Abstract
In this study, we describe the isolation and characterization of previously unreported Y280-lineage H9N2 viruses from two live bird markets in Korea in June 2020. Genetic analysis revealed that they were distinct from previous H9N2 viruses circulating in Korea and had highest homology to A/chicken/Shandong/1844/2019(H9N2) viruses. Their genetic constellation showed they belonged to genotype S, which is the predominant genotype in China since 2010, where genotype S viruses have infected humans and acted as internal gene donors to H5 and H7 zoonotic influenza viruses. Active surveillance and control measures need to be enhanced to protect the poultry industry and public health.
Keywords: Avian influenza, H9N2, live bird market, phylogenetic analysis, surveillance
INTRODUCTION
The H9N2 subtype avian influenza virus (AIV), which is prevalent in poultry worldwide, causes low pathogenic respiratory symptoms. The poultry-adapted H9N2 virus has diverged into several HA lineages: G1, Y280, and Y439. The G1 lineage viruses (represented by A/quail/Hong Kong/G1/1997) have been detected in Asia, the Middle East, and Africa. The Y280 lineage viruses (represented by A/duck/Hong Kong/Y280/1997) circulate mainly in China, but have been reported in Asian countries such as Vietnam, Cambodia, Myanmar, and the Russian Far East. The Y439 lineage viruses (represented by A/duck/Hong Kong/Y439/1997) have been isolated from wild birds and poultry in Europe, Asia, and South Africa [1].
Infection of humans with H9N2 viruses had been reported since 1999 [2]. All sequenced human H9N2 isolates belong to the G1 or Y280 lineages. Both lineages are thought to have donated internal genes to the H5NX and H7N9 AI viruses, which have caused severe human infections and casualties [1,3,4]. However, human infection by Y439 lineage viruses has never been reported.
After the first outbreak of H9N2 AI in poultry in Korea in 1996, the HA gene evolved into a Korean sub-lineage within the Y439 lineage, while the internal genes were reassorted with AIVs of duck and wild waterfowl origin [5]. The use of inactivated H9N2 AIV vaccines in layers and breeders on commercial farms since 2007 greatly reduced the outbreak of H9N2 AIVs [6]. Meanwhile, H9N2 viruses were still detected in live bird markets (LBMs) selling Korean native chickens and ducks from various sources [7]. Intensive control measures were implemented in LBMs and on poultry farms during the national 2016/2017 epidemics caused by H5Nx highly pathogenic AI. As a result of these control measures, the incidence of H9N2 AIVs in LBMs fell sharply over time, and circulation of H9N2 AIV in LBMs stopped in June 2018 (Supplementary Table 1).
However, 2 years later, the national AI surveillance program identified H9N2 AIVs at LBMs in Korea. Surprisingly, the viruses had genetic features distinct from previous indigenous H9N2 AIVs in Korea. Here, we analyzed the genetic characteristics of these viruses and assessed the risk to public health in comparison with the previous Y439 endemic.
MATERIALS AND METHODS
Poultry in every LBM registered in Korea are tested quarterly to detect AIVs; tests are conducted using oropharyngeal swabs, cloacal swabs, or fresh feces. From April to June 23, 2020, 12,867 samples were collected from 337 LBM stores in 11 provinces and screened by real-time-reverse transcription polymerase chain reaction to detect M, H5, and H7; positive samples were inoculated into embryonated chicken eggs. Virus isolates with HA activity were subtyped using gene specific primers, and their whole genomes were sequenced using the Miseq NGS platform (Illumina). Data were analyzed using a de novo assembly program (CLC genomics workbench 10.1) as previously described [8]. For phylogenetic analysis, maximum likelihood trees were constructed in MEGA 6 (https://megasoftware.net) using the general time-reversible nucleotide substitution model with a gamma distribution of among-site rate variations. Reference sequences were acquired from a Global Initiative on Sharing All Influenza Data (GISAID) and GenBank.
