ABSTRACT
Histone deacetylases (HDACs) are important for global gene expression and contribute to numerous physiological events. Deacetylase Rpd3 in yeast and its conserved homolog HDAC1 in mammals oppositely regulate autophagy; however, how Rpd3/HDAC1 is regulated to mediate autophagy remains unclear. Here, we showed autophagy occurrence in silkworm (Bombyx mori) required BmRpd3, wherein steroid hormone 20-hydroxyecdysone (20E) signaling regulated its protein level and nuclear localization negatively. Inhibition of MTOR led to dephosphorylation and nucleo-cytoplasmic translocation of BmRpd3/HsHDAC1. Besides, cholesterol, 20E, and 27-hydroxycholesterol could all induce massive dephosphorylation and cytoplasmic localization of BmRpd3/HsHDAC1, and thus autophagy by affecting MTORC1 activity. In addition, three phosphorylation sites (Ser392, Ser421, and Ser423) identified in BmRpd3 were conserved in HsHDAC1. Single or triple phosphorylation-site mutation attenuated the phosphorylation levels of BmRpd3/HsHDAC1, leading to their cytoplasmic localization and autophagy activation. In general, cholesterol derivatives, especially hydroxylated cholesterol, caused dephosphorylation and nucleo-cytoplasmic shuttling of BmRpd3/HsHDAC1 through inhibition of MTOR signaling to facilitate autophagy in B. mori and mammals. These findings improve our understandings of BmRpd3/HsHDAC1-mediated autophagy induced by cholesterol derivatives and shed light on their potential as a therapeutic target for neurodegenerative diseases and autophagy-related studies.
Abbreviations: 20E: 20-hydroxyecdysone; 27-OH: 27-hydroxycholesterol; ACTB: actin beta; AMPK: AMP-activated protein kinase; Atg: autophagy-related; BmSqstm1: Bombyx sequestosome 1; CQ: chloroquine; HDAC: histone deacetylase; LMNB: Lamin B1; MTOR: mechanistic target of rapamycin kinase; PE: phosphatidylethanolamine; SQSTM1/p62: sequestosome 1; TUBA1A: tubulin alpha 1a.
KEYWORDS: Autophagy, BmRpd3/HsHDAC1, cholesterol derivatives, dephosphorylation, nucleo-cytoplasmic translocation, MTOR
Introduction
Macroautophagy/autophagy is a critical process responsible for the degradation of intracellular components, including damaged organelles, long-lived proteins, and lipids, widely conserved from yeast to mammals [1,2]. The initiation, nucleation, elongation, and maturation steps of autophagosome formation require a series of autophagy-related (Atg) proteins [2,3]. Nutrients and energy regulate the initiation of autophagosome formation via phosphorylation of the ULK1/Atg1-ATG13/Atg13 protein kinase complex through MTOR (mechanistic target of rapamycin kinase) signaling in coordination with the AMP-activated protein kinase (AMPK) pathway [1,4,5]. Unique to insects, the steroid hormone 20-hydroxyecdysone (20E) predominantly triggers the global expression of Atg genes and simultaneously inhibits MTOR signaling to induce autophagy initiation, highlighting the differential regulation of autophagy from yeast to mammals [4,5].
Moreover, post-translational modifications of Atg proteins, including acetylation, SUMOylation, and phosphorylation, all affect autophagy occurrence [6]. In addition to phosphorylation, acetylation of lysine sites in Atg proteins is a universal mechanism that regulates their autophagic functions. Starvation reduces acetyl coenzyme A activity and cytoplasmic acetylation level, which consequently results in the inactivation of histone acetyltransferase KAT8/hMOF/MYST1 and hence the induction of autophagy in mammals [7]. Besides, knockdown of AcCoAS/acetyl-coenzyme A synthetase in the Drosophila brain enhances autophagy, indicating a similar regulatory mechanism of autophagy by deacetylation in both Drosophila and mammals [7,8].
In mammals, deacetylation of ATG5, ATG7, LC3, and ATG12 by SIRT1 (sirtuin 1) and HDAC6 (histone deacetylase 6) promotes, while acetylase EP300/p300 inhibits autophagy [6,9,10]. In yeast, Atg3, Atg5, and Atg8 are acetylated in nutrient-rich conditions; however, Atg3 is further acetylated by acetylase Esa1 during starvation, and then deletion of deacetylase Rpd3 promotes starvation-induced autophagy [11]. Dysfunction of mammalian HDACs is associated with tumorigenesis and metastasis; mammalian HDACs are classified as I, II, III, and IV types, and class I type HDACs contribute to autophagic flux and skeletal muscle homeostasis in mice [12-14]. Knockout of Hdac1 or Hdac2, which are members of mammalian class I type HDACs, inhibits autophagy, and accordingly, their overexpression upregulates autophagy in mouse skeletal muscle [12,13,14]. Therefore, there appear to be gaps in our understanding between lower and higher evolutionary-status organisms concerning the autophagic function of HDAC homologs. This discrepancy in autophagy regulation by acetylation in mammals and yeast has not been investigated in detail in other species. Their post-translational modifications regulate the activity of HDACs, and the subcellular localization of class I and II HDACs requires phosphorylation. Serine-site phosphorylation determines nucleo-cytoplasmic translocation of HDAC1 and occurs in inflammatory demyelinating lesions [14,15]. Two phosphorylation sites at Ser421 and Ser423 mediate the subcellular localization of HDAC1 in mice; in comparison, phosphorylation at Ser278 and Ser279 leads to the nuclear localization of HDAC5, while other HDAC homologs are not well documented [15–17].
Cholesterol is a cell membrane structural component essential for membrane fluidity and permeability, and also acts as a precursor for steroid hormones and bile acid biosynthesis [18–20]. The lack of cholesterol results in various inborn defects in humans, whereas excessive cholesterol causes a high risk of cardiovascular diseases [21]. Cholesterol is highly concentrated in the brain in its free state and participates in neural growth, regeneration, and repair [21,22]. However, high concentrations of cholesterol in the brain are usually associated with neurodegenerative diseases such as Alzheimer and Parkinson diseases [22,23]. Moreover, 27-hydroxycholesterol (27-OH), the major metabolite of cholesterol, increases SNCA/α-synuclein level in human dopaminergic neurons and the risk of Parkinson disease [23]. 20E is biosynthesized from cholesterol absorbed from food digestion in insects, and responsible for multiple physiological changes, including autophagy during larval molting and metamorphosis [24–26]. We found that the Bombyx histone deacetylase BmRpd3 was highly conserved with mammalian homologs and essential for autophagy occurrence. 20E signaling induced the dephosphorylation of BmRpd3 and its nucleo-cytoplasmic shuttling to promote autophagy. Moreover, cholesterol and 27-OH regulate the dephosphorylation of BmRpd3/HsHDAC1 homologs, which were similar to 20E by affecting MTOR activity in organisms such as B. mori and mammals. These results document the roles of cholesterol derivatives in BmRpd3/HsHDAC1-mediated autophagy and provide potential therapeutic targets for neurodegenerative diseases.
Results
Deacetylase BmRpd3 is highly conserved and required for protein deacetylation
The homolog of mammalian HDAC1 has not been functionally identified in B. mori. We found BmRpd3 by blasting the amino acid sequence of HsHDAC1 (BAG70111.1) against the Bombyx genome. We also performed a phylogenetic analysis of the BmRpd3 homologs between more than 30 species from Saccharomyces cerevisiae to Homo sapiens, mainly including Insecta. Results showed that BmRpd3 was more conserved with the homologs in Pieris rapae and other lepidopteran species but still harbored around 80% identity compared to HsHDAC1 (Figure 1A).
Figure 1.
