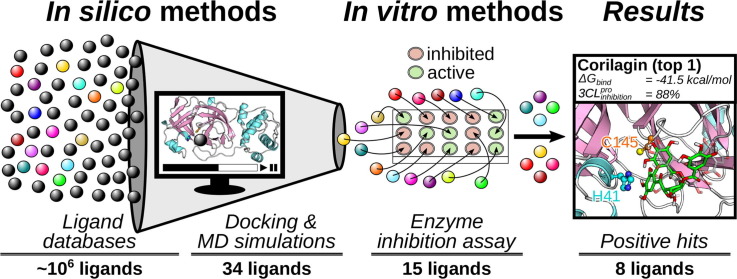
Graphical abstract
Keywords: COVID-19, 3CLpro, Viral replication inhibition, MD simulations, Enzyme inhibition assay, Natural products, Drug repurposing
Abstract
For the COVID-19 pandemic caused by SARS-CoV-2, there are currently no effective drugs or vaccines to treat this coronavirus infection. In this study, we focus on the main protease enzyme of SARS-CoV-2, 3CLpro, which is critical for viral replication. We employ explicit solvent molecular dynamics simulations of about 150 compounds docked into 3CLpro’s binding site and that had emerged as good main protease ligands from our previous in silico screening of over 1.2 million compounds. By incoporating protein dynamics and applying a range of structural descriptors, such as the ability to form specific contacts with the catalytic dyad residues of 3CLpro and the structural fluctuations of the ligands in the binding site, we are able to further refine our compound selection. Fourteen compounds including estradiol shown to be the most promising based on our calculations were procured and screened against recombinant 3CLpro in a fluorescence assay. Eight of these compounds have significant activity in inhibiting the SARS-CoV-2 main protease. Among these are corilagin, a gallotannin, and lurasidone, an antipsychotic drug, which emerged as the most promising natural product and drug, respectively, and might thus be candidates for drug repurposing for the treatment of COVID-19. In addition, we also tested the inhibitory activity of testosterone, and our results reveal testosterone as possessing moderate inhibitory potency against the 3CLpro enzyme, which may thus provide an explanation why older men are more severely affected by COVID-19.
1. Introduction
COVID-19 infection begins with the exposure of a human host to the recently discovered SARS-CoV-2, a positive-sense RNA virus belonging to the coroviridae family. The first critical stage in the development of a COVID-19 diseased state involves viral entry whereby the viral pathogen employs its surface spike glycoprotein in binding to the host’s angiotensin converting enzyme 2 (ACE2) [1], [2], [3], [4]. Following invasion of the host cells, the next critical stage that is central to a successful colonization of host cellular processes and establishment of the infection is viral replication. SARS-CoV-2 employs a multiplex array of independently functional enzymatic units called the replication-transcription complex (RTC) for its replication and transcription functions. The host’s ribosome is hijacked and appropriated for translating the infecting SARS-CoV-2’s mRNA from which crucial viral enzymes, structurally important protein units, as well as new viral genomes needed to create functional viral units, are produced.
SARS-CoV-2’s genome in its full assembly involves a replication complex featuring 14 open reading frames (ORFs) of which ORF1a encoding for polyprotein 1a (PP1a) and ORF1ab that encodes for PP1ab are integral. The genome assembly runs from an ORF1a segment at the 5′ end, followed immediately by an ORF1b section, and terminating with a segment of about 8 kb units that contains genetic codes for critical structural proteins and accessory factors at the 3′ end [5]. The ORF1ab is thus a linear arrangement of the separate ORF1a and ORF1b units. Important structural proteins, including the spike protein employed in viral entry, the membrane and envelope proteins, as well as the nucleocapsid are encoded at the 3′ end. At the 5′ end of the genome, non-structural proteins (NSPs) involved in various enzymatic functions are encoded. A total of sixteen such NSPs (named nsp1 through nsp16) have been identified in SARS-CoV-2 and known to be responsible for different specific catalytic functions, which are crucial for a successful viral replication and transcription. The autocatalytic activity of the two important RTC proteases, the papain-like protease PLpro and the main protease 3CLpro, on the two large polyproteins is responsible for the generation of functional enzymes without which the entire replication architecture becomes nonfunctional. PLpro cleaves the polyprotein PP1ab at three positions producing nsp1, nsp2, and nsp3, while 3CLpro processes PP1ab and PP1a at eleven points to generate nsp4 to nsp16.
This makes the main protease the most important enzyme in the RTC and therefore it represents an attractive target for the development of therapeutics for the treatment of COVID-19. And indeed, since the outbreak of the disease in late 2019 and the recognition of its pandemic status, hundreds of research articles have been published exploring the possibility of evolving small molecules inhibitors of 3CLpro. Computational design involving ligand docking protocols is a significant percentage of these publications. In our first work, we conducted large-scale virtual screening of diverse compound libraries against the three-dimensional structure of 3CLpro [6]. In total we screened over 1.2 million compounds, where we incorporated protein dynamics via so-called ensemble docking [7]. Using this approach we identified important structural factors within the enzyme substrate site as well as encoded in the docked compounds that we believe are crucial for substrate recognition.
In the present work we use all-atom, explicit-solvent molecular dynamics (MD) simulations of the 147 best binding compounds docked to 3CLpro to further delineate good 3CLpro binders from poor ones regardless of initially computed affinities. As the present pandemic mandates fast action, we decided to focus our current attention on existing drugs with possible drug repurposing for the treatment of COVID-19 in mind as well as natural products, which exhibit a wide range of pharmacophores and a high degree of stereochemistry creating a great source of possible hits. A detailed analysis of the MD simulations involving geometric and energetic arguments identified 34 ligands with binding to 3CLpro predicted to be superior compared to the other compounds investigated here. From this list, 14 compounds were chosen and, for comparison with estradiol, augmented by testosterone, which were then tested for their 3CLpro inhibition capabilities using an in vitro assay. This step revealed eight novel non-covalent inhibitors comprising five existing drugs, four of them being approved by the FDA (U.S. Food and Drug Administration) and one being under investigation, and three natural products. Our analysis provides important information about the inhibition of the SARS-CoV-2 main protease as well as new leads for the development of treatment options against COVID-19.
2. Results and discussion
2.1. Selection procedure
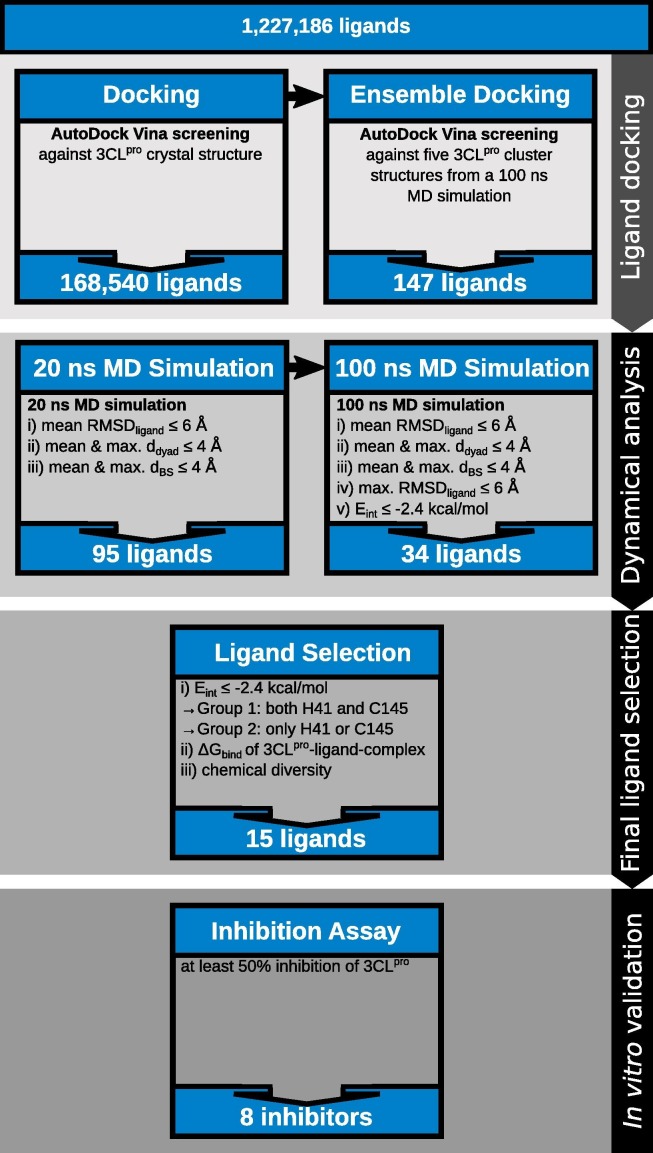
This study addresses the dynamical aspects of prospective inhibitors of the 3CLpro enzyme of SARS-CoV-2, previously identified using ensemble docking [6]. A scheme of our selection procedure employed in our previous and current work is given in Fig. 1 . The current work starts with 147 independent 20 ns MD simulations for ligand-main protease complexes involving the top-performing compounds identified by our previous virtual screening approach. Of this number 61 are FDA-approved drugs, 38 are drugs approved by other countries’ national regulatory agencies (non-FDA) and investigational drugs (INV), 39 are natural products (NP), while the remaining nine compounds are steroids (ST), most of which also have FDA approval status. The steroids were included because some of them were identified as good binders by the initial study [6]. In consideration of the observed gender-related differences in the clinical presentation of COVID-19 [8] we additionally aim to understand differences (if any) at the steroids’ level of interaction with the virus’ main protease. Apart from these, we also simulated different reference compounds (REF) involving previously identified inhibitors of the SARS-CoV-2 enzyme, including eight ligands from Jin et al. [9], and four recently identified inhibitory -ketoamides [10]. In total, 160 compounds were considered in this work, which are listed in Table S1 and for which 20 ns MD simulations were performed. To progress the predicted inhibitors to the next simulation phase, we employed the following three selection criteria:
-
1.
