Abstract
Programmed cell death (PCD) is one of the most intensively researched fields in modern mammalian biology with roles in cancer, aging, diabetes and numerous neurodegenerative diseases. It is becoming increasingly clear that PCD also plays significant roles in plant defence and responses to the environment. Given their unique ability to tolerate desiccation (cells remain viable even after they’ve lost 95% of their water), resurrection plants make ideal models to study the regulation of plant PCD pathways. Previously, we showed that the Australian resurrection plant, Tripogon loliiformis, suppresses plant PCD, via trehalose-mediated activation of autophagy pathways, during drying. In the present study, we created a full-length T. loliiformis cDNA library, performed a large-scale Agrobacterium screen for improved salinity tolerance and identified Stachyose synthase (TlStach) as a potential candidate for improving stress tolerance. Tripogon loliiformis shoots accumulate stachyose synthase transcripts and stachyose during drying. Attempts to generate transgenic plants expressing TlStach failed and were consistent with previous reports in mammals that demonstrated stachyose-mediated induction of apoptosis. Using a combination of transcriptomics, metabolomics and cell death assays (TUNNEL and DNA laddering), we investigated whether stachyose induces apoptotic-like cell death in T. loliiformis. We show that stachyose triggers the formation of the hallmarks of plant apoptotic-like cell death in the desiccation sensitive Nicotiana benthamiana but not the resilient T. loliiformis. These findings suggest that T. loliiformis suppresses stachyose-mediated apoptotic-like cell death and provides insights on the role of sugar metabolism and plant PCD pathways. A better understanding of how resilient plants regulate sugar metabolism and PCD pathways may facilitate future targeting of plant metabolic pathways for increased stress tolerance.
Subject terms: Genetics, Molecular biology, Plant sciences
Introduction
Rainfall projections predict more frequent and severe droughts and other effects of climate change in agricultural regions worldwide. Currently, almost half of the world’s population live in areas that experience severe water scarcity at least one month a year1. Plants are particularly sensitive to changing environments and as such, have developed/evolved a range of tolerance strategies. Despite these adaptations, the flowering plants (angiosperms) that comprise the majority of globally cultivated crops are sensitive to adverse environmental conditions, particularly water deficit and die when their vegetative relative water content (RWC) falls from 60 to 30%. A small group of flowering plants termed resurrection plants can tolerate extreme levels of environmental stresses including excessive heat, salinity and drying (desiccation). Desiccation tolerance is the capacity of a cell or organism to equilibrate their water potential with relatively dry air (< 10% cellular relative water content (RWC) at 20 °C and a humidity of 50%)2,3. In nature, desiccation tolerance is widespread in orthodox seeds. Orthodox seeds tolerate drying through the reversible suspension of metabolic activity and select and controlled activation of specific plant PCD pathways. Recent studies suggest that the vegetative desiccation tolerance observed in resurrection plants is the reactivation of seed desiccation tolerance mechanisms4–6. Resurrection plants can ‘resurrect’ from a dry and seemingly mordant state (RWC 1%) upon watering and are found on rocky outcrops subjected to wet and dry cycles in tropical and sub-tropical climates7,8. An understanding of the water-use efficiency, desiccation tolerance mechanisms and how resurrection plants maintain the vitality of their cells, even when desiccated, could lead to the improvement of these traits in economically important grasses such as rice, sorghum and wheat. Tripogon loliiformis, a diploid, Australian native resurrection plant provides an ideal system for testing numerous hypotheses about the ecology of desiccation-tolerant plants.
Many stress tolerance strategies are either genetically coded or regulated at the molecular level, thus making resilient species not only attractive models but potential genetic sources for the dissection of stress tolerance mechanisms3,9. Researchers have significant knowledge of the drought responses of economically important crops; however, far less information is available on the molecular and metabolic strategies used by resurrection plants to cope with desiccation. The lack of efficient transformation systems for desiccation-tolerant plants has further hindered efforts to characterise the molecular strategies used by this small group of plants to tolerate desiccation.
Transformational mutagenesis is not possible, and although random mutagenesis using chemicals and radiation is possible, identification of the mutated gene that is responsible for the phenotype is near impossible. One method with the potential to identify molecular mechanisms driving desiccation tolerance in non-model plants is the use of gain of function mutants in model plant systems. The Full‐length cDNA Over‐eXpressing (FOX) gene hunting system is one of these methods10. In FOX gene hunting, libraries of cDNAs are inserted into expression cassettes and transformed into a model plant system such as Arabidopsis where they are screened directly for the trait of interest10.
Using a FOX screen of a T. loliiformis cDNA in Agrobacterium subjected to salinity stress, we identified a stress-induced stachyose synthase. Stachyose is a member of the raffinose family of oligosaccharides and a potential stress osmolyte. The raffinose family oligosaccharides (RFOs) are a group of sugars that play roles in transport, carbon storage and protection against abiotic stress11. Two RFOs that play vital roles in stress responses are raffinose and stachyose12. In most plants, raffinose accumulates in vegetative tissues during abiotic stress while stachyose accumulates predominantly during seed development/desiccation11,13. Some resurrection plants, however, accumulate stachyose in their vegetative tissues during drying. Why non-resurrection species mostly limit stachyose accumulation to seeds is unclear. Studies in mammalian cells linking stachyose to the induction of programmed cell death pathways (PCD), specifically, apoptosis, may provide clues14. We show that stachyose accumulates in T. loliiformis shoots during drying; however, we could not generate N. benthamiana plants expressing the T. loliiformis stachyose synthase. Exogenous application of stachyose induced typical apoptotic hallmarks, including DNA laddering and nuclear fragmentation in N. benthamiana plants but not in T. loliiformis plants. These results suggest that T. loliiformis encodes, and implements, measures that suppress stachyose-induced apoptotic cell death and may highlight potential gene targets for the suppression of plant PCD pathways and the production of stress-tolerant crops.
