallite domain size perpendicular to the reflecting planes, λ is the X-ray wavelength, β is the full width at half maximum (FWHM), and θ is the diffraction angle. The average crystallite size was found to be 16.8 nm which was in close agreement with DLS analysis. No secondary peaks were observed, hence the crystalline stable AgNP formation from the endophytic extract is justified with the previous XRD data green synthesized from endophytic bacteria, fungi and plant extracts [36, 51, 52].
Fig. 6.
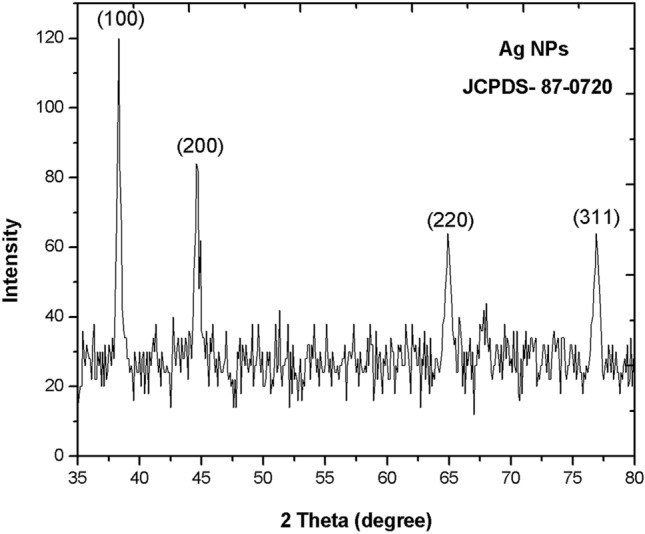
X-ray Diffraction analysis of crystal structure of Pa-AgNPs
TEM Measurement
The antimicrobial activity of AgNPs is greatly influenced by their shape and size [53]. The TEM micrographs (Fig. 7) exposed that Pa-AgNPs have a spherical shape with an average size of 20 ± 1.1 nm. They were found to be circular aggregates with smooth edges that are not in direct contact with each other, which indicates the Pa-AgNPs may be stabilized by capping agents from the endophytic extract. Figure 8 shows the selected area electron diffraction (SAED) pattern of Pa-AgNPs with 100, 200, 220 and 311 crystal alignments matching with diffraction rings that were in agreement with XRD data. The presence of bright spots within the diffraction rings of crystal orientations confirms the FCC structure Pa-AgNP with polycrystalline nature. The average size of the particle was also nearly in congruence with DLS analysis. The results obtained were identical to earlier reports of green synthesized AgNPs [31, 54]. The small size of nanoparticles may enhance the reactivity and catalytic activity in various applications.
Fig. 7.
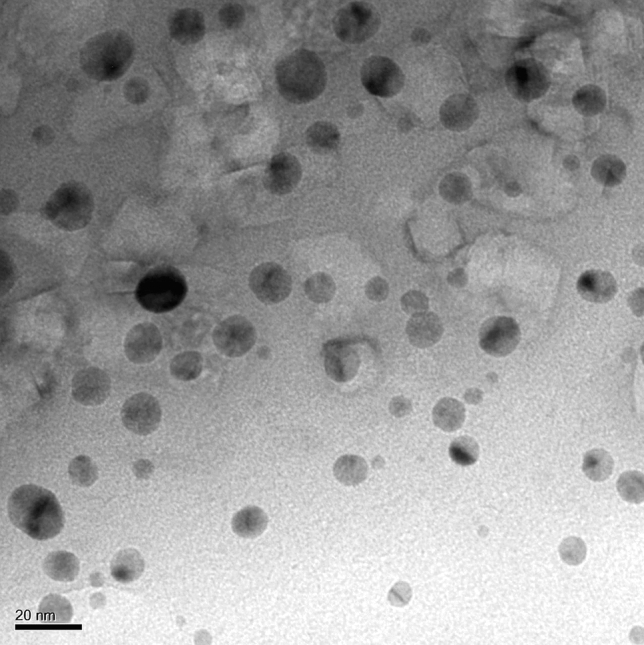
Transmission Electron Microscopy pattern of Pa-AgNPs
Fig. 8.

Selected area electron diffraction pattern of Pa-AgNPs
SEM and EDAX
The SEM micrograph (Fig. 9) showed that the Pa-AgNPs were spherical shaped, nonuniform polydispersed with an average size of 50 nm. The smaller particles may aggregate to give the large-size appearance of nanoparticles [54]. The large particle size depicted in SEM compared to DLS, XRD and TEM may be due to the difference in sample preparation and the presence of various forces of interaction in the solution [55, 56]. The elemental analysis of Pa-AgNPs done using EDAX (Fig. 10) showed a sharp absorption peak between 3–4 keV that is typical of the presence of metallic AgNPs. The additional peak for Fe were also observed that may be due to the trapping of proteins and other metabolites from the bacterial extract on AgNPs [35, 55]. The weight of silver was found to be 41.77%, thus confirms Pa-AgNPs was successfully biosynthesized from endophytic bacterial extracts.
Fig. 9.