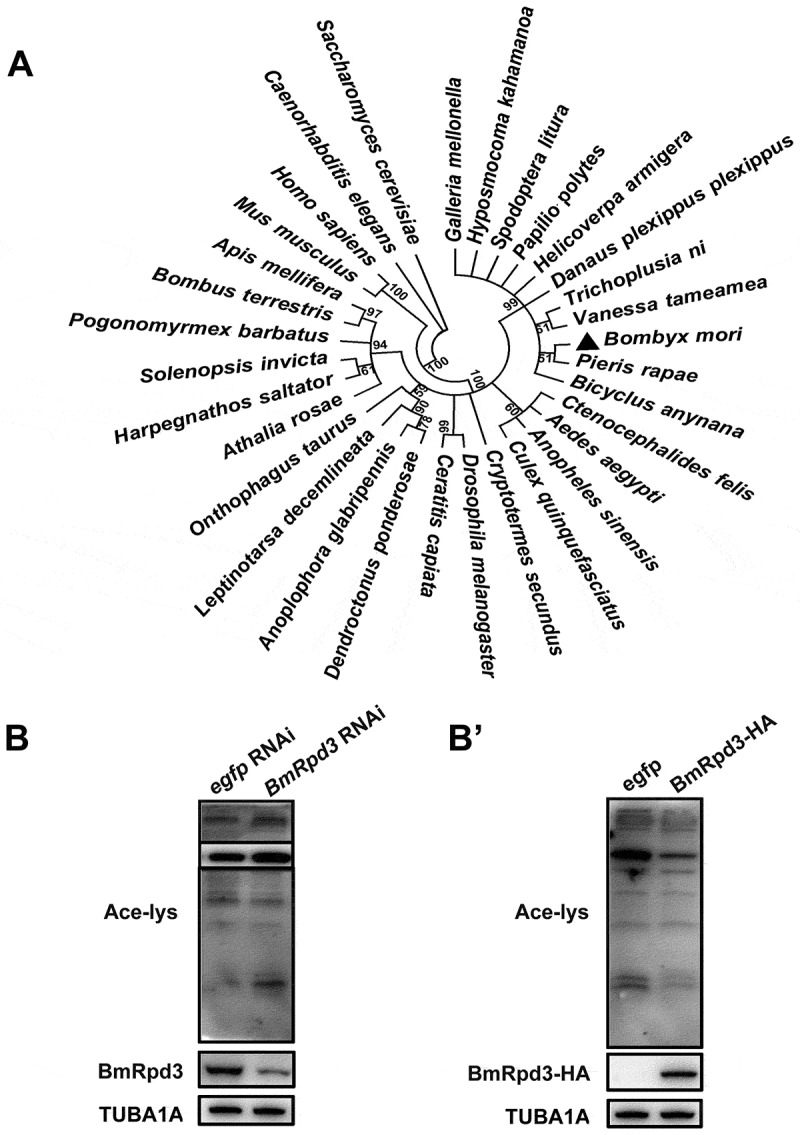
Phylogenetic analysis of the HDAC1 homologs and functional analysis of BmRpd3. (A) The phylogenetic tree of HDAC1 homologs constructed from 32 species, numbers indicate bootstrap value. (B-B’) Acetylation levels of total proteins at 24 h after BmRpd3 RNAi treatment in B. mori fat body (B) and at 48 h after BmRpd3-HA overexpression in BmN cells (B’)
We further verified whether BmRpd3 possessed the catalytic activity for deacetylation using BmRpd3 RNAi in B. mori larvae. After BmRpd3 RNAi for 24 h, we observed a significant reduction of BmRpd3 protein levels; consequently, the acetylation levels of total proteins were increased in the B. mori larval fat body as indicated by the pan-acetyl-lysine western blot detection (Figure 1B). Conversely, overexpression of BmRpd3 decreased the acetylation levels of total proteins in Bombyx BmN cells (Figure 1B’). Thus, BmRpd3 was identified as a functional deacetylase to mediate protein deacetylation in B. mori.
BmRpd3 is indispensable for autophagy in B. mori
Deletion of HDAC1 reduces autophagy, and its overexpression upregulates autophagy in mice [12]. Thus, we investigated the involvement of BmRpd3 in autophagy in B. mori. RNAi knockdown experiments showed that the transcriptional and protein levels of BmRpd3 in the B. mori fat body were reduced by approximately 50% at 24 h after BmRpd3 RNAi treatment (Figure 2A,B). The protein levels of autophagic substrate Bombyx sequestosome 1 (BmSqstm1) accumulated and, conversely, BmAtg8–phosphatidylethanolamine (PE) formation was inhibited, indicating an autophagy block after BmRpd3 RNAi treatment (Figure 2B,B’). Immunofluorescent staining of BmRpd3 indicated that the protein expression reduced both in the nucleus and cytoplasm after RNAi treatment. Moreover, BmAtg8 was mainly arrested in the nucleus after BmRpd3 RNAi, followed by a decrease of BmAtg8 puncta in the cytoplasm (Figure 2C,C’).
Figure 2.
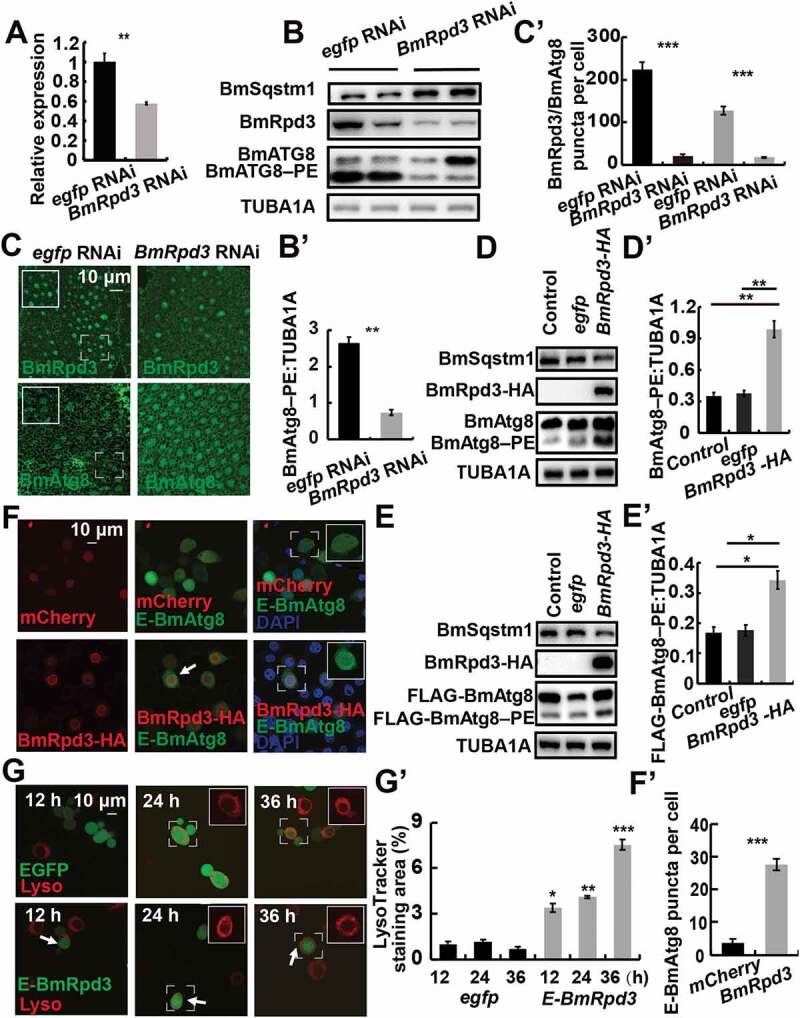
BmRpd3 is indispensable for autophagy in B. mori. (A-C’) mRNA levels of BmRpd3 (A), protein levels of BmSqstm1, BmAtg8, and BmRpd3 (B), immunofluorescent staining of BmRpd3 and BmAtg8 (C) at 24 h after BmRpd3 RNAi treatment in the fat body. Quantification of BmAtg8–PE in B (B’). Quantification of fluorescent BmRpd3 and BmAtg8 in C (C’). Black columns: BmRpd3, gray columns: BmAtg8–PE, wireframes indicate the magnified fields. (D-D’) Protein levels of BmSqstm1, BmAtg8, and BmRpd3-HA after BmRpd3-HA overexpression for 48 h (D). Quantification of BmAtg8–PE in D (D’), egfp overexpression is used as the control. (E-E’) Protein levels of BmSqstm1, FLAG-BmAtg8, and BmRpd3-HA after co-overexpression of BmRpd3 and FLAG-BmAtg8 for 48 h (E). Quantification of FLAG-BmAtg8–PE in E (E’). (F-F’) Punctation of EGFP-BmAtg8 in BmRpd3 overexpressing cells, mCherry overexpression is used as the control (F). Quantification of BmAtg8 puncta in F (F’). Arrows: the typically treated cells, E-BmAtg8: EGFP-BmAtg8. (G-G’) LysoTracker Red staining after overexpression of egfp-BmRpd3 compared to the egfp-overexpressed control (G). Quantification of LysoTracker Red staining in G (G’), E-BmRpd3: EGFP-BmRpd3, Lyso: LysoTracker Red staining
In contrast, the BmSqstm1 protein level decreased, while BmAtg8–PE conjugation and co-overexpressed FLAG-BmAtg8–PE formation increased in BmN cells overexpressing BmRpd3, showing a direct promotion of autophagy by BmRpd3 (Figure 2D–E’). Moreover, EGFP-BmAtg8 puncta and LysoTracker Red staining were induced in cells overexpressing BmRpd3, which was consistent with autophagy occurrence as indicated by BmSqstm1 degradation and BmAtg8–PE formation (Figure 2F–G’). These results show that autophagy occurrence requires BmRpd3 in B. mori.
BmRpd3 protein levels and nuclear localization are negatively related by 20E during larval-pupal metamorphosis
The fat body is a key site for material storage or utilization and insect innate immunity, and 20E regulates multiple physiological processes in this tissue [27,28]. Developmental profiles showed that BmRpd3 protein levels attenuated in the B. mori fat body from day 2 of the 5th instar (5L-2) to day 2 of the prepupa (PP2), which was inversely proportional to 20E titer (Figure 3A,A’). Immunofluorescent staining of BmRpd3 co-stained with DAPI indicated that BmRpd3 almost completely localized in the nuclei of the fat body during larval feeding stage; however, it shuttled from the nucleus to the cytoplasm at wandering (W), PP1, and PP2 stages (Figure 3B,B’), similar to the mammalian HDAC1 that also presents nucleo-cytoplasmic shuttling [15–17]. Overall, BmRpd3 is a nuclear-localized protein that is negatively associated with 20E titer in the hemolymph during larval-pupal metamorphosis.
Figure 3.
