Abstract
Background
Bladder urothelial carcinoma (BLCA) is one of the most frequently appearing, lethal and aggressive malignancies of the genitourinary system with growing morbidity and mortality, which affects human health seriously. Protein glycosylation, catalyzed by specific glycosyltransferase, has been found to be abnormal in several diseases, especially cancer. Fucosyltransferase VII (FUT7), one of the fucosyltransferases, was observed abnormally expressed in various cancers, however, the role of FUT7 in BLCA, and the association between its expression and clinical outcomes or immune infiltration remains unclear.
Methodology
FUT7 expression in BLCA was analyzed in Oncomine database, which was further confirmed with immunohistochemistry and ELISA. The prognostic value of FUT7 for BLCA was evaluated with PrognoScan database, and its genetic alteration was examined in cBioPortal database. The proliferation, migration, invasion and epithelial–mesenchymal transition (EMT) changes of bladder cancer cells after FUT7 siRNA or cDNA transfection were determined by CCK8, colony formation, transwell and Western blot, respectively. The correlation between FUT7 expression and immune infiltration levels was analyzed in TIMER and TISIDB databases, and the methylation level of FUT7 was detected in UALCAN database.
Results
The results showed that the expression of FUT7 was increased in BLCA, and patients with high FUT7 level were predicted to have lower overall survival and disease-specific survival rates, which were not influenced by FUT7 genetic alterations. Downregulation FUT7 inhibited the proliferation, migration, invasion and EMT of bladder cancer cells, whereas upregulation of FUT7 showed the opposite effects. We found that FUT7 was positively correlated with immune cell infiltration levels (CD8+ T cells, CD4+T cells, macrophage, neutrophil and dendritic cells), and also the expression of gene markers of immune cells. The negative correlation between FUT7 expression and FUT7 methylation level was observed, among which FUT7 expression was positively correlated with the abundance of 28 kinds of tumor-infiltrating lymphocytes (TILs), while FUT7 methylation level was negatively correlated with TILs.
Conclusion
Altogether, these findings suggested that FUT7 possessed the potential to serve as a detection biomarker or immunotherapeutic target for BLCA.
Keywords: bladder urothelial carcinoma, FUT7, EMT, diagnosis, prognosis, immune infiltration
Introduction
Bladder urothelial carcinoma (BLCA), one of the most frequently appearing, lethal and aggressive malignancies of the genitourinary system, displays growing morbidity and mortality each year and ranks the 7th leading cause of cancer-related deaths.1–3 It can be classified into two major categories: non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC), of which 75% of the patients represent as NMIBC and 25% represent as MIBC.4,5 Patients with NMIBC have a high recurrence rate which is 50%-70%, and their 5-year survival rate is more than 90%.6 After progressing to MIBC, the 5-year survival rate of the patients will drop to less than 50%.7,8 Despite the improvements in multimode therapeutics and surgical techniques, the 5-year survival rate for BLCA patients has not been promoted significantly due to the lack of sensitive biomarkers, obvious clinical symptoms at the early stage and high recurrence rate.9–11 Therefore, it is imperative to explore the underlying mechanism responsible for cancer progression, as well as novel effective biomarkers for the diagnosis and prognosis prediction, and also therapeutic targets for BLCA to ameliorate patients’ survival rate.
Protein glycosylation, a critical and the most common post-translational modification form of proteins that involves in various physiological and pathological processes,12,13 is an enzymatic reaction catalyzed by specific glycosyltransferases.14,15 Abnormal glycosylation has been found to be closely associated with tumor proliferation, metastasis, and angiogenesis.16–18 Fucosylation, including N-fucosylation and O-fucosylation, is one major type of protein glycosylation and regulates by specific fucosyltransferases. The glycobiology of cancer indicates that fucosylation, a characteristic alteration of cancer, participates in cancer proliferation and metastasis.19 Until now, 13 fucosyltransferases (FUTs) have been identified, and they can be divided into four subfamilies according to the glycosidic bonds, namely α1,2-, α1,3/4-, α1,6- and protein O-fucosyltransferase. Fucosyltransferase VII (FUT7) belongs to the α1,3/4-fucosyltransferase family, which includes eight members and catalyzes the final step in the synthesis of fucosylated glycoconjugates.20,21 Accumulating evidence demonstrated that the expression of FUT7 was elevated in kinds of cancers, such as hepatocellular carcinoma,22 and lung cancer,23,24 etc. It is one of the contributors to the malignant phenotype and plays functional roles in cancer development and progression. For instance, Qin et al found that FUT7 expression was upregulated in follicular thyroid carcinoma (FTC), and it promoted the migration and invasion of FTC cells through activating MAPK and PI3K/Akt signaling pathways.25 Liang et al reported that FUT7 expression was elevated in A549 cells, and it promoted cell proliferation via triggering EGFR/AKT/mTOR signaling pathway.24 Based on the research and the analysis of online databases, we found FUT7 expression was elevated in BLCA, that was why we speculated the abnormally expressed FUT7 might be a latent contributor to BLCA. Here, we aimed to explore the role of FUT7 in BLCA, as well as its association between FUT7 and immune infiltration levels as the underlying mechanism leading to cancer progression.
To identify novel biomarkers with diagnostic and prognostic values and also therapeutic targets for BLCA, we performed the bioinformatics analysis of FUT7 in multiple online databases. We explored the expression profile of FUT7 and evaluated its prognostic value for BLCA. And then investigated the effects of FUT7 on the proliferation, migration, invasion and EMT of bladder cancer cells. At last, we investigated DNA methylation level of FUT7 in BLCA, as well as the association between FUT7 and immune infiltration levels to elucidate the potential mechanism of FUT7 in bladder cancer progression.
Materials and Methods
Ethics Statement
This study was approved by the Clinical Ethics Review Board of the First Affiliated Hospital of Dalian Medical University according to the guidelines of the Declaration of Helsinki. Clinical samples were obtained, and the informed consent forms were acquired from all participants. The clinical investigation was carried out in accordance with the humane and ethical research principles of the First Affiliated Hospital of Dalian Medical University.
Clinical Samples
The serum samples from 45 patients diagnosed with bladder cancer and 44 healthy controls from January 2018 to July 2019 were collected from the First Affiliated Hospital of Dalian Medical University. The samples were got after written consent was obtained from each patient, and stored at −80°C before further analysis. Paraffin-embedded tissue samples were obtained from 30 patients diagnosed with bladder cancer from January 2018 to July 2019 at the First Affiliated Hospital of Dalian Medical University. Informed consent was obtained from each patient. The use of clinical samples was approved by the Clinical Ethics Review Board of the First Affiliated Hospital of Dalian Medical University.
Expression and Prognostic Analysis of FUT7
Oncomine (https://www.oncomine.org/resource/login.html) is an online tumor microarray database compiled 86,733 samples and 715 gene expression, which is used to analyze gene expression profiling for various cancers. FUT7 expression in bladder cancer was analyzed by searching “FUT7”, and selected the catalog “Bladder Cancer vs Normal Analysis” under the module of “Cancer vs Normal Analysis”. The threshold was set as “P-value<0.05”. PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/) is a powerful platform that includes a wide range of publicly available tumor microarray datasets with corresponding clinical information, which is widely used to explore survival information of interested genes in cancers, such as overall survival (OS), relapse-free survival (RFS), disease-specific survival (DSS), and distant metastasis-free survival (DMFS). The prognostic value of FUT7 for BLCA was estimated with PrognoScan database. OS and DSS were assessed by entering gene identifier “FUT7”, and chose cohort “Aarhus (1995–2004)” with probe “217696_at” for OS and “CNUH” with probe “ILMN_1715286” for DSS.
Immunohistochemistry
The tissue sections were deparaffinized in xylol and rehydrated in descending concentrations of ethanol following with antigen retrieve in citrate buffer (pH6.0). Endogenous peroxidases were removed with 0.3% H2O2, and non-specific binding sites were blocked with complete serum. Then, the sections were incubated with primary antibody (FUT7: 1:100, Proteintech, Wuhan, China) at 4°C overnight. The second antibody was used after washing with PBS. The sections were visualized with diaminobenzidine (DAB) and counterstained with hematoxylin. Images were taken under an inverted microscope (Olympus, Tokyo, Japan), and yellowish-brown staining indicated a positive result. The staining intensity was evaluated in comparison with normal tissues, and scored as follows: 0: no staining; 1: weak staining; 2: moderate staining; 3: strong staining.
