Abstract
The National Health Service (NHS) Pathways triage system collates data on enquiries to 111 and 999 services in England. Since the 18th of March 2020, these data have been made publically available for potential COVID-19 symptoms self-reported by members of the public. Trends in such reports over time are likely to reflect behaviour of the ongoing epidemic within the wider community, potentially capturing valuable information across a broader severity profile of cases than hospital admission data. We present a fully reproducible analysis of temporal trends in NHS Pathways reports until 14th May 2020, nationally and regionally, and demonstrate that rates of growth/decline and effective reproduction number estimated from these data may be useful in monitoring transmission. This is a particularly pressing issue as lockdown restrictions begin to be lifted and evidence of disease resurgence must be constantly reassessed. We further assess the correlation between NHS Pathways reports and a publicly available NHS dataset of COVID-19-associated deaths in England, finding that enquiries to 111/999 were strongly associated with daily deaths reported 16 days later. Our results highlight the potential of NHS Pathways as the basis of an early warning system. However, this dataset relies on self-reported symptoms, which are at risk of being severely biased. Further detailed work is therefore necessary to investigate potential behavioural issues which might otherwise explain our conclusions.
Subject terms: Epidemiology, Infectious diseases
Introduction
The coronavirus 2019 disease (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus originally identified in Wuhan (China) in December 20191. As of the beginning of May 2020, more than 3 million confirmed COVID-19 cases and 200,000 deaths have been reported worldwide1. To mitigate epidemic spread and reduce the risk of healthcare systems being overrun, countries have implemented different measures to slow down transmission, such as travel restrictions and various degrees of social distancing2.
More than 120,000 confirmed COVID-19 cases and 25,000 deaths have been reported in England as of the beginning of May 20203. However, laboratory confirmation has largely been restricted to hospitalised cases3. The majority of milder cases, which likely represent a large fraction of the total are unaccounted for4. We should therefore explore alternative, community-driven surveillance data to help characterise the progression of the COVID-19 epidemic in the country.
The National Health Service (NHS) Pathways is a triage system for public calls and online reports for medical care5. This system is currently being used throughout England to assist individuals reporting potential COVID-19 symptoms. Since the 18th of March 2020, data on daily phone calls and completed online assessments which have received a potential COVID-19 final disposition are openly available. These assessments are either completed via calls to 111 and 999 (which are respectively for non-urgent, and urgent medical problems), or through 111-online self-completed reports. The fraction of assessments corresponding to actual COVID-19 cases is unknown, but in the absence of wide-scale testing, the NHS Pathways dataset may be one of the best available proxies for COVID-19 incidence in the community. While prone to self-reporting biases, it is likely to better reflect milder cases and be less biased by different severity profiles than hospital admission data, which by definition reflect the most acute cases.
Here, we analyse NHS Pathways data until 14th May 2020 to assess the temporal dynamics of COVID-19 in England. Specifically, we investigated potential changes in the growth rate of the epidemic over time, and compared the observed patterns across NHS regions. We derived time-varying estimates of the growth rates, halving time and effective reproduction numbers for the different regions. We also assessed the potential correlation between NHS Pathways data with COVID-19 daily deaths in England, to gain an initial understanding of its possible value within an early detection system.
Methods
Data extraction
We extracted the NHS Pathways data from 18th March 2020 up to 14th May 2020 through the NHS Digital website5, where they are updated daily, every weekday. This dataset contains daily numbers of calls to 111 and 999, as well as 111-online reports that were classified as potential COVID-19 cases by the triage algorithm. This algorithm follows a symptom-based approach, where the respondents are asked a series of questions to self-report their symptoms until an end point is reached, or the call is handed to a clinician6. Some individuals might report symptoms multiple times through the system, therefore the number of reports are not directly equivalent to the underlying number of cases. The number of reports are stratified by Clinical Commissioning Group (CCG), gender and age group (0–18, 19–69 and 70–120 years old) of the patients. We mapped the CCGs to their corresponding NHS regions using publicly available CCG data7, and used this geographic resolution for our analysis. All dates indicated refer to the date of reporting.
