Abstract
The sensing of small molecules poses the challenge of developing devices able to discriminate between compounds that may be structurally very similar. Here, attention has been paid to the use of self-assembled monolayer (SAM)-protected gold nanoparticles since they enable a modular approach to tune single-molecule affinity and selectivity simply by changing functional moieties (i.e., covering ligands), along with multivalent molecular recognition. To date, the discovery of monolayers suitable for a specific molecular target has relied on trial-and-error approaches, with ligand chemistry being the main criterion used to modulate selectivity and sensitivity. By using molecular dynamics, we showcase that either individual molecular characteristics and/or collective features such as ligand flexibility, monolayer organization, ligand local ordering, and interfacial solvent properties can also be exploited conveniently. The knowledge of the molecular mechanisms that drive the recognition of small molecules on SAM-covered nanoparticles will critically expand our ability to manipulate and control such supramolecular systems.
Introduction
Sensitive, selective chemical and biological sensors are highly demanded in a broad range of applications in chemistry, biology, healthcare, medicine, and environmental protection. Nevertheless, the development of more efficient, low-cost, versatile, and miniaturized sensors requires continuous advancements in technology, coupled with fundamental knowledge in chemistry, biology, and materials science.1−3 In 2012, on recognizing the considerable potential for nanotechnology to facilitate the development of sensitive, adaptable devices for detection, identification, and quantification of substances, the National Nanotechnology Initiative launched its fifth Nanotechnology Signature Initiative (NSI), entitled “Nanotechnology for Sensors and Sensors for Nano-technology: Improving and Protecting Health, Safety, and the Environment” (or the Sensors NSI).4 Engineered nanomaterials possess characteristics that might advance both the recognition and transduction steps of a probing event, as well as the signal-to-noise ratio, thanks to the miniaturization of the sensor elements.5,6 Thus, sensing at the nanoscale may be viewed as a natural fit. Nanomaterials with a high surface-to-volume ratio offer inherently high sensitivity to surface processes and lead to enhanced chemical reactivity, which can be modulated by the particle type, shape, and surface topography.1 Then, only a small number of analyte molecules are needed to produce a measurable signal, allowing both a reduction of sample volumes and a miniaturization of sensors.7 Moreover, the possibility to tailor nanomaterials with functional moieties confers precise sensitivity and specificity.8
Among other nanosensing platforms,9−15 gold nanoparticles (AuNPs) have inspired intensive efforts in the scientific community. Besides offering highly controllable sizes, shapes, and optical or electrical properties, they can be functionalized with a large variety of molecules involved in (bio)recognition with, for instance, oligonucleotides, antibodies, peptides, proteins, microorganisms, drugs, and other small molecules.16−19 In this regard, AuNPs capped with organic thiols are emerging as appealing chemical sensing tools.20 Thiolated ligands are known to bind strongly to gold surfaces and form self-assembled monolayers (SAMs). SAM-protected AuNPs (SAM-AuNPs) are thus stable multivalent systems, able to operate multiple molecular recognition events simultaneously at their surface.21−27
By changing ligands in the nanoparticle capping layer, it is possible to impart different chemical selectivities and sensitivities toward target analytes or groups of target analytes.28−30 Rotello and Bunz used cationic gold nanoparticles coated with different ammonium thiol derivatives to generate sensor arrays and polyanionic fluorescent polymers or proteins as indicators.28 This method was later expanded by Prins et al. to sense small polyanionic molecules.29 Based on a newly developed “NMR chemosensing” analytical approach, Mancin et al. demonstrated the ability of small gold nanoparticles passivated by a monolayer of amphiphilic thiols to detect salicylate molecules in a selective way. They could distinguish among a set of isomers, which differed only in the relative position of two functional groups, even when present in a mixture.31 By a combination of molecular dynamics (MD) calculations and magnetization-transfer NMR protocols, the authors proved the existence of transient binding pockets (for salicylate) in the monolayer with molecular features mimicking drug–protein recognition processes.32,33
Very recently, Gabrielli et al. have reported on a set of alkyl thiols bearing different terminal groups.34 If self-assembled on a ∼2 nm size gold core, they could detect and discriminate among a series of phenethylamine derivatives (designer drugs) in water, with estimated binding constants falling in the range of 1 × 105–1.3 × 106 M–1 for the most efficient system. An interesting point of this study is that it indicates the ability of rather nonspecific monolayers to discriminate chemically similar analytes, a sign of the complexity of noncovalent phenomena taking place on the monolayer.
To date, discovery of monolayers suitable for a specific sensing application is typically through trial and error, based on a handful of candidates, where ligand chemistry is the only criterion commonly adopted to modulate selectivity and sensitivity.35 Thus, deepening the knowledge of the basic principles governing molecular sensing at the monolayer surface has the potential to critically expand our ability to manipulate SAM-AuNP-based devices.
To this purpose, taking advantage of the molecular view offered by MD calculations, we show how recognition occurs at the surface of three differently designed SAM-AuNPs, and we decipher which molecular features of both the coating ligand and monolayer affect the identification and discrimination of six small amphiphilic molecules (Scheme 1). Several of the thiols and compounds we consider here were previously tested experimentally by Gabrielli et al.34 for sensing phenethylamine derivatives (designer drugs) in water, and this also offers us the opportunity to dissect the influence of electrostatic and hydrophobic interactions, two major driving forces in supramolecular recognition. Moreover, for the first time, this study takes explicitly into account the role of monolayer organization and the solvent in mediating the interaction between SAM-AuNPs and small molecules. Even though the multivalent nature of these systems is an important feature and synergistic effects between binding sites could arise through cooperative recognition, we focus here on profiling molecular forces and ligand properties regulating the recognition of small molecules on SAM-AuNPs, leaving the detailed investigation of multivalency for a further study.
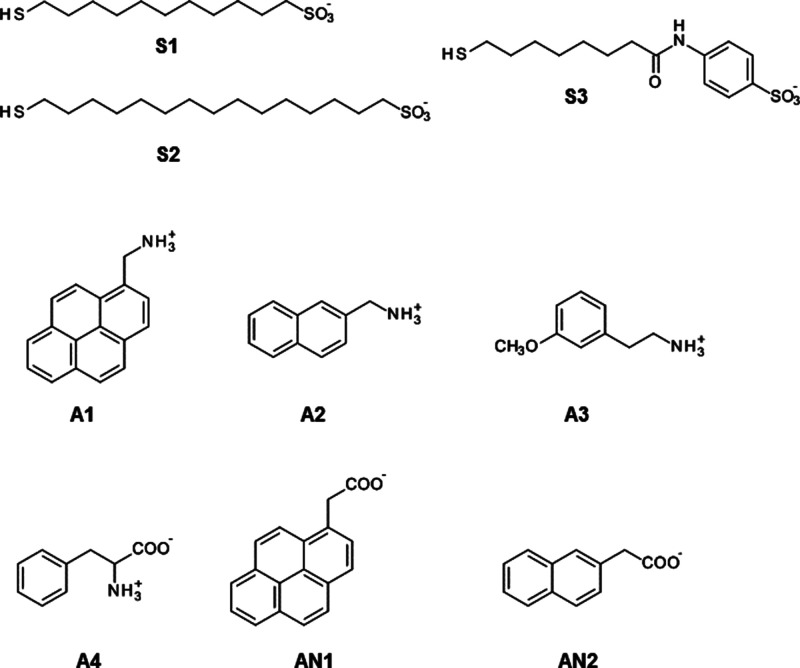
Scheme 1. Structure of Nanoparticle-Coating Thiols (S1, S2, and S3) and Small Molecules Considered in This Work.