Enzyme-Linked Immunosorbent Assay
FUT7 (Langton, Shanghai, China) level in serum of bladder cancer patients was determined by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions. The samples (50µL) and biotinylated human FUT7 antibody (50µL) were incubated at 37°C for 1h. The plate was washed for five times, and 50µL streptavidin-horseradish peroxidase conjugates were added for 1h. The absorbance value at 450nm (OD450nm) was detected and recorded after washing and colored. The concentration of FUT7 in serum was calculated according to the standard curve with curve expert software.
Genetic Alteration Analysis
The cBioPortal database (https://www.cbioportal.org/) is a useful web-based on TCGA database which provides visualization, analysis, and downloads of large-scale tumor genome data for multiple kinds of cancer. It was used to investigate the genetic alterations of FUT7 with “OncoPrint module”, and the prognostic value of FUT7 alteration with “Comparison/Survival module” in bladder cancer (TCGA, Cell 2017, 413 samples). The correlation between FUT7 expression and gene marker levels of immune cells was evaluated in cBioPortal database with “Co-expression module”.
Cell Culture
Human bladder cancer cell lines, T24 and 5637, were purchased from the American Culture Collection (ATCC, Manassas, VA, USA), and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Invitrogen, Grand Island, NY, USA), 100U/mL penicillin and 100µg/mL streptomycin at 37°C with 5% CO2. The culture medium was renewed every 2–3 days.
Transfection
T24 and 5637 cells were plated into 6-well plates and transfected with FUT7 siRNA (50nM, Genepharma, Shanghai, China) and FUT7 cDNA (4µg, Genepharma, Shanghai, China) with lipofectamine 2000 reagent (Invitrogen, Grand Island, NY, USA) when cell density reached 70% confluence. The culture medium was renewed after 6h transfection. The cell protein and RNA samples were collected after 48h of transfection for further analysis. FUT7 siRNA: sense: 5ʹ-GCGCGACUCGG ACAUCUUUTT-3ʹ, anti-sense: 5ʹ-AAAGAUGUCC GAGUCGCGCTT-3ʹ.
Cell Proliferation Assay
The alteration of cell proliferation rate was determined by Cell Counting Kit-8 (CCK8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Cells were seeded into 96-well plates with a density of 3000 cells/well. After transfection, CCK8 solution was added into FBS-free culture medium (1:9) and incubated at 37°C for 2h. Absorbance at 450nm (OD450nm) was detected by using a microplate reader, and data were recorded and analyzed. The experiments were carried out in triplicate for each group.
Colony Formation Assay
Cells were seeded into 6-well plates with a density of 1000 cells/well after FUT7 siRNA or cDNA transfection. After culturing for two weeks, the surviving colonies were fixed with methanol at room temperature for 20min, following with crystal violet staining for 20min. Images were taken under an inverted microscope after washing with PBS for three times. Colonies that contained more than 50 cells were counted, and data were recorded and analyzed.
Transwell Assay
For migration assay, the inserts (8µm, Costar, Cambridge, MA) were used without matrigel. For invasion assay, the inserts were coated with 50µL matrigel (1:9, BD Bioscience, San Jose, CA, USA). Cells were harvested and placed into the upper chamber with 1×105 cells in 0.2mL serum-free medium, and 800µL RPMI-1640 with 10% FBS was added into the lower chamber. After 24h incubation, the inserts were fixed with methanol and stained with crystal violet for 20min at room temperature. The images were captured under an inverted microscope, and the cell number was counted in ten random fields. The representative images and statistical analysis were shown. The experiments were conducted in triplicate for each group.
Western Blot
Cell lysates were prepared with RIPA lysis buffer containing protease inhibitor. The protein concentration was measured with the BCA kit (Beyotime Biotechnology, Beijing, China), and separated by 12% SDS-PAGE gel. After transferring onto nitrocellulose membrane (Millipore, Billerica, MA, USA), 5% non-fat dry milk was used for blocking for 2h at room temperature. Then, the membranes were incubated at 4°C with primary antibodies: FUT7 (1:1000, Proteintech, Wuhan, China), E-cadherin (1:1000, Proteintech, Wuhan, China), N-cadherin (1:1000, Proteintech, Wuhan, China), GAPDH (1:4000, Proteintech, Wuhan, China). After washing with TBST for three times, the membranes were incubated with horseradish peroxidase-conjugated second antibody (1:4000, ZSGB-BIO, Beijing, China) for 45min. The protein bands were visualized with ECL detection system and quantified by using Image J software (NIH, Bethesda, MD, USA).
Immune Infiltration Analysis
TIMER database (https://cistrome.shinyapps.io/timer/) is an effective web interface designed to explore the abundance of tumor-infiltrating immune cells in cancers, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. There are six subsets of tumor-immune interactions which are being pre-calculated in 32 types of cancer and TCGA data. The relationship between FUT7 expression and immune infiltration levels was analyzed in TIMER with “Gene module”. The relationship between somatic copy number changes of FUT7 and immune infiltration level was analyzed with “SCNA module”. The correlation between FUT7 level and expression of immune cell gene markers was analyzed with “Correlation module”.
Promoter Methylation Level of FUT7 Analysis
UALCAN database (http://ualcan.path.uab.edu/analysis.html) is an interactive web resource, which provides comprehensive transcriptome data of cancer from TCGA (31 types of cancer). The methylation level of FUT7 in bladder cancer, as well as its correlation with clinicopathological features were evaluated in UALCAN by entering gene symbol “FUT7” with TCGA dataset for “bladder urothelial carcinoma”. The correlation between FUT7 methylation level and FUT7 expression was analyzed with cBioPortal database.
Tumor Immune-Interaction Analysis
TISIDB database (http://cis.hku.hk/TISIDB/index.php) is a useful and integrated repository web portal that contains 4176 records from 2530 publications and 988 genes correlated to oncoimmunology, which is used to investigate the functions and roles of interested genes in tumor-immune interplay via high-throughput data analysis and literature mining. The correlations between FUT7 expression or FUT7 methylation level with the abundance of tumor-infiltrating lymphocytes were analyzed by using TISIDB database with “Lymphocyte module”.
Statistical Analysis
All of the data got from three independent experiments were expressed as mean ± S.E.M, and the differences between groups were analyzed with Student’s t-test by using SPSS 22.0 (SPSS Inc, Chicago, IL). The receiver operating characteristic curve (ROC) was conducted to evaluate the diagnostic value of FUT7 for bladder cancer. P<0.05 was considered as statistical significance.
Results
The Expression and Prognostic Value of FUT7 for BLCA
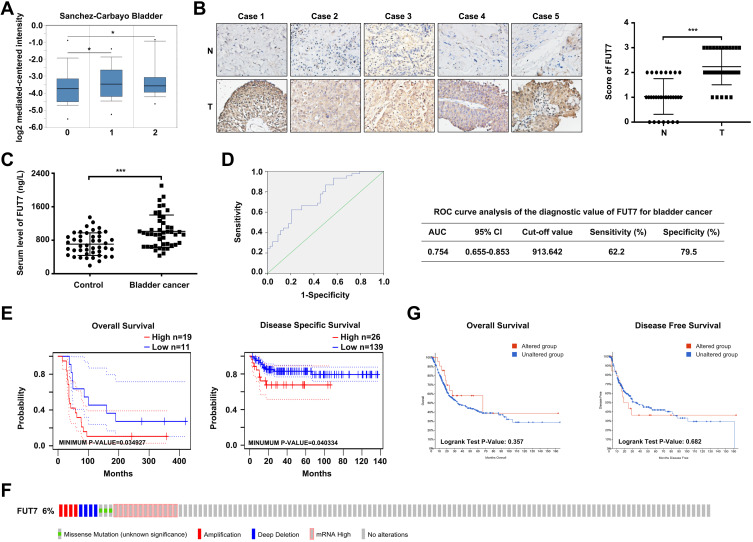
To explore the role of FUT7 in BLCA, we first analyzed its expression profile in Oncomine database. The result showed that FUT7 expression was higher in bladder cancer tissues than in normal tissues (Figure 1A). We further collected clinical bladder cancer tissues and serum samples to confirm its upregulation. The immunohistochemical staining results indicated that FUT7 expression was increased in bladder cancer tissues compared to normal bladder tissues (Figure 1B). Also, we used ELISA to detect serum level alteration of FUT7, which found that FUT7 was significantly elevated in bladder cancer patients (Figure 1C). ROC curve analysis indicated FUT7 could be served as a potential biomarker for bladder cancer detection with the area under the curve (AUC) 0.754, sensitivity 62.2% and specificity 79.5% (Figure 1D). In addition, we investigated the link between FUT7 expression and the outcome of bladder cancer patients by using PrognoScan database. It was found that FUT7 was closely associated with patients’ prognosis. Bladder cancer patients with low FUT7 level indicated better prognosis than patients with high FUT7 level, including overall survival and disease-specific survival (Figure 1E). We then used the cBioPortal database to detect the types and frequencies of FUT7 genetic alterations in BLCA (TCGA, Cell 2017). As shown, FUT7 was altered in 24 out of 413 (6%) BLCA patients. These alterations included missense mutation, amplification, deep deletion, and mRNA high, among which mRNA high was the major modification (Figure 1F). However, the genetic alterations of FUT7 showed no significant influence on overall survival and disease-free survival (Figure 1G) of BLCA patients. These findings demonstrated that FUT7 expression was increased in bladder cancer, and significantly correlated with poorer outcome of BLCA patients.