A reporting change in the NHS Pathways data occured over the period between the 9th and 23rd of April, during which the number of 111-online reports for individuals aged between 0 and 18 years old was not available. However, due to the relatively small proportion of total reports for that age group, this likely does not affect our results (see Figures S3-S5 in Supplementary Material for further details). We also note that towards the end of March 2020, patients with potential COVID-19 symptoms were directed away from health practitioner (HP) services, which may have increased 111 reporting.
Temporal analysis
Total numbers of reports (including all three data sources: 111 calls, 999 calls, and 111-online reports) were modelled using quasi-Poisson generalised linear models (GLM) with log links, to account for exponential trends as well as over-dispersion of the data8. Predictors included time (in days since the first data point (18th March 2020) with interaction terms for varying slopes and intercepts between NHS regions, and day of the week (weekend, Monday, or rest of the week) to account for potential differences in reporting over the weekend and at the start of the week. For a number of reports on day t in NHS region i, the equation for this GLM is therefore:
where θ is the over-dispersion parameter.
Growth rates (r) for each NHS region and their 95% confidence intervals were directly deduced from the corresponding coefficients. All models were fitted using maximum-likelihood.
To assess potential changes of the growth rate over time, analyses were performed over rolling windows of 14 days from the earliest available date (18th March 2020) to the latest available one (14th May 2020; see Figure S1 in Supplementary Material for a sensitivity analysis). Growth rates and associated confidence intervals were calculated for each time window. Whenever the upper bound of r was negative, corresponding halving times were calculated as log(0.5)/r. Halving times correspond to the time period in days that it takes for the number of reports to reduce by half, should it continue to decline following the corresponding negative growth rate r. Growth rates were converted to effective reproduction numbers Re using the approach described in Wallinga and Lipsitch9 and implemented in the epitrix package10, with a serial interval modelled as a gamma distribution with mean 4.7 days and standard deviation 2.9 days11 (see Figure S2 in Supplementary Material for further details on the choice of distribution). The equation used for this calculation is:
where aj are possible values from the serial interval distribution and yj are their corresponding relative frequencies.
Correlation with reported deaths
To test the validity of the NHS Pathways dataset as an early detection system, we compared daily total counts of reports (including all three data sources: 111 calls, 999 calls, and 111-online reports) with publicly available NHS data on COVID-19 daily deaths12. This dataset includes daily counts of COVID-19 deaths in hospitals in England NHS regions since 1st March 2020. All dates refer to the date of death. However, the data are subject to bias from reporting delays, with more recent counts excluding a proportion of deaths which have not yet been reported. To account for this, we excluded data from the last 3 weeks (i.e. from the 23rd April 2020) from this analysis.
We calculated Pearson’s correlation between the daily time series of deaths and NHS Pathways reports, lagging the reports from zero to thirty days. Approximate 95% confidence intervals for each correlation estimate were calculated by bootstrapping with 1,000 replicates. From this we identified an optimal lag at which the reports correlate most strongly with subsequent deaths. We then further evaluated the potential of NHS Pathways reports lagged at this value as a predictor, assuming a quasi-Poisson distribution for daily deaths.
All analyses were performed using the R software13, and the code is publicly available from https://github.com/qleclerc/nhs_pathways_report and distributed under the MIT license.
Results
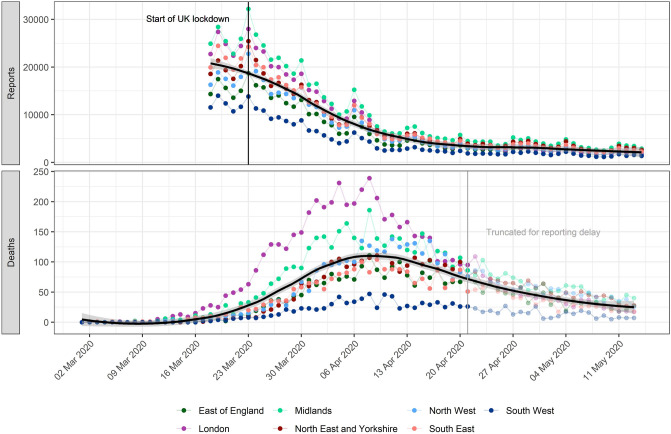
Overall reports of potential COVID-19 cases through NHS Pathways have been clearly decreasing in all NHS regions since 18th March 2020 and until approximately 13th April 2020, after which the trend seems to plateau (Fig. 1). Weekly spikes were consistently observed for all NHS regions, with increased reports on Mondays likely reflecting less reporting over weekends. The NHS COVID-19 daily deaths dataset shows daily deaths increased until approximately 10th April 2020, and have been decreasing since.