Results
Expression of a T. loliiformis cDNAs protect Agrobacterium from salinity
Before taking advantage of the genetic potential encoded by resurrection plants in crops for improved stress tolerance, molecular biologists must have a thorough understanding of the strategies used to confer stress tolerance. To provide further insights into the molecular mechanisms used by T. loliiformis to tolerate desiccation, we generated a full-length, mixed-dehydration enriched, cDNA library. The resultant library contained approximately 1.62 × 106 cDNAs with an average size of 1.45 Kbp and was cloned into a plant binary vector driven by the Cauliflower mosaic virus 35S promoter. Previous studies demonstrated that the CaMV35S promoter drives the expression of plant cDNAs in Agrobacterium, thus presenting a potentially rapid system for screening stress tolerance before the generation of plants. To assess whether/which T. loliiformis cDNAs provide stress tolerance, we performed a preliminary salinity assay of recombinant Agrobacterium cultures harbouring the cDNA library.
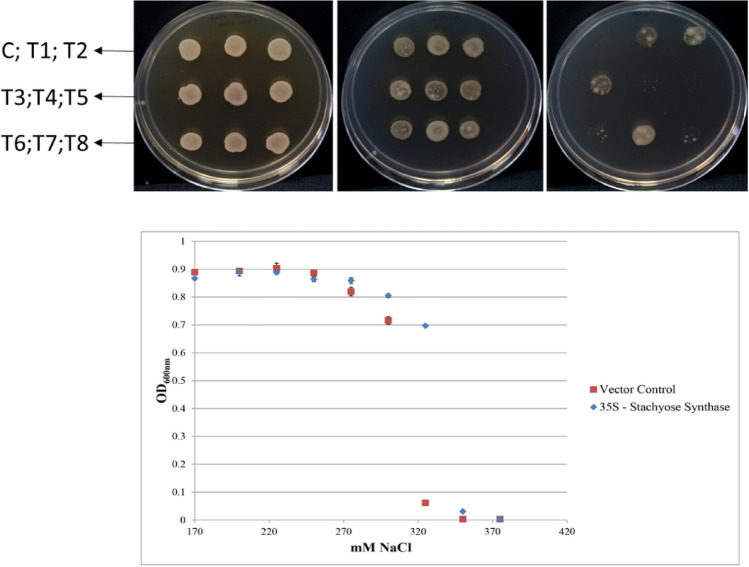
Several Agrobacterium cultures displayed improved growth compared to controls harbouring an empty cloning vector. As shown in Fig. 1, one culture in particular (Tl-7), demonstrated vigorous growth on media containing 370 mM NaCl. In contrast, no growth was observed for Agrobacterium harbouring the empty vector control. Sequence analysis of the cDNA revealed an open reading frame of 758 amino acids with significant homology (81%) to Zea mays and Sorghum bicolour encoded stachyose synthases. Protein modelling displayed 100% homology to the crystal structure of an alpha-galactosidase thus, the cDNA was renamed Tl-Stach.
Figure 1.
FOX screening of Stachyose Synthase genes from Tripogon loliiformis cDNA library in 320 mM; 345 mM and 370 Mm NaCl respectively at 48 h. Plates contain C:Control; T1–T7 Tripogon loliiformis stachyose synthase. Graph shows T7 (TlStach) growth in LB broth supplemented with NaCl. Optical density of bacterial growth presented as means at p < 0.05 using Minitab software version 17 (https://www.minitab.com/en-us/products/minitab/free-trial/).
Dehydrating and desiccated T. loliiformis shoots express stachyose synthase transcripts and accumulate stachyose sugar
Stachyose is a tetrasaccharide member of the family of raffinose oligosaccharides that does not accumulate in the vegetative tissue of most plants but is implicated in the desiccation tolerance of seeds15. Conversely, at least some resurrection plants accumulate stachyose in vegetative tissues16,17. To determine whether stachyose accumulates in drying and desiccated T. loliiformis shoots, we analysed the T. loliiformis transcriptome and metabolome for increased accumulation of stachyose synthase, the enzyme that catalyses the synthesis of stachyose, transcripts and stachyose respectively18,19. The transcriptome data coupled with RT-PCR showed that dehydrating T.loliiformis shoots accumulate stachyose synthase transcripts (Table 1). The increased number of stachyose synthase transcripts in dehydrating shoots produced elevated stachyose levels (Fig. 2).
Table 1.
RT-PCR and transcriptome RNA-Seq data showing fold 2 change of stachyose synthase genes expressed during dehydration in Tripogon loliiformis shoots.
| Hydrated | DS60 | DS40 | DS24 | |
|---|---|---|---|---|
| Ssynthase | ||||
| RT-PCR | 1 | 86.698 ± 3.523 | 45.682 ± 4.287 | 5.915 ± 0.533 |
| RNA-Seq | 1 | 24.797 ± 7.07E−31 | 50.091 ± 1.17E−34 | 7.062 ± 1.96E−34 |
Analysis done in three replicates and analysed at p < 0.05 using R Studio DESeq 2 version 3.11 https://bioconductor.org/packages/release/bioc/html/DESeq2.html. Cells denoted in red and green represent increase and decrease in transcript accumulation respectively in comparison to hydrated controls.
