Abstract
Mycobacterium bovis is the causative agent of bovine tuberculosis (bTB) in wildlife. Confirmation of M. bovis infection relies on mycobacterial culture, which is time-consuming. Collection and transportation of infectious material also pose a human health risk. PrimeStore Molecular Transport Medium (MTM) has been shown to effectively inactivate infectious organisms, making it a safe method for handling infectious samples. This study investigated an in-field sampling technique for rapid, safe detection of M. bovis in buffalo tissues. Potentially infected tissues from bTB test-positive buffaloes were swabbed at post-mortem examination and stored in PrimeStore MTM at ambient temperature until Xpert MTB/RIF Ultra testing was performed. Additionally, tissue samples were frozen and transported before homogenisation for culture and Ultra testing. Oral swabs were collected from M. bovis-unexposed buffaloes as a negative control cohort. Mycobacterium tuberculosis complex (MTBC) DNA was detected by Ultra in 13/16 tissue swabs and 9/16 matched tissue homogenates from culture-confirmed M. bovis-positive buffalo tissues. MTBC DNA was not detected in swabs from M. bovis-unexposed animals, showing the potentially high specificity of Ultra with PrimeStore swabs. PrimeStore MTM sample processing, in combination with the Ultra assay, has the potential to provide a safe, rapid post-mortem screening test for M. bovis in buffaloes.
Subject terms: Molecular biology, Tuberculosis, Translational research
Introduction
Bovine tuberculosis (bTB) is an important zoonosis caused by Mycobacterium bovis (M. bovis) and is responsible for severe economic losses and disruption of conservation programs when livestock and wildlife are affected1,2. African buffaloes (Syncerus caffer) are important wildlife maintenance hosts of M. bovis and serve as a source of infection for other susceptible mammals sharing the same habitat2. This highlights the potential disease risk and importance of its control1,2. Diagnosis of M. bovis infection in African buffaloes typically relies on the measurement of cell-mediated immune (CMI) responses to M. bovis antigens3. Buffalo TB tests include in vitro interferon-gamma (IFN-γ) release assays (IGRAs) and IFN-γ inducible protein 10 (IP-10) release assays (IPRA) that use a modified QuantiFERON-TB Gold Plus (QFT) In-tube antigen stimulation platform, and the in vivo single comparative intradermal tuberculin test (SCITT)4,5. However, confirmation of M. bovis infection usually requires mycobacterial culture and genetic speciation of mycobacterial isolates cultured from tissue specimens collected during post-mortem examination. Conventional culture methods, however, have suboptimal sensitivity and are time-consuming, taking up to 56 days before results are available6. In addition, post-mortem examination of potentially M. bovis-infected buffaloes, and the collection, transport and storage of tissue specimens are considered a potential human health risk7,8. Infectious pathogens, other than M. bovis, may also be present during sample collection and transport, such as Foot-and-Mouth Disease (FMD) virus, which may lead to sample movement restrictions to laboratories due to the risk of spreading the disease. Movement restrictions placed on buffalo samples from locations with reportable diseases to laboratories may result in inadequate bTB surveillance in these areas9.
PrimeStore Molecular Transport Medium (MTM), used in combination with PrimeStore swabs, has been shown to effectively inactivate infectious organisms, while stabilising the nucleic acids, through lysis of the cell membranes, destruction of proteins and enzymes, and inactivation of nucleases10. PrimeStore MTM therefore provides a safer option for collection and transportation of samples that potentially pose a risk to human or animal health. In addition, this method may facilitate sample transport from areas that may be restricted due to the presence of reportable diseases (such as FMD), since it inactivates all pathogens.
The rapid Xpert MTB/RIF Ultra (Ultra) assay is an automated cartridge-based semi-quantitative nested real-time PCR assay that detects Mycobacterium tuberculosis complex (MTBC) DNA and rifampicin resistance in clinical specimens11. The assay targets the multicopy sequences, IS6110 and IS1081, which greatly improves the detection of DNA in paucibacillary samples. IS1081 and IS6110 are exclusively found in MTBC members, which makes it ideal for samples collected from animals that may be infected with members of the MTBC12,13. Although the Ultra is typically used for detection of MTBC DNA in human samples, it has been shown to accurately detect M. bovis in wildlife specimens from white rhinoceros (Ceratotherium simum) and elephants (Loxodonta africana)13, and several other wildlife species14,15.