Figure 1.
Daily potential COVID-19 cases reported through NHS Pathways and reported COVID-19-related deaths, by NHS region. Pathways data include calls to 111 and 999, as well as 111-online reports. Dates correspond to the date of case report and death report, respectively, with x-axis labels corresponding to Mondays. The solid black line and grey ribbon correspond to a lowess smoother and its 95% confidence interval, calculated across all data points and NHS regions. The start of the lockdown in England (23rd March 2020) and date at which death data were truncated to avoid bias from reporting delay (21st April 2020) are highlighted by vertical lines.
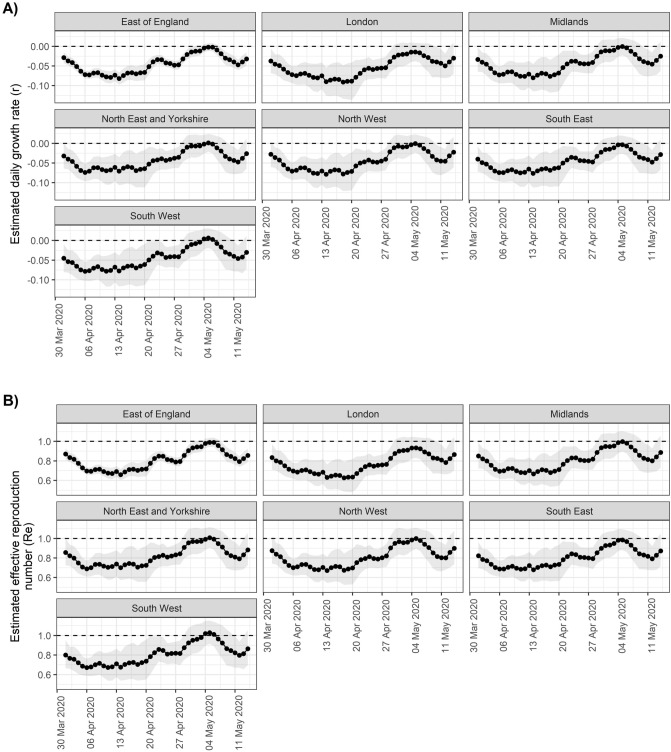
As epidemics are expected to decrease exponentially, a plateau in case incidence would be expected even if the decline rate (i.e., negative growth rate r) remained constant over the time period considered. However, this could also reflect a genuine change in r over time. Analyses over sliding time windows show that daily decline rates have likely been changing substantially during this period. Results show a marked decrease in r and the corresponding effective reproduction numbers (Re) until the 6th April 2020, after which these numbers remained low in all NHS regions for a period of about two weeks (Fig. 2). The lowest r estimated was for Saturday 18 April 2020 in the London region at -0.08 (95% CI: − 0.10– − 0.06), corresponding to a halving time of 8.45 days (95% CI: 6.77–11.20) and an Re of 0.66 (95% CI: 0.59–0.73). Similar trends were observed across all NHS regions, with the exception of London which showed lower r (and Re) after the 13th of April.
Figure 2.
Estimates of (A) daily growth rates (r) and (B) effective reproduction numbers (Re) for potential COVID-19 cases reported through NHS Pathways. Dotted lines indicate the central estimate, and ribbons their 95% confidence intervals. Estimates are indicated at the end of the time window used for estimation, so that values of r and Re provided on a given day correspond to the 2 weeks leading up to that day.