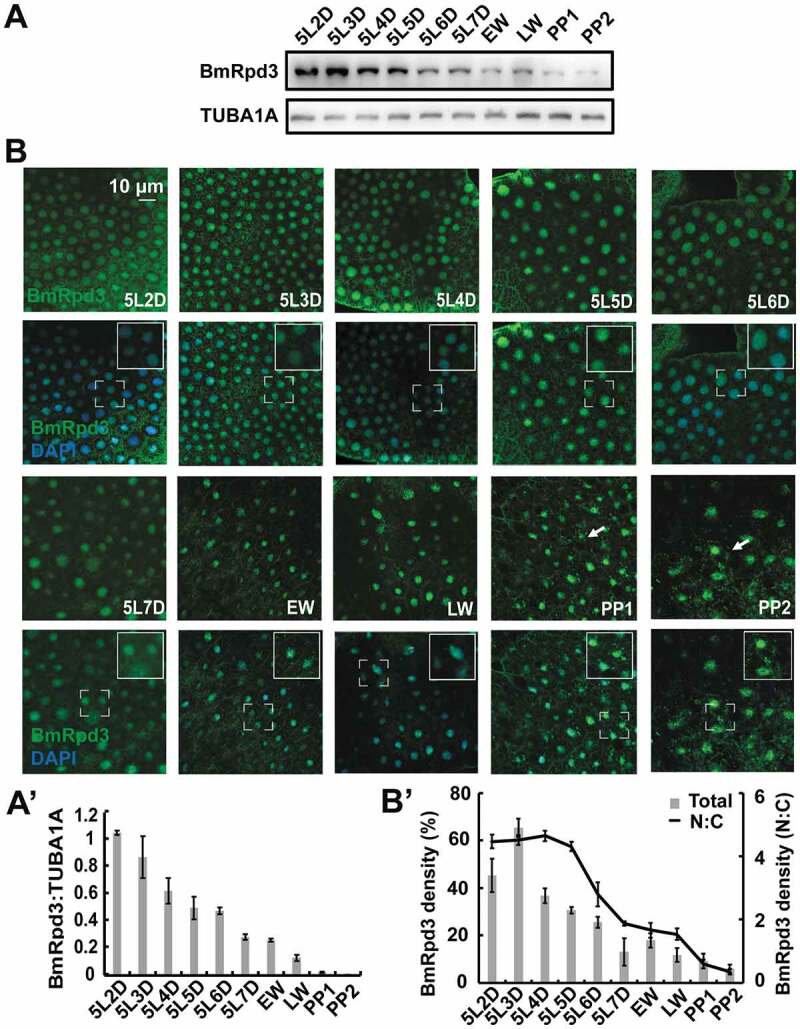
Developmental profiles of BmRpd3 protein levels and subcellular localization in the fat body. (A-A’) Protein levels of BmRpd3 from 5L-2 to PP2. EW: early wandering stage, LW: late wandering stage, PP1: day 1 of the prepupa (A). Quantification of BmRpd3 compared to the TUBA1A in A (A’). (B-B’) Immunofluorescent staining of BmRpd3 from 5L-2 to PP2 (B). Quantification of the total (columns) and nuclear/cytoplasmic (line chart) BmRpd3 in B (B’). N: nucleus, C: cytoplasm, arrows indicate the typical cell with BmRpd3 localized in the cytoplasm
20E signaling induces BmRpd3 nucleo-cytoplasmic translocation and autophagy in B. mori
According to the developmental profiles, 20E is implicated in the negative regulation of BmRpd3 protein levels and its nuclear localization (Figure 3). Thus, we further investigated the regulatory mechanism of 20E on BmRpd3. Three-dimensional structure analysis revealed that there were high binding affinities and close interactions between 20E/cholesterol/27-OH and BmRpd3/HsHDAC1 (Figure S1A–B”). We injected 20E into 5L-3 larvae at a low titer when BmRpd3 mainly localized in the nucleus [4,27]. BmE75a is the primary response gene of 20E, and its mRNA levels increased > 10-fold in the fat body at 24 h after 20E injection. Subsequently, BmSqstm1 and BmRpd3 protein levels decreased, whereas BmAtg8–PE conjugation was promoted (Figure 4A, A’ and S2A), and then BmRpd3 was shuttled from the nucleus to the cytoplasm in the fat body (Figure 4A’’, A’’’). Moreover, BmSqstm1 and the overexpressed BmRpd3-HA protein levels decreased, while BmAtg8–PE formation and nucleo-cytoplasmic translocation of BmRpd3-HA were enhanced by 20E treatment in BmN cells (Figure 4B–B’’’ and S2B).
Figure 4.
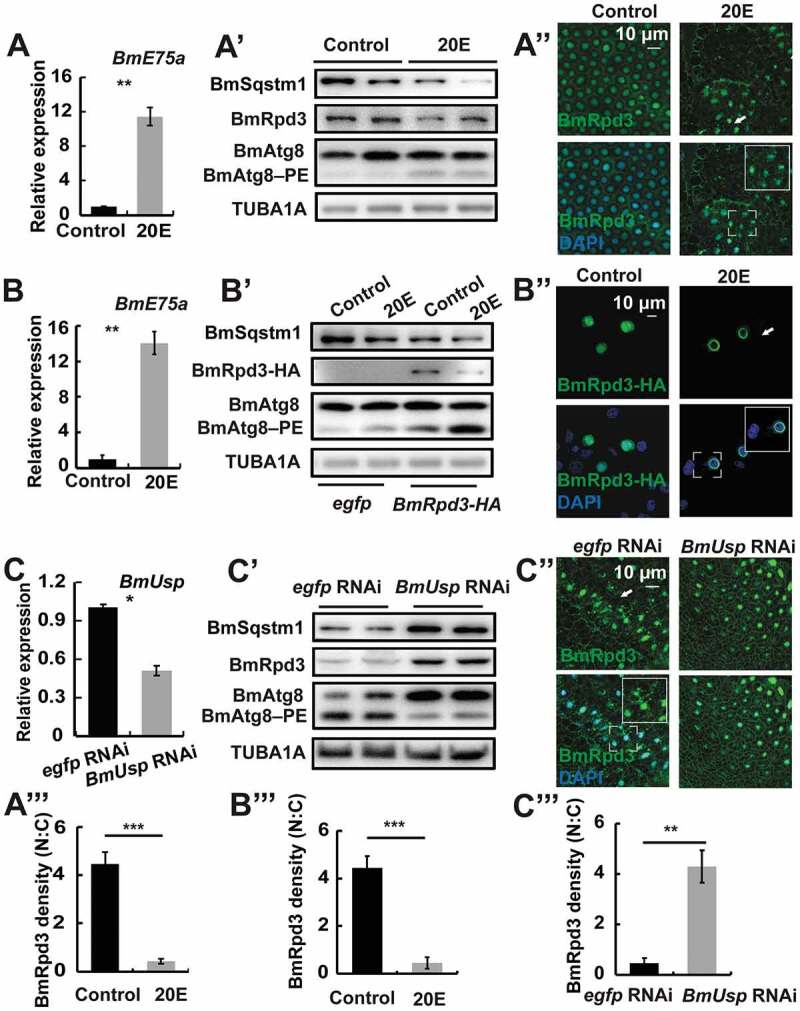
20E signaling induces nucleo-cytoplasmic translocation of BmRpd3 and autophagy in B. mori. (A-A”’) mRNA levels of BmE75a (A). Protein levels of BmSqstm1, BmRpd3, and BmAtg8 (A’). Immunofluorescent staining of BmRpd3 (A”) at 24 h after 20E treatment in the fat body. Quantification of fluorescent BmRpd3 in the nucleus and cytoplasm in A” (A”’). N: nucleus, C: cytoplasm. (B-B”’) mRNA levels of BmE75a (B). Protein levels of BmSqstm1, BmRpd3-HA, and BmAtg8 (B’). Subcellular localization of the overexpressed BmRpd3-HA at 6 h after 20E treatment in BmN cells (B”). Quantification of fluorescent BmRpd3-HA in B” (B”’). (C-C”’) mRNA levels of BmUsp (C). Protein levels of BmSqstm1, BmRpd3, and BmAtg8 (C’). Immunofluorescent staining of BmRpd3 (C”) at 24 h after BmUsp RNAi in the fat body. Quantification of fluorescent BmRpd3 in C” (C”’)
20E signaling transduction occurs through its heterodimer EcR/USP complex [4,27]. RNAi-mediated knockdown of BmUsp significantly downregulated BmUsp mRNA level in the Bombyx fat body (Figure 4C). In contrast to 20E treatment, BmUsp RNAi caused accumulation of BmSqstm1, BmRpd3, and full-length BmAtg8, as well as a decrease in BmAtg8–PE formation (Figure 4C’ and S2C). In addition, fluorescent BmRpd3 decreased in the cytoplasm after BmUsp RNAi treatment (Figure 4C’’,C’’’). These results suggest that 20E regulates autophagy occurrence by affecting BmRpd3 in B. mori.
Although 20E signaling induced nuclear export and consequent degradation of BmRpd3 protein, the mechanisms of these processes remain unclear. As predicted by GPS-SUMO software, BmRpd3 may undergo SUMOylation modification (Table S1). An antibody against SUMOylation detected that the immunoprecipitated BmRpd3 was SUMOylated 6 h after 20E treatment in BmN cells (Figure S2D). Overall, 20E signaling induces the nuclear export and degradation of BmRpd3 in B. mori.
Cholesterol promotes the nuclear export of BmRpd3 and autophagy
Cholesterol is the precursor of 20E, and then we investigated whether cholesterol regulated the subcellular localization of BmRpd3. The degradation of BmSqstm1 and the increase of BmAtg8–PE conjugation indicated that autophagy was induced 24 h after cholesterol treatment in B. mori fat body (Figure S3A,A’). Immunofluorescent staining revealed that the nucleo-cytoplasmic shuttling of BmRpd3 occurred 12 h and 24 h after cholesterol treatment (Figure S3A’ and B). Moreover, BmSqstm1 and BmRpd3 protein levels decreased, whereas BmAtg8–PE formation increased after overexpression of BmRpd3 for 48 h and then treated with different doses of cholesterol (10 to 500 μM) (Figure S3C,C’). In addition, immunofluorescent staining indicated that the overexpressed BmRpd3-HA exported from the nucleus to the cytoplasm after treatment with different doses of cholesterol (Figure S3D,D’). In general, cholesterol-induced nucleo-cytoplasmic translocation of BmRpd3 and autophagy are similar to that of insect molting hormone 20E in B. mori.