Figure 1.
The expression and prognostic value of FUT7 in BLCA. (A) FUT7 expression in bladder cancer was analyzed by Oncomine database. 0: No value (n=48); 1: Superficial bladder cancer (n=28); 2: Infiltrating bladder urothelial carcinoma (n=81). (B) Immunohistochemical staining of FUT7 in bladder cancer tissues and normal bladder tissues. The representative images and statistical analysis were shown. (C) ELISA assay of FUT7 level in serum samples from patients with bladder cancer and healthy controls. (D) ROC curve analysis of the diagnostic value of FUT7 for bladder cancer. (E) Overall survival curve of FUT7 for probe 217696_at in bladder cancer cohort (GSE5287) and disease-specific survival curve of FUT7 for probe ILMN_1715286 in bladder cancer cohort (GSE13507) from PrognoScan database. (F) Analysis of the genetic alteration frequency of FUT7 in BLCA by cBioPortal, including missense mutation, amplification, deep deletion and mRNA high. (G) The overall survival curve and disease-free survival curve of FUT7 for BLCA in FUT7-altered and unaltered group. *P<0.05, ***P<0.001.
The Effects of FUT7 on Cell Proliferation, Migration, Invasion, and EMT of Bladder Cancer
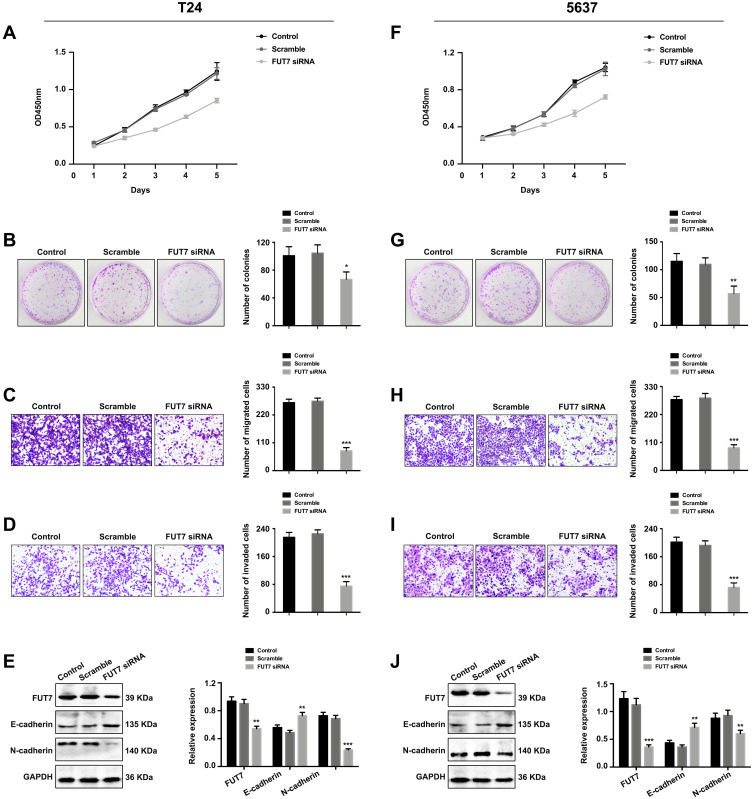
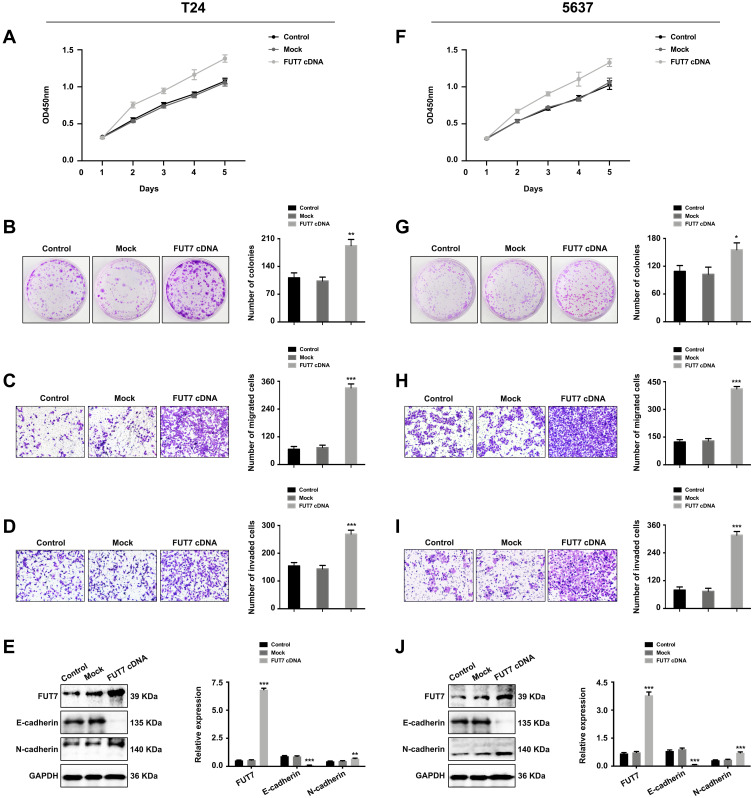
After confirming the expression and prognostic value of FUT7 for bladder cancer, we further investigated the effects of FUT7 on cell proliferation, migration, invasion, and EMT of bladder cancer cells. FUT7 siRNA and cDNA were transfected into T24 and 5637 bladder cancer cell lines. CCK8 assay and colony formation assay were applied to evaluate the effect of FUT7 on cell proliferation, which showed that downregulation of FUT7 with FUT7 siRNA inhibited cell proliferation (Figure 2A, B, F, and G), while upregulation of FUT7 with FUT7 cDNA promoted cell proliferation with increased proliferation rate and colony number (Figure 3A, B, F, and G). When compared with scramble or mock group, bladder cancer cells treated with FUT7 siRNA demonstrated decreased migration (Figure 2C and H) and invasion abilities (Figure 2D and I), while cells treated with FUT7 cDNA showed increased migration (Figure 3C and H) and invasion capabilities (Figure 3D and I). The expression changes of EMT-related markers were analyzed with Western blot after FUT7 siRNA and cDNA transfection. In FUT7 siRNA group, E-cadherin expression was upregulated while N-cadherin expression was downregulated (Figure 2E and J). And in FUT7 cDNA group, the expression changes of EMT markers were opposite (Figure 3E and J). These results illustrated that FUT7 siRNA inhibited cell proliferation, migration, invasion and EMT, while FUT7 cDNA represented the opposite effects, which indicated its active roles on cell functions of bladder cancer.
Figure 2.
Downregulation of FUT7 inhibited the proliferation, migration, invasion and EMT of bladder cancer cells. (A and F) CCK8 assay of cell proliferation rate after FUT7 siRNA transfection. (B and G) Colony formation assay of cell proliferation ability after transfection with FUT7 siRNA. (C and H) Transwell assay of cell migration ability after treated with FUT7 siRNA. (D and I) Transwell assay of cell invasion capability after FUT7 siRNA transfection. (E and J) Western blot analysis of the expression changes of EMT-related markers after FUT7 siRNA transfection. The statistical analysis was shown. *P<0.05, **P<0.01, ***P<0.001.
Figure 3.