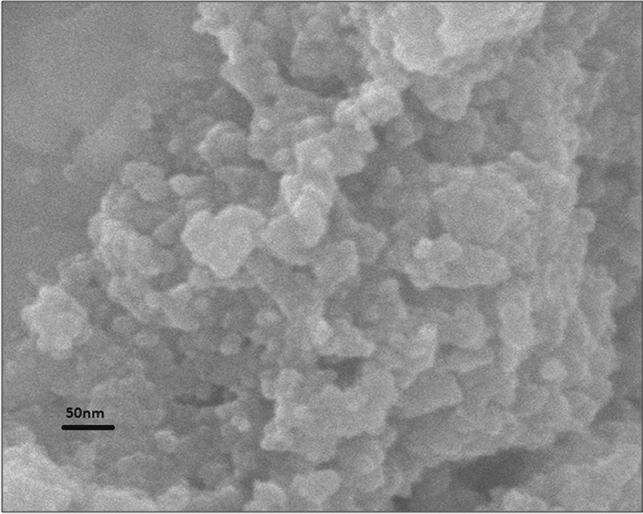
Scanning Electron Microscopy image of Pa-AgNPs
Fig. 10.
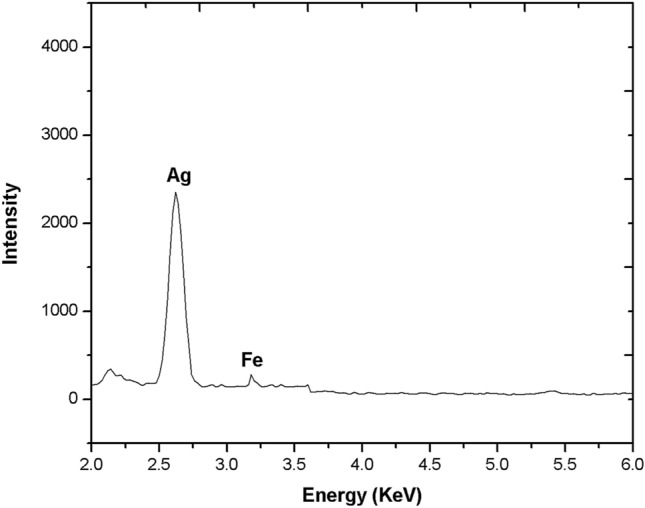
Elemental composition of Pa-AgNPs using EDAX profile
Antimicrobial Screening of Pa-AgNPs
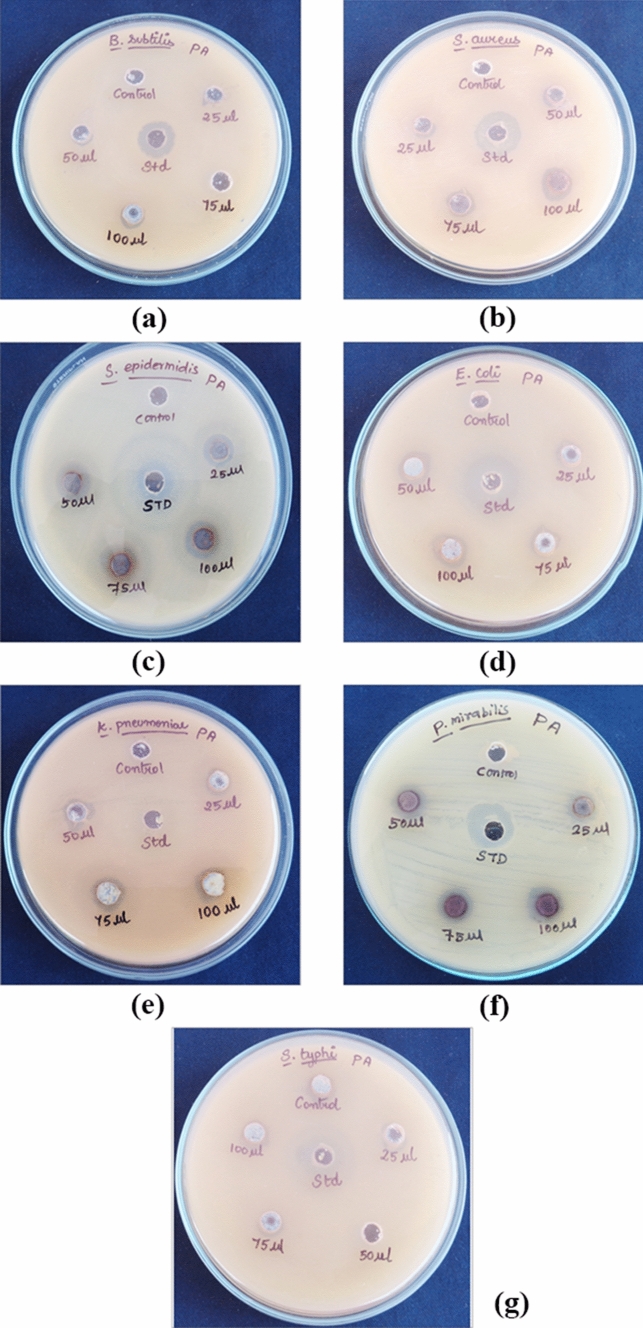
Pa-AgNPs showed promising antimicrobial activity in a dose-dependent manner. Significant antibacterial activity with minimum ZOI of 4.83 ± 0.21 mm against S. aureus, 4.93 ± 0.21 mm against S. epidermidis, P. mirabilis, and 4.03 ± 0.06, 4.03 ± 0.21, 4.03 ± 0.15 mm against E. coli, S. typhi and K. pneumonia at 25 µg/ml was observed. The maximum ZOI between 6.83 ± 0.29 to 7.03 ± 0.06 mm was observed at 100 µg/ml for all pathogens, Comparatively B. Subtilis exhibited low activity of 5 ± 0.10 mm at 100 µg/ml (Fig. 11). The diameters of the ZOI developed by Pa-AgNPs compared to the tetracycline standard for the bacterial pathogens (Table 1) are illustrated in Fig. 12. Hu et al. [57] observed similar ZOI by AgNPs synthesized from endophytic fungi Talaromyces purpureogenus against S. aureus, B. cereus, S. enterica, P. aeruginosa and E. coli. Khatoon et al. [58] chemically synthesized AgNPs using tri-sodium citrate that had higher antibacterial activity against E. coli, followed by B. subtilis which was similar to our results. The results obtained confirm the broad-spectrum activity of Pa-AgNPs against both Gram-positive and Gram-negative bacteria.