Therefore, the aim of this study was to investigate the combined use of the Ultra assay with PrimeStore MTM swab samples as a new screening technique for the detection of MTBC DNA in buffalo tissue specimens, which could provide a rapid and safer method for sample collection and transport, or in-field testing.
Results
Sixteen tissue samples collected from 13 buffaloes were confirmed to be M. bovis positive by mycobacterial culture and PCR-based speciation (Table 1). Each of these 16 culture-confirmed M. bovis positive tissue samples had a matching PrimeStore swab sample collected during post-mortem examination and stabilised in PrimeStore MTM, and a pre-culture tissue homogenate aliquot. Both the swab and tissue homogenate were tested with the Ultra assay prior to mycobacterial culture. The Ultra assay detected MTBC DNA in 13 of 16 (81.3%) tissue swab samples, while MTBC DNA was detected directly in 9 of 16 (56.3%) tissue homogenates from culture-confirmed M. bovis-positive specimens. The difference between the proportion of positive Ultra assay results from tissue homogenates compared to tissue swabs was not significant using the McNemar’s test (p > 0.05). The agreement between these tests were regarded as “fair” on the kappa agreement scale, with κ = 0.259 (95% confidence interval (CI) − 0.2 to 0.72 and standard error (SE) of 0.23).
Table 1.
Ultra assay results from PrimeStore MTM tissue swab samples and tissue homogenates from culture-confirmed M. bovis positive African buffaloes (n = 13). Positive results are shown in bold.
| Buffalo number | Culture-confirmed M. bovis positive tissue samples | Ultra result for PrimeStore MTM tissue swabs | Ultra result for tissue homogenates |
|---|---|---|---|
| A107 | Left retropharyngeal LN | MTB trace detected | MTB detected low |
| A113 | Tonsils | MTB trace detected | MTB not detected |
| A20 | Tonsils | MTB detected very low | MTB not detected |
| A98 | Retropharyngeal LN | MTB trace detected | MTB trace detected |
| A98 | Tonsils | MTB not detected | MTB not detected |
| B15 | Lung lesion | MTB detected low | MTB detected very low |
| B19 | Retropharyngeal LN | MTB trace detected | MTB detected very low |
| B30 | Lung lesion | MTB not detected | MTB not detected |
| B30 | Mediastinal LN | MTB trace detected | MTB not detected |
| B48 | Right retropharyngeal LN lesion | MTB detected very low | MTB detected low |
| B64 | Abdominal serosa | MTB detected very low | MTB detected low |
| B65 | L/R tracheobronchial LN | MTB detected very low | MTB detected very low |
| B8 | L/R tracheobronchial LN | MTB detected very low | MTB detected low |
| C22 | Scalpel | MTB not detected | MTB detected low |
| C28 | Retropharyngeal LN | MTB trace detected | MTB not detected |
| C28 | Tonsil lesion | MTB detected very low | MTB detected very low |
Ultra—Xpert MTB/RIF Ultra; LN—Lymph node; L/R—Left and right; MTM—molecular transport medium.
Additionally, Ultra assay did not detect MTBC DNA in any oral swabs collected from the 12 randomly selected M. bovis unexposed buffaloes from historically bTB free buffalo herds (data not shown).
Discussion
The findings from this study show that the Ultra assay combined with tissue swabs that were inactivated in PrimeStore MTM can accurately identify culture-confirmed M. bovis positive tissues from African buffaloes. The swabs taken directly from dissected tissues and later tested on the Ultra assay performed similar to the Ultra on tissue homogenates prepared prior to culture. Recent studies have shown that Ultra provides accurate, rapid diagnosis of MTBC infection in livestock and wildlife when performed on tissue homogenates13–15. However, the combined use of PrimeStore MTM and tissue swabs with the Ultra assay have the added advantage of providing more rapid test results, since it can directly be used in the Ultra assay without the extra steps of tissue processing; results may even be acquired in-field if portable Ultra equipment is available (such as Cepheid’s GeneXpert Edge system), providing same day results. Typically, tissue samples are kept frozen at − 20 °C and transported to a biosafety level 3 (BSL-3) facility before being homogenised to perform the Ultra test. PrimeStore MTM incubated swabs do not require refrigeration and since PrimeStore MTM inactivates pathogens, samples could even be transported through standard postal services without biosafety concerns. Furthermore, a BSL-3 facility is not required for sample processing of swabs in PrimeStore MTM and samples can be handled safely without the risk of infection to other animals and humans.