Upregulation of FUT7 promoted the proliferation, migration, invasion and EMT of bladder cancer cells. (A and F) CCK8 assay of cell proliferation rate after FUT7 cDNA transfection. (B and G) Colony formation assay of cell proliferation ability after transfection with FUT7 cDNA. (C and H) Transwell assay of cell migration ability after treated with FUT7 cDNA. (D and I) Transwell assay of cell invasion capability after FUT7 cDNA transfection. (E and J) Western blot analysis of the expression of EMT-related markers after FUT7 cDNA transfection. The statistical analysis was shown. *P<0.05, **P<0.01, ***P<0.001.
Association Between FUT7 Expression and Immune Cell Infiltration Levels
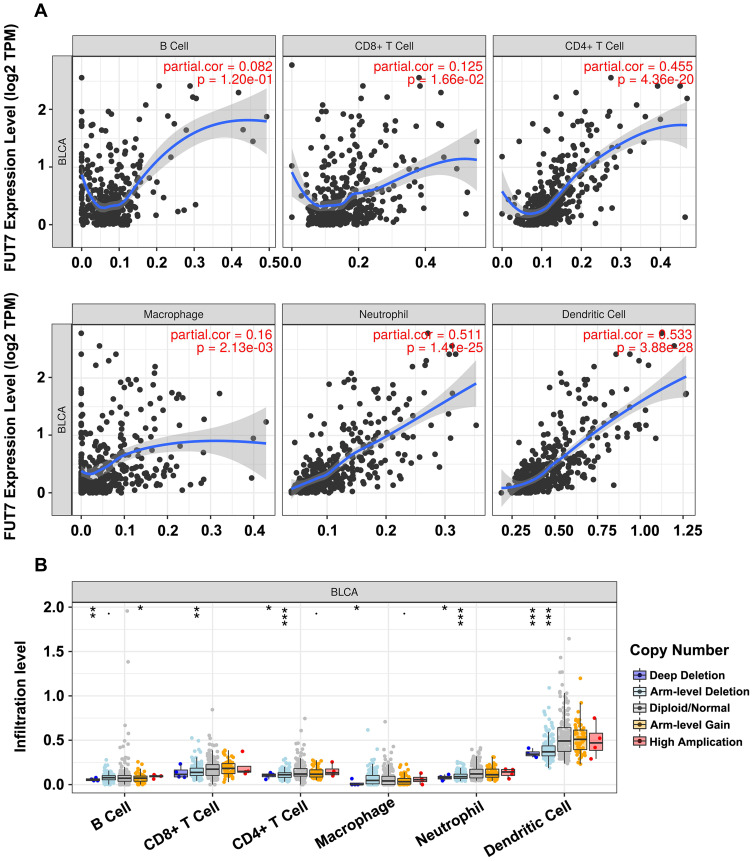
TIMER database was conducted to illustrate the relationship between FUT7 expression and infiltration levels of immune cells in BLCA. The results revealed that high FUT7 expression was closely correlated with dominant immune cell infiltration, including CD8+ T cells (cor=0.125, p=1.66e-02), CD4+ T cells (cor=0.455, p=4.36e-20), macrophages (cor=0.16, p=2.13e-03), neutrophils (cor=0.511, p=1.41e-25) and dendritic cells (cor=0.533, p=3.88e-28) (Figure 4A). We also investigated the relationships between somatic copy number alterations (SCNA) of FUT7 and the abundance of immune infiltration, which showed its significant correlations with infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells (Figure 4B). After observing the close relationship between FUT7 expression and immune infiltration, we further investigated its correlations with gene marker levels of diverse immune cells (Table 1). Specifically, the analysis results showed that FUT7 expression was significantly positive correlated with immune cell gene marker expression, such as B cell markers (BLK: r=0.587, p=5.15e-39; CD19: r=0.564, p=1.74e-35; CD79A: r=0.644, p=3.93e-49); T cell markers (CD3D: r=0.795, p=4.16e-90; CD3E: r=0.844, p=2.06e-111; CD3G: r=0.71, p=9.41e-64; CD2: r=0.86, p=1.35e-120); CD4+ T cell marker (CD4: r=0.796, p=2.03e-90); CD8+ T cell markers (CD8A: r=0.726, p=7.12e-68; CD8B: r=0.567, p=5.13e-36; TBX21: r=0.772, p=1.24e-81; IFNG: r=0.651, p=1.86e-50; CXCL9: r=0.723, p=3.66e-67; CXCL10: r=0.65, p=3.22e-50); Th1 cell markers (TNF: r=0.474, p=3.89e-24; STAT4: r=0.74, p=8.25e-72; STAT1: r=0.556, p=2.18e-34); Th2 cell markers (STAT5A: r=0.43, p=9.41e-20; IL13: r=0.369, p=1.48e-14); Th17 cell markers (IL17A: r=0.187, p=1.50e-04; STAT3: r=0.359, p=7.64e-14); TAM markers (CCL2: r=0.565, p=1.24e-35; CD68: r=0.537, p=1.03e-31; IL10: r=0.596, p=2.02e-40; CSF2: r=0.382, p=1.31e-15); M1 macrophage cell marker (NOS2: r=0.22, p=7.231e-06); M2 macrophage cell markers (CD163: r=0.705, p=2.10e-62; VSIG4: r=0.697, p=1.46e-60; MS4A4A4: r=0.717, p=2.19e-65); NK cell markers (KIR2DL1: r=0.392, p=2.27e-16; KIR2DL3: r=0.473, p=4.24e-24; KIR2DL4: r=0.55, p=1.54e-33; KIR3DL1: r=0.447, p=2.18e-21; KIR3DL2: r=0.46, p=1.16e-22; KIR3DL3: r=0.25, p=3.24e-07); neutrophil cell markers (CCR7: r=0.209, p=2.228e-05; ITAGM: r=0.748, p=5.58e-74). These data highlighted that elevated FUT7 was positively associated with immune infiltration levels, as well as immune exhaustion, and FUT7 might serve as one potential immunotherapeutic target for BLCA treatment.
Figure 4.
Correlations between the expression of FUT7 and immune cell infiltration levels. (A) The correlations between FUT7 expression and infiltrating levels of immune cells, including B cells (p=1.20e-01), CD8+ T cells (p=1.66e-02), CD4+ T cells (p=4.36e-20), macrophage (p=2.13e-03), neutrophil (p=1.41e-25), and dendritic cell (p=3.88e-28). (B) The comparison of tumor infiltrating levels in BLCA with different somatic copy number alterations for FUT7 from TIMER database. *P<0.05, **P<0.01, ***P<0.001.
Table 1.
Correlation Analysis Between FUT7 Expression and Related-Makers of Immune Cells
| Cell Name | Immune Cell Marker | Spearman Correlation | P-value |
|---|---|---|---|
| B cell | BLK | 0.587 | 5.15e-39*** |
| CD19 | 0.564 | 1.74e-35*** | |
| CD79A | 0.644 | 3.93e-49*** | |
| T cell | CD3D | 0.795 | 4.16e-90*** |
| CD3E | 0.844 | 2.06e-111*** | |
| CD3G | 0.71 | 9.41e-64*** | |
| CD2 | 0.86 | 1.35e-120*** | |
| CD4+ T cell | CD4 | 0.796 | 2.03e-90*** |
| CD8+ T cell | CD8A | 0.726 | 7.12e-68*** |
| CD8B | 0.567 | 5.13e-36*** | |
| TBX21 | 0.772 | 1.24e-81*** | |
| IFNG | 0.651 | 1.86e-50*** | |
| CXCL9 | 0.723 | 3.66e-67*** | |
| CXCL10 | 0.65 | 3.22e-50*** | |
| Th1 | TNF | 0.474 | 3.89e-24*** |
| STAT4 | 0.74 | 8.25e-72*** | |
| STAT1 | 0.556 | 2.18e-34*** | |
| Th2 | STAT5A | 0.43 | 9.41e-20*** |
| IL13 | 0.369 | 1.48e-14*** | |
| Th17 | IL17A | 0.187 | 1.50e-04*** |
| STAT3 | 0.359 | 7.64e-14*** | |
| TAMs | CCL2 | 0.565 | 1.24e-35*** |
| CD68 | 0.537 | 1.03e-31*** | |
| IL10 | 0.596 | 2.02e-40*** | |
| CSF2 | 0.382 | 1.31e-15*** | |
| M1 macrophage | NOS2 | 0.22 | 7.231e-06*** |
| M2 macrophage | CD163 | 0.705 | 2.10e-62*** |
| VSIG4 | 0.697 | 1.46e-60*** | |
| MS4A4A | 0.717 | 2.19e-65*** | |
| NK | KIR2DL1 | 0.392 | 2.27e-16*** |
| KIR2DL3 | 0.473 | 4.24e-24*** | |
| KIR2DL4 | 0.55 | 1.54e-33*** | |
| KIR3DL1 | 0.447 | 2.18e-21*** | |
| KIR3DL2 | 0.46 | 1.16e-22*** | |
| KIR3DL3 | 0.25 | 3.24e-07*** | |
| Neutrophil | CCR7 | 0.209 | 2.23e-05*** |
| ITGAM | 0.748 | 5.58e-74*** |
Note: ***P<0.001.