Fig. 11.
Antibacterial activity of Pa-AgNPs against a B. subtilis, b S. aureus, c S. epidermidis, d E. coli, e K. pneumonia, f P. mirabilis, g S. typhi
Table 1.
Antibacterial activity of Pa-AgNPs against Gram-positive and Gram-negative bacterial strains
| S. no | Test organisms | ZOI of Pa-AgNPs | |||||
|---|---|---|---|---|---|---|---|
| Standard (mm) | Control (mm) | 25 µl (mm) | 50 µl (mm) | 75 µl (mm) | 100 µl (mm) | ||
| 1. | B. subtilis | 7.77 ± 0.25 | – | – | – | 4 ± 0.20 | 5 ± 0.10 |
| 2. | S. aureus | 9.90 ± 0.36 | – | 4.83 ± 0.21 | 5.77 ± 0.25 | 5.97 ± 0.06 | 6.93 ± 0.12 |
| 3. | S. epidermidis | 9.07 ± 0.12 | – | 4.93 ± 0.21 | 6.13 ± 0.15 | 5.90 ± 0.10 | 7.03 ± 0.06 |
| 4. | E. coli | 13.03 ± 0.25 | – | 4.03 ± 0.06 | 6 ± 0.20 | 7.03 ± 0.15 | 6.97 ± 0.06 |
| 5. | S. typhi | 11.03 ± 0.25 | – | 4.03 ± 0.06 | 4.80 ± 0.26 | 6.07 ± 0.12 | 6.93 ± 0.12 |
| 6. | P. mirabilis | 8.07 ± 0.12 | – | 4.93 ± 0.21 | 6.10 ± 0.10 | 6.03 ± 0.06 | 6.83 ± 0.29 |
| 7. | K. pneumoniae | 8.93 ± 0.21 | – | 4.03 ± 0.15 | 5 ± 0.10 | 6.03 ± 0.06 | 6.97 ± 0.06 |
Fig. 12.
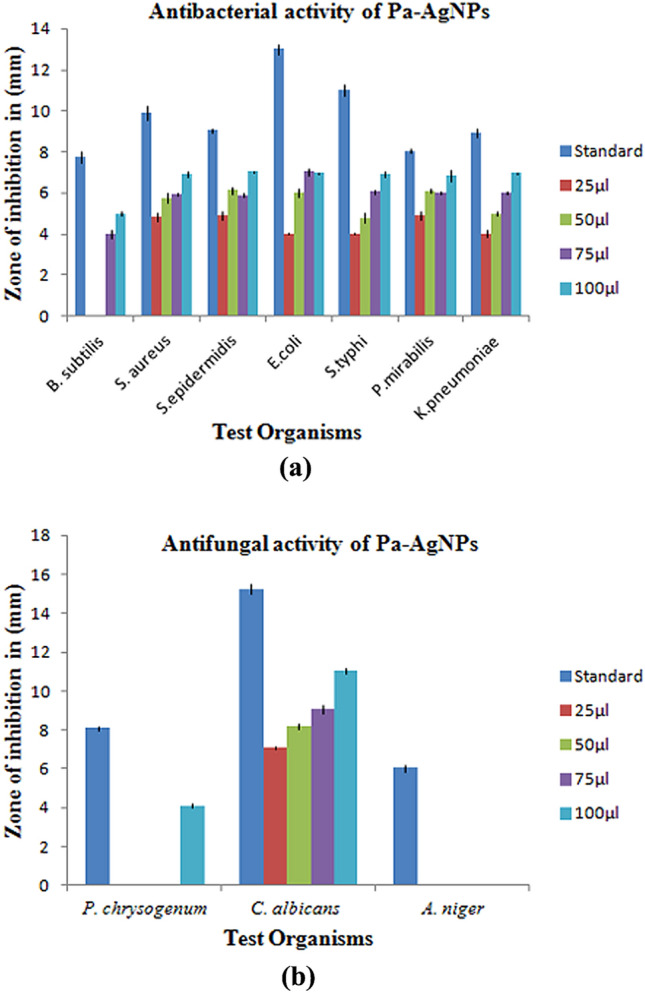
Graphical representation of ZOI produced by Pa-AgNPs against a Bacterial pathogens, b Fungal pathogens
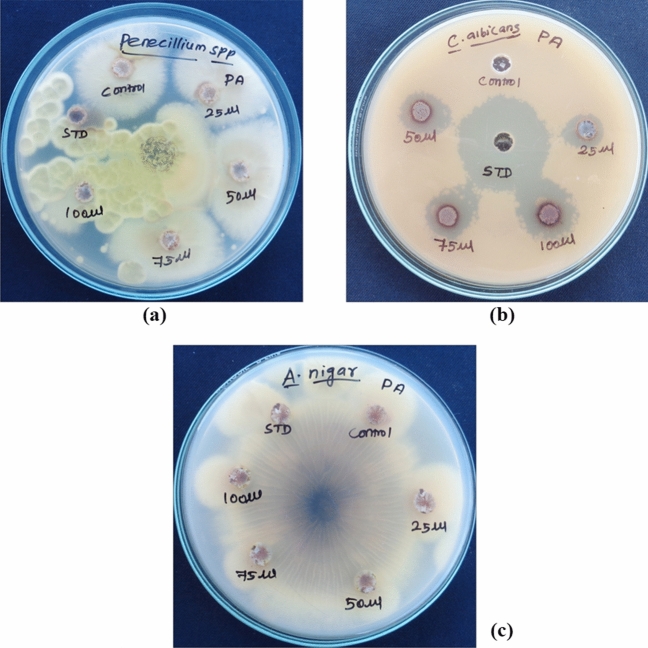
Penicillium chrysogenum and Candida albicans had ZOI of 4.10 ± 0.10 mm (100 µg/ml) and 7.10 ± 0.10 mm to 11.03 ± 0.15 mm (dose-dependent) (Fig. 13) respectively, whereas there is no ZOI for A. Niger (Table 2). Hence, the growth of C. albicans was greatly controlled than Penicillium and Aspergillus. The antimicrobial effect of Pa-AgNPs biosynthesized from endophytic bacteria isolated from W. indica against C. albicans was strong when compared to the crude W. indica leaf extracts reported by Koma et al. [28].
Fig. 13.
Antifungal activity of Pa-AgNPs against a P. chrysogenum, b C. Albicans, c A. niger
Table 2.
Antifungal activity of Pa-AgNPs against selected fungal strains
| S. no | Test organisms | ZOI of Pa-AgNPs | |||||
|---|---|---|---|---|---|---|---|
| Standard (mm) | Control (mm) | 25 µl (mm) | 50 µl (mm) | 75 µl (mm) | 100 µl (mm) | ||