There was a greater number of positive Ultra results using swabs than tissue homogenates. However, the difference was not significant and there was only fair agreement between the two methods. There could be various reasons for the discordant Ultra results between tissue swabs and matching tissue homogenate samples. PrimeStore MTM has been shown to inactivate nucleases in samples. Since the tissue homogenate samples were not incubated in stabilising media (PrimeStore MTM), there may have been degradation of DNA in some samples during freeze–thaw cycles, which could result in a negative Ultra result, especially if the samples were paucibacillary. Tissue swabs, however, did not undergo a freeze–thaw cycle prior to testing with Ultra. Also, since Ultra results are influenced by bacillary load16, swab samples from tissues with visible lesions are more targeted than taking an aliquot of the tissue homogenate for Ultra testing, which may result in some dilution of mycobacteria in a larger homogenate volume than the swabs.
Studies performed with human specimens have shown that Ultra results provide a measure of the bacillary load in a specimen, with a positive correlation between the semi-quantitative Ultra results and detection of acid-fast bacilli with smear-microscopy17. The “MTB trace detected” and “MTB not detected” Ultra results have been shown to be correlated with no acid-fast bacilli detected, whereas Ultra results “MTB detected high” and “medium” typically correlate with positive smear-microscopy results17. The Ultra test results in this study included “MTB detected low”, “MTB detected very low”, “MTB trace detected” and “MTB not detected”. Since the “MTB trace detected” Ultra results from tissue swabs and homogenates were seen in culture-confirmed M. bovis positive specimens, it may indicate greater sensitivity of this method compared to smear microscopy. However, varying copy numbers of IS6110 and IS1081 between MTBC members and between strains have also been shown to influence the sensitivity and may not entirely relate to the performance of the Ultra assay11,13. The bacillary load in samples in which no MTBC DNA were detected (“MTB not detected”) was likely below the threshold for detection by the Ultra assay, although the homogenates from the same tissue sample had a positive M. bovis result by mycobacterial culture and PCR speciation. This could be due to a low but viable bacillary load that would have increased to a detectable level during the 56 days of mycobacterial culture, which could further explain the discordant results between mycobacterial culture (all M. bovis positive) and Ultra results from tissue homogenates. Difference in sample volume between mycobacterial culture and Ultra from tissue swabs may also contribute to discordant results between these two methods.
The oral swabs from the M. bovis-unexposed buffaloes were all negative on the Ultra assay. These results are consistent with the reported high specificity of the Ultra assay11. Interestingly, although these swabs were used as negative controls, the swabs would have had a greater likelihood of contamination than those collected post-mortem, since these animals were sampled in a dusty, natural environment, where they were likely exposed to various environmental organisms.
The study had several limitations. Firstly, there was a small sample size of culture-confirmed M. bovis-positive buffaloes and oral swab samples available from the M. bovis-unexposed buffaloes, which did not allow for direct comparison of the same sample type tested in the Ultra. Secondly, since the Ultra assay was only performed in the laboratory, we were unable to compare laboratory and in-field testing with the Ultra assay and PrimeStore sampling platform. Finally, since tissue samples for Ultra were specifically selected based on visible lesions, it is unknown how the Ultra would perform using M. bovis culture-positive tissue samples with no visible lesions and therefore presumed low bacillary load. Future studies should include a larger sample size with buffaloes at various stages of infection. The application of field testing should also be investigated, which would allow more rapid sample processing and the use of fresh samples. Additionally, pooling swab samples from each buffalo should be investigated as a method to decrease the number of samples tested.
The Ultra assay with samples stored in PrimeStore MTM shows promise as a rapid post-mortem screening test, which could be used to identify MTBC DNA in tissue samples and ultimately identify infected herds more rapidly than mycobacterial culture. This test can also be used to decrease the number of samples sent for mycobacterial culture by identifying those that contain mycobacterial DNA. In addition, the use of PrimeStore MTM to safely store and transport infectious samples will safeguard human and animal health as well as permit greater use of samples that may otherwise be restricted from being transported to diagnostic laboratories.