Data availability statement
The data that support the findings of this study are available in GISIAD at http://www.GISAID.org, accession number [EPI_ISL_492088, EPI_ISL_492104 ~ EPI_ISL_492108].
Ethical statement
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. All applicable national and institutional guidelines for the care and use of animals were followed. The article does not contain any studies involving human participants performed by any of authors.
RESULTS
We isolated two H9N2 AIVs from Korean native chickens at two LBMs: one in Yeongcheon city, Gyeongsangbuk province (June 17, 2020) and one in Hwasoon city, Jeollanam province (June 23, 2020). The virus from Gyeongsangbuk province was designated as A/chicken/Korea/LBM261/2020(H9N2), and the one from Jeollanam province was designated as A/chicken/Korea/LBM314/2020(H9N2); the sequence of each virus was submitted to GISAID (Supplementary Table 1).
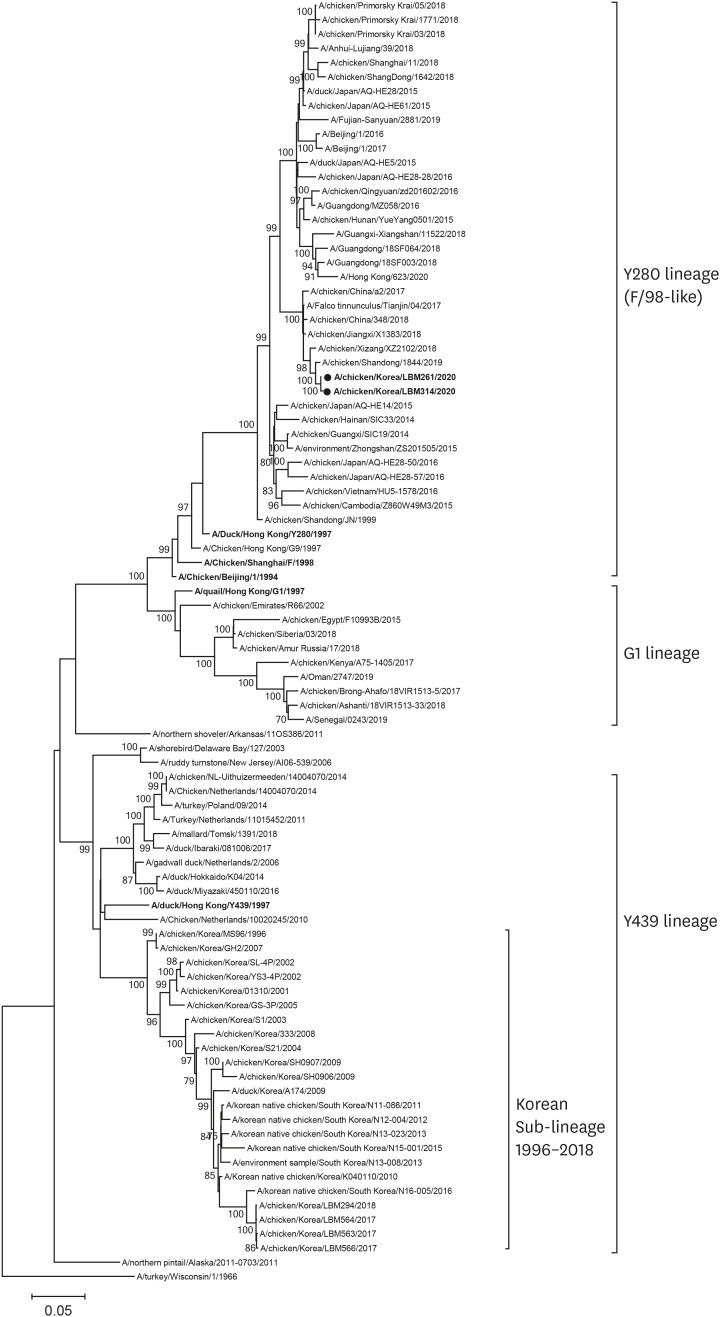
The two viruses were genetically very closely related (nucleotide identity: 99.04%–99.90%), although the LBMs are over 200 km apart. The HA genes from the two isolates were < 82% homologous with those of the indigenous Korean sub-lineage H9N2 virus which had been circulating from 1996 to 2018. The HA genes of the two new isolates showed highest similarity to A/chicken/Shandong/1844/2019(H9N2) belonging to the Y280 lineage, which was recently isolated from China (Fig. 1). All eight segments of the genomes of the two viruses showed highest homology to those of A/chicken/Shandong/1844/2019(H9N2) (98.63%–99.38%) (Fig. 1; Supplementary Fig. 1). Annual intensive surveillance of over 150,000 poultry samples did not detect any AIVs since the last detection of H9N2 AIV in June 2018 (data not shown). All of the internal genes of these two new viruses are distinct from previously identified indigenous Korean H9N2 AIVs (Supplementary Fig. 1).
