Abstract
Background
Pseudorabies (PR), caused by the pseudorabies virus (PRV), is an endemic disease in some regions of China. Although there are many reports on epidemiological investigations into pseudorabies, information on PRV gI antibody dynamics in one pig farm is sparse.
Objectives
To diagnose PR and analyze the course of PR eradication in one pig farm.
Methods
Ten brains and 1,513 serum samples from different groups of pigs in a pig farm were collected to detect PRV gE gene and PRV gI antibody presence using real-time polymerase chain reaction and enzyme-linked immunosorbent assay, respectively.
Results
The July 2015 results indicated that almost all brain samples were PRV gE gene positive, but PRV gI antibody results in the serum samples of the same piglets were all negative. In the boar herd, from October 2015 to July 2018 three positive individuals were culled in October 2015, and the negative status of the remaining boars was maintained in the following tests. In the sow herd, the PRV gI antibody positive rate was always more than 70% from October 2015 to October 2017; however, it decreased to 27% in January 2018 but increased to 40% and 52% in April and July 2018, respectively. The PRV gI antibody positive rate in 100-day pigs markedly decreased in October 2016 and was maintained at less than 30% in the following tests. For 150-day pigs, the PRV gI antibody positive rate decreased notably to 10% in April 2017 and maintained a negative status from July 2017. The positive trend of PRV gI antibody with an increase in pig age remarkably decreased in three tests in 2018.
Conclusions
The results indicate that serological testing is not sensitive in the early stage of a PRV infection and that gilt introduction is a risk factor for a PRV-negative pig farm. The data on PRV gI antibody dynamics can provide reference information for pig farms wanting to eradicate PR.
Keywords: Pseudorabies, diagnosis, antibody, real-time PCR, ELISA
INTRODUCTION
Pseudorabies (PR), also known as Aujeszky's disease, is caused by the pseudorabies virus (PRV). The PRV can cause infection in a broad spectrum of mammals, including ruminants and rodents; however, domestic and wild pigs are the only hosts capable of surviving PRV infection and can serve as reservoirs for the virus [1]. Clinical manifestations in pigs infected by PRV vary from subclinical signs to death. For example, PRV infections may result in nervous system disorders and high mortality in piglets, respiratory symptoms and growth retardation in finishing pigs, and abortions and/or stillbirths in sows [2]. Like other herpesviruses, PRV usually establishes lifetime latent infections in pigs by inducing specific host cells to retain the viral genome. Latency is established mainly in nervous ganglia cells such as the trigeminal ganglia in the head and the sacral ganglia in the lumbar region. PRV replication can reactivate from latently infected cells after host stress or immune system depression; for example, during gestation. Reactivation increases PRV shedding and promotes transmission; thus, latently infected pigs can be a source of reinfection [3,4].
Because of the threat from PRV latent infection and the benefits of an absence of PR [5], the best approach for PR is eradication. In North America and several European countries, PR has been eradicated from domestic pigs due to the implementation of effective eradication programs, which include enhanced biosecurity measures and nationwide compulsory vaccination with gE-deleted vaccines [6,7,8,9]. Both gI and gE are type I transmembrane proteins that form a heterodimer in the endoplasmic reticulum through noncovalent interactions between their ectodomains, and this protein complex is involved in virion assembly, cell-to-cell spread, species-specific binding of IgG as an Fc receptor, and mediating full virulence in animal infections [10]. However, the coding region for gI and gE is deleted in the genome of the attenuated PRV strain Bartha compared to that of virulent PRV [11]. Thus, the development of DIVA vaccines to differentiate infected animals from vaccinated animals and gI-enzyme-linked immunosorbent assay (ELISA) kits has an important role in PR eradication. In China, PR was well controlled before 2010 because of the nationwide vaccination with the Bartha-K61 vaccine strain [12]. However, in late 2011, PR outbreaks were reported in some Bartha-K61-vaccinated pig farms in China, resulting in significant economic losses [13]. A national survey indicated that the PRV gE antibody positive rate was between 22.17 and 13.14% during 2013–2016 [14].
PR has been listed within the National Middle-to-Long Term Plan for Animal Disease Control (2012–2020) as one of the priority swine diseases to be controlled. One of the tasks of that program is to eradicate PR in breeding pig farms in China by the end of 2020 [15]. There was a PR outbreak in a previously PR-negative breeding pig farm in Hei Longjiang Province, China in July 2015. Subsequently, the farm undertook a series of measures to eradicate PR. The main aim of the present study was to describe and analyze PRV gI antibody dynamics in this farm by examining time series data. The authors hope the results of this study will provide reference information that will be useful for farms that want to carry out a PR eradication program.