Figure 2.
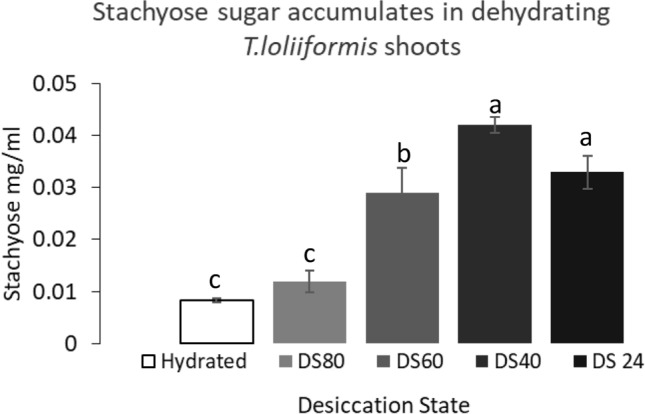
Stachyose sugar accumulation in in dehydrating T.loliiformis shoots. All samples measured using triplicate biological replicates. Stachyose accumulation presented as means at p < 0.05 using Minitab software version 17 (https://www.minitab.com/en-us/products/minitab/free-trial/). Samples denoted with the same letter were not significantly different from each other using p value < 0.05.
Stachyose may induce apoptotic-like cell death in plant cells
The agrobacterium screen, transcriptome and metabolome data suggest that accumulation of stachyose supports stress tolerance. To determine whether increasing stachyose content could improve stress tolerance, we attempted to generate transgenic arabidopsis and Nicotiana benthamiana plants expressing Tl_Stach. Despite numerous attempts and the generation of transgenic plants containing control constructs, we could not generate plants harbouring the TlStach expression vector. The inability to regenerate plants expressing a stachyose synthase raised the possibility that stachyose accumulation may be toxic. Mammalian studies have linked stachyose accumulation with the induction of apoptosis pathways in a caspase-dependant manner14. To determine whether we could not generate transgenic plants because of stachyose-mediated induction of plant apoptotic-like cell death pathways, we performed transient assays. Nicotiana benthamiana leaf tissues were infiltrated with stachyose and assessed for the presence of the apoptotic hallmarks, DNA laddering and TUNEL positive nuclei. Plants infiltrated with oxalate, an established inducer of apoptotic-like PCD, or Murashige and Skoog media (MS), were used as positive and negative controls, respectively20. Previously, we showed that plants display apoptotic-like features between 36 and 48 h post-stress21,22. As shown in Fig. 3, DNA laddering was visible in samples infiltrated with KOX and stachyose after 48 h, with more pronounced laddering observed after 36 h. TUNEL Assays detected the presence of nuclear fragmentation in stachyose treated plants (36 h/48 h), an indication of apoptotic-like cell death (Fig. 4). Despite evident laddering in trehalose treated plants at 48 h, nuclear fragmentation was absent in the trehalose treated TUNEL samples.
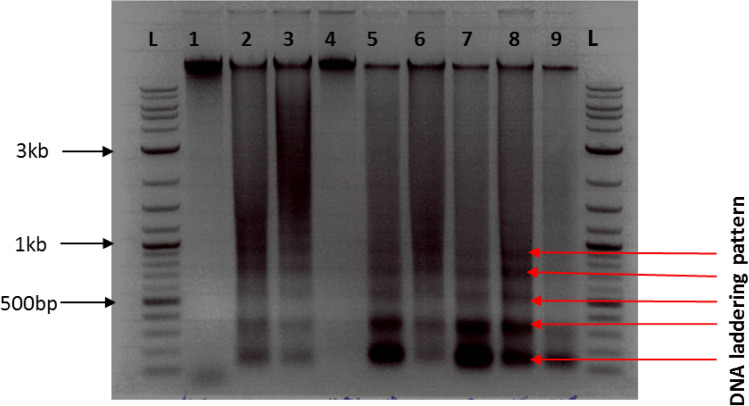
Figure 3.
Stachyose and Trehalose induce time-dependent DNA laddering in Nicotiana benthamiana. DNA was extracted on N. benthamiana leaves treated with stachyose, trehalose and a combination of stachyose and trehalose sugars at 36 h and 48 h. 20 ng of the DNA was run on a 1% agarose gel. Potassium oxalate (KOX) treated sample was the positive control. L: 100 bp ladder; 1: untreated sample; 2/3: 20 mM KOX (36/48 h); 4/5: 5 mM Trehalose samples (36/48 h); 6/7: 3.2 mg/ml Stachyose treated samples (36/48 h); 8/9: 5 mM Trehalose + 3.2 mg/ml Stachyose (36/48 h).
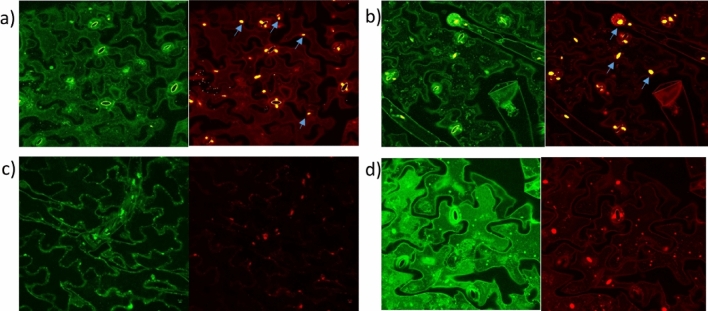
Figure 4.