the root mean square deviation (RMSD) of the ligand (mean RMSDligand Å),
-
2.
the distance to the binding site (mean and maximal d BS Å),
-
3.
the distance to the catalytic dyad residues H41 and C145 (mean and maximal d dyad Å).
Fig. 1.
Scheme of the selection procedure..
These selection parameters were computed for the last 5 ns of the 20 ns MD simulations to make allowance for structural adaption in the first part of the trajectories. Ninety-five of the 147 selections from our previous work as well as three of the 12 reference compounds satisfied the filtering parameters, leading to 98 compounds for which the MD simulations were subsequently extended to 100 ns. In addition, testosterone, which did not meet the selection criteria, was also simulated for 100 ns to have a representative of the male hormone system for comparison to estradiol, the only steroid satisfying the filtering parameters. The last 25 ns of each 100 ns trajectory was subjected to the same filtering criteria 1.–3. listed above, which were augmented with two additional criteria: the requirement of maximal RMSDligand Å and an energy parameter that reflects the interaction between the catalytic dyad residues (H41 and C145) and the ligand requiring kcal/mol. For the initial 20 ns MD simulations, only the average RMSDligand was required to be lower than 6 Å to allow the ligands to adapt to a more energetically favorable binding pose, a phenomenon expected to temporarily increase the RMSDligand values. However, after 75 ns of MD simulations we expect the strong binders to have finally adopted a stable binding pose, which is why a maximal RMSDligand of 6 Å was additionally employed as a criterion in the second set of MD simulations. Applying these five selection criteria yielded 34 compounds of the selection library as well as one reference compound meeting the filtering conditions, which were grouped based on the number of interactions with the catalytic dyad residues. More precisely, ligands interacting with both H41 and C145 were considered the preferred binders as opposed to those interacting with just one catalytic dyad residue. Finally, the total binding free energy was computed and 14 compounds with lower values as well as testosterone were selected for in vitro testing using a 3CLpro inhibition assay, which revealed eight ligands with strong inhibitory activity.
2.2. Filtering based on 20 ns MD simulations
2.2.1. Ligand detachment
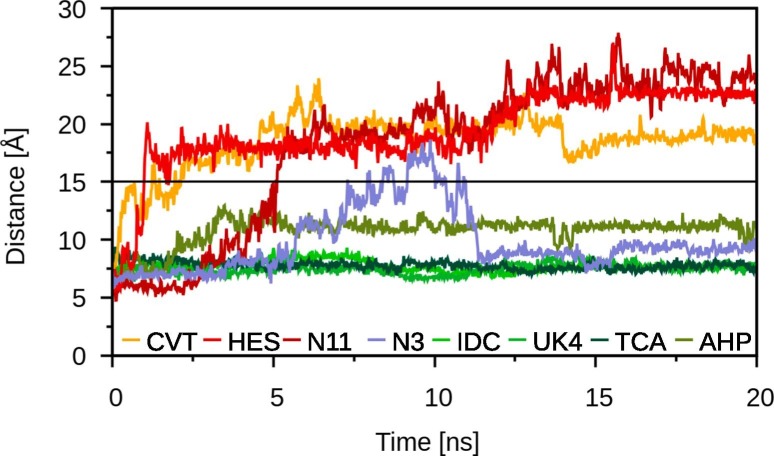
Docking studies allow a good estimation if a compound fits well into the catalytic site of a potential drug target and therefore might be a good binder. To properly differentiate between good and poor binders using computational methods, not only a static analysis but also a dynamic investigation like MD simulations should be conducted [11]. It is possible that only weak interactions between the ligand and the binding site exist, which cause a ligand to leave the binding site under dynamical force. Indeed, during the 20 ns MD simulations some of the ligands display very high distances to the catalytic dyad, which raises the question of whether these ligands remain bound to the binding site. To answer this question, we calculated the distance between the center of mass (COM) of the ligand with respect to the COM of the binding site residues, where the binding site of 3CLpro was defined as all amino acids within 10 Å of the covalently bound ligand N3 in the structure of the 3CLpro-N3 complex [9], yielding a total of 72 residues. Based on this quantity denoted , the following detachment criterion was formulated: a ligand is detached when it resides at a above 15 Å for at least 2 ns. This criterion is applied to the whole trajectories, instead of focusing on only the last 5 ns. This analysis reveals three ligands of different classes as detaching from the binding site, for which is plotted in Fig. 2 . Conivaptan (FDA) and 2-hydroxyestrone (ST) are leaving the binding site already after around 1 ns, while the of the natural product with the ID ZINC000008764269 remains below 15 Å for the first 5 ns before increasing beyond the cutoff value. Once detached, these compounds did not reenter the binding site during the 20 ns MD simulations. Interestingly, also the reference ligand N3 moves away from the binding site, even though not for a sufficiently long duration to have been considered a detachment event. It is important to note that N3 is a Michael acceptor inhibitor, which inactivates 3CLpro irreversibly by forming a covalent bond with C145 [9]. In contrast, N3 was not covalently bound in our MD simulation setup and for this reason the nonbonded interactions seem not to be strong enough resulting in reorientation of the ligand.
Fig. 2.
The distance between the COMs of the ligand and the binding site residues,, of selected ligands during 20 ns MD simulations. The distance is shown for the detaching ligands conivaptan (CVT, orange), 2-hydroxyestrone (HES, red) and ZINC000008764269 (N11, dark red). The cutoff distance at 15 Å is indicated by a black line. The reference compound N3 (blue) does not detach despite transient motions away from the binding site. For comparison, the distances of the ligands displaying the smallest in each ligand class are shown (from dark to light green): indocyanine (IDC, FDA), UK-432,097 (UK4, INV), theacitrin A (TCA, NP) and 17--hydroxypregnanolone (AHP, ST).
For each compound class the ligand exhibiting the smallest values is also shown in Fig. 2. Interestingly, UK-432,097 (INV), theacitrin A (NP), and 17--hydroxypregnanolone (ST) already appeared among the ligands with the smallest distances to the catalytic dyad residues in their respective class in our previous docking study [6]. Only indocyanine (FDA) did not stand out in the previous study, where it even displayed a higher distance to the catalytic dyad than conivaptan which detached from the binding site within 5 ns of the MD simulation.
2.2.2. Ligand flexibility
To further quantify the reorientation and flexibility of the ligands that remained in the binding site within the 20 ns MD simulations, we computed the RMSDligand values. High RMSD values indicate unsatisfactory binding with low affinity towards the binding site. An RMSD value over 2 Å suggests a pose that is significantly different from the reference structure, which in our case was set to the starting structure of each MD production run. However, we recognize that subject to dynamical forces, there may be need for readjustment of the docking-generated binding poses to adapt to conformational changes in the binding site of 3CLpro; because of this we chose a relatively high and slightly forgiving cutoff of 6 Å for the mean RMSDligand. The values obtained for the selection library range from 1.3 Å to 25.2 Å, and from 3.3 Å to 9.7 Å for the reference ligands. Nine ligands display RMSDligand mean values over 10 Å, which include the three ligands that detached, namely conivaptan (FDA, 19.5 Å), ZINC000008764269 (NP, 25.2 Å) and 2-hydroxyestrone (ST, 21.7 Å), as well as dolutegravir (FDA, 11.0 Å), tucatinib (INV, 11.2 Å), telomestatin (INV, 11.5 Å), bavacoumestan-A (NP, 11.1 Å), allopregnanolone (ST, 10.8 Å), and androstenedione (ST, 11.2 Å). It is important to point out that all these compounds feature a rigid chemical structure with few or no rotatable bonds present. This feature most likely renders the compounds incapable of employing intramolecular readjustment for adapting to the dynamically adjusting 3CLpro binding site. Instead, the entire ligand molecules have to be involved in adjusting to the enzyme binding site with consequential increase in RMSDligand.