MATERIALS AND METHODS
Pig farm background and sample collection
The studied pig farm was built in 2013, and about 1000 gilts were introduced in August 2014. All pigs were fed by automatic equipment. The sows started to farrow in March 2015. The farm's PR vaccination schedule was three times per year for sows and boars, and once at 70 d and 100 d for nursery and fattening pigs, respectively. The live PR vaccine used at the farm was produced by HIPRA, Spain.
All samples in this study were collected from the above pig farm. From May 2015 to July 2018, 1513 serum samples from different pigs were randomly collected according to a systematic sampling scheme (Tables 1 and 2) to detect the PRV gI antibody. Ten brain tissues were collected in July 2015 to detect the PRV gE gene; those samples were from 3 suckled piglets with clinical signs, 2 suckled piglets without clinical signs, 3 weaned piglets with clinical signs, and 2 weaned piglets without clinical signs. The serum samples were obtained after centrifugation of coagulated blood samples, and all samples were placed in tubes and stored at −20°C prior to testing.
Table 1. Pig farm serum sampling scheme in May and July 2015.
| May | Jul | ||
|---|---|---|---|
| Pig type | Number of serum samples | Pig type | Number of serum samples |
| Boars | 18 | Boars | 10 |
| Gilts | 21 | Gilts | 10 |
| Sows | 30 | Sows in pregnancy house | 20 |
| 10-day pigs | 5 | Sows in farrowing house | 8 |
| 40-day pigs | 5 | Suckled pigs with clinical signs | 10 |
| Suckled pigs without clinical signs | 10 | ||
| Weaned pigs with clinical signs | 10 | ||
| Weaned pigs without clinical signs | 8 | ||
Table 2. Pig farm serum sampling scheme from October 2015 to July 2018.
| Pig type | Number of serum samples | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2015 | 2016 | 2017 | 2018 | |||||||||
| Oct | Jan | Apr | Jul | Oct | Jan | Apr | Jul | Oct | Jan | Apr | Jul | |
| Boars | 18 | 27 | 24 | 21 | 18 | 17 | 18 | 18 | 16 | 25 | 20 | 20 |
| Sows | 30 | 30 | 30 | 30 | 30 | 28 | 30 | 30 | 30 | 30 | 30 | 29 |
| 40-day pigs | 30 | 30 | 30 | |||||||||
| 100-day pigs | 20 | 20 | 30 | 29 | 29 | 19 | 30 | 30 | 29 | 30 | 30 | 29 |
| 150-day pigs | 20 | 20 | 30 | 30 | 30 | 25 | 29 | 30 | 30 | 30 | 30 | 30 |
The detection of PRV gI antibody by ELISA
All serum samples were assessed using PRV gI Ab test kits (IDEXX Laboratories, USA) according to the manufacturer's protocol. For testing, a diluted sample (100 μL; 1:2) was added to the ELISA plate and incubated for 60 min at 18°C–26°C. The plate was then washed 4 times with 300 μL of wash solution, after which 100 μL of the conjugate was added, and the mixture was incubated for 20 min at 18°C–26°C followed by washing as described above. Then, 100 μL of the substrate was added, and the mixture incubated for 15 min at 18°C–26°C. After incubation, 50 μL of stop solution was added to each well, and the plate was immediately read at 650 nm. The reaction result was read as an optical density (OD) by using an ELISA plate reader (BioTek Instruments Inc., USA). Sample to negative (S/N) ratios were calculated from the ODs according to the kit manufacturer's formula. Cut-off S/N values ≤ 0.6 or ≥ 0.7 were used to classify samples as positive or negative, respectively, for PRV gI antibody presence. It should be noted that the PRV-suspected sample in this study was recognized as a PRV-positive sample when the antibody positive rate was calculated. Additionally, the gilts sampled in May 2015 were different gilts from those sampled in July 2015.
The detection of PRV gE gene by real-time polymerase chain reaction (PCR)
The brain was ground with liquid nitrogen to powder and diluted with five volumes of PBS. The 1 mL homogenate was centrifuged at 3,000 × g for 10 min at 4°C, and the supernatant was transferred to another 1.5 mL tube. The DNA extraction was performed with a commercial TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd, China) according to the manufacturer’s instructions. All DNA extracts were stored at −40°C until assayed by real-time PCR.