Computational Methods
Ligands (S1, S2, and S3) and analytes (A1, A2, A3, A4, AN1, and AN2) were parametrized using antechamber, assigning gaff2 atom types,36,37 and their partial charges were derived by applying the RESP method provided by the RED server.38 The ligand protonation state was assigned based on a report of Gabrielli et al.34 Au–Au interactions were described with the parameters of the INTERFACE39 force field for metals. The Nanoparticle Builder module of OpenMD40 was used to generate an icosahedral gold cluster of 144 atoms, which models nanoparticles with an average core size of 1.6–1.8 nm.41 To preserve the geometry during simulation, all gold atoms within a distance of 2.90 Å were bonded to each other.42 Fifty34 sulfur headgroups and attached ligands were uniformly distributed on the gold surface;43 a harmonic bond was created between each sulfur atom and a gold atom within 3.3 Å.42 All Au–S and Au–Au bonds were modeled with a harmonic potential with a spring constant of 50.000 kJ/mol nm2.42 The interface structure disregards possible gold–sulfur binding motifs (e.g., staples, trimeric motifs, etc.); it has been shown recently42 that this simplified treatment yields a description of the structure of self-assembled alkanethiols of various lengths (n = 3–15) on a 2–6 nm size gold core in agreement with experiments.
Each solvated model (e.g., nanoparticles, analytes, and nanoparticle–analyte complexes) was prepared as described in the following paragraphs. Using the tleap program,44 the system was solvated with TIP3P water molecules, extending at least 20 Å from each solute atom; counterions were added to neutralize the system and match the experimental concentration.34 A combination of the steepest descent (10,000 cycles) and conjugate gradient methods (10,000 cycles) followed by a heating phase of 100 ps in the NVT ensemble (integration step = 1 fs) was carried out to reach the production temperature of 300 K. Then, density was brought to its final value with at least 50 ns in NPT conditions (integration step = 2 fs, pressure 1 atm), and pressure was maintained using a Berendsen barostat.45 Finally, we switched to the Monte Carlo barostat implemented in Amber for production run of which the first part was discarded until the steady state of the ligand RMSD was reached. The trajectory for final ensemble averages (400 ns) was stored from this point on. Temperature was controlled by the Langevin method (damping coefficient of 5 ps–1) throughout all simulations. Electrostatic interactions were computed by means of the Particle Mesh Ewald (PME)46 algorithm, and calculations were carried out using the AMBER 1844,47 suite of programs running on our hybrid CPU (minimization and heating) and GPU (all other steps) cluster48,49 (mixed precision). Each analyte–nanoparticle complex was built with 30 analyte molecules (as estimated experimentally34) placed randomly in the simulation box. Structural and energetic analysis was performed via AMBER programs pytraj, cpptraj, and MM-PBSA.py and by several in-house developed python scripts. Specifically, the SAM-AuNP/analyte free energy of binding ΔGb was derived following the Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) approach.50 It estimates the average interaction energy based on the solute molecular mechanics internal energy change (ΔEMM), solvation energy (ΔGsolv), and conformational entropy change of the solute upon binding (−TΔS). ΔEMM consists of changes in the internal energies (ΔEint), electrostatic energy (ΔEele), and van der Waals energy (ΔEvdW). The solvation energy term ΔGsolv includes two components: the electrostatic term (ΔGp_solv) and the nonpolar term (ΔGnp_solv). The sum of ΔEMM and ΔGsolv accounts for the enthalpy change associated with the binding (ΔH). Details on the calculation of each term are provided in the Supporting Information. The results were ensemble-averaged on three repeated and converged simulations.
Results and Discussion
As a first step, we considered S1-functionalized AuNPs (S1-AuNPs) (Scheme 1). S1-AuNPs showed the highest affinities and remarkable selectivity in the experimental tests.34 MD simulations in water revealed that S1 ligands self-organized around the gold core mainly into opposite oriented bundles (Figure 1), and only a limited amount of chains moved freely (see Table S1). Thus, the shell was elongated, and its shape was far from being spherical (see Table S1).
Figure 1.
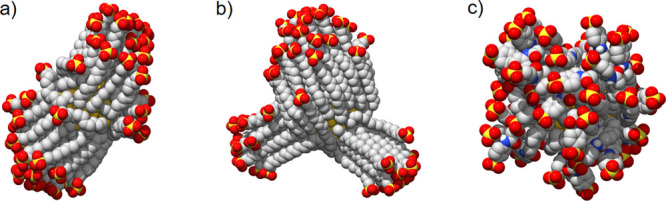
Monolayer organization as predicted by MD calculations for the three shells: (a) S1-AuNP, (b) S2-AuNP, and (c) S3-AuNP. Water molecules and ions are not displayed for clarity.
Spatially heterogeneous surfaces impact overall the NP behavior, especially their interfacial properties.51−56 We investigated the ability of S1-AuNP to bind and distinguish among three positively charged analytes (A1, A2, and A3) and one zwitterionic compound (A4) (see Scheme 1) having decreasing lipophilicity.
In addition, we considered compounds AN1 and AN2 (see Scheme 1). The negatively charged carboxylic group should make the interaction with the monolayer unfavorable, which could in turn be counterbalanced by the aromatic portion of the molecule. This wide spectrum range of analytes also allowed us to span the response of the computational approach in describing NP–small molecule recognition. In fact, despite the undoubted potential of these systems, to date, computational studies have been limited due to the complexity of sampling a multibinding event.57
The simulations showed that S1-AuNP was able to associate effectively with the positively charged compounds (Figure 2). The contact was not permanent, but we observed binding and unbinding events. On average, all A1 molecules interacted with S1-AuNP. The number of contacting molecules (i.e., at a distance lower than 0.5 nm from any heavy atom of the monolayer for times longer than 10 ns) decreased to 29 and 27 for A2 and A3, respectively.
Figure 2.
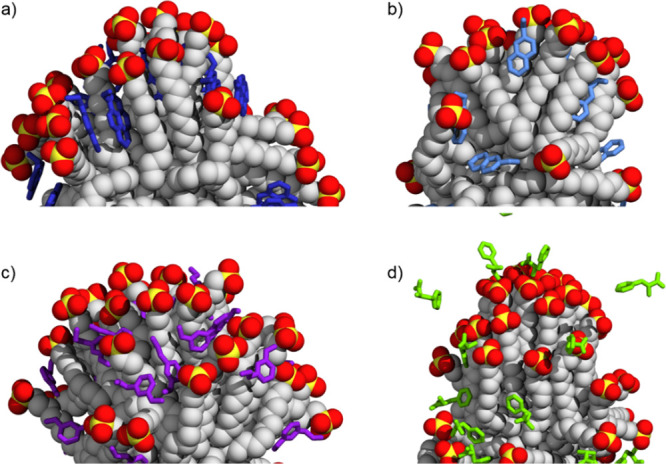
Selected configurations of S1-AuNP association with (a) A1, (b) A2, (c) A3, and (d) A4 as obtained by MD calculations. Water and ions are not shown for the sake of clarity.