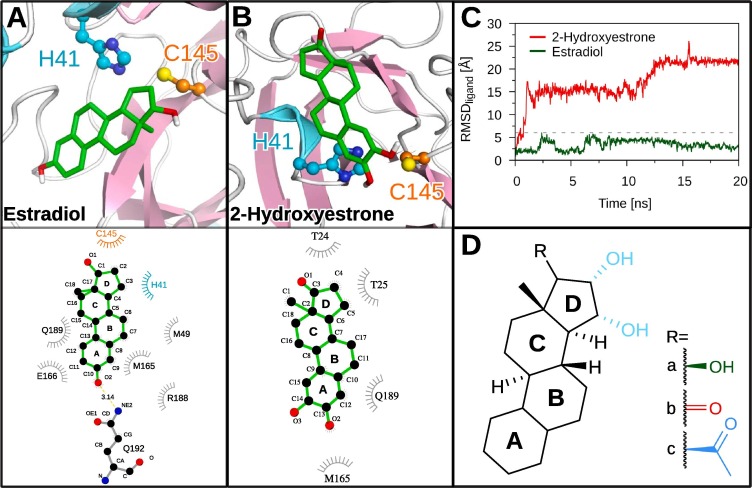
A significant number of the steroids (three out of nine) also show high fluctuations at the binding site, most likely due to weaker interactions compared to steroids with low RMSDligand such as estradiol (3.0 Å) or estetrol (3.4 Å) (Fig. 3 ). The main difference between these steroids lies in their moieties at the D ring. Estradiol and estetrol contain one or three hydroxy groups at the D ring, respectively. It is possible that binding site interactions involving the D ring moieties might be impaired in androstenedione and 2-hydroxyestrone harboring carbonyl moieties or allopregnanolone having an acetyl group in their D ring. Surprisingly, cortisol, which was identified as one of the best binders among the FDA-approved compounds previously [6], is not falling below the catchment RMSD cutoff with an RMSDligand of 7.7 Å. Similar to the steroids with extremely high RMSD, the D ring of cortisol contains not only a hydroxy group but additionally an hydroxyacetone group which might hinder interactions under dynamical forces.
Fig. 3.
The binding pose and 3CLpro-steroid interactions for estradiol and 2-hydroxyestrone. Cartoon representation of the binding site of 3CLpro (-sheets in lilac, -helices in light blue) with (A, top) estradiol and (B, top) 2-hydroxyestrone bound to it. The ligands are shown as green sticks, the side chains of H41 and C145 are shown in ball-and-stick representation in cyan and orange, respectively, with the N atoms (blue), O atoms (red), and the S atom of C145 (yellow) being highlighted. (A/B, bottom) The interactions of the two ligands with the binding site were analyzed and plotted with LigPlot+ [12], [13]. Hydrogen bonds are indicated by yellow dashed lines between the atoms involved and the donor–acceptor distance is given in Å, while hydrophobic contacts are represented by gray arcs with spokes radiating towards the ligand atoms they contact. The contacted atoms are shown with spokes radiating back. (C) The RMSDligand of estradiol (green) and 2-hydroxyestrone (red) is shown over simulation time. The cutoff value at 6 Å used to distinguish between good (e.g., estradiol) and poor (e.g., 2-hydroxyestrone) binders is marked as a gray dotted line. (D) The steroid scaffold is shown with functional moieties at the D ring of some of the steroids analyzed in this study highlighted: estradiol (R = a, green), 2-hydroxyestrone and androstenedione (R = b, red), estetrol (R = a plus hydroxy groups shown in light-blue), and allopregnanolone (R = c, blue).
Besides cortisol, there are various ligands that showed great binding affinity in the initial virtual screening [6] but poor dynamical characteristics with respect to their flexibility at the binding site. These include naldemedine (6.3 Å) and enasidenib (7.8 Å) that ranked among the top binding FDA-approved drugs as well as ritonavir (6.6 Å), which is currently under investigation for a potential treatment of COVID-19 [14]. Furthermore, several non-FDA and INV ligands, which were able to surpass the best-binding FDA drug nilotinib (5.2 Å) in the docking studies [6], display here an RMSDligand above the cutoff, especially the previously mentioned INV drug telomestatin (11.5 Å), a compound with an inflexible macrocyclic core. These findings highlight the importance of fully incorporating structural dynamics while searching for prospective inhibitors of 3CLpro. We also observed that more than half of the reference compounds surpass the RMSDligand cutoff, namely carmofur, N3, PX-12, shikonin, tideglusib and two -ketoamides, as well as testosterone (8.5 Å).
2.2.3. Distance between ligand and binding site
Inhibition of 3CLpro requires interaction of the ligand with the enzyme’s binding site as a sine qua non regardless of the nature of inhibition, competitive or irreversible. Preferably, such interactions should involve direct contacts with the catalytic dyad residues H41 and C145. To identify the compounds in close binding proximity, we set the cutoff for the mean and the maximum distance to both the binding site (d BS) and the dyad (d dyad) at 4 Å. These distances are defined as minimum distances between ligand and the respective group of residues over the last 5 ns of the MD simulations, yielding a maximum and a mean for them. All compounds that were not already identified as detaching before, remain associated with the enzyme with d BS mean values ranging between 1.6 Å and 3.1 Å. In contrast, several of the ligands violate the averaged and/or maximal d dyad cutoff values. These include the antibiotic ertapenem (FDA, maximal = 4.7 Å), epigallocatechin gallate (EGCG, NP, maximal = 5.1 Å), and adozelesin (INV, maximal = 7.5 Å) that had represented the top drugs in their respective categories in the previous docking study [6]. This again underpins the importance of analyzing dynamical features beyond docking.
2.3. Filtering based on 100 ns MD simulations
2.3.1. Ligand detachment
After applying the various cutoffs using the last 5 ns of the initial 20 ns MD simulations, the remaining 95 ligands of our compound selection, 3 remaining reference compounds, as well as testosterone for comparison with estradiol and estetrol were simulated for 100 ns to detect the most strongly binding ligands. During these longer simulations another ligand (the FDA approved drug crizotinib) detached from the 3CLpro binding site after around 85 ns. The dichlorfluorphenyl moiety of this compound was highly mobile and flexible in the binding site during the MD simulation. Being solvent-exposed it was observed to leave the binding site first, following which the entire molecule also detached. For this reason, crizotinib was exlcuded in subsequent analysis. The fact that ligand detachment could be observed even after around 85 ns highlights the importance of a more detailed screening of the most promising ligands by extending the initial MD simulations to at least 100 ns.
2.3.2. Ligand flexibility and distance to the catalytic dyad
To identify the best binding ligands based on the 100 ns MD simulations, the same selection criteria as before were applied using the last 25 ns of these longer simulations. In addition, for the RMSDligand also the maximal values are now considered. In the following, the statistics provided is in relation to the total amount of ligands in the second MD selection round without the reference ligands (i.e., 100% = 95 ligands). The mean RMSDligand ranges between 1.4 Å and 12.0 Å, d BS between 1.7 Å and 2.1 Å, and d dyad between 1.9 Å and 8.8 Å. All ligands stay close to the binding site but four ligands (corresponding to 4% of 95 ligands) – olsalazine (FDA, 8.2 Å), entospletinib (INV, 13.3 Å), amrubicin (non-FDA, 8.5 Å), and estetrol (ST, 12.7 Å) – seem to loose contact with the catalytic dyad as they display maximal d dyad values above 8 Å. The criterion of a maximum d dyad below 4 Å is not met by 22 ligands (23%), most of them belonging to the FDA-approved drugs (12 ligands, 13%). Sixty ligands (63%), including 21 of the 22 ligands surpassing the maximum d dyad cutoff, are very flexible as indicated by maximal RMSDligand values above 6 Å. Only deferasirox (FDA) displays a maximal d dyad above the cutoff (4.6 Å) without simultaneously exceeding the maximum RMSDligand cutoff (5.6 Å). Deferasirox harbors four rotatable bonds, which enable rotation of the three aromatic rings at the central triazole and therewith adjusting the position of the hydroxy and carboxy moieties at the rings. However, the overall structure is quite rigid, so that the movement of the compound away from the catalytic dyad is mainly accompanied by overall translation.