| 1. | P. chrysogenum | 8.10 ± 0.10 | – | – | – | – | 4.10 ± 0.10 |
| 2. | C. albicans | 15.27 ± 0.25 | – | 7.10 ± 0.10 | 8.17 ± 0.15 | 9.07 ± 0.21 | 11.03 ± 0.15 |
| 3. | A. niger | 6.03 ± 0.15 | – | – | – | – | – |
Though different rational mechanisms elucidate the antimicrobial activity of AgNPs, the interpretations are uncertain and researchers are still working to identify the exact mechanism. Khatoon et al. [43] were able to obtain the highest antifungal activity with AgNPs of 24 nm size against S. cerevisiae and C.albicans than AuNPs of 35 nm. Hence the small particle size of about 16.8 nm may attribute to the antimicrobial activity of Pa-AgNPs. At higher concentration, the Pa-AgNPs were able to break the thick peptidoglycan layer of Gram-positive as well as thin layered Gram-negative bacterial cell wall strongly than at the lower concentrations. The much thicker cell wall of fungal species compared to bacteria make a difference in the antifungal activity of Pa-AgNPs [58]. Hence the concentration of the AgNPs and microbial species type influence the antimicrobial activity. According to Levard et al. [59] the positive surface charge of Pa-AgNPs may contribute to the inhibitory effects of bacterial species. Ag+ ions released from Pa-AgNPs binds with the cell wall, alters the membrane potential and transmembrane electrochemical gradient. The AgNPs also binds with the DNAthat leads to DNAdamage and ROS generation ultimately causes cell death. Hence our results justify the plausible mechanisms (Fig. 14) for potent microbicidal activity of Pa-AgNPs that were in accordance with the reported literature [60–62] of AgNPs synthesized by biogeneic approach.
Fig. 14.
Rational mechanisms for antimicrobial activity of Pa-AgNPs
To the best of our knowledge, the present study is the first scientific report on the potential of Pa-AgNPs against human pathogenic microbes. The study can be extended against MDR bacteria, other clinical pathogens and pandemic viruses to overcome the current challenging situations. The low cost biosynthesized AgNPs can also be used to reduce the secondary risks caused in COVID-19 patients.