Ser392, Ser421, and Ser423 mediate the phosphorylation, nuclear export, and autophagic function of BmRpd3
Phosphorylation mediates the subcellular localization of HDAC5 and HDAC1 in mammals [15,16]; therefore, we next investigated whether BmRpd3 exhibited phosphorylation modification. Three phosphorylation sites, Ser392, Ser421, and Ser423, were identified in BmRpd3-HA by liquid chromatography-mass spectrometry (LC-MS/MS). These serine residues were conserved from insects to humans but not in Caenorhabditis elegans and S. cerevisiae (Figure S4A-C). Single phosphorylation-site mutations significantly affected BmRpd3-HA phosphorylation, and triple phosphorylation-site mutation abolished phosphorylation of BmRpd3-HA as detected using an antibody against phosphoserine/threonine/tyrosine-phosphorylation, and led to its cytoplasmic localization (Figure 5A–B’). Moreover, BmRpd3 completely distributed in the cytoplasm after the triple phosphorylation-site mutation (Figure 5C).
Figure 5.
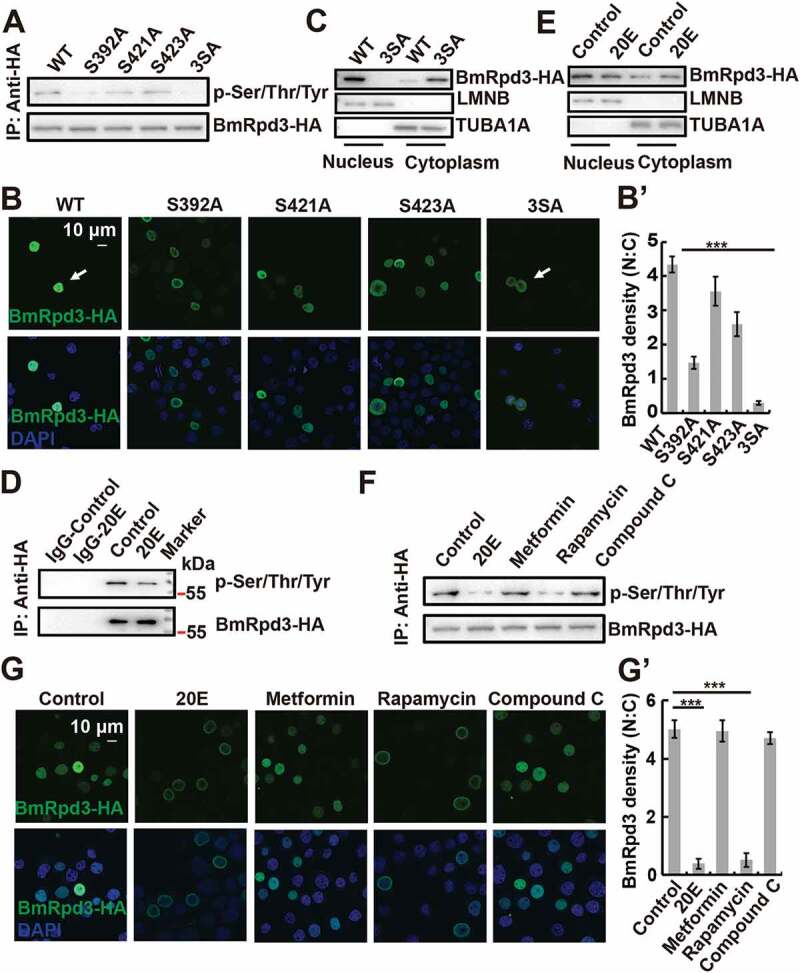
Phosphorylation levels and subcellular localization of BmRpd3 after its phosphorylation sites mutation, 20E treatment, and MTORC1 inhibition in BmN cells. (A) Phosphorylation levels of BmRpd3-HA with a mutation at S392A (Ser392 to Ala392), S421A (Ser421 to Ala421), S423A (Ser423 to Ala423) or 3SA under nutrient-rich conditions, 3SA: triple phosphorylation-site mutation, IP: immunoprecipitation. (B-B’) Subcellular localization of BmRpd3-HA with a mutation at S392A, S421A, S423A or 3SA under nutrient-rich conditions (B). Quantification of fluorescent BmRpd3-HA in the nucleus and the cytoplasm in B (B’), N: nucleus, C: cytoplasm. (C) BmRpd3-HA protein levels in the nuclear and cytoplasmic proteins after the triple phosphorylation-site mutation, LMNB and TUBA1A are respectively used as nuclear and cytoplasmic reference proteins. (D) BmRpd3-HA phosphorylation levels after 20E treatment for 6 h, IgG immunoprecipitation is used as the negative control. (E) BmRpd3-HA protein levels in the nuclear and cytoplasmic proteins after 20E treatment for 6 h. (F-G’) Phosphorylation levels (F), and immunofluorescent staining of BmRpd3-HA (G) after 20E, metformin, rapamycin or compound C treatment for 6 h. Quantification of fluorescent BmRpd3-HA in the nucleus and the cytoplasm in G (G’)
Phosphorylation modification also regulated the autophagic function of BmRpd3. BmSqstm1 protein became significantly degraded, while EGFP-BmAtg8–PE formation was induced by the overexpression of single or triple phosphorylation-site mutation in BmRpd3 under nutrient-rich conditions, which became further enhanced after 20E treatment (Figure 6A and S2G). Accordingly, overexpression of phosphorylation-site mutants, especially the triple phosphorylation-site mutated BmRpd3 induced EGFP-BmAtg8 puncta in the cytoplasm, which was further promoted by 20E addition (Figure 6B,B’). Overall, dephosphorylation at Ser392, Ser421, and Ser423 residues in BmRpd3 causes its nucleo-cytoplasmic translocation and promotes autophagy in B. mori.
Figure 6.
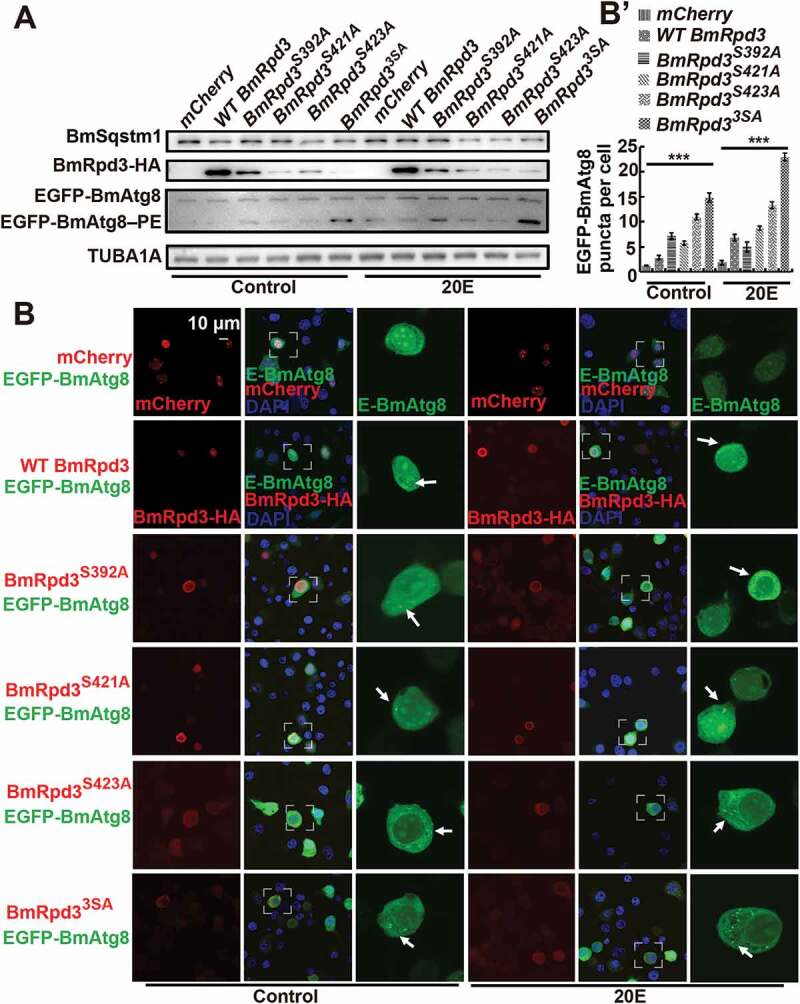
Mutation of S392, S421, and S423 in BmRpd3 promotes autophagy. (A-B’) Protein levels of BmSqstm1, EGFP-BmAtg8, and BmRpd3-HA (A), and fluorescent observation of EGFP-BmAtg8 and BmRpd3-HA, BmRpd3S392A-HA, BmRpd3S421A-HA, BmRpd3S423A-HA or BmRpd33SA-HA after their respective co-overexpression with egfp-BmAtg8 followed by 20E treatment for 6 h (B). Quantification of EGFP-BmAtg8 puncta in B (B’), mCherry overexpression is used as the control. E-BmAtg8: EGFP-BmAtg8
Cholesterol derivatives inhibit MTOR signaling to dephosphorylate BmRpd3
Similar to cholesterol, 20E treatment led to the dephosphorylation of immunoprecipitated BmRpd3-HA from BmN cells (Figure 5D and S3E). We observed that 20E increased the cytoplasmic and decreased the nuclear BmRpd3 protein levels compared to the untreated controls (Figure 5E), indicating the essential role of 20E in regulating the nucleo-cytoplasmic translocation of BmRpd3 via dephosphorylation modification. To address how 20E interacts with signaling pathways that regulate the phosphorylation status of BmRpd3, we individually injected 20E, the AMPK-specific activator metformin, the MTORC1-specific inhibitor rapamycin, and the AMPK-specific inhibitor compound C into 5L-3 larvae. Both 20E and rapamycin treatments led to a significant decrease in BmRpd3 protein and induced its cytoplasmic localization in the fat body (Figure S2E-F). Furthermore, the phosphorylation levels and nuclear localization of BmRpd3-HA attenuated after 20E and rapamycin treatments in BmN cells (Figure 5F-G’). We then investigated whether the derivatives of cholesterol interacted with MTOR signaling and found that 20E, cholesterol, and 27-OH treatment all inhibited nutrient signaling, indicated by changes in p-EIF4EBP1 levels (Figure 7A–C’). Moreover, BmMTORC1 RNAi decreased the phosphorylation levels and nuclear localization of BmRpd3-HA, suggesting cooperation between 20E and MTOR signaling in regulating the phosphorylation modification of BmRpd3 (Figure 7D–F).