The criterion for the maximal RMSDligand cutoff is accomplished by 36 (38%) ligands. This leads to 35 ligands (37%) that meet all three criteria, while out of the 3 reference drugs and testosterone only disulfiram possesses RMSDligand, d BS and d dyad values below the respective cutoff. Interestingly, the two FDA-approved drugs nilotinib (maximal RMSDligand = 13.3 Å) and afatinib (maximal RMSDligand = 10.3 Å) as well as the non-FDA drug R-343 (maximal RMSDligand = 9.4 Å), which belong to the class of tyrosine kinase inhibitors that were shown to be able to inhibit related coronaviruses [15], display a maximal RMSDligand above 6 Å. These three, among others, were considered as top-performance ligands due to low binding free energies obtained from docking in the previous study [6].
2.3.3. Interaction of ligands with catalytic dyad residues
For further selection of the top ligands, the interaction energy composed of Coulomb and Lennard-Jones (LJ) interactions between ligand and either catalytic dyad residue was calculated and used as an additional criterion. The requirement is that a ligand needs to interact with one or both catalytic dyad residues, H41 and C145, using the criterion kcal/mol to be selected for further consideration. This removes tubocurarine (non-FDA) from the list since it does not interact strong enough with the dyad residues. Although it stays closely buried in the binding site with its benzylisoquinoline ring facing the dyad residues, the methyl ether and the tertiary nitrogen are unable to constitute strong interaction partners for H41 and C145, respectively. From the remaining 34 compounds, 15 ligands interact with only one of the two catalytic dyad residues and the other 19 ligands form contacts with both H41 and C145. The only reference compound left, disulfiram, interacts with H41 and is therefore also included in the final ligand selection. To get a better view on the class composition of the final ligand selection, statistics is provided in relation to the total amount of ligands in the respective class and to the total amount of ligands subjected to MD simulation (100% = 160 ligands, including steroids and reference compounds). Accordingly, 34 ligands (21%) are identified as top binders, including 10 out of 61 FDA-approved drugs (16%, 6%), 8 out of 38 non-FDA-approved drugs (21%, 5%), 15 out of 39 natural products (38%, 9%), 1 out of 9 steroids (11%, 1%). The binding poses of these ligands in the active site of 3CLpro are shown in Figs. S1–S3.
2.3.4. Best performing 35 compounds
After narrowing down the tremendous amount of potential SARS-CoV-2 main protease 3CLpro ligands to a sufficiently small library of 35 compounds (34 from our selection list [6] plus disulfiram as the only remaining reference compound), we analyzed the top binders further based on binding free energies, , calculated with the molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) method [16]. Here, we divided the ligands into two groups, those interacting with both catalytic dyad residues, which are considered better binders, and those interacting with only H41 or C145. Each group was then ranked using the values, which in comparison to not only consider the Coulomb- and LJ interactions, but also include solvent effects. For ligands interacting with both dyad residues, ranges from kcal/mol to kcal/mol, while values between kcal/mol to kcal/mol are obtained for the compounds interacting with only one of the dyad residues (Table 1 ).
Table 1.
Characteristics of the 35 top ligands of 3CLproidentified by MD simulations. The compounds selected for validation in a 3CLpro inhibition assay are highlighted in bold.
| No. |
Compound name |
Class |
ddyad [Å] |
dBS [Å] |
[kcal/mol] |
|---|---|---|---|---|---|
| Interaction with both catalytic dyad residues (H41 and C145) | |||||
| 1 | Cilostazol | FDA | |||
| 2 | Proanthocyanidin A1 | NP | |||
| 3 | Rhoifolin | NP | |||
| 4 | Apixaban | FDA | |||
| 5 | Theacitrin C | NP | |||
| 6 | Corilagin | NP | |||
| 7 | Dihydroergotoxine | non-FDA | |||
| 8 | Telcagepant | INV | |||
| 9 | ZINC000002114470 | NP | |||
| 10 | ZINC000011865175 | NP | |||
| 11 | Lurasidone | FDA | |||
| 12 | ZINC000049888572 | INV | |||
| 13 | Hypericin | INV | |||
| 14 | Pimozide | FDA | |||
| 15 | Sotrastaurin | INV | |||
| 16 | Proanthocyanidin A2 | NP | |||
| 17 | Enzastaurin | INV | |||
| 18 | ZINC000008297065 | NP | |||
| 19 | Telmisartan | FDA | |||
|
Interaction with one catalytic dyad residues (H41 or C145) | |||||
| 20 | Isocorilagin | NP | |||
| 21 | Dasatinib | FDA | |||
| 22 | Teniposide | FDA | |||
| 23 | Palbociclib | FDA | |||
| 24 | Tadalafil | FDA | |||
| 25 | ZINC000012881832 | NP | |||
| 26 | TMC647055 | INV | |||
| 27 | Fenoverine | INV | |||
| 28 | Zeylanone | NP | |||
| 29 | Remdesivir | NP | |||
| 30 | Estradiol | ST | |||
| 31 | Disulfiram | REF | |||
| 32 | Epitaraxerol | NP | |||
| 33 | Theaflavin | NP | |||
| 34 | Daidzein | NP | |||
| 35 | Montelukast | FDA | |||
Statistics is generated in relation to the total number of ligands in each class as well as for all ligands selected in the top binders list (100% = 35 ligands). 14 ligands interact with only H41, while epitaraxerol and fenoverine interact with C145 only. These include five FDA compounds (8%, 14%), two non-FDA and INV ligands (5%, 6%), seven NP (18%, 20%), one ST (11%, 3%), and one REF (8%, 3%). The group of ligands binding both dyad residues includes 19 ligands in total, among these are five FDA compounds (8%, 14%), six non-FDA and INV ligands (16%, 17%), and eight NP (21%, 23%). It is particularly noticeable that many NP compounds rank among the top binders, even six exist among the top ten (Table 1). They resemble each other structurally by harboring a lot of oxygen atoms mostly in hydroxy functional groups and involving multiple ring systems. These hydroxy groups were observed to mostly orient towards the dyad residues allowing the establishment of hydrogen bonds with the catalytic dyad. Furthermore, estradiol representing the female hormonal system is found to be the only steroid present among the top 35 binders, while testosterone as a prominent male hormone violates the selection criteria.
2.4. Enzyme inhibition assay
Of the 35 compounds in the selection list (Table 1), 15 were procured and a 20 μM concentration of each was employed in an enzyme-based fluorescence assay to determine their inhibitory activities against the SARS-CoV-2 main protease enzyme at 20 μM concentration in the presence of a peptide substrate. The choice which of the compounds to test in vitro was to a large extent based on , i.e., ligands with smaller binding free energy values were given preference. Moreover, compounds forming contacts with both H41 and C145 were also preferentially considered, resulting in 10 compounds from this group (cilostazol, rhoifolin, apixaban, corilagin, dihydroergotoxine, telcagepant, ZINC000011865175, lurasidone, hypericin, and proanthocyanidin A2), while dasatinib, teniposide, palbociclib, and estradiol, which were also chosen, interact with only one of the catalytic dyad residues. Other selection criteria were chemical diversity among the compounds tested as well as availability. For instance, theacitrin C, which we wanted to include in the enzyme inhibition assay, is currently not commercially available. One out of the 15 compounds tested is testosterone, which we included as comparison to estradiol despite it not making it onto the final selection list that resulted from the in silico screening.