Fig. 1. Phylogenetic analysis of the HA genes of H9 subtype avian influenza viruses isolated from poultry in Korea. The maximum likelihood tree was constructed using a general time-reversible nucleotide substitution model, with a gamma distribution of among-site rate variations and bootstrap analysis (n = 1,000). A/turkey/Wisconsin/1/1966(H9N2) was used as an out-group. All positions containing gaps and missing data were eliminated. There were a total of 1,549 positions in the final dataset. Representative sequences of viruses are listed in bold. The reported isolates are marked with a black dot. Bootstrap scores < 70% are hidden.
The new isolates have the amino acid sequence, PSRSSR/GLF, within the HA cleavage site, which is a dominant feature among representative H9N2 viruses circulating in China between 2013 and 2016. Several key amino acid residues, including 183(N) and 226(L) (H3 numbering), were identified in the HA receptor binding site, which has high binding affinity for α2,6-linked sialic acid receptors that are predominant in humans [9]. The majority of human isolates of H9N2 harbor a leucine residue at position 226 of the HA gene; however, the H9N2 isolate of the Korean sub-lineage within the Y439 lineage harbors a glutamine residue at this position (Table 1) [1,4].
Table 1. Molecular characteristics of H9N2 viruses isolated from live bird markets in Korea between 2017 and 2020.
| Strain name | Cleavage site | HA* | NA† | PB2 | PB1 | PA | M1 | M2 | NS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I155T | H183N | Q226L | G228S | Amino acid deletion | E119D | R292K | A588V | E627K | D701N | D622G | K356R | N30D | S31N/G | P42S | ||
| A/chicken/Korea/LBM261/2020 | PSRSSR/GLF | T | N | L | G | 63–65 | E | R | V | E | D | G | R | D | N | S |
| A/chicken/Korea/LBM314/2020 | PSRSSR/GLF | T | N | L | G | 63–65 | E | R | V | E | D | G | R | D | N | S |
| A/chicken/Korea/LBM294/2018 | PATTGR/GLF | T | S | Q | G | Not deleted | E | R | V | E | D | G | K | D | S | S |
| A/chicken/Korea/LBM567/2017 | PATTGR/GLF | T | S | Q | G | Not deleted | E | R | V | E | D | G | K | D | S | S |
*Position H3 numbering relative to A/Aichi/2/1968(H3N2); †Position N2 numbering relative to A/Aichi/2/1968(H3N2).
In addition, the new isolates have a three-amino-acid deletion in the NA stalk region at positions 63 to 65, which is related to virus adaptation from wild birds to poultry [10]. The E119D and R292K mutations in the NA gene, which are associated with the resistance to neuraminidase inhibitors such as oseltamivir [11], were not observed. Point mutations that increase polymerase activity in mammalian hosts (i.e., E627K and D701N in PB2 gene) were not found (Table 1) [12].