Stachyose treatment induced TUNEL positive nuclei in N. benthamiana. TUNEL Assays were done on sugar treated Nicotiana benthamiana triplicate biological replicates to detect apoptotic-like cell death. N. benthamiana samples treated with DNAse were used as positive controls as DNAse induces nuclei fragmentation. (a) 5 U/μl DNase treated samples (positive control); (b) 3.2 mg/ml Stachyose treated samples; (c) No enzyme control (Negative control); (d) 5 mM Trehalose treated samples (48 h). Arrows represent DNA fragmentation.
Tripogon loliiformis prevents apoptosis caused by stachyose accumulation
Autophagy, another form of cell death is antagonistic to apoptosis. Previously, we showed that trehalose treatment induces the formation of autophagosomes, even in hydrated T. loliiformis leaves18,19. To determine whether T. loliiformis suppresses stachyose-mediated apoptosis, we infiltrated T. loliiformis leaves with stachyose and KOX and looked for the presence of DNA laddering. N. benthamiana samples treated with stachyose served as positive controls. No laddering was detected in T. loliiformis leaves infiltrated with stachyose within the same timeframe as N. benthamiana plants (Fig. 5). The lack of DNA laddering in T. loliiformis indicates that the grass has already put in place mechanisms that suppress nuclear fragmentation and DNA laddering.
Figure 5.
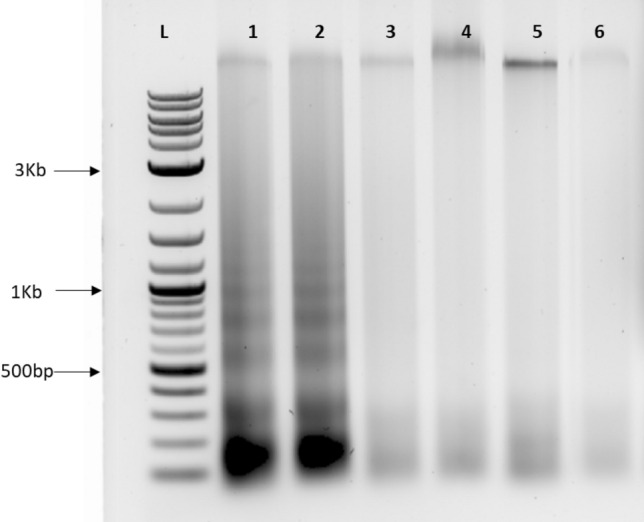
Stachyose does not induce DNA laddering in T. loliiformis shoots. DNA was extracted on both N. benthamiana and T. loliiformis shoots treated with stachyose at 36 h and 48 h. T. loliiformis shoots were also treated with 20 mM Potassium Oxalate (KOX) known to induce DNA laddering. 20 ng of the DNA was run on a 1% agarose gel. N. benthamiana samples treated with stachyose were positive controls. Lane 1/2: 3.2 mg/ml Stachyose treated Nicotiana benthamiana (36 h/48 h); Lane 3/4: 20 mM KOX treated T. loliiformis (36 h/48 h); Lane 5/6: 3.2 mg/ml Stachyose treated T. loliiformis (36 h/48 h).
Discussion
Environmental stresses impact plants and animals in several ways. Plants, due to their sessile nature, are particularly sensitive and severely affected by environmental stresses. To combat stress, plants use a combination of strategies at the molecular, biochemical and physiological level. Accumulation of osmolytes and initiation of PCD are just a few of the physiological and metabolic changes employed by plants to protect cells against stress. Stachyose induces apoptosis in mammals. Similarly, we show that stachyose treatments induce DNA laddering and TUNEL positive nuclei, hallmark features of apoptotic-like cell death, in plants. No such features were observed following stachyose treatment of T. loliiformis which also accumulates stachyose in its vegetative tissue upon dehydration. We speculate that stachyose acts similarly in plant and animal cells, causing apoptotic-like cell death; however, desiccation-tolerant T. loliiformis utilises molecular and physiological strategies coupled with sugar metabolism to inhibit apoptosis.
Stachyose is a member of the Raffinose family Oligosaccharides (RFOs) that plays a significant role in seed desiccation16,23. Raffinose family oligosaccharides, including stachyose, accumulate in the late stages of soybean seed maturation and desiccation16. Soybean seeds induce stress tolerance through gradual drying correlating with an increase in stachyose content24. Arabidopsis and smaize also accumulate stachyose during seed maturation but not in vegetative tissues24,25. Conversely, stachyose accumulates in vegetative tissues of the resurrection plants Craterostigma plantagineum, Xerophyta viscosa and Myrothamnus flabellifolia during dehydration26–28. T. loliiformis shoots also accumulate stachyose during dehydration. Whether the accumulated stachyose sacts as an osmoprotectant in T. loliiformis desiccation tolerance in remains unclear.