Confidence intervals suggest r values remained lower than 0 (and Re lower than 1) in all regions until 15th April, consistent with a decrease in COVID-19 related reports. After this, values of r and Re seem to have gradually increased, to a point where there was no longer any strong evidence of a decrease in COVID-19 reports in any NHS region as of 2nd May 2020, as the 95% CIs of all growth rates include 0. The most recent estimate of r averaged over all NHS regions is -0.03 (95% CI: − 0.06–0.01), corresponding to an Re of 0.88 (95% CI: 0.73–1.04).
The strongest correlation between NHS Pathways reports and deaths was obtained with a lag of 16 days (Pearson’s correlation = 0.94; 95% CI: 0.78–0.98). Figure 3 illustrates the observed trend in correlation across all tested lags. Estimates become increasingly unstable for lags above 25 days as the number of points within the overlapping time window becomes small (n = 5 at 30 days lag). There is however a clear, initially upward trend and subsequent plateau between 16 and 19 days, after which the strength of correlation appears to decrease. Further analysis suggests that this pattern is also observed when looking at NHS regions separately, albeit with some variation in the optimal lag (see Figure S6 in Supplementary Material for further details).
Figure 3.
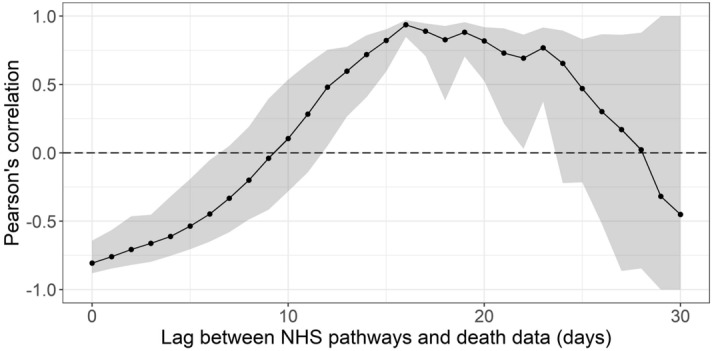
Pearson’s correlation between deaths and potential COVID-19 cases reported through NHS Pathways, lagged between 0 and 30 days. 95% confidence intervals are calculated by bootstrapping with 1000 replicates.
Fitting a quasi-Poisson GLM, we found that over 85.8% of the deviance in daily reported deaths could potentially be explained by NHS Pathways reports 16 days prior, with an average of 1.91 (bootstrap 95% CI: 1.70–2.07) additional deaths for every 1,000 potential COVID-19 cases reported in NHS Pathways 16 days before (intercept = 397, 95% CI: 357–442; % increase per 1000 notifications = 0.48, 95% CI: 0.39–0.57; Fig. 4).
Figure 4.
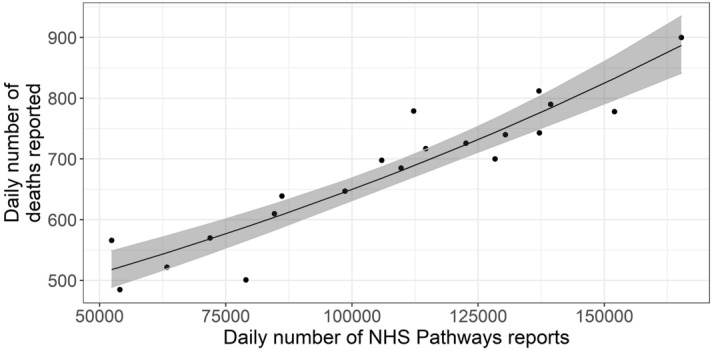
Daily total COVID-19 deaths reported in England between 3rd and 22nd April 2020, against the number of potential COVID-19 cases reported through NHS Pathways with a lag of 16 days (between 18th March and 6th April 2020). The black line and grey ribbon correspond to predictions from the regression model and associated 95% confidence intervals. The coefficient of determination indicates that 85.8% of the deviance in reported deaths is linearly explained by NHS Pathways reports.