Antioxidant Efficacy of Pa-AgNPs
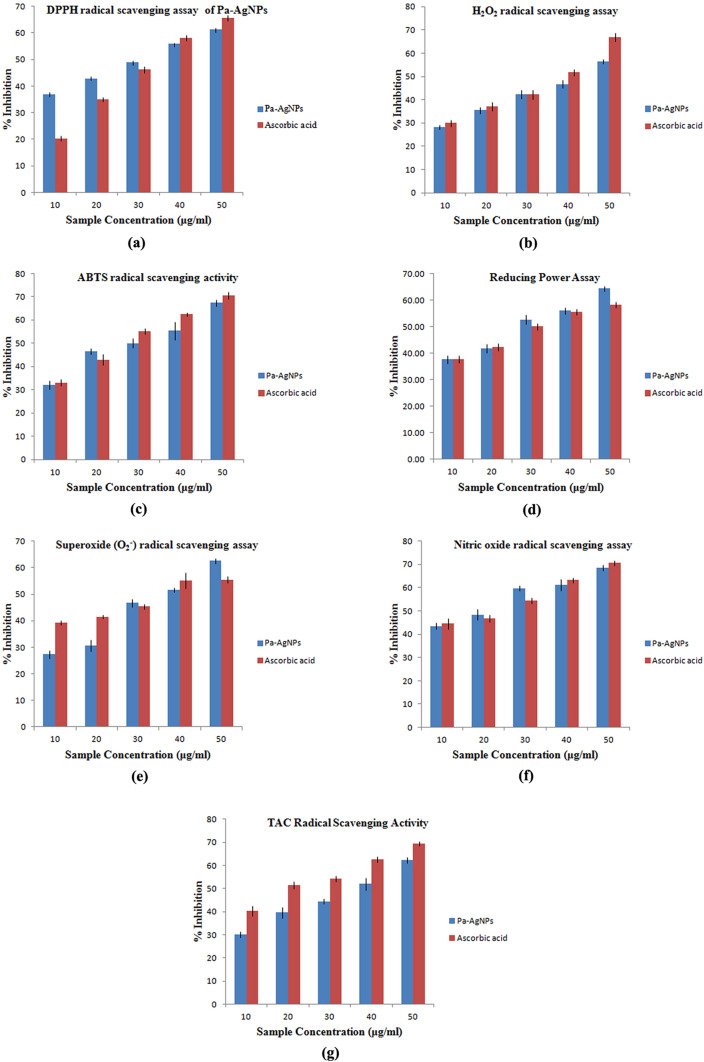
The DPPH radical, H2O2 radical and ABTS radical scavenging activity of Pa-AgNPs increased in a dose-dependent manner from 36.99 ± 0.61% to 61.31 ± 0.47%, 28.36 ± 0.90 to 56.37 ± 0.91% and 31.94 ± 1.84% to 67.44 ± 1.39% respectively at the concentration from 10 to 50 μg/ml, which is comparable with standard ascorbic acid that had 20.97 ± 0.89% to 64.64 ± 1.06%, 29.95 ± 1.27 to 66.96 ± 1.85%, 33.11 ± 1.40 to 70.47 ± 1.49% of inhibition respectively (Fig. 15a, b, c).
Fig. 15.
Antioxidant efficacy of Pa-AgNPs a DPPH radical scavenging assay, b H2O2 radical scavenging assay, c ABTS radical scavenging assay, d Reducing power assay, e Superoxide radical scavenging assay, f NO radical scavenging assay, g Total antioxidant capacity assay
The reducing power of compounds also increased analogous to their antioxidant ability. The reductive capability of the Pa-AgNPs increased from 37.71 ± 1.51 to 64.51 ± 0.94%, while it is 37.81 ± 1.45% to 58.39 ± 1.12% at the same concentration for standard (Fig. 15d).
Pa-AgNPs scavenges the superoxide and nitric oxide radical up to 62.57 ± 0.87% and 68.59 ± 1.21% at 50 μg/ml that was comparable to the scavenging effect of ascorbic acid 55.54 ± 1.23% and 70.54 ± 0.96% at 50 μg/ml (Fig. 15e, f). The total antioxidant capacity of the Pa-AgNPs showed the highest inhibition of 62.22 ± 1.75% at 50 μg/ml, while the reference showed 69.41 ± 1.16% at the same concentration (Fig. 15g).
Thus Pa-AgNPs have strong DPPH radical scavenging activity, nitric oxide radical scavenging activity and reducing power activity when compared to the standard. The H2O2 radical scavenging assay, total antioxidant capacity assay and ABTS radical scavenging assay, standard ascorbic acid showed strong antioxidant activity than Pa-AgNPs. The superoxide radical scavenging activity was similar for both Pa-AgNPs and standard. Netala et al. [63] observed 55.62 ± 0.25% of inhibition in DPPH radical scavenging assay at 50 μg/ml by AgNPs synthesized by endophytic fungi that were lower than that obtained by Pa-AgNPs. Also, the nitric oxide radical, H2O2 radical scavenging and reducing assay of biogenic AgNPs reported at 50 μg/ml was lower than that obtained by Pa-AgNPs [64]. The results were also in line with the previous reports [65, 66]. The comparison of IC50 in each assay is illustrated in Table 3 and the lower IC50 values show the higher antioxidant potency of the Pa-AgNPs.
Table 3.