DISCUSSION
The Y280 lineage H9N2 has prevailed in China since 2010, where it has caused significant losses in the poultry industry. Since mid-2010, spillover of the H9N2 Y280 lineage has been reported in Asian countries, including Vietnam and Cambodia [1]. Anecdotal evidence for this includes isolation of viruses of the genotype S from imported poultry meat products in Japan in 2015 and 2016 [14]. Also, there was an outbreak of H9N2 with this genotype on chicken farms in the Russian Far East in 2018 [15].
In China, Y280 lineage H9N2 has evolved, with gene constellations showing diverse genotypes from A to W [1]. The two new isolates belong to genotype S and possess two G1/97-like genes (PB2 and M) and six F/98-like genes (PB1, PA, NP, HA, NA, and NS) (Fig. 1, Supplementary Fig. 1, and Supplementary Table 1). Genotype S (also known as G57) has caused human infections and is suspected to be an internal gene donor to zoonotic H5NX and H7N9 HPAI viruses [13]. In addition, Y280 lineage H9N2 is known to have a HA motif that confers higher binding affinity for α2,6-linked sialic acid receptors, as observed in this study, which increases zoonotic risk. Human cases of Y280 lineage H9N2 infection have been reported in poultry-related facilities including LBMs in China [1]. To monitor AIV prevalence and genetic evolution, particularly with respect pathogenicity, we have been performing comprehensive AI surveillance on domestic poultry since 2008. In this study, we identified the introduction of the new Y280 lineage H9N2 in 2020, which has zoonotic potential, and disclose the genetic makeup of the new Korean isolates to facilitate AI preparedness in the veterinary community.
Although the route of introduction is unknown, the new viruses are likely to be recent introductions to Korea because no AIVs have been detected in the last 2 years despite intensive active surveillance. Thus, we speculate that the virus is likely to have been introduced across the border by human activities such as illegal importation of raw meat products in luggage or mail because infection of wild birds with Y280 HA lineage H9N2 virus is very rare, and viruses of genotype S have been isolated from luggage intercepted by border control in Japan [14].
To protect the poultry industry, biosecurity and active surveillance must be strengthened and maintained. Stronger control measures are needed to reduce the risk of human infection.
ACKNOWLEDGMENTS
We thank Eun-bi Noh, Jeong-Eui Lee, In-Kyeong Kim, and Byeong-suk Jeon for excellent technical assistance. We also thank the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, and the Regional Office for Animal Disease Control for their efforts to control AIV. We thank our colleagues worldwide for their laboratory contributions, which were made available through GISAID and GenBank.
Footnotes
Funding: This work was supported by the Animal and Plant Quarantine Agency, Republic of Korea (grant numbers B-1543418-2019-20-01).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Heo GB.
- Data curation: Heo GB, Sagong M, Lee EK.
- Formal analysis: Heo GB, Kye SJ.
- Funding acquisition: Lee EK.
- Investigation: Sagong M, Lee EK, Lee YN.
- Methodology: Heo GB, Kye SJ.
- Project administration: Lee MH.
- Resources: Sagong M, Lee YN.
- Supervision: Lee MH.
- Validation: Lee YJ.
- Visualization: Heo GB, Kye SJ.
- Writing - original draft: Heo GB, Kye SJ.
- Writing - review & editing: Lee YJ, Lee KN, Choi KS, Lee MH.
SUPPLEMENTARY MATERIALS
Complete genome sequences of H9N2 viruses isolated from live bird markets in Korea between 2017 and 2020
Phylogenetic analysis of the internal genes of H9 subtype avian influenza viruses isolated from poultry in Korea. (A) PB2, (B) PB1, (C) PA, (D) NP, (E) NA, (F) M, and (G) NS. The phylogenetic trees were rooted to A/turkey/Wisconsin/1/1966(H9N2). Maximum likelihood trees were constructed using a general time-reversible nucleotide substitution model, with a gamma distribution of among-site rate variations using bootstrap analysis (n = 1,000). All positions containing gaps and missing data were eliminated. Representative sequences of viruses are listed in bold. The reported isolates are marked by a black dot. Bootstrap scores < 70% are hidden.