Figure 7.
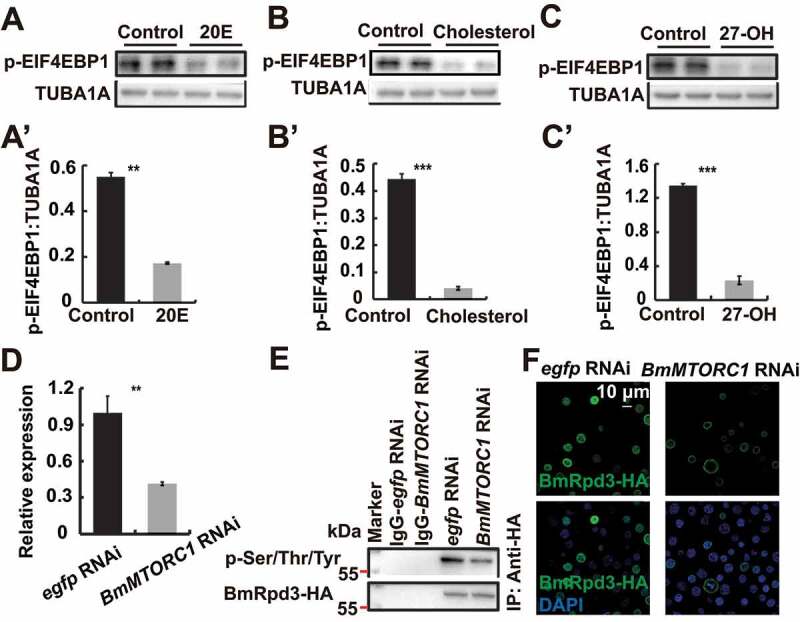
Cholesterol derivatives cooperate with nutrient signaling to regulate phosphorylation modification of BmRpd3. (A-C’) Protein levels of p-EIF4EBP1 after 20E (A), cholesterol (B) or 27-OH (C) treatment for 6 h. Quantification of p-EIF4EBP1, respectively in A (A’), B (B’), and C (C’). (D-F) mRNA level of BmMTORC1 (D), phosphorylation levels of immunoprecipitated BmRpd3-HA (E), and immunofluorescent staining of BmRpd3-HA (F) after BmMTORC1 RNAi treatment for 48 h in BmN cells
Cholesterol, 20E, and 27-OH administrations result in the dephosphorylation and nuclear export of HsHDAC1 and then promote autophagy
We next investigated whether cholesterol, 20E, and 27-OH could similarly regulate the phosphorylation and subcellular localization of H. Sapiens HDAC1. As indicated by a decrease in human SQSTM1/p62 protein levels, and an increase in LC3-II formation, HsHDAC1 overexpression in HEK 293 cells significantly induced autophagy (Figure 8A and S5A). Chloroquine (CQ) treatment inhibits the fusion between autophagosome and lysosome, thus determining inhibition of the autophagic flux [1]. Also, we detected that CQ inhibited HsHDAC1-induced autophagic flux. Firstly, SQSTM1/p62 and LC3-II protein levels linearly accumulated in response to an increasing dose of CQ (20, 40, 60, 80 μM) in HEK 293 cells. We used 40 μM CQ for subsequent experiments (Figure S4D and D’). After HsHDAC1 overexpression and treatment with 40 μM CQ, the SQSTM1/p62 and LC3-II protein levels both significantly increased compared to HsHDAC1 overexpression, showing inhibition of HsHDAC1-induced autophagic flux by CQ treatment (Figure 8A). Moreover, HsHDAC1 overexpression also induced the presence of EGFP-LC3 puncta, further indicating autophagy induction (Figure 8A’,A”). Notably, degradation of both human SQSTM1/p62 and overexpressed HsHDAC1-HA, and LC3-II formation were induced 6 h after 250 μM cholesterol treatment; in contrast, they were all accumulated after 40 μM CQ addition (Figure 8B and S5B). Besides, the phosphorylation levels and nuclear localization of overexpressed HsHDAC1-HA simultaneously and significantly decreased after cholesterol treatment (Figure 8B’,B” and S5C).
Figure 8.
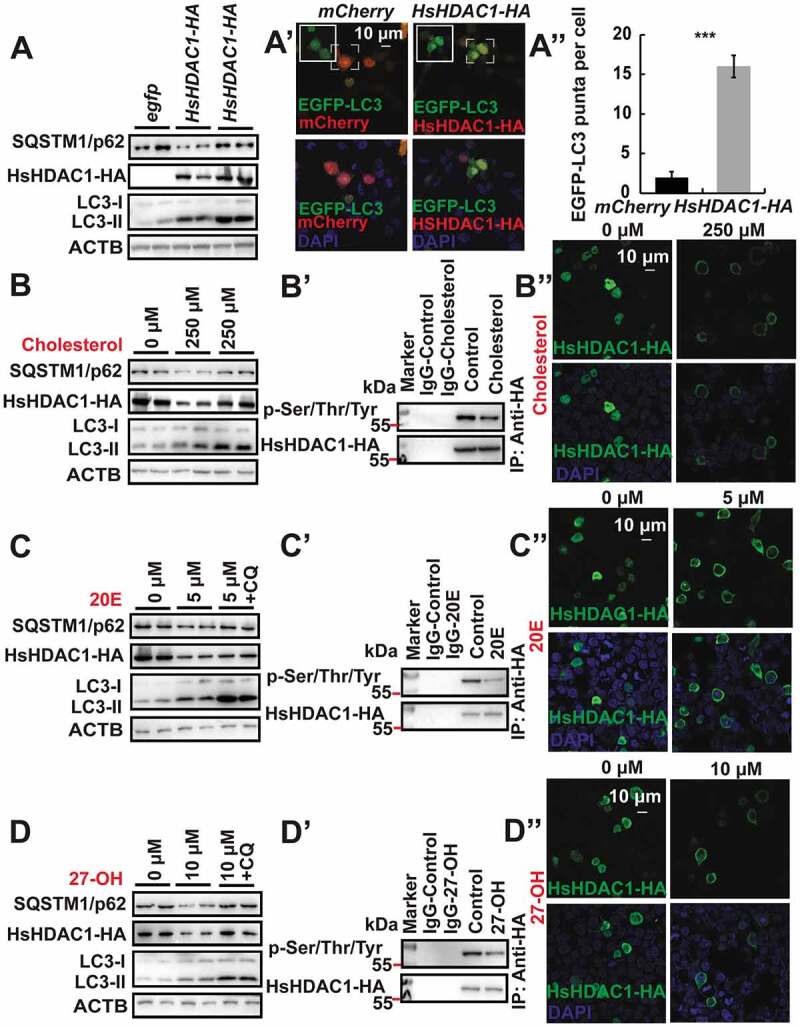
Cholesterol derivatives induce dephosphorylation and cytoplasmic localization of HsHDAC1, and autophagy in HEK 293 cells. (A-A”) Protein levels of SQSTM1/p62 and LC3 after HsHDAC1 overexpression or HsHDAC1 overexpression followed by 40 μM CQ treatment (A). Punctation of EGFP-LC3 after HsHDAC1 overexpression (A’). Quantification of EGFP-LC3 puncta in A’ (A”). (B-B”) Protein levels of SQSTM1/p62, HsHDAC1-HA, and LC3 after 250 µM cholesterol treatment for 6 h, or 3 h followed by 40 μM CQ treatment for further 3 h (B). Phosphorylation levels of immunoprecipitated HsHDAC1-HA (B’), and immunofluorescent staining of HsHDAC1 at 6 h after 250 µM cholesterol treatment (B”). (C-C”) Protein levels of SQSTM1/p62, HsHDAC1-HA, and LC3 after 5 µM 20E treatment for 6 h, or 3 h followed by 40 μM CQ treatment for further 3 h (C). Phosphorylation levels of immunoprecipitated HsHDAC1-HA (C’), and immunofluorescent staining of HsHDAC1 after 5 µM 20E treatment for 6 h (C”). (D-D”) Protein levels of SQSTM1/p62, HsHDAC1-HA, and LC3 after 10 µM 27-OH treatment for 6 h, or 3 h followed by 40 μM CQ treatment for further 3 h (D). Phosphorylation levels of immunoprecipitated HsHDAC1-HA (D’), and immunofluorescent staining of HsHDAC1 after 10 µM 27-OH treatment for 6 h (D”)
We hypothesized that 20E would have a similar function as cholesterol in regulating HsHDAC1 in HEK 293 cells. The results showed that 5 μM 20E was able to induce autophagic flux as well as dephosphorylation and nucleo-cytoplasmic translocation of HsHDAC1 (Figure 8C–C”, S5D and S5E). Treatment with 10 μM 27-OH had similar effects compared to 20E and cholesterol treatment, demonstrating a conserved regulatory mechanism of 27-OH on HsHDAC1 dephosphorylation and autophagy (Figure 8D–D”, S5F and S5G). Overall, cholesterol and its hydroxylated derivatives are able to induce dephosphorylation and nucleo-cytoplasmic shuttling of BmRpd3/HsHDAC1, and then autophagy.