The three analytes associated with S1-AuNP in the same region of the monolayer (see Figure S1a).
Comparing the distribution of the sulfonate groups carried by the S1 ligand with that of the amine groups A1–A3, we saw that they almost overlapped, suggesting a local interaction (see Figure S1b–d) that promotes the association. This likely arises from ion pairing and hydrogen bonding between SO3– and NH3+ moieties as can be visually inferred from Figure S2. The complexation was also stabilized by the presence of a few water bridges between the aforementioned functional groups. The total number of salt and water bridges between S1 and the three positively charged compounds was comparable in all systems (i.e., 26 on average), indicating that it is not specific to each analyte.
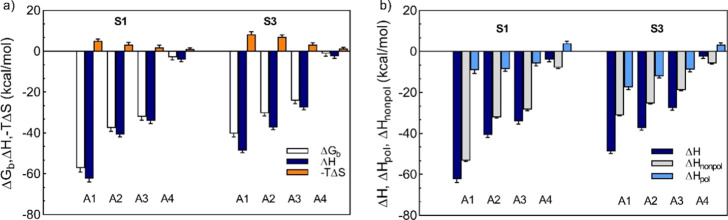
Binding affinity is a straightforward measure of molecular recognition58 and can be computed by MD simulations. The MM/PBSA approach50 was used here to sample the bound states and evaluate the Gibbs binding energy ΔGb of each analyte toward S1-AuNP. It is an end-point free energy method commonly used to compute the binding free energies of small molecules to large biomolecule receptors as well as to describe large interbiomolecular recognitions.59−64 Moreover, it allows to decompose ΔGb in its enthalpic (ΔH) and entropic (−TΔS) terms and to reveal the molecular forces that drive the binding. This thermodynamic signature is reported in Figure 3a.
Figure 3.
Thermodynamic binding signature of S1-AuNP and S3-AuNP to A1, A2, A3, and A4. (a) Binding free energy (ΔGb, white), enthalpy (ΔH, blue), and entropy (−TΔS, orange) variation on an analyte basis. (b) Decomposition of total binding enthalpy (ΔH, blue) into nonpolar (ΔHnonpol, gray) and polar (ΔHpol, light blue) interaction changes. ΔHpol accounts for electrostatics forces (ΔEele) and polar contribution to solvation (ΔGp_solv)); ΔHnonpol is the sum of van der Waals energy (ΔEvdW), nonpolar solvation (ΔEnp_solv), and internal energy variation (ΔEint) terms.
For A1–A4 compounds, the enthalpy change was favorable (ΔH < 0), whereas entropy variation opposed binding (−TΔS > 0). However, the entropic penalty paid was outweighed by the enthalpy gain, thus suggesting that enthalpy is the major driving force for complex formation. This is also a hallmark of the so-called enthalpy–entropy compensation mechanism observed widely in (bio)molecular complexes.65,66
Pleasingly, the predicted affinity trend agrees well with the experimental counterpart34 (see also Table S2 and Figure S3), with A1 outperforming A2 and A3 also in the calculations.
Polar contribution (ΔHpol) to enthalpy, which arises from Coulombic interactions between S1-AuNP and each analyte and polar solvation energy, was always favorable for binding (see Figure 3b). Thus, the unfavorable desolvation of polar groups was compensated for by favorable intermolecular electrostatic interactions. For all positively charged analytes, ΔHpol yielded contribution in the range 4–9 kcal/mol and accounted for only a small fraction of the total binding enthalpy. Strong intermolecular van der Waals interactions and hydrophobic forces are instead required for boosting the molecular recognition. Indeed, ΔHnonpol was the dominant energetic contribution in association and the main interaction responsible for the observed selectivity of S1-AuNP.
The complementarity of electrostatic and hydrophobic interactions in driving complex formation on SAMs is even more evident, including the binding of the zwitterionic (A4) and negatively charged compounds (AN2, AN1) in the discussion. A4 is close to A3 in terms of log D values (−1.46 and −1.04, respectively) but bears a negatively charged carboxylic group besides a positively charged amine. This had a dramatic effect on the affinity and led to a decrease (less negative value) of ΔGb from −32.2 ± 1.6 kcal/mol for A3 to −3.0 ± 1.2 kcal/mol for A4. The reduced affinity of A4 was also seen by Gabrielli et al. in the NOE pumping spectra, where A4 did not produce any signal.34 The polar contribution (ΔHpol) became positive (unfavorable); at the same time, the nonpolar term (ΔHnonpol) reduced significantly, overall accounting for a much less effective enthalpic stabilization. As a consequence, the number of A4 molecules temporarily making contact with S1-AuNP dropped down to 19.
At the same time, none of the negatively charged analytes (AN1 and AN2) bound significantly to S1-AuNP. Both transiently approached the nanoparticle on the surface (see Figure S4), but their free energy of binding was positive (ΔGb = 1.40 ± 0.8 kcal/mol for AN1 and 5.3 ± 1.2 kcal/mol for AN2), evidencing that the association with anionic amphiphilic molecules is not favored by thermodynamics.
Taken together, these findings provide clues that electrostatic interactions between oppositely charged species are needed to drive analytes toward their optimal binding mode; hydrophobic forces that originate from the interaction of aromatic units in the hydrophobic portion of the shell stabilize the complex and modulate the affinity of S1-AuNP toward the binding partner. A precise combination of these two forces thus appears as a way to control the overall affinity and specificity.
Among positively charged analytes, the binding mode of A1 deserves a specific discussion. At a closer look, while some molecules interacted with the monolayer at the water–bundle interface, the majority of A1 resided inside the bundles with the aromatic rings oriented parallel to the hydrophobic backbone of S1 (see Figure S5a). This specific placement allows optimization of the van der Waals forces and may explain the highest affinity (ΔGb) of this compound due to a more favorable nonpolar contribution (ΔHnonpolar) to association (see Figure 3a). For comparison, A2, which features only a naphthalene moiety, is not able to create a pattern of hydrophobic interactions with the extent similar to that observed for A1 (see Figure S5b). To test the efficiency of this recognition mechanism, we then considered S2-AuNP (see Scheme 1). S2-AuNP has the same core size of S1-AuNP, but the alkyl chain in S2 is longer than that in S1 (16 vs 11 carbon atoms, respectively). This endows ligands with a higher degree of freedom and the nanosensor with greater hydrophobic potential. S2-AuNP presented a monolayer organized in bundles (see Figure 1b) with a few free chains (see Table S1). Binding of A1 altered the monolayer organization: due to their inherent flexibility and small NP core size, the chains fluctuate and bend over the gold core to optimize ligand–ligand and ligand–A1 multiple interactions (see Figure S6). This reduced the number of bundles in the monolayer upon binding (see Table S3). Still, A1 resided among ligands as seen for S1-AuNP, thus confirming the strength and role of ligand–analyte parallel pairing in leading the molecular interaction.