Discussion
We analysed publicly available NHS Pathways data to assess the temporal dynamics of the COVID-19 epidemic in England until 14th May 2020. Trends in NHS Pathways reports are similar across all England NHS regions, based on casual observation. Our results suggest after a sharp initial decrease up until early April 2020, transmissibility may have slowly increased until early May. After this, it becomes unclear if cases are still declining as confidence intervals of r and Re include the threshold values for growth / decline (respectively, 0 and 1) in all NHS regions. Trends in COVID-19 associated deaths in England showed more variation between NHS regions than the Pathways reports, with one potential explanation being differing fatality risk amongst cases due to age and comorbidities. As a national total, the number of reported deaths was found to be strongly correlated with the number of reported 16 days prior. However, if these data are to serve as an early warning for potential disease resurgence, further investigation will be required to ascertain whether this correlation holds beyond this period of decline in both trends which we have analysed here.
A number of caveats may affect our results. Firstly, the data we considered are at best a proxy for the true incidence of COVID-19 in the country as they rely on self-reported symptoms interpreted by an algorithm, and were not confirmed by virological tests. This is further exacerbated by the fact that individuals might access NHS Pathways more than once to report their symptoms, which could artificially increase the numbers of potential COVID-19 cases. Unfortunately, the degree to which this occurs is not measurable and we are unable to apply a correction to our data to account for this.
As NHS Pathways is based on self-reporting, several biases could affect the data, such as changes in service availability and delays in the uptake of the 111-online reporting system. When estimating growth rates over time and geographic regions, we implicitly assume that self-reporting behaviours have not substantially changed over time, and have been similar across different NHS regions. In reality, self-reporting could be strongly biased by behavioural issues, such as the effect of news coverage which might lead individuals to pay more attention to their symptoms and report them. Inversely, individuals could reduce their perception of the risk of the disease over time as they become used to hearing about it daily, which would decrease their likelihood of noticing and reporting symptoms. Similarly, differences in self-reporting behaviours across various age groups would likely bias the age composition of the potential COVID-19 cases reported here.
While NHS Pathways data may better capture the epidemic in the community than hospitalisation data, these data do not only reflect community cases. In fact, a fraction of cases reported through 999 are likely to be hospitalised, as well as smaller proportions of cases reported through 111 calls and 111-online. Therefore, the data we considered here most likely reflect the epidemic as a whole, rather than just in the community. We also note that all results presented rely on dates of calls or online reports, so that estimates of transmissibility are likely lagged by a few days compared to the true, underlying epidemic. Unfortunately, the delays from exposure to notification through NHS Pathways cannot be estimated from current publicly available data.
The analysis of lagged correlation between the NHS Pathways and deaths data is limited by the observed time windows. With a longer window, correlation estimates for higher lags would be more stable and perhaps the optimal lag would be clearer. However, a recent review found that estimates of hospital length of stay from admission to death were mostly below 10 days14, therefore a potential lag of 16 days between potential COVID-19 cases reported through NHS Pathways and COVID-19 deaths would be coherent with the timelines for patients observed so far. Future work will aim to better exploit the temporal correlation in these data by regressing on a series of lags.
The initial sharp decrease in potential COVID-19 cases could suggest that social distancing measures put in place have had a strong impact on reducing transmission and brought the effective reproduction number under 1 across all NHS regions. However, the more recent estimates of Re suggest that transmissibility might have increased again since, such that it is no longer clear that case incidence is declining. This aligns with other estimates from the same time period based on confirmed cases in England15, which adds strength to the result. The main limitation in interpreting this result is that as true incidence reaches low levels in the population, the relative proportion of false positives among potential COVID-19 cases reported through NHS Pathways is likely to increase. In fact, if there were no more cases but an approximately constant number of false positives, the analysis of NHS Pathways data would estimate Re to be around 1. Nevertheless, the observed changes in estimated transmissibility over time, together with the potentially strong correlation between NHS Pathways reports and COVID-19 deaths time series suggest that future changes in Re could possibly be reflected in NHS Pathways data. Moreover, we found that, for the time period covered by our analysis, the NHS Pathways data were a good predictor of COVID-19 deaths reported 16 days later, making this data source a good candidate for designing early warning systems in the upcoming weeks as lockdown restrictions are progressively lifted in England. Further work is now needed to test whether this correlation holds outside a declining epidemic, and to investigate potential behavioural issues which might otherwise explain the trends we have highlighted in our analysis.