Materials and methods
Animals and post-mortem sample collection
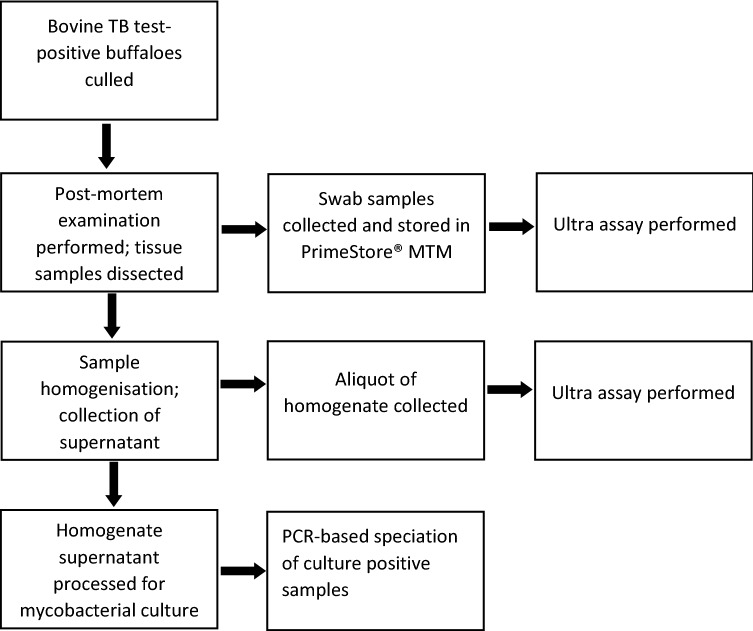
The Hluhluwe-iMfolozi Park (HiP) in KwaZulu-Natal, South Africa, is a game park that is endemic for M. bovis-infection in the wildlife population. During the 2019 annual test-and-slaughter bTB control program, African buffaloes were captured, immobilised, sampled and tested for bTB, using the testing regime previously described18. Bovine TB test-positive buffaloes (n = 22) underwent post-mortem examination as previously described18. Various tissues with gross pathological lesions consistent with bTB18 were dissected and swabbed during post-mortem examination using PrimeStore swabs (Longhorn Vaccines and Diagnostics, San Antonio, Texas, USA) and stored in PrimeStore MTM (Longhorn Vaccines and Diagnostics) at ambient temperature (which may reach temperatures up to 25 °C during winter in HiP) until further processing. Tissues included retropharyngeal lymph nodes, tonsils, lung, mediastinal lymph nodes, abdominal serosa, and tracheobronchial lymph nodes. In addition, the scalpel blade used to cut tissues for a particular animal was also sampled on some occasions (Fig. 1). Samples with visible lesions were stored in separate containers and frozen at − 20 °C until processed for mycobacterial culture. Since tissue samples were not available from M. bovis-unexposed buffaloes, oropharyngeal swabs were collected from 12 randomly selected buffaloes that were tested for bTB in historically bTB-negative private game parks, as previously described18. PrimeStore swab samples were stored in PrimeStore MTM at ambient temperature until further processing. All samples were transported to Stellenbosch University for further analyses.
Figure 1.
Study method flow chart for African buffaloes from the bTB-endemic game reserve, HiP.
Ethical approval for this study was granted by Stellenbosch University Animal Care and Use Committee (ACU-2019-9081). Permission to perform animal research in terms of section 20 of the Animal Diseases Act was granted by the South African Department of Agriculture, Land Reform and Rural Development (DALRRD), formerly the Department of Agriculture, Forestry and Fisheries (DAFF), South Africa (12/11/1/7/2). All buffaloes were handled by the Ezemvelo KZN Wildlife services’ veterinarians and game capture teams according to their guidelines. ARRIVE guidelines for reporting animal research have been followed as much as possible (https://arriveguidelines.org/).