Antioxidant activity of Pa-AgNPs
| S. no | Parameters | IC50 value (μg/ml) | |
|---|---|---|---|
| Pa-AgNPs | Ascorbic acid | ||
| 1. | DPPH radical scavenging assay | 31.29 ± 0.73 | 34.83 ± 0.32 |
| 2. | NO radical scavenging assay | 19.83 ± 1.57 | 21.36 ± 1.75 |
| 3. | Total antioxidant capacity assay | 35.64 ± 0.94 | 21.78 ± 2.22 |
| 4. | H2O2 radical scavenging assay | 42.07 ± 1.30 | 34.94 ± 0.15 |
| 5. | ABTS radical scavenging assay | 29.70 ± 2.26 | 26.96 ± 0.15 |
| 6. | Reducing power assay | 29.10 ± 0.82 | 32.11 ± 0.63 |
| 7. | Superoxide radical scavenging assay | 36.80 ± 0.63 | 35.76 ± 1.56 |
Thus Pa-AgNPs was proved to have strong antioxidant activity that may occur due to the adsorption of functional groups from the endophytic extract on the surface of Pa-AgNPs [67]. The excellent radical scavenging activity of Pa-AgNPs admit them to be used as potent antioxidants or as a valuable component of antioxidant formulations in biomedical and pharmaceutical fields [68]. The antioxidant property can be evaluated in in vivo models prior to human applications for the treatment of various oxidative stress-related degenerative diseases.
Conclusions
In the present study, the endophytic bacterium P. anthophila isolated from the stem of W. indica was used for extracellular biosynthesis of AgNPs. Structural characterization showed the optimal formation of Pa-AgNPs with crystalline nature at 410 nm in the UV–visible spectrum, 16 to 20 nm size with positive surface potential. The bacterial metabolites formed a stable capping moiety around AgNPs and the elemental composition of Pa-AgNPs was also confirmed. They exhibited potential antimicrobial activity against 10 different bacterial and fungal pathogens. Higher antioxidant activity of Pa-AgNPs was also confirmed by DPPH, NO, H2O2, ABTS, superoxide radical scavenging assay, total antioxidant capacity assay and reducing power assay. Thus endophytic P. anthophila mediated biosynthesis of AgNPs prevails as an eco-friendly, cost-effective and substantial method with increased potency of applications in nanomedicine. Further, the efficiency and mechanism of Pa-AgNPs to establish as anticancer and antiviral agents will be evaluated in future to ascertain that green synthesized AgNPs are an optimistic therapeutic agent in the trending era of biomedicines.
Acknowledgements
The authors gratefully acknowledge the Department of Biotechnology, Vinayaka Mission's Kirupananda Variyar Engineering College, Salem and Acme ProGen Biotech (India) Private Limited, Salem for providing the essential facilities to carry out the research.
Abbreviations
- AgNPs
Silver nanoparticles
- Pa-AgNPs
Silver nanoparticles biosynthesized from Pantoea anthophila
- rpm
Revolution per minute
- mM
Milli molar
- DLS
Dynamic light scattering analysis
- FTIR
Fourier Transform Infrared Spectroscopy
- XRD
X-ray diffraction
- SEM
Scanning Electron Microscopy
- TEM
Transmission Electron Microscopy
- EDAX
Energy dispersive of X-Ray spectrum
- Ag+
Silver ions
- DMSO
Dimethyl sulfoxide
- DPPH
1, 1-Diphenyl-2-picrylhydrazyl
- NO
Nitric oxide
- H2O2
Hydrogen peroxide
- ABTS
2,2′-Azino-bis-3-ethyl benzothiozoline-6-sulfonic acid
- NBT
Nitro blue tetrazolium
- PMS
Phenazinemethosulfate
- COVID
Corona Virus Disease
- IC50
50% Inhibition
Author Contributions
CN had collected all the literature, samples and undergone the research work, analyzed, and prepared the complete manuscript under the guidance of MS, who has also critically reviewed the article for improvement. All authors have read and approved the final manuscript.
Funding
No funding was received for conducting this study.
Data Availability
All data generated or analyzed during this study are available upon request.
Declarations
Conflict of interest
The authors declare that there are no Competing interests.
Research Involving Human and Animal Rights
This article does not contain any studies with human participants or animals performed by the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
C. Nirmala, Email: nirmalabt@gmail.com
M. Sridevi, Email: drsridevimuruhan@gmail.com
References
- 1.Ivanov AV, Bartosch B, Isaguliants MG. Oxid. Med. Cell Longev. 2017 doi: 10.1155/2017/3496043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta P, Authimoolam SP, Hilt JZ, Dziubla TD. Actabiomaterialia. 2015 doi: 10.1016/j.actbio.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim YY, Lim TT, Tee JJ. Food Chem. 2007 doi: 10.1016/j.foodchem.2006.08.038. [DOI] [Google Scholar]