The Promoter Methylation Level of FUT7 in BLCA
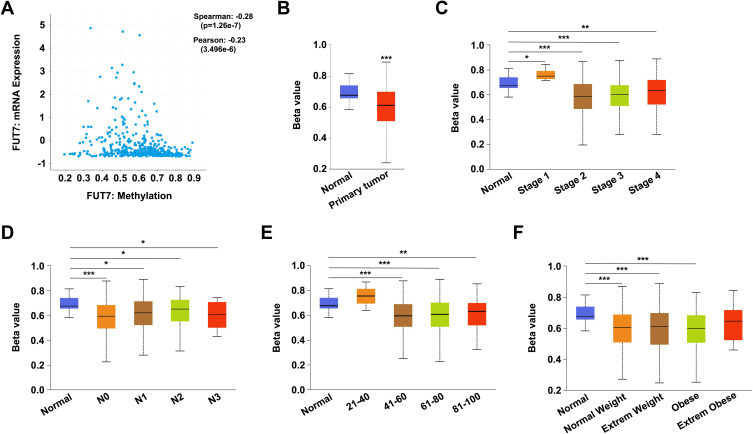
To explore the potential mechanism of FUT7 involving in the carcinogenesis of BLCA, we examined the methylation level of FUT7 by using cBioPortal and UALCAN databases. As shown, the methylation level of FUT7 was negatively correlated with FUT7 expression in bladder cancer (Spearman: −0.28, p=1.26e-7; Pearson: −0.23, p=3.496e-6) (Figure 5A). We also detected the relationships between FUT7 methylation level and clinicopathological features of BLCA patients. Compared with normal tissues, the promoter methylation level of FUT7 was decreased in bladder cancer tissues (Figure 5B) and also downregulated in patients with high stage status (stage 2, 3, 4; Figure 5C) and nodal metastasis (Figure 5D). Besides, it was also negatively correlated with patients’ age (Figure 5E) and weight (Figure 5F). Therefore, decreased FUT7 methylation level might be an underlying indicator, which reflected clinical characteristics of BLCA.
Figure 5.
Correlations between promoter methylation level of FUT7 and clinicopathological features of BLCA patients. (A) The negative correlation between the promoter methylation level of FUT7 and the expression of FUT7 in BLCA analyzed by cBioPortal database. (B–F) The relative promoter methylation level of FUT7 was analyzed with UALCAN database based on sample type (B), cancer stage (C), nodal metastasis status (D), patient’s age (E), and weight (F). *P<0.05, **P<0.01, ***P<0.001.
Associations Between FUT7 Expression, Methylation Level and the Abundance of Tumor-Infiltrating Lymphocytes
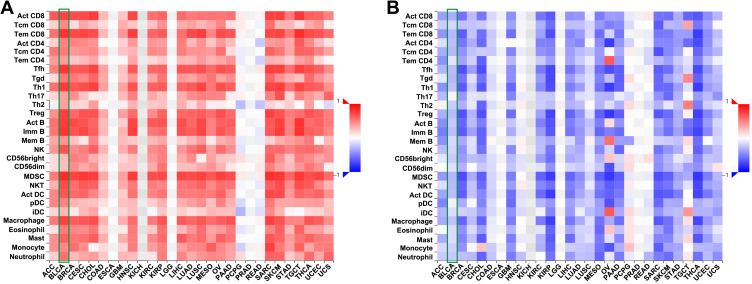
In TISIDB database, we found that FUT7 expression was positively correlated with the abundance of 28 kinds of tumor-infiltrating lymphocytes (Figure 6A), including Act CD8 cells (rho=0.721, p<2.2e-16), Tcm CD8 cells (rho=0.454, p<2.2e-16), Tem CD8 cells (rho=0.801, p<2.2e-16), Act CD4 cells (rho=0.48, p<2.2e-16), Tcm CD4 cells (rho=0.529, p<2.2e-16), Tem CD4 cells (rho=0.388, p=1.49e-16), Tfh cells (rho=0.798, p<2.2e-16), Tgd cells (rho=0.529, p<2.2e-16), Th1 cells (rho=0.778, p<2.2e-16), Th17 cells (rho=0.376, p=3.72e-15), Th2 cells (rho=0.372, p=9.57e-15), Treg cells (rho=0.751, p<2.2e-16), Act B cells (0.771, p<2.2e-16), Imm B cells (rho=0.797, p<2.2e-16), Mem B cells (rho=0.415, p<2.2e-16), NK cells (rho=0.683, p<2.2e-16), CD56bright cells (rho=0.265, p=6.42e-08), CD56dim cells (rho=0.256, p=1.76e-07), MDSC cells (rho=0.836, p<2.2e-16), NKT cells (rho=0.702, p<2.2e-16), Act DC cells (rho=0.668, p<2.2e-16), pDC cells (rho=0.493, p<2.2e-16), iDC cells (rho=0.277, p=1.5e-08), macrophages (rho=0.745, p<2.2e-16), eosinophil (rho=0.513, p<2.2e-16), Mast cells (rho=0.621, p<2.2e-16), monocytes (rho=0.468, p<2.2e-16) and neutrophils (rho=0.549, p<2.2e-16) (Table 2). We further detected the relationship between FUT7 methylation level and the abundance of tumor-infiltrating lymphocytes, and found it was significantly negatively correlated with tumor-infiltrating lymphocytes’ abundance (Figure 6B), including Act CD8 cells (rho=−0.323, p=2.86e-11), Tcm CD8 cells (rho=−0.163, p=0.000948), Tem CD8 cells (rho=−0.276, p=1.57e-08), Act CD4 cells (rho=−0.26, p=1.06e-07), Tcm CD4 cells (rho=−0.283, p=7.18e-09), Tem CD4 cells (rho=−0.107, p=0.0307), Tfh cells (rho=−0.273, p=2.28e-08), Tgd cells (rho=−0.287, p=4.35e-09), Th1 cells (rho=−0.251, p=3.05e-07), Th2 cells (rho=−0.208, p=2.48e-05), Treg cells (rho=−0.274, p=2.23e-08), Act B cells (rho=−0.22, p=7.65e-06), Imm B cells (rho=−0.262, p=9.06e-08), Mem B cells (rho=−0.153, p=0.00193), NK cells (rho=−0.168, p=0.000665), CD56bright cells (rho=−0.213, p=1.49e-05), CD56dim cells (rho=−0.125, p=0.0116), MDSC cells (rho=−0.287, p=4.1e-09), NKT cells (rho=−0.275, p=1.89e-08), Act DC cells (rho=−0.263, p=7.42e-08), iDC cells (rho=−0.132, p=0.00741), macrophages (rho=−0.248, p=4.28e-07), eosinophil (rho=−0.136, p=0.00614), mast cells (rho=−0.139, p=0.00484), monocytes (rho=−0.184, p=0.000196) and neutrohpils (rho=−0.215, p=1.26e-05) (Table 3). These results revealed that the expression of FUT7 was positively, while the methylation level of FUT7 was negatively associated with the abundance of tumor-infiltrating lymphocytes, and it had the potential to serve as one of the immunotherapeutic targets for BLCA treatment.
Figure 6.
The relationships between FUT7 expression, methylation level and the abundance of tumor-infiltrating lymphocytes. (A) Relations between the expression of FUT7 and the abundance of 28 kinds of tumor-infiltrating lymphocytes (TILs) in various cancers. (B) Relations between the methylation level of FUT7 and the abundance of 28 kinds of tumor-infiltrating lymphocytes in various cancers.
Table 2.
