Abstract
Oxalis corymbosa DC. is an important medicinal and edible perennial herb belonging to the wood-sorrel family Oxalidaceae. In this study, we report the complete chloroplast (cp) genome sequence of O. corymbosa. The assembled chloroplast genome was 151,351 bp in length, containing two inverted repeated (IR) regions of 24,587 bp each, a large single copy (LSC) region of 85,476 bp, and a small single copy (SSC) region of 16,701 bp. The genome encodes 128 genes, consisting of 82 protein-coding genes, 37 tRNA genes, eight rRNA genes, and one pseudogene (ycf1). The 82 protein-coding genes encode 25,751 amino acids in total, most of which use the initiation codon ATG, except rps19 and psbC genes start with GTG. The lengths of the tRNA genes range from 71 bp to 93 bp, with the highest GC content of 62.16% in tRNA-Arg (ACG). The overall GC content of O. corymbosa is 36.47%, with the highest GC content of 42.64% in IR region. In addition, a total of 74 simple sequence repeats were identified in the cp genome of O. corymbosa. Phylogenetic analysis indicated a sister relationship between O. corymbosa and O. drummondii, suggesting a close genetic relationship between the two Oxalis species. This work provides basic genetic resources for investigating the evolutionary status and population genetics diversities for this medicinal species.
Keywords: Oxalis corymbosa, complete chloroplast genome, phylogenetic analysis, simple sequence repeats
Oxalis corymbosa DC. is an important medicinal and edible perennial herb and widely distributed throughout the world. This species was introduced into China as an ornamental plant in the mid-19th century and is now abundantly found in agricultural farms, gardens, and lawns (Tsai et al. 2020). Oxalis corymbosa was recorded as Tongchuicao in a variety of Local Records of Chinese Traditional Medicine with effects of removing blood stasis and detumescence, clearing heat, and removing dampness (Gao et al. 2011). The extracts of O. corymbosa have been demonstrated as a valuable natural source of antioxidants, suggesting potential applications in both medicinal and food industries (Liao et al. 2019). The antioxidant capacity of hydrophilic extracts from O. corymbosa was positively related with the total phenolic contents (Tukun et al. 2014). In addition, Oxalis is the largest genus of family Oxalidaceae, comprising of more than 500 species (Vaio et al. 2013). The taxonomy has been revised with similarities in phenotypes across different species (Lubna et al. 2020). The chloroplast genome (cp) has been proven to be a valuable resource for species identification and plant phylogenetic analysis (Wang et al. 2020). Furthermore, it is necessary to develop effective molecular identification strategy for O. corymbosa to ensure the safety of clinical application. The aim of this study is to analyze chloroplast genome sequence of O. corymbosa, which could contribute to the development of molecular markers and phylogenetic relationship investigation.
The sample of Oxalis corymbosa was collected from Fuyang area of Zhejiang Province (30°05′2.4″N, 119°53′20.4″E). The leaf specimen was deposited at Medicinal Herbarium Center of Zhejiang Chinese Medical University, Hangzhou, China (Voucher Identifying Number CPC-02). Total genomic DNA was extracted and sequenced using the Illumina Hiseq Platform according to the previous report (Dong et al. 2020; Gao et al. 2020). The chloroplast genome of O. corymbosa was assembled by metaSPAdes with the chloroplast genome sequence of Oxalis drummondii as reference (Nurk et al. 2017). Chloroplast genome sequence was annotated using GeSqe and further confirmed by BLAST (Tillich et al. 2017). The complete cp genome of O. corymbosa was submitted to GenBank with the accession number of MW057776.
The length of the complete chloroplast genome sequence of O. corymbosa was 151,351 bp, with a large single copy (LSC) region of 85,476 bp, a small single copy (SSC) region of 16,701 bp, and two separated inverted repeated (IR) regions of 24,587 bp each. A total of 128 genes were identified in the cp of O. corymbosa, including 82 protein-coding genes, 37 tRNA genes, eight rRNA genes, and one pseudogene (ycf1). The overall GC content was 36.47%, and the corresponding contents for LSC, SSC, and IR regions were 34.19%, 29.97%, and 42.64%, respectively. The genome included 15 duplicated genes in the IR region, including seven tRNAs, four rRNAs and four protein-coding genes. The proportion of coding sequences with a total length of 77,499 bp is 51.2%, which encodes 25,751 amino acids. The most frequently used amino acids were Leu (10.6%), followed by Ile (8.8%), Ser (7.7%), Gly (6.6%), and Phe (5.9%), respectively. Most of the protein-coding genes in O. corymbosa started with a typical ATG codon, except for rps19 and psbC genes that used the initiation codon GTG. For the stop codon, 44 of 82 genes ended with TAA, 20 protein-coding genes ended with TAG, while the other 18 genes terminated with TGA. It is interestingly to note that one of ycf1 is a pseudogene that has only partial fragment. Similar to most chloroplast genomes, the space gap of nucleotides are very common in O. corymbosa. The tRNA genes of O. corymbosa vary from 71 to 93 nucleotides, with the GC content range from 39.73% to 62.16%. Moreover, a total of 74 small single repeats (SSR) are identified in the cp of O. corymbosa, ranging from 10 bp to 131 bp.