- 4.Anagnostopoulou MA, Kefalas P, Papageorgiou VP, Assimopoulou AN, Boskou D. Food Chem. 2006 doi: 10.1016/j.foodchem.2004.09.047. [DOI] [Google Scholar]
- 5.Verma AK. J. Pharm. Res. 2014;8:7. [Google Scholar]
- 6.Ahmed S, Ahmad M, Swami BL, Ikram S. J. Adv. Res. 2016 doi: 10.1016/j.jare.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saifuddin N, Wong CW, Yasumira AA. J. Chem. 2009;6:1. [Google Scholar]
- 8.Wang L, Hu C, Shao L. Int. J. Nanomed. 2017 doi: 10.2147/IJN.S121956. [DOI] [Google Scholar]
- 9.Pandey S, Goswami GK, Nanda KK. Sci. Rep. 2013 doi: 10.1038/srep02082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey S, Nanda KK. ACS Sens. 2016 doi: 10.1021/acssensors.5b00013. [DOI] [Google Scholar]
- 11.Pandey S, Do JY, Kim J, Kang M. Carbohydr. Polym. 2020 doi: 10.1016/j.carbpol.2019.115597. [DOI] [PubMed] [Google Scholar]
- 12.Nair LS, Laurencin CT. J. Biomed. Nanotech. 2007 doi: 10.1166/jbn.2007.041. [DOI] [Google Scholar]
- 13.Patra JK, Baek KH. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coe S, Woo WK, Bawendi M, Bulović V. Nature. 2002 doi: 10.1038/nature01217. [DOI] [PubMed] [Google Scholar]
- 15.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998 doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 16.Singh PK, Bhardwaj K, Dubey P, Prabhune A. RSC Adv. 2015 doi: 10.1039/C4RA17233G. [DOI] [Google Scholar]
- 17.Mishra A, Kaushik NK, Sardar M, Sahal D. Colloid Surf. B. 2013 doi: 10.1016/j.colsurfb.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Krishnaraj RN, Berchmans S. RSC Adv. 2013 doi: 10.1039/C3RA41246F. [DOI] [Google Scholar]
- 19.Castro-Aceituno V, Ahn S, Simu SY, Singh P, Mathiyalagan R, Lee HA, Yang DC. Biomed. Pharmacother. 2016 doi: 10.1016/j.biopha.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Rigo C, Ferroni L, Tocco I, Roman M, Munivrana I, Gardin C, Cairns WR, Vindigni V, Azzena B, Barbante C, Zavan B. Int. J. Mol. Sci. 2013 doi: 10.3390/ijms14034817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baptista PV, McCusker MP, Carvalho A, Ferreira DA, Mohan NM, Martins M, Fernandes AR. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei L, Lu J, Xu H, Patel A, Chen ZS, Chen G. Drug Discov. Today. 2015 doi: 10.1016/j.drudis.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazem A, Mansoori GA. J. Alzheimer's Dis. 2008 doi: 10.3233/JAD-2008-13210. [DOI] [PubMed] [Google Scholar]
- 24.Hari N, Thomas TK, Nair AJ. J. Nanosci. 2014 doi: 10.1155/2014/201482. [DOI] [Google Scholar]
- 25.Chen JC, Lin ZH, Ma XX. Lett. Appl. Microbiol. 2003 doi: 10.1046/j.1472-765X.2003.01348.x. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad A, Mukherjee P, Senapati S, Mandal D, Khan IM, Kumar R, Sastry M. Colloid Surf. B. 2003 doi: 10.1016/S0927-7765(02)00174-1. [DOI] [Google Scholar]
- 27.Rónavári A, Kovács D, Igaz N, Vágvölgyi C, Boros IM, Kónya Z, Pfeiffer I, Kiricsi M. Int. J. Nanomed. 2017 doi: 10.2147/IJN.S122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koma OS, Fatokun OA, Theophilus OA. Biomed. Sci. 2017 doi: 10.11648/j.bs.20170305.11. [DOI] [Google Scholar]
- 29.Musa Y, Musah M, Yerima H, Erena NB. J. Adv. Res. Appl. Sci. 2016;3:4. [Google Scholar]
- 30.Nirmala C, Sridevi M. Future J. Pharm. Sci. 2021 doi: 10.1186/s43094-020-00174-3. [DOI] [Google Scholar]
- 31.Devi LS, Joshi SR. J. Microsc. Ultrastruct. 2015 doi: 10.1016/j.jmau.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monowar T, Rahman M, Bhore SJ, Raju G, Sathasivam KV. Mole. 2018 doi: 10.3390/molecules23123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalimuthu K, Babu RS, Venkataraman D, Bilal M, Gurunathan S. Colloid Surf. B. 2008 doi: 10.1016/j.colsurfb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Das VL, Thomas R, Varghese RT, Soniya EV, Mathew J, Radhakrishnan EK. 3 Biotech. 2014 doi: 10.1007/s13205-013-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh T, Jyoti K, Patnaik A, Singh A, Chauhan R, Chandel SS. J. Genet. Eng. Biotechnol. 2017 doi: 10.1016/j.jgeb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh D, Rathod V, Ninganagouda S, Hiremath J, Singh AK, Mathew J. Bioinorg. Chem. Appl. 2014 doi: 10.1155/2014/408021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.PatilShriniwas P. Biochem. Biophys. Rep. 2017;10:76. [Google Scholar]