Mycobacterial culture
All previously frozen tissue samples collected during post-mortem examination were homogenised in phosphate buffer (Becton Dickinson, Franklin Lakes, NJ, USA) with 4.8 mm stainless steel beads using a Bullet Blender 50 (Next Advance, Averill Park, NY, USA) for 15 min at maximum speed in a BSL-3 laboratory at Stellenbosch University, as previously described3. A 1 ml aliquot of the liquid fraction of each tissue homogenate was used for quantitative polymerase chain reaction (qPCR) analysis by the Xpert MTB/RIF Ultra (Ultra) assay (Cepheid Inc., Sunnyvale, California, United States). Tissue homogenates were then further processed for mycobacterial culture using the standard mycobacterial culture protocol with the BBL MycoPrep specimen decontamination kit (Becton Dickinson) and BACTEC MGIT 960 Mycobacterial Detection System (Becton Dickinson), as previously described3. Speciation by region of difference PCR was performed on all culture positive samples to confirm the presence of M. bovis19 (Fig. 1).
Xpert MTB/RIF ultra assay
The Ultra assay (Cepheid) was performed using the oropharyngeal swabs collected from M. bovis-unexposed buffaloes and tissue swab samples from HiP buffaloes, according to the Ultra alternative sample assay instructions, as previously described13. Briefly, Xpert MTB/RIF sample lysis reagent (Cepheid) was mixed in a 1:1 ratio with each PrimeStore MTM stabilised swab sample and incubated for 10 min at room temperature (RT). Hereafter it was vortexed for 10 s before incubation at RT for 5 min followed by a 5 s vortex step. Approximately 2 ml of the prepared sample was dispensed into the sample chamber of the Ultra cartridge for automated processing. Aliquots of tissue homogenate processed for mycobacterial culture were also tested with the Ultra assay, with sample preparation as described above. The Ultra assay classifies MTBC DNA detection as high, medium, low, very low or trace, based on the quantity of MTBC DNA that was detected, and all of these classifications were considered positive in this study. When the Ultra could not detect MTBC DNA (“MTB not detected”), it was considered a negative result. Rifampicin resistance was also reported but is irrelevant in the context of this study (Fig. 1).
Statistical analysis
Agreement between tests was calculated as Cohen’s kappa coefficient (κ) using the calculator on the GraphPad Prism Software (https://www.graphpad.com/quickcalcs/kappa2/). The McNemar’s test was used to compare Ultra results from tissue swabs and tissue homogenates using the GraphPad QuickCalcs software https://www.graphpad.com/quickcalcs/McNemar1.cfm). A p value < 0.05 was considered statistically significant.
Acknowledgements
We thank Dr. Gwynn Stevens and Dipti Lallubhai (Cepheid) for their support of this project, Alicia and Warren McCall, Dr Rowan Leeming, Dumisani Zwane, JP van Heerden, and the Game Capture staff from KwaZulu-Natal Ezemvelo Wildlife for their assistance with this study.
Author contributions
C.C., M.A.M., and W.J.G. conceived the experiments. W.J.G. and C.C. conducted the experiments. D.V.C. provided access to immobilized buffaloes; W.J.G., K.S., S.J.G., and M.A.M. performed sample collection. C.H. provided swabs and MTM media as well as provided input on study design. M.A.M., R.M.W., and P.D.H. provided funding. C.C., M.A.M., T.J.K., L.K., W.J.G., R.M.W. and P.D. v H. analysed the data. All authors reviewed the manuscript.