The PRV gE gene was detected by using a PRV gE real-time PCR diagnostic kit (Beijing Anheal Laboratories Co., Ltd, China). The amplification reaction was conducted in a 20 μL reaction volume containing 10 μL of PCR master mix, 2.1 μL primers and probe mix, 2 μL of DNA extract, and 5.9 μL of nuclease-free water. The real-time PCR was performed under the following conditions: 95°C for 2 min, and 40 cycles of 95°C for 5 sec and 60°C for 35 sec using an Applied Biosystems ABI 7500 real-time PCR system (Thermo Fisher Scientific, USA). Analysis of the results was carried out using ABI 7500 software, version 2.3. A sample with a Ct value ≤ 35 in the gE real-time PCR assay result was considered positive.
Statistical analysis
All statistical analyses were performed using GraphPad Prism® 7.00 software (GraphPad Software, Inc., USA). Descriptive statistics were used to define the pattern of the PRV gI antibody detection in serum. The S/N ratio data are presented as mean ± SD.
RESULTS
Detection and analysis of PRV gI antibody in May 2015
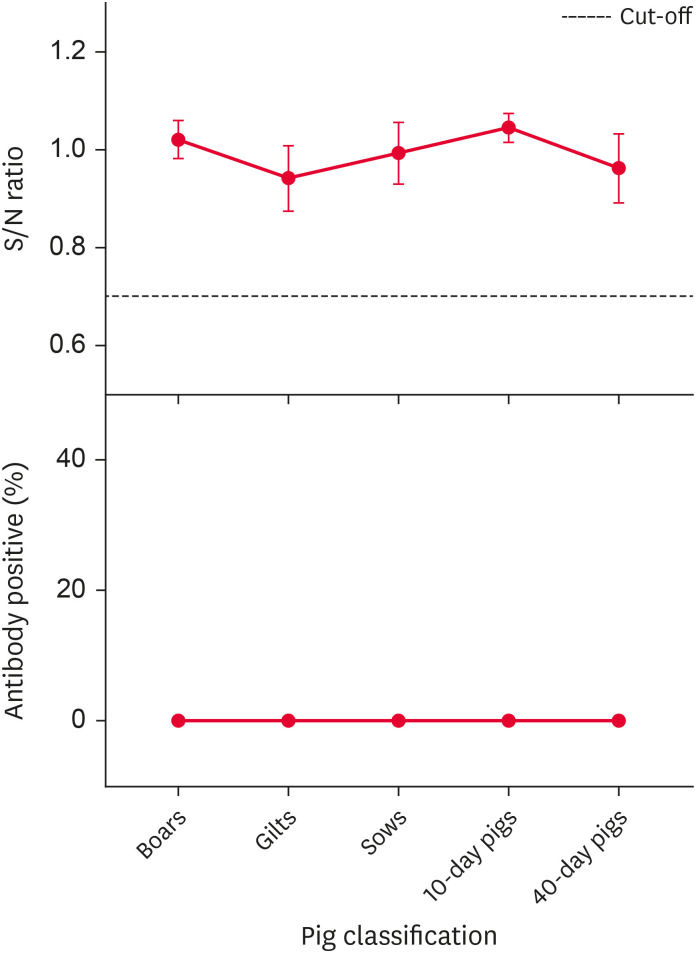
The May 2015 PRV gI antibody results are shown in Fig. 1. The S/N values for the PRV gI antibody from the different groups of pigs were all above the suggested cut-off, so there were no PRV-positive individuals among the sampled pigs. The results indicated that this pig farm was free of PR.
Fig. 1. Pseudorabies virus gI antibody S/N ratio and positive rate (%) in different groups of pigs in May 2015.
S/N, sample to negative.