- 38.Jain PK, Agrawal RK. Asian J. Exp. Sci. 2008;22:3. [Google Scholar]
- 39.Nabavi SM, Ebrahimzadeh MA, Nabavi SF, Hamidinia A, Bekhradnia AR. Pharmacology. 2008;2:560–567. [Google Scholar]
- 40.Prieto P, Pineda M, Aguilar M. Anal. Biochem. 1999 doi: 10.1006/abio.1999.4019. [DOI] [PubMed] [Google Scholar]
- 41.Yen GC, Duh PD. J. Am. Oil Chem. Soc. 1993 doi: 10.1007/BF02552711. [DOI] [Google Scholar]
- 42.Liu F, Ooi VE, Chang ST. Life Sci. 1997 doi: 10.1016/S0024-3205(97)00004-0. [DOI] [PubMed] [Google Scholar]
- 43.Wrótniak-Drzewiecka W, Gaikwad S, Laskowski D, Dahm H, Niedojadło J, Gade A, Rai M. Austin J. Biotechnol. Bioeng. 2014;1:1. [Google Scholar]
- 44.Khatoon UT, Rao GN, Mohan MK, Ramanaviciene A, Ramanavicius A. J. Environ. Chem. Eng. 2018 doi: 10.1016/j.jece.2018.08.009. [DOI] [Google Scholar]
- 45.Attia SM, Abdelfatah MS, Mossad MM, Phys J. Conf. Ser. 2017 doi: 10.1088/1742-6596/869/1/012036. [DOI] [Google Scholar]
- 46.Ali ZA, Yahya R, Sekaran SD, Puteh R. Adv. Mater. Sci. Eng. 2016 doi: 10.1155/2016/4102196. [DOI] [Google Scholar]
- 47.Prakasham RS, Kumar BS, Kumar YS, Shankar GG. J. Microbiol. Biotechnol. 2012 doi: 10.4014/jmb.1107.07013. [DOI] [PubMed] [Google Scholar]
- 48.Roseline TA, Murugan M, Sudhakar MP, Arunkumar K. Environ Technol. Innov. 2019 doi: 10.1016/j.eti.2018.10.005. [DOI] [Google Scholar]
- 49.Pandey S, Goswami GK, Nanda KK. Int. J. Biol. Macromol. 2012 doi: 10.1016/j.ijbiomac.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 50.Cullity BD. Elements of X-ray Diffraction. 2. Reading: Addison-Wesley Publishing Company Inc; 1978. [Google Scholar]
- 51.Gnanadesigan M, Anand M, Ravikumar S, Maruthupandy M, Vijayakumar V, Selvam S, Dhineshkumar M, Kumaraguru AK. Asian Pac. J. Trop. Med. 2011 doi: 10.1016/S1995-7645(11)60197-1. [DOI] [PubMed] [Google Scholar]
- 52.Dong ZY, NarsingRao MP, Xiao M, Wang HF, Hozzein WN, Chen W, Li WJ. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumari M, Pandey S, Giri VP, Bhattacharya A, Shukla R, Mishra A, Nautiyal CS. Microb Pathogen. 2017 doi: 10.3390/molecules23123220. [DOI] [PubMed] [Google Scholar]
- 54.Devaraj P, Kumari P, Aarti C, Renganathan A. J. Nanotechnol. 2013 doi: 10.1155/2013/598328. [DOI] [Google Scholar]
- 55.Banerjee P, Nath D. Nanosci. Technol. 2015;2:1. [Google Scholar]
- 56.Otunola GA, Afolayan AJ. Biotechnol. Biotechnol. Equip. 2018 doi: 10.1080/13102818.2018.1448301. [DOI] [Google Scholar]
- 57.Hu X, KandasamySaravanakumar TJ, Wang MH. Int. J. Nanomed. 2019 doi: 10.2147/IJN.S200817. [DOI] [Google Scholar]
- 58.Khatoon UT, Rao GN, Mohan KM, Ramanaviciene A, Ramanavicius A. Vacuum. 2017 doi: 10.1016/j.vacuum.2017.10.003. [DOI] [Google Scholar]
- 59.Levard C, Hotze EM, Lowry GV, Brown GE., Jr Environ. Sci. Technol. 2012 doi: 10.1021/es2037405. [DOI] [PubMed] [Google Scholar]
- 60.Pandey S, Klerk CD, Kim J, Kang M, Fosso-Kankeu E. Polymers. 2020 doi: 10.3390/polym12061418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandey S, Ramontja J. Int. J. Biol. 2016 doi: 10.1016/j.ijbiomac.2016.09.033. [DOI] [Google Scholar]
- 62.Mishra AK, Tiwari KN, Saini R, Kumar P, Mishra SK, Yadav VB, Nath G. J. Inorg. Organomet. Polym. 2020 doi: 10.1007/s10904-019-01392-w. [DOI] [Google Scholar]
- 63.Netala VR, Kotakadi VS, Bobbu P, Gaddam SA, Tartte V. 3 Biotech. 2016 doi: 10.1007/s13205-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhakya S, Muthukrishnan S, Sukumaran M, Muthukumar M. Appl. Nanosci. 2016 doi: 10.1007/s13204-015-0473-z. [DOI] [Google Scholar]
- 65.Hemashekhar B, Chandrappa C, Govindappa M, Chandrasekhar N, Ganganagappa N, Ramachandra Y. Int. J. Eng. Res. Appl. 2017 doi: 10.9790/9622-0708011724.14. [DOI] [Google Scholar]
- 66.Ramamurthy CH, Padma M, Mareeswaran R, Suyavaran A, Kumar MS, Premkumar K, Thirunavukkarasu C. Colloids Surf. B. 2013 doi: 10.1016/j.colsurfb.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 67.Keshari AK, Srivastava R, Singh P, Yadav VB, Nath G. J. Ayurveda Integr. Med. 2020 doi: 10.1016/j.jaim.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chandrappa CP, Govindappa M, Chandrasekar N, Sarkar S, Ooha S, Channabasava R. Adv. Nat. Sci. Nanosci. 2016 doi: 10.1088/2043-6262/7/2/025016. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are available upon request.