The Association Between the Expression of FUT7 and the Abundance of Tumor-Infiltrating Lymphocytes
| Cell Name | Spearman Correlation Test: Rho | P-value |
|---|---|---|
| Act CD8 | 0.721 | <2.2e-16*** |
| Tcm CD8 | 0.454 | <2.2e-16*** |
| Tem CD8 | 0.801 | <2.2e-16*** |
| Act CD4 | 0.48 | <2.2e-16*** |
| Tcm CD4 | 0.529 | <2.2e-16*** |
| Tem CD4 | 0.388 | 1.49e-16*** |
| Tfh | 0.798 | <2.2e-16*** |
| Tgd | 0.529 | <2.2e-16*** |
| Th1 | 0.778 | <2.2e-16*** |
| Th17 | 0.376 | 3.72e-15*** |
| Th2 | 0.372 | 9.57e-15*** |
| Treg | 0.751 | <2.2e-16*** |
| Act B | 0.771 | <2.2e-16*** |
| Imm B | 0.797 | <2.2e-16*** |
| Mem B | 0.415 | <2.2e-16*** |
| NK | 0.683 | <2.2e-16*** |
| CD56bright | 0.265 | 6.42e-08*** |
| CD56dim | 0.256 | 1.76e-07*** |
| MDSC | 0.836 | <2.2e-16*** |
| NKT | 0.702 | <2.2e-16*** |
| Act DC | 0.668 | <2.2e-16*** |
| pDC | 0.493 | <2.2e-16*** |
| iDC | 0.277 | 1.5e-08*** |
| Macrophage | 0.745 | <2.2e-16*** |
| Eosinophil | 0.513 | <2.2e-16*** |
| Mast | 0.621 | <2.2e-16*** |
| Monocyte | 0.468 | <2.2e-16*** |
| Neutrophil | 0.549 | <2.2e-16*** |
Note: ***P<0.001.
Table 3.
The Association Between the Methylation Level of FUT7 and the Abundance of Tumor-Infiltrating Lymphocytes
| Cell Name | Spearman Correlation Test: Rho | P-value |
|---|---|---|
| Act CD8 | −0.323 | 2.86e-11*** |
| Tcm CD8 | −0.163 | 0.000948*** |
| Tem CD8 | −0.276 | 1.57e-08*** |
| Act CD4 | −0.26 | 1.06e-07*** |
| Tcm CD4 | −0.283 | 7.18e-09*** |
| Tem CD4 | −0.107 | 0.0307* |
| Tfh | −0.273 | 2.28e-08*** |
| Tgd | −0.287 | 4.35e-09*** |
| Th1 | −0.251 | 3.05e-07*** |
| Th17 | 0.042 | 0.398 |
| Th2 | −0.208 | 2.48e-05*** |
| Treg | −0.274 | 2.23e-08*** |
| Act B | −0.22 | 7.65e-06*** |
| Imm B | −0.262 | 9.06e-08*** |
| Mem B | −0.153 | 0.00193** |
| NK | −0.168 | 0.000665*** |
| CD56bright | −0.213 | 1.49e-05*** |
| CD56dim | −0.125 | 0.0116* |
| MDSC | −0.287 | 4.1e-09*** |
| NKT | −0.275 | 1.89e-08*** |
| Act DC | −0.263 | 7.42e-08*** |
| pDC | −0.049 | 0.326 |
| iDC | −0.132 | 0.00741** |
| Macrophage | −0.248 | 4.28e-07*** |
| Eosinophil | −0.136 | 0.00614** |
| Mast | −0.139 | 0.00484** |
| Monocyte | −0.184 | 0.000196*** |
| Neutrophil | −0.215 | 1.26e-05*** |
Notes: *P<0.05; **P<0.01; ***P<0.001.
Discussion
Bladder cancer is one of the leading causes of cancer death worldwide, which can occur at any age. So there is a pressing demand to search for the sensitive and specific biomarkers of BLCA to improve the diagnosis, treatment, and prognosis evaluation. Glycosylation, a key regulatory mechanism, is involved in kinds of cancer-related processes.26,27 Abnormal glycosylation is a common phenomenon that has been observed in cancer, and it can determine cancer stage, grade, and the fate of cancer progression.28–30 It is a contributor to the occurrence and development of cancers, including bladder cancer. Deb et al carried out LC-MS/MS-based quantitative proteomic and glycoproteomic profiling analysis of glycosite-containing peptides and found aberrant protein glycosylation would provide additional avenues to employ glycan-based therapies and may lead to the identification of novel therapeutic targets.31 Li et al found that the glycotransferase GALNT1 was highly expressed and correlated with clinicopathological features of bladder cancer, and its mediated O-linked glycosylation was essential for the self-renewal maintenance of BCSCs and bladder tumorigenesis.32 Wahby et al reported that the expression of poFUT1 was upregulated in bladder cancer, and patients with low poFUT1 level predicted poor overall survival, cancer-specific survival, and disease-free survival, which demonstrated the importance of altered glycosylation for the progress of bladder cancer.33 FUT7 is a member of the fucosyltransferase family that is known to play a regulatory role in multivariate physiological and pathological processes, such as the formation of endometrial receptivity,34 embryo implantation,35 as well as cancer development and metastasis.36 The overexpression of FUT7 was found in follicular thyroid carcinoma,25 acute myeloid leukemia,37 and hepatocarcinoma,22 etc. To determine the role of FUT7 in BLCA, we first evaluated its expression profile and prognostic value by using Oncomine and PrognoScan databases. It was found that FUT7 expression was increased both in infiltrating bladder urothelial carcinoma and superficial bladder cancer, which was further confirmed with immunohistochemical staining and ELISA assay. Then, we explored the association of FUT7 expression and the survival rate of BLCA patients. The results showed that lower FUT7 level was correlated with better overall survival rate and disease-free survival rate in bladder cancer. As the progression of cancer is a series of histopathological processes influenced by genetic or proteomic alterations, we utilized cBioPortal web tool to check its genetic alterations, mutations, and survival rate changes in BLCA. Several genetic alterations of FUT7 were observed, including missense mutation, amplification, deep deletion, and mRNA high. These alterations might have played critical roles in BLCA progression. However, the alterations represented no impact on the survival of BLCA patients (Figure 1). After confirming the diagnostic and prognostic value of FUT7 for BLCA, we further explored the effects of FUT7 on bladder cancer cell functions. Metastasis is the leading cause of cancer death globally.38 It is a complicated pathological process that involves multiple steps, including epithelial to mesenchymal transition, in which cancer cells transit between epithelial and mesenchymal states that facilitating cell dissemination.39 Our results demonstrated that FUT7 siRNA inhibited the migration, invasion, and EMT of bladder cancer cells, while FUT7 cDNA promoted these cell behaviors (Figures 2 and 3). Overall, our study provided novel and valuable insights into FUT7 as a diagnostic biomarker, prognosis predictor, and therapeutic target for BLCA.
The enhanced inflammatory milieu is reported to be closely related to tumor progression and prognosis, which is recognized as one of the hallmarks in the pathogenesis of malignancies.40 The inflammatory tumor microenvironment eventually leads to tumor progression and metastasis.41–43 To investigate the role of tumor milieu with FUT7, we analyzed the relationship between FUT7 expression and immune cell infiltration. Growing evidence demonstrated that the infiltration and activation of immune cells were believed to be closely associated with tumor processes, such as breast cancer,44 lung cancer,45 uterine corpus endometrial carcinoma,46 and bladder cancer.47 Nowadays, more researchers began to pay attention to the role of immune infiltration in cancers. Wang et al found that elevated expression of CtBP1 correlated positively with tumor-associated macrophage infiltration into non-small cell lung cancer (NSCLC) tissues, induced EMT and modulated the activated NF-κB signaling pathway leading to the increase in CCL2 secretion to promote tumor-associated macrophages recruitment and polarization. These findings showed a novel insight into the process of CtBP1-regulated tumor-associated macrophages infiltration in NSCLC.48 Tekin et al reported that PAR1 expression was correlated with macrophage infiltration in primary pancreatic ductal adenocarcinoma (PDAC), which macrophages induced mesenchymal transition through PAR1 activation. This macrophage-induced mesenchymal transition supported the tumor-promoting role of macrophage influx, explaining the dichotomous contributions of these immune cells to tumor growth.49 Thus, we hypothesized that the correlation between FUT7 expression and immune infiltration might act as a potential contributor to BLCA. Pearson analysis was conducted between FUT7 and immune cell infiltration in both TIMER and cBioPortal databases. We found that FUT7 expression was positively correlated with tumor heterogeneity regarding the infiltration levels of immune cells, particularly with CD8+ T cells, CD4+ T cells, B cells, Th1, Th2, macrophages, neutrophils, dendritic cells, NK, and MDSC. Moreover, it was also positively correlated with most gene markers of immune cells significantly (Table 1). These findings suggested that FUT7 exerted its functional roles in the recruitment and regulation of immune cell infiltration in BLCA microenvironment (Figures 4 and 6, Table 2). To further elucidate the underlying mechanism of FUT7 in BLCA, we examined the methylation level of FUT7, which was a hallmark of cancer and had been recognized as an essential biological process of tumorigenesis, cancer development and progression.50,51 Yang et al found that CLDN10 expression was remarkably reduced in clear cell renal cell carcinoma (ccRCC), which was negatively correlated with the methylation levels of cg10305311 and cg16275739, and DNA hypermethylation played an important role in decreased CLDN10 expression and its association with immune cell infiltration.52 In this study, we found that the methylation level of FUT7 was decreased in BLCA and negatively correlated with FUT7 overexpression. In addition, it was further decreased in a high stage, or with nodal metastasis, or high age and obese BLCA patients (Figure 5). These indicated that the methylation level was closely associated with the clinicopathological characteristics of patients with BLCA. Moreover, it was greatly correlated with the abundance of CD8+ T cells, CD4+ T cells, Th1, Th2, B cells, NK, MDSC, macrophages, and neutrophils, etc. (Table 3). These correlations indicated the potential mechanism of FUT7 in regulating immune cell functions and providing a basis that immunotherapy might be considered based on FUT7 expression. In conclusion, the dysregulation of tumor immune microenvironment was thought to be responsible for BLCA development and progression.