In mammalian cells, stachyose plays a role in controlling various diseases like cancer by initiating apoptosis29. For instance, stachyose-induced apoptosis and expression of caspases 3 and 9 in a dose-dependent manner leading to the release of Cytochrome C (Cyt C) from the mitochondria to the cytosol. The effector caspase, caspase 3, is activated by the initiator caspase, caspase 9, via the release of Cyt C, leading to cell death14. A key feature in apoptotic cell death is nuclear fragmentation and DNA laddering. The laddering observed in stachyose treated N. benthamiana cells potentially show that despite the lack of true caspases in plant cells, parts of the pathways are conserved and stachyose acts in a similar manner to mammalian cells by inducing apoptotic-like cell death.
In yeast, trehalose acts as a chemical chaperone that prevents protein denaturation30. The role of trehalose in plants is less clear; however, studies suggest that it has a role in stress signalling pathways. Previously, treatment of T. loliiformis and Nicotiana benthamiana shoots with trehalose showed induction of autophagy18,31. Thus, suggesting that, in respect to autophagy, sensitive and tolerant plants react similarly to trehalose. Autophagy helps restore cellular homeostasis during stress sequestering cellular material for degradation and recycling of nutrients18,32. Due to its role in maintaining homeostasis and removing unwanted and cellular toxins, autophagy increases lifespan in mammals and delays senescence in plant cells33–35. By removing damaged and misfolded proteins as well as other apoptotic signals, autophagy suppresses cell death36,37. For instance, trehalose application induces autophagy and protects human neuroblastoma cells against Bax-induced cell death38. Additionally, mammalian cells displaying trehalose triggered m-TOR independent autophagy, have fewer cells showing features of apoptotic-like cell death39. In plants, trehalose application suppresses petal senescence and promotes the accumulation of autophagosomes in T. loliiformis18,40.
When faced with prolonged stress, some sensitive plants like N. benthamiana respond by accelerating their life cycle and prematurely promoting senescence. The funneling of resources in reproduction, however, renders these plants susceptible to pro-apoptotic signals and cell death. In contrast, T. loliiformis sutilises autophagy via regulation of the snRK1 pathway to cause an immediate shift from anabolism to catabolism. This shift is crucial and helps prevent kernel abortion and increase smaize yields41. T. loliiformis cells treated with stachyose did not undergo apoptosis as shown by lack of laddering and nuclear fragmentation. The absence of laddering is because T. loliiformis is primed for stress tolerance in the hydrated state and therefore activates signals that trigger autophagy early. Autophagy through the sremobilisation of nutrients provides alternative energy sources that T. loliiformis uses to combat stress. However, prolonged autophagy (overeating) causes the depletion of cellular machinery and may lead to cell death. In our work, the presence of DNA laddering at 48, but not 36 h post trehalose treatment may be indicative of such as case.
A key feature of resurrection plants is the suppression of apoptosis and senescence during desiccation18,42. One strategy that helps resurrection plants survive desiccation is transcriptional and metabolic priming in the hydrated state to enable rapid responses to stress. The early onset of dehydration adaptive strategies like autophagy may facilitate desiccation tolerance19,43. The cell cycle also plays a vital role in growth and development and is driven by energy metabolism40,44–46. The transition from the G2 to Mitosis phase is crucial and therefore acts as an essential stop-check by the cell for the presence of genetic deformities before duplication. In case the deformity cannot be repaired, PCD eliminates the cell from the organism47. Sugar signals regulate the major cell cycle components in meristematic cells for G2/M transition48. For instance, a reduction in cellular sucrose pushes the cells to the G2 phase rendering them susceptible to signals causing apoptotic-like cell death. Since the cells have been transitioned quickly to the G2 phase, they are at a high risk of acquiring abnormalities. In mammalian cells, cell death caused by cadmium application in the G2 phase displayed apoptotic-like cell death features potentially aided by changes to sugar metabolism44,49. Low energy levels affect plant and mammalian cells, forcing them to accelerate their life cycles. Contrary, high glucose levels promote senescence and nutrient recycling to provide alternative energy reserves required for growth and development50. Autophagy is employed to meet additional energy needs through sremobilisation of resources51. This was possibly the case with T. loliiformis, as a switch from anabolism to catabolism, aided by autophagy, provides the additional energy resources required to suppress apoptotic signals.
In summary, stachyose can function either as an osmoprotectant or as an inducer of apoptotic-like cell death in plant cells. However, T. loliiformis potentially sutilises autophagy to suppress apoptotic-like cell death. Induction of autophagy through the snRK1 pathway, as shown in T. loliiformis18 provides an alternative energy source required by the cells to maintain cellular and metabolic homeostasis during stress. Contrary, stachyose treated cells were unable to maintain cellular homeostasis and triggered apoptosis. TUNEL assays showed that stachyose induces apoptotic-like cell death in N. benthamiana plants. These results provide much-needed insight into the effect of stachyose on plant cells and the importance of energy metabolism in determining cell fate. How energy metabolism affects the plant cell can dictate whether a cell lives or dies when faced with developmental or environmental stresses. However, how T. loliiformis plants survive accumulation of stachyose and whether autophagy functions to induce signals stronger than apoptosis in dehydrating T. loliiformis plants warrants further research. Future work on the signalling pathways activated by stachyose application and their potential role in plant PCD should be reviewed. Additionally, further research is needed to ascertain the molecular mechanisms by which T. loliiformis suppresses plant PCD, including stachyose-induced PCD observed in sensitive plants.