The behavior of S2-AuNP also raises another important issue commonly neglected when designing new ligands for supramolecular sensors based on SAM-AuNPs: the role of ligand flexibility and collective/individual loss (or gain) in entropy upon the recognition and the consequent negative (or positive) contribution to the total free energy of binding. The computed entropy change (−TΔS) for all S1-AuNP complexes (see Figure 3a) was positive, indicating a loss of conformational flexibility of the binding partners, which is unfavorable for binding. This cost was lower for A3 and A4 than for A1 and A2 complexes. A3/A4 compounds had a lower affinity toward S1-AuNP, and A4 also showed a decreased number of bound molecules, overall resulting in a smaller effect on chain mobility. Consistently, the structural features of S1-AuNP were closer to those of the unbound nanoparticle for S1-AuNP/A3 and S1-AuNP/A4 systems (see Table S4).
This first set of calculations provides us with unprecedented molecular details into factors affecting the ability of a self-assembled monolayer to discriminate between small target molecules on spherical surfaces. Yet, molecular recognition is a two-player game, and acquiring a complete picture is possible only by exploring the binding also from the nanoparticle perspective, i.e., changing its covering ligands. Fluorescence titration experiments performed by Gabrielli et al.34 assessed that modification of the coating thiol with an aromatic head group (S3, see Scheme 1) results in a reduction of the affinity for each analyte, compared to S1-AuNP (see Table S2). However, no comprehensive molecular rationale was attempted at that time.
Our MD calculations predicted an essentially spherical organization of S3 ligands around the gold core (see Figure 1c and Table S1) devoid of any chain bundling (contrary to S1 and S2 ligands), which can be ascribed to the presence of a bulkier headgroup and shorter length of the alkyl chain that restrict ligand association.67 Considering again the three positively charged (A1, A2, and A3) and zwitterionic (A4) compounds (see Figure 4), the average number of molecules bound was comparable to that found for S1-AuNP as well as the total number of ion pairs and water bridges among S3 chains and each analyte (see Table S5), suggesting that these features are only modestly influenced by the ligand chemistry in the systems investigated here.
Figure 4.
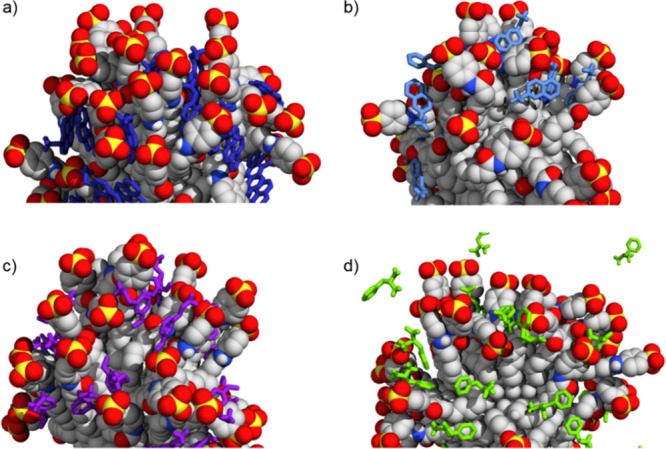
Selected configurations of S3-AuNP association with (a) A1, (b) A2, (c) A3, and (d) A4 as obtained from MD calculations. Water and ions are not shown for the sake of clarity.
The binding thermodynamics (see Figure 3) indicated that the entropic term still caused an energetic penalty; its value was larger than that found for the corresponding S1-AuNP systems. In the unbound state, the absence of bundles endowed thiols with a higher degree of mobility and flexibility; when analytes approach the monolayer, they hinder ligand natural conformational fluctuations more than those in S1-AuNP, leading to an increased entropic cost. Reasonably, this decreased with the affinity toward each analyte. At the same time, we observed a significant reduction in the enthalpy contribution to binding. Coupling these two effects led to a reduced affinity of S3-AuNP toward each compound if compared to S1-AuNP, which matches the experimental findings32 (see also Table S2 and Figure S3). A summary of the structural characterization of S3-AuNP upon A1, A2, A3, and A4 binding can be found in Table S6 and Figure S7. Again, none of the negatively charged analytes (AN2 and AN1) was detected proficiently by S3-AuNP, showing positive ΔGb values of 4.1 ± 1.0 and 1.1 ± 0.5 kcal/mol, respectively, highlighting the contribution of electrostatic interactions in recognition processes involving SAMs and small charged molecules.
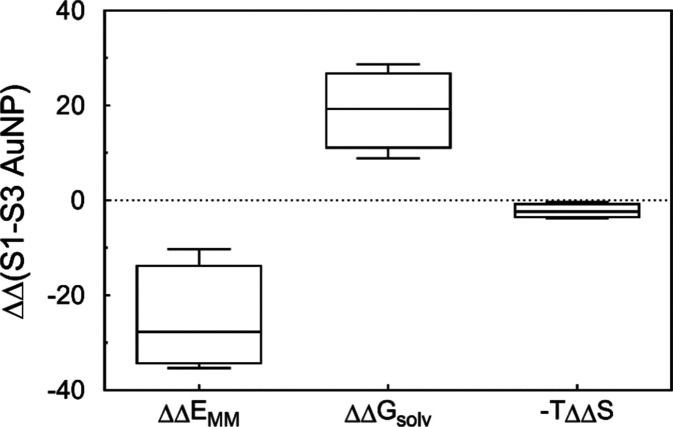
In contrast to S1-AuNP, both polar and nonpolar terms contributed to ΔH, and we sought where these differences may arise. Figure 5 shows the difference in terms of ΔEMM, ΔGsolv, and −TΔS between S1-AuNP and S3-AuNP bound to A1, A2, A3, and A4. Although the change in entropy was obviously dissimilar between the two systems, it did not represent the major contribution to recognition. Intermolecular (and intramolecular) interactions (ΔEMM) were mainly responsible for the differing affinity of these systems toward the same analyte. Clearly, the chemical structure of the coating ligands determines the ability to establish more (or less) favorable interactions with a binding partner. At the same time, solvent-mediated forces (ΔGsolv) were equally responsible for the different interaction abilities.
Figure 5.
Box plot of the difference in terms of ΔEGMM, ΔGsolv, and −TΔS between S1-AuNP and S3-AuNP once bound to A1, A2, A3, and A4.