In the current study, we systematically explored the role of FUT7 in BLCA. By analyzing the underlying mechanism of FUT7, we found that FUT7 had the potential to serve as a detection and prognostic biomarker for BLCA. Moreover, FUT7 expression was positively correlated with infiltration levels and gene markers levels of immune cells, which might be due to the alterations of its methylation level. To date, there are relatively few studies on FUT7 in BLCA. Our comprehensive analysis of FUT7, as well as its potential mechanism, may lay a foundation for future research.
Acknowledgments
We would like to thank Oncomine, PrognoScan, cBioPortal, TIMER, TISIDB and UALCAN databases for the availability of the data.
Funding Statement
This study was supported by the Natural Science Foundation of Liaoning Province (grant number 2020-MS-253).
Abbreviations
BLCA, Bladder urothelial carcinoma; EMT, Epithelial–mesenchymal transition; TILs, Tumor-infiltrating lymphocytes; NMIBC, Non-muscle-invasive bladder cancer; MIBC, Muscle-invasive bladder cancer; FUTs, Fucosyltransferases; FTC, Follicular thyroid carcinoma; OS, Overall survival; RFS, Relapse-free survival; DSS, Disease-specific survival; DMFS, Distant metastasis-free survival; DAB, Diaminobenzidine; ELISA, Enzyme-linked immunosorbent assay; TAMs, Tumor-associated macrophages; NK, Natural killer; DC, Dendritic cells; ATCC, American Culture Collection; FBS, Fetal bovine serum; ROC, Receiver operating characteristic curve; AUC, Area under the curve; Act CD8, Activated CD8 T cell; Tcm CD8, Central memory CD8 T cell; Tem CD8, Effector memory CD8 T cell; Act CD4, Activated CD4 T cell; Tcm CD4, Central memory CD4 T cell; Tem CD4, Effector memory CD4 T cell; Tfh, T follicular helper cell; Tgd, Gamma delta T cell; Th1, Type 1 T helper cell; Th17, Type 17 T helper cell; Th2, Type 2 T helper cell; Treg, Regulatory T cell; Act B, Activated B cell; Imm B, Immature B cell; Mem B, Memory B cell; CD56bright, CD56bright natural killer cell; CD56dim, CD56dim natural killer cell; MDSC, Myeloid derived suppressor cell; NKT, Natural killer T cell; Act DC, Activated dendritic cell; pDC, Plasmacytoid dendritic cell; iDC, Immature dendritic cell; NSCLC, Non-small cell lung cancer; PDAC, Pancreatic ductal adenocarcinoma; ccRCC, Clear cell renal cell carcinoma.
Data Sharing Statement
All figures and tables adopted in this study to support the findings are included in the article.
Disclosure
The authors declare no conflict of interests.
References
- 1.van Wilpe S, Gerretsen ECF, van der Heijden AG, de Vries IJM, Gerritsen WR, Mehra N. Prognostic and predictive value of tumor-infiltrating immune cells in urothelial cancer of the bladder. Cancers (Basel). 2020;12(9):E2692. doi: 10.3390/cancers12092692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zangouei AS, Barjasteh AH, Rahimi HR, Mojarrad M, Moghbeli M. Role of tyrosine kinases in bladder cancer progression: an overview. Cell Commun Signal. 2020;18(1):127. doi: 10.1186/s12964-020-00625-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4.Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70(5):404–423. doi: 10.3322/caac.21631 [DOI] [PubMed] [Google Scholar]
- 5.Witjes JA, Bruins HM, Cathomas R, et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur Urol. 2020;79(1):82–104. [DOI] [PubMed] [Google Scholar]
- 6.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41. doi: 10.1038/nrc3817 [DOI] [PubMed] [Google Scholar]
- 7.Alfred witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475. [DOI] [PubMed] [Google Scholar]
- 8.Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. doi: 10.1038/nrdp.2017.22 [DOI] [PubMed] [Google Scholar]
- 9.Ng K, Stenzl A, Sharma A, Vasdev N. Urinary biomarkers in bladder cancer: a review of the current landscape and future directions. Urol Oncol. 2020;39(1):41–51. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Liu C, Yang X. Endoscopic molecular imaging plus photoimmunotherapy: a new strategy for monitoring and treatment of bladder cancer. Mol Ther Oncolytics. 2020;18:409–418. doi: 10.1016/j.omto.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afonso J, Santos LL, Longatto-Filho A, Baltazar F. Competitive glucose metabolism as a target to boost bladder cancer immunotherapy. Nat Rev Urol. 2020;17(2):77–106. doi: 10.1038/s41585-019-0263-6 [DOI] [PubMed] [Google Scholar]
- 12.Rasheduzzaman M, Kulasinghe A, Dolcetti R, et al. Protein glycosylation in head and neck cancers: from diagnosis to treatment. Biochim Biophys Acta Rev Cancer. 2020;1874(2):188422. doi: 10.1016/j.bbcan.2020.188422 [DOI] [PubMed] [Google Scholar]
- 13.Moffett S, Shiao TC, Mousavifar L, Mignani S, Roy R. Aberrant glycosylation patterns on cancer cells: therapeutic opportunities for glycodendrimers/metallodendrimers oncology. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;13(1):e1659. doi: 10.1002/wnan.1659 [DOI] [PubMed] [Google Scholar]
- 14.Council CE, Kilpin KJ, Gusthart JS, Allman SA, Linclau B, Lee SS. Enzymatic glycosylation involving fluorinated carbohydrates. Org Biomol Chem. 2020;18(18):3423–3451. doi: 10.1039/D0OB00436G [DOI] [PubMed] [Google Scholar]
- 15.Rudman N, Gornik O, Lauc G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019;593(13):1598–1615. doi: 10.1002/1873-3468.13495 [DOI] [PubMed] [Google Scholar]
- 16.Du T, Jia X, Dong X, et al. Cosmc disruption-mediated aberrant O-glycosylation suppresses breast cancer cell growth via impairment of CD44. Cancer Manag Res. 2020;12:511–522. doi: 10.2147/CMAR.S234735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Ten Dijke P, Wuhrer M, Zhang T. Role of glycosylation in TGF-β signaling and epithelial-to-mesenchymal transition in cancer. Protein Cell. 2020. doi: 10.1007/s13238-020-00741-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler KB, Costello CE, Rahimi N. Glycosylation in the tumor microenvironment: implications for tumor angiogenesis and metastasis. Cells. 2019;8(6):544. doi: 10.3390/cells8060544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J, Hu Y, Sun S, et al. Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer. Nat Commun. 2020;11(1):6139. doi: 10.1038/s41467-020-19976-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu ZL, Whitaker M, Person AD, Kalabokis V. Detecting substrate glycans of fucosyltransferases with fluorophore-conjugated fucose and methods for glycan electrophoresis. Glycobiology. 2020;30(12):970–980. doi: 10.1093/glycob/cwaa030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pink M, Ratsch BA, Mardahl M, et al. Imprinting of skin/inflammation homing in CD4+ T cells is controlled by DNA methylation within the Fucosyltransferase 7 gene. J Immunol. 2016;197(8):3406–3414. doi: 10.4049/jimmunol.1502434 [DOI] [PubMed] [Google Scholar]
- 22.Li D, Sun H, Bai G, et al. α-1,3-Fucosyltransferase-VII siRNA inhibits the expression of SLex and hepatocarcinoma cell proliferation. Int J Mol Med. 2018;42(5):2700–2708. doi: 10.3892/ijmm.2018.3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jassam SA, Maherally Z, Ashkan K, Pilkington GJ, Fillmore HL. Fucosyltransferase 4 and 7 mediates adhesion of non-small cell lung cancer cells to brain-derived endothelial cells and results in modification of the blood-brain-barrier: in vitro investigation of CD15 and CD15s in lung-to-brain metastasis. J Neurooncol. 2019;143(3):405–415. doi: 10.1007/s11060-019-03188-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang JX, Gao W, Cai L. Fucosyltransferase VII promotes proliferation via the EGFR/AKT/mTOR pathway in A549 cells. Onco Targets Ther. 2017;10:3971–3978. doi: 10.2147/OTT.S140940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin H, Liu J, Yu M, et al. FUT7 promotes the malignant transformation of follicular thyroid carcinoma through α1,3-fucosylation of EGF receptor. Exp Cell Res. 2020;393(2):112095. doi: 10.1016/j.yexcr.2020.112095 [DOI] [PubMed] [Google Scholar]