BmRpd3 phosphorylation sites are functionally conserved in the regulation of HsHDAC1 phosphorylation and subcellular localization
As the three phosphorylation sites (Ser392, Ser421, and Ser423) were conserved in BmRpd3 and HsHDAC1, we asked if these sites were involved in HsHDAC1 phosphorylation regulation. Results showed that single phosphorylation-site mutation partly attenuated, while triple phosphorylation-site mutation completely impaired phosphorylation of HsHDAC1 and led to the linear decrease of its nuclear localization (Figure 9A–A”). Autophagy was gradually induced after the overexpression of the single and triple phosphorylation-site mutated HsHDAC1, as shown by SQSTM1/p62 and LC3-II protein levels, further enhanced by 27-OH treatment (Figure 9B,B’). Moreover, triple phosphorylation-site mutation of HsHDAC1-HA induced the formation of mCherry-LC3 puncta, which was further promoted by 27-OH treatment (Figure S5H,H’). Overall, the phosphorylation sites identified on BmRpd3 are functionally conserved in B. moriand humans and mediate the phosphorylation and nuclear localization of its mammalian homolog HsHDAC1.
Figure 9.
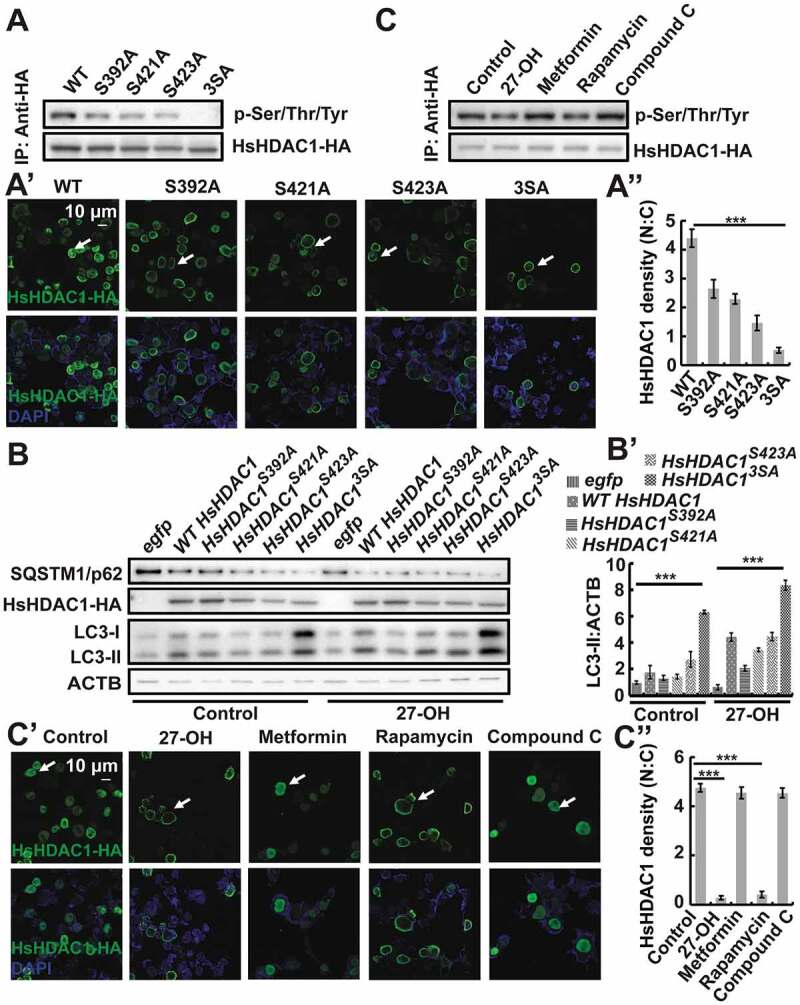
BmRpd3 phosphorylation sites are functionally conserved and regulate phosphorylation levels, subcellular localization of HsHDAC1, and autophagy in HEK 293 cells. (A-A”) Phosphorylation levels (A) and immunofluorescent staining (A’) of HsHDAC1-HA mutated with a single phosphorylation site at S392A (Ser392 to Ala392), S421A (Ser421 to Ala421) or S423A (Ser423 to Ala423), or mutated with triple phosphorylation sites (3SA) under nutrient-rich conditions. Quantification of fluorescent HsHDAC1-HA in the nucleus and the cytoplasm in A’ (A”). (B-B’) Protein levels of SQSTM1/p62, HsHDAC1-HA, and LC3 after the overexpression of single or triple phosphorylation-site mutation of HsHDAC1-HA under nutrient-rich conditions or further treated with 27-OH, ACTB is used as the reference protein (B). Quantification of LC3-II in B (B’). (C-C”) Phosphorylation levels (C) and immunofluorescent staining (C’) of HsHDAC1-HA after 27-OH, metformin, rapamycin or compound C treatment for 6 h. Quantification of fluorescent HsHDAC1-HA in the nucleus (N) and cytoplasm (C) in C’ (C”)
Cholesterol derivatives regulate the phosphorylation of HsHDAC1 through their interaction with HsMTORC1 signaling
Similar to BmMTORC1, the HsMTORC1 was hypothesized to regulate the phosphorylation of HsHDAC1. As expected, treatments with 27-OH and rapamycin both reduced HsHDAC1 phosphorylation levels and nuclear localization in HEK 293 cells (Figure 9C–C”). Cholesterol, 20E, and 27-OH treatments all led to the inhibition of HsMTORC1 activity as indicated by a decrease of p-EIF4EBP1 protein levels (Figure 10A–C’). In HEK 293 cells, HsMTORC1 RNAi significantly reduced HsMTORC1 transcripts after 48 h, and then the phosphorylation of HsHDAC1-HA was almost abolished, which consequently led to the nucleo-cytoplasmic translocation of fluorescent HsHDAC1-HA (Figure 10D–F). In conclusion, cholesterol derivatives are functionally conserved and are able to dephosphorylate BmRpd3/HsHDAC1 through the inhibition of MTORC1 activity, leading to the cytoplasmic localization of BmRpd3/HsHDAC1, and thus promoting autophagy (Figure 11).
Figure 10.
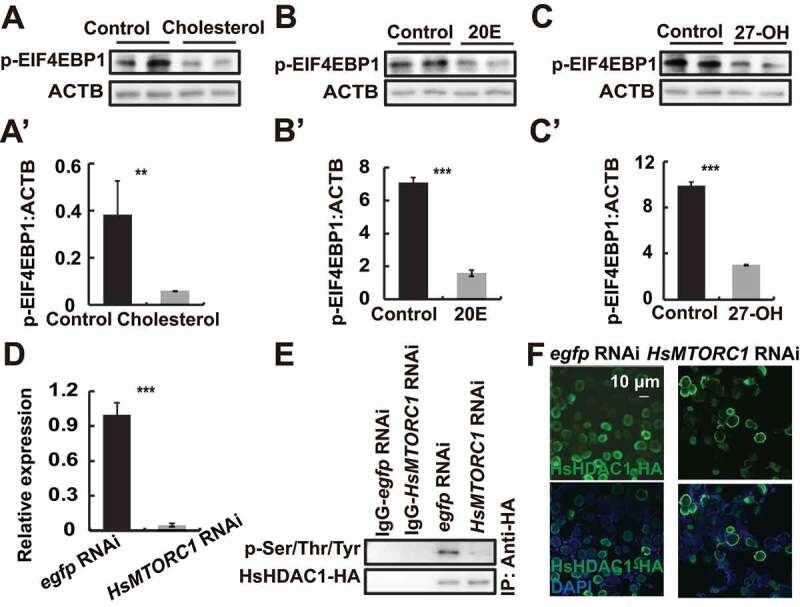
HsMTORC1 mediates the phosphorylation of HsHDAC1. (A-A’) p-EIF4EBP1 levels after cholesterol treatment for 6 h (A). Quantification of p-EIF4EBP1 in A (A’). (B-B’) p-EIF4EBP1 levels after 20E treatment for 6 h (B). Quantification of p-EIF4EBP1 in B (B’). (C-C’) p-EIF4EBP1 levels after 27-OH treatment for 6 h (C). Quantification of p-EIF4EBP1 in C (C’). (D-F) mRNA level of HsMTORC1 (D), phosphorylation levels of immunoprecipitated HsHDAC1-HA (E), and immunofluorescent staining of HsHDAC1-HA (F) after HsMTORC1 RNAi treatment for 48 h in HEK 293 cells
Figure 11.