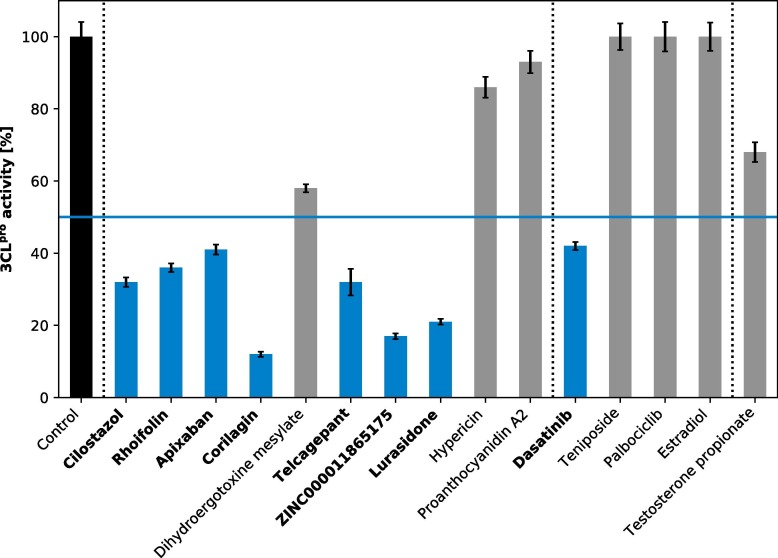
In Fig. 4 the results from the enzyme inhibition assay show various degrees of 3CLpro inhibition for the 15 compounds. The three compounds having zero inhibitory activity at a ligand concentration of 20 μM against the proteolytic properties of the main protease (teniposide, estradiol, and palbociclib) are from the selection group that interacts with only one catalytic dyad residue. This represents 75% of those screened from this group in the enzyme inhibitory assay. The only ligand efficiently inhibiting 3CLpro out of this group is dasatinib, which is an FDA-approved drug interacting with H41 only, provoking an activity loss of 58%. Out of the ten compounds from the selection group shown to interact with both catalytic dyad residues, seven of them (corresponding to 70%) were found to inhibit greater than 50% of the enzyme function. Of these, the natural product corilagin produces an 88% loss of the SARS-CoV-2 main protease activity, followed by 83% for ZINC000011865175, another natural product. Trailing closely behind these two are three drugs: lurasidone, an antipsychotic FDA-approved drug with 79% inhibition, and cilostazol and telcagepant both displaying 68% main protease inhibition. Cilostazol inhibits the phosphodiesterase 3 and platelet aggregation and has been approved for peripheral vascular disease, while telcagepant was initially being developed as a treatment for migraine. After these, rhoifolin, a natural product and apixaban, an FDA-approved drug, were found to inhibit the enzyme by 64% and 59%, respectively. Interestingly, the anticoagulant apixaban was found to reduce the mortality from COVID-19 by about 50% in a therapeutic dose (data from Mount Sinai Hospital, New York, USA) and the flavonoid rhoifolin has been previously proposed to be an efficient inhibitor of 3CLpro [17]. Comparing the 3CLpro activity of estradiol and testosterone, one can see that 3CLpro looses about 30% of its activity in the presence of testosterone but is fully active in the presence of estradiol. On account of this, we can exclude a potential 3CLpro inhibition activity of the female hormone estradiol as the cause behind the different COVID-19 progression between men and women [8]. In total, eight compounds could successfully be identified as potent 3CLpro inhibitors with more than 50% inhibition at 20 μM concentration comprising five FDA-approved drugs, two natural products, and one investigational drug. The binding poses of the top eight inhibitors are shown in Fig. 5 , and their chemical structures can be seen in Fig. S4.
Fig. 4.
Activity of 3CLproin the presence of the 15 tested compounds for their inhibition capacity. The activity of 3CLpro is provided relative to the control sample (black) without additional ligand. The group of compounds displaying interactions with both catalytic dyad residues is separated from the one with only one such interaction by a dashed vertical line and the compounds in each group are sorted by their values. Testosterone not belonging to the top 35 ligands is shown separately. The bars for compounds with a residual 3CLpro activity above 50% are colored in gray, while those with a residual activity below 50% are highlighted in blue with the compound names written in bold. A ligand concentration of 20 μM was used for the inhibition test, which was performed in triplicate per ligand. The standard deviation is shown as error bar.
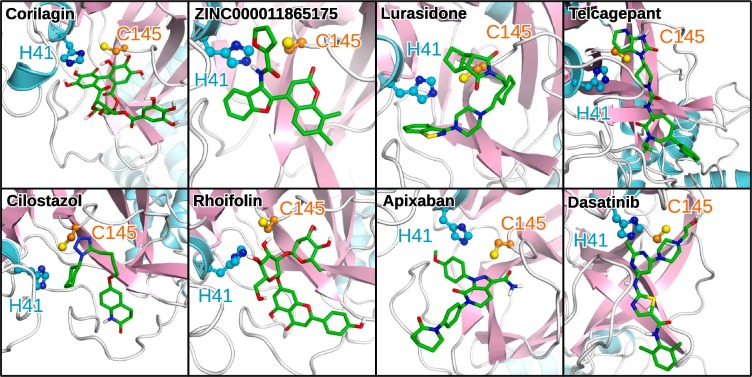
Fig. 5.
The binding poses of the top eight binders. The top eight compounds identified by an enzyme inhibition assay following in silico screening are shown in the order of decreasing inhibitory activity: corilagin (NP), ZINC000011865175 (NP), lurasidone (FDA), telcagepant (INV), cilostazol (FDA), rhoifolin (NP), apixaban (FDA), and dasatinib (FDA). The same protein and ligand representations as in Fig. 3 are used.
2.5. Top eight inhibitors
2.5.1. Interactions with 3CLpro
It is interesting to note that all eight compounds found to inhibit 3CLpro in our in vitro assay passed all employed filtering criteria and were additionally found to be capable of forming specific interactions with both catalytic dyad residues as well as establishing hydrogen bonding networks with other binding site amino acids (Fig. S5). A difference in the classes can be observed as the three natural products corilagin, rhoifolin and ZINC000011865175 contain many oxygen containing functional groups, while the FDA-approved drugs apixaban, dasatinib, cilostazol and lurasidone as well as the investigational compound telcagepant contain more functional moieties comprising nitrogen. We analyzed the interactions with binding site residues using LigPlot+ [12], [13] and extracted all residues exhibiting contacts, accounting specifically for hydrogen bonding with one or several of the eight 3CLpro inhibitors (Table 2 ). Here, residues interacting with several ligands and those being involved in hydrogen bonding are considered as more important for inhibition than those not fulfilling these two criteria.
Table 2.
Binding site residues interacting with the top eight inhibitors. The number of ligands interacting with the different binding site residues via hydrogen bonds exclusively or via any kind of interactions (including hydrogen bonds) is listed.
| Number of ligands involved | Hydrogen bonds | All interactions |
|---|---|---|
| 1 | H41, C145, E166, V186 | L27, S46, L50, Y54,L167, T169, V186 |
| 2 | L141, N142, G143,H164, Q189, T190 | T25, F140, L141,H172, A191 |
| 3 | R188, Q192 | T26, H163, Q192 |
| 4 | S144 | M49, S144, P168, T190 |
| 5 | — | N142,D187 |
| 6 | — | G143, H164 |
| 7 | — | H41, E166 |
| 8 | — | C145, M165, R188, Q189 |
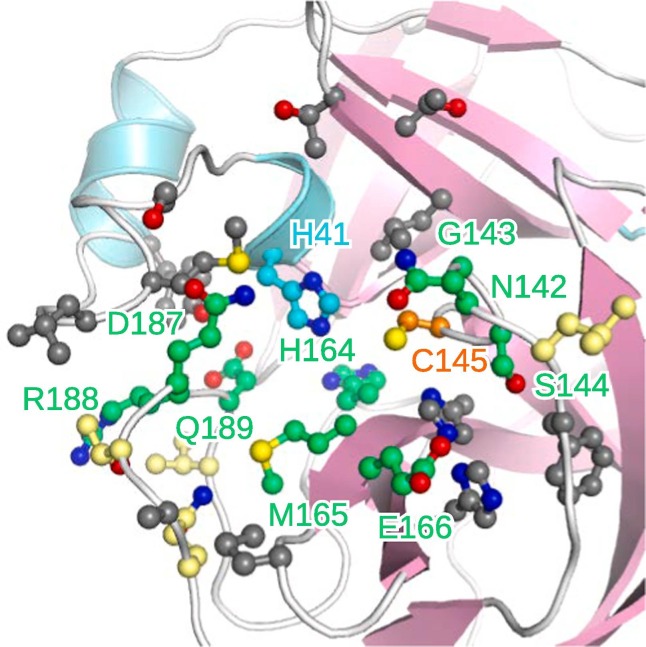
The catalytic dyad residues H41 and C145 form hydrogen bonds exclusively with ZINC000011865175 but have less strong interactions with the other inhibitors (Fig. S5). While C145 interacts with all ligands, no interaction can be observed between H41 and rhoifolin. Besides C145, also M165, R188, and Q189 interact with all eight ligands. The latter two form hydrogen bonds with three ligands each, but similar to the catalytic dyad mainly weaker interactions with the other ligands. No hydrogen bond formation but other contacts with all eight ligands is recorded for M165. Also G143, H164, E166, and N142 are suggested as important residues for inhibitor binding forming both hydrogen bonds and other contacts with the latter as the main interaction type. D187 establishes contacts but no hydrogen bonds with five ligands, while S144 exclusively forms hydrogen bonds with half of the ligands. Except from G143 and M165, these residues are all polar or charged and located either at the entrance to or at the inside of the binding site (Fig. 6 ). The latter might be responsible for tight inhibitor binding and the former might hinder ligand detachment due to a closure of the binding pocket, together accounting for the high 3CLpro inhibition activity. Hydrophobic residues interacting with few of the ligands are located rather at the surface close to the binding site and might be responsible for initial recruitment of binders to the active site.
Fig. 6.