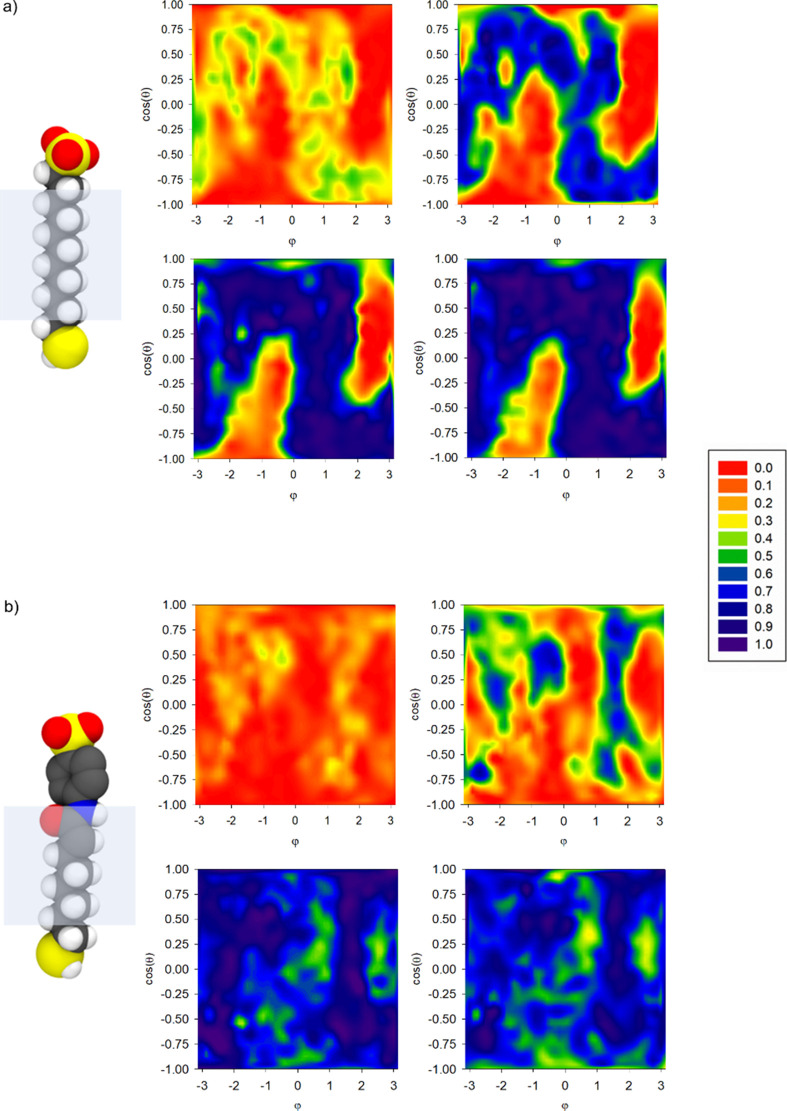
Figure 6 shows the comparison of averaged density distributions of water around the core for S1-AuNP and S3-AuNP at several distances from the gold surface. It is evident that different monolayer morphologies led to dissimilar nanoparticle hydration within the monolayer. For S1-AuNP, the aggregation of ligands made the distribution of water molecules around the nanoparticle spatially heterogeneous, with areas less hydrated or not accessible to the solvent, for instance, inside the bundles (red areas in Figure 6a).
Figure 6.
Normalized water distribution at increasing distance from the gold surface (∼4 to 10 Å up-left to right-down panels) for (a) S1-AuNP and (b) S3-AuNP. The graphs plot the distribution of the atom (oxygen of water or carbon of thiols) closest to spherical surfaces (centered on the gold core and placed at increasing distances from the NP core) shown as a two-dimensional projection of the sphere surface (x axis, the azimuthal angle φ; y axis, the cosine of the polar angle θ). A value of 1 indicates that an oxygen atom of a water molecule is always the closest; if it is equal to 0, it indicates that a carbon atom of a chain is always the closest.
Vice versa, when the organization of the monolayer is disordered (as in S3-AuNP, see Figure 6b), the penetration of the solvent within the shell was higher and more uniform. Thus, the presence of bundles induces a hydrophobic environment that favors the recognition of small amphiphilic molecules.
This observation is consistent with recent evidence by van Lehn et al. on planar SAMs.68−70 The authors proved that spatially disordered SAMs affect the interfacial properties of the water solvent and decrease the interfacial hydrophobicity with respect to ordered surfaces. Such a phenomenon not only explains the marked positive difference in the solvation term ΔGsolv between S1-AuNP and S3-AuNP (see Figure 5) but also brings out the active contribution of the solvent in the recognition mechanism between SAMs and small amphiphilic molecules on curved surfaces.
Summing all up, the reduced affinity of S3-AuNP with respect to S1-AuNP stems from several concomitant factors: first, the different ligand chemistry responsible for less effective interactions as evidenced by ΔΔEMM; second, the dissimilar ligand flexibility, which modulates the binding affinity toward an analyte through different entropy costs, higher for S3-AuNPs; and last, the different monolayer organization—the disordered shell in S3-AuNP offers a less hydrophobic solvation microenvironment, which disfavors the partition of amphiphilic analytes.
Conclusions
Sensing platforms based on self-assembled monolayers (SAMs) of organic thiols on gold nanoparticles are multivalent and cooperative systems whose strength and selectivity toward selected substrates can be tailored by designing ad hoc the monolayer constituents. To that end, mastering the basic principles that regulate recognition at the monolayer surface is needed. In this paper, we have investigated three different SAMs and by means of molecular dynamics calculations have analyzed their ability to detect and discriminate a set of small amphiphilic charged molecules from a molecular perspective. The chosen SAMs are deprived of any structural and chemical feature that would permit specific interactions, allowing us to explore the underlying forces and molecular attributes that shape the formation of such supramolecular complexes.
Our comprehensive investigation reveals that probing small molecules with SAMs on curved surfaces is a complex, multidimensional phenomenon distinct from that occurring on planar SAMs. It is regulated by either single-molecule properties (such as ligand chemistry and flexibility) or collective features (such as SAM organization and presence of interfaces). Moreover, the same binding event may significantly alter the monolayer structure, thus adding another level of complexity. We also showcase that the shell structure influences the solvation interfacial microenvironment through combined hydrophobic interactions, which may be tuned to tailor the affinity.
We believe that the acquired knowledge of the intimate mechanisms driving sensing at the SAM surface will expand our ability to manipulate and computationally design nanodevices with enhanced recognition ability toward small molecules, such as drugs, metabolites, or small molecular markers for cancer.
Acknowledgments
This research was supported by the Italian Ministry of University Research (MIUR) through the project “Structure and function at the nanoparticle biointerface” (RBSI14PBC6) and by the Czech Ministry of Education, Youth and Sports through the Large Infrastructures for Research, Experimental Development and Innovations project “IT4Innovations National Supercomputing Center – LM2015070”. Z. P. would also like to acknowledge the internal grant of J. E. Purkyne University (UJEP-IGA-TC-2019-53-02-2).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.9b03686.