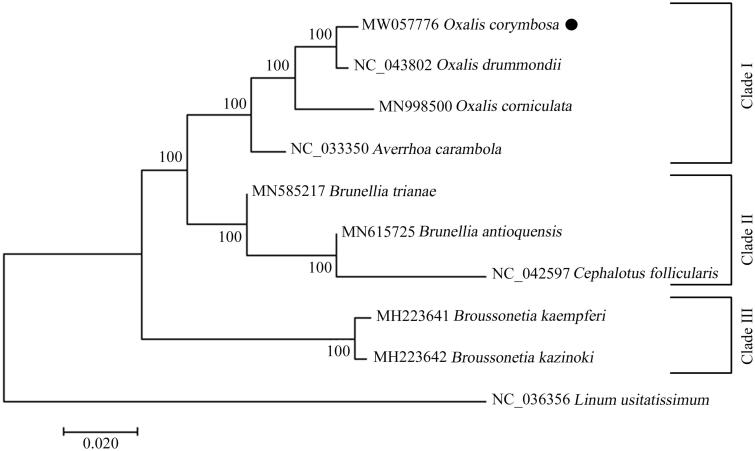
The ML tree was inferred by MEGA 7.0 using the newly determined complete genome sequence of O. corymbosa as well as the cp sequences of other eight representative species from the family Oxalidaceae. The result demonstrated that O. corymbosa was clustered together with O. drummondii, suggesting a close genetic relationship between the two species (Figure 1). In addition, the three species from genus Oxalis formed a monophyletic group, and exhibited a sister relationship with Averrhos carambola. The four species combined together to form the group of Clade I (Figure 1). Our results would contribute to the development of molecular markers and further investigation on the population genetics and phylogenetics of the genus Oxalis.
Figure 1.
Phylogenetic relationships of the newly sequenced Oxalis corymbosa and other representative species from family Oxalidaceae based on the complete chloroplast genomes. The tree was generated by MEGA 7.0 with maximum-likelihood (ML) method. The species of Linum usitatissimum was chosen as the out-group. The newly determined genome of O. corymbosa was indicated with black circle. Numbers on the nodes represent bootstrap values from ML analysis with 100 replicates. The GenBank accession numbers were listed before the species name.
Funding Statement
This work was supported by the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University [No. ZYAOX2018033] and Project of Quality Guarantee System of Chinese Herbal Medicines [Grant No. 201507002-4].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW057776. The associated BioProject, SRA, and BioSample numbers are PRJNA690071, SRR13370090 and SAMN17227902, respectively.
References
- Dong S, Gao C, Wang Q, Ge Y, Cheng R.. 2020. Characterization of the complete chloroplast genome of Macleaya cordata and its phylogenomic position within the subfamily papaveroideae. Mitochondrial DNA Part B. 5(2):1714–1715. [Google Scholar]
- Gao C, Wang Q, Ying Z, Ge Y, Cheng R.. 2020. Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang Province. Mitochondrial DNA Part B. 5(1):1077–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao PL, Guo SL, Lou YX.. 2011. Adaptation characteristics of Oxalis corymbosa and O. triangularis to light factor based on Chlorophyll fluorescence parameters. J Shanghai Normal Univ (Nat Sci). 40(5):528–532. [Google Scholar]
- Liao XY, Xu HM, Feng P, Wang YX, Huang JY.. 2019. Evaluation of Oxalis Corymbosa extracts from different plant parts and seasons as a potential source of antioxidants. Curr Top Nutraceutical Res. 17:50–55. [Google Scholar]
- Lubna Asaf S., Jan R, Khan AL, Lee IJ.. 2020. Complete chloroplast genome characterization of Oxalis Corniculata and its comparison with related species from family Oxalidaceae. Plants. 9:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA.. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MY, Jane WN, Chen SH, Kao WY.. 2020. Female sterility and ovule malformation of Oxalis corymbosa, an exotic plant in Taiwan. Flora. 265:151571. [Google Scholar]
- Tukun AB, Shaheen N, Banu CP, Mohiduzzaman M, Islam S, Begum M.. 2014. Antioxidant capacity and total phenolic contents in hydrophilic extracts of selected Bangladeshi medicinal plants. Asian Pac J Trop Med. 7:S568–S573. [DOI] [PubMed] [Google Scholar]
- Wang Q, Yu S, Gao C, Ge Y, Cheng R.. 2020. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Rubus chingii Hu. Mitochondrial DNA Part B. 5(2):1307–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaio M, Gardner A, Emshwiller E, Guerra M.. 2013. Molecular phylogeny and chromosome evolution among the creeping herbaceous Oxalis species of sections Corniculatae and Ripariae (Oxalidaceae). Mol Phylogenet Evol. 68(2):199–211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW057776. The associated BioProject, SRA, and BioSample numbers are PRJNA690071, SRR13370090 and SAMN17227902, respectively.