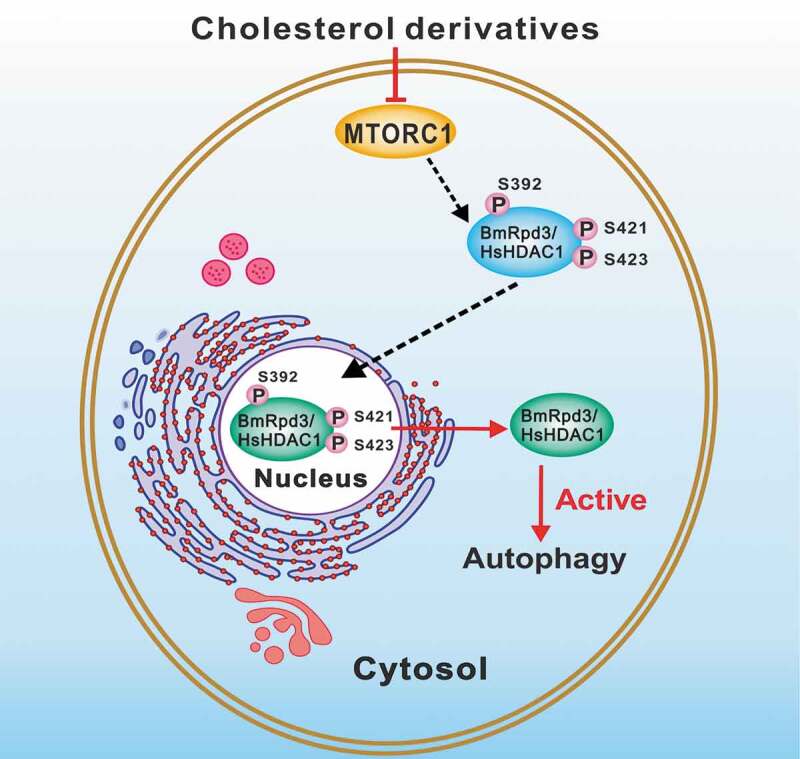
A model for cholesterol derivatives in regulating BmRpd3/HsHDAC1 dephosphorylation and subsequent autophagy occurrence. MTORC1 signaling phosphorylates BmRpd3/HsHDAC1 and sequesters them in the nucleus. After being dephosphorylated by cholesterol derivatives signaling through inhibiting MTORC1 activity, BmRpd3/HsHDAC1 shuttles from the nucleus to the cytoplasm and promotes autophagy
Discussion
Oxysterols exhibit an effective and evolutionarily conserved ability to regulate Rpd3/HDAC1 function in B. mori and humans
Members of class I HDACs in mammals promote autophagy, whereas Rpd3, the homolog of HsHDAC1 in yeast, inhibits autophagy induction that is not documented in other species [11,12]. Knockdown of AcCoAS/acetyl-coenzyme A synthetase in the Drosophila brain enhances autophagy, which is consistent with the induction of autophagy by the reduction of acetyl coenzyme A activity and cytoplasmic acetylation levels in mammals [7,8]. In the present study, we demonstrate that HsHDAC1 and its homolog BmRpd3 in B. mori could deacetylate proteins and are required for autophagy occurrence. Their similar functions imply that the regulatory mechanism of BmRpd3/HsHDAC1 is likely highly conserved in B. mori and humans, and potentially, from insects to mammals.
It is well known that cholesterol has a close relationship with deacetylases involved in the regulation of cholesterol biosynthesis and catabolism [21,29]. However, whether the cholesterol and its derivatives inversely regulate the functions of deacetylases remains unclear. Cholesterol induces autophagic and apoptotic features in mammalian insulin-secreting βTC-6 and INS-1 cells, although the precise mechanism of this regulation is not well defined [30]. Here we show that cholesterol and oxysterols such as 20E and 27-OH are all able to induce the dephosphorylation of BmRpd3/HDAC1 and their cytoplasmic localization through inactivation of MTORC1 signaling, revealing a conserved regulation of HsHDAC1 homologs by cholesterol and oxysterol (Figures 5, 7, 9 and 10). Notably, oxysterols that freely pass through the blood-brain barrier are more efficient at inducing dephosphorylation and nucleo-cytoplasmic translocation of BmRpd3/HDAC1 and autophagy, indicating that the hydroxylated metabolites of cholesterol may play more important roles in some physiological processes.
Oxysterols are involved in several events underlying Alzheimer pathogenesis. Certain oxysterols cause neuron dysfunction and degeneration; conversely, some display neuroprotective effects. 24-hydroxylcholesterol increases the transcription and translation of the deacetylase SIRT1, which prevents the neurotoxic accumulation of the hyperphosphorylated tau protein [31]. However, 27-OH decreases the proteasomal degradation of SNCA and results in memory deficits and synucleinopathy risk [23]. Whether class I type HDACs have similar functions to class II type SIRTs in neurodegenerative diseases and whether other oxysterols functionally differ from 24-hydroxycholesterol in nervous system diseases are worthy of investigation.
20E regulates autophagy occurrence via multiple pathways in B. mori
In insects, 20E signaling controls programmed cell death, mainly including autophagy and apoptosis [4,32,33]. 20E upregulates autophagy occurrence via multiple pathways, several studies have shown that 20E induces massive autophagy during larval-pupal metamorphosis through EcR-USP signaling by upregulating the expression of Atg genes in Drosophila and Bombyx [4,5]. Moreover, 20E interacts with the nutrient pathway to inhibit the activity of MTORC1 and initiate the formation of autophagosomes through the phosphorylation of the Atg1-Atg13 protein complex [4,5]. Besides, 20E signaling upregulates the transcription of most genes encoding subunits of the vacuolar-type H+-adenosine triphosphatases (BmV-ATPases) from V0 and V1 subcomplexes and the subunit assembly in B. mori, mainly through the inhibition of BmMTORC1 activity and induction of BmTFEB (Bombyx transcription factor EB) transcription and nuclear translocalization [34]. In addition, 20E also induces starvation-like conditions in Bombyx by reducing food consumption [35].
Herein, we describe that BmMTOR1 signaling mediates BmRpd3 phosphorylation and nuclear localization; whereas, 20E inhibits BmMTORC1 signaling and then dephosphorylates BmRpd3, resulting in its nucleo-cytoplasmic translocation and consequent promotion of autophagy (Figure 11). Notably, 20E signaling simultaneously induced SUMOylation and degradation of BmRpd3, although the function and mechanism of this process are still unclear.
Materials and methods
Animals and cell culture
Silkworms (Dazao) were provided by the Sericultural and Agri-Food Research Institute of the Guangdong Academy of Agricultural Sciences (Guangzhou, China). Insects were reared on fresh mulberry leaves in the laboratory at 25°C under a 14 h light/10 h dark cycle [34,36].
BmN cells were maintained in Grace’s insect medium (Sigma-Aldrich, G9771) supplemented with 10% fetal bovine serum; Human HEK 293 cells (ATCC, CRL-1573TM) were cultured in Dulbecco’s Modified Eagle Medium (ThermoFisher, 10569044) supplemented with 5% fetal bovine serum (AusGeneX, FBS500-S).
Plasmid construction and transfection
BmRpd3 (XM_028180486.1) cDNAs were cloned from B. mori total cDNA and inserted into the pIEx4 overexpression vector fused with HA tag [27,34]. HsHDAC1 was inserted into the pCMV3 overexpression vector (Sino Biological, HG11486-CY). BmRpd3 and HsHDAC1 were mutated accordingly. BmN (ATCC, CRL-8910TM) or HEK 293 cells were transfected with BmRpd3- or HsHDAC1-containing plasmids using the FuGENE® HD Transfection Reagent (Promega, E2311) or TransIntroTM EL Transfection Reagent (TransGen Biotech, FT201-01) according to the manufacturer’s instructions.
Chemical treatments
Metformin (Selleck, S1950; 30 μg/larva), rapamycin (Selleck,S1039; 10 μg/larva), compound C (Sigma-Aldrich, P5499; 5 μg/larva), 20E (Sigma-Aldrich, H5142; 5 μg/larva) and cholesterol (Yuanye Biology, 57885; 8 μg/larva) were individually injected into silkworm larva. Control groups were injected with the same volume of solvent [4,34]. Metformin (5 μM), rapamycin (10 μM), compound C (10 μM), cholesterol (5, 10, 100, 250 or 500 μM) and 20E (5 μM) were used in BmN cells. Cholesterol (250 μM), 20E (5 μM) and 27-OH (Yuanye Biology, B21770; 10 μM) were used in HEK 293 cells. Different concentrations of CQ (Sigma-Aldrich, C6628; 20, 40, 60 or 80 μM) were added to HEK 293 cells for 3 h. Ten animals were included in each group, and three biological replicates were performed.
RNAi
The double-stranded RNA (dsRNA) of BmRpd3 and egfp were generated using T7 RiboMAX™ Express RNAi system (Promega, P1700) according to the manufacturer’s instruction. Each larva was injected with BmRpd3 dsRNA (50 μg) at 12 h before initiation of wandering (IW), and egfp dsRNA (50 μg) was injected as a control. BmMTORC1 and HsMTORC1 siRNA were synthesized (Sangon Biotech) and transfected into BmN or HEK 293 cells for 48 h, egfp siRNA transfection was used as the negative control. All primers used in this paper are listed in Table S2.
Quantitative real-time PCR (qPCR)
Total RNA from fat body tissue and cells was extracted using TRIzol reagent (TaKaRa, RR037A), and cDNA was generated using the PrimeScriptTM RT reagent Kit (TaKaRa, RR047A). qPCR was performed as previously described [36]. The BmRp49 and ACTB genes were used as the reference genes.
Immunoprecipitation and western blot
BmN cells overexpressing different forms of BmRpd3-HA and HEK 293 cells overexpressing different forms of HsHDAC-HA were harvested and lysed in Nonidet P-40 lysis buffer (Beyotime Biotechnology Co., Ltd., P0013F) supplemented with a protease inhibitor cocktail (ThermoFisher Scientific, 78429). The supernatant of lysed cells was pre-incubated with the HA antibody (Santa Cruz Biotechnology, sc-7392; 1:200) at 4°C for 2 h. Then, the mixtures were incubated with agarose beads (Thermo Fisher Scientific, 20421) at 4°C overnight according to a standard immunoprecipitation procedure.