Details on MM-PB/SA methodology and additional results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Zhang S.; Geryak R.; Geldmeier J.; Kim S.; Tsukruk V. V. Synthesis, Assembly, and Applications of Hybrid Nanostructures for Biosensing. Chem. Rev. 2017, 117, 12942–13038. 10.1021/acs.chemrev.7b00088. [DOI] [PubMed] [Google Scholar]
- Paulovich F. V.; De Oliveira M. C. F.; Oliveira O. N. A Future with Ubiquitous Sensing and Intelligent Systems. ACS Sensors 2018, 3, 1433–1438. 10.1021/acssensors.8b00276. [DOI] [PubMed] [Google Scholar]
- Justus K. B.; Hellebrekers T.; Lewis D. D.; Wood A.; Ingham C.; Majidi C.; LeDuc P. R.; Tan C. A Biosensing Soft Robot: Autonomous Parsing of Chemical Signals through Integrated Organic and Inorganic Interfaces. Sci. Robot. 2019, 4, eaax0765 10.1126/scirobotics.aax0765. [DOI] [PubMed] [Google Scholar]
- Fadel T. R.; Farrell D. F.; Friedersdorf L. E.; Griep M. H.; Hoover M. D.; Meador M. A.; Meyyappan M. Toward the Responsible Development and Commercialization of Sensor Nanotechnologies. ACS Sensors 2016, 1, 207–216. 10.1021/acssensors.5b00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani L.; Li F. Mechanistic Challenges and Advantages of Biosensor Miniaturization into the Nanoscale. ACS Sensors 2017, 2, 458–467. 10.1021/acssensors.7b00069. [DOI] [PubMed] [Google Scholar]
- Quesada-González D.; Merkoçi A. Nanomaterial-based Sevices for Point-Of-Care Diagnostic Applications. Chem. Soc. Rev. 2018, 47, 4697–4709. 10.1039/C7CS00837F. [DOI] [PubMed] [Google Scholar]
- Zhang A.; Lieber C. M. Nano-Bioelectronics. Chem. Rev. 2016, 116, 215–257. 10.1021/acs.chemrev.5b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkaew N.; Simsek M.; Griesche C.; Baeumner A. J. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem. Rev. 2019, 119, 120–194. 10.1021/acs.chemrev.8b00172. [DOI] [PubMed] [Google Scholar]
- Liu X.; Wang F.; Aizen R.; Yehezkeli O.; Willner I. Graphene Oxide/Nucleic-Acid-Stabilized Silver Nanoclusters: Functional Hybrid Materials for Optical Aptamer Sensing and Multiplexed Analysis of Pathogenic DNAs. J. Am. Chem. Soc. 2013, 135, 11832–11839. 10.1021/ja403485r. [DOI] [PubMed] [Google Scholar]
- Howes P. D.; Chandrawati R.; Stevens M. M. Colloidal Nanoparticles as Advanced Biological Sensors. Science 2014, 346, 1247390. 10.1126/science.1247390. [DOI] [PubMed] [Google Scholar]
- Barnard A.; Posocco P.; Fermeglia M.; Tschiche A.; Calderon M.; Pricl S.; Smith D. K. Double-degradable Responsive Self-assembled Multivalent Arrays – Temporary Nanoscale Recognition between Dendrons and DNA. Org. Biomol. Chem. 2014, 12, 446–455. 10.1039/C3OB42202J. [DOI] [PubMed] [Google Scholar]
- Meyyappan M. Carbon Nanotube-Based Chemical Sensors. Small 2016, 12, 2118–2129. 10.1002/smll.201502555. [DOI] [PubMed] [Google Scholar]
- Xue T.; Liang W.; Li Y.; Sun Y.; Xiang Y.; Zhang Y.; Dai Z.; Duo Y.; Wu L.; Qi K.; Shivananju B. N.; Zhang L.; Cui X.; Zhang H.; Bao Q. Ultrasensitive Detection of miRNA with an Antimonene-based Surface Plasmon Resonance Sensor. Nat. Commun. 2019, 10, 28. 10.1038/s41467-018-07947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Li H.; Wu Z.; Wang M.; Luo J.; Torun H.; Hu P.; Yang C.; Grundmann M.; Liu X.; Fu Y. Advances in Designs and Mechanisms of Semiconducting Metal Oxide Nanostructures for High-precision Gas Sensors Operated at Room Temperature. Mater. Horiz. 2019, 6, 470–506. 10.1039/C8MH01365A. [DOI] [Google Scholar]
- Cai R.; Du Y.; Yang D.; Jia G.; Zhu B.; Chen B.; Lyu Y.; Chen K.; Chen D.; Chen W.; Yang L.; Zhao Y.; Chen Z.; Tan W. Free-standing 2D Nanorafts by Assembly of 1D Nanorods for Biomolecule Sensing. Nanoscale 2019, 11, 12169–12176. 10.1039/C9NR02636C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K.; Agasti S. S.; Kim C.; Li X.; Rotello V. M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779. 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Li X.; Dong Y.; Wang B.; Li D.; Shi Y.; Wu Y. Colorimetric Sensor Array Based on Gold Nanoparticles with Diverse Surface Charges for Microorganisms Identification. Anal. Chem. 2017, 89, 10639–10643. 10.1021/acs.analchem.7b02594. [DOI] [PubMed] [Google Scholar]
- Cantarutti C.; Bertoncin P.; Posocco P.; Hunashal Y.; Giorgetti S.; Bellotti V.; Fogolari F.; Esposito G. The Interaction of β2-microglobulin with Gold Nanoparticles: Impact of Coating, Charge and Size. J. Mater. Chem. B 2018, 6, 5964–5974. 10.1039/C8TB01129J. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Poss G.; Weng Y.; Qi R.; Zheng H.; Nianias N.; Kay E. R.; Guldin S. Probing the Interaction of Nanoparticles with Small Molecules in Real Time via Quartz Crystal Microbalance Monitoring. Nanoscale 2019, 11, 11107–11113. 10.1039/C9NR03162F. [DOI] [PubMed] [Google Scholar]
- Prins L. J. Emergence of Complex Chemistry on an Organic Monolayer. Acc. Chem. Res. 2015, 48, 1920–1928. 10.1021/acs.accounts.5b00173. [DOI] [PubMed] [Google Scholar]
- Liu X.; Hu Y.; Stellacci F. Mixed-ligand Nanoparticles as Supramolecular Receptors. Small 2011, 7, 1961–1966. 10.1002/smll.201100386. [DOI] [PubMed] [Google Scholar]
- Boccalon M.; Bidoggia S.; Romano F.; Gualandi L.; Franchi P.; Lucarini M.; Pengo P.; Pasquato L. Gold Nanoparticles as Drug Carriers: a Contribution to the Quest for Basic Principles for Monolayer Design. J. Mater. Chem. B 2015, 3, 432–439. 10.1039/C4TB01536C. [DOI] [PubMed] [Google Scholar]
- Yapar S.; Oikonomou M.; Velders A. H.; Kubik S. Dipeptide Recognition in Water Mediated by Mixed Monolayer Protected Gold Nanoparticles. Chem. Commun. 2015, 51, 14247–14250. 10.1039/C5CC05909G. [DOI] [PubMed] [Google Scholar]