Funding
This work was supported by (1) the Harry Crossley Foundation, (2) South African government through the South African Medical Research Council and the National Research Foundation South African Research Chair Initiative (Grant #86949), (3) Cepheid, Inc., and (4) American Association of Zoo Veterinarians Wild Animal Health Fund (S005651). The content is the sole responsibility of the authors and does not necessarily represent the official views of the funders.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arnot LF, Michel A. Challenges for controlling bovine tuberculosis in South Africa. Onderstepoort J. Vet. Res. 2020;87(1):a1690. doi: 10.4102/ojvr.v87i1.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vos V, et al. The epidemiology of tuberculosis in free-ranging African buffalo (Syncerus caffer) in the Kruger national park, South Africa. Onderstepoort J. Vet. Res. 2001;68:119–130. [PubMed] [Google Scholar]
- 3.Goosen WJ, et al. Agreement between assays of cell-mediated immunity utilizing Mycobacterium bovis-specific antigens for the diagnosis of tuberculosis in African buffaloes (Syncerus caffer) Vet. Immunol. Immunopathol. 2014;160:133–138. doi: 10.1016/j.vetimm.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Goosen WJ, Cooper D, Miller MA, Van Helden PD, Parsons SDC. IP-10 is a sensitive biomarker of antigen recognition in whole-blood stimulation assays used for the diagnosis of Mycobacterium bovis infection in African buffaloes (Syncerus caffer) Clin. Vaccine Immunol. 2015;22:974–978. doi: 10.1128/CVI.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernitz N, et al. Detection of Mycobacterium bovis infection in African buffaloes (Syncerus caffer) using QuantiFERON-TB Gold (QFT) tubes and the Qiagen cattletype IFN-gamma ELISA. Vet. Immunol. Immunopathol. 2018;196:48–52. doi: 10.1016/j.vetimm.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Stewart LD, McNair J, McCallan L, Gordon A, Grant IR. Improved Detection of Mycobacterium bovis infection in bovine lymph node tissue using immunomagnetic separation (IMS)-based methods. PLoS ONE. 2013;8:e58374. doi: 10.1371/journal.pone.0058374.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vayr F, Martin-blondel G, Savall F, Soulat J, Herin F. Occupational exposure to human Mycobacterium bovis infection: a systematic review. PLoS Negl. Trop. Dis. 2018;12(1):e0006208. doi: 10.1371/journal.pntd.0006208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khattak I, Mushtaq MH, Ahmad MUD, Khan MS, Haider J. Zoonotic tuberculosis in occupationally exposed groups in Pakistan. Occup. Med. 2016;66:371–376. doi: 10.1093/occmed/kqw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Department of Agriculture, Forestry & Fisheries, 2016. The South African Veterinary Strategy (2016–2026). [online] pp. 1–60. https://www.daff.gov.za/vetweb/Animal%20Identification/Veterinary%20Strategy%20%202016-09-08_%20(Final).pdf (Accessed 8 December 2020).
- 10.Daum LT, et al. A clinical specimen collection and transport medium for molecular diagnostic and genomic applications. Epidemiol. Infect. 2011;139:1764–1773. doi: 10.1017/S0950268810002384. [DOI] [PubMed] [Google Scholar]
- 11.Chakravorty S, et al. The new Xpert MTB/RIF Ultra: Improving detection of Mycobacterium tuberculosis and resistance to Rifampin in an assay suitable for point-of-care testing. MBio. 2017;8:e00812-17. doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Soolingen D, Hermans PWM, De Haas PEW, Van Embden JDA. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J. Clin. Microbiol. 1992;30:1772–1777. doi: 10.1128/JCM.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goosen WJ, et al. The Xpert MTB / RIF Ultra assay detects Mycobacterium tuberculosis complex DNA in white rhinoceros (Ceratotherium simum) and African elephants (Loxodonta africana) Sci. Rep. 2020 doi: 10.1038/s41598-020-71568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hlokwe TM, Mogano RM. Utility of Xpert MTB/RIF Ultra assay in the rapid diagnosis of bovine tuberculosis in wildlife and livestock animals from South Africa. Prev. Vet. Med. 2020 doi: 10.1016/j.prevetmed.2020.104980. [DOI] [PubMed] [Google Scholar]
- 15.Kerr TJ, et al. Novel techniques for detection of Mycobacterium bovis infection in a cheetah. Emerg. Infect. Dis. 2020;26:630–631. doi: 10.3201/eid2603.191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Risco D, Rodriguez-Temporal D, Valledor-Sanchez I, Alcaide F. Evaluation of the Xpert MTB/RIF ultra assay for direct detection of Mycobacterium tuberculosis complex in smear-negative extrapulmonary samples. J. Clin. Microbiol. 2018;56:e00659-18. doi: 10.1128/JCM.00659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opota O, Mazza-Stalder J, Greub G, Jaton K. The rapid molecular test Xpert MTB/RIF ultra: towards improved tuberculosis diagnosis and rifampicin resistance detection. Clin. Microbiol. Infect. 2019;25:1370–1376. doi: 10.1016/j.cmi.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Bernitz N, et al. Impact of Mycobacterium bovis -induced pathology on interpretation of QuantiFERON-TB Gold assay results in African buffaloes (Syncerus caffer) Vet. Immunol. Immunopathol. 2019;217:109923. doi: 10.1016/j.vetimm.2019.109923. [DOI] [PubMed] [Google Scholar]
- 19.Warren RM, et al. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 2006;10:818–822. [PubMed] [Google Scholar]