Detection and analysis of PRV gI antibody and PRV gE gene in July 2015
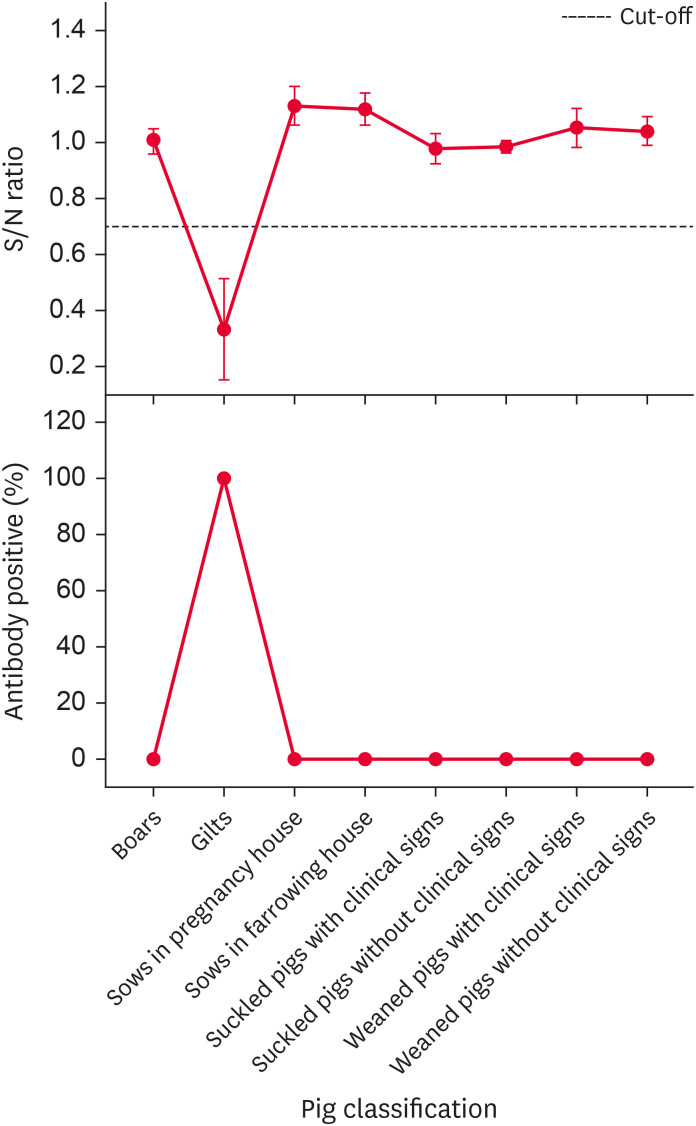
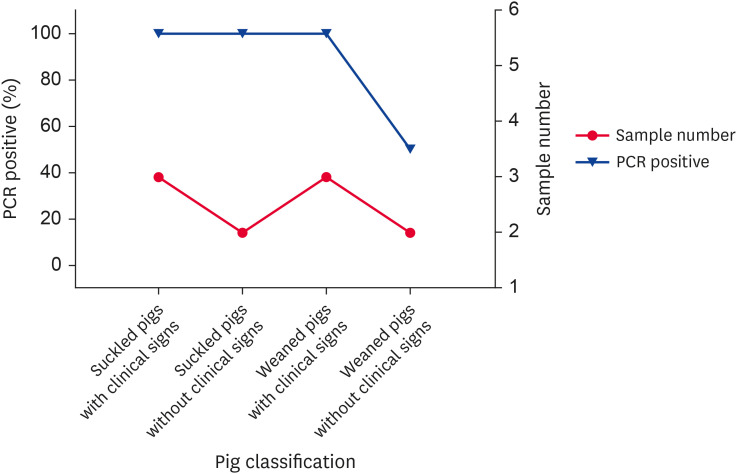
In July 2015, suckled piglets manifested vomiting, diarrhea, and nervous system disorders, and weaned piglets manifested fever, lethargy, and anorexia. Based on those observations, PR was the first suspected disease. Subsequently, serum and brain samples were collected to determine PRV gI antibody and gE gene presence. The results of the serological and etiological test are respectively given in Figs. 2 and 3. The serological results indicated that only the gilts' PRV gI antibody was positive, and the PRV gI antibody results were negative from all other pig samples. The etiological results indicated that the PRV gE gene was positive in 9 piglets, and their Ct values were all less than 30. The serological and etiological results indicated a PR outbreak at this pig farm, with the infectious source probably being the introduced gilts. Notably, the brain sample PRV gE gene results were mostly positive, but the serum PRV gI antibody results from the same piglets were all negative.
Fig. 2. Pseudorabies virus gI antibody S/N ratio and positive rate (%) in different groups of pigs in July 2015.
S/N, sample to negative.
Fig. 3. Detection of Pseudorabies virus gE genes by real time PCR in the suckled and weaned pigs in July 2015.
PCR, polymerase chain reaction.