- Lucarini M.; Franchi P.; Pedulli G. F.; Gentilini C.; Polizzi S.; Pengo P.; Scrimin P.; Pasquato L. Effect of Core Size on the Partition of Organic Solutes in the Monolayer of Water-Soluble Nanoparticles: An ESR Investigation. J. Am. Chem. Soc. 2005, 127, 16384–16385. 10.1021/ja0560534. [DOI] [PubMed] [Google Scholar]
- Sun X.; Liu P.; Mancin F. Sensor Arrays made by Self-organized Nanoreceptors for Detection and Discrimination of Carboxylate Drugs. Analyst 2018, 143, 5754–5763. 10.1039/C8AN01756E. [DOI] [PubMed] [Google Scholar]
- Cho E. S.; Kim J.; Tejerina B.; Hermans T. M.; Jiang H.; Nakanishi H.; Yu M.; Patashinski A. Z.; Glotzer S. C.; Stellacci F.; Grzybowski B. A. Ultrasensitive Detection of Toxic Cations through Changes in the Tunnelling Current across Films of Striped Nanoparticles. Nat. Mater. 2012, 11, 978–985. 10.1038/nmat3406. [DOI] [PubMed] [Google Scholar]
- Li Y.; Wang Y.; Huang G.; Gao J. Cooperativity Principles in Self-Assembled Nanomedicine. Chem. Rev. 2018, 118, 5359–5391. 10.1021/acs.chemrev.8b00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz U. H. F.; Rotello V. M. Gold Nanoparticle–Fluorophore Complexes: Sensitive and Discerning “Noses” for Biosystems Sensing. Angew. Chem. Int. Ed. 2010, 49, 3268–3279. 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- Pezzato C.; Maiti S.; Chen J. L. Y.; Cazzolaro A.; Gobbo C.; Prins L. J. Monolayer Protected Gold Nanoparticles with Metal-ion Binding Sites: Functional Systems for Chemosensing Applications. Chem. Commun. 2015, 51, 9922–9931. 10.1039/C5CC00814J. [DOI] [PubMed] [Google Scholar]
- Ertem E.; Diez-Castellnou M.; Ong Q. K.; Stellacci F. Novel Sensing Strategies Based on Monolayer Protected Gold Nanoparticles for the Detection of Metal Ions and Small Molecules. Chem. Rec. 2018, 18, 819–828. 10.1002/tcr.201700065. [DOI] [PubMed] [Google Scholar]
- Perrone B.; Springhetti S.; Ramadori F.; Rastrelli F.; Mancin F. “NMR Chemosensing” Using Monolayer-Protected Nanoparticles as Receptors. J. Am. Chem. Soc. 2013, 135, 11768–11771. 10.1021/ja406688a. [DOI] [PubMed] [Google Scholar]
- Riccardi L.; Gabrielli L.; Sun X.; De Biasi F.; Rastrelli F.; Mancin F.; De Vivo M. Nanoparticle-Based Receptors Mimic Protein-Ligand Recognition. Chem 2017, 3, 92–109. 10.1016/j.chempr.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Riccardi L.; De Biasi F.; Rastrelli F.; De Vivo M.; Mancin F. Molecular-Dynamics-Simulation-Directed Rational Design of Nanoreceptors with Targeted Affinity. Am. Ethnol. 2019, 58, 7702–7707. 10.1002/anie.201902316. [DOI] [PubMed] [Google Scholar]
- Gabrielli L.; Rosa-Gastaldo D.; Salvia M.-V.; Springhetti S.; Rastrelli F.; Mancin F. Detection and Identification of Designer Drugs by Nanoparticle-based NMR Chemosensing. Chem. Sci. 2018, 9, 4777–4784. 10.1039/C8SC01283K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubble L. J.; Cooper J. S.; Sosa-Pintos A.; Kiiveri H.; Chow E.; Webster M. S.; Wieczorek L.; Raguse B. High-Throughput Fabrication and Screening Improves Gold Nanoparticle Chemiresistor Sensor Performance. ACS Comb. Sci. 2015, 17, 120–129. 10.1021/co500129v. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wang W.; Kollman P. A.; Case D. A. Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graph. Model. 2006, 25, 247–260. 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Vanquelef E.; Simon S.; Marquant G.; Garcia E.; Klimerak G.; Delepine J. C.; Cieplak P.; Dupradeau F.-Y. R.E.D. Server: A Web Service for Deriving RESP and ESP Charges and Building Force Field Libraries for New Molecules and Molecular Fragments. Nucleic Acids Res. 2011, 39, W511–W517. 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz H.; Lin T.-J.; Kishore Mishra R.; Emami F. S. Thermodynamically Consistent Force Fields for the Assembly of Inorganic, Organic, and Biological Nanostructures: The INTERFACE Force Field. Langmuir 2013, 29, 1754–1765. 10.1021/la3038846. [DOI] [PubMed] [Google Scholar]
- http://openmd.org/download/.
- Jensen K. M. Ø.; Juhas P.; Tofanelli M. A.; Heinecke C. L.; Vaughan G.; Ackerson C. J.; Billinge S. J. L. Polymorphism in magic-sized Au144(SR)60 clusters. Nat. Commun. 2016, 7, 11859. 10.1038/ncomms11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew A. K.; Van Lehn R. C. Effect of Core Morphology on the Structural Asymmetry of Alkanethiol Monolayer-Protected Gold Nanoparticles. J. Phys. Chem. C 2018, 122, 26288–26297. 10.1021/acs.jpcc.8b09323. [DOI] [Google Scholar]
- Wong O. A.; Heinecke C. L.; Simone A. R.; Whetten R. L.; Ackerson C. J. Ligand Symmetry-Equivalence on Thiolate Protected Gold Nanoclusters Determined by NMR Spectroscopy. Nanoscale 2012, 4, 4099–4102. 10.1039/c2nr30259d. [DOI] [PubMed] [Google Scholar]
- Case D.A.; Ben-Shalom I.Y.; Brozell S.R.; Cerutti D.S.; Cheatham T.E.I.; Cruzeiro V.W.D.; Darden T.A.; Duke R.E.; Ghoreishim D.; Gilson M.K.; Gohlke H.; Goetz A.W.; Greene D.; Harris R.; Homeyer N.; Izadi S.; Kovalenko A.; Kurtzman T.; Lee T.S.; LeGrand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Mermelstein D.J.; Merz K.M.; Miao Y.; Monard G.; Nguyen C.; Nguyen H.; Omelyan I.; Onufriev A.; Pan F.; Qi R.; Roe D.R.; Roitberg A.; Sagui C.; Schott-Verdugo S.; Shen J.; Simmerling C.L.; Smith J.; Salomon-Ferrer R.; Swails J.; Walker R.C.; Wang J.; Wei H.; Wolf R.M.; Wu X.; Xiao L.; York D.M.; Kollman P. A.. AMBER 2018, University of California, San Francisco.