The binding site with the residues predominantly interacting with the top eight inhibitors highlighted. The residues showing the most pronounced interactions are colored in green and those forming hydrogen bonds are emphasized by a yellow color, while all other interacting residues are shown in gray.
In order to better quantify the hydrogen bonding between the ligands and 3CLpro, the average number of hydrogen bonds formed with all the binding site residues during the 100 ns MD simulations were calculated for the top 35 ligands identified in silico (Fig. S6). Except from ZINC000011865175, lurasidone, and telcagepant, the top eight inhibitors belong to the ligands developing on average the most hydrogen bonds, between 2.75 and 5.00. From the other ligands interacting with both catalytic dyad residues, four ligands – proanthocyanidin A1, theacitrin C, dihydroergotoxine, and proanthocyanidin A2 – form a similar amount of hydrogen bonds, while of those interacting solely with one catalytic dyad residue only two ligands – isocorilagin and remdesivir – exceed 2.75 hydrogen bonds on average. It is noticable that besides the top eight inhibitors, the compounds considered as the best binders as judged by show the most pronounced hydrogen bond formation with binding site residues. Thus, hydrogen bond formation considerably contributes to the binding free energy. Ligands, which were excluded from the list of potential 3CLpro inhibitors by the activity assay such as hypericin, teniposide, palbociclib, and estradiol are found to form less than 2.75 hydrogen bonds on average. Therefore we can conclude that a ligand being able to develop many hydrogen bounds with binding site residues is sought to be important for its strong binding and potential inhibition activity. However, some of the ligands like ZINC000011865175, lurasidone, and telcagepant as well as testosterone form on average less than 2.75 hydrogen bonds (Fig. S6), yet nonetheless are able to inhibit 3CLpro, in the case of the former three even strongly. Testosterone formed on average only 0.63 hydrogen bonds and also failed all other selection criteria, but surprisingly reduces the activity of 3CLpro by 32%. Testesterone may thus be an endogenous inhibitor of the SARS-CoV-2 main protease and help to attenuate a COVID-19 infection. This would contribute to an explanation for the age-dependent severity of COVID-19, given the positive correlation between serum testosterone levels, disease progression and clinical outcomes in male Covid-19 patients, independent of patient age and comorbidities [18].
2.5.2. Distance to the catalytic dyad residues
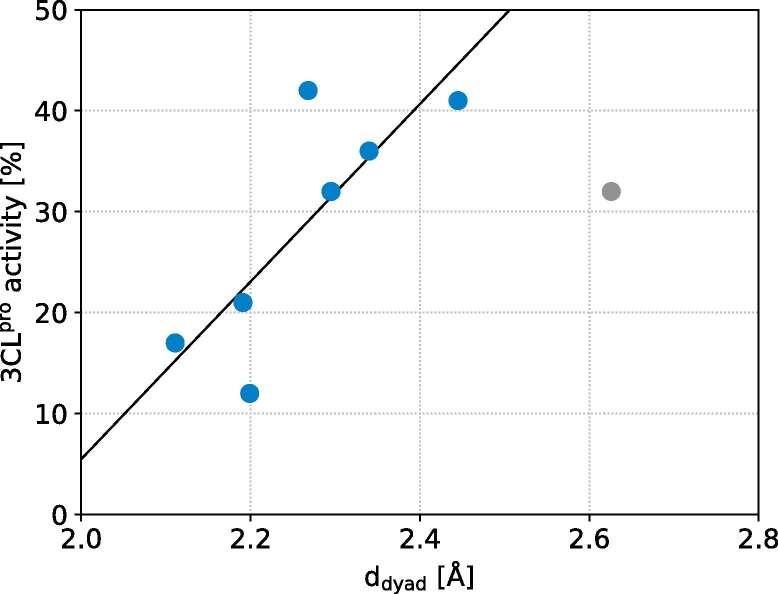
In addition to hydrogen bonds, we analyzed various other properties of the ligands, such as the number of aromatic rings and rotatable bonds, as well as their interactions with 3CLpro and plotted the resulting values against the experimentally obtained inhibition data to identify further characteristics contributing to the inhibitory activities. A fairly good correlation is obtained between experimental data and d dyad suggesting higher protease inhibition at closer distances from the catalytic dyad (Fig. 7 ). Comparing the chemical structures of the top eight inhibitors (Fig. S4) and the other experimentally addressed ligands, a decent number of ring systems can be found as a common feature among the ligands. Furthermore, good inhibitors seem to need a certain flexibility to adapt a strong binding position, which is not the case for e.g. the large and rigid hypericin and the tested steroids. It would be interesting to extend the experimental testing to ligands exhibiting descriptors found to be important, i.e. those with small d dyad values and those with pronounced hydrogen bonding like proanthocyanidin A1 and theacitrin C, which were not experimentally investigated in the first testing round. This includes remdesivir, which according to our in silico screening could act as a 3CLpro inhibitor, while based on cell assays it is suggested to interfere with the RNA polymerase, i.e., nsp12 of SARS-CoV-2 [19], [20].
Fig. 7.
Correlation between the distance to the catalytic dyad (ddyad) and the experimentally determined residual activity of 3CLprofor the top eight inhibitors. A linear regression without the outlier telcagepant colored in gray shows that lower ddyad correlates with stronger 3CLpro inhibition.
3. Conclusion
By carefully combining computational investigation methods with an enzyme inhibition assays we have identified eight chemical compounds that showed inhibitory activity in the low micromolar range against the main protease enzyme central to viral replication in SARS-CoV-2. With functional groups known for covalent attack absent in the molecular structure of these eight compounds, a non-covalent inhibitory mechanism is strongly implicated. Of the eight identified inhibitors, four are approved drugs, one an investigational drug, and three are natural products, a feature that will likely shorten the time to clinical availability following successful completion of relevant testing proceedings. We expect that the findings presented in this work will be of importance in the development of novel therapeutics for managing COVID-19. In addition, we have presented important structural features that appear to underly 3CLpro enzyme inhibition as well as surprising inhibition features for hormonal steroids involved in gender-based response to COVID-19 infection. While estradiol was found to be devoid of inhibitory activity against SARS-CoV-2’s main protease enzyme, testosterone was discovered to possess relatively strong inhibitory activity against the enzyme. This observation may explain, at least in parts, why obese and/or elderly men with expected reduced testosterone titres appear to be more vulnerable to the infection compared with males with higher serum testesterone levels. Further analyses are required to explore the full implications and ramifications of these findings both for the development of COVID-19 treatment and for understanding hormonal involvements in host response to SARS-CoV-2 infection.
4. Methods and materials
4.1. Molecular dynamics simulations
4.1.1. Simulation flow
In our previous study [6], we docked FDA-approved drugs, non-FDA and investigational drugs as well as natural products to the main protease 3CLpro binding site of the SARS-CoV-2 virus. In the first screening step, we screened 1,227,186 ligands against the 3CLpro crystal structure (PDB code 6LU7) [9]. To consider protein flexibility, ensemble docking [7] was performed with the 168,540 best performing compounds from the first screening (Fig. 1). To this end, five different 3CLpro conformers were taken from a 500 ns MD simulation of the protein with the N3 ligand bound to it. In order to select the ligands for the MD simulations for testing their stability in the binding site, we sorted the ligands based on the binding free energies predicted by docking and using the cutoff kcal/mol and the distances to the catalytic dyad residues with cutoff Å. For each of the resulting 147 compounds we performed a 20 ns MD simulation of the 3CLpro-ligand complex using the crystal structure of the enzyme. Additionally, 20 ns MD simulations were performed for 13 other ligands of interest in connection with 3CLpro (Table S1) [9], [10]. For 99 of these ligands the MD simulations were extended to 100 ns using the selection criteria explained in Section 2.1.
4.1.2. Parameterization of the ligands
Before the MD simulations could be started, generalized AMBER force field (GAFF) parameters [21] for the 160 ligands considered had to be derived. To this end, quantum mechanics calculations at the HF6-31G* level were performed using Gaussian 09 [22], followed by restrained electrostatic potential (RESP) calculations for determining partial charges [23], [24] via Antechamber [21], [25] as available in AmberTool 19 [26]. The GROMACS input files we then generated with the ACPYPE tool [27].