- 26.Song X, Zhou Z, Li H, et al. Pharmacological suppression of B7-H4 glycosylation restores antitumor immunity in immune-cold breast cancers. Cancer Discov. 2020;10(12):1872–1893. doi: 10.1158/2159-8290.CD-20-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Li Q, Niu L, et al. Suppression of G6PD induces the expression and bisecting GlcNAc-branched N-glycosylation of E-Cadherin to block epithelial-mesenchymal transition and lymphatic metastasis. Br J Cancer. 2020;123(8):1315–1325. doi: 10.1038/s41416-020-1007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahradnikova M, Ihnatova I, Lattova E, et al. N-Glycome changes reflecting resistance to platinum-based chemotherapy in ovarian cancer. J Proteomics. 2020;230:103964. doi: 10.1016/j.jprot.2020.103964 [DOI] [PubMed] [Google Scholar]
- 29.Legler K, Rosprim R, Karius T, et al. Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer. Br J Cancer. 2018;118(6):847–856. doi: 10.1038/bjc.2017.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, Lu J, Yang M, Wang Y, Liu H, Xu C. Elevated GALNT10 expression identifies immunosuppressive microenvironment and dismal prognosis of patients with high grade serous ovarian cancer. Cancer Immunol Immunother. 2020;69(2):175–187. doi: 10.1007/s00262-019-02454-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deb B, Patel K, Sathe G, Kumar P. N-glycoproteomic profiling reveals alteration in extracellular matrix organization in non-type bladder carcinoma. J Clin Med. 2019;8(9):1303. doi: 10.3390/jcm8091303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Du Y, Yang Z, et al. GALNT1-mediated glycosylation and activation of sonic hedgehog signaling maintains the self-renewal and tumor-initiating capacity of bladder cancer stem cells. Cancer Res. 2016;76(5):1273–1283. doi: 10.1158/0008-5472.CAN-15-2309 [DOI] [PubMed] [Google Scholar]
- 33.Wahby S, Jarczyk J, Fierek A, et al. POFUT1 mRNA expression as an independent prognostic parameter in muscle-invasive bladder cancer. Transl Oncol. 2021;14(1):100900. doi: 10.1016/j.tranon.2020.100900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Q, Zhang D, Yang YU, et al. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and α1,3-fucosylation. Cell Death Differ. 2017;24(12):2161–2172. doi: 10.1038/cdd.2017.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YM, Zhang YY, Bulbul A, Shan X, Wang XQ, Yan Q. Baicalin promotes embryo adhesion and implantation by upregulating fucosyltransferase IV (FUT4) via Wnt/beta-catenin signaling pathway. FEBS Lett. 2015;589(11):1225–1233. doi: 10.1016/j.febslet.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 36.Lu HH, Lin SY, Weng RR, et al. Fucosyltransferase 4 shapes oncogenic glycoproteome to drive metastasis of lung adenocarcinoma. EBioMedicine. 2020;57:102846. doi: 10.1016/j.ebiom.2020.102846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai Y, Cheng Z, Pang Y, et al. Prognostic value of the FUT family in acute myeloid leukemia. Cancer Gene Ther. 2020;27(1–2):70–80. doi: 10.1038/s41417-019-0115-9 [DOI] [PubMed] [Google Scholar]
- 38.Yang H, Kuo YH, Smith ZI, Spangler J. Targeting cancer metastasis with antibody therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2021;e1698. doi: 10.1002/wnan.1698 [DOI] [PubMed] [Google Scholar]
- 39.He C, Li A, Lai Q, et al. The DDX39B/FUT3/TGFbetaR-I axis promotes tumor metastasis and EMT in colorectal cancer. Cell Death Dis. 2021;12(1):74. doi: 10.1038/s41419-020-03360-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mola S, Pandolfo C, Sica A, Porta C. The macrophages-microbiota interplay in colorectal cancer (CRC)-related inflammation: prognostic and therapeutic significance. Int J Mol Sci. 2020;21(18):E6866. doi: 10.3390/ijms21186866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M, Hu C, Gao Q, et al. Study on metastasis inhibition of Kejinyan decoction on lung cancer by affecting tumor microenvironment. Cancer Cell Int. 2020;20:451. doi: 10.1186/s12935-020-01540-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabold K, Aschenbrenner A, Thiele C, et al. Enhanced lipid biosynthesis in human tumor-induced macrophages contributes to their protumoral characteristics. J Immunother Cancer. 2020;8(2):e000638. doi: 10.1136/jitc-2020-000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trimaglio G, Tilkin-Mariamé AF, Feliu V, et al. Colon-specific immune microenvironment regulates cancer progression versus rejection. Oncoimmunology. 2020;9(1):1790125. doi: 10.1080/2162402X.2020.1790125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oshi M, Asaoka M, Tokumaru Y, et al. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21(18):E6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang J, Ramis-Cabrer D, Curull V, et al. B cells and tertiary lymphoid structures influence survival in lung cancer patients with resectable tumors. Cancers (Basel). 2020;12(9):E2644. doi: 10.3390/cancers12092644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li BL, Wan XP. Prognostic significance of immune landscape in tumour microenvironment of endometrial cancer. J Cell Mol Med. 2020;24(14):7767–7777. doi: 10.1111/jcmm.15408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang Z, Zhou Q, Zeng H, et al. Intratumoral CCR5 + neutrophils identify immunogenic subtype muscle-invasive bladder cancer with favorable prognosis and therapeutic responses. Oncoimmunology. 2020;9(1):1802176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Zhao Y, Xu H, et al. CtBP1 promotes tumour-associated macrophage infiltration and progression in non-small-cell lung cancer. J Cell Mol Med. 2020;24(19):11445–11456. doi: 10.1111/jcmm.15751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tekin C, Aberson HL, Waasdorp C, et al. Macrophage-secreted MMP9 induces mesenchymal transition in pancreatic cancer cells via PAR1 activation. Cell Oncol (Dordr). 2020;43(6):1161–1174. doi: 10.1007/s13402-020-00549-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Khoury LY, Fu S, Hlady RA, et al. Identification of DNA methylation signatures associated with poor outcome in lower-risk stage, size, grade and necrosis (SSIGN) score clear cell renal cell cancer. Clin Epigenetics. 2021;13(1):12. doi: 10.1186/s13148-020-00998-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin L, Cai Q, Wang S, Wang S, Wang J, Quan Z. Long noncoding RNA PVT1 promoted gallbladder cancer proliferation by epigenetically suppressing miR-18b-5p via DNA methylation. Cell Death Dis. 2020;11(10):871. doi: 10.1038/s41419-020-03080-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Li L, Zhang K, et al. CLDN10 associated with immune infiltration is a novel prognostic biomarker for clear cell renal cell carcinoma. Epigenomics. 2020. doi: 10.2217/epi-2020-0256 [DOI] [PubMed] [Google Scholar]