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Salomon-Ferrer R.; Case D. A.; Walker R. C. An Overview of the Amber Biomolecular Simulation Package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. 10.1002/wcms.1121. [DOI] [Google Scholar]
- Salomon-Ferrer R.; Götz A. W.; Poole D.; Le Grand S.; Walker R. C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory 2013, 9, 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Le Grand S.; Götz A. W.; Walker R. C. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. 10.1016/j.cpc.2012.09.022. [DOI] [Google Scholar]
- Wang E.; Sun H.; Wang J.; Wang Z.; Liu H.; Zhang J. Z. H.; Hou T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- Bauer C. A.; Stellacci F.; Perry J. W. Relationship between Structure and Solubility of Thiol-Protected Silver Nanoparticles and Assemblies. Top. Catal. 2008, 47, 32–41. 10.1007/s11244-007-9032-5. [DOI] [Google Scholar]
- Centrone A.; Penzo E.; Sharma M.; Myerson J. W.; Jackson A. M.; Marzari N.; Stellacci F. The Role of Nanostructure in the Wetting Behavior of Mixed-monolayer-protected Metal Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9886–9891. 10.1073/pnas.0803929105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuna J. J.; Voïtchovsky K.; Singh C.; Jiang H.; Mwenifumbo S.; Ghorai P. K.; Stevens M. M.; Glotzer S. C.; Stellacci F. The Effect of Nanometre-scale Structure on Interfacial Energy. Nat. Mater. 2009, 8, 837–842. 10.1038/nmat2534. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Basak S.; Wunsch B. H.; Kumar R.; Stellacci F. Effect of Composition on the Catalytic Properties of Mixed-ligand-coated Gold Nanoparticles. Angew. Chem. Int. Ed. 2011, 50, 7900–7905. 10.1002/anie.201101821. [DOI] [PubMed] [Google Scholar]
- Huang R.; Carney R. P.; Stellacci F.; Lau B. L. T. Colloidal Stability of Self-assembled Monolayer-coated Gold Nanoparticles: The Effects of Surface Compositional and Structural Heterogeneity. Langmuir 2013, 29, 11560–11566. 10.1021/la4020674. [DOI] [PubMed] [Google Scholar]
- Huang R.; Carney R. P.; Stellacci F.; Lau B. L. T. Protein-nanoparticle Interactions: The Effects of Surface Compositional and Structural Heterogeneity are Scale Dependent. Nanoscale 2013, 5, 6928–6935. 10.1039/c3nr02117c. [DOI] [PubMed] [Google Scholar]
- Nash J. A.; Kwansa A. L.; Peerless J. S.; Kim H. S.; Yingling Y. G. Advances in Molecular Modeling of Nanoparticle–Nucleic Acid Interfaces. Bioconjugate Chem. 2016, 28, 3–10. 10.1021/acs.bioconjchem.6b00534. [DOI] [PubMed] [Google Scholar]
- Baron R.; McCammon J. A. Molecular Recognition and Ligand Association. Annu. Rev. Phys. Chem. 2013, 64, 151–175. 10.1146/annurev-physchem-040412-110047. [DOI] [PubMed] [Google Scholar]
- Tonelli M.; Boido V.; Colla P. L.; Loddo R.; Posocco P.; Paneni M. S.; Fermeglia M.; Pricl S. Pharmacophore Modeling, Resistant Mutant Isolation, Docking, and MM-PBSA analysis: Combined Experimental/Computer-Assisted Approaches to Identify New Inhibitors of the Bovine Viral Diarrhea Virus (BVDV). Bioorg. Med. Chem. 2010, 18, 2304–2316. 10.1016/j.bmc.2010.01.058. [DOI] [PubMed] [Google Scholar]
- Bromfield S. M.; Posocco P.; Fermeglia M.; Pricl S.; Rodríguez-López J.; Smith D. K. A Simple New Competition Assay for Heparin Binding in Serum Applied to Multivalent PAMAM Dendrimers. Chem. Commun. 2013, 49, 4830–4832. 10.1039/c3cc41251b. [DOI] [PubMed] [Google Scholar]
- Bromfield S. M.; Posocco P.; Chan C. W.; Calderon M.; Guimond S. E.; Turnbull J. E.; Pricl S.; Smith D. K. Nanoscale Self-assembled Multivalent (SAMul) Heparin Binders in Highly Competitive, Biologically Relevant, Aqueous Media. Chem. Sci. 2014, 5, 1484–1492. 10.1039/c4sc00298a. [DOI] [Google Scholar]
- Kong X.; Sun H.; Pan P.; Zhu F.; Chang S.; Xu L.; Li Y.; Hou T. Importance of Protein Flexibility in Molecular Recognition: A Case Study on Type-I1/2 Inhibitors of ALK. Phys. Chem. Chem. Phys. 2018, 20, 4851–4863. 10.1039/C7CP08241J. [DOI] [PubMed] [Google Scholar]
- Chen C.; Posocco P.; Liu X.; Cheng Q.; Laurini E.; Zhou J.; Liu C.; Wang Y.; Tang J.; Col V. D.; Yu T.; Giorgio S.; Fermeglia M.; Qu F.; Liang Z.; Rossi J. J.; Liu M.; Rocchi P.; Pricl S.; Peng L. Mastering Dendrimer Self-Assembly for Efficient siRNA Delivery: From Conceptual Design to In Vivo Efficient Gene Silencing. Small 2016, 12, 3667–3676. 10.1002/smll.201503866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurini E.; Harel D.; Marson D.; Schepmann D.; Schmidt T. J.; Pricl S.; Wünsch B. Identification, pharmacological evaluation and binding mode analysis of novel chromene and chromane based σ1 receptor ligands. Eur. J. Med. Chem. 2014, 83, 526–533. 10.1016/j.ejmech.2014.06.054. [DOI] [PubMed] [Google Scholar]
- Laurini E.; Marson D.; Posocco P.; Fermeglia M.; Pricl S. Structure and Binding Thermodynamics of Viologen-phosphorous Dendrimers to Human Serum Albumin: A Combined Computational/Experimental Investigation. Fluid Phase Equilib. 2016, 422, 18–31. 10.1016/j.fluid.2016.02.014. [DOI] [Google Scholar]
- Fox J. M.; Zhao M.; Fink M. J.; Kang K.; Whitesides G. M. The Molecular Origin of Enthalpy/Entropy Compensation in Biomolecular Recognition. Annu. Rev. Biophys. 2018, 47, 223–250. 10.1146/annurev-biophys-070816-033743. [DOI] [PubMed] [Google Scholar]
- Giri A. K.; Spohr E. Influence of Chain Length and Branching on the Structure of Functionalized Gold Nanoparticles. J. Phys. Chem. C 2018, 122, 26739–26747. 10.1021/acs.jpcc.8b08590. [DOI] [Google Scholar]
- Yeon H.; Wang C.; Van Lehn R. C.; Abbott N. L. Influence of Order within Nonpolar Monolayers on Hydrophobic Interactions. Langmuir 2017, 33, 4628–4637. 10.1021/acs.langmuir.7b00226. [DOI] [PubMed] [Google Scholar]
- Dallin B. C.; Yeon H.; Ostwalt A. R.; Abbott N. L.; Van Lehn R. C. Molecular Order Affects Interfacial Water Structure and Temperature-Dependent Hydrophobic Interactions between Nonpolar Self-Assembled Monolayers. Langmuir 2019, 35, 2078–2088. 10.1021/acs.langmuir.8b03287. [DOI] [PubMed] [Google Scholar]
- Dallin B. C.; Van Lehn R. C. Spatially Heterogeneous Water Properties at Disordered Surfaces Decrease the Hydrophobicity of Nonpolar Self-Assembled Monolayers. J. Phys. Chem. Lett. 2019, 10, 3991–3997. 10.1021/acs.jpclett.9b01707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.