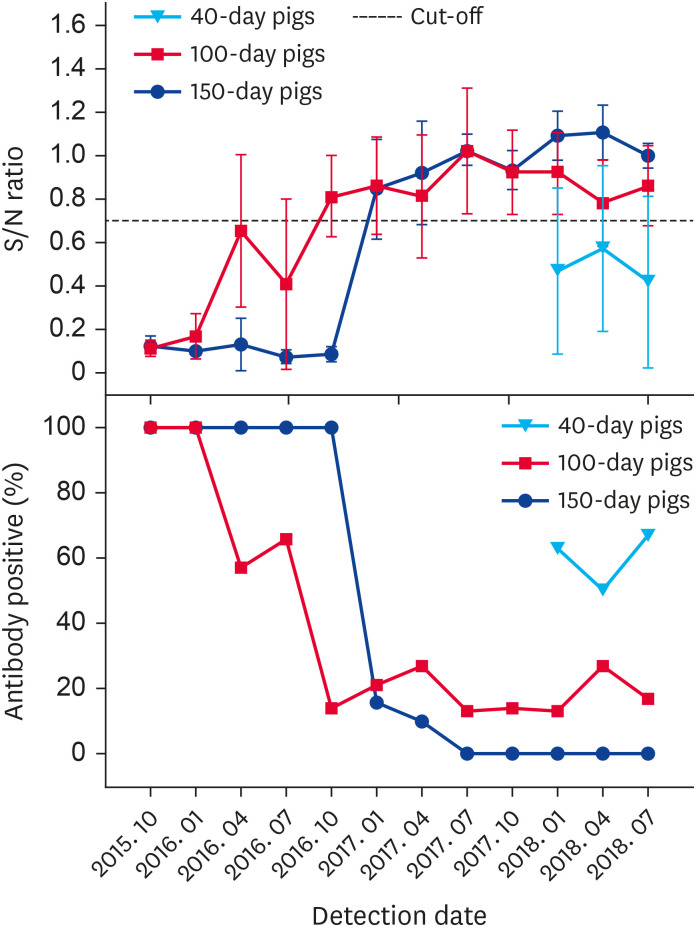
Dynamics of PRV gI antibody from October 2015 to July 2018
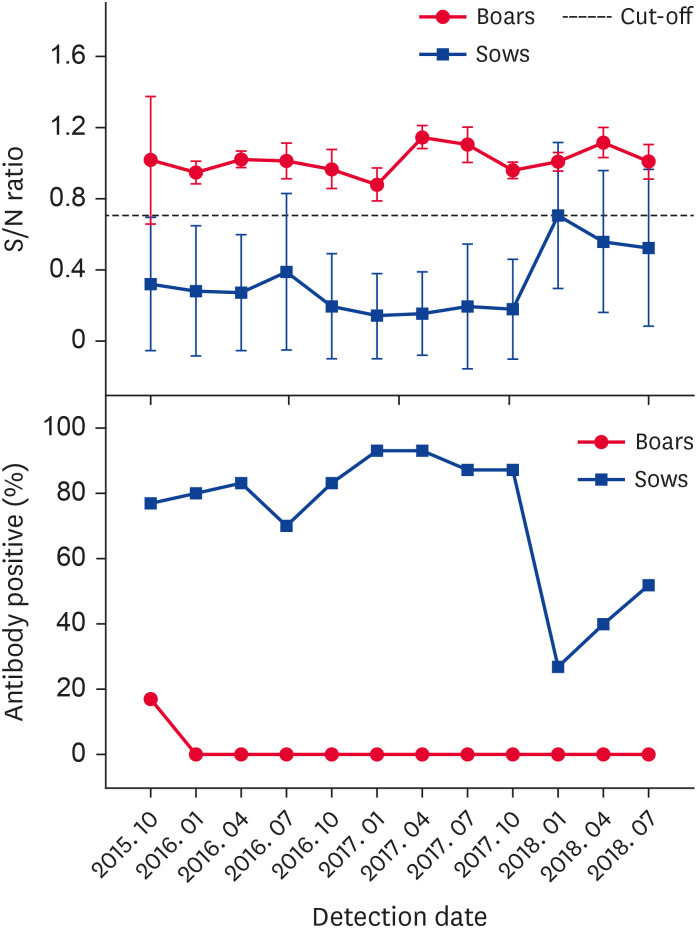
Most pigs in the assessed farm quickly became PRV-positive, and a series of measures, including a routine surveillance plan and improvement of biosecurity measures, were implemented to eradicate PR from the farm. From October 2015 to July 2018, 1,348 serum samples from boars, sows, and nursery and fattening pigs were collected to determine PRV gI antibody presence. The dynamic results for the PRV gI antibody from the different groups of pigs are presented in Figs. 4 and 5. Within the boar herd, 3 PRV-positive individuals were culled in October 2015 and the PRV-negative status of boars was maintained in subsequent tests. The PRV gI antibody positive rate in the sow herd was very high and ranged between 70% and 93% from October 2015 to October 2017. In January 2018, the sow positive rate decreased to 27%, but it rose to 40% and 52% in April and July 2018, respectively. Among the 100-day pigs sampled, the PRV gI antibody positive rate markedly decreased in October 2016 and remained less than 30% in subsequent tests. For the 150-day pigs, the PRV gI antibody positive rate decreased notably to 10% in April 2017, and the results remained negative from July 2017. Sampling of 40-day pigs was temporarily added to the assessment program in January 2018 to detect a trend in PRV gI antibody-positive results related to an increase in pig age. The results indicated that the PRV gI antibody positive rates for pigs of increasing age were all remarkably decreased in three tests performed in 2018.