References
- 1.Peacock TH, James J, Sealy JE, Iqbal M. A global perspective on H9N2 avian influenza virus. Viruses. 2019;11(7):11. doi: 10.3390/v11070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, et al. Human infection with influenza H9N2. Lancet. 1999;354(9182):916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 3.Carnaccini S, Perez DR. H9 influenza viruses: an emerging challenge. Cold Spring Harb Perspect Med. 2020;10(6):10. doi: 10.1101/cshperspect.a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pusch EA, Suarez DL. The multifaceted zoonotic risk of H9N2 avian influenza. Vet Sci. 2018;5(4):82. doi: 10.3390/vetsci5040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youk SS, Lee DH, Jeong JH, Pantin-Jackwood MJ, Song CS, Swayne DE. Live bird markets as evolutionary epicentres of H9N2 low pathogenicity avian influenza viruses in Korea. Emerg Microbes Infect. 2020;9(1):616–627. doi: 10.1080/22221751.2020.1738903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho HK, Kang YM, Kim HM, Lee CH, Kim DY, Choi SH, et al. Sales and immunogenicity of commercial vaccines to H9N2 low pathogenic avian influenza virus in Korea from 2007 to 2017. Vaccine. 2020;38(16):3191–3195. doi: 10.1016/j.vaccine.2020.02.083. [DOI] [PubMed] [Google Scholar]
- 7.Lee EK, Kang HM, Song BM, Lee YN, Heo GB, Lee HS, et al. Surveillance of avian influenza viruses in South Korea between 2012 and 2014. Virol J. 2017;14(1):54. doi: 10.1186/s12985-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83(19):10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281(2):156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 10.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73(2):1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baek YH, Song MS, Lee EY, Kim YI, Kim EH, Park SJ, et al. Profiling and characterization of influenza virus N1 strains potentially resistant to multiple neuraminidase inhibitors. J Virol. 2015;89(1):287–299. doi: 10.1128/JVI.02485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin X, Zha Y, Hu J, Li X, Chen J, Xie S, et al. New molecular evolutionary characteristics of H9N2 avian influenza virus in Guangdong Province, China. Infect Genet Evol. 2020;77:104064. doi: 10.1016/j.meegid.2019.104064. [DOI] [PubMed] [Google Scholar]
- 14.Shibata A, Hiono T, Fukuhara H, Sumiyoshi R, Ohkawara A, Matsuno K, et al. Isolation and characterization of avian influenza viruses from raw poultry products illegally imported to Japan by international flight passengers. Transbound Emerg Dis. 2018;65(2):465–475. doi: 10.1111/tbed.12726. [DOI] [PubMed] [Google Scholar]
- 15.Sharshov K, Kurskaya O, Sobolev I, Leonov S, Kabilov M, Tatyana A, et al. First detection of a G1-like H9N2 virus in Russia, 2018. Korean J Vet Res. 2019;59(1):37–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete genome sequences of H9N2 viruses isolated from live bird markets in Korea between 2017 and 2020
Phylogenetic analysis of the internal genes of H9 subtype avian influenza viruses isolated from poultry in Korea. (A) PB2, (B) PB1, (C) PA, (D) NP, (E) NA, (F) M, and (G) NS. The phylogenetic trees were rooted to A/turkey/Wisconsin/1/1966(H9N2). Maximum likelihood trees were constructed using a general time-reversible nucleotide substitution model, with a gamma distribution of among-site rate variations using bootstrap analysis (n = 1,000). All positions containing gaps and missing data were eliminated. Representative sequences of viruses are listed in bold. The reported isolates are marked by a black dot. Bootstrap scores < 70% are hidden.
Data Availability Statement
The data that support the findings of this study are available in GISIAD at http://www.GISAID.org, accession number [EPI_ISL_492088, EPI_ISL_492104 ~ EPI_ISL_492108].