4.1.3. Simulation details
All MD simulations were performed with GROMACS 2018 [28]. We used AMBER14SB [29] with Parmbsc1 parameters [30] as protein force field combined with the TIP3P water model [31] to explicitly simulate water. The, 3CLpro-ligand complexes were centered in a cubic box of size Å3, solvated with water, and Na+ and Cl− added at a concentration of 150 mM while at the same time neutralizing the system. This results in a system size of 51,000 atoms in total. The energy of the systems was minimized via the steepest descent algorithm [32]. Afterwards, the systems were equilibrated, first in the NVT ensemble (i.e., with a constant number of molecules, volume, and temperature) for 0.1 ns and second for 1 ns in the NpT ensemble at 310 K (37 °C, Nosé-Hoover thermostat [33], [34]) and 1.0 bar (Parrinello-Rahman barostat [35]). The production runs of 20 ns or 100 ns lengths used the same settings as the NpT equilibration runs. Electrostatic interactions were processed with the particle-mesh Ewald method [36], [37] in combination with periodic boundary conditions and a real-space cutoff of 12 Å. The Lennard-Jones (LJ) interactions were also cut at 12 Å. For the integration of the equations of motion, a leapfrog stochastic dynamics integrator was used with a time step of 2 fs. The LINCS algorithm [38] was applied to constrain all bond lengths during the MD simulations. The coordinates were saved every 20 ps.
4.1.4. Analysis
As explained in the Results and Discussion section, various quantities were calculated for each ligand to determine the stability of the compounds in the binding site of 3CLpro. One of these quantities is the distance d dyad, which defines the minimum distance between the catalytic dyad residues (H41 and C145) and ligand and was calculated with the GROMACS tool gmx mindist. Another distance that was determined is the distance d BS, which is the minimum distance between the binding site and ligand. Here, the binding site was defined as the collection of 72 residues that reside within 10 Å around the ligand N3 in the crystal structure of the 3CLpro-N3 complex (PDB code 6LU7) [9]. This distance was also calculated with gmx mindist. To identify the ligands detaching from the binding site, we defined a third distance called which measures the distance of the center of mass of the binding site residues to the center of mass of the ligand in question. This distance was computed with gmx distance. We further determined the root mean square deviation (RMSD) of each ligand, called RMSDligand here, which indicates how flexible a ligand is in the binding site. To this end, we aligned the protein structures sampled during the MD simulations to the MD starting structure (excluding the ligand during the alignment) and then calculated the RMSD for the ligand using the gmx rms tool.
To quantify the strength of the 3CLpro-ligand interactions, their interaction energy consisting of Coulomb and LJ contributions was determined, which was accomplished by rerunning the simulation using gmx mdrun -rerun to obtain the energies, which were processed using gmx energy to calculate and between the ligand and the catalytic dyad residues H41 and C145. For the 35 best ligands identified in silico, we also computed the binding free energy using the MM/PBSA method as implemented in g_mmpbsa (https://rashmikumari.github.io/g_mmpbsa/) [16]. This analysis was applied to 626 MD snapshots sampled every 40 ps between 75 ns and 100 ns of the MD simulations. Within the MM/PBSA scheme the binding free energy is defined as
| (1) |
where indicates the average over the 626 snapshots in the current case. The free energy for each of these three entities is given as
| (2) |
where describes the bonded interactions, which is like and obtained from the force field, and are the polar and nonpolar contributions to the solvation free energy, and the last term is the absolute temperature, T, multiplied by the configurational entropy, S, which can be estimated by a normal-mode analysis of the vibrational frequencies. However, this entropy term is not calculated by g_mmpbsa. The polar energy term is obtained by solving the Poisson–Boltzmann equation, whereas the nonpolar term is estimated from a linear relation to the solvent accessible surface area (SASA). The parameters for the calculation of were set as K (37 °C), for the dielectric constant of the solvent (corresponding to water), for the dielectric constant of the solute (corresponding to a globular protein), kJ/(molÅ2) for the surface tension, Å as probe radius for the SASA calculation. was further decomposed into its per-residue contributions to determine the interaction strength with the catalytic dyad (H41/C145) or other residues in the binding site. This was accomplished with the Python script MmPbSaDecomp.py, while the script MmPbSaStat.py was used for the calculation of . Both scripts are provided via the g_mmpbsa website.
The energetic analysis of the 3CLpro-ligand interactions was augmented by an analysis of the hydrogen bonds formed between both entities. This was accomplished with gmx hbond applied to the binding site and ligand as interaction partners and using a cutoff of 3.5 Å for the distance between hydrogen donor and acceptor and a maximal allowed deviation from linearity of 30°.
4.1.5. Visualization
The protein–ligand systems were visualized using the PyMOL software [39]. For the selection of the binding poses shown in this manuscript, the ligand structures and orientations were clustered using the algorithmus of Daura et al. [40] as implemented in gmx cluster. A cutoff of 2 Å was used for the clustering and the most populated cluster per ligand chosen for visualization. Interactions between protein and ligands were analyzed and plotted using LigPlot+ [12], [13]. Results were plotted using the Gnuplot software [41] and Python3 [42].
4.1.6. Data availability
The AMBER force field (GAFF) parameters of the 160 organic molecules including drugs, natural products, and steroids that were derived as part of this work are available on Mendeley Data (https://doi.org/10.17632/phxtv76n5s.3). These parameters can be employed without further processing in MD simulations using GROMACS. Moreover, the Gaussian and GROMACS input files used for generating the force field parameters, along with bash scripts for automating the parameterization procedure are also provided at Mendeley Data. In [46] this dataset and how the data were obtained are described in detail.
4.2. In vitro testing
4.2.1. Cloning, expression and purification of SARS-CoV-2 3CLpro
The codon optimized cDNA encoding SARS-CoV-2 3CLpro (Uniprot entry: P0DTD1) was synthesized and implemented in the ampicillin resistant vector pGEX-6P-3 (BioCat GmbH, Heidelberg, Germany). The construct contains an N-terminal GST-tag and a PreScission protease cleavage site (LEFLFQGP).
SARS-CoV-2 3CLpro-pGEX-6P-3 vectors were transformed into E. coli Lemo21 (DE3) (New England BioLabs, USA) competent cells and grown overnight at 37 °C in LB-medium. This pre-culture was added to fresh LB-medium (Ampicillin and Chloramphenicol) and grew at 37 °C until the cells reached an OD600 of 0.6. Gene expression was induced with IPTG at final concentration of 0.5 mM (1 mM Rhamnose was added) and incubated for 3 h, at 37 °C and 120 rpm. Subsequently, the culture was harvested by centrifugation (4,000 rpm) at 5 °C for 20 min (Sorvall RC-5B Plus Superspeed Centrifuge, Thermo Fisher Scientific, USA; GSA rotor). The supernatant was discarded and the cells containing the recombinant SARS-CoV-2 3CLpro_GST were resuspended in 50 mM Tris–HCl pH 8.0, 200 mM NaCl (lysis buffer) and stored at −20 °C for subsequent purification.
For purification, the cell-suspension was incubated on ice for 1 h with addition of lysozyme, subsequently it was lysed by sonication in four pulses of 30 s each with amplitude of 30% interspersed by intervals of 10 s. The crude cell extract obtained was centrifuged (7,000 rpm for 90 min at 6 °C). The supernatant containing SARS-CoV-2 3CLpro_GST was loaded onto a GSH-Sepharose matrix which was previously equilibrated with the lysis buffer and was extensively washed with the same buffer. The protein was eluted with the same buffer plus addition of 10 mM GSH. The eluted fractions were concentrated and dialyzed against PreScission protease cleavage buffer (50 mM Tris pH 7.0, 200 mM NaCl, 1 mM DTT and 1 mM EDTA). PreScission protease was used to cleave the GST-tag from the SARS-CoV-2 3CLpro_GS°CT protein. For 100 μg target protein, 10 μg PreScission protease was added and the sample incubated for 36 h at 4 °C. Separation of the target protein, the GST-tag and the PreScission protease was achieved using GSH-Sepharose. Further, to remove aggregated fraction, size exclusion chromatography was used (Superdex 200 10/300 GL GE Healthcare, USA), the column was equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl. Sample purity after each purification step was assessed by 15% SDS–PAGE gels. The corresponding protein fraction was concentrated up to 2 mg/mL and stored at °C.
4.2.2. Activity assay of SARS-CoV-2 3CLpro
SARS-CoV-2 3CLpro activity assay was performed as described earlier using a fluorogenic substrate DABCYL-KTSAVLQSGFRKME-EDANS (Bachem, Switzerland) in a buffer containing 20 mM Tris pH 7.2, 200 mM NaCl, 1 mM EDTA and 1 mM TCEP [43], [10], [44]. The reaction mixture was pipetted in a Corning 96-Well plate (Sigma Aldrich) consisting of 0.5 μM protein and the assay was initiated with the addition of the substrate at a final concentration of 50 μM.
The inhibitory potential against the SARS-CoV-2 3CLpro activity of the best compounds identified in the virtual screening was investigated using the activity assay described above. 20 μM of the compounds was used for the screening tests. The mixtures were incubated for 30 min at RT. When the substrate with a final concentration of 50 μM was added to the mixture, the fluorescence intensities were measured at 60 s intervals over 30 min using an Infinite 200 PRO plate reader (Tecan, Männedorf, Switzerland). The temperature was set to 37 °C. The excitation and emission wavelengths were 360 nm and 460 nm, respectively. Inhibition assays were performed as triplicates.