Fig. 4. Pseudorabies virus gI antibody S/N ratio and positive rate (%) in boars and sows from October 2015 to July 2018.
S/N, sample to negative.
Fig. 5. Pseudorabies virus gI antibody S/N ratio and positive rate (%) in nursery and fattening pigs from October 2015 to July 2018.
S/N, sample to negative.
DISCUSSION
PR is an endemic disease in some regions of China, even though gene-deleted live vaccines have been widely used for decades. The epidemiology investigation reported by Liu et al. [14] indicated that PRV gE positive rates differed among different geographical regions in China, with results from Northeast China showing increasing average PRV gE positive rates from 2013 to 2016. The pig farm assessed in the present study was located in Hei Longjiang Province, Northeast China, and in May 2015, results indicated the pig farm was PR-free. But in July 2015, observations indicated a PR outbreak at this pig farm, with sampling results indicating that the infectious source was probably the introduced gilts. Previous studies have identified that the introduction of gilts is one of the risk factors related to PRV seropositivity in pig farms [16,17,18]. After observing an outbreak, a PR eradication program, including PRV vaccine replacement, increased immune times and disinfection frequencies, implementing an all-in/all-out practice, and improving biosecurity measures, was initiated. From October 2015, routine surveillance was undertaken to monitor the dynamics of the PRV gI antibody results in different groups of pigs.
Maintenance of a PRV-free status involves surveillance of susceptible swine, and serological detection of PRV antibodies is the most common method used for herd diagnosis. Such detection is quite efficient and sensitive once the animal has seroconverted. However, serological detection is not sensitive during the early infection period, leading to false-negative results [19,20]. In this study, the PRV gE gene results from brain samples were mostly positive, while the serum PRV gI antibody results from the same piglets were all negative. These results further indicate that serological detection is not sufficiently sensitive in an early infection because the specific gI antibody against PRV was either not produced or produced at a low level. Our study could not confirm whether the PRV gE gene was positive in serum samples because detection of PRV gE genes in serum samples was not performed. Regardless, the PRV gE gene has been widely used in the detection of PRV DNA from many specimens, including serum, nasal swabs, brain, tonsil, and other sources, and these studies have suggested that serum sampling is not sufficiently sensitive for detecting PRV [21,22,23,24]. In conclusion, in the early stage of a PRV infection, serological detection and serum sampling for PCR detection are not suitably sensitive.
There are many reports on epidemiological investigations of PR [14,16,25,26], but there are few reports on PRV gI antibody dynamics in a single farm. In this study, 1348 serums from the different groups of pigs in a single farm were collected to analyze PRV gI antibody dynamics. In the boar herd, 3 PRV-positive individuals were detected in October 2015; the apparent reason for the infections was that the boars were temporarily housed in the sow pen during renovation of the boar pen. Subsequently, the PRV-positive boars were culled, and based on further testing, a PRV-negative status was maintained in the boar herd. In nursery and fattening pigs, the 150-day pigs were always negative after July 2017, and the positive PRV gI antibody results with increasing pig age were all remarkably decreased in three tests performed in 2018. These results indicate that the PRV gI antibodies present in 40-day and 100-day pigs after April 2017 were probably from the maternal antibodies, not from infection. Moreover, the results also indicate that a positive sow cannot infect the fetus through the placenta. Based on the above results and conclusion, the farm decided to introduce PRV-negative gilts into the sow herd in November 2017. The PRV gI antibody positive rate in the sow herd in 2018 was notably lower than that in 2016 and 2017, although the PRV gI antibody positive rate in 2018 wavered in accord with the number of serum samples from negative gilts. Additionally, the results showed no seroconversion among the introduced gilts, this further indicating that the positive sows did not shed the virus.
