As indicated in the letter by Rissiek et al., our paper characterizing P2X5-dependent elements of the immune response to Listeria monocytogenes (Lm) [1] employed a P2X5 KO mouse strain (B6;129S5-P2rx5tm1Lex/Ieg; MGI:5430769) that was generated by Lexicon Genetics using 129S5/SvEvBrd (129) background embryonic stem (ES) cells. While we backcrossed P2X5 KO mice to the C57BL/6 background at least 10 times before using them for experiments, the very close proximity of the P2rx5 locus to Nlrp1a, Nlrp1b, and Nlrp1c loci, suggests a high probability that mice carrying the P2X5 KO allele continue to harbor Nlrp1 passenger mutations associated with the 129 background [2]. Indeed, having now analyzed RNA-Seq data generated from WT and P2X5 KO bone marrow-derived macrophages (BMMs), we find that P2X5 KO samples show dramatically reduced expression levels of Nlrp1a and Nlrp1c, compared to WT samples, but normal levels of Nlrp1b (Figure 1). These findings are consistent with the presence of Nlrp1 passenger mutations from 129 ES cells in our backcrossed P2X5 KO mice [3]. Therefore, observations we made with P2X5 KO mice/cells should be considered to have been made in the context of potential accompanying differences in NLRP1 function compared to experimentally-paired WT control mice/cells. However, to be clear, we made no claim in our paper that P2X5 regulates NLRP1 function or Nlrp1 gene expression, and while Lm may trigger NLRP1 activation, it is remains unclear whether NLRP1 plays any significant role in anti-Lm immune responses [4, 5]. It is possible that NLRP1 contributes to Lm-induced immune responses, though we are not currently in a position to address whether differences in Nlrp1 genes impact susceptibility of P2X5 KO mice infected with Lm. These questions will need to be addressed in the future, ideally with the use of tools that enable independent assessment of contributions by NLRP1 versus P2X5. In this regard, we recommend that future studies addressing the role of P2X5 in anti-Lm immunity be performed using P2X5 KO mice newly derived from C57BL/6 background ES cells in order to dissect whether P2X5 acts alone or in concert with NLRP1.
Figure 1. P2X5 WT and P2X5 KO BMMs exhibit differential expression profiles of Nlrp1 paralogs.
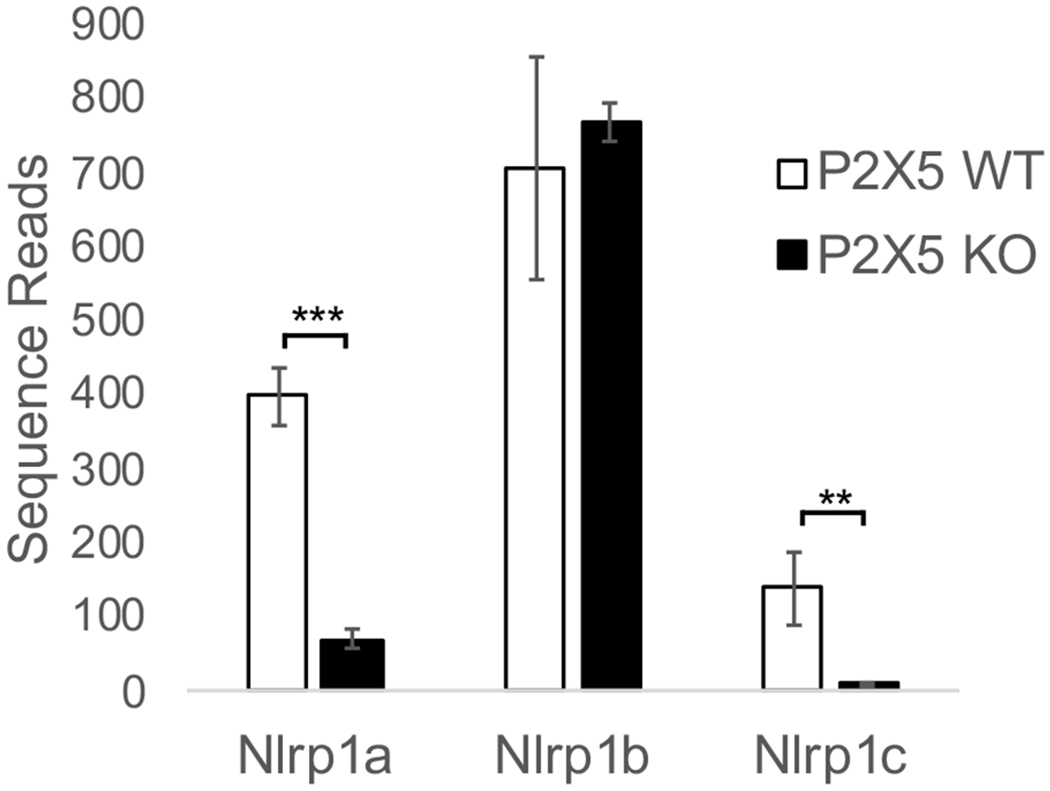
cDNAs from P2X5 WT (C57BL/6 background) and P2X5 KO (mixed 129 and C57BL/6 background) BMMs were subjected to total RNA sequencing (RNAseq) analysis and Nlrp1 expression quantitated. **p<0.01, ***p<0.0005.
REFERENCES
- 1.Jeong YH, Walsh MC, Yu J, Shen H, Wherry EJ, and Choi Y, Mice Lacking the Purinergic Receptor P2X5 Exhibit Defective Inflammasome Activation and Early Susceptibility to Listeria monocytogenes. J Immunol, 2020. 205(3): p. 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanden Berghe T, Hulpiau P, Martens L, Vandenbroucke RE, Van Wonterghem E, Perry SW, Bruggeman I, Divert T, Choi SM, Vuylsteke M, Shestopalov VI, Libert C, and Vandenabeele P, Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice. Immunity, 2015. 43(1): p. 200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sastalla I, Crown D, Masters SL, McKenzie A, Leppla SH, and Moayeri M, Transcriptional analysis of the three Nlrp1 paralogs in mice. BMC Genomics, 2013. 14: p. 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meixenberger K, Pache F, Eitel J, Schmeck B, Hippenstiel S, Slevogt H, N’Guessan P, Witzenrath M, Netea MG, Chakraborty T, Suttorp N, and Opitz B, Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J Immunol, 2010. 184(2): p. 922–30. [DOI] [PubMed] [Google Scholar]
- 5.Neiman-Zenevich J, Stuart S, Abdel-Nour M, Girardin SE, and Mogridge J, Listeria monocytogenes and Shigella flexneri Activate the NLRP1B Inflammasome. Infect Immun, 2017. 85(11). [DOI] [PMC free article] [PubMed] [Google Scholar]