Materials and methods
Plant materials and cultivation
Tripogon loliiformis (F.Muell.) C.E.Hubb. plants (Queensland Herbarium voucher accession number: Williams 01) were identified and collected under licence from Charleville (GPS: –26.42686 S, 146.25002E), Queensland, Australia, by Stephen Peck (QLD Parks and Wildlife) and Dr Brett Williams. The plants were germinated from seeds collected from a single plant and grown in a chamber at 27 °C and 16 h photoperiod. Twenty-one, 65 mm pots, each containing several tufts of T. loliiformis, were grown for two months. Before dehydration, all plants were watered to saturation. Hydrated controls were randomly collected in triplicate, 1-day post-watering. The remaining plants were subjected to water deficit by withholding water until they reached an air-dry state and a relative water content (RWC) less than 10%. Triplicate samples were collected at 80, 60, 40, and < 10% RWC throughout the dehydration. The plants were watered and rehydrated samples were collected after 48 h. The RWC was determined on T. loliiformis shoots and roots and was calculated according to Barrs and Weatherley (1962) using the formula (RWC (%) = ((Fresh weight − Dry weight)/(Turgid weight − Dry weight)) × 100). All the shoot and root samples were snap-frozen in liquid nitrogen and stored at − 80 °C until analysed19. For exogenouse application so stachyose, T. loliiformis shoots were infiltrated with 1.6 mg/ml and 3.2 mg/ml stachyose before subjection to analysis for DNA laddering.
Non-transgenic Nicotiana benthamiana seeds, collected from a single plant, were germinated in soil and grown in a chamber at 27 °C under a 16/8-h light/dark cycles at 100 μE/m2/s. Leaves were infiltrated with 20 mM Potassium oxalate, 5 mM trehalose, 1.6 and 3.2 mg/ml stachyose and a combination of 5 mM trehalose and 3.2 mg/ml stachyose. Samples were collected 36hrs and 48 h post-infiltration and used for either DNA laddering or TUNEL Assays.
Total RNA extraction and high throughput sequencing
For sequencing analysis, total RNA was isolated from the shoot tissues of triplicate; hydrated, dehydrating, dehydrated and rehydrated T. loliiformis plants using a modified Trizol (Invitrogen) and spin column (Qiagen) method. The Bioanalyzer (Agilent technologies) was used to verify RNA integrity and quality. For library preparation, polyadenylated RNA was enriched and chemically fragmented; cDNA was ssynthesised using an Illumina RNA-seq kit according to the manufacturer’s instructions. Sequencing of the cDNA libraries was performed at Texas A&M AgriLife Genomics and Bioinformatics service, United States using an Illumina HiSeq 2500 Sequencer (Illumina, Inc.). 100 bp single-read sequences were collected18,19.
RNA-seq analysis
Quality control of the sequences was performed. Primer and barcode sequences were removed by trimming. A de novo assembled and blast annotated T. loliiformis transcriptome assembly served as the reference for RNA-Seq profiling of the independent cDNA libraries18. Over 80% of the reads from each sample were mapped. All data sets were paired and used in an in silico microarray experiment using CLC genomics workbench. Using the hydrated samples as a reference for unstressed gene expression, each data set was enriched for genes that had a fold change > 2 or < − 2. Prior to analysis, the data were subjected to quantile normalisation. The distributions of the expression values for each replicate were used to create a common target distribution. The targeted distribution was then used to calculate normalised expression values18,19. Fold 2 change and statisticical analysis at p < 0.05 was done using R Studio DESeq 2.
Quantitative real-time PCR analysis
cDNA was generated from 0.8 μg of Total RNA using an oligo (dT) (100 ρmol) primer and Superscript III Reverse Transcriptase (Invitrogen). Quantitative PCR was done using a ViiA7 Real-Time PCR System and the SYBR Green PCR Master Mix kit (Applied Biosystems) according to the manufacturer’s instructions using 300 mM primer and 1/100 dilution of cDNA and standard cycling parameters. Stachyose synthase primers were designed using Primer3 bioinformatic software (MIT) and are listed in Supplementary Table 1. The data analysis was completed using ExpressionSuite Software (Life Technologies). The Tripogon loliiformis homologue of Arabidopsis Actin identified from the annotated transcriptome were used for quantitative normalisation. Fold changes were calculated against hydrated tissue18,19.
GCMS analysis
Tripogon loliiformis shoots at the various dehydration points described above, were sampled and analysed for sugar content by Gas Chromatography-Mass Spectroscopy (GCMS). Approximately, 100 mg of dry tissue was analysed using a Waters Acquity UPLC coupled to a mass spectrometer. For quantitation purposes, stachyose standards were analysed alongside the samples. Chromatographic separation was performed using a Waters BEH Amide UPLC column (1.7 mm, 2.1 mm 3 150 mm; 30C). The mobile phase used a mixture of water (Fisher Optima) containing 0.1% formic acid (solvent A; Fisher) and acetonitrile (Fisher Optima) containing 0.1% formic acid (solvent B) in a linear gradient from 80% B to 50% B at a flow rate of 0.35 ml/min. For statistical significance, triplicate samples were measured19.
Salinity assays of recombinant Agrobacterium harbouring the T. loliiformis cDNA library
Recombinant Agrobacterium tumefaciens cultures harbouring expression cassettes of individual T. loliiformis cDNAs were grown in Luria Bertani (LB) broth containing 50 mg/l Kanamycin. Recombinant cultures containing an empty cloning vector were used as controls. The recombinant cultures were grown on LB solid media with 170, 225, 300 and 400 mM sodium chloride and incubated at 28 °C for 48 h. Comparisons were made by observing differences in growth between the recombinant cultures grown on the salinity media, as mentioned above.