Except for the sow herd, the other pigs became PRV-negative; thus, this pig farm could probably eradicate PR by introducing PRV-negative gilts in the next two years. Unfortunately, all pigs in this farm were culled because African Swine Fever was detected in a nearby pig farm at the end of 2018. Regardless, the following summarized the farm's experience with PR eradication. First, creating an eradication program is important, but practicing the components of that eradication program is even more important. Second, effective vaccines, improved biosecurity measures, and strict production management are significant aspects of such an eradication program. Third, the eradication program may differ at different farms or in different regions, and it also should be adjusted according to the monitoring results and practical situations at the farm. Fourth, it is difficult but essential to detect subclinical infections in a timely manner; thus, a pig farm should have a suitable detection plan for diagnosis, eradication, or other purposes. Fifth, based on our results from the boar herd, isolation effectively controls PRV transmission. Sixth, positive gI antibody results can probably come from a maternal antibody source, not from infection within a herd of 100-day pigs. Seventh, an introduction of negative gilts must be on the premise that PRV-positive sows do not shed the virus, and there is no virus in the farm’s environment. Eighth, the farm owner should believe the validity of scientific virus detection and not rely on clinical manifestations during PR eradication.
PR has been listed in the National Middle-to-Long Term Plan for Animal Disease Control (2012–2020) as one of the priority swine diseases that need to be controlled [15]. Based on our results, it is possible to control and eradicate this disease in China in the near future via the use of PRV gE-deletion vaccines and the concomitant use of ELISA kits.
ACKNOWLEDGMENTS
We thank the pig farm owner for assistance in sample collection.
Footnotes
Funding: This work was funded by Heilongjiang Bayi Agricultural University Support Program for the introduced talent (Award Number: XYB201913).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Wang J.
- Data curation: Han H, Liu W.
- Formal analysis: Han H, Li S.
- Funding acquisition: Wang J.
- Investigation: Wang J, Han H, Liu W, Li S.
- Methodology: Wang J, Guo D.
- Writing - original draft: Wang J.
- Writing - review & editing: Guo D.
References
- 1.Müller T, Hahn EC, Tottewitz F, Kramer M, Klupp BG, Mettenleiter TC, et al. Pseudorabies virus in wild swine: a global perspective. Arch Virol. 2011;156(10):1691–1705. doi: 10.1007/s00705-011-1080-2. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis--state of the art, June 1999. Vet Res. 2000;31(1):99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 3.Casades-Martí L, González-Barrio D, Royo-Hernández L, Díez-Delgado I, Ruiz-Fons F. Dynamics of Aujeszky's disease virus infection in wild boar in enzootic scenarios. Transbound Emerg Dis. 2020;67(1):388–405. doi: 10.1111/tbed.13362. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Luo Y, Wang CH, Yuan J, Li N, Song K, et al. Control of swine pseudorabies in China: opportunities and limitations. Vet Microbiol. 2016;183:119–124. doi: 10.1016/j.vetmic.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Chen Q, Rao X, Diao X, Yang L, Fang X, et al. An economic assessment of pseudorabies (Aujeszky' disease) elimination on hog farms in China. Prev Vet Med. 2019;163:24–30. doi: 10.1016/j.prevetmed.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Elbers AR, Braamskamp J, Dekkers LJ, Voets R, Duinhof T, Hunneman WA, et al. Aujeszky's disease virus eradication campaign successfully heading for last stage in The Netherlands. Vet Q. 2000;22(2):103–107. doi: 10.1080/01652176.2000.9695034. [DOI] [PubMed] [Google Scholar]
- 7.Müller T, Bätza HJ, Schlüter H, Conraths FJ, Mettenleiter TC. Eradication of Aujeszky's disease in Germany. J Vet Med B Infect Dis Vet Public Health. 2003;50(5):207–213. doi: 10.1046/j.1439-0450.2003.00666.x. [DOI] [PubMed] [Google Scholar]
- 8.Smith G. Preferential sexual transmission of pseudorabies virus in feral swine populations may not account for observed seroprevalence in the USA. Prev Vet Med. 2012;103(2-3):145–156. doi: 10.1016/j.prevetmed.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stegeman A. Aujeszky's disease (pseudorabies) virus eradication campaign in The Netherlands. Vet Microbiol. 1997;55(1-4):175–180. doi: 10.1016/s0378-1135(96)01301-6. [DOI] [PubMed] [Google Scholar]