Supplementary Information
Author contributions
Q.J.L. and T.J. conceptualised the analysis and T.J. supervised the work. Q.J.L. and E.S.N. performed the analyses and prepared all figures. S.A. gave feedback on methodology and presentation of results, and reviewed the code. Q.J.L. and E.S.N. wrote the manuscript. S.A. and T.J. reviewed and edited the manuscript. Q.J.L. produced the remote code repository. All co-authors provided feedback on the analyses and the manuscript, and agreed with the final submitted version. Authors who were part of the Centre for Mathematical Modelling of Infectious Disease COVID-19 working group each contributed in processing, cleaning and interpretation of data, interpreted findings, contributed to the manuscript, and approved the work for publication.
Funding
The following funding sources are acknowledged as providing funding for the named authors. This research was partly funded by the Bill & Melinda Gates Foundation (OPP1183986: ESN). This research was partly funded by the Global Challenges Research Fund (GCRF) project 'RECAP' managed through RCUK and ESRC (ES/P010873/1: TJ). This research was partly funded by the National Institute for Health Research (NIHR) using UK aid from the UK Government to support global health research. The views expressed in this publication are those of the author(s) and not necessarily those of the NIHR or the UK Department of Health and Social Care (Health Protection Research Unit for Modelling Methodology HPRU-2012-10096: TJ). UK MRC (LID DTP MR/N013638/1: QJL; MC_PC_19065: TJ). Authors of this research receive funding from UK Public Health Rapid Support Team funded by the United Kingdom Department of Health and Social Care (TJ). Wellcome Trust (210758/Z/18/Z: SA). The following funding sources are acknowledged as providing funding for the working group authors. Alan Turing Institute (AE). BBSRC LIDP (BB/M009513/1: DS). This research was partly funded by the Bill & Melinda Gates Foundation (INV-003174: KP, MJ, YL; NTD Modelling Consortium OPP1184344: CABP, GM; OPP1180644: SRP; OPP1191821: KO'R, MA). DFID/Wellcome Trust (Epidemic Preparedness Coronavirus research programme 221303/Z/20/Z: CABP, KvZ). Elrha R2HC/UK DFID/Wellcome Trust/This research was partly funded by the National Institute for Health Research (NIHR) using UK aid from the UK Government to support global health research. The views expressed in this publication are those of the author(s) and not necessarily those of the NIHR or the UK Department of Health and Social Care (KvZ). ERC Starting Grant (#757688: CJVA, KEA; #757699: JCE, MQ, RMGJH). This project has received funding from the European Union's Horizon 2020 research and innovation programme—project EpiPose (101003688: KP, MJ, PK, WJE, YL). This research was partly funded by the Global Challenges Research Fund (GCRF) project 'RECAP' managed through RCUK and ESRC (ES/P010873/1: AG, CIJ). HDR UK (MR/S003975/1: RME). Nakajima Foundation (AE). NIHR (16/137/109: BJQ, CD, FYS, MJ, YL; Health Protection Research Unit for Modelling Methodology HPRU-2012-10096: NGD; PR-OD-1017-20002: AR). Royal Society (Dorothy Hodgkin Fellowship: RL; RP\EA\180004: PK). UK DHSC/UK Aid/NIHR (ITCRZ 03010: HPG). UK MRC (LID DTP MR/N013638/1: EMR; MC_PC 19065: RME; MR/P014658/1: GMK). Wellcome Trust (206250/Z/17/Z: AJK, TWR; 208812/Z/17/Z: SC, SFlasche; 210758/Z/18/Z: JDM, JH, NIB, SFunk, SRM). No funding (AKD, AMF, DCT, SH).