DNA laddering
Leaf tissues of N. benthamiana treated with 20 mM Potassium oxalate, 5 mM trehalose, 1.6 and 3.2 mg/ml stachyose and a combination of 5 mM trehalose and 3.2 mg/ml stachyose respectively and T. loliiformis treated with 1.6 mg/ml and 3.2mgml of stachyose as described above were snap-frozen in liquid nitrogen and ground to a fine powder. Extraction of DNA was performed using a modified CTAB protocol52. For analysis of DNA laddering, equal amounts of DNA (20 µg) were treated with 25 μg/ml DNase-free RNase A (Boehringer Mannheim) and incubated for one h at 37 °C. Following RNA digestion, the DNA was separated by electrophoresis on a 1% (w/v) agarose gel, and visualised by staining with 0.5 μg/ml Gel Red (Biotium) and observation under UV light.
TUNEL assays
Nicotiana benthamiana samples were infiltrated with stachyose and trehalose and harvested for TUNEL assays. As a positive control, samples were treated with 5 U/μl DNase I for 10 min at 37 °C. In situ nick end labelling of nuclear DNA fragmentation was performed at 37 °C in a humid chamber for 1 h in the dark using the fluorescein, in situ Cell Death Detection Kit (Boehringer Mannheim, Mannheim, Germany) at a dilution of 1:2 in reaction buffer. For negative controls, stachyose and trehalose treated samples were labelled in parallel as above, except for the absence of TdT enzyme. After labelling, the samples were mounted on slides and rinsed in 1X PBS and counterstained with propidium iodide (PI). Confocal microscopy was performed using a Nikon A1 microscope. Seven samples were viewed under the microscope for each treatment.
Data analysis
Data on stachyose concentration and optical density of bacterial growth were collected and analysed using Minitab software Version 17 https://www.minitab.com/en-us/products/minitab/free-trial/. ANOVA was carried out to determine the significance at p < 0.05.
Supplementary Information
Acknowledgements
PA was supported by a QUT PhD scholarship. BW was supported by an Advance QLD fellowship (AQRF04016-17RD2). The confocal microscopy data reported in this paper were generated at the Central Analytical Research Facility (CARF) operated by QUT’s Institute for Future Environments (QUT).
Author contributions
All authors contributed equally to this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86559-7.
References
- 1.Boretti A, Rosa L. Reassessing the projections of the World Water Development Report. NPJ Clean Water. 2019;2:1–6. doi: 10.1038/s41545-019-0039-9. [DOI] [Google Scholar]
- 2.Gaff DF. Desiccation tolerant vascular plants of southern Africa. Oecologia. 1977;31:95–109. doi: 10.1007/BF00348713. [DOI] [PubMed] [Google Scholar]
- 3.Proctor MCF, Tuba Z. Poikilohydry and homoihydry: Antithesis or spectrum of possibilities? New Phytol. 2002;156:327–349. doi: 10.1046/j.1469-8137.2002.00526.x. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths CA, Gaff DF, Neale AD. Drying without senescence in resurrection plants. Front. Plant Sci. 2014;5:36. doi: 10.3389/fpls.2014.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa MCD, Cooper K, Hilhorst HWM, Farrant JM. Orthodox seeds and resurrection plants: Two of a kind? Plant Physiol. 2017;175:589–599. doi: 10.1104/pp.17.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa MD, et al. A footprint of desiccation tolerance in the genome of Xerophyta viscosa Maria-Cecília. Nat. Plants. 2017;3:17038. doi: 10.1038/nplants.2017.38. [DOI] [PubMed] [Google Scholar]
- 7.Alpert P. Constraints of tolerance: Why are desiccation-tolerant organisms so small or rare? J. Exp. Biol. 2006;209:1575–1584. doi: 10.1242/jeb.02179. [DOI] [PubMed] [Google Scholar]
- 8.Bewley JD. Physiological aspects +7670 of desiccation tolerance. Annu. Rev. Plant Biol. 1979;30:195–238. doi: 10.1146/annurev.pp.30.060179.001211. [DOI] [Google Scholar]
- 9.Deeba, F. & Pandey, V. Adaptive Mechanisms of Desiccation Tolerance in Resurrection Plants. (Springer Nature Singapore PTE Ltd, 2017).