- 10.Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch'ng TH, Cristea IM, et al. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J Virol. 2013;87(17):9431–9440. doi: 10.1128/JVI.01317-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, et al. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011;7(10):e1002282. doi: 10.1371/journal.ppat.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong GZ, Chen HC. Pseudorabies epidemic status and control measures in China. Chin J Vet Med. 1999;19:1–2. [Google Scholar]
- 13.Tong W, Li G, Liang C, Liu F, Tian Q, Cao Y, et al. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antiviral Res. 2016;130:110–117. doi: 10.1016/j.antiviral.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Zhang S, Xu Q, Wu J, Zhai X, Li S, et al. Investigation on pseudorabies prevalence in Chinese swine breeding farms in 2013–2016. Trop Anim Health Prod. 2018;50(6):1279–1285. doi: 10.1007/s11250-018-1555-1. [DOI] [PubMed] [Google Scholar]
- 15.The State Council of the People's Republic of China. National Middle-to-Long Term Plan for Animal Disease Control (2012–2020). The Bulletin of the State Council of the People's Republic of China. Beijing: The State Council of the People's Republic of China; 2012. [Google Scholar]
- 16.Hu D, Lv L, Zhang Z, Xiao Y, Liu S. Seroprevalence and associated risk factors of pseudorabies in Shandong province of China. J Vet Sci. 2016;17(3):361–368. doi: 10.4142/jvs.2016.17.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel AM, Weigel RM. Herd factors affecting the selection and success of intervention strategies in the program for eradication of pseudorabies (Aujeszky's disease) virus from Illinois swine farms. Prev Vet Med. 1999;40(3-4):243–259. doi: 10.1016/s0167-5877(99)00026-4. [DOI] [PubMed] [Google Scholar]
- 18.Tamba M, Calabrese R, Finelli E, Cordioli P. Risk factors for Aujeszky's-disease seropositivity of swine herds of a region of northern Italy. Prev Vet Med. 2002;54(3):203–212. doi: 10.1016/s0167-5877(02)00028-4. [DOI] [PubMed] [Google Scholar]
- 19.Kinker DR, Swenson SL, Wu LL, Zimmerman JJ. Evaluation of serological tests for the detection of pseudorabies gE antibodies during early infection. Vet Microbiol. 1997;55(1-4):99–106. doi: 10.1016/s0378-1135(96)01308-9. [DOI] [PubMed] [Google Scholar]
- 20.Oren SL, Swenson SL, Kinker DR, Hill HT, Hu HL, Zimmerman J. Evaluation of serological pseudorabies tests for the detection of antibodies during early infection. J Vet Diagn Invest. 1993;5(4):529–533. doi: 10.1177/104063879300500405. [DOI] [PubMed] [Google Scholar]
- 21.Balasch M, Pujols J, Segalés J, Plana-Durán J, Pumarola M. Study of the persistence of Aujeszky's disease (pseudorabies) virus in peripheral blood mononuclear cells and tissues of experimentally infected pigs. Vet Microbiol. 1998;62(3):171–183. doi: 10.1016/s0378-1135(98)00208-9. [DOI] [PubMed] [Google Scholar]
- 22.Ma W, Lager KM, Richt JA, Stoffregen WC, Zhou F, Yoon KJ. Development of real-time polymerase chain reaction assays for rapid detection and differentiation of wild-type pseudorabies and gene-deleted vaccine viruses. J Vet Diagn Invest. 2008;20(4):440–447. doi: 10.1177/104063870802000405. [DOI] [PubMed] [Google Scholar]
- 23.Panyasing Y, Kedkovid R, Kittawornrat A, Ji J, Zimmerman J, Thanawongnuwech R. Detection of Aujeszky's disease virus DNA and antibody in swine oral fluid specimens. Transbound Emerg Dis. 2018;65(6):1828–1835. doi: 10.1111/tbed.12961. [DOI] [PubMed] [Google Scholar]
- 24.Zanella EL, Miller LC, Lager KM, Bigelow TT. Evaluation of a real-time polymerase chain reaction assay for Pseudorabies virus surveillance purposes. J Vet Diagn Invest. 2012;24(4):739–745. doi: 10.1177/1040638712447279. [DOI] [PubMed] [Google Scholar]
- 25.Gu J, Hu D, Peng T, Wang Y, Ma Z, Liu Z, et al. Epidemiological investigation of pseudorabies in Shandong Province from 2013 to 2016. Transbound Emerg Dis. 2018;65(3):890–898. doi: 10.1111/tbed.12827. [DOI] [PubMed] [Google Scholar]
- 26.Xia L, Sun Q, Wang J, Chen Q, Liu P, Shen C, et al. Epidemiology of pseudorabies in intensive pig farms in Shanghai, China: herd-level prevalence and risk factors. Prev Vet Med. 2018;159:51–56. doi: 10.1016/j.prevetmed.2018.08.013. [DOI] [PubMed] [Google Scholar]