Data and code availability
The data and code used in this work can be accessed at: https://github.com/qleclerc/nhs_pathways_report.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Quentin J. Leclerc and Emily S. Nightingale.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Emily S. Nightingale, Email: emily.nightingale@lshtm.ac.uk
CMMID COVID-19 Working Group:
Eleanor M. Rees, Rosalind M. Eggo, Matthew Quaife, Fiona Yueqian Sun, Carl A. B. Pearson, Kiesha Prem, James D. Munday, Sophie R. Meakin, Graham Medley, Kevin van Zandvoort, W. John Edmunds, Alicia Rosello, Sebastian Funk, Kathleen O’Reilly, Billy J. Quilty, Simon R. Procter, Amy Gimma, Adam J. Kucharski, Arminder K. Deol, Jon C. Emery, Nikos I. Bosse, Hamish P. Gibbs, David Simons, Stéphane Hué, Christopher I. Jarvis, Petra Klepac, Yang Liu, Anna M. Foss, Charlie Diamond, C. Julian Villabona-Arenas, Akira Endo, Rein M. G. J. Houben, Stefan Flasche, Timothy W. Russell, Samuel Clifford, Gwenan M. Knight, Joel Hellewell, Nicholas G. Davies, Katherine E. Atkins, Damien C. Tully, Megan Auzenbergs, Mark Jit, and Rachel Lowe
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86266-3.
References
- 1.World Health Organization. Q&A on coronaviruses (COVID-19). Available from: https://www.who.int/news-room/q-a-detail/q-a-coronaviruses.
- 2.A third of the global population is on coronavirus lockdown—here’s our constantly updated list of countries locking down and opening up [cited 2020 May 5]. Available from: https://www.businessinsider.fr/us/countries-on-lockdown-coronavirus-italy-2020-3.
- 3.Coronavirus (COVID-19) cases in the UK [cited 2020 May 5]. Available from: https://coronavirus.data.gov.uk/.
- 4.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potential Coronavirus (COVID-19) symptoms reported through NHS Pathways and 111 online - NHS Digital [Internet]. [cited 2020 May 5]. Available from: https://digital.nhs.uk/data-and-information/publications/statistical/mi-potential-covid-19-symptoms-reported-through-nhs-pathways-and-111-online/latest.
- 6.NHS Pathways - NHS Digital. NHS Digital [Internet]. [cited 2020 May 13]. Available from: https://digital.nhs.uk/services/nhs-pathways.
- 7.Clinical Commissioning Group to NHS England (Region, Local Office) and NHS England (Region) (April 2019) Lookup in England | Open Geography portal [cited 2020 May 5]. Available from: http://geoportal.statistics.gov.uk/datasets/clinical-commissioning-group-to-nhs-england-region-local-office-and-nhs-england-region-april-2019-lookup-in-england?page=6.
- 8.McCullagh, P., Nelder, J. A. Generalized Linear Models, Second Edition. (CRC Press, 1989).
- 9.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. R. Soc. B Biol. Sci. 2007;274(1609):599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jombart, T., Cori, A. Epitrix: Small helpers and tricks for epidemics analysis (2019). Available from: https://CRAN.R-project.org/package=epitrix.
- 11.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int. J. Infect. Dis. 2020;93:284–286. doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NHS UK. COVID-19 Daily Deaths. [cited 2020 May 13]. Available from: https://www.england.nhs.uk/statistics/statistical-work-areas/covid-19-daily-deaths/.
- 13.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2020). Available from: https://www.R-project.org/.
- 14.Rees, E. M., Nightingale, E. S., Jafari, Y., Waterlow, N. R., Clifford, S., Pearson, C. A., Jombart, T., Procter, S. R., Knight, G. M., CMMID Working Group. COVID-19 length of hospital stay: a systematic review and data synthesis. BMC Med. 18, 270 (2020). [DOI] [PMC free article] [PubMed]
- 15.Abbott, S., Hellewell, J., Thompson, R. N., Sherratt, K., Gibbs, H. P., Bosse, N. I., Munday, J. D., Meakin, S., Doughty, E. L., Chun, J. Y., Chan, Y. W. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts [version 1; peer review: awaiting peer review]. Wellcome Open Research (2020). 10.12688/wellcomeopenres.16006.1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code used in this work can be accessed at: https://github.com/qleclerc/nhs_pathways_report.