Western blot was performed according to the standard procedure using antibodies against BmRpd3/HsHDAC1 (Bioss, bs-1414R; 1:1000), HA (Beijing Biodragon, B1021; 1:2000), SQSTM1/p62 (Cell Signaling Technology, 5114; 1:1000), LC3B (Abcam, ab192890; 1:2000), LMNB/Lamin B1 (Bioworld, AP6001; 1:5000), BmAtg8 (1:3000) [4,34], phosphoserine/threonine/tyrosine (Abcam, ab15556; 1:2000), acetylated-lysine (Cell Signaling Technology, 9441; 1:1000), p-EIF4EBP1-T37/46 (ABclonal Technology, AP0030; 1:1000), SUMOylation (PTM Biolabs, PTM-1110; 1:2000), and BmSqstm1 (ABclonal Technology, A18679; 1:3000). TUBA1A/tubulin alpha 1a (Beyotime Biotechnology Co., Ltd, AT819; 1:5000) and ACTB/actin beta (ProMab, 20270; 1:1000) were used as the reference proteins. Images of western blots were taken under a series of exposure time, and the moderately exposed blots without over-saturation were used for quantification and figure presentation. ImageJ software (National Institutes of Health, USA) was used to quantify the protein blots. Three independent biological replicates were performed.
Isolation of nuclear and cytoplasmic proteins
BmN cells were treated with 5 μM 20E for 6 h, or overexpressed with wild type or triple phosphorylation-site mutated BmRpd3-HA. Cells were then harvested for nuclear and cytoplasmic extraction using a NE-PERTM Nuclear Cytoplasmic Extraction Reagent Kit (Pierce, 78833) according to the manufacturer’s instruction. The separated proteins were evaluated with western blots.
Immunofluorescent staining
The fat body and cells were collected for immunofluorescent staining with BmRpd3 or HA tag primary antibodies (Santa Cruz Biotechnology, sc-7392; 1:200), then further co-stained with DAPI (Beyotime Biotechnology Co., Ltd, C1005) as previously described [4,36]. Fluorescence observation was conducted under a confocal microscope equipped with an Olympus digital camera (Olympus FV3000, Tokyo). To quantify Alexa Fluor 488-labeled BmRpd3, BmRpd3-HA, HsDAC1-HA, or GFP-LC3/BmAtg8 puncta, 150–200 fat body cells or a total of 30 overexpressed BmN cells from three independent biological repeats were recorded and analyzed using ImageJ software.
LysoTracker staining
Sterilized coverslips were placed in 6-well plates (Guangzhou Jet Bio-Filtration Co., Ltd, TCP-010-006) during cell plating. After pre-incubated for 24 h, the BmN cells were transfected with pIEx-4-BmRpd3-HA for 12, 24, and 36 h before being stained with LysoTracker Red DND-99 (Thermo Fisher Scientific, L7528) [4]. Observation was performed under an Olympus FV300 confocal microscope.
LC-MS/MS
The phosphorylation sites of immunoprecipitated BmRpd3 were determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry using LC-MS (Applied Biosystems, Foster City). Gel-digested BmRpd3 protein was loaded onto a reversed-phase analytical column (75 µm i.d. × 150 mm, packed with Acclaim PepMap RSLC C18, ThermoFisher; 2 µm, 100Å, nanoViper) and separated by a gradient elution buffer. The MS analysis of peptides was performed on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher, USA). Protein identification was performed with MASCOT software (Mascot SoftWeb Solution Pvt. Ltd., Uttarakhand) by searching Uniprot for B. mori.
Bioinformatics analysis
Phylogenetic analysis was constructed using the MEGA7.0 software (Molecular Evolutionary Genetics Anlysis, USA) and the Neighbor-Joining method. Only values > 50% were indicated. The three-dimensional structures between BmRpd3/HsHDAC1 and cholesterol derivatives were generated using AutoDock Tool software (The Scripps Research Institute, USA).
Statistical analysis
Experimental data were analyzed by Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001. Values are presented as the means ± standard deviation of three independent biological replicates.
Supplementary Material
Acknowledgments
This study was supported by the National Science Foundation of China to LT (NSFC; 31672368, 31970463), the Natural Science Foundation of Guangdong Province to LT (2017A030311024), and NSFC to KL (31702053).
Funding Statement
This work was supported by the the National Science Foundation of China [31672368 and 31970463];Natural Science Foundation of Guangdong Province (CN) [2017A030311024]; NSFC to KL [31702053].
Disclosure statement
The authors have no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lee CY, Cooksey BA, Baehrecke EH.. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250(1):101–111. [DOI] [PubMed] [Google Scholar]
- [3].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tian L, Ma L, Guo EE, et al. 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy. 2013;9(8):1172–1187.23674061 [Google Scholar]
- [5].Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FFBS Lett. 2010;584(7):1342–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Popelka H, Klionsky DJ. Post-translationally-modified structures in the autophagy machinery: an integrative perspective. Febs J. 2015;282(18):3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wani WY, Boyer-Guittaut M, Dodson M, et al. Regulation of autophagy by protein post-translational modification. Lab Invest. 2015;95(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Eisenberg T, Schroeder S, Buttner S, et al. A histone point mutation that switches on autophagy. Autophagy. 2014;10(6):1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huang R, Xu Y, Wan W, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57(3):456–466. [DOI] [PubMed] [Google Scholar]
- [10].Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284(10):6322–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336(6080):474–477. [DOI] [PubMed] [Google Scholar]
- [12].Moresi V, Carrer M, Grueter CE, et al. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci U S A. 2012;109(5):1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev . 2007;21(14):1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhu Y, Vidaurre OG, Adula KP, et al. Subcellular distribution of HDAC1 in neurotoxic conditions is dependent on serine phosphorylation. J Neurosci. 2017;37(31):7547–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Greco TM, Yu F, Guise AJ, et al. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10(2):M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. [DOI] [PubMed] [Google Scholar]
- [18].Chang TY, Chang CC, Ohgami N, et al. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. [DOI] [PubMed] [Google Scholar]
- [19].Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. [DOI] [PubMed] [Google Scholar]
- [20].Head BP, Patel HH, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta. 2014;1838(2):532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kirkegaard P, Edwards A, Risor MB, et al. Risk of cardiovascular disease? A qualitative study of risk interpretation among patients with high cholesterol. BMC Fam Pract. 2013;14:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kwiterovich PJ, Chen SC, Virgil DG, et al. Response of obligate heterozygotes for phytosterolemia to a low-fat diet and to a plant sterol ester dietary challenge. J Lipid Res. 2003;44(6):1143–1155. [DOI] [PubMed] [Google Scholar]
- [23].Schommer J, Marwarha G, Schommer T, et al. 27-Hydroxycholesterol increases alpha-synuclein protein levels through proteasomal inhibition in human dopaminergic neurons. BMC Neurosci. 2018;19(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomo. 2002;47:883–916. [DOI] [PubMed] [Google Scholar]
- [25].Tettamanti G, Casartelli M. Cell death during complete metamorphosis. Philos Trans R Soc Lond B Biol Sci. 2019;374(1783):20190065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ryoo HD, Baehrecke EH. Distinct death mechanisms in Drosophila development. Curr Opin Cell Biol. 2010;22(6):889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li K, Guo E, Hossain MS, et al. Bombyx E75 isoforms display stage- and tissue-specific responses to 20-hydroxyecdysone. Sci Rep. 2015;5:12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li S, Yu X, Feng Q. Fat body biology in the last decade. Annu Rev Entomol. 2019;64:315–333. [DOI] [PubMed] [Google Scholar]
- [29].Pipalia NH, Subramanian K, Mao S, et al. Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J Lipid Res. 2017;58(4):695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu J, Kong F, Pan Q, et al. Autophagy protects against cholesterol-induced apoptosis in pancreatic beta-cells. Biochem Biophys Res Commun. 2017;482(4):678–685. [DOI] [PubMed] [Google Scholar]
- [31].Testa G, Staurenghi E, Giannelli S, et al. A silver lining for 24-hydroxycholesterol in Alzheimer’s disease: the involvement of the neuroprotective enzyme sirtuin 1. Redox Bio. 2018;17:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xie K, Tian L, Guo XY, et al. BmATG5 and BmATG6 mediate apoptosis following autophagy induced by 20-hydroxyecdysone or starvation. Autophagy. 2016;12(2):381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Romanelli D, Casartelli M, Cappellozza S, et al. Roles and regulation of autophagy and apoptosis in the remodelling of the lepidopteran midgut epithelium during metamorphosis. Sci Rep. 2016;6:32939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dai YC, Li K, Wu WM, et al. Steroid hormone 20-hydroxyecdysone induces the transcription and complex assembly of V-ATPases to facilitate autophagy in Bombyx mori. Insect Biochem Mol Biol. 2020;116:103255. [DOI] [PubMed] [Google Scholar]
- [35].Wang S, Liu S, Liu H, et al. 20-hydroxyecdysone reduces insect food consumption resulting in fat body lipolysis during molting and pupation. J Mol Biol. 2010;2(3):128–138. [DOI] [PubMed] [Google Scholar]
- [36].Guo SY, Wu WM, Li SY, et al. 20-Hydroxyecdysone-upregulated proteases involved in Bombyx larval fat body destruction. Insect Mol Biol. 2018;27(6):724–738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