- 10.Ichikawa T, et al. The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- 11.Taji T, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 12.Peterbauer T, et al. Analysis of the raffinose family oligosaccharide pathway in pea seeds with contrasting carbohydrate composition. Plant Physiol. 2001;127:1764–1772. doi: 10.1104/pp.010534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizawa-yokoi A, Yabuta Y, Shigeoka S. The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal. Behav. 2008;3:1016–1018. doi: 10.4161/psb.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang G, Mao J, Ji Z, Ailati A. Stachyose-induced apoptosis of Caco-2 cells via the caspase-dependent mitochondrial pathway. Food Funct. 2015;6:765–771. doi: 10.1039/C4FO01017E. [DOI] [PubMed] [Google Scholar]
- 15.González-morales SI, Chávez-montes RA, Hayano-kanashiro C, Alejo-jacuinde G. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. PNAS. 2016 doi: 10.1073/pnas.1610985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saravitz DM, Pharr DM, Carter TE. Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiol. 1987;83:185–189. doi: 10.1104/pp.83.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver MJ, et al. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell. 2011;23:1231–1248. doi: 10.1105/tpc.110.082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams B, et al. Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet. 2015;11:1–17. doi: 10.1371/journal.pgen.1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asami P, et al. Roots of the resurrection plant Tripogon loliiformis survive desiccation without the activation of autophagy pathways by maintaining energy reserves. Front. Plant Sci. 2019;10:459. doi: 10.3389/fpls.2019.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KS, Min J, Dickman MB. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 2008;21:605–612. doi: 10.1094/MPMI-21-5-0605. [DOI] [PubMed] [Google Scholar]
- 21.Kabbage M, Williams B, Dickman MB. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013;9:e1003287. doi: 10.1371/journal.ppat.1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y. Programmed cell death during Petunia petal senescence. Plant Physiol. 2000;122:266. doi: 10.1104/pp.122.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillo EM, de Lumen BO, Reyes PS, de Lumen HZ. Raffinose synthase and galactinol synthase in developing seeds and leaves of legumes. J. Agric. Food Chem. 1990;38:351–355. doi: 10.1021/jf00092a003. [DOI] [Google Scholar]
- 24.Blackman SA, Obendorf RL, Leopold AC. Maturation proteins and sugars in desiccation tolerance of developing soybean seeds. Plant Physiol. 1992;100:225–230. doi: 10.1104/pp.100.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenac P, et al. Raffinose accumulation related to desiccation tolerance during maize (Zea mays L.) seed development and maturation. J. Plant Physiol. 1997;150:481–488. doi: 10.1016/S0176-1617(97)80102-2. [DOI] [Google Scholar]
- 26.Norwood M, Truesdale MR, Richter A, Scott P. Photosynthetic carbohydrate metabolism in the resurrection plant Craterostigma plantagineum. J. Exp. Bot. 2000;51:159–165. doi: 10.1093/jexbot/51.343.159. [DOI] [PubMed] [Google Scholar]
- 27.Peters S, Mundree SG, Thomson JA, Farrant JM, Keller F. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): Both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J. Exp. Bot. 2007 doi: 10.1093/jxb/erm056. [DOI] [PubMed] [Google Scholar]
- 28.Moore JP, Lindsey GG, Farrant JM, Brandt WF. An overview of the biology of the desiccation-tolerant resurrection plant Myrothamnus flabellifolia. Ann. Bot. 2007;99:211–217. doi: 10.1093/aob/mcl269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cervera-Tison M, et al. Functional analysis of family GH36 α-galactosidases from Ruminococcus gnavus E1: Insights into the metabolism of a plant oligosaccharide by a human gut symbiont. Appl. Environ. Microbiol. 2012;78:7720–7732. doi: 10.1128/AEM.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: A multifunctional molecule. Glycobiology. 2003;13:17–27. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, et al. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Sci. Rep. 2016;6:28423. doi: 10.1038/srep28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N. The role of mammalian autophagy in protein metabolism. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007;83:39–46. doi: 10.2183/pjab.83.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat. Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 34.Vellai T, Takács-Vellai K, Sass M, Klionsky DJ. The regulation of aging: Does autophagy underlie longevity? Trends Cell Biol. 2009;19:487–494. doi: 10.1016/j.tcb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584:1350–1358. doi: 10.1016/j.febslet.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 37.Andrew T. Apoptosis and Autophagy: Regulatory connections between two supposedly different processes. Methods. 2008;13:23–29. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and ??-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;10:588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asami P, Mundree S, Williams B. Saving for a rainy day: Control of energy needs in resurrection plants. Plant Sci. 2018;271:62–66. doi: 10.1016/j.plantsci.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Bledsoe SW, et al. The role of Tre6P and SnRK1 in maize early kernel development and events leading to stress-induced kernel abortion. BMC Plant Biol. 2017;17:74. doi: 10.1186/s12870-017-1018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radermacher AL, du Toit SF, Farrant JM. Desiccation-driven senescence in the resurrection plant Xerophyta schlechteri (Baker) N.L. Menezes: Comparison of anatomical, ultrastructural, and metabolic responses between senescent and non-senescent tissues. Front. Plant Sci. 2019;10:1–16. doi: 10.3389/fpls.2019.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez MCS, et al. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010;63:212–228. doi: 10.1111/j.1365-313X.2010.04243.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Ruan Y-L. Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 2013;4:163. doi: 10.3389/fpls.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabbage, M., Kessens, R., Bartholomay, L. C. & Williams, B. The life and death of a plant cell. Annu. Rev. Plant Biol.68, annurev-arplant-043015-111655 (2017). [DOI] [PubMed]
- 46.Fogarty CE, Bergmann A. Killers creating new life: Caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ. 2017;24:1390–1400. doi: 10.1038/cdd.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pucci B, Kasten M, Giordano A. Cell cycle and apoptosis. Nature. 2000;2:291–299. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azzopardi M, Farrugia G, Balzan R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech. Ageing Dev. 2016;161:211–224. doi: 10.1016/j.mad.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Chuan ZC, Lu LY, Min LY, He LM. Cadmium exposure enhances bisphenol A-induced genotoxicity through inhibition in NIH3T3 fibroblast cells. Cell. Physiol. Biochem. 2016;400038:961–974. doi: 10.1159/000447804. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Chang C, Tucker ML. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015;6:1–7. doi: 10.3389/fpls.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izumi M, Hidema J, Ishida H. Deficiency of autophagy leads to significant changes of metabolic profiles in Arabidopsis. Plant Signal. Behav. 2013;8:e25023. doi: 10.4161/psb.25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.