Funding sources
The authors gratefully acknowledge the computing time granted through JARA-HPC (project COVID19MD) on the supercomputer JURECA at Forschungszentrum Jülich [45], the hybrid computer cluster purchased from funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) project number INST 208/704-1 FUGG, and the Centre for Information and Media Technology at Heinrich Heine University Düsseldorf. R.J.E. recognizes with appreciation funding from FAPESP [Grant Nos. 2018/07572-3, 2019/05614-3].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.bioorg.2021.104862.
Supplementary material
The following are the Supplementary data to this article:
References
- 1.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ace2 predicts the host range and an optimized ace2 for sars-cov-2 infection. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wrapp D., Wang N., Corbett K., Goldsmith J., Hsieh C., Abiona O., Graham B., McLellan J. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science. 2002;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-ncov and fusion inhibitors targeting hr1 domain in spike protein. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T., Herrler G., Wu N., Nitsche A., et al. Sars-cov-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of sars-cov-2 rna replication machinery: Rna synthesis, proofreading and final capping. Cell. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olubiyi O., Olagunju M., Keutmann M., Loschwitz J., Strodel B. High throughput virtual screening to discover inhibitors of the main protease of the coronavirus sars-cov-2. Molecules. 2020;3193 doi: 10.3390/molecules25143193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaro R., Baudry J., Chodera J., Demir O., McCammon J., Miao Y., Smith J. Ensemble docking in drug discovery. Biophys. J. 2018;114:2271–2278. doi: 10.1016/j.bpj.2018.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. Gender differences in patients with covid-19: Focus on severity and mortality. Front. in Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 11.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace A., Laskowski R., Thornton J. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 13.Laskowski R., Swindells M. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 14.Hung I., Lung K., Tso E., Liu R., Chung T., Chu M., Ng Y., Lo J., Chan J., Tam A., Shum H., Chan V., Wu A., Sin K., Leung W., Law W., Lung D., Sin S., Yeung P., Yip C., Zhang R., Fung A., Yan E., Leung K., Ip J., Chu A., Chan W., Ng A., Lee R., Fung K., Yeung A., Wu T., Chan J., Yan W., Chan W., Chan J., Lie A., Tsang Q., Cheng V., Que T., Lau C., Chan K., To K., Yue K. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norman P. A novel syk kinase inhibitor suitable for inhalation: R-343(?) – wo-2009031011. Expert. Opin. Ther. Pat. 2009;19:1469–1472. doi: 10.1517/13543770903059281. [DOI] [PubMed] [Google Scholar]
- 16.Kumari R., Kumar R., Lynn A. g_mmpbsa–A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Comp. Sci. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 17.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowland S., O’Brien Bergin E. Screening for low testosterone is needed for early identification and treatment of men at high risk of mortality from covid-19. Crit. Care. 2020;24:367. doi: 10.1186/s13054-020-03086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (gs-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tchesnokov E.P., Gordon C.J., Woolner E., Kocincova D., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Template-dependent inhibition of coronavirus rna-dependent rna polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2004 doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Wolf R., Caldwell J., Kollman P., Case D. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 22.M. Frisch, G. Trucks, H. Schlegel, G. Scuseria, M. Robb, J. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. Hratchian, A. Izmaylov, J. Bloino, G. Zheng, J. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. Montgomery, J. Peralta, F. Ogliaro, M. Bearpark, J. Heyd, E. Brothers, K. Kudin, V. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. Burant, S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. Millam, M. Klene, J. Knox, J. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. Stratmann, O. Yazyev, A. Austin, R. Cammi, C. Pomelli, J. Ochterski, R. Martin, K. Morokuma, V. Zakrzewski, G. Voth, P. Salvador, J. Dannenberg, S. Dapprich, A. Daniels, Ö. Farkas, J. Foresman, J. Ortiz, J. Cioslowski, D. Fox, Gaussian 09 Revision E.01, 2009. Gaussian Inc., Wallingford CT.
- 23.Bayly C., Cieplak P., Cornell W., Kollman P. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 24.Cornell W., Cieplak P., Bayly C., Kollman P. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Phys. Chem. 1993;115:9620–9631. [Google Scholar]
- 25.Wang J., Wang W., Kollman P., Case D. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graphics Modell. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Case D., Ben-Shalom I., Brozell S., Cerutti D., Cheatham T., Cruzeiro V., III, Darden T., Duke R., Ghoreishi D., Giambasu G., Giese T., Gilson M., Gohlke H., Goetz A., Greene D., Harris R., Homeyer N., Huang Y., Izadi S., Kovalenko A., Krasny R., Kurtzman T., Lee T., LeGrand S., Li P., Lin C., Liu J., Luchko T., Luo R., Man V., Mermelstein D., Merz K., Miao Y., Monard G., Nguyen C., Nguyen H., Onufriev A., Pan F., Qi R., Roe D., Roitberg A., Sagui C., Schott-Verdugo S., Shen J., Simmerling C., Smith J., Swails J., Walker R., Wang J., Wei H., Wilson L., Wolf R., Wu X., Xiao L., Xiong Y., York D., Kollman P. University of California; San Francisco: 2019. Amber 2019. [Google Scholar]
- 27.Sousa da Silva A., Vranken W. ACPYPE – AnteChamber PYthon Parser interfacE. BMC Res. Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham M., Murtola T., Schulz R., Páll S., Smith J., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 29.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14sb: Improving the accuracy of protein side chain and backbone parameters from ff99sb. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivani I., Dans P.D., Noy A., Pérez A., Faustino I., Hospital A., Walther J., Andrio P., Goñi R., Balaceanu A., Portella G., Battistini F., Gelpí J., González C., Vendruscolo M., Laughton C., Harris S., Case D., Orozco M. Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2016;13:55. doi: 10.1038/nmeth.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen W., Chandrasekhar J., Madura J., Impey R., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Theory Comput. 1983;79:926–935. [Google Scholar]
- 32.Cauchy M.A. Méthode généerale pour la résolution des systèmes d’ éequations simultanéees. CR Hebd. Acad. Sci. 1847;25:536–538. [Google Scholar]
- 33.Nosé S. Molecular-dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 34.Hoover W. Canonical dynamics – equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 35.Parrinello M., Rahman A. Polymorphic transitions in single-crystals - a new molecular-dynamics method. Mol. Phys. 1981;52:7182–7190. [Google Scholar]
- 36.Darden T., York D., Pedersen L. Particle Mesh Ewald - an N. Log(N) method for ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 37.Essmann U., Perera L., Berkowitz M. A smooth particle mesh ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 38.Hess B., Bekker H., Berendsen H., Fraaije J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 39.The PyMOL Molecular Graphics System, Version 1.8, 2015. Schrödinger LLC, 2015.
- 40.Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 1999;38:236–240. [Google Scholar]
- 41.T. Williams, C. Kelley, many others, Gnuplot 4.6: an interactive plotting program, http://gnuplot.sourceforge.net/, 2013.
- 42.Van Rossum G., Drake F.L. CreateSpace; Scotts Valley, CA: 2009. Python 3 Reference Manual. [Google Scholar]
- 43.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.C. Ma, M.D. Sacco, B. Hurst, J.A. Townsend, Y. Hu, T. Szeto, X. Zhang, B. Tarbet, M.T. Marty, Y. Chen, J. Wang, Boceprevir, gc-376, and calpain inhibitors ii, xii inhibit sars-cov-2 viral replication by targeting the viral main protease, bioRxiv: the preprint server for biology (2020) 678–692. [DOI] [PMC free article] [PubMed]
- 45.Krause D., Thörnig P. JURECA: Modular supercomputer at Jülich Supercomputing Centre. JLSRF. 2018;4:A132. [Google Scholar]
- 46.Loschwitz Jennifer, Jäckering Anna, Keutmann Monika, Olagunju Maryam, Olubiyi Olujide O., Strodel Birgit. Dataset of AMBER force field parameters of drugs, natural products and steroids for simulations using GROMACS. Data in Brief. 2021;35 doi: 10.1016/j.dib.2021.106948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AMBER force field (GAFF) parameters of the 160 organic molecules including drugs, natural products, and steroids that were derived as part of this work are available on Mendeley Data (https://doi.org/10.17632/phxtv76n5s.3). These parameters can be employed without further processing in MD simulations using GROMACS. Moreover, the Gaussian and GROMACS input files used for generating the force field parameters, along with bash scripts for automating the parameterization procedure are also provided at Mendeley Data. In [46] this dataset and how the data were obtained are described in detail.